Abstract
Anaplastic lymphoma kinase (ALK) is a validated molecular target in several ALK-rearranged malignancies, including non-small-cell lung cancer (NSCLC). However, the clinical benefit of targeting ALK using tyrosine kinase inhibitors (TKIs) is almost universally limited by the emergence of drug resistance. Diverse mechanisms of resistance to ALK TKIs have now been discovered, and these basic mechanisms are informing the development of novel therapeutic strategies to overcome resistance in the clinic. In this Review, we summarize the current successes and challenges of targeting ALK.
Keywords: ALK, anaplastic lymphoma kinase, TKI, resistance, NSCLC
INTRODUCTION
The discovery of anaplastic lymphoma kinase (ALK) dates back to 1994 when a chromosomal rearrangement t(2;5), resulting in a nucleophosmin (NPM1)-ALK fusion, was described in anaplastic large-cell lymphoma (ALCL) (1). Subsequent work over the next two decades identified ALK fusion proteins as the oncogenic driver in numerous different malignancies. Perhaps most widely recognized is the echinoderm microtubule-associated protein-like 4 (EML4)-ALK fusion detected in non-small-cell lung cancer (NSCLC) in 2007 (2). ALK-rearranged cancers have since emerged as a salient example of the paradigm of “oncogene addiction.”
In the span of less than a decade, collaborative efforts among basic and clinical scientists in academia and the pharmaceutical industry have led to the development of numerous ALK tyrosine kinase inhibitors (TKIs). To date, three ALK TKIs (crizotinib, ceritinib, and alectinib) have received approval by the US Food and Drug Administration (FDA) for treatment of advanced “ALK-positive” NSCLC (i.e., NSCLC harboring an ALK rearrangement), with clinical trials demonstrating remarkable responses in this patient population (3–8). Yet, as with any targeted therapy, tumor cells evolve and invariably acquire resistance to ALK TKIs, leading to clinical relapse. Since the first demonstration of crizotinib’s activity in ALK-positive NSCLC, a multitude of studies have focused on elucidating mechanisms of resistance to ALK inhibition.
The rapid pace of ALK-targeted drug development, and the knowledge gained in parallel on resistance mechanisms, illustrate the power of an iterative, systematic discovery process—at the bench and the bedside—to advance cancer research and transform patients’ lives. This Review will provide an up-to-date, mechanistic framework for understanding how ALK, and the mechanisms of resistance to ALK TKIs, may be effectively targeted in order to extend and improve the lives of patients with ALK-driven cancers.
ALK BIOLOGY
The Function of Native ALK
ALK encodes a highly conserved receptor tyrosine kinase (RTK) within the insulin receptor superfamily, comprised of an extracellular domain, a single-pass transmembrane region, and an intracellular kinase domain (9). In adult humans, ALK expression is limited to the nervous system, testes, and small intestines (1). Activation of endogenous ALK requires ligand-dependent receptor dimerization and autophosphorylation. Recent work has established Augmentor α and β (FAM150) as the ligand for ALK (10–12). Although little is known about the biological function of ALK in humans, insights may be gleaned from studying model systems.
In Drosophila melanogaster, Alk is activated by its ligand Jelly belly (Jeb) and regulates the development of the gut musculature and neuronal circuitry within the visual system (13–16). In mice, Alk is expressed in the nervous system at the embryonic and neonatal stage, but minimally expressed in adults (9). Alk knockout mice notably achieve a normal life span, exhibiting mild abnormalities involving the frontal cortex and hippocampus and hypogonadotropic hypogonadism (17–19). Interestingly, visual disturbance and hypogonadism have been reported in patients treated with the first-generation, multitargeted ALK inhibitor crizotinib (3, 4). These toxicities, however, are unlikely to be ALK-related, given their decreased incidence with more selective ALK TKIs such as ceritinib and alectinib (5–8).
ALK Rearrangements in Cancer
With the advent of next generation sequencing (NGS)-based diagnostics, more than 20 different ALK fusion partner genes have been reported across multiple malignancies. In each cancer type, the full spectrum of chimeric ALK proteins and their individual frequency varies (Table 1). For example, in ALCL where ALK rearrangements are seen in ~55% of adult patients (and nearly universally in pediatric patients), NPM1-ALK resulting from t(2;5)(p23;q35) is the most common fusion, accounting for up to 80% of cases (1, 20–29). Inflammatory myofibroblastic tumor (IMT) was the first solid tumor found to harbor ALK rearrangements, which occur in up to 50% of cases (30). In ALK-positive IMT, NPM1-ALK has not been reported, but a variety of other fusion partners not seen in ALCL have instead been identified (Table 1) (30–37).
Table 1.
ALK rearrangements in cancer.
| Cancer type | Frequency of ALK rearrangements | ALK fusion partner gene | Location of fusion partner | References |
|---|---|---|---|---|
| NSCLC | 3–7% |
TPR CRIM1 EML4* STRN TFG HIP1 PTPN3 KIF5B KLC1 CLTC |
1q25 2p21 2p21 2p22.2 3q12.2 7q11.23 9q31 10p11.22 14q32.3 17q23.1 |
2, 40–45 |
| ALCL | ~55% (in adults) |
TPM3 ATIC TFG NPM1* TRAF1 CLTC RNF213 TPM4 MYH9 MSN |
1q21.2 2q35 3q12.2 5q35.1 9q33-q34 17q23.1 17q25.3 19p13.1 22q13.1 Xq11.1 |
1, 21–29 |
| IMT | Up to 50% |
TPM3 RANBP2 ATIC SEC31A CARS PPFIBP1 CLTC TPM4 |
1q21.2 2q12.3 2q35 4q21.22 11p15.5 12p12.1 17q23.1 19p13.1 |
30–37 |
| DLBCL | < 1% |
RANBP2 EML4 SEC31A SQSTM1 NPM1 CLTC |
2q12.3 2p21 4q21.22 5q35 5q35.1 17q23.1 |
164–171 |
| Colorectal cancer | < 1% |
EML4 WDCP |
2p21 2p23.3 |
172–175 |
| Breast cancer | N.D. | EML4 | 2p21 | 173 |
| RCC | < 1% |
TPM3 EML4 STRN VCL |
1q21.2 2p21 2p22.2 10q22.2 |
176–179 |
| RMC | N.D. | VCL | 10q22.2 | 180 |
| Esophageal cancer | N.D. | TPM4 | 19p13.1 | 181, 182 |
| Ovarian cancer | N.D. | FN1 | 2q34 | 183 |
Abbreviations: ALK, anaplastic lymphoma kinase; NSCLC, non-small-cell lung cancer; TPR, translocated promoter region, nuclear basket protein; CRIM1, cysteine rich transmembrane BMP regulator 1; EML4, echinoderm microtubule associated protein like 4; STRN, striatin; TFG, TRK-fused gene; HIP1, huntingtin interacting protein 1; PTPN3, protein tyrosine phosphatase, non-receptor type 3; KIF5B, kinesin family member 5B; KLC1, kinesin light chain 1; CLTC, clathrin heavy chain; ALCL, anaplastic large cell lymphoma; TPM3, tropomyosin 3; NPM1, nucleophosmin; TRAF1, TNF receptor associated factor 1; RNF213, ring finger protein 213; TPM4, tropomyosin 4; MYH9, myosin, heavy chain 9, non-muscle; MSN, moesin; IMT, inflammatory myofibroblastic tumor; RANBP2, RAN binding protein 2; SEC31A, SEC31 homolog A; CARS, cysteinyl-tRNA synthetase; PPFIBP1, PTPRF interacting protein, binding protein 1; DLBCL, diffuse large B-cell lymphoma; SQSTM1, sequestosome 1; WDCP, WD repeat and coiled coil containing; N.D., not determined; RCC, renal cell carcinoma; VCL, vinculin; RMC, renal medullary carcinoma; FN1, fibronectin 1.
EML4 and NPM1 are the most common fusion partner genes in NSCLC and ALCL, respectively.
NSCLC was the second solid tumor in which oncogenic ALK fusions were detected. Soda and colleagues reported the identification of EML4-ALK in a small cohort of Japanese NSCLC patients (2). Since then, ALK fusions have been detected in 3–7% of NSCLCs, and have been associated with absence of smoking, younger age, and adenocarcinoma histology (38). Even though the relative proportion of NSCLCs harboring an ALK rearrangement is significantly lower than that of ALCL or IMT, NSCLC patients constitute the largest subset of patients with an ALK-rearranged cancer due to the high incidence of lung cancer worldwide (39). Notably, studies in NSCLC have identified several additional ALK fusion proteins (40–45), which collectively occur less frequently than EML4-ALK. Furthermore, a number of breakpoint variants may be seen for a given fusion protein. The classic example is EML4-ALK, with over 10 distinct variants (46). In most cases, the breakpoint in ALK at intron 19, just preceding exon 20, is conserved.
At low frequency, ALK rearrangements have been detected in other cancers, including colorectal, breast, renal cell, esophageal, ovarian, and anaplastic thyroid carcinoma, and diffuse large B-cell lymphoma (Table 1). Several common themes have emerged based on the identified ALK rearrangements (47). First, in all ALK fusions, the entire ALK kinase domain is preserved. Second, the N-terminal partner contributes its promoter and oligomerization domain to the ALK fusion protein, leading to aberrant expression and constitutive activation of ALK. Consequently, the level of ALK fusion expression and the degree of signaling may vary depending on the partner gene. Indeed, in vitro studies using NIH3T3 cells have suggested differential effects of ALK fusion proteins on cell proliferation and invasion depending on the exact fusion (48). These findings have not yet been validated in patients, but could be clinically relevant if they translate into differential sensitivity of ALK fusions to TKIs in the clinic, as described below (49). Lastly, ALK fusion proteins interact with a complex network of proteins and signal via multiple downstream pathways, including JAK/STAT, PI3K/AKT, and MEK/ERK, driving aberrant proliferation and survival (Figure 1) (50, 51). In the setting of chronic TKI exposure, dysregulation of these signaling nodes may enable acquired resistance to ALK inhibition (see bypass signaling section below).
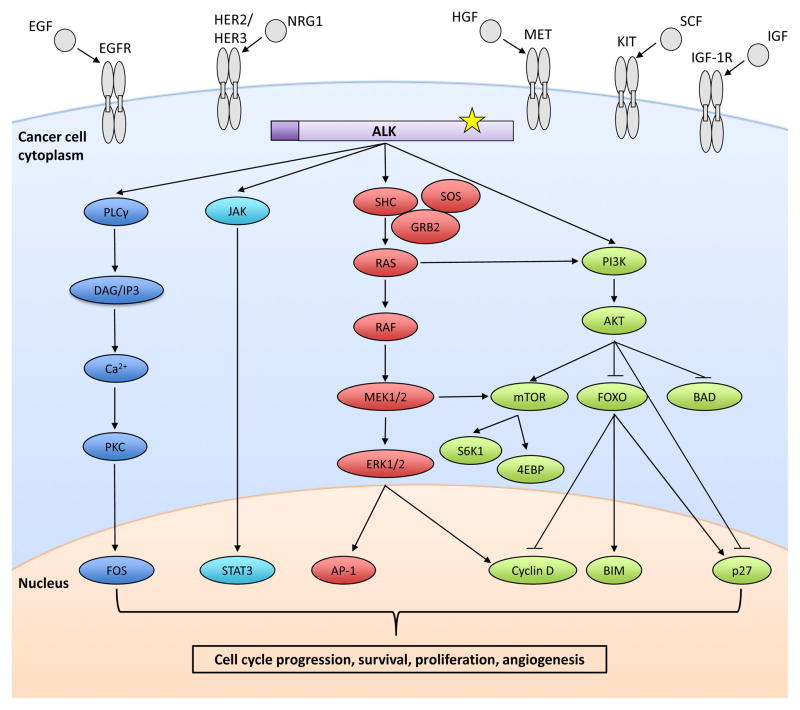
Figure 1. Oncogenic ALK signaling.
The ALK fusion protein is constitutively active and signals via phospholipase Cγ (PLCγ), JAK-STAT, RAS-RAF-MEK-ERK, and PI3K-AKT-mTOR pathways (13). This signaling results in the aberrant regulation of a number of genes (some of which are represented here), ultimately driving cell cycle progression, survival, proliferation, and angiogenesis (50). Secondary mutations in the ALK kinase domain (starred) cause acquired resistance to ALK TKIs. Several of the bypass signaling tracks implicated in ALK TKI resistance are also shown here —EGFR, HER2/HER3, MET, KIT, and IGF-1R with their respective ligands.
Additional questions remain regarding the biology of ALK. For example, it is unclear why certain fusion partners dominate in certain cancers (e.g., NPM1-ALK in ALCL and EML4-ALK in NSCLC). The etiology of ALK rearrangements, and their predilection for younger patients, also remains unknown. Finally, activating point mutations in ALK have been identified in a subset of patients with familial or sporadic neuroblastoma, and investigations into the biology of mutant ALK are ongoing. ALK mutations outside the context of TKI resistance are beyond the scope of this Review, and are covered elsewhere (13, 47).
TARGETING ONCOGENIC ALK
Within months of the identification of EML4-ALK in NSCLC, preclinical studies established the transforming potential of this ALK fusion (2, 52). Transgenic mice expressing EML4-ALK in lung alveolar epithelial cells developed innumerable adenocarcinoma nodules in their lungs soon after birth (52). The dependency of ALK-rearranged lung cancer cells on ALK signaling for survival and growth (i.e., “addiction” to ALK) was demonstrated in cell lines and mouse models (52, 53), spurring the development of TKIs targeting ALK.
Although originally developed as a potent MET inhibitor, crizotinib was the first ALK-directed TKI to enter the clinic (53). In 2011, crizotinib was granted accelerated approval by the FDA based on the results of phase I/II studies demonstrating robust clinical activity in advanced ALK-rearranged NSCLC (54). Soon thereafter, two phase III trials showed that crizotinib was superior to first- and second-line cytotoxic chemotherapy in advanced ALK-rearranged NSCLC (3, 4). Across all crizotinib trials, the objective response rate (ORR) in ALK-positive NSCLC patients ranged from 60–74%, with median progression-free survival (PFS) of 8–11 months. Subsequently, two second-generation ALK inhibitors, ceritinib and alectinib, received accelerated approval for patients with ALK-positive NSCLC previously treated with crizotinib. In the phase 1 ASCEND-1 trial, ceritinib demonstrated an ORR of 56% and median PFS of 6.9 months in crizotinib-pre-treated ALK-rearranged NSCLC patients (5, 6). In two phase II studies, alectinib induced objective responses in ~50% of crizotinib-resistant ALK-rearranged NSCLCs (7, 8); median PFS in both studies was 8–9 months. Another second-generation inhibitor, brigatinib, has been associated with a confirmed ORR of 62% and a median PFS of 13.2 months in crizotinib-pre-treated patients in one phase I/II study (55). These second-generation inhibitors are more potent than crizotinib, can overcome the most common crizotinib-resistant mutations including the L1196M gatekeeper mutation, are active in crizotinib-resistant tumors without ALK resistance mutations, and are more effective than crizotinib against central nervous system (CNS) metastases (5–8).
ALK inhibitors have not yet been approved for use in other, non-lung ALK-driven cancers, although reports have been published of patients’ responses to ALK TKIs in ALK-positive ALCL and IMT (56–58). Multiple clinical trials are underway evaluating the activity of ALK TKIs across different tumors. In particular, the National Cancer Institute (NCI)-Molecular Analysis for Therapy Choice (MATCH) trial is enrolling adult patients with any advanced solid tumors and lymphomas refractory to standard therapy (NCT02465060); those patients found to have ALK-rearranged cancers (other than NSCLC and ALCL) will be eligible to receive treatment with crizotinib. Hence, the NCI-MATCH protocol will allow for systematic analysis of response to ALK inhibition in patients with ALK-rearranged cancers, irrespective of the cancer type.
Despite initial responses, patients treated with ALK TKIs inevitably progress within 1–2 years due to acquired resistance (3–8). This resistance is an expected consequence of tumor evolution (59) and is similar in principle to what has been observed with other targeted therapies (e.g., KIT inhibitors in gastrointestinal stromal tumors (GISTs) and epidermal growth factor receptor (EGFR) inhibitors in EGFR-mutant NSCLC).
MECHANISMS OF RESISTANCE TO ALK TKIs
Current Approaches to Study TKI Resistance
Numerous ALK TKI resistance mechanisms have been identified since the discovery of ALK as a therapeutic target in NSCLC. These discoveries have relied on the use of diverse experimental systems and approaches (Figure 2). Traditionally, sequencing analyses of pre-TKI and post-TKI tumor biopsies, followed by functional validation of candidate resistance mechanisms, have been a common approach (60–63). As an example, many of the secondary resistance mutations within the ALK tyrosine kinase domain were discovered in this manner.
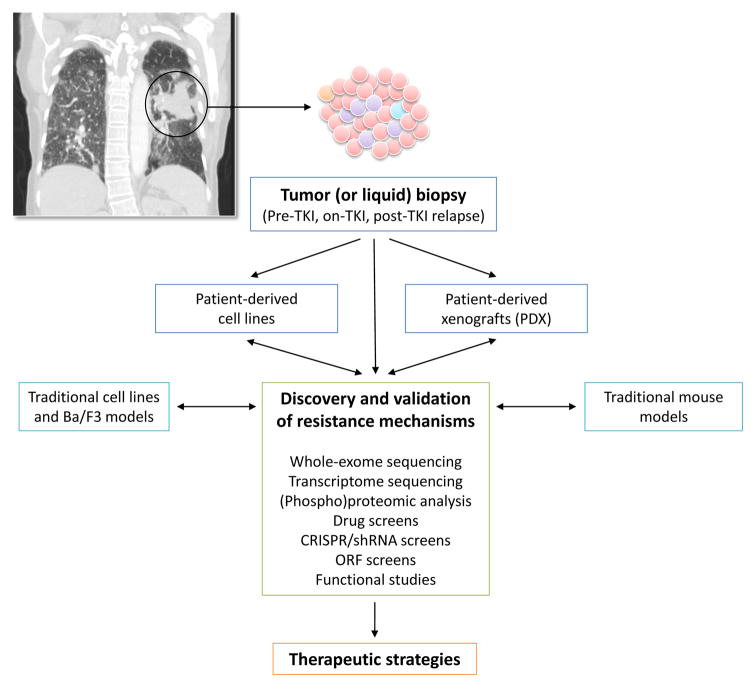
Figure 2. Experimental platforms for discovery and validation of TKI resistance mechanisms.
Tumor biopsy specimens from patients obtained at baseline (pre-TKI), on TKI, and after relapse on a TKI (post-TKI) serve as the gold standard model for studying resistance mechanisms. Liquid biopsies with circulating tumor DNA (ctDNA) analyses are being increasingly utilized. Generation of patient-derived cell lines and xenografts (PDX), if successful, can help facilitate the execution of systematic functional assays. Based on the identified resistance mechanisms, novel therapeutic strategies may be developed and tested preclinically, prior to entering clinical trials.
To overcome the inherent limitations in biopsy specimens with regards to tumor quantity and quality, more recent studies have employed patient-derived cell lines and xenograft (PDX) models (62, 64). These models can serve as a platform for systematic loss-of-function (e.g., CRISPR, shRNA, or pharmacologic) and gain-of-function (e.g., open reading frame (ORF) library) screens. For example, compound drug screens in patient-derived cell lines led to the identification of MEK and SRC activation as novel ALK TKI resistance mechanisms (further discussed below) (64). Liquid biopsies can also overcome some of the challenges of tumor biopsies (discussed below).
Together with traditional in vitro generated cell line models (63, 65) and random mutagenesis screens (66, 67), these experimental approaches have led to the identification of two major classes of ALK TKI resistance mechanisms: (i) ALK-dependent, “on-target” mechanisms including ALK secondary resistance mutations or amplification, where the tumor cell dependency on ALK signaling persists; and (ii) ALK-independent, “off-target” mechanisms including activation of bypass tracks and lineage changes, where the tumor cells effectively escape dependency on ALK. While pharmacological properties of TKIs can also limit ALK TKI efficacy, particularly for CNS disease, these issues are beyond the scope of this Review and have been discussed elsewhere (68). Below, we explore the biological mechanisms of acquired ALK TKI resistance primarily based on data obtained from studies of ALK-positive NSCLC.
ALK-Dependent Resistance: Secondary Mutations in the ALK Tyrosine Kinase Domain
In general, secondary mutations within the target kinase cause drug resistance by re-inducing kinase activation and signaling despite the presence of the TKI. These resistance mutations can directly hinder TKI binding to the target kinase, alter the kinase’s conformation, and/or alter the ATP-binding affinity of the kinase. Unlike in EGFR-mutant NSCLC, where the T790M gatekeeper mutation is the predominant, clinically-observed EGFR mutation causing resistance to first- and second-generation EGFR TKIs, a much broader spectrum of on-target mutations has been identified in ALK-positive NSCLC treated with ALK TKIs (Figure 3) (69). This situation is reminiscent of the wide array of resistance mutations observed after treatment with the ABL inhibitor imatinib in patients with chronic myelogenous leukemia (CML). The difference in spectrum of resistance mutations may be attributable to the genetic mechanism of oncogene activation (i.e., gene rearrangements involving ALK or ABL, versus activating point mutations within the EGFR kinase domain) and/or the mode of TKI binding (i.e., to the inactive kinase conformation for imatinib and crizotinib, versus the active conformation for EGFR TKIs erlotinib and gefitinib) (70, 71). Both of these factors may influence the spectrum of drug-resistant mutations that arise in TKI-resistant patients.
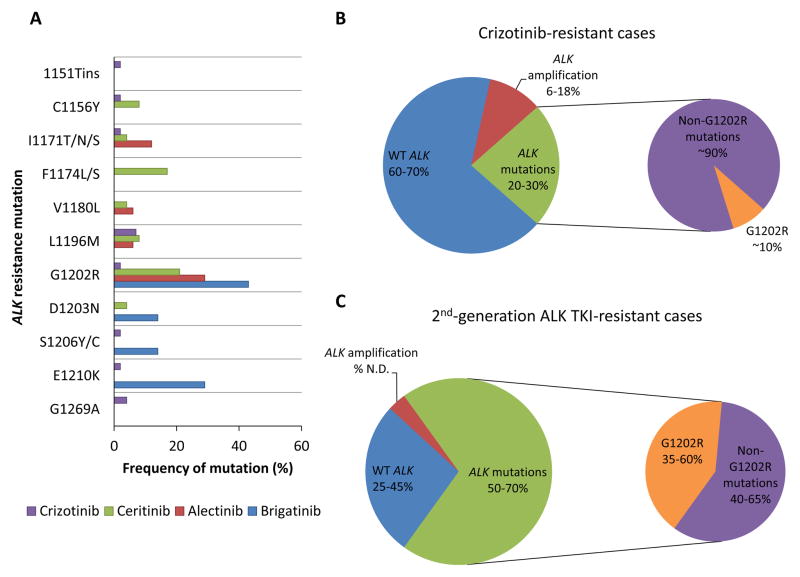
Figure 3. Acquired resistance mechanisms to ALK TKIs.
(A) The reported frequency of each secondary resistance mutation in the ALK kinase domain is depicted for post-crizotinib (n = 55), post-ceritinib (n = 24), post-alectinib (n = 17), and post-brigatinib (n = 7) cases, based on reference 78. The absence of a colored bar representing an ALK TKI (e.g., a blue bar representing brigatinib) indicates that the particular ALK mutation was not detected in the tested cases progressing after that specific ALK TKI (78). In (B) and (C), the differential frequency of ALK secondary mutations in crizotinib-resistant versus second-generation ALK TKI (e.g., ceritinib, alectinib, brigatinib)-resistant cases (20–30% versus 50–70%, respectively) is highlighted. Notably, the frequency of the G1202R mutation is significantly higher after relapse on a second-generation ALK TKI compared to crizotinib. ALK amplification appears to be an infrequent mechanism of resistance to second-generation ALK TKIs, although the exact frequency has not been determined (N.D.). WT = wild-type.
The first ALK resistance mutation reported was the L1196M gatekeeper mutation, analogous to EGFR T790M (61). This mutation, also identified in a cell line model of crizotinib resistance (65), alters the gatekeeper residue at the bottom of the ATP-binding pocket and impairs TKI binding. The G1269A mutation also lies in the ATP-binding pocket and hinders crizotinib binding (72). Other resistance mutations affect residues adjacent to the N-terminus (C1156Y, L1152R, and I1151Tins) and C-terminus of the αC helix (F1174C/L/V) (61–63, 73). The exact structural mechanisms by which these mutations cause resistance are unclear, although they may enhance the kinase’s ATP-binding affinity and increase its enzymatic activity (63, 74). The I1171T/N/S mutation may distort the αC helix to interfere with TKI binding (75–78). Solvent-front mutations (G1202R, G1202del, D1203N, and S1206Y/C) represent another class of ALK resistance mutations that impair drug binding likely through steric hindrance (63, 73, 76–78). Numerous studies have revealed that G1202R in particular confers high level resistance to first- and second-generation ALK TKIs (63, 76, 78). It remains to be seen whether the same spectrum of mutations will be detected in other ALK-positive cancers treated with ALK TKIs; to date, several of the same mutations have been identified in ALCL cell lines (e.g., I1171T/N, F1174C, L1196Q) and an IMT patient post-crizotinib (e.g., F1174L) (79–81).
Given the structural differences among the available ALK TKIs, it is perhaps not surprising that each ALK TKI appears to be associated with a specific profile of secondary ALK resistance mutations (Figure 3A). A notable example involves the general difference in resistance mutations that arise on the first-generation ALK TKI crizotinib versus second-generation ALK TKIs (e.g., ceritinib, alectinib, brigatinib) (Figure 3). Recently, over 100 repeat biopsies from patients with ALK-positive NSCLC progressing on first- and second-generation AKI TKIs were analyzed (78). Secondary ALK mutations were observed in 20–30% of patients progressing on crizotinib, versus 56% of patients progressing on second-generation ALK TKIs (Figures 3B, 3C) (63, 72, 78). In line with prior studies, L1196M and G1269A were the most common resistance mutations detected in post-crizotinib samples. Interestingly, G1202R was found in only 2% of post-crizotinib samples, whereas it was the predominant resistance mechanism post-ceritinib, -alectinib, and -brigatinib (frequency ranging between 21–43%) (78). This distinction likely reflects the greater potency of second-generation ALK TKIs versus crizotinib, resulting in the suppression of other, less potent resistance mutations that are seen with crizotinib. Of note, the third-generation pan-inhibitory ALK TKI lorlatinib has been shown to effectively inhibit G1202R in cell lines and in patients (78, 82).
Several other ALK resistance mutations also show differential sensitivity/resistance to distinct ALK TKIs. For example, I1171 mutations are frequently identified in alectinib-resistant specimens, but not in ceritinib-resistant cases (75–78). Cell lines harboring I1171 mutations are resistant to alectinib, but sensitive to ceritinib. Conversely, F1174 mutations confer resistance to ceritinib, but are sensitive to alectinib, both in preclinical and clinical studies (73, 76, 78). Defining the resistance mutation profiles associated with each ALK TKI will be critical to the rational sequencing of ALK TKIs in patients.
In the setting of failure of a second-generation ALK TKI, the development of a secondary ALK resistance mutation implies that ALK may still be functioning as the oncogenic driver. In patient-derived NSCLC cell lines with resistance to second-generation ALK TKIs, the third-generation ALK TKI lorlatinib could inhibit the growth of cell lines harboring ALK resistance mutations, but was inactive against those lines without ALK resistance mutations (78). This preclinical work suggests that defining the ALK status is particularly crucial in patients who have progressed on a second-generation ALK TKI. In this working model (Figure 4), those patients whose ALK TKI-resistant tumors harbor secondary mutations should be treated with another ALK TKI tailored to that resistance mutation, whereas those patients whose resistant tumors do not harbor an ALK resistance mutation should consider ALK-based combinatorial strategies (discussed below), rather than monotherapy with another ALK TKI.
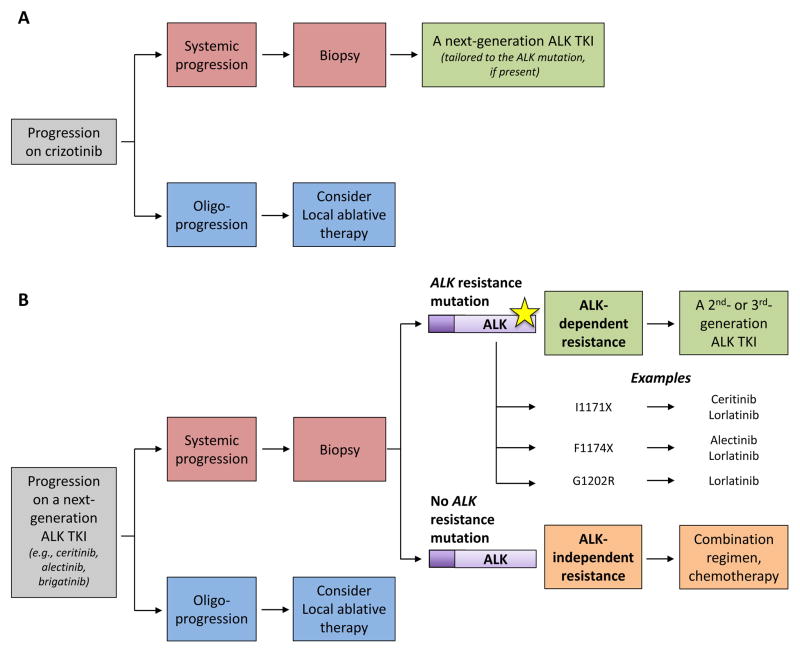
Figure 4. Guidelines for selecting treatment after progression on an ALK TKI.
When patients have oligoprogression on an ALK TKI, local ablative therapies can be considered. (A) When patients have systemic relapse on crizotinib, repeat biopsies should ideally be pursued if feasible, as the detection of particular ALK resistance mutations in a small number of patients (e.g., I1171, F1174, or G1202R mutations) may impact the choice of next-generation ALK TKI. However, the absence of an ALK resistance mutation after crizotinib usually does not translate into ALK independence, and these patients should go on to be treated with a next-generation ALK inhibitor. (B) After relapse on a second- or later-generation ALK TKI, we strongly recommend repeat biopsies at the time of progression to determine the resistance mechanism. The detection of a secondary ALK resistance mutation is suggestive of continued ALK dependency of tumor cells, and therefore, sensitivity to another ALK TKI that has activity against the mutant kinase. If an ALK resistance mutation is not identified, resistance may have arisen due to bypass signaling, lineage changes, or other ALK-independent mechanisms. In these cases, the tumor cells are likely no longer ALK-dependent, and combinatorial approaches or standard chemotherapy may be more effective.
As patients are treated with sequential ALK TKIs, compound resistance mutations can emerge, which adds another layer of complexity. Examples include C1156Y/I1171N after progression on sequential crizotinib, ceritinib, and alectinib; and E1210K/D1203N after sequential crizotinib and brigatinib (78). Functional studies in Ba/F3 cells suggest that compound mutations generally confer increased resistance to ALK TKIs (78). Similar examples exist in other oncogene-driven cancers treated using sequential TKIs. For example, in EGFR-mutant NSCLC treated with a first-generation followed by a third-generation EGFR TKI, compound T790M/C797S mutations arise, and when in cis, confer resistance to all available EGFR TKIs (83, 84). In the case of ALK TKI-resistant cells with compound mutations, however, in vitro studies suggest that the highly potent TKI lorlatinib may remain a potentially effective option (78).
It should be noted that the presence of compound mutations does not always translate into increased drug resistance. As an illustrative example, a compound C1156Y/L1198F mutation was recently discovered in a patient with ALK-positive NSCLC who had relapsed on crizotinib due to C1156Y, followed by sequential ceritinib and lorlatinib (74). While C1156Y is sensitive to lorlatinib, the addition of L1198F disrupts binding of the drug and leads to lorlatinib resistance. Interestingly, biochemical and cellular assays demonstrated that the L1198F mutation paradoxically leads to re-sensitization to the less potent and selective inhibitor crizotinib. Based on the in vitro findings, the patient was re-treated with crizotinib, and had a durable response (74). This case underscores the emerging complexity of ALK resistance mutations and the importance of serial biopsies. It also serves as proof of principle that timely collaborative research at the bench and the bedside can reveal important biological insights into tumor evolution and translate into immediate clinical benefit for patients.
ALK-Dependent Resistance: Amplification of ALK
ALK amplification occurs less frequently than secondary mutations, but it is a recognized cause of acquired resistance to crizotinib. In one series of crizotinib-resistant NSCLC cases, high-level ALK amplification was identified in 1 of 15 (6.7%), without an accompanying ALK mutation (63). Another series found new ALK copy number gain in 2 of 11 post-crizotinib cases (18.2%), although one of these also had a concomitant ALK resistance mutation (72). Notably, a study using an ALK-positive cell line made resistant to crizotinib in vitro demonstrated that ALK amplification resulted in partial resistance, whereas the addition of the L1196M gatekeeper mutation resulted in high level resistance (65). ALK amplification has not yet been detected as a resistance mechanism after next-generation ALK TKIs (78), and therefore, may not be a clinically relevant resistance mechanism in the face of more potent ALK inhibition.
ALK-Independent Resistance: Activation of Bypass Signaling Pathways
Secondary ALK mutations and/or amplification account for ~30% of crizotinib resistance in ALK-positive NSCLC, yet most crizotinib-resistant tumors—including those without an identifiable on-target mechanism—remain ALK-dependent with sensitivity to next-generation ALK TKIs (78). In contrast, 40–50% of cases resistant to second-generation ALK TKIs do not harbor on-target resistance mechanisms (Figures 3B, 3C), and these are no longer ALK-dependent based on studies described above (78, 85). One important category of ALK-independent, or off-target, resistance mechanism is the activation of bypass signaling track(s) through genetic alterations, autocrine signaling, or dysregulation of feedback signaling, resulting in the reactivation of downstream effectors required for tumor cell growth and survival.
Numerous examples of bypass signaling activation have been discovered as a cause of ALK TKI resistance (Figure 1). EGFR activation was the first identified bypass mechanism (62, 63, 86). Phospho-RTK array analysis of ALK-rearranged lung cancer cell lines revealed increased EGFR phosphorylation in crizotinib-resistant cell lines lacking secondary ALK alterations, compared to parental crizotinib-sensitive cells (63). This was associated with higher EGFR mRNA expression and persistent activation of downstream ERK and AKT signaling. Interestingly, these cells did not harbor any EGFR mutations or amplification, suggesting that EGFR activation may result from receptor and/or ligand upregulation (63, 86). In patients, assessment of paired pre- and post-crizotinib biopsy samples identified increased EGFR activation in 4 of 9 cases (44.4%) (63).
Subsequent work has implicated additional members of the HER receptor family in ALK TKI resistance (63, 87, 88). In a lentiviral ORF library screen designed to identify mediators of ALK TKI resistance, neuregulin-1 (NRG1), the ligand for ERBB3 (HER3) and ERBB4 (HER4) tyrosine kinases, emerged as the strongest driver of resistance (87). The resistant phenotype induced by NRG1 overexpression was abrogated by combined inhibition of ALK and HER2, the dimerization partner for HER3. Consistent with these findings, gene expression profiling of crizotinib-resistant versus crizotinib-naïve NSCLC tumor samples using RNA sequencing (RNA-seq) followed by single-sample gene set enrichment analysis (ssGSEA) identified EGFR and HER2 signatures as two of the most enriched gene expression signatures in resistant tumors (87). It is unknown whether dual blockade of EGFR/HER2 and ALK could be effective in treating patients with acquired resistance to ALK TKIs, but toxicities of the combination may be a major hurdle (further discussed below).
MET amplification is an example of bypass signaling that highlights the importance of considering the full spectrum of kinase targets for a given TKI in anticipating resistance mechanisms. MET activation is a known bypass signaling track in EGFR-mutant NSCLC (89, 90), but does not cause resistance to crizotinib, a potent ALK and MET inhibitor, in ALK-positive lung cancer. However, most next-generation ALK TKIs in development do not have anti-MET activity, and indeed, MET has recently been reported as a bypass mechanism in patients who have failed second-generation inhibitors like alectinib. In one case report, a post-alectinib biopsy did not reveal genetic alterations in ALK or EGFR, but demonstrated MET amplification by fluorescence in situ hybridization (FISH). This patient subsequently responded to crizotinib (91).
Direct reactivation of downstream effector proteins can also enable acquired resistance. MEK reactivation is a key example of this resistance mechanism (64, 92). In a recent study using a patient-derived ALK-rearranged lung cancer cell line post-ceritinib, a compound drug screen identified the MEK inhibitor selumetinib (AZD6244) as a potent hit combined with ceritinib (64). Subsequent NGS of this cell line revealed a MAP2K1 K57N activating mutation as the underlying genetic alteration leading to MEK activation. More importantly, a separate study demonstrated that ALK/MEK dual blockade may be effective not only in overcoming but also delaying ALK TKI resistance (92). In ALK-positive lung adenocarcinoma cell lines and mouse xenograft models, the RAS-MEK pathway was found to be the critical downstream effector of EML4-ALK. Upfront combination therapy with ALK and MEK inhibitors led to an increase in the magnitude and duration of treatment responses, and suppression of TKI resistance. Based on these findings, multiple combination regimens of ALK and MEK inhibitors may soon enter early-phase clinical testing.
Other examples of bypass signaling tracks clinically implicated in ALK TKI resistance include PIK3CA mutations (1 of 27 samples (3.7%), post-alectinib; a case post-ceritinib) (64, 78), KIT amplification (1 of 6 samples (16.7%), post-crizotinib) (63), IGF-1R activation (4 of 5 samples (80%), post-crizotinib) (93), and SRC activation (64) (Figure 1). Parallel use of different experimental platforms has been pivotal to the discovery and validation of off-target resistance mechanisms. With the growing use of genomic sequencing, proteomic, and phospho-proteomic technologies, more candidate off-target mechanisms will likely be discovered. In one series utilizing targeted NGS, mutations were detected post-second-generation ALK TKI in TP53 (in 56% of 27 specimens examined), DDR2, BRAF, FGFR2, MET, NRAS, and PIK3CA (each in 1 specimen) (78). Systematic assessment of pre- and post-TKI paired biopsies together with rigorous functional validation of candidate resistance mechanisms will be critical to identify those bypass tracks that represent therapeutic targets in resistant ALK-positive patients.
ALK-Independent Resistance: Lineage Changes
Phenotypic changes such as epithelial-to-mesenchymal transition (EMT) and small cell lung cancer (SCLC) transformation can contribute to the development of ALK TKI resistance. With EMT, tumor cells acquire a mesenchymal morphology along with migratory and invasive capacities. At the molecular level, they gain the expression of mesenchymal markers (e.g., vimentin) and lose epithelial markers (e.g., E-cadherin). This phenomenon has been reported in ALK TKI-resistant cell lines and tumor samples (78, 94, 95). In one series, immunohistochemical (IHC) staining on ceritinib-resistant biopsies revealed evidence of EMT in 5 of 12 cases, although in 3 of these, concurrent ALK resistance mutations were detected. Therefore, the relative contribution of EMT to the ALK TKI resistant phenotype remains to be established. The molecular mechanism by which EMT mediates ALK TKI resistance is also not known. At least in EGFR-mutant NSCLCs that acquire EGFR TKI resistance via EMT, preliminary studies suggest a role for AXL and IGF-1R activation (96, 97). In another study, the SRC/FAK pathway emerged as a key signaling node in mesenchymal EGFR TKI-resistant cancer cells, whose growth was inhibited by the multitargeted SRC inhibitor dasatinib (98).
Histologic change of tumor cells from adenocarcinoma to SCLC has been observed in 3–10% of EGFR TKI-resistant NSCLCs (90), and a small number of recent case reports have now described resistance to crizotinib and alectinib mediated by this mechanism (99–102). Conceptually, this lineage change may contribute to ALK TKI resistance in a manner similar to EGFR TKI resistance, which has been associated with the acquisition of RB loss and genetic or epigenetic features of classical SCLC (103). As successful strategies are developed to target ALK resistance mutations and bypass signaling tracks (see below), lineage changes may emerge as an increasingly important mechanism of TKI resistance. Therefore, a better biologic understanding of this process is needed.
ALK-Independent Resistance: Drug Efflux Pump
P-glycoprotein (P-gp) is a highly conserved ATP-dependent efflux pump encoded by the multidrug resistance 1 (MDR1) gene, also known as ATP-binding cassette sub-family B member 1 (ABCB1) (104). Recent work has identified P-gp overexpression as a potential resistance mechanism in 3 of 11 ALK-positive, crizotinib- or ceritinib-resistant NSCLC patients (105). In a patient-derived cell line overexpressing P-gp, shRNA-mediated knockdown of ABCB1 or pharmacologic inhibition of P-gp using verapamil re-sensitized the resistant cells to crizotinib and ceritinib (105). These findings need to be validated in larger patient cohorts. Importantly, P-gp can limit the CNS penetration of crizotinib and ceritinib (106, 107); alectinib, by comparison, is not a P-gp substrate and is able to achieve higher CNS levels (105, 108). Rational design of novel TKIs that are not substrates of P-gp or other ABC efflux transporters may help enhance penetration into the brain.
Primary ALK TKI Resistance
In addition to secondary (acquired) resistance, primary (intrinsic) resistance—defined as de novo lack of treatment response—can be seen after treatment with a TKI. For example, a small number (~5%) of ALK-positive NSCLC patients treated with first-line crizotinib have progressive disease as their best response (4). Mechanisms of intrinsic resistance are poorly understood, and this represents an important gap in the field of ALK TKI resistance. In theory, any of the acquired resistance mechanisms outlined above could cause primary resistance if they were pre-existing in TKI-naïve tumors. Indeed, in the case of EGFR-mutant lung cancers, primary resistance can arise due to a pre-existing T790M gatekeeper mutation (109, 110). However, the occurrence of de novo ALK resistance mutations appears to be rare in ALK-positive lung cancers (111). Similarly, pre-existing genetic alterations affecting bypass signaling could cause primary resistance (112), but have not yet been reported in ALK-positive NSCLC.
A number of studies have examined whether the specific ALK fusion variant may influence de novo sensitivity to ALK inhibitors. Differential ALK TKI sensitivity of EML4-ALK fusion variants was initially suggested by an in vitro study using a Ba/F3 cell line, in which variant v3a showed the least sensitivity to crizotinib and the tool compound TAE684, followed by v1 and v3b with intermediate sensitivity, and v2 with the greatest sensitivity (113). Drug sensitivity was found to correlate with the overall protein stability of each fusion variant. More recently, a retrospective analysis of 55 EML4-ALK-positive NSCLC patients treated with crizotinib demonstrated a higher disease control rate and longer median PFS among those with EML4-ALK variant 1 compared to those with non-variant 1, suggesting that the precise ALK fusion variant may be clinically relevant (114). However, given the small number of patients in this retrospective analysis, these findings will need to be validated in larger studies (49).
It should be noted that a fraction of primary resistant cases may in fact be due to false-positive genotyping. The registration trials of the currently approved ALK TKIs required ALK positivity by the break-apart FISH assay, which can yield false-positive results due to technical issues and variability in pathologist interpretation (115). On the other hand, ALK FISH may also yield false-negative results. The alternative FDA-approved diagnostic, IHC, has several advantages over FISH, but also has imperfect sensitivity and specificity, highlighting the importance of using multiple diagnostic methods including targeted NGS when clinically warranted (116, 117).
TUMOR HETEROGENEITY AND RESISTANCE
Polyclonal Resistance
A number of studies have demonstrated that heterogeneous resistance mechanisms may exist in tumor sites within one patient, or in tumor cells within one site, complicating efforts to therapeutically target resistant tumors. For example, ALK kinase domain resistance mutations have been reported to co-occur with ALK amplification (72) and with bypassing signaling activation such as c-KIT amplification (63). In another study, sequencing of 11 resistant tumor lesions acquired at autopsy from an ALK-positive, ceritinib-resistant NSCLC patient revealed a MAP2K1 activating mutation and a PIK3CA activating mutation in separate tumor sites (64). These findings collectively demonstrate the phenomenon of “polyclonal resistance,” whereby tumors acquire drug resistance through the simultaneous development of multiple resistance mechanisms. Over time, with exposure to the selective pressures of different TKIs, the clonal composition of these tumors can continue to evolve (68, 74, 118).
Liquid Biopsies
The ability to detect and characterize polyclonal resistance in patients is critical, as targeting only one subclonal cell population can potentially lead to therapy failures (118). As multiple tissue biopsies are usually not feasible, non-invasive liquid biopsies and analyses of circulating tumor DNA (ctDNA) represent a promising avenue to identify polyclonal resistance. The potential of liquid biopsies was recently highlighted in the context of colorectal cancer. In a patient progressing on the anti-EGFR monoclonal antibody cetuximab, discrete metastatic lesions were found to harbor different resistance mechanisms (e.g., a MAP2K1 mutation and a KRAS mutation), modulating treatment responses in a lesion-specific manner. While single-lesion tumor biopsies failed to capture this heterogeneity, both resistance mutations were detected in the patient’s serial plasma ctDNA (118).
In lung cancer, significant inroads have been made in using plasma-based assays to detect EGFR mutations, including the T790M gatekeeper mutation (119, 120). Indeed, a plasma-based EGFR mutation test was recently FDA-approved based on data from a phase IV single-arm study of gefitinib (121), serving as a proof of concept that this diagnostic approach can be clinically implemented. Studies are now assessing whether liquid biopsies can similarly detect ALK fusions and resistance mutations in patients treated with ALK TKIs. Preliminary results appear promising (122, 123). In one study, cell-free DNA (cfDNA) isolated from plasma could be used to monitor the emergence and temporal evolution of ALK resistance mutations, including G1202R, during treatment with alectinib (122). However, more data is required before plasma-based assays can be routinely used in the clinic for monitoring ALK-positive patients. First, the ability of liquid biopsies to detect oncogenic fusions in general needs to be optimized. Second, their utility in real-time treatment monitoring and therapeutic stratification also needs to be further demonstrated through prospective and clinical intervention studies; this is one goal of the NCI-sponsored ALK Master Protocol. Ultimately, liquid biopsies have the potential to become a powerful diagnostic tool that complements repeat tumor biopsies, and at the same time, allows for more precise understanding at the genetic level of the longitudinal evolution and global landscape of resistance mechanisms.
DEVELOPING STRATEGIES TO OVERCOME RESISTANCE
Our understanding of the molecular mechanisms of ALK TKI resistance continues to mature, informing the development of new therapeutic strategies. The aim of targeting mechanisms of TKI resistance is two-fold: (i) effectively treat the relapsed disease by overcoming the dominant resistance mechanism, and (ii) enhance the depth and duration of tumor response upfront by preventing the emergence of resistance (60). While most efforts thus far have focused on the former, strategies to prevent resistance upfront are likely to have greater impact in the clinic.
Novel ALK TKIs: Many Options
Given the wide array of resistance mutations that can arise, multiple structurally distinct ALK inhibitors are needed. ALK TKIs that are currently available or being developed are listed in Table 2. As with the approved inhibitors crizotinib, ceritinib and alectinib, we anticipate that these emerging ALK TKIs will: (i) possess different potencies against resistant ALK mutants, (ii) differ in target kinase selectivity, and (iii) give rise to a distinct spectrum of ALK resistance mutations.
Table 2.
Pharmacologic properties of ALK inhibitors approved by the US FDA or in clinical testing.
| ALK TKI | Crizotinib (PF-02341066) |
Ceritinib (LDK378) |
Alectinib (RO/CH5424802) |
Brigatinib (AP26113) |
Lorlatinib (PF-06463922) |
Entrectinib (RXDX-101) |
Ensartinib (X-396) |
|---|---|---|---|---|---|---|---|
| Manufacturer | Pfizer | Novartis | Genentech | Ariad | Pfizer | Ignyta | Xcovery |
| Targets other than ALK | ROS1MET |
ROS1 IGF-1R IR |
GAK LTK RET |
ROS1 | ROS1 |
NTRK1 NTRK2 NTRK3 ROS1 |
ROS1 MET AXL |
| Resistance mutations known to be targeted by TKI | L1198F | I1171T/N L1196M S1206C/Y G1269A/S |
L1152P/R C1156Y/T F1174C/L/V L1196M S1206C/Y G1269A/S |
I1151Tins L1152P/R C1156Y/T F1174C/L/V L1196M G1202Ra G1269A/S |
I1151Tins L1152P/R C1156Y/T I1171T/N/S F1174C/L/V L1196M G1202Rb S1206C/Y E1210K G1269A/S |
C1156Y/T L1196M |
C1156Y/T L1196M |
| Reported resistance mutations to the TKI | I1151Tins L1152P/R C1156Y/T I1171T/N/S F1174C/L/V V1180L L1196M G1202R S1206C/Y E1210K G1269A/S |
I1151Tins L1152P/R C1156Y/T F1174C/L/V G1202R |
I1171T/N/S V1180L G1202R |
G1202Ra E1210K + S1206C E1210K + D1203N |
L1198F + C1156Yc | G1202R | N.D. |
| Regulatory approval | Approved for 1L and beyond | Approved for crizotinib-pretreated | Approved for crizotinib-pretreated; Breakthrough therapy designation for 1L | Breakthrough therapy designation for crizotinib-pretreated | N/A | N/A | N/A |
| Phase of testing | Phase III complete | III | III | III | III | II | III |
| References | 3, 4, 74, 78 | 73, 78 | 78, 137, 184 | 78, 124–130 | 74, 78, 131 | 132–134 | 135, 136 |
Abbreviations: ALK, anaplastic lymphoma kinase; US, United States; FDA, Food and Drug Administration; TKI, tyrosine kinase inhibitor; IGF-1R, insulin-like growth factor 1 receptor; IR, insulin receptor; GAK, cyclin G-associated kinase; LTK, leukocyte receptor tyrosine kinase; RET, rearranged during transfection; TRK, tropomyosin receptor kinase; N.D., not determined; 1L, first-line; N/A, not applicable.
Brigatinib has been reported to have some activity against the ALK G1202R mutation (128–130), but G1202R has also been detected in biopsy specimens from patients with ALK-rearranged NSCLC who relapsed on brigatinib (78), suggesting that its potency may be compromised against this mutation.
G1202R is a highly refractory mutation resistant to all first- and second-generation inhibitors, but targeted by lorlatinib (74, 78).
L1198F has been found to confer resistance to lorlatinib in the context of another mutation, C1156Y (74).
For example, brigatinib (AP26113) is a potent ALK inhibitor that inhibits most crizotinib-resistant ALK mutants (124–129). In the phase II ALTA trial, 222 patients with advanced ALK-positive NSCLC who progressed on crizotinib were randomized to two different dosing schedules of brigatinib. The confirmed ORR ranged from 45–54%, with median PFS of 9.2–12.9 months (127), similar to the phase II data seen with alectinib. A phase III trial (ALTA-1L; NCT02737501) is ongoing to assess the efficacy of brigatinib versus crizotinib in TKI-naïve ALK-positive NSCLC. In the interim, brigatinib has received breakthrough therapy designation by the FDA for the treatment of crizotinib-resistant, ALK-rearranged NSCLC. Some preclinical studies of brigatinib suggest activity against the G1202R mutation in cell lines and mouse models (128, 130), and a confirmed response to brigatinib has been reported in a patient with G1202R-mutant NSCLC (129). However, G1202R has also been detected in 43% (3/7) of ALK-positive NSCLC biopsies post-brigatinib (78). More importantly, a patient who relapsed after brigatinib with documented ALK G1202R in a repeat biopsy went on to have a durable response to lorlatinib (131), suggesting that while brigatinib may have some activity against G1202R, its potency is compromised by this mutation.
Entrectinib (RXDX-101) and ensartinib (X-396) are notable for their additional kinase targets. Entrectinib inhibits TRK in addition to ROS1 and ALK (132, 133). Updated results from a phase I study of entrectinib showed significant responses in TKI-naïve patients with NTRK-, ROS1-, and ALK1-rearranged solid tumors (confirmed ORR of 100%, 86%, and 57%, respectively) (134). A phase II basket study of entrectinib is currently recruiting participants (NCT02568267). Similar to crizotinib, ensartinib has activity against ROS1 and MET in addition to ALK, and also targets AXL (135, 136). Preliminary analysis of ensartinib’s activity in ALK-rearranged NSCLC demonstrated responses in 7 of 8 patients who were crizotinib-naïve and in 11 of 19 patients who were previously treated with crizotinib (136). A phase III randomized trial (eXalt3; NCT02767804) will compare ensartinib to crizotinib in TKI-naïve ALK-positive NSCLC.
Lastly, lorlatinib (PF-06463922) is a third-generation ALK TKI that offers several advantages over second-generation TKIs. Lorlatinib has activity against all of the known ALK resistance mutations including G1202R, and is highly selective for ALK/ROS1 (78, 82, 137). In the phase I portion of an ongoing phase I/II study (NCT01970865), lorlatinib was associated with an ORR of 46% among 41 patients with ALK-positive NSCLC, many of whom had progressed after two or more ALK TKIs (85). Responses to lorlatinib have also been reported in patients with ALK G1202R-positive NSCLC (78, 85, 131). Notably, lorlatinib was developed to evade P-gp-mediated efflux, and thus can achieve excellent CNS penetration (105). In the phase I study, the intracranial ORR to lorlatinib was 39% among all patients (85), and CNS responses have been noted after failure of prior crizotinib, ceritinib and alectinib, including dramatic improvements in leptomeningeal disease. Despite its promising activity, resistance to lorlatinib also emerges. A compound C1156Y/L1198F mutation was detected in a patient relapsing on lorlatinib (74). Structural studies suggest that this mutation causes steric hindrance to the binding of lorlatinib, and when superimposed on another ALK mutation, C1156Y, it confers high level resistance to lorlatinib (74). Studies are ongoing to uncover additional lorlatinib resistance mechanisms.
Sequencing of ALK TKIs
The recent advances reviewed above underscore the critical need for repeat biopsies to guide therapeutic strategies. We strongly recommend pursuing repeat biopsies—when feasible and safe—in patients progressing on an ALK TKI. Particularly for patients who relapse on a second-generation ALK TKI, the detection of on-target versus off-target resistance mechanisms in repeat biopsy specimens is an essential factor in selecting the next therapy. The former generally indicates persistent ALK dependency and potential sensitivity to another ALK TKI, while the latter suggests the need to pursue an alternative approach (e.g., clinical trial of an ALK-based combination regimen or standard cytotoxic chemotherapy) (Figure 4). In addition, the detection of a particular ALK resistance mutation may inform the choice of the next ALK TKI, as discussed above, both in the crizotinib-resistant setting as well as after failure of a second-generation ALK TKI.
While general guidelines are emerging for determining the optimal later-line ALK TKI, which TKI to use in the first-line setting remains an ongoing controversy. Currently, crizotinib is the only FDA-approved agent for use in TKI-naïve ALK-positive NSCLC (4); a second-generation ALK inhibitor (e.g., ceritinib, alectinib) can be used once patients relapse on, or are intolerant of, crizotinib. However, the upfront use of a more potent and selective second-generation ALK inhibitor may substantially delay disease progression through various mechanisms. First, next-generation ALK TKIs have activity against multiple crizotinib-resistant mutations including the most common L1196M and G1269A mutations (82, 138), and may thus suppress the outgrowth of any pre-existing clones that harbor these mutations and also prevent them from emerging de novo. Additionally, most second-generation TKIs have greater CNS activity than crizotinib, and would hence delay the development of brain and leptomeningeal metastases which are commonly seen in crizotinib-treated patients (7, 8, 139, 140).
The first reported results comparing a second-generation ALK TKI to crizotinib in the TKI-naïve setting came from the J-ALEX study, comparing alectinib to crizotinib in Japanese patients with ALK-positive NSCLC. Preliminary results suggest that front-line alectinib may be superior to crizotinib (141). The median PFS in the crizotinib arm was 10.2 months (95% confidence interval (CI), 8.2–12.0 months), while the median PFS was not reached in the alectinib arm (95% CI, 20.3 months-not estimated) (141). Based on these results, alectinib was recently granted FDA breakthrough therapy designation for first-line treatment of ALK-positive NSCLC. It is worth noting that there are a number of important limitations to the J-ALEX study, including an imbalance of patients with baseline brain metastases and a higher than expected rate of toxicities with crizotinib. A similar but global phase III trial (ALEX) comparing first-line alectinib to crizotinib in ALK-positive NSCLC is ongoing (NCT02075840), and results of this study will likely be practice-changing. The ALEX trials have not allowed patient crossover at the time of progression. Therefore, they will not address the more relevant comparison of first-line alectinib versus sequential crizotinib followed by alectinib.
Combination Regimens
The knowledge of bypass signaling tracks that can foster ALK TKI resistance has fueled efforts to develop combinatorial approaches for use in patients who relapse on TKI therapy, or in the upfront setting to delay resistance and potentially enable more durable responses than those achieved using monotherapy alone. Similarly, the discovery that each ALK TKI is associated with a unique spectrum of ALK resistance mutations suggests that combinations of ALK TKIs could be beneficial in overcoming or preventing on-target resistance mechanisms. Different combinations of targeted agents, chemotherapies, and/or immunotherapies with ALK TKIs are currently being evaluated. However, many of the ongoing studies are not biomarker-driven.
Clinical trials to test the efficacy of dual ALK and MEK blockade are being developed in light of the compelling preclinical data discussed above (64, 92). Combinations of ceritinib and LEE011, a CDK4/6 inhibitor, and of ceritinib and everolimus, an mTOR inhibitor, are in early-phase testing in NSCLC (NCT02292550 and NCT02321501, respectively); preclinical data for these approaches are limited. The combination of alectinib with bevacizumab, an anti-angiogenesis agent targeting vascular endothelial growth factor (VEGF), is being tested in patients with ALK-positive NSCLC with at least one CNS target lesion (NCT02521051). The rationale is that bevacizumab may help augment the systemic and intracranial drug activity by modulating the tumor vasculature (142). Findings from the ALK TKI resistance studies described above would also support efforts to therapeutically co-target ALK with MET, EGFR, KIT, or SRC (Figure 1).
Several studies are investigating the efficacy and tolerability of an ALK TKI combined with immunotherapy in lung cancer (e.g., crizotinib with nivolumab or ipilimumab (NCT01998126) or pembrolizumab (NCT02511184); ceritinib with nivolumab (NCT02393625); alectinib with atezolizumab (NCT02013219); lorlatinib with avelumab (NCT02584634)). However, there is limited preclinical data thus far to support this combination strategy. Although PD-1/PD-L1 inhibitors have demonstrated durable activity in a subset of NSCLC, responses are limited to ~20% of patients, and have been associated with high PD-L1 expression, high tumor mutational load, and smoking history (143–150). ALK-positive NSCLC patients tend to be never-smokers with a low tumor mutational load (151). Moreover, a recent study demonstrated that ALK-positive NSCLCs tend to lack concurrent PD-L1 expression and CD8+ tumor-infiltrating lymphocytes in the tumor microenvironment—an important component of response to immunotherapy (152). Indeed, no responses were seen among 6 ALK-positive NSCLC patients treated with checkpoint inhibitors in a small retrospective analysis (152), and in large randomized studies of previously treated NSCLC, subgroup analyses demonstrated no survival benefit to checkpoint inhibitors over chemotherapy in never-smokers and/or EGFR-mutant patients (144, 145). Therefore, the potential benefit of combining immunotherapies with ALK TKIs is unclear at this time, and the optimal sequencing and timing of such an approach warrants careful investigation. In addition, future studies will need to define the level of PD-L1 expression in a larger cohort of ALK-positive (and other oncogene-driven) NSCLCs, and explore whether additional biomarkers predictive of response to immunotherapy may be present in a subset of these patients.
Ultimately, any successful combination regimen will need to have demonstrated not only superior efficacy compared to monotherapy, but also tolerability and feasible dosing in patients. Toxicities are often exacerbated when two drugs are given in combination, and even unanticipated toxicities may arise. For example, combination of the third-generation EGFR TKI osimertinib plus the PD-L1 inhibitor durvalumab (MEDI4736) was found to cause interstitial lung disease in 38% of the patients, compared to 2–3% in patients receiving either osimertinib or durvalumab as monotherapy (153). Similarly, combined pan-HER and ALK/ROS1/MET inhibition using dacomitinib and crizotinib, respectively, led to grade 3 or 4 treatment-related adverse events in 43% of NSCLC patients, most common of which were diarrhea, rash, and fatigue (154). Design of alternative dosing schedules, such as the use of submaximal doses of each individual drug, or pulsed or intermittent dosing, may help mitigate toxicities.
Targeting Oligoprogressive or Oligopersistent Disease
Clinically, the course and pace of disease progression on a TKI can be variable among patients. While most patients develop systemic, multisite progression requiring a change in systemic therapy, a subset of patients develop progression limited to only one or a few anatomic sites, with the remaining disease sites continuing to be controlled by the TKI. This phenomenon has been termed “oligoprogression” (155, 156). In the case of oligoprogressive disease, local ablative therapy (LAT) using surgery or radiation may serve as a strategy that offers several advantages (Figure 4). First, LAT can eradicate the TKI-resistant clone(s) at the progressing sites and potentially delay the evolution of more heterogeneous resistant tumor cell populations. Second, it permits the continued, maximal use of the TKI that is otherwise active against sensitive tumor clones in sites that are not progressing.
The feasibility and utility of the LAT approach has been suggested by a limited number of single-institution, retrospective studies. In one study including 25 EGFR-mutant and ALK-positive NSCLC patients with oligoprogressive disease on erlotinib and crizotinib, respectively, LAT followed by recommencement of the TKI led to an additional 6.2 months of median PFS (157). In an updated analysis of 14 ALK-positive NSCLC patients with extracranial oligoprogression on crizotinib, LAT led to 6- and 12-month actuarial local lesion control rate of 100% and 86%, respectively. The median time to second progression was 5.5 months, and median duration on crizotinib was 28 months (158). These results provide the rationale for larger, prospective studies of the local ablative strategy.
Taken one step further, patients may achieve an overall tumor response to a TKI but continue to have a few sites of persistent, residual disease (“oligopersistent disease”) (155). Similar to the oligoprogressive situation, these sites of oligopersistent disease may serve as a reservoir of residual, TKI-insensitive tumor cells that can eventually drive systemic therapy failure (159). In one randomized phase II study, the efficacy of adding local consolidative therapy after induction systemic therapy was compared to maintenance systemic therapy or surveillance in oligometastatic NSCLC (160). Among 49 patients including 8 with EGFR-mutant or ALK-positive NSCLC who achieved disease control after the induction systemic therapy (e.g., platinum-doublet chemotherapy, erlotinib, or crizotinib), the addition of local therapy versus no local therapy led to an improved median PFS (11.9 months versus 3.9 months), suggesting that aggressive treatment of oligopersistent sites of disease may help improve patient outcomes. Another phase II trial is assessing the efficacy of treating up to 5 sites of oligopersistent disease using stereotactic body radiation therapy (SBRT) within 6 months of initiating TKI therapy, specifically in patients with EGFR-mutant, ALK-rearranged, or ROS1-rearranged NSCLC (NCT02314364).
Targeting Persister Cells
An emerging question in ALK-rearranged cancers involves the role of drug-tolerant persister cells in driving residual disease on TKIs and ultimately therapy failure. Recent work suggests that persister cells, which survive the initial TKI exposure through adaptive mechanisms, can eventually acquire overt genetic alterations leading to full-fledged resistance and clinical relapse (161, 162).
In EGFR-mutant NSCLC, persister cells have been identified as an important nidus for the de novo development of heterogeneous EGFR TKI resistance mechanisms including EGFR T790M (162, 163). The persister cell-derived, de novo EGFR T790M-mutant cells emerge later than, and are biologically distinct from, the early-resistant, pre-existing EGFR T790M-mutant cells (163). Compared to early-arising T790M-mutant cells, late-arising T790M-mutant cells exhibit diminished apoptotic response to the T790M-mutant-selective EGFR TKI WZ4002. However, this can be overcome by the addition of navitoclax, a BCL2 inhibitor. Based on these preclinical findings, the combination of osimertinib and navitoclax is now being evaluated in EGFR-mutant lung cancer patients after progression on a first-generation EGFR TKI (NCT02520778).
A role for persister cells in mediating ALK TKI resistance has yet to be determined. It will be important to characterize the biology of persister cells and identify their therapeutic vulnerabilities which could be targeted to prevent de novo resistance. More broadly, there is a growing need to focus research efforts on designing and validating strategies to eradicate residual disease in order to suppress the emergence of resistance upfront. This anticipatory approach—rather than treating resistance once it has already developed—has the greatest potential to truly transform patient outcomes.
CONCLUSIONS
ALK is an established therapeutic target in lung cancer and several other hematologic and solid malignancies, including ALCL and IMT. Since its discovery as a fusion oncogene in 1994, much insight has been gained into the biology of both native and oncogenic ALK. In parallel, numerous ALK inhibitors have entered the clinic, and to date, three have become standard therapies for advanced ALK-positive lung cancer.
Despite the remarkable responses seen with ALK TKIs, patients invariably relapse due to acquired resistance, and therefore, developing strategies to overcome or prevent resistance is an urgent priority. With the growing knowledge of resistance mechanisms, new treatment approaches can be rationally designed. These new approaches hold the promise of more effectively overcoming and suppressing drug resistance, translating into deeper and more prolonged responses in patients with ALK-driven cancers.
Significance.
Effective long-term treatment of ALK-rearranged cancers requires a mechanistic understanding of resistance to ALK TKIs so that rational therapies can be selected to combat resistance. This Review underscores the importance of serial biopsies in capturing the dynamic therapeutic vulnerabilities within a patient’s tumor, and offers a perspective into the complexity of on-target and off-target ALK TKI resistance mechanisms. Therapeutic strategies that can successfully overcome, and potentially prevent, these resistance mechanisms will have the greatest impact on patient outcome.
Acknowledgments
GRANT SUPPORT
This work was supported by grants from the National Cancer Institute (5R01CA164273, to ATS) and the National Foundation for Cancer Research (to ATS).
We apologize to the numerous colleagues whose important contributions could not be cited in this Review due to space constraints. This work was supported by grants from the National Cancer Institute (5R01CA164273, to A.T.S.), and the National Foundation for Cancer Research (to A.T.S), and by Be a Piece of the Solution and LungStrong.
Footnotes
Disclosures: GJR has served as a compensated consultant to Genentech/Roche, and his institution receives clinical research support from Pfizer, Novartis, Genentech/Roche, Ariad, and Millennium. ATS has served as a compensated consultant or received honoraria from Pfizer, Novartis, Genentech/Roche, Ariad, Ignyta, Daiichi-Sankyo, Taiho, Blueprint Medicines, Loxo, EMD Serono, and Foundation Medicine. JJL has no financial interests to declare.
References
- 1.Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–4. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- 2.Soda M, Choi YL, Enomoto M, Takada S, Yamashita Y, Ishikawa S, et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature. 2007;448:561–6. doi: 10.1038/nature05945. [DOI] [PubMed] [Google Scholar]
- 3.Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn MJ, et al. Crizotinib versus chemotherapy in advanced ALK-positive lung cancer. N Engl J Med. 2013;368:2385–94. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 4.Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa K, Mekhail T, et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N Engl J Med. 2014;371:2167–77. doi: 10.1056/NEJMoa1408440. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AT, Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N Engl J Med. 2014;370:1189–97. doi: 10.1056/NEJMoa1311107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim DW, Mehra R, Tan DS, Felip E, Chow LQ, Camidge DR, et al. Activity and safety of ceritinib in patients with ALK-rearranged non-small-cell lung cancer (ASCEND-1): updated results from the multicentre, open-label, phase 1 trial. Lancet Oncol. 2016;17:452–63. doi: 10.1016/S1470-2045(15)00614-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou SH, Ahn JS, De Petris L, Govindan R, Yang JC, Hughes B, et al. Alectinib in crizotinib-refractory ALK-rearranged non-small-cell lung cancer: a phase II global study. J Clin Oncol. 2016;34:661–8. doi: 10.1200/jco.2015.63.9443. [DOI] [PubMed] [Google Scholar]
- 8.Shaw AT, Gandhi L, Gadgeel S, Riely GJ, Cetnar J, West H, et al. Alectinib in ALK-positive, crizotinib-resistant, non-small-cell lung cancer: a single-group, multicentre, phase 2 trial. Lancet Oncol. 2016;17:234–42. doi: 10.1016/S1470-2045(15)00488-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwahara T, Fujimoto J, Wen D, Cupples R, Bucay N, Arakawa T, et al. Molecular characterization of ALK, a receptor tyrosine kinase expressed specifically in the nervous system. Oncogene. 1997;14:439–49. doi: 10.1038/sj.onc.1200849. [DOI] [PubMed] [Google Scholar]
- 10.Reshetnyak AV, Murray PB, Shi X, Mo ES, Mohanty J, Tome F, et al. Augmentor α and β (FAM150) are ligands of the receptor tyrosine kinases ALK and LTK: Hierarchy and specificity of ligand-receptor interactions. Proc Natl Acad Sci U S A. 2015;112:15862–7. doi: 10.1073/pnas.1520099112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murray PB, Lax I, Reshetnyak A, Ligon GF, Lillquist JS, Natoli EJ, Jr, et al. Heparin is an activating ligand of the orphan receptor tyrosine kinase ALK. Sci Signal. 2015;8:ra6. doi: 10.1126/scisignal.2005916. [DOI] [PubMed] [Google Scholar]
- 12.Lemke G. Adopting ALK and LTK. Proc Natl Acad Sci U S A. 2015;112:15783–4. doi: 10.1073/pnas.1521923113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hallberg B, Palmer RH. Mechanistic insight into ALK receptor tyrosine kinase in human cancer biology. Nat Rev Cancer. 2013;13:685–700. doi: 10.1038/nrc3580. [DOI] [PubMed] [Google Scholar]
- 14.Bazigou E, Apitz H, Johansson J, Lorén CE, Hirst EM, Chen PL, et al. Anterograde Jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell. 2007;128:961–75. doi: 10.1016/j.cell.2007.02.024. [DOI] [PubMed] [Google Scholar]
- 15.Englund C, Lorén CE, Grabbe C, Varshney GK, Deleuil F, Hallberg B, et al. Jeb signals through the Alk receptor tyrosine kinase to drive visceral muscle fusion. Nature. 2003;425:512–6. doi: 10.1038/nature01950. [DOI] [PubMed] [Google Scholar]
- 16.Lee HH, Norris A, Weiss JB, Frasch M. Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature. 2003;425:507–12. doi: 10.1038/nature01916. [DOI] [PubMed] [Google Scholar]
- 17.Bilsland JG, Wheeldon A, Mead A, Znamenskiy P, Almond S, Waters KA, et al. Behavioral and neurochemical alterations in mice deficient in anaplastic lymphoma kinase suggest therapeutic potential for psychiatric indications. Neuropsychopharmacology. 2008;33:685–700. doi: 10.1038/sj.npp.1301446. [DOI] [PubMed] [Google Scholar]
- 18.Lasek AW, Lim J, Kliethermes CL, Berger KH, Joslyn G, Brush G, et al. An evolutionary conserved role for anaplastic lymphoma kinase in behavioral responses to ethanol. PLoS One. 2011;6:e22636. doi: 10.1371/journal.pone.0022636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Witek B, El Wakil A, Nord C, Ahlgren U, Eriksson M, Vernersson-Lindahl E, et al. Targeted disruption of ALK reveals a potential role in hypogonadotropic hypogonadism. PLoS One. 2015;10:e0123542. doi: 10.1371/journal.pone.0123542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amin HM, Lai R. Pathobiology of ALK+ anaplastic large-cell lymphoma. Blood. 2007;110:2259–67. doi: 10.1182/blood-2007-04-060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamant L, Dastugue N, Pulford K, Delsol G, Mariamé B. A new fusion gene TPM3-ALK in anaplastic large cell lymphoma created by a (1;2)(q25;p23) translocation. Blood. 1999;93:3088–95. [PubMed] [Google Scholar]
- 22.Meech SJ, McGavran L, Odom LF, Liang X, Meltesen L, Gump J, et al. Unusual childhood extramedullary hematologic malignancy with natural killer cell properties that contains tropomyosin 4-anaplastic lymphoma kinase gene fusion. Blood. 2001;98:1209–16. doi: 10.1182/blood.v98.4.1209. [DOI] [PubMed] [Google Scholar]
- 23.Hernández L, Beà S, Bellosillo B, Pinyol M, Falini B, Carbone A, et al. Diversity of genomic breakpoints in TFG-ALK translocations in anaplastic large cell lymphomas: identification of a new TFG-ALK(XL) chimeric gene with transforming activity. Am J Pathol. 2002;160:1487–94. doi: 10.1016/S0002-9440(10)62574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Colleoni GW, Bridge JA, Garicochea B, Liu J, Filippa DA, Ladanyi M. ATIC-ALK: a novel variant ALK gene fusion in anaplastic large cell lymphoma resulting from the recurrent cryptic chromosomal inversion, inv(2)(p23q35) Am J Pathol. 2000;156:781–9. doi: 10.1016/S0002-9440(10)64945-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cools J, Wlodarska I, Somers R, Mentens N, Pedeutour F, Maes B, et al. Identification of novel fusion partners of ALK, the anaplastic lymphoma kinase, in anaplastic large-cell lymphoma and inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2002;34:354–62. doi: 10.1002/gcc.10033. [DOI] [PubMed] [Google Scholar]
- 26.Touriol C, Greenland C, Lamant L, Pulford K, Bernard F, Rousset T, et al. Further demonstration of the diversity of chromosomal changes involving 2p23 in ALK-positive lymphoma: 2 cases expressing ALK kinase fused to CLTCL (clathrin chain polypeptide-like) Blood. 2000;95:3204–7. [PubMed] [Google Scholar]
- 27.Abate F, Todaro M, van der Krogt JA, Boi M, Landra I, Machiorlatti R, et al. A novel patient-derived tumorgraft model with TRAF1-ALK anaplastic large-cell lymphoma translocation. Leukemia. 2015;29:1390–401. doi: 10.1038/leu.2014.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tort F, Pinyol M, Pulford K, Roncador G, Hernandez L, Nayach I, et al. Molecular characterization of a new ALK translocation involving moesin (MSN-ALK) in anaplastic large cell lymphoma. Lab Invest. 2001;81:419–26. doi: 10.1038/labinvest.3780249. [DOI] [PubMed] [Google Scholar]
- 29.Lamant L, Gascoyne RD, Duplantier MM, Armstrong F, Raghab A, Chhanabhai M, et al. Non-muscle myosin heavy chain (MYH9): a new partner fused to ALK in anaplastic large cell lymphoma. Genes Chromosomes Cancer. 2003;37:427–32. doi: 10.1002/gcc.10232. [DOI] [PubMed] [Google Scholar]
- 30.Lovly CM, Gupta A, Lipson D, Otto G, Brennan T, Chung CT, et al. Inflammatory myofibroblastic tumors harbor multiple potentially actionable kinase fusions. Cancer Discov. 2014;4:889–95. doi: 10.1158/2159-8290.CD-14-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, et al. TPM3-ALK and TPM4-ALK oncogenes in inflammatory myofibroblastic tumors. Am J Pathol. 2000;157:377–84. doi: 10.1016/S0002-9440(10)64550-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bridge JA, Kanamori M, Ma Z, Pickering D, Hill DA, Lydiatt W, et al. Fusion of the ALK gene to the clathrin heavy chain gene, CLTC, in inflammatory myofibroblastic tumor. Am J Pathol. 2001;159:411–5. doi: 10.1016/S0002-9440(10)61711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Debiec-Rychter M, Marynen P, Hagemeijer A, Pauwels P. ALK-ATIC fusion in urinary bladder inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;38:187–90. doi: 10.1002/gcc.10267. [DOI] [PubMed] [Google Scholar]
- 34.Ma Z, Hill DA, Collins MH, Morris SW, Sumegi J, Zhou M, et al. Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;37:98–105. doi: 10.1002/gcc.10177. [DOI] [PubMed] [Google Scholar]
- 35.Debelenko LV, Arthur DC, Pack SD, Helman LJ, Schrump DS, Tsokos M. Identification of CARS-ALK fusion in primary and metastatic lesions of an inflammatory myofibroblastic tumor. Lab Invest. 2003;83:1255–65. doi: 10.1097/01.lab.0000088856.49388.ea. [DOI] [PubMed] [Google Scholar]
- 36.Panagopoulos I, Nilsson T, Domanski HA, Isaksson M, Lindblom P, Mertens F, et al. Fusion of the SEC31L1 and ALK genes in an inflammatory myofibroblastic tumor. Int J Cancer. 2006;118:1181–6. doi: 10.1002/ijc.21490. [DOI] [PubMed] [Google Scholar]
- 37.Takeuchi K, Soda M, Togashi Y, Sugawara E, Hatano S, Asaka R, et al. Pulmonary inflammatory myofibroblastic tumor expressing a novel fusion, PPFIBP1-ALK: reappraisal of anti-ALK immunohistochemistry as a tool for novel ALK fusion identification. Clin Cancer Res. 2011;17:3341–8. doi: 10.1158/1078-0432.CCR-11-0063. [DOI] [PubMed] [Google Scholar]
- 38.Shaw AT, Yeap BY, Mino-Kenudson M, Digumarthy SR, Costa DB, Heist RS, et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J Clin Oncol. 2009;27:4247–53. doi: 10.1200/JCO.2009.22.6993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 40.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 41.Takeuchi K, Choi YL, Togashi Y, Soda M, Hatano S, Inamura K, et al. KIF5B-ALK, a novel fusion oncokinase identified by an immunohistochemistry-based diagnostic system for ALK-positive lung cancer. Clin Cancer Res. 2009;15:3143–9. doi: 10.1158/1078-0432.CCR-08-3248. [DOI] [PubMed] [Google Scholar]
- 42.Togashi Y, Soda M, Sakata S, Sugawara E, Hatano S, Asaka R, et al. KLC1-ALK: a novel fusion in lung cancer identified using a formalin-fixed paraffin-embedded tissue only. PLoS One. 2012;7:e31323. doi: 10.1371/journal.pone.0031323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jung Y, Kim P, Jung Y, Keum J, Kim SN, Choi YS, et al. Discovery of ALK-PTPN3 gene fusion from human non-small cell lung carcinoma cell line using next generation RNA sequencing. Genes Chromosomes Cancer. 2012;51:590–7. doi: 10.1002/gcc.21945. [DOI] [PubMed] [Google Scholar]
- 44.Choi YL, Lira ME, Hong M, Kim RN, Choi SJ, Song JY, et al. A novel fusion of TPR and ALK in lung adenocarcinoma. J Thorac Oncol. 2014;9:563–6. doi: 10.1097/JTO.0000000000000093. [DOI] [PubMed] [Google Scholar]
- 45.Tan DS, Kim DW, Thomas M, Pantano S, Wang Y, Szpakowski SL, et al. Genetic landscape of ALK+ non-small cell lung cancer (NSCLC) patients (pts) and response to ceritinib in ASCEND-1. J Clin Oncol. 2016;34(suppl) doi: 10.1016/j.lungcan.2021.11.007. abstr 9064. [DOI] [PubMed] [Google Scholar]
- 46.Shaw AT, Engelman JA. ALK in lung cancer: past, present, and future. J Clin Oncol. 2013;31:1105–11. doi: 10.1200/JCO.2012.44.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katayama R, Lovly CM, Shaw AT. Therapeutic targeting of anaplastic lymphoma kinase in lung cancer: a paradigm for precision cancer medicine. Clin Cancer Res. 2015;21:2227–35. doi: 10.1158/1078-0432.CCR-14-2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Armstrong F, Duplantier MM, Trempat P, Hieblot C, Lamant L, Espinos E, et al. Differential effects of X-ALK fusion proteins on proliferation, transformation, and invasion properties of NIH3T3 cells. Oncogene. 2004;23:6071–82. doi: 10.1038/sj.onc.1207813. [DOI] [PubMed] [Google Scholar]
- 49.Lin JJ, Shaw AT. Differential sensitivity to crizotinib: does EML4-ALK fusion variant matter? J Clin Oncol. 2016;34:3363–5. doi: 10.1200/JCO.2016.68.5891. [DOI] [PubMed] [Google Scholar]
- 50.Chiarle R, Voena C, Ambrogio C, Piva R, Inghirami G. The anaplastic lymphoma kinase in the pathogenesis of cancer. Nat Rev Cancer. 2008;8:11–23. doi: 10.1038/nrc2291. [DOI] [PubMed] [Google Scholar]
- 51.Zhang G, Scarborough H, Kim J, Rozhok AI, Chen YA, Zhang X, et al. Coupling an EML4-ALK-centric interactome with RNA interference identifies sensitizers to ALK inhibitors. Sci Signal. 2016;9:rs12. doi: 10.1126/scisignal.aaf5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soda M, Takada S, Takeuchi K, Choi YL, Enomoto M, Ueno T, et al. A mouse model for EML4-ALK-positive lung cancer. Proc Natl Acad Sci U S A. 2008;105:19893–7. doi: 10.1073/pnas.0805381105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zou HY, Li Q, Lee JH, Arango ME, McDonnell SR, Yamazaki S, et al. An orally available small-molecule inhibitor of c-Met, PF-2341066, exhibits cytoreductive antitumor efficacy through antiproliferative and antiangiogenic mechanisms. Cancer Res. 2007;67:4408–17. doi: 10.1158/0008-5472.CAN-06-4443. [DOI] [PubMed] [Google Scholar]
- 54.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N Engl J Med. 2010;363:1693–703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gettinger SN, Bazhenova LA, Langer CJ, Salgia R, Gold KA, Rosell R, et al. Activity and safety of brigatinib in ALK-rearranged non-small-cell lung cancer and other malignancies: a single-arm, open-label, phase 1/2 trial. Lancet Oncol. 2016 doi: 10.1016/S1470-2045(16)30392-8. [DOI] [PubMed] [Google Scholar]
- 56.Gambacorti Passerini C, Farina F, Stasia A, Redaelli S, Ceccon M, Mologni L, et al. Crizotinib in advanced, chemoresistant anaplastic lymphoma kinase-positive lymphoma patients. J Natl Cancer Inst. 2014;106:djt378. doi: 10.1093/jnci/djt378. [DOI] [PubMed] [Google Scholar]
- 57.Richly H, Kim TM, Schuler M, Kim DW, Harrison SJ, Shaw AT, et al. Ceritinib in patients with advanced anaplastic lymphoma kinase-rearranged anaplastic large-cell lymphoma. Blood. 2015;126:1257–8. doi: 10.1182/blood-2014-12-617779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Butrynski JE, D’Adamo DR, Hornick JL, Dal Cin P, Antonescu CR, Jhanwar SC, et al. Crizotinib in ALK-rearranged inflammatory myofibroblastic tumor. N Engl J Med. 2010;363:1727–33. doi: 10.1056/NEJMoa1007056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greaves M. Evolutionary determinants of cancer. Cancer Discov. 2015;5:806–20. doi: 10.1158/2159-8290.CD-15-0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garraway LA, Jänne PA. Circumventing cancer drug resistance in the era of personalized medicine. Cancer Discov. 2012;2:214–26. doi: 10.1158/2159-8290.CD-12-0012. [DOI] [PubMed] [Google Scholar]
- 61.Choi YL, Soda M, Yamashita Y, Ueno T, Takashima J, Nakajima T, et al. EML4-ALK mutations in lung cancer that confer resistance to ALK inhibitors. N Engl J Med. 2010;363:1734–9. doi: 10.1056/NEJMoa1007478. [DOI] [PubMed] [Google Scholar]
- 62.Sasaki T, Koivunen J, Ogino A, Yanagita M, Nikiforow S, Zheng W, et al. A novel ALK secondary mutation and EGFR signaling cause resistance to ALK kinase inhibitors. Cancer Res. 2011;71:6051–60. doi: 10.1158/0008-5472.CAN-11-1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Katayama R, Shaw AT, Khan TM, Mino-Kenudson M, Solomon BJ, Halmos B, et al. Mechanisms of acquired crizotinib resistance in ALK-rearranged lung cancers. Sci Transl Med. 2012;4:120ra17. doi: 10.1126/scitranslmed.3003316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Crystal AS, Shaw AT, Sequist LV, Friboulet L, Niederst MJ, Lockerman EL, et al. Patient-derived models of acquired resistance can identify effective drug combinations for cancer. Science. 2014;346:1480–6. doi: 10.1126/science.1254721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Katayama R, Khan TM, Benes C, Lifshits E, Ebi H, Rivera VM, et al. Therapeutic strategies to overcome crizotinib resistance in non-small cell lung cancers harboring the fusion oncogene EML4-ALK. Proc Natl Acad Sci U S A. 2011;108:7535–40. doi: 10.1073/pnas.1019559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang S, Wang F, Keats J, Zhu X, Ning Y, Wardwell SD, et al. Crizotinib-resistant mutants of EML4-ALK identified through an accelerated mutagenesis screen. Chem Biol Drug Des. 2011;78:999–1005. doi: 10.1111/j.1747-0285.2011.01239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heuckmann JM, Hölzel M, Sos ML, Heynck S, Balke-Want H, Koker M, et al. ALK mutations conferring differential resistance to structurally diverse ALK inhibitors. Clin Cancer Res. 2011;17:7394–401. doi: 10.1158/1078-0432.CCR-11-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Camidge DR, Pao W, Sequist LV. Acquired resistance to TKIs in solid tumours: learning from lung cancer. Nat Rev Clin Oncol. 2014;11:473–81. doi: 10.1038/nrclinonc.2014.104. [DOI] [PubMed] [Google Scholar]
- 69.Lin JJ, Shaw AT. Resisting resistance: targeted therapies in lung cancer. Trends Cancer. 2016;2:350–64. doi: 10.1016/j.trecan.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Balak MN, Gong Y, Riely GJ, Somwar R, Li AR, Zakowski MF, et al. Novel D761Y and common secondary T790M mutations in epidermal growth factor receptor-mutant lung adenocarcinomas with acquired resistance to kinase inhibitors. Clin Cancer Res. 2006;12:6494–501. doi: 10.1158/1078-0432.CCR-06-1570. [DOI] [PubMed] [Google Scholar]
- 71.Cui JJ, Tran-Dubé M, Shen H, Nambu M, Kung PP, Pairish M, et al. Structure based drug design of crizotinib (PF-02341066), a potent and selective dual inhibitor of mesenchymal-epithelial transition factor (c-MET) kinase and anaplastic lymphoma kinase (ALK) J Med Chem. 2011;54:6342–63. doi: 10.1021/jm2007613. [DOI] [PubMed] [Google Scholar]
- 72.Doebele RC, Pilling AB, Aisner DL, Kutateladze TG, Le AT, Weickhardt AJ, et al. Mechanisms of resistance to crizotinib in patients with ALK gene rearranged non-small cell lung cancer. Clin Cancer Res. 2012;18:1472–82. doi: 10.1158/1078-0432.CCR-11-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Friboulet L, Li N, Katayama R, Lee CC, Gainor JF, Crystal AS, et al. The ALK inhibitor ceritinib overcomes crizotinib resistance in non-small cell lung cancer. Cancer Discov. 2014;4:662–73. doi: 10.1158/2159-8290.CD-13-0846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shaw AT, Friboulet L, Leshchiner I, Gainor JF, Bergqvist S, Brooun A, et al. Resensitization to crizotinib by the lorlatinib ALK resistance mutation L1198F. N Engl J Med. 2016;374:54–61. doi: 10.1056/NEJMoa1508887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Katayama R, Friboulet L, Koike S, Lockerman EL, Khan TM, Gainor JF, et al. Two novel ALK mutations mediate acquired resistance to the next-generation ALK inhibitor alectinib. Clin Cancer Res. 2014;20:5686–96. doi: 10.1158/1078-0432.CCR-14-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ignatius Ou SH, Azada M, Hsiang DJ, Herman JM, Kain TS, Siwak-Tapp C, et al. Next-generation sequencing reveals a novel NSCLC ALK F1174V mutation and confirms ALK G1202R mutation confers high-level resistance to alectinib (CH5424802/RO5424802) in ALK-rearranged NSCLC patients who progressed on crizotinib. J Thorac Oncol. 2014;9:549–53. doi: 10.1097/JTO.0000000000000094. [DOI] [PubMed] [Google Scholar]
- 77.Toyokawa G, Hirai F, Inamasu E, Yoshida T, Nosaki K, Takenaka T, et al. Secondary mutations at I1171 in the ALK gene confer resistance to both crizotinib and alectinib. J Thorac Oncol. 2014;9:e86–7. doi: 10.1097/JTO.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 78.Gainor JF, Dardaei L, Yoda S, Friboulet L, Leshchiner I, Katayama R, et al. Molecular mechanisms of resistance to first- and second-generation ALK inhibitors in ALK-rearranged lung cancer. Cancer Discov. 2016;6:1118–33. doi: 10.1158/2159-8290.CD-16-0596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ceccon M, Mologni L, Bisson W, Scapozza L, Gambacorti-Passerini C. Crizotinib-resistant NPM-ALK mutants confer differential sensitivity to unrelated ALK inhibitors. Mol Cancer Res. 2013;11:122–32. doi: 10.1158/1541-7786.MCR-12-0569. [DOI] [PubMed] [Google Scholar]
- 80.Zdzalik D, Dymek B, Grygielewicz P, Gunerka P, Bujak A, Lamparska-Przybysz M, et al. Activating mutations in ALK kinase domain confer resistance to structurally unrelated ALK inhibitors in NPM-ALK positive anaplastic large-cell lymphoma. J Cancer Res Clin Oncol. 2014;140:589–98. doi: 10.1007/s00432-014-1589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Sasaki T, Okuda K, Zheng W, Butrynski J, Capelletti M, Wang L, et al. The neuroblastoma-associated F1174L ALK mutation causes resistance to an ALK kinase inhibitor in ALK-translocated cancers. Cancer Res. 2010;70:10038–43. doi: 10.1158/0008-5472.CAN-10-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zou HY, Friboulet L, Kodack DP, Engstrom LD, Li Q, West M, et al. PF-06463922, an ALK/ROS1 inhibitor, overcomes resistance to first and second generation ALK inhibitors in preclinical models. Cancer Cell. 2015;28:70–81. doi: 10.1016/j.ccell.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thress KS, Paweletz CP, Felip E, Cho BC, Stetson D, Dougherty B, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nat Med. 2015;21:560–2. doi: 10.1038/nm.3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Niederst MJ, Hu H, Mulvey HE, Lockerman EL, Garcia AR, Piotrowska Z, et al. The allelic context of the C797S mutation acquired upon treatment with third-generation EGFR inhibitors impacts sensitivity to subsequent treatment strategies. Clin Cancer Res. 2015;21:3924–33. doi: 10.1158/1078-0432.CCR-15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Solomon BJ, Bauer TM, Felip E, Besse B, James LP, Clancy JS, et al. Safety and efficacy of lorlatinib (PF-06463922) from the dose-escalation component of a study in patients with advanced ALK+ or ROS1+ non-small cell lung cancer (NSCLC) J Clin Oncol. 2016;34(suppl) abstr 9009. [Google Scholar]
- 86.Miyawaki M, Yasuda H, Tani T, Hamamoto J, Arai D, Ishioka K, et al. Overcoming EGFR bypass signal-induced acquired resistance to ALK tyrosine kinase inhibitors in ALK-translocated lung cancer. Mol Cancer Res. 2016 doi: 10.1158/1541-7786.MCR-16-0211. [DOI] [PubMed] [Google Scholar]
- 87.Wilson FH, Johannessen CM, Piccioni F, Tamayo P, Kim JW, Van Allen EM, et al. A functional landscape of resistance to ALK inhibition in lung cancer. Cancer Cell. 2015;27:397–408. doi: 10.1016/j.ccell.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tanizaki J, Okamoto I, Okabe T, Sakai K, Tanaka K, Hayashi H, et al. Activation of HER family signaling as a mechanism of acquired resistance to ALK inhibitors in EML4-ALK-positive non-small cell lung cancer. Clin Cancer Res. 2012;18:6219–26. doi: 10.1158/1078-0432.CCR-12-0392. [DOI] [PubMed] [Google Scholar]
- 89.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. doi: 10.1126/science.1141478. [DOI] [PubMed] [Google Scholar]
- 90.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. doi: 10.1126/scitranslmed.3002003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gouji T, Takashi S, Mitsuhiro T, Yukito I. Crizotinib can overcome acquired resistance to CH5424802: is amplification of the MET gene a key factor? J Thorac Oncol. 2014;9:e27–8. doi: 10.1097/JTO.0000000000000113. [DOI] [PubMed] [Google Scholar]
- 92.Hrustanovic G, Olivas V, Pazarentzos E, Tulpule A, Asthana S, Blakely CM, et al. RAS-MAPK dependence underlies a rational polytherapy strategy in EML4-ALK-positive lung cancer. Nat Med. 2015;21:1038–47. doi: 10.1038/nm.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lovly CM, McDonald NT, Chen H, Ortiz-Cuaran S, Heukamp LC, Yan Y, et al. Rationale for co-targeting IGF-1R and ALK in ALK fusion-positive lung cancer. Nat Med. 2014;20:1027–34. doi: 10.1038/nm.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kim HR, Kim WS, Choi YJ, Choi CM, Rho JK, Lee JC. Epithelial-mesenchymal transition leads to crizotinib resistance in H2228 lung cancer cells with EML4-ALK translocation. Mol Oncol. 2013;7:1093–102. doi: 10.1016/j.molonc.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gower A, Hsu WH, Hsu ST, Wang Y, Giaccone G. EMT is associated with, but does not drive resistance to ALK inhibitors among EML4-ALK non-small cell lung cancer. Mol Oncol. 2016;10:601–9. doi: 10.1016/j.molonc.2015.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhou J, Wang J, Zeng Y, Zhang X, Hu Q, Zheng J, et al. Implication of epithelial-mesenchymal transition in IGF1R-induced resistance to EGFR-TKIs in advanced non-small cell lung cancer. Oncotarget. 2015;6:44332–45. doi: 10.18632/oncotarget.6293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wilson C, Nicholes K, Bustos D, Lin E, Song Q, Stephan JP, et al. Overcoming EMT-associated resistance to anti-cancer drugs via Src/FAK pathway inhibition. Oncotarget. 2014;5:7328–41. doi: 10.18632/oncotarget.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Takegawa N, Hayashi H, Iizuka N, Takahama T, Ueda H, Tanaka K, et al. Transformation of ALK rearrangement-positive adenocarcinoma to small-cell lung cancer in association with acquired resistance to alectinib. Ann Oncol. 2016;27:953–5. doi: 10.1093/annonc/mdw032. [DOI] [PubMed] [Google Scholar]
- 100.Fujita S, Masago K, Katakami N, Yatabe Y. Transformation to SCLC after treatment with the ALK inhibitor alectinib. J Thorac Oncol. 2016;11:e67–72. doi: 10.1016/j.jtho.2015.12.105. [DOI] [PubMed] [Google Scholar]
- 101.Cha YJ, Cho BC, Kim HR, Lee HJ, Shim HS. A case of ALK-rearranged adenocarcinoma with small cell carcinoma-like transformation and resistance to crizotinib. J Thorac Oncol. 2016;11:e55–8. doi: 10.1016/j.jtho.2015.12.097. [DOI] [PubMed] [Google Scholar]
- 102.Miyamoto S, Ikushima S, Ono R, Awano N, Kondo K, Furuhata Y, et al. Transformation to small-cell lung cancer as a mechanism of acquired resistance to crizotinib and alectinib. Jpn J Clin Oncol. 2016;46:170–3. doi: 10.1093/jjco/hyv173. [DOI] [PubMed] [Google Scholar]
- 103.Niederst MJ, Sequist LV, Poirier JT, Mermel CH, Lockerman EL, Garcia AR, et al. RB loss in resistant EGFR mutant lung adenocarcinomas that transform to small-cell lung cancer. Nat Commun. 2015;6:6377. doi: 10.1038/ncomms7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nat Rev Cancer. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 105.Katayama R, Sakashita T, Yanagitani N, Ninomiya H, Horiike A, Friboulet L, et al. P-glycoprotein mediates ceritinib resistance in anaplastic lymphoma kinase-rearranged non-small cell lung cancer. EBioMedicine. 2015;3:54–66. doi: 10.1016/j.ebiom.2015.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kort A, Sparidans RW, Wagenaar E, Beijnen JH, Schinkel AH. Brain accumulation of the EML4-ALK inhibitor ceritinib is restricted by P-glycoprotein (P-GP/ABCB1) and breast cancer resistance protein (BCRP/ABCG2) Pharmacol Res. 2015;102:200–7. doi: 10.1016/j.phrs.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 107.Tang SC, Nguyen LN, Sparidans RW, Wagenaar E, Beijnen JH, Schinkel AH. Increased oral availability and brain accumulation of the ALK inhibitor crizotinib by coadministration of the P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2) inhibitor elacridar. Int J Cancer. 2014;134:1484–94. doi: 10.1002/ijc.28475. [DOI] [PubMed] [Google Scholar]
- 108.Kodama T, Hasegawa M, Takanashi K, Sakurai Y, Kondoh O, Sakamoto H. Antitumor activity of the selective ALK inhibitor alectinib in models of intracranial metastases. Cancer Chemother Pharmacol. 2014;74:1023–8. doi: 10.1007/s00280-014-2578-6. [DOI] [PubMed] [Google Scholar]
- 109.Yu HA, Arcila ME, Hellmann MD, Kris MG, Ladanyi M, Riely GJ. Poor response to erlotinib in patients with tumors containing baseline EGFR T790M mutations found by routine clinical molecular testing. Ann Oncol. 2014;25:423–8. doi: 10.1093/annonc/mdt573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Su KY, Chen HY, Li KC, Kuo ML, Yang JC, Chan WK, et al. Pretreatment epidermal growth factor receptor (EGFR) T790M mutation predicts shorter EGFR tyrosine kinase inhibitor response duration in patients with non-small-cell lung cancer. J Clin Oncol. 2012;30:433–40. doi: 10.1200/JCO.2011.38.3224. [DOI] [PubMed] [Google Scholar]
- 111.Lucena-Araujo AR, Moran JP, VanderLaan PA, Dias-Santagata D, Folch E, Majid A, et al. De novo ALK kinase domain mutations are uncommon in kinase inhibitor-naïve ALK rearranged lung cancers. Lung Cancer. 2016;99:17–22. doi: 10.1016/j.lungcan.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heuckmann JM, Balke-Want H, Malchers F, Peifer M, Sos ML, Koker M, et al. Differential protein stability and ALK inhibitor sensitivity of EML4-ALK fusion variants. Clin Cancer Res. 2012;18:4682–90. doi: 10.1158/1078-0432.CCR-11-3260. [DOI] [PubMed] [Google Scholar]
- 114.Yoshida T, Oya Y, Tanaka K, Shimizu J, Horio Y, Kuroda H, et al. Differential crizotinib response duration among ALK fusion variants in ALK-positive non-small-cell lung cancer. J Clin Oncol. 2016;34:3383–9. doi: 10.1200/JCO.2015.65.8732. [DOI] [PubMed] [Google Scholar]
- 115.Sholl LM, Weremowicz S, Gray SW, Wong KK, Chirieac LR, Lindeman NI, et al. Combined use of ALK immunohistochemistry and FISH for optimal detection of ALK-rearranged lung adenocarcinomas. J Thorac Oncol. 2013;8:322–8. doi: 10.1097/JTO.0b013e31827db604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Marchetti A, Di Lorito A, Pace MV, lezzi M, Felicioni L, D’Antuono T, et al. ALK protein analysis by IHC staining after recent regulatory changes: a comparison of two widely used approaches, revision of the literature, and a new testing algorithm. J Thorac Oncol. 2016;11:487–95. doi: 10.1016/j.jtho.2015.12.111. [DOI] [PubMed] [Google Scholar]
- 117.Ali SM, Hensing T, Schrock AB, Allen J, Sanford E, Gowen K, et al. Comprehensive genomic profiling identifies a subset of crizotinib-responsive ALK-rearranged non-small cell lung cancer not detected by fluorescence in situ hybridization. Oncologist. 2016;21:762–70. doi: 10.1634/theoncologist.2015-0497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Russo M, Siravegna G, Blaszkowsky LS, Corti G, Crisafulli G, Ahronian LG, et al. Tumor heterogeneity and lesion-specific response to targeted therapy in colorectal cancer. Cancer Discov. 2016;6:147–53. doi: 10.1158/2159-8290.CD-15-1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chabon JJ, Simmons AD, Lovejoy AF, Esfahani MS, Newman AM, Haringsma HJ, et al. Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nat Commun. 2016;7:11815. doi: 10.1038/ncomms11815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Oxnard GR, Thress KS, Alden RS, Lawrance R, Paweletz CP, Cantarini M, et al. Association between plasma genotyping and outcomes of treatment with osimertinib (AZD9291) in advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:3375–82. doi: 10.1200/JCO.2016.66.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Douillard JY, Ostoros G, Cobo M, Ciuleanu T, McCormack R, Webster A, et al. First-line gefitinib in Caucasian EGFR mutation-positive NSCLC patients: a phase-IV, open-label, single-arm study. Br J Cancer. 2014;110:55–62. doi: 10.1038/bjc.2013.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Puig O, Yang JC, Ou SI, Chiappori A, Chao BH, Belani CP, et al. Pooled mutation analysis for the NP28673 and NP28761 studies of alectinib in ALK+ non-small-cell lung cancer (NSCLC) J Clin Oncol. 2016;34(suppl) abstr 9061. [Google Scholar]
- 123.Horn L, Wakelee H, Reckamp KL, Blumenschein GR, Infante JR, Carter CA, et al. Plasma genotyping of patients in the eXalt2 trial: ensartinib (X-396) in ALK+ non-small cell lung cancer (NSCLC) J Clin Oncol. 2016;34(suppl) abstr 9056. [Google Scholar]
- 124.Huang WS, Liu S, Zou D, Thomas M, Wang Y, Zhou T, et al. Discovery of brigatinib (AP26113), a phosphine oxide-containing, potent, orally active inhibitor of anaplastic lymphoma kinase. J Med Chem. 2016;59:4948–64. doi: 10.1021/acs.jmedchem.6b00306. [DOI] [PubMed] [Google Scholar]
- 125.Zhang S, Wang F, Keats J, Ning Y, Wardwell SD, Moran L, et al. AP26113, a potent ALK inhibitor, overcomes mutations in EML4-ALK that confer resistance to PF-02341066 (PF1066) [abstract] Cancer Res. 2014;70:LB-298. [Google Scholar]
- 126.Squillace RM, Anjum R, Miller D, Vodala S, Moran L, Wang F, et al. Abstract 5655: AP26113 possesses pan-inhibitory activity versus crizotinib-resistant ALK mutants and oncogenic ROS1 fusions. Cancer Res. 2014;73:5655. [Google Scholar]
- 127.Kim DW, Tiseo M, Ahn MJ, Reckamp KL, Hansen KH, Kim SW, et al. Brigatinib (BRG) in patients (pts) with crizotinib (CRZ)-refractory ALK+ non-small cell lung cancer (NSCLC): First report of efficacy and safety from a pivotal randomized phase (ph) 2 trial (ALTA) J Clin Oncol. 2016;34(suppl) abstr 9007. [Google Scholar]
- 128.Gettinger SN, Bazhenova L, Salgia R, Langer CJ, Gold KA, Rosell R, et al. Updated efficacy and safety of the ALK inhibitor AP26113 in patients (pts) with advanced malignancies, including ALK+ non-small cell lung cancer (NSCLC) J Clin Oncol. 2014;32(suppl):5s. abstr 8047. [Google Scholar]
- 129.Gettinger SN, Zhang S, Hodgson JG, Bazhenova L, Burgers S, Kim DW, et al. Activity of brigatinib (BRG) in crizotinib (CRZ) resistant patients (pts) according to ALK mutation status. J Clin Oncol. 2016;34(suppl) abstr 9060. [Google Scholar]
- 130.Zhang S, Anjum R, Squillace R, Nadworny S, Zhou T, Keats J, et al. The potent ALK inhibitor brigatinib (AP26113) overcomes mechanisms of resistance to first- and second-generation ALK inhibitors in preclinical models. Clin Cancer Res. 2016 doi: 10.1158/1078-0432.CCR-16-0569. [DOI] [PubMed] [Google Scholar]
- 131.Shaw AT, Bauer TM, Felip E, Besse B, James LP, Clancy JS, et al. Clinical activity and safety of PF-06463922 from a dose escalation study in patients with advanced ALK+ or ROS1+ NSCLC. J Clin Oncol. 2015;33(suppl) abstr 8018. [Google Scholar]
- 132.Menichincheri M, Ardini E, Magnaghi P, Avanzi N, Banfi P, Bossi R, et al. Discovery of entrectinib: a new 3-aminoindazole as a potent anaplastic lymphoma kinase (ALK), c-ros oncogene 1 kinase (ROS1), and pan-tropomyosin receptor kinases (pan-TRKs) inhibitor. J Med Chem. 2016;59:3392–408. doi: 10.1021/acs.jmedchem.6b00064. [DOI] [PubMed] [Google Scholar]
- 133.Ardini E, Menichincheri M, Banfi P, Bosotti R, De Ponti C, Pulci R, et al. Entrectinib, a pan-TRK, ROS1, and ALK inhibitor with activity in multiple molecularly defined cancer indications. Mol Cancer Ther. 2016;15:628–39. doi: 10.1158/1535-7163.MCT-15-0758. [DOI] [PubMed] [Google Scholar]
- 134.Drilon A, De Braud FG, Siena S, Ou SH, Patel M, Ahn MJ, et al. Entrectinib, an oral pan-Trk, ROS1, and ALK inhibitor in TKI-naïve patients with advanced solid tumors harboring gene rearrangements: Updated phase I results [abstract] Cancer Res. 2016;76:CT007. [Google Scholar]
- 135.Lovly CM, Heuckmann JM, de Stanchina E, Chen H, Thomas RK, Liang C, et al. Insights into ALK-driven cancers revealed through development of novel ALK tyrosine kinase inhibitors. Cancer Res. 2011;71:4920–31. doi: 10.1158/0008-5472.CAN-10-3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lovly CM, Infante JR, Blumenschein GR, Reckamp K, Wakelee H, Carter CA, et al. Phase I/II trial of X-396, a novel anaplastic lymphoma kinase (ALK) inhibitor, in patients with ALK+ non-small cell lung cancer (NSCLC) [abstract] Cancer Res. 2016;76:CT088. [Google Scholar]
- 137.Johnson TW, Richardson PF, Bailey S, Brooun A, Burke BJ, Collins MR, et al. Discovery of (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]-benzoxadiazacyclotetradecine-3-carbonitrile (PF-06463922), a macrocyclic inhibitor of anaplastic lymphoma kinase (ALK) and c-ros oncogene 1 (ROS1) with preclinical brain exposure and broad-spectrum potency against ALK-resistant mutations. J Med Chem. 2014;57:4720–44. doi: 10.1021/jm500261q. [DOI] [PubMed] [Google Scholar]
- 138.Sakamoto H, Tsukaguchi T, Hiroshima S, Kodama T, Kobayashi T, Fukami TA, et al. CH5424802, a selective ALK inhibitor capable of blocking the resistant gatekeeper mutant. Cancer Cell. 2011;19:679–90. doi: 10.1016/j.ccr.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 139.Gadgeel SM, Gandhi L, Riely GJ, Chiappori AA, West HL, Azada MC, et al. Safety and activity of alectinib against systemic disease and brain metastases in patients with crizotinib-resistant ALK-rearranged non-small-cell lung cancer (AF-002JG): results from the dose-finding portion of a phase 1/2 study. Lancet Oncol. 2014;15:1119–28. doi: 10.1016/S1470-2045(14)70362-6. [DOI] [PubMed] [Google Scholar]
- 140.Gainor JF, Sherman CA, Willoughby K, Logan J, Kennedy E, Brastianos PK, et al. Alectinib salvages CNS metastases in ALK-positive lung cancer patients previously treated with crizotinib and ceritinib. J Thorac Oncol. 2015;10:232–6. doi: 10.1097/JTO.0000000000000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Nokihara H, Hida T, Kondo M, Kim YH, Azuma K, Seto T, et al. Alectinib (ALC) versus crizotinib (CRZ) in ALK-inhibitor naïve ALK-positive non-small cell lung cancer (ALK+ NSCLC): Primary results from the J-ALEX study. J Clin Oncol. 2016;34(suppl) abstr 9008. [Google Scholar]
- 142.Kato T, Seto T, Nishio M, Goto K, Atagi S, Hosomi Y, et al. Erlotinib plus bevacizumab (EB) versus erlotinib alone (E) as first-line treatment for advanced EGFR mutation-positive nonsquamous non-small cell lung cancer (NSCLC): An open-label randomized trial. J Clin Oncol. 2014;32(suppl) abstr 8005. [Google Scholar]
- 143.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WE, Poddubskaya E, et al. Nivolumab versus docetaxel in advanced squamous-cell non-small-cell lung cancer. N Engl J Med. 2015;373:123–35. doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373:1627–39. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Herbst RS, Baas P, Kim DW, Felip E, Pérez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1 positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet. 2016;387:1540–50. doi: 10.1016/S0140-6736(15)01281-7. [DOI] [PubMed] [Google Scholar]
- 146.Gettinger S, Rizvi NA, Chow LQ, Borghaei H, Brahmer J, Ready N, et al. Nivolumab monotherapy for first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:2980–7. doi: 10.1200/JCO.2016.66.9929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Rizvi NA, Hellmann MD, Brahmer JR, Juergens RA, Borghaei H, Gettinger S, et al. Nivolumab in combination with platinum-based doublet chemotherapy for first-line treatment of advanced non-small-cell lung cancer. J Clin Oncol. 2016;34:2969–79. doi: 10.1200/JCO.2016.66.9861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, et al. Pembrolizumab versus chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl J Med. 2016 doi: 10.1056/NEJMoa1606774. [DOI] [PubMed] [Google Scholar]
- 149.Langer CJ, Gadgeel SM, Borghaei H, Papadimitrakopoulou VA, Patnaik A, Powell SF, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol. 2016;17:1497–508. doi: 10.1016/S1470-2045(16)30498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Govindan R, Ding L, Griffith M, Subramanian J, Dees ND, Kanchi KL, et al. Genomic landscape of non-small cell lung cancer in smokers and never-smokers. Cell. 2012;150:1121–34. doi: 10.1016/j.cell.2012.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gainor JF, Shaw AT, Sequist LV, Fu X, Azzoli CG, Piotrowska Z, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer (NSCLC): a retrospective analysis. Clin Cancer Res. 2016;22:4585–93. doi: 10.1158/1078-0432.CCR-15-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Ahn MJ, Yang J, Yu H, Saka H, Ramalingam S, Goto K, et al. Osimertinib combined with durvalumab in EGFR-mutant non-small cell lung cancer: Results from the TATTON phase 1b trial. J Thorac Oncol. 2016;11:S115. [Google Scholar]
- 154.Jänne PA, Shaw AT, Camidge DR, Giaccone G, Shreeve SM, Tang Y, et al. Combined pan-HER and ALK/ROS1/MET inhibition with dacomitinib and crizotinib in advanced non-small cell lung cancer: results of a phase I study. J Thorac Oncol. 2016;11:737–47. doi: 10.1016/j.jtho.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 155.Campo M, Al-Halabi H, Khandekar M, Shaw AT, Sequist LV, Willers H. Integration of stereotactic body radiation therapy with tyrosine kinase inhibitors in stage IV oncogene-driven lung cancer. Oncologist. 2016;21:964–73. doi: 10.1634/theoncologist.2015-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Castellanos EH, Horn L. Re-evaluating progression in an era of progress: a review of first- and second-line treatment options in anaplastic lymphoma kinase-positive non-small cell lung cancer. Oncologist. 2016;21:755–61. doi: 10.1634/theoncologist.2015-0396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Weickhardt AJ, Scheier B, Burke JM, Gan G, Lu X, Bunn PA, Jr, et al. Local ablative therapy of oligoprogressive disease prolongs disease control by tyrosine kinase inhibitors in oncogene-addicted non-small-cell lung cancer. J Thorac Oncol. 2012;7:1807–14. doi: 10.1097/JTO.0b013e3182745948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Gan GN, Weickhardt AJ, Scheier B, Doebele RC, Gaspar LE, Kavanagh BD, et al. Stereotactic radiation therapy can safely and durably control sites of extra-central nervous system oligoprogressive disease in anaplastic lymphoma kinase-positive lung cancer patients receiving crizotinib. Int J Radiat Oncol Biol Phys. 2014;88:892–8. doi: 10.1016/j.ijrobp.2013.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bivona TG, Doebele RC. A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nat Med. 2016;22:472–8. doi: 10.1038/nm.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Gomez DR, Blumenschein GR, Jr, Lee JJ, Hernandez M, Ye R, Camidge DR, et al. Local consolidative therapy versus maintenance therapy or observation for patients with oligometastatic non-small-cell lung cancer without progression after first-line systemic therapy: a multicentre, randomised, controlled, phase 2 study. Lancet Oncol. 2016 doi: 10.1016/S1470-2045(16)30532-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ramirez M, Rajaram S, Steininger RJ, Osipchuk D, Roth MA, Morinishi LS, et al. Diverse drug-resistance mechanisms can emerge from drug-tolerant cancer persister cells. Nat Commun. 2016;7:10690. doi: 10.1038/ncomms10690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Hata AN, Niederst MJ, Archibald HL, Gomez-Caraballo M, Siddiqui FM, Mulvey HE, et al. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat Med. 2016;22:262–9. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.De Paepe P, Baens M, van Krieken H, Verhasselt B, Stul M, Simons A, et al. ALK activation by the CLTC-ALK fusion is a recurrent event in large B-cell lymphoma. Blood. 2003;102:2638–41. doi: 10.1182/blood-2003-04-1050. [DOI] [PubMed] [Google Scholar]
- 165.Adam P, Katzenberger T, Seeberger H, Gattenlöhner S, Wolf J, Steinlein C, et al. A case of a diffuse large B-cell lymphoma of plasmablastic type associated with the t(2;5)(p23;q35) chromosome translocation. Am J Surg Pathol. 2003;27:1473–6. doi: 10.1097/00000478-200311000-00012. [DOI] [PubMed] [Google Scholar]
- 166.Onciu M, Behm FG, Raimondi SC, Moore S, Harwood EL, Pui CH, et al. ALK-positive anaplastic large cell lymphoma with leukemic peripheral blood involvement is a clinicopathologic entity with an unfavorable prognosis. Report of three cases and review of the literature. Am J Clin Pathol. 2003;120:617–25. doi: 10.1309/WH8P-NU9P-K4RR-V852. [DOI] [PubMed] [Google Scholar]
- 167.Van Roosbroeck K, Cools J, Dierickx D, Thomas J, Vandenberghe P, Stul M, et al. ALK-positive large B-cell lymphomas with cryptic SEC31A-ALK and NPM1-ALK fusions. Haematologica. 2010;95:509–13. doi: 10.3324/haematol.2009.014761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Bedwell C, Rowe D, Moulton D, Jones G, Bown N, Bacon CM. Cytogenetically complex SEC31A-ALK fusions are recurrent in ALK-positive large B-cell lymphomas. Haematologica. 2011;96:343–6. doi: 10.3324/haematol.2010.031484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.d’Amore ES, Visco C, Menin A, Famengo B, Bonvini P, Lazzari E. STAT3 pathway is activated in ALK-positive large B-cell lymphoma carrying SQSTM1-ALK rearrangement and provides a possible therapeutic target. Am J Surg Pathol. 2013;37:780–6. doi: 10.1097/PAS.0b013e318287791f. [DOI] [PubMed] [Google Scholar]
- 170.Lee SE, Kang SY, Takeuchi K, Ko YH. Identification of RANBP2-ALK fusion in ALK positive diffuse large B-cell lymphoma. Hematol Oncol. 2014;32:221–4. doi: 10.1002/hon.2125. [DOI] [PubMed] [Google Scholar]
- 171.Sakamoto K, Nakasone H, Togashi Y, Sakata S, Tsuyama N, Baba S, et al. ALK-positive large B-cell lymphoma: identification of EML4-ALK and a review of the literature focusing on the ALK immunohistochemical staining pattern. Int J Hematol. 2016;103:399–408. doi: 10.1007/s12185-016-1934-1. [DOI] [PubMed] [Google Scholar]
- 172.Lipson D, Capelletti M, Yelensky R, Otto G, Parker A, Jarosz M, et al. Identification of new ALK and RET gene fusions from colorectal and lung cancer biopsies. Nat Med. 2012;18:382–4. doi: 10.1038/nm.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lin E, Li L, Guan Y, Soriano R, Rivers CS, Mohan S, et al. Exon array profiling detects EML4-ALK fusion in breast, colorectal, and non-small cell lung cancers. Mol Cancer Res. 2009;7:1466–76. doi: 10.1158/1541-7786.MCR-08-0522. [DOI] [PubMed] [Google Scholar]
- 174.Aisner DL, Nguyen TT, Paskulin DD, Le AT, Haney J, Schulte N, et al. ROS1 and ALK fusions in colorectal cancer, with evidence of intratumoral heterogeneity for molecular drivers. Mol Cancer Res. 2014;12:111–8. doi: 10.1158/1541-7786.MCR-13-0479-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Medico E, Russo M, Picco G, Cancelliere C, Valtorta E, Corti G, et al. The molecular landscape of colorectal cancer cell lines unveils clinically actionable kinase targets. Nat Commun. 2015;6:7002. doi: 10.1038/ncomms8002. [DOI] [PubMed] [Google Scholar]
- 176.Debelenko LV, Raimondi SC, Daw N, Shivakumar BR, Huang D, Nelson M, et al. Renal cell carcinoma with novel VCL-ALK fusion: new representative of ALK-associated tumor spectrum. Mod Pathol. 2011;24:430–42. doi: 10.1038/modpathol.2010.213. [DOI] [PubMed] [Google Scholar]
- 177.Sugawara E, Togashi Y, Kuroda N, Sakata S, Hatano S, Asaka R, et al. Identification of anaplastic lymphoma kinase fusions in renal cancer: large-scale immunohistochemical screening by the intercalated antibody-enhanced polymer method. Cancer. 2012;118:4427–36. doi: 10.1002/cncr.27391. [DOI] [PubMed] [Google Scholar]
- 178.Sukov WR, Hodge JC, Lohse CM, Akre MK, Leibovich BC, Thompson RH, et al. ALK alterations in adult renal cell carcinoma: frequency, clinicopathologic features and outcome in a large series of consecutively treated patients. Mod Pathol. 2012;25:1516–25. doi: 10.1038/modpathol.2012.107. [DOI] [PubMed] [Google Scholar]
- 179.Kusano H, Togashi Y, Akiba J, Moriya F, Baba K, Matsuzaki N, et al. Two cases of renal cell carcinoma harboring a novel STRN-ALK fusion gene. Am J Surg Pathol. 2016;40:761–9. doi: 10.1097/PAS.0000000000000610. [DOI] [PubMed] [Google Scholar]
- 180.Mariño-Enríquez A, Ou WB, Weldon CB, Fletcher JA, Pérez-Atayde AR. ALK rearrangement in sickle cell trait-associated renal medullary carcinoma. Genes Chromosomes Cancer. 2011;50:146–53. doi: 10.1002/gcc.20839. [DOI] [PubMed] [Google Scholar]
- 181.Jazii FR, Najafi Z, Malekzadeh R, Conrads TP, Ziaee AA, Abnet C, et al. Identification of squamous cell carcinoma associated proteins by proteomics and loss of beta tropomyosin expression in esophageal cancer. World J Gastroenterol. 2006;12:7104–12. doi: 10.3748/wjg.v12.i44.7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Du XL, Hu H, Lin DC, Xia SH, Shen XM, Zhang Y, et al. Proteomic profiling of proteins dysregulated in Chinese esophageal squamous cell carcinoma. J Mol Med. 2007;85:863–75. doi: 10.1007/s00109-007-0159-4. [DOI] [PubMed] [Google Scholar]
- 183.Ren H, Tan ZP, Zhu X, Crosby K, Haack H, Ren JM, et al. Identification of anaplastic lymphoma kinase as a potential therapeutic target in ovarian cancer. Cancer Res. 2012;72:3312–23. doi: 10.1158/0008-5472.CAN-11-3931. [DOI] [PubMed] [Google Scholar]
- 184.Lin JJ, Kennedy E, Sequist LV, Brastianos PK, Goodwin KE, Stevens S, et al. Clinical activity of alectinib in advanced RET-rearranged non-small-cell lung cancer. J Thorac Oncol. 2016;11:2027–32. doi: 10.1016/j.jtho.2016.08.126. [DOI] [PubMed] [Google Scholar]