Abstract
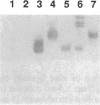
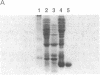
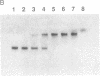
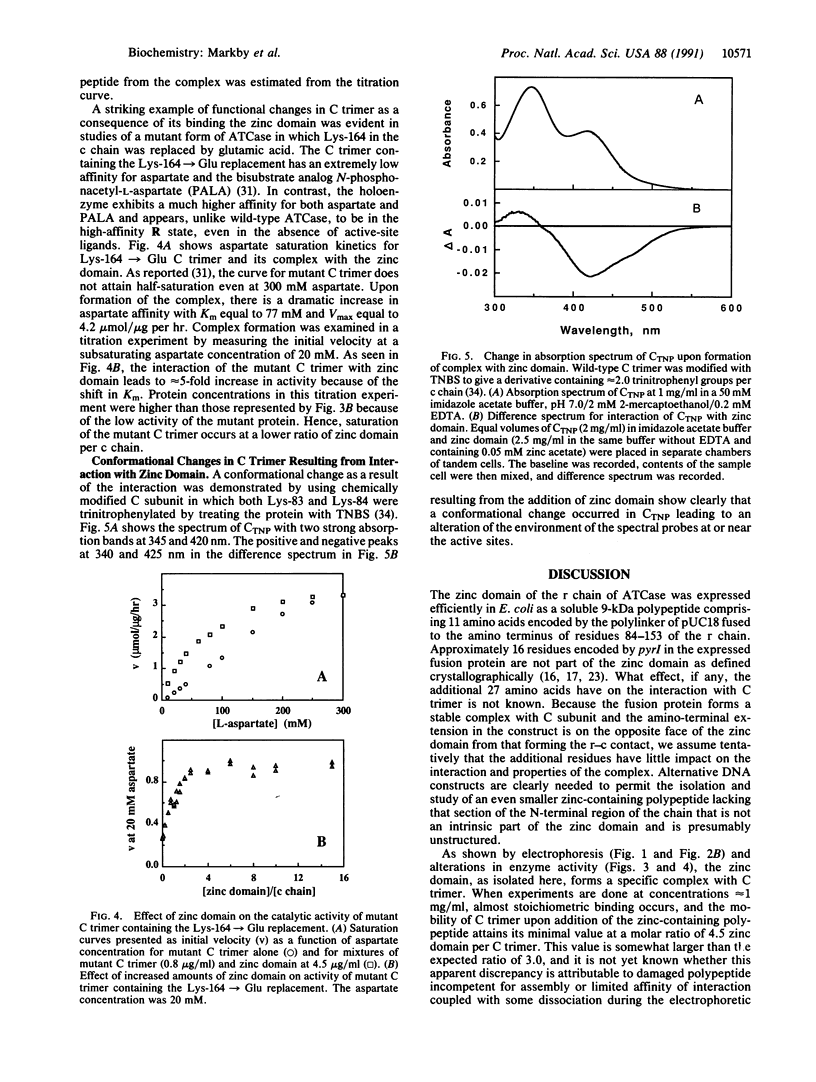
In an effort to clarify effects of specific protein-protein interactions on the properties of the dodecameric enzyme aspartate transcarbamoylase (carbamoyl-phosphate:L-aspartate carbamoyltransferase, EC 2.1.3.2), we initiated studies of a simpler complex containing an intact catalytic trimer and three copies of a fragment from the regulatory chain. The partial regulatory chain was expressed as a soluble 9-kDa zinc-binding polypeptide comprising 11 amino acids encoded by the polylinker of pUC18 fused to the amino terminus of residues 84-153 of the regulatory chain; this polypeptide includes the zinc domain detected in crystallographic studies of the holoenzyme. In contrast to intact regulatory chains, the zinc-binding polypeptide is monomeric in solution because it lacks the second domain responsible for dimer formation and assembly of the dodecameric holoenzyme. The isolated 9-kDa protein forms a tight, zinc-dependent complex with catalytic trimer, as shown by the large shift in electrophoretic mobility of the trimer in nondenaturing polyacrylamide gels. Enzyme assays of the complex showed a hyperbolic dependence of initial velocity on aspartate concentration with Vmax and Km for aspartate approximately 50% lower than the values for free catalytic subunit. A mutant catalytic subunit containing the Lys-164----Glu substitution exhibited a striking increase in enzyme activity at low aspartate concentrations upon interaction with the zinc domain because of a large reduction in Km upon complex formation. These changes in functional properties indicate that the complex of the zinc domain and catalytic trimer is an analog of the high-affinity R ("relaxed") state of aspartate transcarbamoylase, as proposed previously for a transiently formed assembly intermediate composed of one catalytic and three regulatory subunits. Conformational changes at the active sites, resulting from binding the zinc-containing polypeptide chains, were detected by difference spectroscopy with trinitrophenylated catalytic trimers. Isolation of the zinc domain of aspartate transcarbamoylase provides a model protein for study of oligomer assembly, communication between dissimilar polypeptides, and metal-binding motifs in proteins.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bothwell M. A., Schachman H. K. A model for the assembly of aspartate transcarbamoylase from catalytic and regulatory subunits. J Biol Chem. 1980 Mar 10;255(5):1971–1977. [PubMed] [Google Scholar]
- Bothwell M., Schachman H. K. Pathways of assembly of aspartate transcarbamoylase from catalytic and regulatory subunits. Proc Natl Acad Sci U S A. 1974 Aug;71(8):3221–3225. doi: 10.1073/pnas.71.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan W. W. Further studies on aspartate transcarbamoylase: molecular weight of the c3r6 complex and analysis of succinate inhibition in the native enzyme. Can J Biochem. 1976 Dec;54(12):1061–1068. doi: 10.1139/o76-155. [DOI] [PubMed] [Google Scholar]
- Chan W. W., Mort J. S. A complex between the catalytic and regulatory subunits of aspartate transcarbamylase. J Biol Chem. 1973 Nov 10;248(21):7614–7616. [PubMed] [Google Scholar]
- Cohlberg J. A., Pigiet V. P., Jr, Schachman H. K. Structure and arrangement of the regulatory subunits in aspartate transcarbamylase. Biochemistry. 1972 Aug 29;11(18):3396–3411. doi: 10.1021/bi00768a013. [DOI] [PubMed] [Google Scholar]
- Davies G. E., Vanaman T. C., Stark G. R. Aspartate transcarbamylase. Stereospecific restrictions on the binding site for L-aspartate. J Biol Chem. 1970 Mar 10;245(5):1175–1179. [PubMed] [Google Scholar]
- Foltermann K. F., Shanley M. S., Wild J. R. Assembly of the aspartate transcarbamoylase holoenzyme from transcriptionally independent catalytic and regulatory cistrons. J Bacteriol. 1984 Mar;157(3):891–898. doi: 10.1128/jb.157.3.891-898.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GERHART J. C., PARDEE A. B. The enzymology of control by feedback inhibition. J Biol Chem. 1962 Mar;237:891–896. [PubMed] [Google Scholar]
- Gerhart J. C., Schachman H. K. Allosteric interactions in aspartate transcarbamylase. II. Evidence for different conformational states of the protein in the presence and absence of specific ligands. Biochemistry. 1968 Feb;7(2):538–552. doi: 10.1021/bi00842a600. [DOI] [PubMed] [Google Scholar]
- Gerhart J. C., Schachman H. K. Distinct subunits for the regulation and catalytic activity of aspartate transcarbamylase. Biochemistry. 1965 Jun;4(6):1054–1062. doi: 10.1021/bi00882a012. [DOI] [PubMed] [Google Scholar]
- Hensley P., Schachman H. K. Communication between dissimilar subunits in aspartate transcarbamoylase: effect of inhibitor and activator on the conformation of the catalytic polypeptide chains. Proc Natl Acad Sci U S A. 1979 Aug;76(8):3732–3736. doi: 10.1073/pnas.76.8.3732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett G. J., Blackburn M. N., Compton J. G., Schachman H. K. Allosteric regulation of aspartate transcarbamoylase. Analysis of the structural and functional behavior in terms of a two-state model. Biochemistry. 1977 Nov 15;16(23):5091–5100. doi: 10.1021/bi00642a023. [DOI] [PubMed] [Google Scholar]
- Ke H. M., Lipscomb W. N., Cho Y. J., Honzatko R. B. Complex of N-phosphonacetyl-L-aspartate with aspartate carbamoyltransferase. X-ray refinement, analysis of conformational changes and catalytic and allosteric mechanisms. J Mol Biol. 1988 Dec 5;204(3):725–747. doi: 10.1016/0022-2836(88)90365-8. [DOI] [PubMed] [Google Scholar]
- Kim K. H., Pan Z. X., Honzatko R. B., Ke H. M., Lipscomb W. N. Structural asymmetry in the CTP-liganded form of aspartate carbamoyltransferase from Escherichia coli. J Mol Biol. 1987 Aug 20;196(4):853–875. doi: 10.1016/0022-2836(87)90410-4. [DOI] [PubMed] [Google Scholar]
- Krause K. L., Volz K. W., Lipscomb W. N. Structure at 2.9-A resolution of aspartate carbamoyltransferase complexed with the bisubstrate analogue N-(phosphonacetyl)-L-aspartate. Proc Natl Acad Sci U S A. 1985 Mar;82(6):1643–1647. doi: 10.1073/pnas.82.6.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahue R. S., Schachman H. K. The influence of quaternary structure on the active site of an oligomeric enzyme. Catalytic subunit of aspartate transcarbamoylase. J Biol Chem. 1984 Nov 25;259(22):13906–13913. [PubMed] [Google Scholar]
- MONOD J., WYMAN J., CHANGEUX J. P. ON THE NATURE OF ALLOSTERIC TRANSITIONS: A PLAUSIBLE MODEL. J Mol Biol. 1965 May;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- Meighen E. A., Pigiet V., Schachman H. K. Hybridization of native and chemically modified enzymes. 3. The catalytic subunits of aspartate transcarbamylase. Proc Natl Acad Sci U S A. 1970 Jan;65(1):234–241. doi: 10.1073/pnas.65.1.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco H. L., Crawford J. L., Lipscomb W. N. Three-dimensional structures of aspartate carbamoyltransferase from Escherichia coli and of its complex with cytidine triphosphate. Proc Natl Acad Sci U S A. 1978 Nov;75(11):5276–5280. doi: 10.1073/pnas.75.11.5276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mort J. S., Chan W. W. Subunit interactions in aspartate transcarbamylase. Characterization of a complex between the catalytic and the regulatory subunits. J Biol Chem. 1975 Jan 25;250(2):653–660. [PubMed] [Google Scholar]
- Navre M., Schachman H. K. Synthesis of aspartate transcarbamoylase in Escherichia coli: transcriptional regulation of the pyrB-pyrI operon. Proc Natl Acad Sci U S A. 1983 Mar;80(5):1207–1211. doi: 10.1073/pnas.80.5.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell J. O., Markby D. W., Schachman H. K. Cooperative binding of the bisubstrate analog N-(phosphonacetyl)-L-aspartate to aspartate transcarbamoylase and the heterotropic effects of ATP and CTP. J Biol Chem. 1989 Feb 15;264(5):2476–2481. [PubMed] [Google Scholar]
- Newell J. O., Schachman H. K. Amino acid substitutions which stabilize aspartate transcarbamoylase in the R state disrupt both homotropic and heterotropic effects. Biophys Chem. 1990 Aug 31;37(1-3):183–196. doi: 10.1016/0301-4622(90)88018-n. [DOI] [PubMed] [Google Scholar]
- Robey E. A., Schachman H. K. Site-specific mutagenesis of aspartate transcarbamoylase. Replacement of tyrosine 165 in the catalytic chain by serine reduces enzymatic activity. J Biol Chem. 1984 Sep 25;259(18):11180–11183. [PubMed] [Google Scholar]
- Rosenbusch J. P., Weber K. Localization of the zinc binding site of aspartate transcarbamoylase in the regulatory subunit. Proc Natl Acad Sci U S A. 1971 May;68(5):1019–1023. doi: 10.1073/pnas.68.5.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbusch J. P., Weber K. Subunit structure of aspartate transcarbamylase from Escherichia coli. J Biol Chem. 1971 Mar 25;246(6):1644–1657. [PubMed] [Google Scholar]
- Schachman H. K. Assembly of aspartate transcarbamoylase in Escherichia coli. Trans N Y Acad Sci. 1983;41:199–211. doi: 10.1111/j.2164-0947.1983.tb02802.x. [DOI] [PubMed] [Google Scholar]
- Schachman H. K. Can a simple model account for the allosteric transition of aspartate transcarbamoylase? J Biol Chem. 1988 Dec 15;263(35):18583–18586. [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Subramani S., Bothwell M. A., Gibbons I., Yang Y. R., Schachman H. K. Ligand-promoted weakening of intersubunit bonding domains in aspartate transcarbamolylase. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3777–3781. doi: 10.1073/pnas.74.9.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber K. New structural model of E. coli aspartate transcarbamylase and the amino-acid sequence of the regulatory polypeptide chain. Nature. 1968 Jun 22;218(5147):1116–1119. doi: 10.1038/2181116a0. [DOI] [PubMed] [Google Scholar]
- Wente S. R., Schachman H. K. Shared active sites in oligomeric enzymes: model studies with defective mutants of aspartate transcarbamoylase produced by site-directed mutagenesis. Proc Natl Acad Sci U S A. 1987 Jan;84(1):31–35. doi: 10.1073/pnas.84.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D. C., Lipscomb W. N. Crystallographic determination of symmetry of aspartate transcarbamylase. Nature. 1968 Jun 22;218(5147):1119–1121. doi: 10.1038/2181119a0. [DOI] [PubMed] [Google Scholar]
- Yang Y. R., Schachman H. K. Communication between catalytic subunits in hybrid aspartate transcarbamoylase molecules: effect of ligand binding to active chains on the conformation of unliganded, inactive chains. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5187–5191. doi: 10.1073/pnas.77.9.5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]