Abstract
APOBEC-catalyzed cytosine-to-uracil deamination of single-stranded (ss)DNA has beneficial functions in immunity and detrimental roles in cancer. APOBEC enzymes have intrinsic dinucleotide specificities that impart hallmark mutation signatures. Despite numerous structures, mechanisms for global ssDNA recognition and local target sequence selection remain unclear. Here, we report crystal structures of human APOBEC3A and a chimera of human APOBEC3B and APOBEC3A bound to ssDNA at 3.1 and 1.7 angstroms resolution, respectively. These structures reveal a U-shaped DNA conformation, with the specificity-conferring −1 thymine flipped out and the target cytosine inserted deep into the zinc-coordinating active site pocket. The −1 thymine base fits between flexible loops in a groove that forms upon binding ssDNA, and it makes direct hydrogen bonds with the protein accounting for the strong 5′-TC preference. These studies explain both conserved and unique properties among APOBEC family members, and provide a basis for the rational design of inhibitors to impede the evolvability of viruses and tumors.
Vertebrates encode variable numbers of active polynucleotide cytosine deaminase enzymes collectively called APOBECs1,2. All vertebrate species have activation-induced deaminase (AID), which is essential for antibody gene diversification through somatic hypermutation and class switch recombination3,4. Most vertebrates also have APOBEC1, which edits cytosine nucleobases in RNA and ssDNA and functions in regulating the transcriptome and likely also in blocking the spread of endogenous and exogenous mobile elements including viruses5,6. The APOBEC3 subfamily of enzymes is specific to mammals, subject to extreme copy number variation, elicits strong preferences for ssDNA, and provides innate immune protection against a wide variety of DNA-based parasites including common retrotransposons L1 and Alu and retroviruses such as HIV-12,7,8. Human cells have the potential to produce up to 7 distinct APOBEC3 enzymes, APOBEC3A through APOBEC3H (A3A-A3H, excluding A3E), though most cells express subsets due to differential gene regulation9–12.
The local substrate preference of each of these enzymes is an intrinsic property that has helped to elucidate multiple biological and pathological functions including the elucidation of AID as an antibody gene DNA deaminase3,4, the delineation of the subset of APOBEC3 enzymes responsible for HIV-1 hypermutation (A3D, A3F, A3G, and A3H)2,7,8 and, recently, the implication of at least one APOBEC family member in mutagenesis in a wide variety of cancers13–15. The nucleobase immediately 5′ of the target cytosine (−1 relative to the target cytosine at position 0) is the most important determinant of each enzyme’s intrinsic substrate preference16–19. AID preferentially deaminates single-stranded DNA cytosine bases preceded by an adenine or guanine (5′-RC), corresponding to cytosine mutation spectra in immunoglobulin gene variable and switch regions. A3G uniquely targets cytosine bases preceded by another cytosine (5′-CC), a pattern that is evident in patient-derived HIV-1 sequences. APOBEC1 and the remaining APOBEC3 enzymes elicit preferences for cytosine bases preceded by a thymine (5′-TC). The −1 nucleobase preference is governed largely by a loop adjacent to the active site (loop 7), such that loop exchanges can convert one enzyme’s intrinsic preference into that of another (e.g., A3G with loop 7 from A3A becomes a 5′-TC preferring enzyme17). The −2 and +1 nucleobases relative to the target cytosine are also likely to be involved in local target selection but are less influential.
The APOBEC mutation signature in cancer has been defined as C-to-T and C-to-G base substitution mutations within 5′-TC dinucleotide motifs and most commonly within 5′-TCA and 5′-TCT trinucleotide contexts (also 5′-TCG if accounting for the genomic under-representation of this motif12,20). Unlike AID in antibody gene diversification and A3G in HIV-1 hypermutation, absolute assignments of cause and effect in cancer cannot be made based on intrinsic ssDNA deamination preferences alone because most APOBEC family members prefer substrates with 5′-TC motifs and the potential for additional specificity-conferring enzyme-nucleobase contacts is poorly understood. Thus, corroborating data sets including expression profiles, biochemical activities, global mutation analysis, animal experiments, and clinical information are being combined to deduce which subset of enzymes is actually responsible for the observed APOBEC signature mutations in cancer. The leading candidates to explain APOBEC mutagenesis in cancer are A3A21–23, A3B12,20,24, and A3H20. However, to date, only A3B has met all experimental criteria including clinical correlations between high expression levels and poor outcomes for multiple cancer types25–29.
Despite the importance of APOBEC3-mediated mutations in a variety of biological and pathological processes, the molecular mechanisms underlying global ssDNA binding activity and local target selectivity are poorly understood. In particular, prior structural studies have yielded numerous apo-enzyme structurese.g.,30–40 as well as two structures with non-substrate nucleotides37,41, but complexes with relevant ssDNA substrates have proven elusive. Here we solve crystal structures of human A3A and a variant of the human A3B catalytic domain in complex with optimal and commonly mutated ssDNA substrates, respectively. Together with supporting biochemical data and comparisons with prior apo-structures, our studies provide robust molecular explanations for why these enzymes selectively bind ssDNA and elicit strong preferences for cytosine nucleobases in specific 5′-TC contexts in viral and cancer genomes.
RESULTS
Optimal target for A3A-catalyzed C-to-U deamination in ssDNA
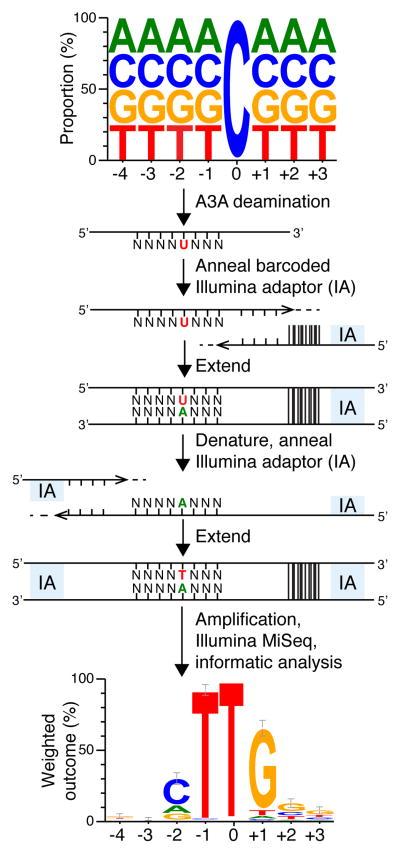
A3A is a globular enzyme with a single zinc-coordinating active site, and it has proven to be the most potent human DNA cytosine deaminase42,43. To elucidate the mechanism for binding ssDNA and preferring 5′-TC dinucleotide targets, we sought to solve the crystal structure of human A3A bound to ssDNA. Although prior studies have examined the next nucleotide preferences of this enzyme44,45, none have determined the extended substrate requirements in an unbiased manner. Therefore, first we determined the optimal substrate for wild-type A3A catalysis by performing deep deamination experiments with ssDNA containing a fixed target cytosine flanked on the 5′ and 3′ sides by 4 and 3 randomized bases, respectively (Fig. 1). Enzymatic reactions were allowed to proceed to ~10% completion (single hit kinetics) and, following conversion to double-stranded DNA with adaptors, deep-sequencing was used to determine the preferred bases flanking C-to-U deamination events (detected as C-to-T events). An analysis of >10,000 reads and 641 mutagenic events revealed a near-complete enrichment for T at the 5′ −1 position relative to the target C (0), a strong preference for a cytosine or purine nucleobase at the −2 position, and an unexpected preference for G at all 3′ positions (+1 to +3) (Fig. 1). This analysis informed the design of an optimal ssDNA substrate for co-crystallization studies, 5′-AAAAAAATCGGGAAA.
Figure 1. Deep-deamination approach to determine an optimal human A3A substrate.
A ssDNA library with a single target C and N’s on the 5′ and 3′ sides was reacted with human A3A (near-single hit kinetics). The resulting pool containing C-to-U deamination products was annealed to a bar-coded Illumina adaptor (IA) and T4 DNA polymerase was used to produce a complementary DNA strand. This intermediate was denatured, annealed to a 5′-IA, and converted to duplex by Phusion thermostable high fidelity DNA polymerase. Illumina Mi-Seq was used to generate reads for subsequent informatics analysis. A weblogo representation of deamination products unique to A3A shows enrichment for −1 T and +1 G that informed the ssDNA sequence for co-crystallization experiments (n=641; error bars are twice the sample correction value). Source data are provided as a spreadsheet (.xlsx) linked to the online legend.
A3A-ssDNA structure reveals a novel U-shaped binding conformation
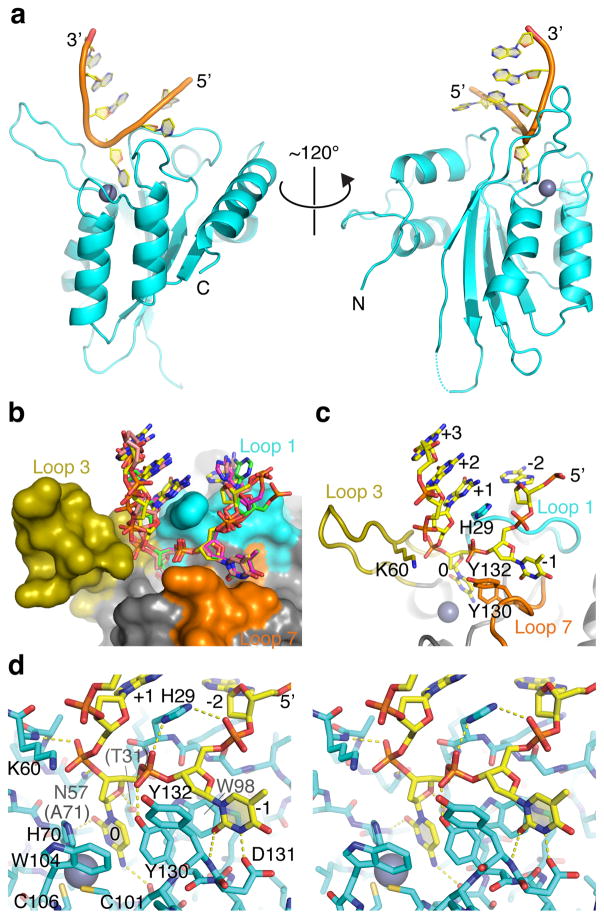
The optimal ssDNA substrate for A3A was co-crystalized with human A3A purified from E. coli. This protein represents the wild-type enzyme apart from a 4 residue C-terminal truncation to improve solubility and a single amino acid substitution of the catalytic glutamate (E72A) to prevent substrate turnover and bacterial genotoxicity (Supplementary Fig. 1). The 3.1 Å resolution A3A-ssDNA structure has 4 monomeric complexes in the asymmetric unit and each shows clear electron density for either 5 (5′-ATCGG) or 6 (5′-ATCGGG) nucleotides centered on the target cytosine (Table 1, Supplementary Fig. 2). A superposition shows near-identical conformations of all 4 proteins as well as the positioning of the −1 T and the target C (0) nucleotides and, as expected, some variation in the locations of bases outside the 5′-TC dinucleotide core (Supplementary Fig. 2).
Table 1.
Data collection and refinement statistics
| A3A-ssDNA (5SWW) | A3Bctd*-ssDNA (5TD5) | |
|---|---|---|
| Data collection | ||
| Space group | P2221 | P6422 |
| Cell dimensions | ||
| a, b, c (Å) | 90.15, 90.20, 167.26 | 96.41, 96.41, 84.88 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å) | 47.42–3.15(3.26–3.15)a | 59.52–1.72(1.78–1.72) |
| Rmerge (%) | 19.1 (112.1) | 5.4 (127.5) |
| Rmeas (%) | 21.1 (123.8) | 5.6 (133.2) |
| Rpim (%) | 8.9 (51.9) | 1.5 (37.8) |
| I/σ(I) | 8.4 (1.4) | 35.3 (2.1) |
| CC1/2 | 0.991 (0.693) | 0.999 (0.783) |
| Completeness (%) | 99.6 (99.0) | 99.7 (97.2) |
| Redundancy | 5.5 (5.6) | 12.7 (11.8) |
| Refinementb | ||
| Resolution (Å) | 47.42–3.15(3.26–3.15) | 59.52–1.72(1.78–1.72) |
| No. reflections | 24189 (2339) | 25281 (2417) |
| Rwork/Rfree (%) | 21.0(30.3)/26.3(35.1) | 18.1(31.9)/21.2(29.4) |
| No. atoms | 6597 | 1712 |
| Protein/DNA | 6553 | 1578 |
| Ligand/ion | 24/4 (GOL/Zn2+) | 24/1/9/2 (EG/Zn2+/I−/Cl−) |
| Water | 16 | 98 |
| B factors (Å2) | 73.54 | 45.97 |
| Protein/DNA | 73.58 | 45.24 |
| Ligand/ion | 82.77 | 76.46 |
| Water | 37.41 | 46.58 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.002 | 0.015 |
| Bond angles (°) | 0.45 | 1.20 |
Values in parentheses are for highest-resolution shell.
Each structure is from one crystal.
The A3A-bound ssDNA adopts a U-shaped conformation anchored by the target cytosine and the −1 thymine, with up- and down-stream ssDNA bent away from the active site (Fig. 2a). At the bottom of the ‘U’, the target cytosine and the 5′ thymine bases are flipped out toward the protein with the sugar-phosphate backbone rotated with respect to those of the flanking nucleotides (Fig. 2a–d, Supplementary Fig. 2). The two flipped-out nucleotides fit between loops 1 and 7 and are stabilized by extensive van der Waals contacts with Trp98 at the base of the groove and hydrogen bonds to backbone phosphates on the 5′ and 3′ sides of the target cytosine, respectively by the side chain of Tyr130 in loop 7 and Asn57 preceding loop 3 (Fig. 2b–d). Across the ssDNA-binding groove and opposite Tyr130, His29 from loop 1 fits inside the ‘U’ and donates hydrogen bonds to the backbone phosphates of both the target cytosine and 5′ thymine. The simultaneous hydrogen-bonding of His29 suggests that this side chain interacts with DNA optimally when doubly protonated, consistent with the reported pH-dependence of A3A and A3G ssDNA deamination activity46,47. The His29 side chain also stacks with the +1 base and makes van der Waals contacts with the nucleotide at −2 position, where the +1 and −2 bases may be close enough to interact. Thus, His29 appears to serve as a scaffold to stabilize ssDNA substrates in the ‘U-shaped’ conformation. The +2 and +3 bases linearly stack on the +1 base analogous to a B-form double-stranded DNA (Fig. 2b–c).
Figure 2. Crystal structure of human A3A bound to ssDNA with preferred 5′-TCG deamination target motif.
(a) Ribbon schematic of A3A-ssDNA complex showing flipped-out target C and −1 T nucleotides, as well as the overall U-shaped binding conformation. (b) Molecular surface of A3A active site with the surrounding loops color-coded, and a superposition of stick-models of ssDNA bound to 4 different molecules in the crystal’s asymmetric unit. (c) A view similar to panel b with a representative ssDNA molecule shown and nucleobases and key amino side chains from the active site loops labeled. (d) A wall-eye stereo view of the A3A active site and the bound ssDNA molecule shown in sticks. Hydrogen bonds are indicated by yellow dashed lines.
A3B-ssDNA structure and the mechanism of local target recognition
To improve crystallographic resolution and increase relevance to cancer, we next sought to solve a crystal structure of the human A3B catalytic domain bound to ssDNA. A3B is a nuclear-localizing enzyme strongly implicated in cancer mutagenesis including associations with poor clinical outcomes for estrogen receptor-positive breast cancer, multiple myeloma, and lung cancer25–29. A3B is double-domain enzyme with an N-terminal pseudo-catalytic domain and a C-terminal catalytic domain2,7,8. The catalytic domain has 92% amino acid sequence identity with A3A and the majority of differences occur in solvent-exposed surfaces including loop regions (Supplementary Fig. 1). We obtained crystals using a catalytic mutant derivative (E255A) of our previously described A3B variant with loop 1 from A3A and near wild-type activity (A3Bctd-QMΔloop3-A3Aloop1, hereafter called A3Bctd*)37 and a 7-mer ssDNA (5′-TTTTCAT) containing the most frequently mutated APOBEC motif in cancer (5′-TCA)13–15.
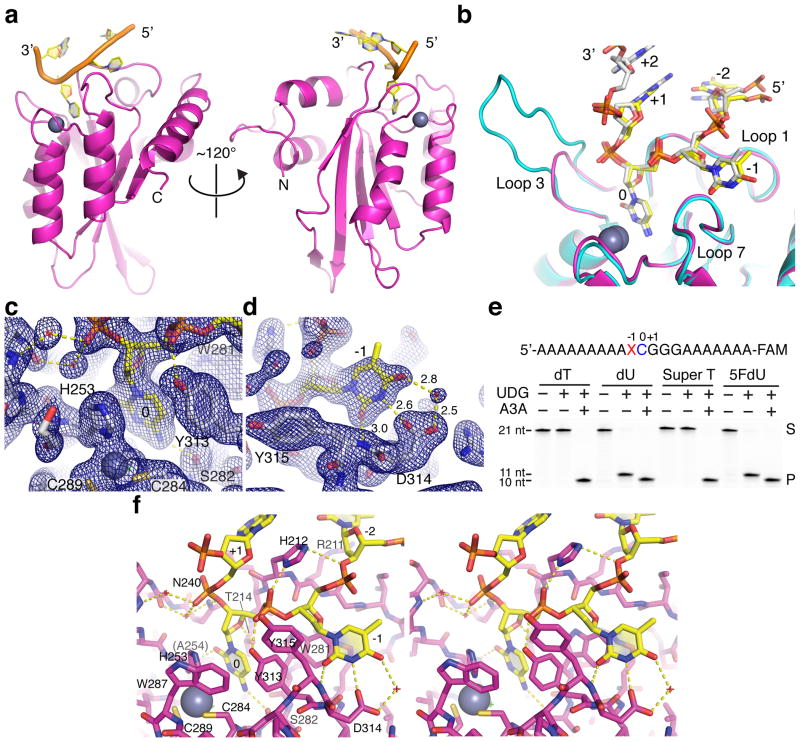
The 1.7 Å resolution structure of this A3Bctd*-ssDNA complex has a single nucleoprotein complex in the asymmetric unit and clear electron density for 4 nucleotides (5′-TTCA) (Fig. 3a, Table 1, Supplementary Fig. 3). Despite the different enzyme-ssDNA combination, the overall DNA conformation is also ‘U-shaped’ and highly similar to that in the DNA-bound A3A structure (Fig. 3b). The target cytosine is inserted deep into the active site pocket, which contains a zinc ion coordinated by His253, Cys284, and Cys289 (equivalent to A3A residues His70, Cys101, and Cys106; A3Bctd*-ssDNA vs. A3A-ssDNA superposition in Fig. 3b and A3Bctd* active site electron density in Fig. 3c). The amino group at the C4 position of the pyrimidine ring donates a hydrogen bond to the backbone carbonyl of Ser282 (Ser99) and is positioned to form the zinc-stabilized tetrahedral intermediate characteristic of the hydrolytic deamination reaction (Fig. 3c). The carbonyl oxygen of the cytosine base accepts a hydrogen bond from the backbone amide group of Ala254 (Ala71). The non-polar edge (C5 and C6) of the cytosine base makes a T-shaped π-stacking interaction with Tyr313 (Tyr130). Cytosine ring positioning is further stabilized by a hydrogen bond between the O4′ of the deoxyribose ring and Thr214 (Thr31) as well as nucleobase sandwiching between Thr214 (Thr31) and His253 (His70).
Figure 3. Crystal structure of a variant of the human A3B catalytic domain bound to ssDNA with a 5′-TCA deamination target motif.
(a) Ribbon schematic of ssDNA-A3Bctd* complex showing the flipped-out target C (0) and −1 T, as well as the overall U-shaped binding conformation. (b) A superposition of the active site region of A3A (cyan) and A3Bctd* (magenta) with relevant ssDNA substrates (opaque from a representative A3A structure and yellow from the A3Bctd* structure) showing the near-identical positioning of the flipped-out target C and −1 T. (c–d) Composite omit 2Fo-Fc map contoured at 1.0σ shown for region surrounding the target cytosine (panel c) or −1 thymine (panel d). (e) Deaminase activity of wild-type A3A on ssDNA substrates containing normal T or the indicated analogs at the −1 position demonstrating that the 5-methyl group is unconstrained structurally. Uracil DNA glycosylase (UDG) readily excises dU and 5FdU from ssDNA and accounts for the 11-nucleotide product in the absence of deamination. However, due to A3A activity on the target C and the 3′-end label, only the shorter 10-nucleotide product is apparent upon deamination and gel fractionation. The uncropped gel image is provided in Supplementary Data Set 1, and the results are representative of 2 independent experiments. (f) A wall-eye stereo view of the A3Bctd* active site and the bound ssDNA molecule shown in sticks. Hydrogen bonds are indicated by yellow dashed lines. Water molecules are represented by small red crosshairs.
The −1 thymine nucleotide fits in a hydrophobic pocket formed by Trp281, Tyr313, and Tyr315 (Trp98, Tyr130, and Tyr132), where the N3 and O2 atoms of the pyrimidine is hydrogen-bonded to the side chain of Asp314 (Asp131) and the backbone amide group of Tyr315 (Tyr132), respectively. These interactions explain the strong selectivity for the 5′-TC motif by A3B and A3A (Fig. 3b). Furthermore, the higher resolution A3Bctd*-ssDNA structure revealed a water-mediated interaction between Asp314 and the O4 carbonyl group of the thymine ring, which may be an additional selectivity factor (Fig. 3d). Interestingly, the 5-methyl group of the −1 T is pointed away from the protein and does not interact with the enzyme or the ssDNA substrate. Consistent with this observation, ssDNA substrates with 5-methyl substituted deoxy-thymidine analogs at −1 position were deaminated efficiently by A3A [2′-deoxyuridine (dU), 5-hydroxybutynl-2′-deoxyuridine (Super T), and 5-fluorodeoxyuridine (5FdU); Fig. 3e]. A3A has indistinguishable activities in dose response experiments with normal T versus Super T at the −1 position, which further validates the observed positioning of the −1 thymine in the bound ssDNA and shows that the methyl group does not contribute to the intrinsic 5′-TC target sequence preference (Supplementary Fig. 4). The stereoview provides a comprehensive visualization of all of these structural features (Fig. 3f).
Interactions with nucleotides flanking the 5′-TC target motif
Outside of the central 5′-TC motif, A3A and A3Bctd* make limited direct interactions with nucleobases of the bound ssDNA, consistent with the degeneracy of these positions in deep deamination experiments (Figs. 2 & 3). Arg28 in loop 1 of A3A interacts with an adenine at the −2 position, whereas the homologous Arg211 in A3Bctd* interacts with a thymine at the same position. This less specific base contact is consistent with A3A R28A and A3B R211A retaining robust ssDNA deaminase activity (data not shown and ref.37) and the occurrence of multiple bases at the −2 position in the A3A deep deamination reaction (Fig. 1). In addition, Lys30 in A3A is positioned close to the major groove edge of the +1 guanine base, potentially contributing to the preference for a purine at this position. The corresponding residue Gln213 of A3B may similarly account for the reported +1 adenine preference12, although our A3Bctd*-ssDNA complex could not show this potential contact due to a likely influence of crystal packing in the positioning of +1 adenine and the necessary engineering of loop 1 residues to facilitate crystallization of the A3Bctd*-ssDNA complex.
To probe whether interactions with any of the linearly stacked nucleobases at +1 to +3 positions are important in ssDNA engagement, we assayed A3A activity using normal versus 5-nitroindole substituted ssDNA substrates. 5-nitroindole is a universal base analog that stacks like a canonical nucleobase but lacks hydrogen-bonding capabilities. A3A has robust DNA cytosine deaminase activity with a ssDNA substrate containing 5-nitroindole substitutions for +1 to +3 positions, only about 2-fold lower than that with an optimal ssDNA substrate with deoxy-guanosines at the same positions (Supplementary Fig. 5). These data indicate that base stacking or hydrophobic interactions between the +1 to +3 nucleotides may be more relevant for the ssDNA deamination mechanism than nucleobase hydrogen-bonding with the enzyme.
The loop 3 region in both the A3A and A3Bctd* complexes makes either direct or water-mediated hydrogen bonds via the peptide main chain atoms with the backbone phosphate of +1 nucleotide (Fig. 2d, Fig. 3b & f). In addition, A3A loop 3 Lys60 points toward a mid-point between the backbone phosphate groups of +1 and +2 guanine nucleotides suggesting a stabilizing interaction, although the ζ amino group is not within direct hydrogen bonding distance to either phosphate. The 5′ and 3′ nucleotides farther from the target cytosine, 5′ of −1 and 3′ of +1 positions, are beyond bonding potential with the enzyme, which is consistent with earlier biochemical footprinting studies48, HIV-1 hypermutation experimentse.g.,49–51, and cancer mutation spectra analysese.g.,24,52,53. However, ssDNA lengths greater than 3 nucleotides appear to be required for full deaminase activity indicating that non-specific contacts may also be important54.
Comparisons of apo and ssDNA-bound structures
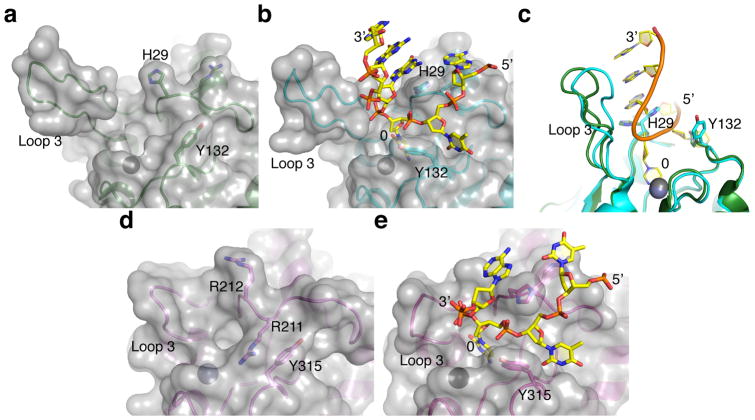
A comparison of the ssDNA-bound structures with previously reported A3A and A3Bctd crystal structures34,37 yields additional mechanistic insights into how the activities of these enzymes are likely to be regulated (Fig. 4). The conserved active site and zinc-coordinating residues are virtually unchanged, consistent with the strong preferences of these enzymes (and other family members) for normal (unmodified) cytosine nucleobases in ssDNA and indicating that the surrounding loop regions govern ssDNA binding activity and local dinucleotide targeting. Indeed, comparisons of the native and ssDNA-bound structures reveal large side chain reorientations for His29 and Tyr132 of A3A, and loop 1 rearrangement and Tyr315 reorientation in A3B. As noted above, A3A His29 provides multiple contacts that enable the ssDNA to adopt the ‘U-shaped’ conformation. The analogous histidine in the A3Bctd*-ssDNA structure has a similar scaffolding conformation, although it should be noted that this residue and most of loop 1 are derived from A3A (necessary alteration for crystallization purposes). Therefore, further structural and biochemical studies will be needed to fully understand the mechanism of ssDNA engagement by wild-type A3B, where one of the three arginines (Arg210, Arg211, or Arg212) in wild-type A3B loop 1 may have a stabilizing function analogous to A3A His29.
Figure 4. Comparison between apo and ssDNA-bound A3A and A3B structures.
(a–b) Molecular surface of A3A around the active site in the DNA-free (panel a, pdb 4XXO) and ssDNA-bound (panel b, this study) states, showing re-orientation of the side chains of His29 and Tyr132. (c) A superposition of the two conformations in panels a and b highlighting the repositioning of His29 and Tyr132 as well as shifting of loop 3 toward the bound ssDNA. (d–e) Molecular surface of A3Bctd* around the active site in the DNA-free (panel d, pdb 5CQH) and ssDNA-bound (panel e, this study) states showing the large transition that is likely to occur between the closed (unbound) and open (ssDNA-bound) conformations.
Importantly, the positioning of a conserved tyrosine in loop 7, Tyr132 and Tyr315, changes upon binding ssDNA in both the A3A and A3Bctd* structures (Fig. 4a–c & d–e for bound and unbound comparisons of A3A and A3B, respectively). For both enzymes, this reorientation is important to confer dinucleotide target specificity through extensive van der Waals contacts with the −1 thymine (above). Indeed, loop 7 swap experiments have shown that the residues including A3A Tyr132 are critical for determining the preference of the −1 nucleobase17. For example, the 5′-CC dinucleotide specificity of A3G skewed toward 5′-TC upon swapping the entirety of loop 7 or by changing Asp317 to a tyrosine in order to mimic this residue in A3A and A3B17. It is also notable that the Tyr315 reorientation converts a closed active site conformation of A3Bctd37 to the more open conformation required for binding ssDNA (Fig. 4d–e). The comparison between the apo and ssDNA-bound A3A structures also shows that loop 3, which is variable among APOBEC3 family members, swings toward the ssDNA and makes a backbone contact (Fig. 4c). However, both the apo and bound A3A structures have relatively open conformations, possibly accounting for the higher catalytic activity of this enzyme in comparison to A3B43,55.
Biochemical analyses corroborate the ssDNA-bound A3A and A3Bctd* structures
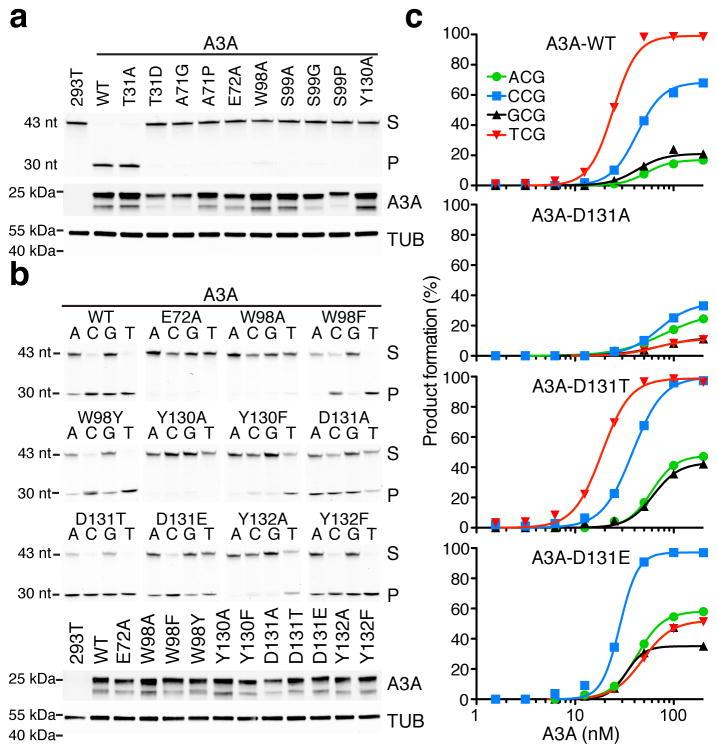
The overall ‘U-shaped’ ssDNA conformation and the positioning of the 5′-TC target dinucleotide are nearly identical between the two independent crystal structures (Fig. 3b). This superposition strongly implies that the observed ssDNA binding and local targeting mechanisms are accurate reflections of the biological and pathological activities of these enzymes in virus and cancer mutagenesis. To further validate these structural results, we compared wild-type A3A and structure-informed mutant derivatives in a series of biochemical experiments with extracts from 293T cells. Active site alanine mutants were inactive in deaminating a 5′-TC-containing ssDNA substrate, as expected, including those with substitutions of the catalytic glutamate (E72A), the zinc-coordinating histidine and cysteines (H70A, C101A, and C106A), and the tryptophan lining a side of the active site pocket (W98A) (upper panel in Fig. 5a and data not shown). The conserved cytosine-contacting residues, Ala71 (A71G, A71P), Ser99 (S99A, S99G, S99P), and Tyr130 (Y130A) also proved essential. The interaction between Thr31 and the 2′-deoxyribose of the target cytosine was only mildly compromised by an alanine substitution (T31A) and fully disrupted by the introduction of a negative charge (T31D), consistent with prior data indicating proximity of this residue to ssDNA56. All constructs expressed similarly in immunoblots indicating that the activity data are not due to poor expression or misfolding (lower panels in Fig. 5a).
Figure 5. Corroborating biochemical data for human A3A.
(a–b) DNA cytosine deamination by human A3A (WT, wild-type) and the indicated mutant derivatives (S, substrate; P, product). The corresponding anti-MYC (A3A) and anti-TUBULIN immunoblots indicate similar levels of A3A and soluble extract in each experiment relative to controls (A3A has two bands due to alternative translation initiation from Met1 or Met13). Reactions in panel a interrogated active site mutants using a 43-nucleotide 5′-TC-containing ssDNA substrate, and reactions in panel b additionally interrogated the identity of the −1 position (A, C, G, or T) relative to the target cytosine. These results are representative of 2 independent experiments, and uncropped gel images are provided in Supplementary Data Set 1. (c) Graphs quantifying product accumulation in dose response experiments for A3A and the indicated D131 derivatives using the same ssDNA substrates as panel b. WT enzyme and D131T show similar activities and local −1 nucleobase preferences, D131A has low activity and relaxed preferences, and D131E has slightly lower activity and a clear preference for −1 C instead of −1 T. These data are representative of 2 independent experiments, and source data are provided as a spreadsheet (.xlsx) linked to the online legend.
Additional ssDNA deamination experiments were done to interrogate A3A contacts with the specificity-conferring −1 thymine nucleobase (upper panel in Fig. 5b). These experiments were performed as above except both activity and selectivity were examined in parallel by systematically varying the −1 position of the ssDNA substrate. As controls, wild-type A3A has the most activity with ssDNA substrates with −1 T, intermediate activity with −1 C, and little activity with −1 A or G, whereas A3A-E72A has no activity. A comparison of Trp98 substitutions indicated that an aromatic residue is sufficient at this position in the structure to stabilize −1 T and the target C, as the W98A mutant was inactive but W98F and W98Y substitution mutants retained robust catalytic activity and near wild-type dinucleotide preferences. The aromatic character of Tyr132 is similarly important in forming the hydrophobic pocket for −1 T, based on near wild-type activity for Y132F but not Y132A constructs. We observed parallel, albeit more severe, effects of phenylalanine and alanine substitutions for Tyr130. The overall greater importance of an aromatic side chain at position 130 is further indicated by contacts with the target C and the ssDNA backbone as well as van der Waals interactions with Tyr132, which help to position the −1 T (Fig. 2c–d). As alluded above, Asp131 strongly influences the −1 preference, with a small non-polar alanine substitution (D131A) loosening selectivity and showing near-equivalent activity with −1 T and −1 C substrates, a shorter hydroxylated residue (D131T) retaining selectivity for −1 T (likely by mimicking the hydrogen bond acceptor role of the aspartate), and a longer and acidic glutamate substitution (D131E) converting the preference at −1 position to C (most likely by creating an opportunity for direct hydrogen-bonding with the amino group of the cytosine ring and simultaneously disrupting the hydrogen-bonding between carboxyl group of the shorter aspartate side chain and the N3 hydrogen of thymine). As above, the mutants expressed similarly in immunoblots with few exceptions indicating that the activity data are not due to poor expression or misfolding (lower panels in Fig. 5b). The D131 results confirmed in quantitative dose response experiments with purified A3A and single amino acid substitution derivatives (Fig. 5c). Overall, these biochemical data strongly support the observed conformation of 5′-TC-containing ssDNA bound to A3A in the crystal structure.
DISCUSSION
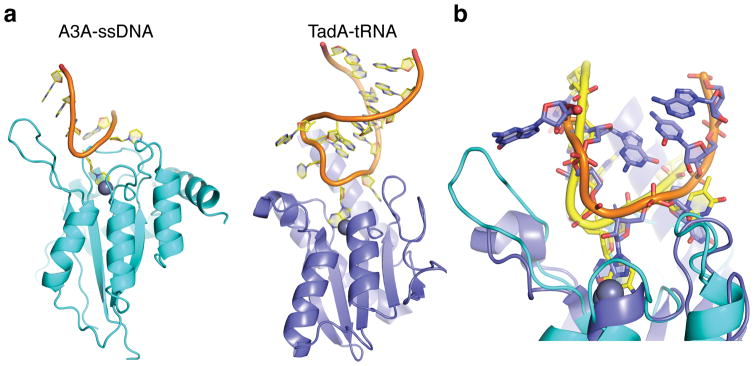
The ‘U-shaped’ ssDNA-binding mechanism, as revealed by co-crystal structures and validated through biochemical analyses, suggests that the loop regions of DNA stem-loop structures may be hotspots for APOBEC mutagenesis. Indeed, loop regions in HIV-1 cDNA have been shown to be preferred sites for A3G-mediated DNA deamination57, and a recent study of 560 breast cancer genomes reported multiple hotspots for APOBEC signature mutations within loop regions of predicated stem-loop structures58. The ‘U-shaped’ ssDNA binding conformation of A3A and A3Bctd* resembles the conformation of RNA bound to the distantly related tRNA adenosine deaminase TadA (Fig. 6). Additional structural studies will be needed to determine whether the common ssDNA-binding mechanism observed here for A3A and A3Bctd* represents that of wild-type A3B with natural loop 1 residues or that of other APOBEC3 family members including A3G. Nonetheless, the similarity between these nucleic acid binding conformations suggests that hairpin or hairpin-like ssDNA or RNA structures may be preferred substrates for many different polynucleotide deaminase family members. However, the vast majority of viral and genomic ssDNA APOBEC mutations are not found in predicted secondary structures and, instead, correlate with properties of DNA replication (single-stranded cDNA in retroviral reverse transcription and lagging-strand DNA in tumor cells) suggesting that the most critical feature may be simply single-strandedness13–15. The mechanism of binding ssDNA by human A3A and A3Bctd* contrasts with prior models including our own30,31,35,37,40, the conformation of a short oligo-dT co-crystalized with A3Gntd41 (Supplementary Fig. 6), and the mechanism of double-stranded RNA binding and adenosine deamination by ADAR259.
Figure 6. Human A3A and S. aureus TadA have similar U-shaped polynucleotide binding conformations.
(a) Ribbon schematics of A3A-ssDNA (this study) and TadA-tRNA (pdb 2B3J61) with the single zinc-coordinating active site regions positioned at similar angles for comparison. (b) Superposition of A3A-ssDNA and TadA-tRNA structures showing similar U-shaped binding conformations (predicted in a commentary62 to the original TadA-tRNA structure report61). A3A-bound ssDNA is shown in yellow and TadA-bound RNA is shown with an orange backbone and magenta nucleobases.
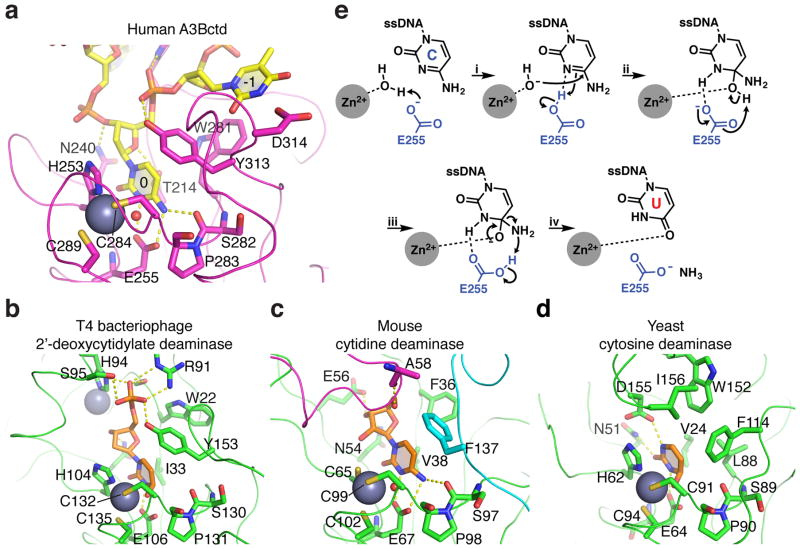
Residues within the A3A and A3B active site pockets are highly conserved within the APOBEC3 family, as evidenced by a close superpositioning of the active site region of crystal structures of several human APOBEC3 enzymes (A3A, A3B, A3C, A3F, and A3G) (Supplementary Fig. 7). The only minor exception is Thr31 of A3A (Thr214 in A3B), which is a Ser216 in A3F. The T31A substitution of A3A is well tolerated but T31D abolishes deaminase activity, consistent with a potential for phospho-regulation at this position (Fig. 5a and refs.56,60). The corresponding residue in more distantly related metabolic cytosine deaminases, which likely catalyze the same hydrolytic chemistry of the cytosine deamination reaction as APOBEC3 enzymes, is either valine or isoleucine and makes van der Waals contact with the target cytosine (Fig. 7). These enzymes also show interesting variations of the residues surrounding the target cytosine base, reflecting substrate-specific interactions. For instance, the aromatic residue stacked over the target cytosine corresponding to Tyr130 of A3A (Tyr313 of A3B) is tyrosine in the 2′-deoxycytidine-5′-monophosphate deaminase of bacteriophage T4, whereas it is phenylalanine in the nucleoside cytidine deaminase and the free cytosine deaminase, consistent with the lack of a 5′-phosphate group.
Figure 7. Structural comparison of the active sites of A3B and distantly related deaminase family members.
Active sites of (a) A3Bctd bound to ssDNA, (b) T4 bacteriophage 2′-deoxycytidylate deaminase bound to a dCMP analog (pdb 1VQ263), (c) murine cytidine deaminase bound to cytidine (pdb 2FR664), and (d) yeast cytosine deaminase bound to an analog of the free nucleobase cytosine (pdb 1P6O65). The catalytic glutamate (Glu255) was modeled into the A3Bctd*-ssDNA structure based on its positioning in the apo structure (pdb 5CQH), closely mimicking conformations of the corresponding residues in the T4, mouse, and yeast enzymes (Glu106, Glu67, and Glu64, respectively). Gray spheres represent zinc ions. Smaller red spheres show the zinc-bound reactive water molecule. Sticks depict key residues contacting bound substrates, and ribbons represent protein backbones. The magenta loop in panel c is from an adjacent subunit in the tetramer. A comparison of the structures shows similar zinc-coordination mechanisms and target cytosine positioning including conservation of surrounding aromatic residues, as well as substrate-specific interactions conferred by unique residues for each class of enzyme. (e) Proposed deamination mechanism (adapted from ref.66): (i) Glu255 deprotonates the Zn2+-coordinated H2O for nucleophilic attack at cytosine with the residual, protonated Glu255 H-bonding to N3 to withdraw electron density from C4 of cytosine, thereby accelerating nucleophilic attack by hydroxide. (ii) Deprotonation of the alcohol on the tetrahedral intermediate by Glu255 ensues, followed by (iii) collapse of the tetrahedral intermediates due to elimination of ammonia, to which protonated Glu255 contributes a hydrogen. (iv) The resulting uracil base is likely stabilized by Zn2+ coordination in the enzyme active site.
In contrast to the strict conservation of the catalytic residues in the APOBEC3 active sites, comparisons of the amino acid sequences and conformations of the loops 1, 3, and 7 suggest that adjacent contacts with ssDNA substrates are either conserved or diverged amongst APOBEC3 family members1,2 (Supplementary Fig. 7). The conservation accounts for the strong preference of the APOBEC3 enzymes for ssDNA, and the divergence provides flexibility to evolve varying catalytic efficiencies and local sequence preferences in order to achieve overlapping but distinct functions in innate immunity. For instance, the closed active site conformation of A3Bctd in the ground state37, in spite of the similar modes of ssDNA interaction between A3A and A3Bctd*, likely reflects a need for a tight regulation of this enzyme’s activity in the nucleus. Given the clear roles of the APOBEC3 enzymes in virus and tumor evolution, most recently demonstrated for A3B driving drug resistance in breast cancer26, the A3A-ssDNA and A3Bctd*-ssDNA crystal structures are expected to provide a foundation for rational design of small molecule inhibitors to impede virus and tumor evolvability.
ONLINE METHODS
Protein purification
Human A3A with single amino acid substitution (E72A) was expressed in the E. coli strain BL21(DE3) as a GST-fusion protein using the pGEX6P-1 vector. Transformed bacteria were grown to mid-log phase in LB medium, then supplemented with 100 μM ZnCl2, and induced by addition of IPTG at a final concentration of 0.5 mM. After overnight incubation at 18°C, bacteria were collected by centrifugation, resuspended in lysis buffer (20 mM Tris-HCl pH 8.0, 0.5 M NaCl, 10 mM β-mercaptoethanol) and lysed by the addition of lysozyme and sonication. After centrifugation, the cleared lysate was passed through a 0.22 μm filter, and the protein was captured with glutathione agarose resin (Pierce), eluted in the lysis buffer supplemented with 10 mM reduced glutathione, cleaved by the human rhinovirus 3C protease to remove GST, and purified over a Superdex75 (GE Healthcare) size-exclusion column. The purified protein contains A3A residues 1–195 (near full-length) and additional vector-derived residues (GPLGSPEF) on the N-terminus. The A3B construct used in this study was the previously reported A3Bctd-QMΔloop3-A3Aloop1, which has substitution of the A3A loop1 residues (GIGRHK) for A3B loop 1 (DPLVLRRRQ) and single serine for the A3B loop 3 residues (spanning Ala242 to Tyr250)37. A3Bctd-QMΔloop3-A3Aloop1 with the additional E255A amino acid substitution (referred to as A3Bctd*) was expressed with a non-cleavable C-terminal His6-tag (LEHHHHHH) in the E. coli strain C41(DE3)pLysS (Lucigen) from a pET24a-based vector and purified as reported37. For both proteins the final size-exclusion chromatography buffer consisted of 20 mM Tris-HCl (pH 7.4), 0.2 M NaCl, and 0.5 mM TCEP. The proteins were >95% pure based on visual inspection of Coomassie Blue-stained polyacrylamide gels. The purified enzymes were concentrated by ultrafiltration for use in the crystallization experiments. The initial A3A and A3B protein sequences match UniProt ID P31941 and Q9UH17, respectively (aligned in Supplementary Fig. 1).
Crystallization and structure determination
Purified A3A and A3Bctd* were mixed with ~1.5 molar excess ssDNA (see below) at a final protein concentration of 25–30 mg ml−1. The A3A-ssDNA complex crystal was obtained in a sitting drop formed by mixing the complex solution with an equal volume of 0.2 M NaF, 20 %(w/v) PEG3350, 0.1 M Bis-Tris propane (pH 6.5), equilibrated via vapor diffusion against the same reservoir solution. The A3Bctd*-ssDNA crystal was obtained similarly, using a reservoir solution consisting of 0.45 M NaI and 20 %(w/v) PEG3350. The crystals were mounted directly from the Intelli-Plate 96 (Art Robbins Instruments) and flash cooled in liquid nitrogen using glycerol or ethylene glycol as cryo-protectant. X-ray diffraction data were collected at the Advanced Photon Source Northeastern Collaborative Access Team beamline 24-ID-C using the selenium K-absorption edge wavelength, and the data were processed with XDS67. The A3A-DNA complex crystal is in the space group of P2221. A Matthews coefficient calculation indicated that there would likely be 4 monomers of A3A in an asymmetric unit. Using monomeric A3A (pdb 4XXO36) as a search model, the molecular replacement calculations by PHASER68 located four copies of A3A in the asymmetric unit. The resulting electron density map clearly showed the presence of ssDNA bound to each A3A molecule (Supplementary Fig. 2). The A3Bctd*-DNA complex crystal is in the space group of P6422 with one monomer in the asymmetric unit and diffracted to ~1.7 Å resolution. As the crystallization condition contained a high concentration of iodide ions, the diffraction data showed strong anomalous signal, and a total of 6 iodine or zinc sites were located using SHELXD69. The resulting single wavelength anomalous dispersion (SAD)-phased electron density map, after density modification using SHELXE69, showed A3B monomer and the presence of a bound ssDNA molecule. The A3Bctd monomer37 (pdb 5CQK) was placed into the electron density by molecular replacement using MOLREP70. Subsequent iterative refinement with PHENIX suite71 and manual model inspection and rebuilding using COOT72 resulted in the final Rwork/Rfree of 20.97/26.30% and 18.11/21.21% for A3A-DNA and A3Bctd*-DNA complexes, respectively. Each A3A or A3Bctd* monomer is bound to one molecule of ssDNA substrate. The summary of x-ray data collection and model refinement statistics is shown in Table 1. Ramachandran analysis shows that 99.5, 0.5, and 0% of the protein residues are in the most favored, allowed, and disallowed region for A3A-ssDNA complex structure and 98.3, 1.7, and 0% for A3Bctd*-ssDNA complex, respectively. The molecular graphics images were produced using PYMOL (www.pymol.org).
Single-stranded DNA (ssDNA) oligonucleotides
The ssDNA oligonucleotides for co-crystallization with A3A and A3Bctd* were 5′-AAAAAAATCGGGAAA and 5′-TTTTCAT, respectively (Integrated DNA Technologies). The unbiased experimental approach for identifying the former sequence is described below, and the latter is based on the fact that 5′-TCA is the most commonly mutated APOBEC signature motif in cancer13–15. 3′-fluorescently labeled ssDNA oligonucleotides for in vitro DNA deamination experiments were obtained from Integrated DNA Technologies or Midland Certified Reagent Company. The ssDNA oligonucleotide substrates used in Fig. 3e are 5′-AAAAAAAAATCGGGAAAAAAA-3′-FAM, 5′-AAAAAAAAA[dU]CGGGAAAAAAA-3′-FAM, 5′-AAAAAAAAA[5-F-dU]CGGGAAAAAAA-3′-FAM, and 5′-AAAAAAAAA[iSuper-dT]CGGGAAAAAAA-3′-FAM (labeled dT, dU, Super T, and 5FdU, respectively). The ssDNA oligonucleotide substrate used in Fig. 5a is 5′-ATTATTATTATTCAAATGGATTTATTTATTTATTTATTTATTT-3′-FAM, the same except (A, C, G, or T)CA as trinucleotide targets in Fig. 5b. 5-Nitroindole containing ssDNA substrates were purchased from Integrated DNA Technologies (Coralville, IA) as desalted oligonucleotides and characterized by LC-MS on an Agilent 1100 series HPLC instrument equipped an Agilent MSD SL ion trap mass spectrometer. A full list of ssDNA oligonucleotides used in site-directed mutation is available upon request.
Deep deamination experiments to determine an optimal A3A target site
We reasoned that deep sequencing of a target ssDNA oligonucleotide with a single cytosine flanked by degenerate Watson-Crick nucleobases could be used to determine an optimal A3A target site. First, a ssDNA substrate oligonucleotide 5′-NNNNCNNN flanked by cytosine-free 22 and 21 nucleotide regions was synthesized (Integrated DNA Technologies). This yielded a pool with 47 unique substrate sequences. Second, wild-type human A3A was purified from semi-confluent 293T cells transfected with a pcDNA4/TO-A3Ai-2xStrep3xFlag (SF) expression vector20. Cells were harvested 48 hours post-transfection and lysed in 50 mM Tris-HCl pH 8.0, 1% (v/v) NP-40, 150 mM NaCl, 0.5% (w/v) deoxycholate, 0.1% (w/v) SDS, 5 mM EDTA, RNase A, 1× EDTA-free Protease Inhibitor Cocktail (Roche), and then further disrupted by sonication. A3A-SF was purified using Strep-tactin resin (IBA). Samples were washed in high salt buffer (20 mM Tris-HCl pH 7.5, 1.5 mM MgCl2, 1 M NaCl, 0.5 mM DTT and 5% glycerol) followed by low salt buffer (same as the high salt buffer except with 150 mM NaCl) and final wash buffer (100 mM Tris-HCl pH 7.5, 150 mM NaCl) and eluted using 2.5 mM desthiobiotin. Purified protein was fractionated using 4–20% SDS-PAGE and quantified by staining with Coomassie brilliant blue (Sigma).
Third, titration experiments were conducted with recombinant A3A-SF to determine single-hit reaction conditions. This amount of enzyme was then incubated with 8 pmol of the substrate oligonucleotide pool for 1 hour at 37 °C in 50 mM Tris pH 7.5, 75 mM NaCl. 2 pmol of the treated pool was annealed to appropriate 3′-barcoded adaptor using T4 DNA Polymerase at 12°C for 20 minutes. Then a universal 3′-adapter was added to the duplex using Phusion DNA polymerase (New England Biolabs, Ipswich, MA). Products were purified using a GeneJET PCR purification kit (ThermoFisher Scientific), analyzed by 20% native PAGE, and diluted to appropriate concentrations for deep sequencing.
The reaction products were analyzed using 2 × 50 nt paired-end reads (Illumina HiSeq 2500, University of Minnesota Genomics Center). Reads were paired using FLASh (http://ccb.jhu.edu/software/FLASH/). Data processing was performed using a locally installed FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). FASTX trimmer was used to trim the 5′ and 3′ constant regions from sequences. Trimmed sequences were then filtered for high-quality reads using FASTQ quality filter. Sequences with a Phred quality score less than 30 (99.9% base calling accuracy) at any position were eliminated. Pre-processed sequences were then further analyzed using the FASTAptamer toolkit (http://burkelab.missouri.edu/fastaptamer.html). FASTAptamer-Count was used to count the number of times each sequence was sampled from the population. Each sequence was then ranked and sorted based on overall abundance, normalized to the total number of reads in each population, and directed into FASTAptamer-Enrich. FASTAptamer-Enrich calculates the fold-enrichment ratios from a starting population (no enzyme control) to a selected population (incubation with A3A or other enzymes). After generating the enrichment file, mutated sequences specific to the A3A reaction versus control reactions were analyzed using WebLogo design software73. Logo error bars are twice the sample correction value74.
A3A expression constructs and DNA deamination activity assays
The pcDNA3.1-A3Ai-myc-His expression construct is reported11,33,42. Derivatives were constructed by site-directed mutagenesis and verified by DNA sequencing. The activities of a subset of the A3A mutant constructs reported here have been described previously35,40,43–45,54,55,75,76. Semi-confluent 293T cells in 6 well plates were transfected with 1 μg plasmid and harvested after 48 hours to allow time for enzyme expression. Soluble whole cell extracts (WCE) were prepared by pelleting the cells and resuspending them in HED buffer (20 mM HEPES pH 7.4, 5 mM EDTA, 100 μg ml−1 RNase A, 1 mM DTT, 10% glycerol, and Roche Complete protease inhibitors). Resuspended cell pellets were freeze-thawed then rotated for 1 hour at room temperature followed by water bath sonication for 20 minutes. Cell debris was pelleted and the clarified lysate was used for DNA deaminase activity assays. 5 μl WCE with desired A3A construct (or control) were mixed with 5 μl HED buffer containing 1.6 μM fluorescently labeled ssDNA (sequences above). Reactions were then allowed to progress for 1 hour at 37°C, followed by treatment with 120 nM recombinant human UNG2 (uracil DNA glycosylase) for 10 minutes at 37°C, and treatment with 100 mM NaOH for 10 minutes at 95°C33,42. Reaction products were separated by 15% denaturing PAGE and scanned on a Typhoon FLA 7000 imager (GE Healthcare). A3A-myc-His expression was verified by immunoblotting using primary rabbit anti-cMYC antibody at 1:3000 (Sigma, C3956) and secondary goat anti-rabbit IgG-Alexa Fluor 680 at 1:10,000 (Life Technologies, A21076). Tubulin expression was used as a loading control, and staining was done with primary mouse anti alpha-Tubulin at 1:80,000 (Sigma, B512) followed by secondary goat anti-mouse IR-Dye 800CW at 1:10,000 (LI-COR, 926-32210). Washed immunoblots were imaged using a LI-COR Odyssey imaging system.
Recombinant A3A-myc-His and mutant derivatives were expressed in 293T cells and purified as described by nickel affinity chromatography11,33,42. Activity assays were conducted as outlined above except reactions were initiated by adding 5 μl of enzyme (2-fold dilutions starting at 200 nM) to 5 μl of 50 mM NaCl, 10 mM HEPES buffer (pH 7.4) containing 1.6 μM fluorescently labeled ssDNA (sequences above). Reactions progressed 1 hour at 37 °C and processed and quantified as described above.
Data availability
The coordinates and structure factors for the A3A-ssDNA and A3Bctd*-ssDNA complexes have been deposited in the Protein Data Bank under accession codes 5SWW and 5TD5, respectively. Source data for Figs. 1 & 3c are available in the online version of the paper. Full gel images of biochemical and immunoblot results in Figs. 3e & 5a–b are shown in Supplementary Data Set 1. Other data are available upon request.
Supplementary Material
Acknowledgments
We thank D. Largaespada and D. Yee for insightful comments, R. Moorthy for oligo sample preparations, and J. Stivers for providing the human UNG2 expression construct and purification protocol. This work was supported by grants from the US National Institutes of Health (NIGMS R01-GM118000 to RSH and HA, NIGMS R35-GM118047 to HA, NIGMS R01-GM110129 to DAH, NCI R21-CA206309 to RSH, DP2-OD007237 and NIGMS P41-GM103426 to REA), the NSF (CHE060073N to REA), the Prospect Creek Foundation (RSH and DAH), and the University of Minnesota Masonic Cancer Center (Spore-Program-Project-Planning Seed Grant to RSH). This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the US National Institutes of Health (NIGMS P41-GM103403). The Pilatus 6M detector on 24-ID-C beamline is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. RSH is the Margaret Harvey Schering Land Grant Chair for Cancer Research and an Investigator of the Howard Hughes Medical Institute.
Footnotes
Accession codes. The coordinates and structure factors for the A3A-ssDNA and A3Bctd*-ssDNA complexes have been deposited in the Protein Data Bank under accession codes 5SWW and 5TD5, respectively.
AUTHOR CONTRIBUTIONS
RSH and HA conceived and designed the studies. KS, NMS, KK, JD, and HA purified proteins and established crystallization conditions. SB collected x-ray diffraction data. KS solved the crystal structures. MAC, DJS, JLM, and GJS performed the deep-deamination studies. MAC performed biochemical experiments. DAH designed modified DNA substrates. OD and REA provided computational and structural insights. KS, MAC, NMS, RSH, and HA drafted the manuscript, and all authors contributed to revisions and figure preparation.
COMPETING FINANCIAL INTERESTS
RSH and DAH are co-founders, shareholders, and consultants of ApoGen Biotechnologies Inc. HA and REA are consultants for ApoGen Biotechnologies Inc. REA is a co-founder of Actavalon Inc. The other authors have no competing financial interests to declare.
BIBLIOGRAPHY
- 1.Conticello SG. The AID/APOBEC family of nucleic acid mutators. Genome Biol. 2008;9:229. doi: 10.1186/gb-2008-9-6-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harris RS, Dudley JP. APOBECs and virus restriction. Virology. 2015;479–480C:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Noia JM, Neuberger MS. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem. 2007;76:1–22. doi: 10.1146/annurev.biochem.76.061705.090740. [DOI] [PubMed] [Google Scholar]
- 4.Robbiani DF, Nussenzweig MC. Chromosome translocation, B cell lymphoma, and activation-induced cytidine deaminase. Annu Rev Pathol. 2013;8:79–103. doi: 10.1146/annurev-pathol-020712-164004. [DOI] [PubMed] [Google Scholar]
- 5.Fossat N, Tam PP. Re-editing the paradigm of cytidine (C) to uridine (U) RNA editing. RNA Biol. 2014;11:1233–1237. doi: 10.1080/15476286.2014.996054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koito A, Ikeda T. Intrinsic immunity against retrotransposons by APOBEC cytidine deaminases. Front Microbiol. 2013;4:28. doi: 10.3389/fmicb.2013.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malim MH, Bieniasz PD. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med. 2012;2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simon V, Bloch N, Landau NR. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol. 2015;16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Refsland EW, Stenglein MD, Shindo K, Albin JS, Brown WL, Harris RS. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 2010;38:4274–4284. doi: 10.1093/nar/gkq174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koning FA, Newman EN, Kim EY, Kunstman KJ, Wolinsky SM, Malim MH. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J Virol. 2009;83:9474–9485. doi: 10.1128/JVI.01089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stenglein MD, Burns MB, Li M, Lengyel J, Harris RS. APOBEC3 proteins mediate the clearance of foreign DNA from human cells. Nat Struct Mol Biol. 2010;17:222–229. doi: 10.1038/nsmb.1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burns MB, Lackey L, Carpenter MA, Rathore A, Land AM, Leonard B, Refsland EW, Kotandeniya D, Tretyakova N, Nikas JB, Yee D, Temiz NA, Donohue DE, McDougle RM, Brown WL, Law EK, Harris RS. APOBEC3B is an enzymatic source of mutation in breast cancer. Nature. 2013;494:366–370. doi: 10.1038/nature11881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Helleday T, Eshtad S, Nik-Zainal S. Mechanisms underlying mutational signatures in human cancers. Nat Rev Genet. 2014;15:585–598. doi: 10.1038/nrg3729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14:786–800. doi: 10.1038/nrc3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swanton C, McGranahan N, Starrett GJ, Harris RS. APOBEC enzymes: mutagenic fuel for cancer evolution and heterogeneity. Cancer Discov. 2015;5:704–712. doi: 10.1158/2159-8290.CD-15-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carpenter MA, Rajagurubandara E, Wijesinghe P, Bhagwat AS. Determinants of sequence-specificity within human AID and APOBEC3G. DNA Repair (Amst) 2010;9:579–587. doi: 10.1016/j.dnarep.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rathore A, Carpenter MA, Demir Ö, Ikeda T, Li M, Shaban NM, Law EK, Anokhin D, Brown WL, Amaro RE, Harris RS. The local dinucleotide preference of APOBEC3G can be altered from 5′-CC to 5′-TC by a single amino acid substitution. J Mol Biol. 2013;425:4442–4454. doi: 10.1016/j.jmb.2013.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohli RM, Maul RW, Guminski AF, McClure RL, Gajula KS, Saribasak H, McMahon MA, Siliciano RF, Gearhart PJ, Stivers JT. Local sequence targeting in the AID/APOBEC family differentially impacts retroviral restriction and antibody diversification. J Biol Chem. 2010;285:40956–40964. doi: 10.1074/jbc.M110.177402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Rada C, Neuberger MS. Altering the spectrum of immunoglobulin V gene somatic hypermutation by modifying the active site of AID. J Exp Med. 2010;207:141–153. doi: 10.1084/jem.20092238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Starrett GJ, Luengas EM, McCann JL, Ebrahimi D, Temiz NA, Love RP, Feng Y, Adolph MB, Chelico L, Law EK, Carpenter MA, Harris RS. The DNA cytosine deaminase APOBEC3H haplotype I likely contributes to breast and lung cancer mutagenesis. Nat Commun. 2016;7:12918. doi: 10.1038/ncomms12918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caval V, Suspene R, Shapira M, Vartanian JP, Wain-Hobson S. A prevalent cancer susceptibility APOBEC3A hybrid allele bearing APOBEC3B 3′UTR enhances chromosomal DNA damage. Nat Commun. 2014;5:5129. doi: 10.1038/ncomms6129. [DOI] [PubMed] [Google Scholar]
- 22.Chan K, Roberts SA, Klimczak LJ, Sterling JF, Saini N, Malc EP, Kim J, Kwiatkowski DJ, Fargo DC, Mieczkowski PA, Getz G, Gordenin DA. An APOBEC3A hypermutation signature is distinguishable from the signature of background mutagenesis by APOBEC3B in human cancers. Nat Genet. 2015;47:1067–1072. doi: 10.1038/ng.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nik-Zainal S, Wedge DC, Alexandrov LB, Petljak M, Butler AP, Bolli N, Davies HR, Knappskog S, Martin S, Papaemmanuil E, Ramakrishna M, Shlien A, Simonic I, Xue Y, Tyler-Smith C, Campbell PJ, Stratton MR. Association of a germline copy number polymorphism of APOBEC3A and APOBEC3B with burden of putative APOBEC-dependent mutations in breast cancer. Nat Genet. 2014;46:487–491. doi: 10.1038/ng.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Burns MB, Temiz NA, Harris RS. Evidence for APOBEC3B mutagenesis in multiple human cancers. Nat Genet. 2013;45:977–983. doi: 10.1038/ng.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sieuwerts AM, Willis S, Burns MB, Look MP, Meijer-Van Gelder ME, Schlicker A, Heideman MR, Jacobs H, Wessels L, Leyland-Jones B, Gray KP, Foekens JA, Harris RS, Martens JW. Elevated APOBEC3B correlates with poor outcomes for estrogen-receptor-positive breast cancers. Horm Cancer. 2014;5:405–413. doi: 10.1007/s12672-014-0196-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Law EK, Sieuwerts AM, LaPara K, Leonard B, Starrett GJ, Molan AM, Temiz NA, Vogel RI, Meijer-van Gelder ME, Sweep FC, Span PN, Foekens JA, Martens JW, Yee D, Harris RS. The DNA cytosine deaminase APOBEC3B promotes tamoxifen resistance in ER-positive breast cancer. Sci Adv. 2016;2:e1601737. doi: 10.1126/sciadv.1601737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cescon DW, Haibe-Kains B, Mak TW. APOBEC3B expression in breast cancer reflects cellular proliferation, while a deletion polymorphism is associated with immune activation. Proc Natl Acad Sci U S A. 2015;112:2841–2846. doi: 10.1073/pnas.1424869112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan S, He F, Gao B, Wu H, Li M, Huang L, Liang J, Wu Q, Li Y. Increased APOBEC3B predicts worse outcomes in lung cancer: a comprehensive retrospective study. J Cancer. 2016;7:618–625. doi: 10.7150/jca.14030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker BA, Boyle EM, Wardell CP, Murison A, Begum DB, Dahir NM, Proszek PZ, Johnson DC, Kaiser MF, Melchor L, Aronson LI, Scales M, Pawlyn C, Mirabella F, Jones JR, Brioli A, Mikulasova A, Cairns DA, Gregory WM, Quartilho A, Drayson MT, Russell N, Cook G, Jackson GH, Leleu X, et al. Mutational spectrum, copy number changes, and outcome: results of a sequencing study of patients with newly diagnosed myeloma. J Clin Oncol. 2015;33:3911–3920. doi: 10.1200/JCO.2014.59.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen KM, Harjes E, Gross PJ, Fahmy A, Lu Y, Shindo K, Harris RS, Matsuo H. Structure of the DNA deaminase domain of the HIV-1 restriction factor APOBEC3G. Nature. 2008;452:116–119. doi: 10.1038/nature06638. [DOI] [PubMed] [Google Scholar]
- 31.Holden LG, Prochnow C, Chang YP, Bransteitter R, Chelico L, Sen U, Stevens RC, Goodman MF, Chen XS. Crystal structure of the anti-viral APOBEC3G catalytic domain and functional implications. Nature. 2008;456:121–124. doi: 10.1038/nature07357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitamura S, Ode H, Nakashima M, Imahashi M, Naganawa Y, Kurosawa T, Yokomaku Y, Yamane T, Watanabe N, Suzuki A, Sugiura W, Iwatani Y. The APOBEC3C crystal structure and the interface for HIV-1 Vif binding. Nat Struct Mol Biol. 2012;19:1005–1010. doi: 10.1038/nsmb.2378. [DOI] [PubMed] [Google Scholar]
- 33.Li M, Shandilya SM, Carpenter MA, Rathore A, Brown WL, Perkins AL, Harki DA, Solberg J, Hook DJ, Pandey KK, Parniak MA, Johnson JR, Krogan NJ, Somasundaran M, Ali A, Schiffer CA, Harris RS. First-in-class small molecule inhibitors of the single-strand DNA cytosine deaminase APOBEC3G. ACS Chem Biol. 2012;7:506–517. doi: 10.1021/cb200440y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bohn MF, Shandilya SM, Albin JS, Kouno T, Anderson BD, McDougle RM, Carpenter MA, Rathore A, Evans L, Davis AN, Zhang J, Lu Y, Somasundaran M, Matsuo H, Harris RS, Schiffer CA. Crystal structure of the DNA cytosine deaminase APOBEC3F: the catalytically active and HIV-1 Vif-binding domain. Structure. 2013;21:1042–1050. doi: 10.1016/j.str.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Byeon IJ, Ahn J, Mitra M, Byeon CH, Hercik K, Hritz J, Charlton LM, Levin JG, Gronenborn AM. NMR structure of human restriction factor APOBEC3A reveals substrate binding and enzyme specificity. Nat Commun. 2013;4:1890. doi: 10.1038/ncomms2883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bohn MF, Shandilya SM, Silvas TV, Nalivaika EA, Kouno T, Kelch BA, Ryder SP, Kurt-Yilmaz N, Somasundaran M, Schiffer CA. The ssDNA mutator APOBEC3A is regulated by cooperative dimerization. Structure. 2015;23:903–911. doi: 10.1016/j.str.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shi K, Carpenter MA, Kurahashi K, Harris RS, Aihara H. Crystal structure of the DNA deaminase APOBEC3B catalytic domain. J Biol Chem. 2015;290:28120–28130. doi: 10.1074/jbc.M115.679951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakashima M, Ode H, Kawamura T, Kitamura S, Naganawa Y, Awazu H, Tsuzuki S, Matsuoka K, Nemoto M, Hachiya A, Sugiura W, Yokomaku Y, Watanabe N, Iwatani Y. Structural insights into HIV-1 Vif-APOBEC3F interaction. J Virol. 2016;90:1034–1047. doi: 10.1128/JVI.02369-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaban NM, Shi K, Li M, Aihara H, Harris RS. 1.92 angstrom zinc-free APOBEC3F catalytic domain crystal structure. J Mol Biol. 2016;428:2307–2316. doi: 10.1016/j.jmb.2016.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Byeon IJ, Byeon CH, Wu T, Mitra M, Singer D, Levin JG, Gronenborn AM. Nuclear magnetic resonance structure of the APOBEC3B catalytic domain: structural basis for substrate binding and DNA deaminase activity. Biochemistry. 2016;55:2944–2959. doi: 10.1021/acs.biochem.6b00382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao X, Li SX, Yang H, Chen XS. Crystal structures of APOBEC3G N-domain alone and its complex with DNA. Nat Commun. 2016;7:12193. doi: 10.1038/ncomms12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carpenter MA, Li M, Rathore A, Lackey L, Law EK, Land AM, Leonard B, Shandilya SM, Bohn MF, Schiffer CA, Brown WL, Harris RS. Methylcytosine and normal cytosine deamination by the foreign DNA restriction enzyme APOBEC3A. J Biol Chem. 2012;287:34801–34808. doi: 10.1074/jbc.M112.385161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Caval V, Bouzidi MS, Suspene R, Laude H, Dumargne MC, Bashamboo A, Krey T, Vartanian JP, Wain-Hobson S. Molecular basis of the attenuated phenotype of human APOBEC3B DNA mutator enzyme. Nucleic Acids Res. 2015;43:9340–9349. doi: 10.1093/nar/gkv935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen H, Lilley CE, Yu Q, Lee DV, Chou J, Narvaiza I, Landau NR, Weitzman MD. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- 45.Logue EC, Bloch N, Dhuey E, Zhang R, Cao P, Herate C, Chauveau L, Hubbard SR, Landau NR. A DNA sequence recognition loop on APOBEC3A controls substrate specificity. PLoS One. 2014;9:e97062. doi: 10.1371/journal.pone.0097062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harjes S, Solomon WC, Li M, Chen KM, Harjes E, Harris RS, Matsuo H. Impact of H216 on the DNA binding and catalytic activities of the HIV restriction factor APOBEC3G. J Virol. 2013;87:7008–7014. doi: 10.1128/JVI.03173-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pham P, Landolph A, Mendez C, Li N, Goodman MF. A biochemical analysis linking APOBEC3A to disparate HIV-1 restriction and skin cancer. J Biol Chem. 2013;288:29294–29304. doi: 10.1074/jbc.M113.504175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rausch JW, Chelico L, Goodman MF, Le Grice SF. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J Biol Chem. 2009;284:7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 50.Yu Q, Konig R, Pillai S, Chiles K, Kearney M, Palmer S, Richman D, Coffin JM, Landau NR. Single-strand specificity of APOBEC3G accounts for minus-strand deamination of the HIV genome. Nat Struct Mol Biol. 2004;11:435–442. doi: 10.1038/nsmb758. [DOI] [PubMed] [Google Scholar]
- 51.Kim EY, Lorenzo-Redondo R, Little SJ, Chung YS, Phalora PK, Maljkovic Berry I, Archer J, Penugonda S, Fischer W, Richman DD, Bhattacharya T, Malim MH, Wolinsky SM. Human APOBEC3 induced mutation of human immunodeficiency virus type-1 contributes to adaptation and evolution in natural infection. PLoS Pathog. 2014;10:e1004281. doi: 10.1371/journal.ppat.1004281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, Harris S, Shah RR, Resnick MA, Getz G, Gordenin DA. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–976. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Borresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjord JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinsk M, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–421. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitra M, Hercik K, Byeon IJ, Ahn J, Hill S, Hinchee-Rodriguez K, Singer D, Byeon CH, Charlton LM, Nam G, Heidecker G, Gronenborn AM, Levin JG. Structural determinants of human APOBEC3A enzymatic and nucleic acid binding properties. Nucleic Acids Res. 2014;42:1095–1110. doi: 10.1093/nar/gkt945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu Y, Ito F, Zhang G, Fernandez B, Yang H, Chen XS. DNA cytosine and methylcytosine deamination by APOBEC3B: enhancing methylcytosine deamination by engineering APOBEC3B. Biochem J. 2015;471:25–35. doi: 10.1042/BJ20150382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Demorest ZL, Li M, Harris RS. Phosphorylation directly regulates the intrinsic DNA cytidine deaminase activity of activation-induced deaminase and APOBEC3G. J Biol Chem. 2011;286:26568–26575. doi: 10.1074/jbc.M111.235721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holtz CM, Sadler HA, Mansky LM. APOBEC3G cytosine deamination hotspots are defined by both sequence context and single-stranded DNA secondary structure. Nucleic Acids Res. 2013;41:6139–6148. doi: 10.1093/nar/gkt246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nik-Zainal S, Davies H, Staaf J, Ramakrishna M, Glodzik D, Zou X, Martincorena I, Alexandrov LB, Martin S, Wedge DC, Van Loo P, Ju YS, Smid M, Brinkman AB, Morganella S, Aure MR, Lingjaerde OC, Langerod A, Ringner M, Ahn SM, Boyault S, Brock JE, Broeks A, Butler A, Desmedt C, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matthews MM, Thomas JM, Zheng Y, Tran K, Phelps KJ, Scott AI, Havel J, Fisher AJ, Beal PA. Structures of human ADAR2 bound to dsRNA reveal base-flipping mechanism and basis for site selectivity. Nat Struct Mol Biol. 2016;23:426–433. doi: 10.1038/nsmb.3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shirakawa K, Takaori-Kondo A, Yokoyama M, Izumi T, Matsui M, Io K, Sato T, Sato H, Uchiyama T. Phosphorylation of APOBEC3G by protein kinase A regulates its interaction with HIV-1 Vif. Nat Struct Mol Biol. 2008;15:1184–1191. doi: 10.1038/nsmb.1497. [DOI] [PubMed] [Google Scholar]
- 61.Losey HC, Ruthenburg AJ, Verdine GL. Crystal structure of Staphylococcus aureus tRNA adenosine deaminase TadA in complex with RNA. Nat Struct Mol Biol. 2006;13:153–159. doi: 10.1038/nsmb1047. [DOI] [PubMed] [Google Scholar]
- 62.Conticello SG, Langlois MA, Neuberger MS. Insights into DNA deaminases. Nat Struct Mol Biol. 2007;14:7–9. doi: 10.1038/nsmb0107-7. [DOI] [PubMed] [Google Scholar]
- 63.Almog R, Maley F, Maley GF, Maccoll R, Van Roey P. Three-dimensional structure of the R115E mutant of T4-bacteriophage 2′-deoxycytidylate deaminase. Biochemistry. 2004;43:13715–13723. doi: 10.1021/bi048928h. [DOI] [PubMed] [Google Scholar]
- 64.Teh AH, Kimura M, Yamamoto M, Tanaka N, Yamaguchi I, Kumasaka T. The 1.48 A resolution crystal structure of the homotetrameric cytidine deaminase from mouse. Biochemistry. 2006;45:7825–7833. doi: 10.1021/bi060345f. [DOI] [PubMed] [Google Scholar]
- 65.Ireton GC, Black ME, Stoddard BL. The 1.14 A crystal structure of yeast cytosine deaminase: evolution of nucleotide salvage enzymes and implications for genetic chemotherapy. Structure. 2003;11:961–972. doi: 10.1016/s0969-2126(03)00153-9. [DOI] [PubMed] [Google Scholar]
- 66.Ko TP, Lin JJ, Hu CY, Hsu YH, Wang AH, Liaw SH. Crystal structure of yeast cytosine deaminase. Insights into enzyme mechanism and evolution. J Biol Chem. 2003;278:19111–19117. doi: 10.1074/jbc.M300874200. [DOI] [PubMed] [Google Scholar]
- 67.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sheldrick GM. Experimental phasing with SHELXC/D/E: combining chain tracing with density modification. Acta Crystallogr D Biol Crystallogr. 2010;66:479–485. doi: 10.1107/S0907444909038360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 71.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 73.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gaborek TJ. Conformational free-energy landscapes for a peptide in saline environments. Biophys J. 2012;103:2513–2520. doi: 10.1016/j.bpj.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Narvaiza I, Linfesty DC, Greener BN, Hakata Y, Pintel DJ, Logue E, Landau NR, Weitzman MD. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bulliard Y, Narvaiza I, Bertero A, Peddi S, Rohrig UF, Ortiz M, Zoete V, Castro-Diaz N, Turelli P, Telenti A, Michielin O, Weitzman MD, Trono D. Structure-function analyses point to a polynucleotide-accommodating groove essential for APOBEC3A restriction activities. J Virol. 2011;85:1765–1776. doi: 10.1128/JVI.01651-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The coordinates and structure factors for the A3A-ssDNA and A3Bctd*-ssDNA complexes have been deposited in the Protein Data Bank under accession codes 5SWW and 5TD5, respectively. Source data for Figs. 1 & 3c are available in the online version of the paper. Full gel images of biochemical and immunoblot results in Figs. 3e & 5a–b are shown in Supplementary Data Set 1. Other data are available upon request.