The crystal structure of the 15-residue peptaibol bergofungin A resembles that of the closely related samarosporin I. The differences in these structures, as well as the significant variation in their respective antibiotic functions, are presented.
Keywords: crystal structure, Emericellopsis donezkii, hydrogen bond, peptaibols, peptide antibiotics, 310-helix, α-helix
Abstract
Bergofungin is a peptide antibiotic that is produced by the ascomycetous fungus Emericellopsis donezkii HKI 0059 and belongs to peptaibol subfamily 2. The crystal structure of bergofungin A has been determined and refined to 0.84 Å resolution. This is the second crystal structure of a natural 15-residue peptaibol, after that of samarosporin I. The amino-terminal phenylalanine residue in samarosporin I is exchanged to a valine residue in bergofungin A. According to agar diffusion tests, this results in a nearly inactive antibiotic peptide compared with the moderately active samarosporin I. Crystals were obtained from methanol solutions of purified bergofungin mixed with water. Although there are differences in the intramolecular hydrogen-bonding scheme of samarosporin I, the overall folding is very similar for both peptaibols, namely 310-helical at the termini and α-helical in the middle of the molecules. Bergofungin A and samarosporin I molecules are arranged in a similar way in both lattices. However, the packing of bergofungin A exhibits a second solvent channel along the twofold axis. This latter channel occurs in the vicinity of the N-terminus, where the natural substitution resides.
1. Introduction
Peptaibols are naturally occurring, microheterogeneous peptides of fungal origin consisting of up to 20 residues and are characterized by the presence of the nonstandard, helix-advancing (Karle & Balaram, 1990 ▸) α-aminoisobutyric acid (Aib) and a 2-amino alcohol at the C-terminus. Usually, they are protected (acetylated) at the N-terminus. Peptaibols are a subgroup of peptaibiotics (Toniolo & Brückner, 2009 ▸) and have been assigned certain types of biological activities such as antibiotic and membrane-modifying. The 14–20-residue peptaibols are synthesized by multisubunit, nonribosomal peptide synthetases representing the largest enzymes known to date (Mukherjee et al., 2011 ▸; Degenkolb et al., 2012 ▸). Peptaibols form transmembrane channels through self-association. These channels enable the translocation of water and ionic species, leading to lysis and thus cell death of the host. Peptaibols have been partitioned into subfamilies (SFs) on the basis of their chain length, sequence and functional properties (Chugh & Wallace, 2001 ▸). Bergofungin belongs to subfamily 2 (SF2). All members of SF2 possess phenylalanine residues at both the amino- and hydroxyl-termini, except for bergofungin and minor components of antiamoebin, namely antiamoebins XIII–XVI. Bergofungin was isolated from the mycelium of Emericellopsis donezkii HKI 0059 (Berg et al., 1996 ▸). Four components of bergofungin have been identified (Neumann et al., 2015 ▸). The main component, bergofungin A (Bf-A), which accounts for 88–93% of the peptide produced, may be considered as a Pro15-deletion peptide of the 16-residue antiamoebin AAM XIV (Ac-V-U-U-U-V-G-L-U-U-O-Q-J-O-U-P-Fol) from various Emericellopsis and Stilbella spp. (Pandey et al., 1977 ▸; Jaworski & Brückner, 2000 ▸) and cephaibols from Acremonium tubakii (Schiell et al., 2001 ▸). To date, crystal structures of the 16-residue SF2 peptaibols antiamoebin and cephaibols A, B and C, and of the 15-residue samarosporin have been determined (Snook et al., 1998 ▸; Karle et al., 1998 ▸; Bunkóczi et al., 2003 ▸; Gessmann, Axford, Evans et al., 2012 ▸). The amino-acid sequences of SF2 members with known crystal structures are as follows.
Here, Ac is acetyl, U is Aib (α-aminoisobutyryl), O is Hyp (trans-4-hydroxy-l-proline), J is Iva (d-isovaline) and Fol is a phenylalaninol residue. All sequences, with the exception of bergofungin, have phenylalanine residues at their termini, which help to anchor the peptide in the membrane (Chugh & Wallace, 2001 ▸). Common to all of these SF2 members are the two Hyp residues at positions 10 and 13, Gly at position 6, Leu at position 7, Gln at position 11 and an N-terminus rich in the achiral residues Aib and Gly. Aib and Gly have been found in both left- and right-handed helical conformations (Gessmann, Brückner, Aivaliotis et al., 2015 ▸; Gessmann, Brückner & Petratos, 2015 ▸). Although Iva is helix-promoting, the configuration of Iva (l or d, equivalent to S or R, respectively) has no influence on the helical sense (De Zotti et al., 2012 ▸). Bf-A has a chiral l-Val at position 5, which augments the number of residues with a preference for right-handed helical conformation to 3/7 among the seven N-terminal residues in the above listed sequences. The best studied member of SF2 remains antiamoebin, which is remarkably less active in channel formation compared with the related SF3 zervamicin IIB (Kropacheva et al., 2005 ▸) with 5/7 N-terminal chiral residues. NMR data of antiamoebin solutions in methanol indicated a left-handed helix for residues 2–7 followed by a right-handed C-terminal helix (Galbraith et al., 2003 ▸). A recent combination of NMR data with CD spectropolarimetry shows that antiamoebin rapidly transforms its N-terminal conformation comprising residues 1–7 between left-handed and right-handed 310-helices, with an equal population of both states (Shenkarev et al., 2007 ▸). This highly dynamic process might explain its relative inefficiency as a channel former compared with the equally well studied zervamicin IIB. Bergofungin and samarosporin, with a chiral l-Val substitution for d-Iva (position 5) in antiamoebin, thus have a smaller probability of forming an N-terminal left-handed helix.
Multimeric assembly has been proposed for bergofungin and confirmed by an assay of the elicitation of the biosynthesis of scents secreted from the leaves of the legume Phaeolus lunatus (Jabs et al., 2001 ▸). A characteristic property of bergofungin is its comparably strong antimicrobial effect against the yeast Sporobolomyces salmonicolor compared with its antimicrobial effect against the fungus Penicillium notatum and Bacillus subtilis (Berg et al., 1996 ▸). Moreover, bergofungin moderately inhibits the activity of prolyl endopeptidase from Flavobacter sp., with inhibition constants K i in the range 0.18–0.34 µM (Christner et al., 1997 ▸).
In the present work, the effect on the structure and function of the substitution of Phe by Val at the amino-terminus of bergofungin A is examined. Its crystal structure has been refined at atomic resolution and the inhibition of bacterial growth has been investigated using agar diffusion assays. The results clearly show a reduction of the biological activity of bergofungin in comparison with the nearly identical samarosporin I. This could be ascribed to a less pronounced embedding of bergofungin in membranes.
2. Materials and methods
2.1. Isolation
Bf-A was isolated from the mycelium and culture broth of a surface culture of E. donezkii HKI 0059 at the former Hans Knöll Institute for Natural Product Research eV, Jena, Germany (Berg et al., 1996 ▸). E. donezkii HKI 0059 was deposited as patent strain Emericellopsis spec. DSM 11079 in the Deutsche Sammlung für Mikroorganismen und Zellkulturen, Braunschweig, Germany and is available on request. The fungus was grown in a liquid medium (pH 6) composed of malt extract (20 g l−1), glucose (10 g l−1), yeast extract (1 g l−1) and ammonium sulfate (5 g l−1). Mycelium and culture broth were extracted with methanol and ethyl acetate, respectively. Bf-A was purified from the combined extracts and isolated by silica-gel column and RP-18 chromatography.
2.2. Crystallization and data collection
The sample used for X-ray analysis was crystallized by slow evaporation in a glass vial at room temperature from a methanol–water (45:55) solution and provided tiny needle-shaped crystals that were barely visible under a stereoscope at 40× magnification. Crystals were transferred to a MiTeGen MicroMesh mount using an ultrasharp needle. Diffraction data were collected at 100 K at the Diamond Light Source in Didcot, UK using the microfocus beamline I24 equipped with a PILATUS3 6M detector (Dectris). The crystals were not clearly visible and a single crystal had to be aligned with the X-ray beam using a raster scan. A data set of 900 images covering 180° rotation was collected from a single bergofungin crystal estimated at less than 10 µm in size in the two smallest dimensions. The data were integrated and scaled using the XDS software package (Kabsch, 2010 ▸). The data-set statistics are summarized in Table 1 ▸.
Table 1. Crystal and diffraction data and structure refinement.
The computer programs XDS (Kabsch, 2010 ▸), SCALA (Evans, 2006 ▸), SHELXS86 (Sheldrick, 2008 ▸), SHELXL2014 (Sheldrick, 2015 ▸), Coot (Emsley et al., 2010 ▸), PyMOL (DeLano, 2002 ▸) and POV-Ray (http://www.povray.org) were used.
| Data collection | |
| Temperature (K) | 100 |
| Radiation wavelength (Å) | 0.7293 |
| Chemical formula | C73H120N16O19·0.5H2O |
| M r | 1533.8 |
| Space group | C2 |
| a, b, c (Å) | 48.275 (10), 8.932 (2), 24.604 (5) |
| β (°) | 119.72 (3) |
| V (Å3) | 9213 (4) |
| Z | 4 |
| μ (mm−1) | 0.08 |
| Crystal size (mm) | 0.2 × 0.02 × 0.02 |
| Resolution range (Å) | 30–0.84 |
| (sinθ/λ)max (Å−1) | 0.595 |
| No. of independent reflections (unmerged Friedel pairs) | 12250 |
| No. of merged reflections | 7347 |
| Completeness for merged reflections (entire range/0.94–0.84 Å) (%) | 83.3/50.5 |
| R int (unmerged Friedel pairs) | 0.076 |
| Refinement | |
| wR [F 2 > 2σ(F 2)], wR (F 2), S | 0.067, 0.172, 0.95 |
| No. of parameters | 985 |
| Δρmax, Δρmin (e Å−3) | 0.35, −0.22 |
2.3. Structure determination and refinement
The structure was solved by direct methods using SHELXS97 (Sheldrick, 2008 ▸) with the options Init 99, Phan and Tref 77777 to obtain more self-consistent phase sets. Thus, all 108 non-H atoms and a well ordered, half-occupied water molecule could be detected. The structure was refined using SHELXL (Sheldrick, 2015 ▸). Unrestrained refinement led to R and R free factors of 15.0 and 15.8%, respectively. Anisotropic temperature-factor refinement and modelling of five disordered water molecules with limited occupancy were performed. Addition of H atoms in calculated positions and their refinement as riding atoms, with the exception of the C-terminal H atom and the H atoms attached to the Oγ atoms of hydroxyprolines, for which the bonding angle was positioned by rotation in the maximum electron density, produced an R factor of 11.3% and an R free of 13% for all reflections. Attempts to describe the disordered solvent region and to interpret the highest peaks in difference Fourier syntheses resulted in more disordered water molecules with limited occupancy. Owing to their close proximity, these water molecules could not coexist in the same unit cell. For better handling of this disordered solvent region the eight disordered water molecules were removed and the PLATON program SQUEEZE (Spek, 2015 ▸) was used to calculate the solvent contribution. The output files were used as input for the last cycles of SHELXL, in which the temperature factors of the H atoms were fixed at a multiple of the equivalent isotropic values of the temperature factors of the atoms that they are riding on: 1.2 times for O and N atoms and 1.5 times for C atoms. R and R free factors of 10.3 and 13.1% were obtained. A total of 985 parameters were refined against 12 250 reflections, and one floating origin restraint was used. The validation server of the Protein Data Bank (Berman et al., 2006 ▸; Laskowski et al., 1993 ▸) was used to examine the quality of the structure. Swiss-PdbViewer (Guex & Peitsch, 1997 ▸; http://spdbv.vital-it.ch/), Coot (Emsley et al., 2010 ▸), POV-Ray (http://www.povray.org) and PyMOL (DeLano, 2002 ▸) were used for geometric analysis, visualization and for the production of figures.
2.4. Agar diffusion test
The antibiotic activity of bergofungin A was determined as the diameter of the inhibition zone produced by 1000 and 50 µg ml−1 bergofungin A in distilled water added to agar wells (9 mm in diameter) in a standardized agar-plate diffusion assay. After 18 h at 37°C the antibiotic effect could be measured (see Supporting Information for more details). All of the microbial strains used for testing Bf-A were obtained from HKI Jena (Leibniz Institute for Natural Product Research and Infection Biology, Hans Knöll Institute). The biological activity assays of the two peptaibols (Bf-A and samarosporin) are admittedly not directly comparable as they were obtained by different protocols using different organisms. Unfortunately, experiments with both peptaibols could not be carried out in a uniform manner owing to a lack of samarosporin samples.
3. Results and discussion
3.1. Molecular structure
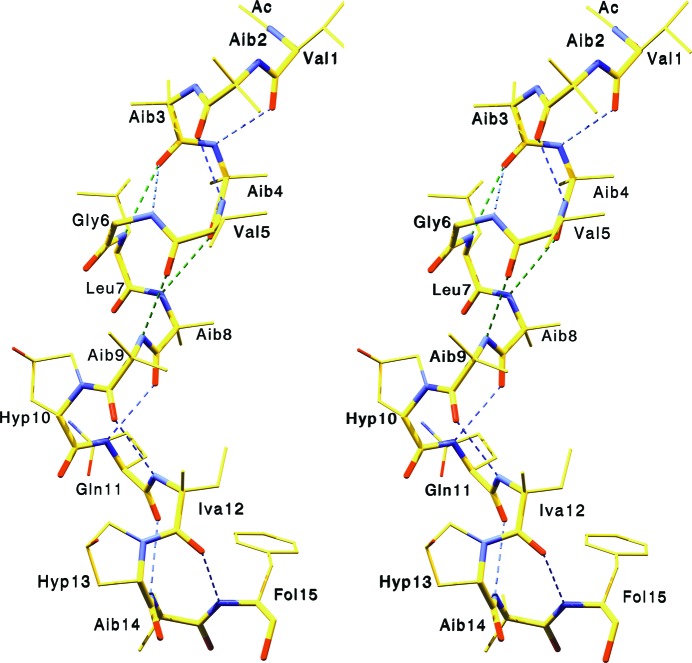
Bergofungin differs from samarosporin only in the first residue (Val versus Phe). Their chemical similarity leads to crystals with the same space-group symmetry (C2) and nearly isomorphous unit-cell parameters (Table 1 ▸ and PDB entry 4g13; Gessmann, Axford, Evans et al., 2012 ▸). The quality of the refined structure at the atomic level is reflected in the electron-density map shown in Fig. 1 ▸. In Fig. 2 ▸, a stereoview of bergofungin is shown. There are ten intramolecular hydrogen bonds. The helix starts with three hydrogen bonds of type 4→1, with the third C=O group of Aib3 also participating in a type 5→1 hydrogen bond to Leu7. The helix continues with two further hydrogen bonds of type 5→1 followed by four 4→1 hydrogen bonds. Thus, the folding of bergofungin A is overwhelmingly 310-helical, forming one kinked helix. The two carbonyl O atoms of Gly6 and Leu7 cannot accept hydrogen bonds from Hyp10 as prolines are imino acids. These O atoms are nevertheless satisfied by intermolecular hydrogen bonding to the side chain of Gln11 and the hydroxyl group of Hyp10. The single water molecule is hydrogen-bonded to the C=O group of Hyp10 and to the carbonyl group of the Gln11 side chain of the same molecule. Val1 lies inside the additionally allowed helical range (Laskowski et al., 1993 ▸), with φ and ψ torsion angles of −120 and −72°, respectively. Torsion angles and hydrogen-bond parameters are listed in Supplementary Tables S1 and S2. The side chains of the two valines and one leucine are in the usual tg+ conformation (Supplementary Table S1; Janin et al., 1978 ▸). The two l-4-hydroxyprolines are in a trans conformation; their pyrrolidine rings assume the Cγ-exo conformation and the asymmetric Cγ atoms are in the d-configuration, thus representing (2S,4R)-4-hydroxyprolines. Iva unambiguously has the d-configuration (Fig. 1 ▸), as also shown by NMR data (De Zotti et al., 2012 ▸).
Figure 1.
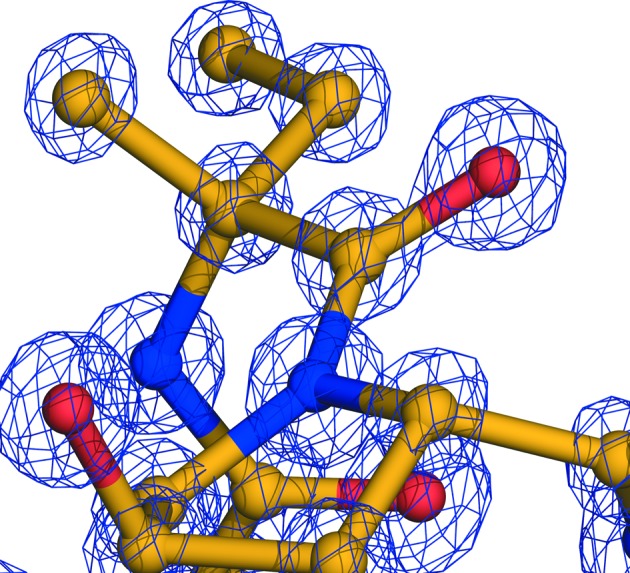
Electron density of the 2F o − F c map contoured at 3σ around Iva12.
Figure 2.
Stereoview of bergofungin A. Hydrogen bonds of type 1←4 are shown in blue and of type 1←5 are shown in green.
3.2. Crystal packing
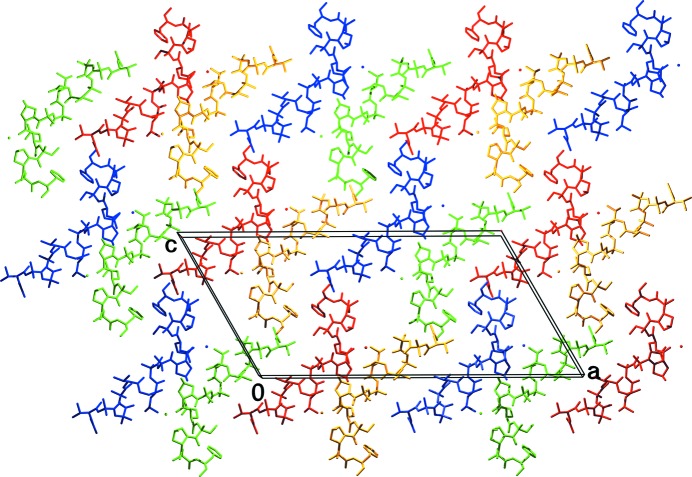
The crystal packing is shown in Fig. 3 ▸. As in the case of the closely related samarosporin I, every two molecules are hydrogen-bonded in the convex middle part of the molecule via three hydrogen bonds (yellow–red and green–blue): the side chain of Hyp10 forms a hydrogen bond to the carbonyl group of Leu7 and to the amino moiety of the side-chain carboxamide group of Gln11. This group is hydrogen-bonded back to the carbonyl group of Gly6 of the first molecule. Thus, a >< shape is adopted by two antiparallelly packed, symmetry-related molecules. The other two groups of every partner molecule (C=O of Leu7 and NH2 of Gln11 for the > molecule and C=O of Gly6 and OH of Hyp10 for the < molecule) which are not involved in this hydrogen bonding, interact with the +1 or −1 y-translated < or > partner, respectively. This partner lies exactly behind or in front of its companion molecule shown in Fig. 3 ▸. Thus, symmetry-related molecules forming the >< shaped ‘dimer’ are connected via hydrogen bonds in the y direction in an alternating zigzag fashion, e.g. one red molecule is hydrogen-bonded to two yellow molecules translated along the b axis. In addition, there is an additional hydrogen bond in the y direction between the side chain of Hyp13 and the carbonyl-group side chain of Gln11.
Figure 3.
Arrangement of the molecules in the crystal viewed along the short b axis. Molecules translated along the unit-cell edges are coloured uniformly, whereas the four molecules of different colour are related by space-group symmetry. A single molecular layer is shown in the ac plane.
Furthermore, there is strong head-to-tail hydrogen bonding between symmetry-related molecules (blue–red and green–yellow in Fig. 3 ▸). There are three hydrogen bonds from every molecule to its counterpart. Moreover, the alcoholic C-terminus connects to the carbonyl of the acetyl protection group of a b-translated molecule in such a way that the planes of the blue–red and green–yellow pairs of molecules lie perpendicular to the ac plane. Together with the above hydrogen bonding of the facing molecules in the ac planes, there are all-four-symmetry-related columns along the short b axis. Neighbours of these planes are packed antiparallel via hydrophobic contacts. All polar groups that can participate in intramolecular or intermolecular hydrogen bonding do so, with the exception of the carbonyl group of Aib13. Further details of hydrogen bonding are provided in Supplementary Table S2.
As in the other peptaibol crystal structures, the calculated space for solvent content is relatively high (Supplementary Table S3). In the two large spaces which are formed between the four symmetry-related molecules (one space at the N-terminus and one at the C-terminus of every molecule in Fig. 3 ▸, looking along the b axis) there are two water molecules with half occupancy per space. Assuming a van der Waals volume of 20.6 Å3 (Li & Nussinov, 1998 ▸) for one water molecule, there is space available for ∼20 water molecules in the asymmetric unit. Nonetheless, only one, half-occupied water molecule was located. Such extensive, practically empty spaces along the short crystal axis and perpendicular to the molecular helical axis that give rise to solvent channels have also been observed in the crystal structures of trichovirin and samarosporin (Gessmann, Axford, Evans et al., 2012 ▸; Gessmann, Axford, Owen et al., 2012 ▸).
3.3. Comparison with other subfamily 2 peptaibols and with samarosporin
Some geometric parameters of peptaibol SF2 crystal structures are listed in Table 2 ▸. The bend angle of bergofungin A is the largest among the members of this group, and this peptaibol is also the shortest member of SF2. The helical parameters lie between the corresponding parameters for a 310-helix and an α-helix, while the value of the dipole moment is the lowest among the members of the group. The low dipole moment may facilitate embedding of Bf-A in the membrane and compensate for the lack of phenylalanine as the first residue.
Table 2. Geometric parameters of peptaibol subfamily 2 structures.
| Peptaibol | No. of residues | Length† (Å) | Bend angle‡ (°) | n § | h § (Å) | Pitch§ | Dipole moment¶ (D) |
|---|---|---|---|---|---|---|---|
| Bergofungin A | 15 | 25.2 | 59.1 | 3.53 | 1.73 | 6.12 | 27.9 |
| Samarosporin I†† | 15 | 26.9 | 56.5 | 3.44 | 1.81 | 6.23 | 28.3 |
| Antiamoebin I‡‡ | |||||||
| Molecule 1 | 16 | 28.3 | 56.0 | 3.55 | 1.65 | 5.86 | 29.1 |
| Molecule 2 | 16 | 28.6 | 48.5 | 3.51 | 1.67 | 5.86 | 28.7 |
| Antiamoebin I§§ | 16 | 28.1 | 53.4 | 3.54 | 1.73 | 6.12 | 32.5 |
| Cephaibol A¶¶ | 16 | 26.8 | 50.3 | 3.54 | 1.72 | 6.08 | 29.8 |
| Cephaibol B¶¶ | |||||||
| Molecule 1 | 16 | 29.0 | 40.2 | 3.52 | 1.74 | 6.12 | 30.4 |
| Molecule 2 | 16 | 30.0 | 47.4 | 3.52 | 1.74 | 6.32 | 31.2 |
| Cephaibol C¶¶ | 16 | 28.4 | 47.6 | 3.56 | 1.71 | 6.09 | 30.8 |
| α-Helix | 3.60 | 1.50 | 5.40 | ||||
| 310-Helix††† | 3.24 | 1.94 | 6.29 | ||||
Distance from the more distant atom (CH3 or O) of the acetyl protection group to the O atom of the C-terminal amino alcohol.
Angle between the planes on either side of Hyp10 (residue n) formed by the N atoms of residues n − 2, n − 4 and n − 6 (plane 1) and residues n + 1, n + 3 and n + 5 (plane 2), as proposed by Snook et al. (1998 ▸).
n is the average number of residues per turn and h is the average unit height in the helix; pitch = n × h; calculated with HELANAL (Bansal et al., 2000 ▸) after substituting all unusual residues with conventional residues.
Calculated at http://dipole.weizmann.ac.il; 1 D = 0.2082 e Å.
Gessmann, Axford, Evans et al. (2012 ▸).
Snook et al. (1998 ▸).
Karle et al. (1998 ▸).
Bunkóczi et al. (2003 ▸).
Toniolo & Benedetti (1991 ▸).
The crystal structures of bergofungin A and samarosporin I are closely related. The r.m.s. deviation of main-chain atoms is 0.65 Å, with maxima of 2.7 and 2.3 Å at the N- and C-termini, respectively.
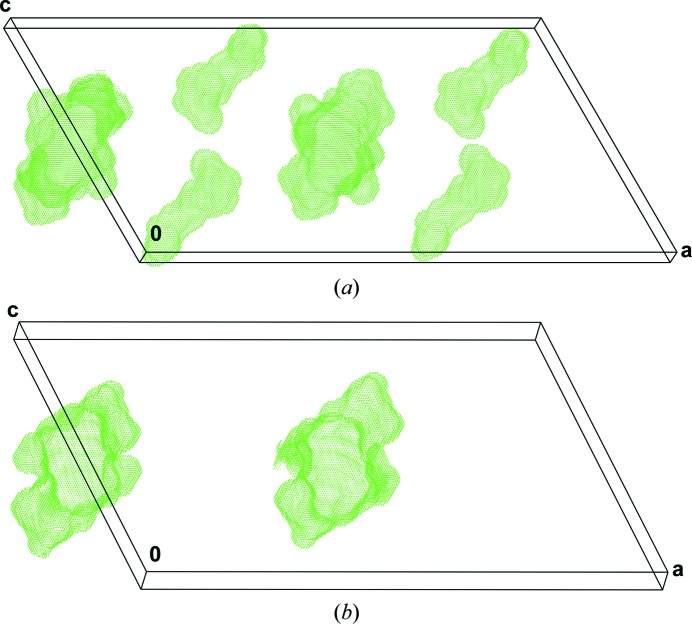
The differences in the intramolecular hydrogen bonding between the above structures are the following: O3←N6 in Bf-A corresponds to O2←N6 in samarosporin, while O4←N8 in Bf-A has a distance (N8 to O4) in samarosporin that is above the limit for hydrogen bonding and the C-terminal O13←O15 hydrogen bond in samarosporin is missing in Bf-A. In the crystal, the aromatic rings of Fol15 pack almost parallel (24°) in bergofungin with a minimum distance of 4.2 Å, while in samarosporin these rings pack with an angle of 58° and approach each other within 3.7 Å. The packing in the crystals of both peptaibols appears quite similar, and extensive empty solvent-accessible channels along the shortest axis exist in both crystals. Nevertheless, the less bulky Val1 side chain leads to an additional solvent-accessible channel per molecule along the short axis, the volume of which is about a third of their common channel (Fig. 4 ▸).
Figure 4.
Solvent-accessible spaces in the crystals of bergofungin A (a) and samarosporin (b) calculated and plotted with PLATON (Spek, 2009 ▸). The parts of the channels corresponding to one unit cell are shown, viewed approximately along the b axis.
The bacterial growth assay in agar diffusion (described in detail in the Supporting Information) shows that bergofungin at 1 mg ml−1 is not active against Staphylococcus aureus, Escherichia coli and Candida albicans, whereas samarosporin at 156 µg ml−1 is active against S. aureus and at 625 µg ml−1 is active against E. coli and C. albicans (data from Inoue et al., 1976 ▸). Furthermore, 1 mg ml−1 bergofungin solution has no effect against Pseudomonas aeruginosa, while samarosporin is active against Gram-positive and Gram-negative bacteria, yeasts, fungi and protozoa at lower concentrations.
Apparently, this lower antibiotic effect can be assigned to the exchange of phenylalanine to valine at the N-terminus of the peptides.
4. Related literature
The following references are cited in the Supporting Information for this article: Chugh et al. (2002 ▸), Fox & Richards (1982 ▸), Matthews (1968 ▸) and Mueller & Hinton (1941 ▸).
Supplementary Material
PDB reference: bergofungin A, 5mas
Supplementary file containing torsion angles, hydrogen bonds and biological data.. DOI: 10.1107/S2053230X17001236/dz5402sup3.pdf
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2053230X17001236/dz5402sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2053230X17001236/dz5402sup2.hkl
CCDC reference: 1529476
Acknowledgments
We thank Dr Kerstin Voigt (HKI Jena) for performing the microbial testing.
References
- Bansal, M., Kumart, S. & Velavan, R. (2000). J. Biomol. Struct. Dyn. 17, 811–819. [DOI] [PubMed]
- Berg, A., Ritzau, M., Ihn, W., Schlegel, B., Fleck, W. F., Heinze, S. & Gräfe, U. (1996). J. Antibiot. 49, 817–820. [DOI] [PubMed]
- Berman, H. M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2006). International Tables for Crystallography, Vol. F, edited by M. G. Rossmann & E. Arnold, pp. 675–684. Dordrecht: Kluwer Academic Publishers.
- Bunkóczi, G., Schiell, M., Vértesy, L. & Sheldrick, G. M. (2003). J. Pept. Sci. 9, 745–752. [DOI] [PubMed]
- Christner, C., Zerlin, M., Gräfe, U., Heinze, S., Küllertz, G. & Fischer, G. (1997). J. Antibiot. 50, 384–389. [DOI] [PubMed]
- Chugh, J. K., Brückner, H. & Wallace, B. A. (2002). Biochemistry, 41, 12934–12941. [DOI] [PubMed]
- Chugh, J. K. & Wallace, B. A. (2001). Biochem. Soc. Trans. 29, 565–570. [DOI] [PubMed]
- Degenkolb, T., Karimi Aghcheh, R., Dieckmann, R., Neuhof, T., Baker, S. E., Druzhinina, I. S., Kubicek, C. P., Brückner, H. & von Döhren, H. (2012). Chem. Biodivers. 9, 499–535. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- De Zotti, M., Biondi, B., Crisma, M., Hjørringgaard, C. U., Berg, A., Brückner, H. & Toniolo, C. (2012). Biopolymers, 98, 36–49. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fox, R. O. & Richards, F. M. (1982). Nature (London), 300, 325–330. [DOI] [PubMed]
- Galbraith, T. P., Harris, R., Driscoll, P. C. & Wallace, B. A. (2003). Biophys. J. 84, 185–194. [DOI] [PMC free article] [PubMed]
- Gessmann, R., Axford, D., Evans, G., Brückner, H. & Petratos, K. (2012). J. Pept. Sci. 18, 678–684. [DOI] [PubMed]
- Gessmann, R., Axford, D., Owen, R. L., Brückner, H. & Petratos, K. (2012). Acta Cryst. D68, 109–116. [DOI] [PubMed]
- Gessmann, R., Brückner, H., Aivaliotis, M. & Petratos, K. (2015). J. Pept. Sci. 21, 476–479. [DOI] [PubMed]
- Gessmann, R., Brückner, H. & Petratos, K. (2015). Acta Cryst. C71, 1114–1117. [DOI] [PubMed]
- Guex, N. & Peitsch, M. C. (1997). Electrophoresis, 18, 2714–2723. [DOI] [PubMed]
- Inoue, N., Inoue, A., Furukawa, M. & Kanda, N. (1976). J. Antibiot. 29, 618–622. [DOI] [PubMed]
- Jabs, T., Ammermann, E., Stierl, R., Korenz, G., Boland, W. & Engelberth, J. (2001). World Patent WO2001067867A3.
- Janin, J., Wodak, S., Levitt, M. & Maigret, B. (1978). J. Mol. Biol. 125, 357–386. [DOI] [PubMed]
- Jaworski, A. & Brückner, H. (2000). J. Pept. Sci. 6, 149–167. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karle, I. L. & Balaram, P. (1990). Biochemistry, 29, 6747–6756. [DOI] [PubMed]
- Karle, I. L., Perozzo, M. A., Mishra, V. K. & Balaram, P. (1998). Proc. Natl Acad. Sci. USA, 95, 5501–5504. [DOI] [PMC free article] [PubMed]
- Kropacheva, T. N., Salnikov, E. S., Nguyen, H. H., Reissmann, S., Yakimenko, Z. A., Tagaev, A. A., Ovchinnikova, T. V. & Raap, J. (2005). Biochim. Biophys. Acta, 1715, 6–18. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Li, A. J. & Nussinov, R. (1998). Proteins, 32, 111–127. [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mueller, J. H. & Hinton, J. (1941). Exp. Biol. Med. 48, 330–333.
- Mukherjee, P. K., Wiest, A., Ruiz, N., Keightley, A., Moran-Diez, M. E., McCluskey, K., Pouchus, Y. F. & Kenerley, C. M. (2011). J. Biol. Chem. 286, 4544–4554. [DOI] [PMC free article] [PubMed]
- Neumann, N. K. N., Stoppacher, N., Zeilinger, S., Degenkolb, T., Brückner, H. & Schuhmacher, R. (2015). Chem. Biodivers. 12, 743–751. [DOI] [PubMed]
- Pandey, R. C., Cook, J. C. & Rinehart, K. L. (1977). J. Am. Chem. Soc. 99, 5205–5206. [DOI] [PubMed]
- Schiell, M., Hofmann, J., Kurz, M., Schmidt, F. R., Vériest, L. S., Vogel, M., Wink, J. & Seibert, G. (2001). J. Antibiot. 54, 220–233. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Shenkarev, Z. O., Paramonov, A. S., Nadezhdin, K. D., Bocharov, E. V., Kudelina, I. A., Skladnev, D. A., Tagaev, A. A., Yakimenko, Z. A., Ovchinnikova, T. V. & Arseniev, A. S. (2007). Chem. Biodivers. 4, 1219–1242. [DOI] [PubMed]
- Snook, C. F., Woolley, G. A., Oliva, G., Pattabhi, V., Wood, S. F., Blundell, T. L. & Wallace, B. A. (1998). Structure, 6, 783–792. [DOI] [PubMed]
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Spek, A. L. (2015). Acta Cryst. C71, 9–18. [DOI] [PubMed]
- Toniolo, C. & Benedetti, E. (1991). Trends Biochem. Sci. 16, 350–353. [DOI] [PubMed]
- Toniolo, C. & Brückner, H. (2009). Peptaibiotics: Fungal Peptides Containing α-Alkyl α-Amino Acids. Weinheim: Wiley-VCH.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: bergofungin A, 5mas
Supplementary file containing torsion angles, hydrogen bonds and biological data.. DOI: 10.1107/S2053230X17001236/dz5402sup3.pdf
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2053230X17001236/dz5402sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2053230X17001236/dz5402sup2.hkl
CCDC reference: 1529476