Abstract
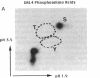
GAL4 protein isolated from yeast in which it is active is phosphorylated predominantly on two different serine residues. One of these was identified as Ser-837; substitution of this residue for alanine has no detectable effect on transcriptional activation by GAL4. Phosphorylation at Ser-837 requires that both the DNA binding and transcriptional activation functions be intact. We propose that some phosphorylations of GAL4, including that at Ser-837, occur concomitantly with activation of transcription.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Carey M., Kakidani H., Leatherwood J., Mostashari F., Ptashne M. An amino-terminal fragment of GAL4 binds DNA as a dimer. J Mol Biol. 1989 Oct 5;209(3):423–432. doi: 10.1016/0022-2836(89)90007-7. [DOI] [PubMed] [Google Scholar]
- Cisek L. J., Corden J. L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature. 1989 Jun 29;339(6227):679–684. doi: 10.1038/339679a0. [DOI] [PubMed] [Google Scholar]
- Flick J. S., Johnston M. Two systems of glucose repression of the GAL1 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Sep;10(9):4757–4769. doi: 10.1128/mcb.10.9.4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz R. D., Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988 Dec 30;74(2):527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Gill G., Sadowski I., Ptashne M. Mutations that increase the activity of a transcriptional activator in yeast and mammalian cells. Proc Natl Acad Sci U S A. 1990 Mar;87(6):2127–2131. doi: 10.1073/pnas.87.6.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelfarb H. J., Pearlberg J., Last D. H., Ptashne M. GAL11P: a yeast mutation that potentiates the effect of weak GAL4-derived activators. Cell. 1990 Dec 21;63(6):1299–1309. doi: 10.1016/0092-8674(90)90425-e. [DOI] [PubMed] [Google Scholar]
- Hoeck W., Groner B. Hormone-dependent phosphorylation of the glucocorticoid receptor occurs mainly in the amino-terminal transactivation domain. J Biol Chem. 1990 Apr 5;265(10):5403–5408. [PubMed] [Google Scholar]
- Jackson S. P., MacDonald J. J., Lees-Miller S., Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990 Oct 5;63(1):155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutational analysis of the GAL4-encoded transcriptional activator protein of Saccharomyces cerevisiae. Genetics. 1988 Sep;120(1):63–74. doi: 10.1093/genetics/120.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature. 1987 Jul 23;328(6128):353–355. doi: 10.1038/328353a0. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laybourn P. J., Dahmus M. E. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J Biol Chem. 1990 Aug 5;265(22):13165–13173. [PubMed] [Google Scholar]
- Long R. M., Mylin L. M., Hopper J. E. GAL11 (SPT13), a transcriptional regulator of diverse yeast genes, affects the phosphorylation state of GAL4, a highly specific transcriptional activator. Mol Cell Biol. 1991 Apr;11(4):2311–2314. doi: 10.1128/mcb.11.4.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- Mylin L. M., Bhat J. P., Hopper J. E. Regulated phosphorylation and dephosphorylation of GAL4, a transcriptional activator. Genes Dev. 1989 Aug;3(8):1157–1165. doi: 10.1101/gad.3.8.1157. [DOI] [PubMed] [Google Scholar]
- Mylin L. M., Johnston M., Hopper J. E. Phosphorylated forms of GAL4 are correlated with ability to activate transcription. Mol Cell Biol. 1990 Sep;10(9):4623–4629. doi: 10.1128/mcb.10.9.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortí E., Mendel D. B., Smith L. I., Munck A. Agonist-dependent phosphorylation and nuclear dephosphorylation of glucocorticoid receptors in intact cells. J Biol Chem. 1989 Jun 15;264(17):9728–9731. [PubMed] [Google Scholar]
- Payne J. M., Laybourn P. J., Dahmus M. E. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989 Nov 25;264(33):19621–19629. [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Sadowski I., Stone J. C., Pawson T. A noncatalytic domain conserved among cytoplasmic protein-tyrosine kinases modifies the kinase function and transforming activity of Fujinami sarcoma virus P130gag-fps. Mol Cell Biol. 1986 Dec;6(12):4396–4408. doi: 10.1128/mcb.6.12.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song D., Dolan J. W., Yuan Y. L., Fields S. Pheromone-dependent phosphorylation of the yeast STE12 protein correlates with transcriptional activation. Genes Dev. 1991 May;5(5):741–750. doi: 10.1101/gad.5.5.741. [DOI] [PubMed] [Google Scholar]
- Sorger P. K., Pelham H. R. Yeast heat shock factor is an essential DNA-binding protein that exhibits temperature-dependent phosphorylation. Cell. 1988 Sep 9;54(6):855–864. doi: 10.1016/s0092-8674(88)91219-6. [DOI] [PubMed] [Google Scholar]
- Sorger P. K. Yeast heat shock factor contains separable transient and sustained response transcriptional activators. Cell. 1990 Aug 24;62(4):793–805. doi: 10.1016/0092-8674(90)90123-v. [DOI] [PubMed] [Google Scholar]