Abstract
Recent studies have shown how volumetric imaging and morphometrics can add significantly to our understanding of morphogenesis, the developmental basis for variation and the etiology of structural birth defects. On the other hand, the complex questions and diverse imaging data in developmental biology present morphometrics with more complex challenges than applications in virtually any other field. Meeting these challenges is necessary in order to understand the mechanistic basis for variation in complex morphologies. This chapter reviews the methods and theory that enable the application of modern landmark-based morphometrics to developmental biology and craniofacial development, in particular. We discuss the theoretical foundations of morphometrics as applied to development and review the basic approaches to the quantification of morphology. Focusing on geometric morphometrics, we discuss the principal statistical methods for quantifying and comparing morphological variation and covariation structure within and among groups. Finally, we discuss the future directions for morphometrics in developmental biology that will be required for approaches that enable quantitative integration across the genotype-phenotype map.
Introduction
Answering the question of how developmental mechanisms result in morphogenesis is a key goal of developmental biology. The study of molecular mechanisms underlying morphogenesis has become increasingly quantitative and integrative with the development of genomic technologies. Yet, the quantitative study of morphology has been fairly peripheral to the mechanistic study of morphogenesis (Hallgrímsson et al. 2009). In recent years, quantitative analysis of morphology has become more prevalent in the study of morphogenesis. New imaging techniques provide increasingly accessible and higher throughput imaging of embryonic morphology. Combined with novel quantitative approaches, this allows for analyses and visualizations that are increasingly intuitive and accessible to developmental biologists and geneticists. Further, studies of the developmental basis for morphogenesis involve increasingly complex analyses of multiple genetic factors or treatments. Finally, there is increased interest in the relationship between the determinants of normal variation and the genetics of structural birth defects, especially in the field of craniofacial biology (Cooper and Albertson 2008; Hallgrímsson et al. 2009; Heuze et al. 2014; Houle 2010; Houle et al. 2010; Young et al. 2010a). Such studies demand more refined quantitative phenotypic assessment than might have sufficed in the past. This chapter reviews the use of 3D imaging and morphometrics for the study of craniofacial development.
Developmental biology is focused on revealing the processes and the interactions among processes that result in embryogenesis and growth (Love 2014). Processes and mechanisms underlying organismal development are the focus of study for developmental biologists and the field assembles approaches and techniques that tackle this basic question and its various components (Burian 2005). As such, processes and mechanisms are the focus of study for developmental biologists. Although morphology is, ultimately, the phenomenon that the field seeks to explain, it is far distant from most developmental biology research that seeks to reveal molecular and cellular mechanisms. Accordingly, phenotypic variation often takes the role of a predicted outcome of an experimental perturbation, and an observation showing morphological difference represents an abnormality that points towards a mechanism of interest. For these reasons, developmental biologists have not generally been interested in phenotypic variation per se. Phenotypic outcomes are usually seen as discrete. Mutants are described as having a phenotype and variation in experimental outcomes at the phenotypic level is more often a nuisance than an object of study as it is thought to obscure the phenotype of interest.
In contrast, phenotypic variation has a central epistemological position within evolutionary biology. Although its coherence can be debated, evolutionary biology has a unifying theory (Sober 1994), within which selection acts on phenotypic variation and evolution occurs through particulate inheritance of genes. Therefore, the quantitative study of morphological variation has been much more prominent within evolutionary biology than within developmental biology (Bookstein et al. 1985). Much of the theory of geometric morphometrics, for example, was developed to answer evolutionary questions. Within this chapter, we discuss some of the ways that the resulting morphometric concepts and methods can be applied to answer questions of development.. However, development also presents huge challenges to morphometrics. Chief among these is the need to integrate quantification across levels from genetic to cellular to morphological. In particular, the need to quantitatively integrate molecular and morphological imaging is a major challenge. Current efforts are being made to meet these challenges. Further, the quantification of morphology across developmental stages often spans ranges of morphological variation that challenge existing techniques, particularly when some attempt is made to identify and preserve homologies, defined as the biological correspondence of structures or location across individuals and ontogeny. Finally, innovation in imaging techniques has led to increasingly rich image datasets that challenge both existing theory and computational power. We discuss these challenges their potential solutions and progress on their development below.
Morphometrics and Morphospaces
Morphometrics is the quantification and statistical analysis of form. Form is the combination of size and shape of a geometric object in an arbitrary orientation and location. Shape is what remains of the geometry of such an object once you standardize for size. Note that in morphometrics, unlike in common parlance, form and shape do not mean the same thing.
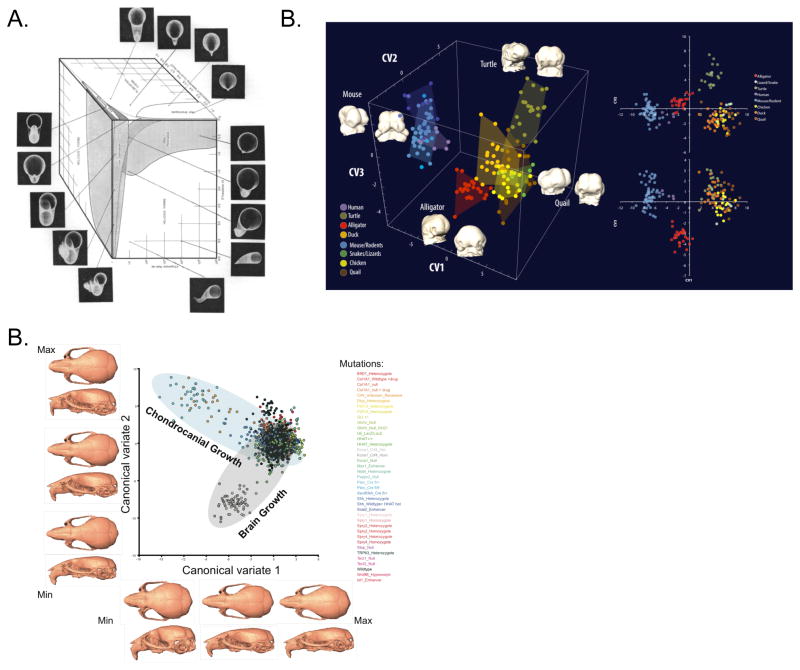
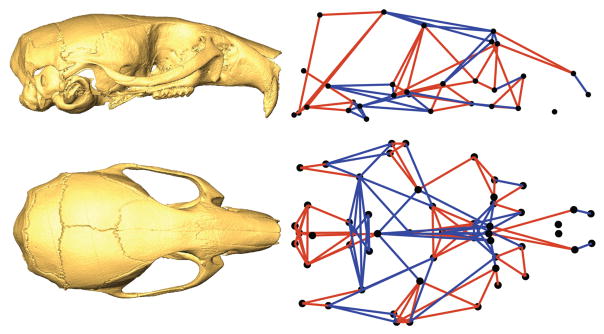
A key concept underlying much of the quantitative analysis of form is the idea that morphology can be mapped in a systematic way, often within a “morphospace”. Morphospaces are maps showing how shapes are defined by quantitative traits. Although the basic idea arguably derives from the deformation grids of D’Arcy Thompson (1942; 1961; 1917), the idea of a quantitative morphospace was first operationalized by David Raup (1966). While the concept of a phenotypic distance between specimens or groups of specimens, based on the measures of multiple traits, was used in numerical taxonomy at that time (Sneath and Sokal 1973; Sokal and Rohlf 1995b), Raup created the visual metaphor of the morphospace. He created a model composed of three parameters that could be used to describe the potential diversity of shell form (Figure 1a), his phenotype of interest. He then plotted the evolutionary diversity of coiled shell animals within the three dimensional morphospace defined by these parameters and found that large areas of potential morphospace were not occupied. This raised the interesting question of why there are not naturally occurring shells that represent all possible regions of morphospace. The proposed explanations included shell function and constraints imposed by developmental processes. In this way, Raup’s morphospace concept was foundational for evolutionary developmental biology because early concepts of developmental constraint were used to explain gaps in the space of possible morphologies (Alberch 1982; Maynard Smith et al. 1985)
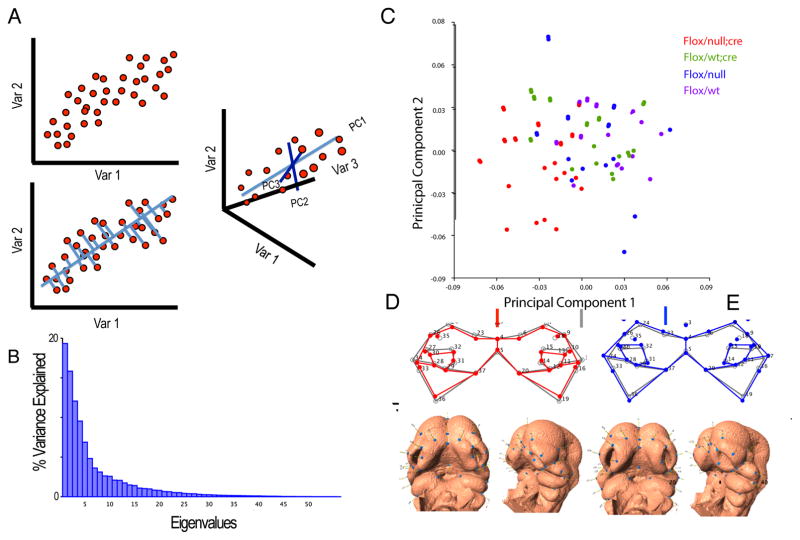
Figure 1.
A) Raup’s morphospace for shell coiling (Raup, 1966). B) Morphology for amniote embryos, including humans, is shown in a space constructed using Principal Components Analysis from 3D landmark data (Young et al. 2014). C) Analysis of mouse mutants with in a Canonical Variates Analysis based space. This plots shows craniofacial effects for mutations affecting chondrocanial growth and brain size forming two distinct common axes of covariation.
Raup’s elegantly simple morphospace provides a surprisingly complete description of shell coiling, but it belies the arbitrariness of such spaces more generally. Morphospaces are defined by their axes. Often such axes are multivariate summaries of form data as in Figure 1B and C. In very few circumstances is it possible to construct a morphospace based on developmental parameters, such as the expression or diffusion gradients of particular morphogens, because developmental systems are rarely understood at that level of granularity. This means that while it is possible to plot measured morphology and quantify morphological variation in accordance with the axes (ie. chosen parameters) of a morphospace, it is rarely possible to capture the all options for potential variation within a plot or single shape-space. Depending on how a morphospace is defined, developmental changes can occur in ways that are not captured. Further, for axes defined on the basis of covariation structure, it must be borne in mind that developmental perturbations can alter covariation structure in ways that can be hard to predict (Hallgrimsson et al. 2009; Jamniczky and Hallgrimsson 2009; Mitteroecker 2009). Morphospaces are generally arbitrary with respect to development, as they are not usually defined by direct developmental parameters. Morphospaces are, nonetheless, essential tools in that they can be used to quantitatively compare and situate morphologies relative to each other. As phenomics gains traction and intersects more meaningfully with developmental biology, careful thought will need to be devoted to the construction of the morphospaces within which phenomic analyses are conducted.
As with all morphometric tools, care must be taken to interpret morphospaces within the context in which they are constructed. To be useful, morphospaces must have a few key properties. The first is that relative locations and distances in such spaces must have biological meaning (Mitteroecker and Huttegger 2009). This means that forms that are similar should cluster together and those that are dissimilar should be far apart and that the distances among them should be proportional to some underlying biological difference. This is critical if a measure of dispersion of a sample within a space is to have any relation to the distance between the mean of two samples or if the distances among various mutant phenotypes are to be compared. Second, directions within the morphospace should have biological meaning. Without biological meaning, it becomes impossible to use the morphospace to predict morphologies based on a continuous relationship or determine whether a group of related mutations or treatments produce effects in the same direction. Closely related to this is the requirement of co-linearity, meaning that parallel trajectories in morphospace should represent comparable axes of phenotypic change. For example, in two samples that differ in mean shape, one should be able to compare the effect of a continuous variable such as age, size or some treatment under the expectation that these effect would produce a displacement in the same direction for both shapes even though they are at different locations in morphospace. Finally, the axes of a morphospace should be independent and consist of commensurate units. For a more fulsome discussion of these properties, see Mitteroecker and Hutteger (2009).
Often, however, morphospaces fail to meet these criteria (Mitteroecker and Hutteger 2009), making the data contained within them difficult to interpret. Raup’s shell coiling space, for instance, uses the parameters of a mathematical model of coiling to create the axes of the space. The problem with this is that there is not a natural scale relationship among the axes. Further, the axes are not all independent as some involve overlapping sets of input parameters. The lack of a natural scale means that angles or directions in Raup’s morphospace are arbitrary. When two phenotypic measures are dependent on one another, then using both measures can lead to a morphospace within which there are regions that cannot be occupied simply because some combination of values is not possible, rather than because of some biological, developmental, or evolutionary constraint. Spaces defined by ratios among traits such as limb proportions or size ratios within structures often produce such artifacts.
Approches to Morphometrics
There are several approaches to doing morphometrics. All of these approaches have advantages and limitations. There is no single correct approach to morphometrics that applies to all problems. Approaches are more or less correct or appropriate only in the context of the question being asked. For this reason, it is critical to understand what different morphometric techniques do and their respective limitations. Much effort has been devoted in morphometrics to debates over the use of one approach or another. Too often such debates are framed within the assumption that there is a single correct approach to the quantification of form. Under this assumption, such debates have sometimes taken on a religious fervor. However, the quantification of form is not an end in itself. Rather, it is a means of answering biological questions. All methods will result in something less than a perfect quantification of the phenomena of interest. One must always ask whether the method used generates useful quantifications and there are occasions when using more than one approach and interpreting the differences among them leads to greater insight into the biological question at hand.
In the age of omics, debates over methods have practical implications because of the increasing need to compare and integrate data or results across studies. This is a good reason to favour some methods over others. On the other hand, maintaining a diversity of approaches in a field can lead to innovation that might otherwise be missed. Balancing these two considerations is important both for reviewers of grants and papers and those who design “omic” resources involving morphometric data.
Landmark based methods
Traditional morphometrics relied on the phenotypic measurements such as linear distances, angles, weights and areas. Most modern morphometric approaches, however, are based on analysis of landmarks. Not all landmarks are equally useful. Bookstein classified landmarks into three types based on their information content and utility (Bookstein 1991). Type 1 landmarks represent discrete identifiable points that usually occur at the intersection of distinct anatomical structures. Intersections of sutures in bones, foramina, vascular branching points are examples of such landmarks. Type 2 landmarks represent points of maximal curvature along definable features. The tip of facial prominence or the inflection point of a cleft would be examples of such points. Type 3 landmarks are defined along extremes that are often defined by other points. These types of landmarks contain progressively less biological information. Type 3 landmarks, specifically, are deficient because they rely on information from other landmarks.
Semi-landmarks are a special case of Type 3 landmarks that are constructed by distributing points across a surface defined by other landmarks (Gunz et al. 2005). Usually, semi-landmarks are placed in an equidistant grid across a surface and then they are “slid” to optimize their position relative to the average shape for the sample. This sliding step is important as it places the landmarks in positions where they correspond better to each other across individuals (Gunz and Mitteroecker 2013). Skipping this step results in artifacts caused by the arbitrary assumption of equidistance among landmarks (Gunz and Mitteroecker 2013).
Mention that some methods for dense landmark quantification of surfaces are semilandmark techniques taken to the extreme?
Geometric Morphometrics
Geometric morphometrics (GMM) is the most established body of morphometric theory for handling landmark-based data.. Paraphrasing Rohlf’s definition of the field (http://life.bio.sunysb.edu/morph/), GMM is the multivariate statistical analysis of form as represented by Cartesian coordinate data. There are many excellent introductions to geometric morphometrics (Mitteroecker and Gunz 2009) and its use in developmental biology (Cooper and Albertson 2008; Hallgrímsson et al. 2009). We will not attempt a thorough review here. GMM methods rely on superimposition of landmark coordinate data in order to place individuals into a common morphospace. In the earliest example of this, Bookstein proposed using two fixed landmarks to register a larger landmark set. After scaling and superimposition of these two landmarks, the locations of the remaining landmarks represent variation in shape between specimens (Bookstein 1986; Bookstein 1991). Currently, the most commonly used form of superimposition is the Generalized Procrustes method (GPA). (Mitteroecker and Gunz 2009). Developed in its more general form for multivariate statistics by Gower (Gower 1975) GPA was adapted to morphometric data by Rohlf and Slice (Rohlf and Bookstein 1990; Rohlf and Slice 1990).
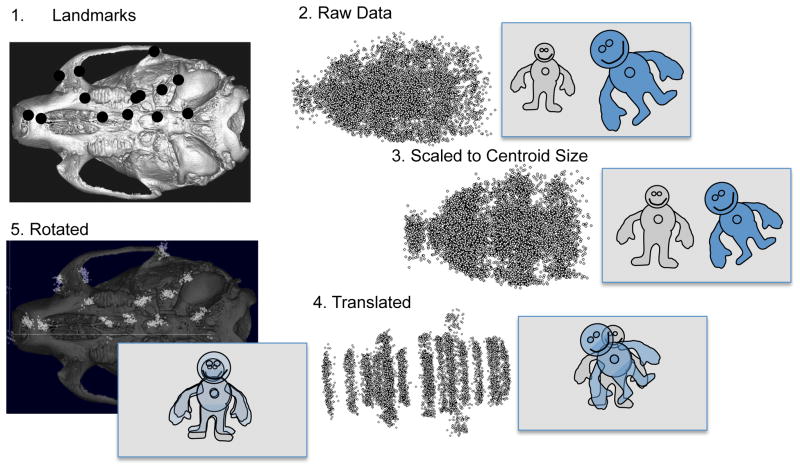
GPA places multiple individual specimens into the same shape space by scaling, translating and rotating the landmark coordinates (Figure 2). To scale the landmark coordinates GPA uses the Centroid Size (CS), a measure of scale representing the overall spread of a specimen’s landmark configuration from its centroid. CS is calculated as the square root of the summed squared deviations from the center (arithmetic mean) of each landmark configuration (Rohlf and Slice 1990). The arithmetic mean of each landmark configuration is the centroid. Each specimen’s landmark configuration is then translated so they share a centroid. Finally, each landmark configuration is rotated around the common centroid such that the deviation of each landmark from the average position for that landmark is minimized. This involves an iterative process. In the first step, the configurations are fitted to an arbitrary rotational angle for which each specimen is rotated to the angle that minimizes the sum of the squared distances from each landmark to its mean. From this first least-squares fit step, an average landmark configuration is generated and the least squares fit is repeated. The GPA process does this repeatedly until further steps produce no improvement in the fit (Dryden and Mardia 1998; Mitteroecker and Gunz 2009; Rohlf and Slice 1990). The result of the superimposition process is a set of modified landmark coordinates, termed Procrustes coordinates, for each specimen. These coordinates preserve the geometric relationship among a specimen’s landmarks and express each specimen as a set of departures from each landmark mean. Preservation of geometry throughout the analysis generates much of the power of geometric morphometric methods.
Figure 2.
General Procrustes Superimposition. A shows a 2D view of a 3D adult mouse landmark set. B. shows the full scatter for the raw landmarks in this dataset. In this particular datasets, the two sides of each specimen are treated as separate individuals. B) The scatter for all individuals (left and right sides) after scaling the LM coordinates for centroid size. C) The scatter after translation so as to center the centroids of each individual. D). The distribution of Procrustes residuals after rotation and, where appropriate, reflection superimposed on a ventral view of a mouse.
Although Procrustes superimposed data correspond to a definable morphospace, that space is not linear (Kendall 1984). For this reason, analyses are often conducted on data in which the Procrustes coordinates have been projected to a tangent space that is approximately linear (Dryden and Mardia 1998; Rohlf 1999). Most commonly, Procrustes coordinates are transformed to partial warp scores. Partial warp scores are based on Bookstein’s thin-plate spline method which uses a bending energy metaphor to calculate the continuous deformation of surfaces (Bookstein 1989; Bookstein 1991). This method is fundamental in computer graphics and image informatics, and is widely used in engineering. Whether to use Procrustes coordinates or partial warp scores does not matter for most morphometric analyses as they tend to involve fairly small amounts of shape variation (Rohlf 1999). In developmental biology, however, analyses often span large shape distances produced by morphogenesis where this choice may well matter for the results of an analysis. In general, it is critical for users of morphometric methods to know and understand the precise form of superimposition to which their data has been subjected, particularly when using comprehensive morphometric analysis software instead of custom-made programs.
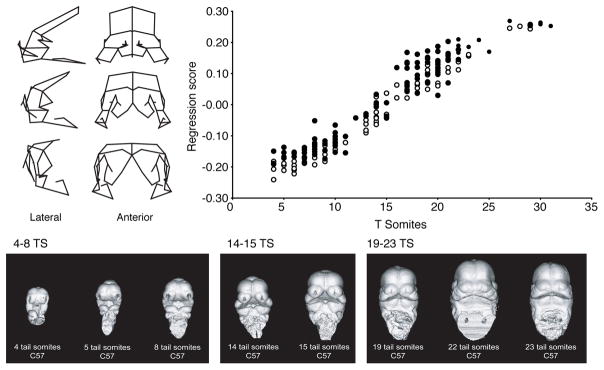
There are several important advantages of geometric morphometric methods over more traditional morphometrics that rely principally on linear distances, ratios or angles. The fact that GMM methods preserve geometry throughout an analysis means that variation within and between groups or the variation that corresponds to covariates of interest can be visualized as displacements of individual landmarks. These displacements can be used to generate 2- or 3-dimensional deformations of an anatomical object, showing how the object changes across the morphospace. Figure 3 shows examples of such analyses applied to microCT scan data for mouse embryos. Here, wireframes show the 3D deformations that correspond to ontogenetic variation in craniofacial shape. A related advantage is that the Procrustes superimposition based methods generate useful morphospaces in which multiple groups can be meaningfully compared. Such spaces have all of the desired properties listed above (Mitteroecker and Huttegger 2009).
Figure 3.
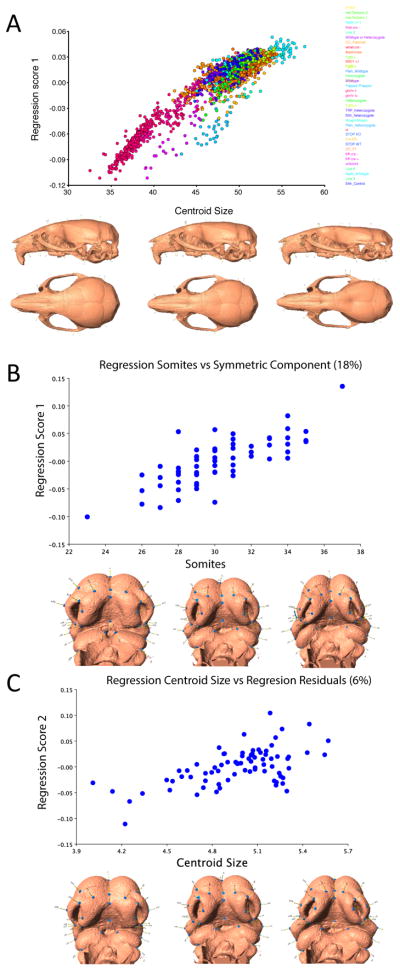
Multivariate regression on Procrustes superimposed landmark data allows calculation of the shape changes that correspond to that regression. Here, tail somite stage is regressed on landmark coordinates that capture craniofacial shape variation during face formation. The wireframes show the deformations that correspond to the regression in frontal and lateral views while the scatter plot shows the regression scores plotted against tail somite stage.
Variation in size and shape and be successfully quantified using GMM methods because they are based on a rigorous definition of shape (Kendall’s shape space and its linearizations). Shape variation can be decomposed into components that relate to biological factors of interest and that can be visualized as deformations of landmark configurations and surfaces, a key advantage in terms of biological relevance. Figure 3B shows an example of this in which the shape variation related to stage and to size are quantified separately within a sample of mouse embryos and this is explained in more detail in the next section.
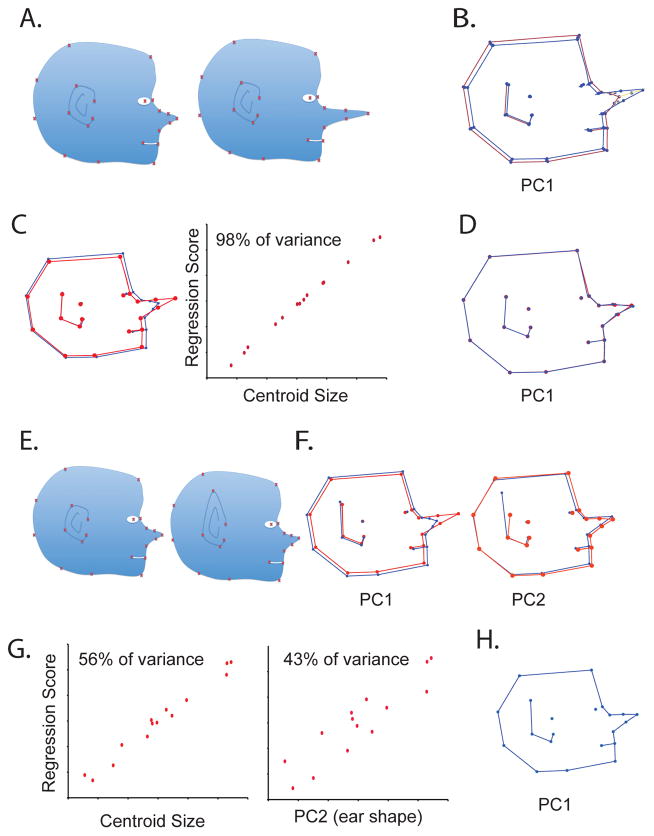
As with all good methods, GMM approaches also have limitations. The most important of these is that superimposition distributes some fraction of variation across landmarks (Marquez et al. 2012). This is known as the Pinocchio effect (Figure 4). The Pinocchio effect is simply a consequence of the GMM definition of shape. Once Pinocchio’s nose has elongated, the shape of his profile has changed and those changes are accurately described by the small differences in landmark positions across the head as well as the point of the nose even if only the nose has actually changed. However, As Klingenberg (Klingenberg 2013) points out, from a GMM perspective there is no a priori reason why this difference in shape should be described in terms of the nose moving forward rather than the head moving backwards. The profile has simple changed in shape. Further, as the Pinocchio’s nose grows, centroid size increases. In a longitudinal analysis of Pinocchio profiles that capture the growth of the nose, the combination of centroid size and shape using GMM does accurately portray what is happening to Pinocchio (Figure 4). Even if you add a second localized shape change such as elongation of the ears, it is still theoretically possible to quantify and remove the artifactual variation. This can be hard in practice, however. The Pinocchio effect does not mean that Procrustes superimposed data contain no information about local variation. However, the fact that variation gets distributed to some degree across landmarks means that care must be taken to ensure that what appears to be local variation isn’t driven by variation somewhere else in a landmark configuration, particularly when dealing with subtle changes in shape.
Figure 4.
Illustration of the Pinocchio effect. In A, a dataset is generated in which a profile of Pinocchio’s head varies only in that the nose gets longer. This only displaces the three landmarks on the nose. However, a PCA based on a Procrustes fit of those data shows small displacements of landmarks that were not actually displaced although the largest displacements involve the nose (B). However, the increase in nose length is perfectly correlated with centroid size and so regressing on CS completely removes both the nose length variation and its associated artifacts (C, D). In D, shape is standardized to the sample average and so shows an intermediate length nose. If we add an additional shape component, ear shape (E) that is uncorrelated with nose length, the Pinocchio effect is complicated slightly. Here, variation is spread across two PCs (F), one which captures nose length and another that captures ear length. These effects and all their associated changes in can be removed from the data as shown in G and H, either by regressing out centroid size and PC2 or PC1 and PC2. The artifactual shape changes, however, are preserved in the shape variation associated with these regressions.
Various methods have been proposed to minimize the distorting effects of variation at particular landmarks. Rohlf and Slice (1990), for example, proposed a resistant fit superimposition for such cases while Marquez et al. (2012) suggest an interpolation based method. There is, however, little agreement on the use of such methods (Klingenberg 2013) and this remains an active area of investigation (Marquez et al. 2012). Although the choice of superimposition method may in practice default to GPA (Richtsmeier 2005), this need not be so. If one suspects that particular landmarks are introducing large amounts of variation that obscures other signals of interest, it is perfectly reasonable to try an alternative superimposition, such as the resistant fit version (Rohlf and Slice 1990), to determine whether this significantly alters the results of an analysis.
Scaling is necessary for superimposition of each specimen’s landmark coordinates into the same shape space. This makes direct comparisons of form across ontogeny impossible. Since variation in size and the associated allometric (age-shape) changes are critical components of phenotypic change across development, researchers may find it useful to add CS or a component of age to a GMM analysis as a covariate (ref to size-shape space analysis and geomorph) or to use a different set of morphometric methods. In general, size and size-related change in shape are important components of any analysis. These are rarely features of shape variation that should simply be removed. Rather, these aspects of form variation need to be quantified carefully and analyzed either separately or conjointly with shape variation (see below).
Euclidean Distance Matrix Analysis
The other major body of landmark-based morphometric methods is Euclidean Distance Matrix Analysis or EDMA (Lele and Richtsmeier 2001). EDMA is a method of morphometric analysis that avoids superimposition and its drawbacks altogether. Instead of comparing variation of superimposed landmark coordinates, EDMA techniques first represent each specimen as a matrix of linear distances between all possible pairs of landmarks (Lele and Richtsmeier 1991), which can be scaled by a chosen scaling factor (Lele and Cole III 1996). Morphological differences between groups can be pinpointed to specific linear distances on an object through pairwise comparisons of mean form or shape matrices, followed by bootstrapping to estimate the significance of these differences (Figure 5) (Lele and Richtsmeier 1991). The ability to conclusively identify specific regions of an object that differ between two samples is not always possible with commonly used multivariate data exploration techniques, including PCA; where a given landmark might be highly weighted on multiple PCs and each PC is associated with many moderately weighted landmarks of potential interest. Additionally, scaling phenotypic values by a measure of size is not automatically performed for EDMA analysis, allowing for comparisons of ontogenetic growth trajectories between groups (Richtsmeier et al. 1993a; Richtsmeier et al. 1993b), without having to reintroduce a previously removed measure of scale. Growth analysis methods can also be applied to other situations, including comparisons of the morphological effects of a specific mutation on two different genetic backgrounds (Percival et al. In Prep)
Figure 5.
The strongest (> 5% difference in means) significantly (α=0.05) different linear dimensions identified in a EDMA SHAPE comparison of skulls from 129 and C57 mouse strains, viewed from the lateral (top) and superior (bottom) aspects. Measurements were standardized by centroid size before analysis. Red lines are dimensions for which C57 is relatively longer than 129, while blue lines are relatively longer in 129 than C57.
The major advantage of EDMA is that it avoids the superimposition step yet allows analysis of morphological variation throughout a morphologically complex object. This is important if an analysis depends critically on the ability to precisely localize variation within a structure. The major disadvantage of EDMA is that since it is based on the analysis of linear distances it does not deal overtly with shape. Methods exist within EDMA to quantify allometry and ontogenetic change in form but they are more difficult to implement within a complex multivariate analysis. Further development of EDMA methods and software may well address these limitations. It is not necessarily the case that one must make a choice between using EDMA or GMM. Similar results from multiple, relatively independent methods of analysis can provide stronger support for subsequent interpretations and conclusions (Martínez-Abadías et al., 2010). Although not currently as popular as GMM methods, EDMA methods continue to be adapted to answer new research questions (Hill et al. 2013; Motch Perrine et al. 2014).
Image Analysis Based Methods
Finally, there is a growing body of methods that are designed to extract morphometric information directly from 2D and 3D image data, without the use of sparse landmarks. Some rely on automated landmarking of a few key features, then evenly distributing landmarks across the remainder of the surface(Heimann and Meinzer 2009). Others rely on analysis of deformation data on a voxel-by-voxel basis (Chakravarty et al. 2011; Heimann and Meinzer 2009; Joshi et al. 2005; Srivastava et al. 2005; Wong et al. 2014). Most applications of such methods have been in neuroimaging but there are examples of applications to craniofacial morphology (Chakravarty et al. 2011; Chinthapalli et al. 2012; Hopman et al. 2014) and embryonic development (Kristensen et al. 2008). For voxel-based methods, thresholding and registration can produce a large amount of error that must be minimized or considered in analyses (Bookstein 2001). Voxel based and dense landmark-based methods involve a superimposition step of some sort and so share the pitfalls of GMM approaches. On the other hand, statistical shape models and voxel based morphometry make use of much more of the image data that is contained in a 2D (eg. photograph) or 3D image (eg. microCT or optical projection tomography image) than approaches based on sparse landmark sets. With further methods development, such approaches may be adaptable to the full range of applications currently available through GMM methods.
Landmarks and Homology
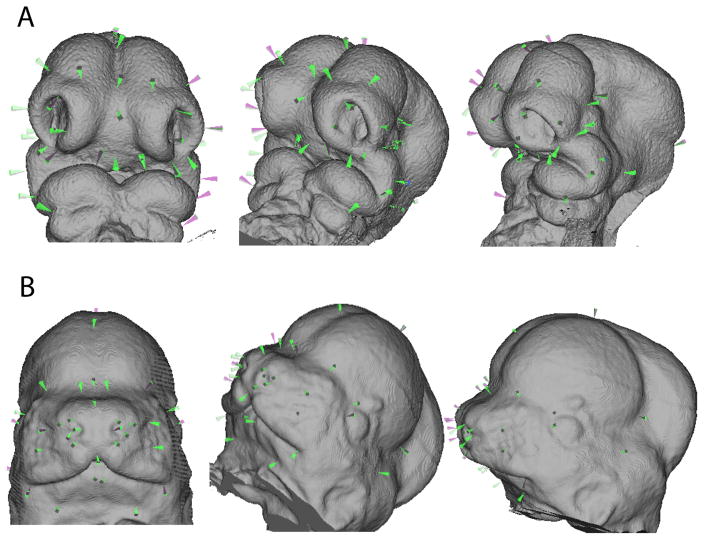
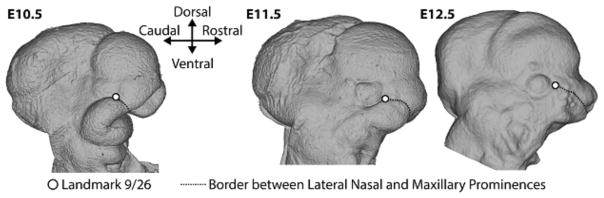
An assumption of most morphometric methods is that landmarks are homologous across individuals, samples and within individuals across ages or stages in the case of longitudinal data (Bookstein 1991; Oxnard and O’Higgins 2009). A particular landmark should effectively correspond to the same point across these situations, but what does landmark homology mean in practice? Leaving aside the debate over homology as representations of discrete, biologically defined, developmental entities versus resemblance caused by continuity of information (Hall 2007; Jamniczky 2008; Roth 1984; Roth 1991; Van Valen 1982; Wagner 2007), homology is a difficult problem for morphometrics (Klingenberg 2008). This is particularly the case for applications of morphometrics to morphogenesis in embryos (Percival et al. 2014). Figure 6 shows a standard landmark set that we use for mouse embryonic craniofacial morphology from E10 to E12.5. While these landmarks are chosen with care, the meaning of homology even within this ontogenetic range is sometimes unclear. The point defined at the caudal extreme of the cleft between the maxillary prominence and the lateral nasal prominence is a good example (Figure 7). This location gives rise eventually to lacrimal duct and so it should be a good homologous point across stages. Yet, as morphogenesis proceeds, this initially obvious anatomical feature moves in relation to the position of the eye, while descendants of the cells found at this landmark at E9.5 are probably buried as the prominences fuse and the whisker field develops by E12.5. Since our landmarking scheme is intended to track the growth and movement of the facial prominences, we define this landmark such that it remains at the border between cells derived from the maxillary and lateral nasal prominences rather than maintaining a standard geometric relationship with other cranial features like the eye (Percival et al. 2014). In this case we have made a conscious choice to attempt to follow the specific cell population rather a point defined by its geometric relationship with other features. Unfortunately, this may not always be possible, particularly across a wide range of developmental stages.
Figure 6.
Standard embryo craniofacial landmark set (Percival et al., 2014).
Figure 7.
Right lateral view of the surface craniofacial morphology of E10.5, E11.5, and E12.5 mouse specimens in a standard orientation. As previously defined (Percival et al., 2014) the homologous location of a landmark between the maxillary and the lateral nasal prominences is shown. We consciously defined it so that it landmark such that it remains at the border between these cell populations rather than maintaining a standard geometric relationship with other cranial features like the eye.
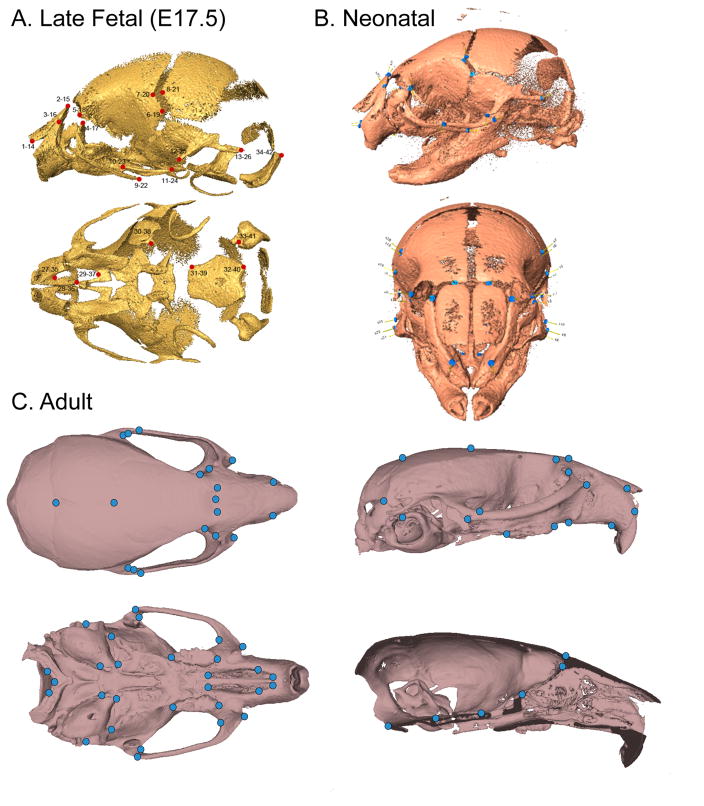
The homology issue becomes much more difficult when one needs to assemble samples across wider ranges of developmental stages. Here there are two issues. The first is that structures come and go. For the quantitative study of face formation, we have used soft tissue landmarks that are mostly located on the epithelial surface. For later fetal craniofacial development, while it is possible to rely on surface features, one is usually interested in other structures such as the skeletal features that are forming at that time. For analysis of post-natal morphology, one may be interested in features of the skull or the brain. Combining such analyses in a coherent way is an unresolved challenge. In practice, we have approached this issue by defining standard landmark sets that are particular to a range of stages and structures. Figure 8 shows standard craniofacial landmark sets for late stage fetuses, neonates and adult mice.
Figure 8.
Late fetal (Gonzalez et al. 2014), Neonatal (Boughner et al. 2008)), and adult mouse landmark sets used in recent studies by our group.
These issues underline the importance of taking care in the definition of landmark sets. Poorly, or improperly defined landmarks can lead to false conclusions. Phenomic analyses that rely on large datasets are particularly vulnerable to drift in landmark position. Reliance on manual landmarking is a significant barrier to the use of image data for phenomic analyses. Manual landmarks vary among and can drift over time for landmarkers. Landmarking protocols are also exceedingly difficult to standardize across labs (Percival et al. 2014). For this reason, development of automated methods that can handle the complexity of real datasets is crucial. Methods exist for 3D landmarking from image data (Brett and Taylor 1999; Douglas 2004) and there are examples of their application in particular contexts (Chakravarty et al. 2011; Subburaj et al. 2009). Due to the complexity and noisiness of most image datasets, however, the majority of morphometric work still relies heavily on manual landmarking.
Quantifying Variation
Comparing Shape and Size Among Groups
Morphometric analyses that address questions in developmental biology seek to quantify variation within samples and to compare variation among samples. Morphometric data based on 3D landmarks are multivariate. The statistical techniques for describing variation within groups and comparing that across groups rely heavily on matrix algebra. A fundamental element of any morphometric analysis is the variance-covariance matrix (VCM). Such matrices consist of the set of landmark by coordinate variances along their diagonal and all of the pairwise covariances in the off-diagonal cells of the matrix. An intuitive way to express the variation in such matrices is to use a multivariate data reduction technique such as Principal Components Analysis (PCA). A PCA uses the variance-covariance matrix to create new variables that correspond to progressively smaller proportions of the total variance in the sample. Imagine a bivariate dataset in which x and y are partially correlated (Figure 9). In this case, one could construct a new variable that captures the covariation among x and y. This would be the first principal component (PC). Every individual point in that dataset can be projected onto this variable, resulting in a set of principal component scores for each individual. In PCA, the second component is defined in the direction that is orthogonal to the first component but contains the second largest amount of covariance. In a two-dimensional dataset, there is only such one such option (Figure 9a), but in a multidimensional dataset, PC2 describes the second largest direction of variation. The variance of all phenotypic measures explained by a PC is its eigenvalue while its direction is its eigenvector. Within GMM, the eigenvector of each PC is its orientation with respect to the original variables. These orientations allow the visualization of the shape transformations that correspond to variation along each PC. Figure 9B–E illustrates this using PCA plots for a mouse embryo craniofacial dataset.
Figure 9.
Principal Components Analysis in Geometric Morphometrics. A) Explanation of PCA (from Zeldicth, (2004)). The 2D plots illustrate PC1 and the projection of points (individuals) on to PC1 to create PC scores. The 3D plot shows how the scatter among three variables would correspond to principal components.
Although PCA is a useful way to approach a multivariate morphometric data, it is important to note that the assumption of orthogonality is one made for convenience and not because it corresponds to biology in any way. Real covariance structures can be very complex, determined by the variances of multiple interacting and overlaid developmental factors (Hallgrimsson et al. 2009; Mitteroecker 2009). This can easily create situations where a biological effect gets distributed across multiple PCs in real datasets. The orthogonility assumption also means that every PC is uncorrelated with every other; each PC is dependent on the definition of the PCs that precede it. PCs capture axes of covariance in the VCM, but they estimate those axes with error (Zelditch 2004). If the identity of a particular PC is important to an interpretation of biological differences, resampling approaches can be used to determine whether that PC is significantly different from its neighbours (Zelditch 2004).
PCA plots are a useful first step to take when examining variation among groups such as genotypes or treatments. Subtle differences among groups, however, may not be readily evident on such plots or the differences may be distributed across several PCs. Canonical Variates Analysis (CVA) orients that data along axes that maximally distinguish groups that are defined a priori (Marcus et al. 2013). CVA is best used as a data exploration technique rather than as a definitive test of the hypothesis that two groups are different. For landmark data, the large numbers of variables and the covariance structure or Procrustes superimposed landmark data produces a situation where CVA may separate groups that actually have the same mean shape when the number of variables is close to the number of specimens (Mitteroecker and Gunz 2009). To put this more simply, use of CVA is not advised when the number of specimens under study is comparable to the number of variables in the analysis (eg. number of landmarks multiplied by number of dimensions), particularly when there are a large number of groups to separate. For the same reason, the Mahalanobis permutation test should not be used to test for differences in mean shape when sample sizes are small. In most cases, tests of significance for differences among groups should rely on permutation tests that are not affected by the ratio of variables to sample size, such as the Procrustes distance permutation test (Gunz and Mitteroecker 2013; Gunz et al. 2005; Mitteroecker and Gunz 2009). The Procrustes distance is also the basis for the parametric Goodall’s F-test, which performs well under most GMM conditions (Rohlf 2000). Finally, there are also EDMA based tests for matrices of linear distances that can be used to test for differences in form before and after scaling for size (Lele and Richtsmeier 1995; Lele and Richtsmeier 2001).
For many analyses, one may wish to quantify the phenotypic variance of a sample and compare it to others. For example, mutations can affect both the phenotypic mean as well the variance about the mean, a phenomenon known as canalization (Wagner et al. 1997). For landmark data, phenotypic variance can be quantified in several ways. The distribution of shape variation at the individual level can be quantified using the Procrustes distance of each individual from the mean. The variance of these distances can also serve as a sample variance for shape (Zelditch et al. 2004). This measure is equivalent to the trace of the variance covariance matrix, or the sum of the diagonal elements in the VCM.
Comparing Covariation Structures
Although covariation structures may seem arcane to most developmental biologists, this is actually fundamental to most advanced morphometrics. The rich datasets that we obtain from 3D images require that we apply data reduction techniques in order to extract meaning from them. Such data reductions techniques (e.g. PCA) are all based on analysis of covariance structure. To compare relationships among complex datasets, we also need to use methods that relate different covariance structures to one another. To study how brain morphology affects the face during face formation, for example, we have used such methods (Parsons et al. 2011). Similarly, if we want to relate variation in cellular dynamics to measures of morphogenesis over time, such analyses are also based on quantification and comparisons of covariance structures. For example, one might wish to determine how a mutation influences morphogenesis via spatially structured changes in gene expression, and cellular dynamics (proliferation, hypertrophy, polarity etc.). To test this, one could obtain external shape data along with data on cellular parameters and gene expression for multiple individuals. The resulting dataset would be large and heterogeneous. Relating such datasets to each other requires sophisticated multivariate approaches, most of which rely on analysis of covariance structure.
In most datasets, PCA reveals a salient feature of morphological variation: multivariate morphological variation tends be structured such that the majority of the variance is captured by the first few PCs. This can be visualized using a histogram of the eigenvalues from a PCA (Figure 9B). This phenomenon, morphological integration (Olson and Miller 1958), reflects the influence of developmental processes that vary during development and produce patterns of coordinated variation among phenotypic traits (Cheverud 1996b; Hallgrimsson et al. 2009; Wagner 1990). The strength of morphological integration can be compared using the tendency for variation to concentrate in the first we PCs. The more variance is biased towards the first few PCs, the higher the variance of the eigenvalues for each PC. Therefore, the variance of eigenvalues (VE) can be used to measure the strength of integration (Wagner 1990). Variation tends to concentrate in the first we PCs and decreases across each subsequent PC. An alteration in the rate of this decrease demonstrates integration. VE can be derived from a covariance matrix and scaled to the mean eigenvalue or simply derived from the correlation matrix.. VE estimates integration more reliably than the older and more intuitive metric of simply using correlation coefficients (Fisher-z transformed for normality), particularly in the case of heterogeneous matrices such as those we usually encounter in biology (Pavlicev et al. 2009). VE is dependent on the number of traits and, for this reason, Pavlicev et al recommend using the relative eigenvalue variance (Pavlicev et al. 2009). Here, VE is scaled to the maximum eigenvalue of a correlation matrix. In some datasets, even with scaling to the mean eigenvalue, there is a persistent correlation between VE and overall variance with the absolute scale of the eigenvalues as demonstrated by resampling the VE and the sample variance (Hallgrimsson et al. 2009; Young et al. 2010b). This artifact is usually small but it is a concern when comparing samples that vary dramatically in phenotypic variance. VE can be compared among groups by resampling.
Covariation structure can be quantified and compared in a multitude of ways. See Steppan et al. (2002) for a high-level review. The simplest and most direct way to compare correlation of covariance matrices is by using the matrix correlation, which is simply the correlation among the corresponding cells in two matrices. Matrix correlations can also be adjusted for matrix repeatability (Cheverud 1996a). To determine the significance of a matrix correlation, one can use a Mantel’s test which permutes the matrices by randomly shuffling their cells, calculating a matrix correlation at each iteration (Sokal and Rohlf 1995a). Klingenberg modified the Mantel’s test for landmark data by shuffling the coordinates for each landmark as a unit rather than treating them as independent cells (Klingenberg and McIntyre 1998). Random skewers is a method that provides for a more flexible analysis of the differences and similarities in covariance structure (Cheverud 1996a). This method has been used successfully for GMM data (Jamniczky and Hallgrimsson 2009).
Covariation among parts of a structure defined by landmarks or among distinct structures defined by landmarks can be analyzed by means of partial least squares (Rohlf and Corti 2000). This approach can provide evidence for interaction between structures during development. This is useful, for instance, if one suspects that a perturbation that directly affects one structure influences the development of another. This approach is preferable to multivariate approaches that have been frequently used in the past, including canonical correlation analysis (Rohlf and Corti 2000). Importantly, the blocks of landmarks that are compared must have independent Procrustes fits so as to remove the possibility that variance from one set of landmarks affects the other. Many studies apply and develop the application of this methods to landmark data (Bastir and Rosas 2005; Bookstein et al. 2003; Klingenberg et al. 2001; Klingenberg et al. 2003; Mitteroecker 2009; Mitteroecker and Bookstein 2007; Mitteroecker et al. 2012) including analysis of embryonic craniofacial structures (Parsons et al. 2011). Bookstein refers to partial least squares as singular warp analysis when applied in GMM (Bookstein et al. 2003).
Allometry - the shape correlates of size and stage
Allometry is a salient feature of the covariance structures of most complex morphologies. Traits are said to exhibit allometric variation when they do not scale isometrically to some measure of size (Gould 1966). Shape and size are inextricably linked in most organisms. Allometry is also a prominent feature of shape variation for complex morphological traits such as craniofacial shape (Figure 10a). For example, allometry represents roughly a third of the total shape variation in baboon skulls and in mice (Frost et al. 2003; Hallgrimsson et al. 2009). In GMM, Procrustes superimposition scales the landmark sets that describe a structure to centroid size, leaving the shape component that is correlated with size. This component represents allometric variation or the tendency for structures of different sizes to be differently shaped. Allometry is often divided into ontogenetic, static or evolutionary (Klingenberg and Zimmermann 1992). Ontogenetic allometry refers to shape changes with ontogenetic stage or age. This is the most important type of allometry for most studies of development. Static allometry refers to the shape correlates of size independently of age. Evolutionary allometry refers to the shape correlates of size among species. The relationships among these three types of allometry has been a significant area of inquiry within evolutionary biology (Lande 1979).
Figure 10.
Allometry in adult mice and in mouse embryos (E1.0–12.5). A shows the allometric component of shape variation in the Parental strains and F1 crosses for the Collaborative Cross Mice. B and C show the results of multiple multivariate regression of shape on both tail somite stage (B) and size (C). B shows the component of shape variation that is related to tail somite stage while C shows the static allometry component that is perpendicular to the variation in C. A challenge in such analyses is the co-linearity among the effects of stage and size.
In some cases, most of the allometric variation is captured by the first component in a PCA (Klingenberg 1998). In others, allometry can be more complex. In such cases, multivariate regression can be used to regress shape on centroid size, age or other relevant measure (Monteiro 1999). Figure 10b shows this in a sample of mouse embryos that vary in age and centroid size. Here, regression of shape on stage quantifies ontogenetic allometry while regression of shape on centroid size quantifies static allometry. This analysis was based on multiple multivariate regression. As there is co-linearity between the effects of stage and size, it is difficult to estimate the independent shape effects of each. These regression based methods can be used more generally to quantify or standardize for the effect of other covariates.
Allometry can complicate comparisons among genotypes or treatments when they vary in size as well as shape. In such cases, it can be necessary to determine what the static allometric shape variation is independent of the treatment or mutation of interest. As an example of this, we have used mice that have reduced growth due to a mutation in the growth hormone releasing hormone receptor to quantify the allometric component of a mutational effect (Boughner et al. 2008). This is a common problem as many mutations affect size with most resulting in a reduction in size. A further complication is that many mutations may be associated with a reduction in developmental rate. For these reasons, quantification of both ontogenetic and static allometry is a critical component of the phenotypic analysis of any mutant. A useful tool in such analyses is to do a combined analysis of shape and size, or form space, by adding centroid size back into a morphometric analysis (Mitteroecker and Bookstein 2008; Smith et al. 2015).
3D Imaging for Morphometrics
A variety of imaging techniques exist for generating data for morphometric analysis and morphometric techniques are broadly applicable across these techniques (Norris et al. 2013). A full review of these methods is beyond the scope of this chapter. Instead, we will briefly describe the three imaging techniques that have been most widely used to create 3D image data for embryos and describe their chief advantages and limitations.
Computed Microtomography
Computed microtomography (μCT) is the technique that has most commonly been used in the quantitative study of craniofacial morphogenesis (Billington et al. 2015; Boughner et al. 2008; Chong et al. 2011; Green et al. 2015; Hu et al. 2015b; Parsons et al. 2008; Percival et al. 2014; Schmidt et al. 2010a; Young et al. 2010a; Young et al. 2014). Although μCT was initially used mostly for visualizing skeletal tissues, it has proven to be an effective modality for 3D volumetric imaging of embryonic soft tissues (Boughner et al. 2008; Metscher 2009; Parsons et al. 2008; Wong et al. 2014). The major issue for μCT imaging of embryos is the distortions that are produced by fixation or dessication (Schmidt et al. 2010b). Scanning in a liquid medium is also problematic because of the low contrast between embryonic tissues and water in a CT scan. We have experimented with a variety of techniques and have found that a 4% formaldehyde + 5% biological grade glutaraldehyde buffered fix produces scans with the least amount of fixation related shrinkage and shape distortion (Schmidt et al. 2010b) (Figure 11). Embryos are scanned in air but carefully positioned for scanning so as to avoid compression or damage that can alter facial morphology.
Figure 11.
MicroCT scans of embryos fixed using different protocols shown at the same scale. Illustrating the effect of different fixation and scanning procedures on morphology. A) 4% formaldehyde + Bouin’s. B) 4% formaldehyde + 1% glutaraldehyde with Iothalamate meglumine for contrast. C) 4% formaldehyde + 5% glutaraldehyde plus contrast agent (Schmidt et al. 2010b).
Optical Projection Tomography
Optical projection tomography (OPT) is a form of optical computed tomography scanning that is based on visible light rather than X-rays (Sharpe 2009; Sharpe et al. 2002). The principal advantage of this technique is one can use optical molecular markers, such as fluorescent tags to image different tissues or regions expressing particular proteins, protein modifications or mRNAs. Current instrumentation allows visualization in several UV or white light channels, allowing simultaneous visualization of several markers as well as generation of 3D morphology in a single scan session (Figure 12). This combination of molecular and morphological imaging makes OPT a very powerful technique for morphometrics in developmental biology. The principal drawback of the technique is that the need to partially clear tissues increases processing artifacts that affect morphology. Further, processing and imaging is more time-consuming than microCT. Finally, the fact that OPT relies on lenses to focus images limits the depth of focus and thus the size of tissue that can be imaged in this way (Sharpe 2009).
Figure 12.
Examples of Optical Projection Tomography Images. A) E10.5 mouse embryo stained with Sytox Green B) E12.5 mouse embryo stained with Sytox Green (blue) and Ser10 phosphohistone H3 (green). C) Hamburger Hamilton Stage 23 chick embryo showing Shh expression in the Frontonasal ectodermal zone (FEZ) highlighted in red.
High-Resolution Magnetic Resonance Imaging
High-resolution magnetic resonance imaging (μMRI) has been used to image embryos for over two decades (Smith 1999; Smith et al. 1992; Smith et al. 1994; Smith et al. 1996). μMRI offers tremendous advantages over other volumetric imaging modalities in terms of soft tissue contrast (Baghdadi et al. 2011). Advances in molecular imaging based on MRI also have tremendous potential to advance the abilities of this imaging technique. μMRI can be performed in unfixed tissues in a liquid medium, which means that specimen distortion is minimal. The drawbacks of μMRI are the availability and cost of scanning time and the specialized expertise required to optimize scanning protocols. Further, the spatial resolution and signal to noise are determined by the strength of the magnet and so high-resolution imaging studies of embryos require powerful and expensive magnets. Even so, μMRI scanning resolutions are generally between 20 and 40 microns, compared to 5 micron or higher for μCT. Although the soft tissue differentiation is much better than with μCT, the large voxel size results in significant pixilation error for small embryos. At higher resolutions, scan times on a per specimen basis can be prohibitively long (Powell and Wilson 2012). For these reasons, morphometric analysis of craniofacial morphogenesis have not to date employed μMRI. Instead, efforts have focused on atlas construction or (Petiet et al. 2008) or automated organism-wide detection of abnormalities (Nieman et al. 2011).
Future Directions - Integrating Molecular and Anatomical Imaging
Recent studies have used morphometrics and 3D imaging to quantify variation in craniofacial morphogenesis (Billington et al. 2015; Green et al. 2015; Hu et al. 2015a; Hu et al. 2015c; Smith et al. 2015; Young et al. 2014). These studies have shown that morphometric analyses can significantly add to mechanistic studies of development and thus that morphometrics and 3D imaging of embryos have significant potential for developmental biology. However, these current studies fall far short of realizing the full potential of the quantitative study of variation.
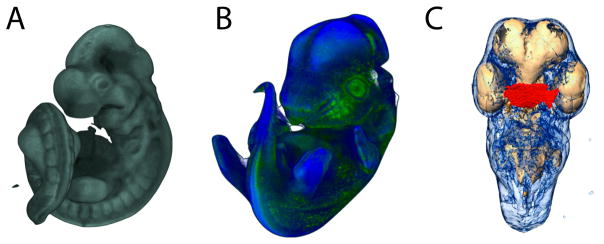
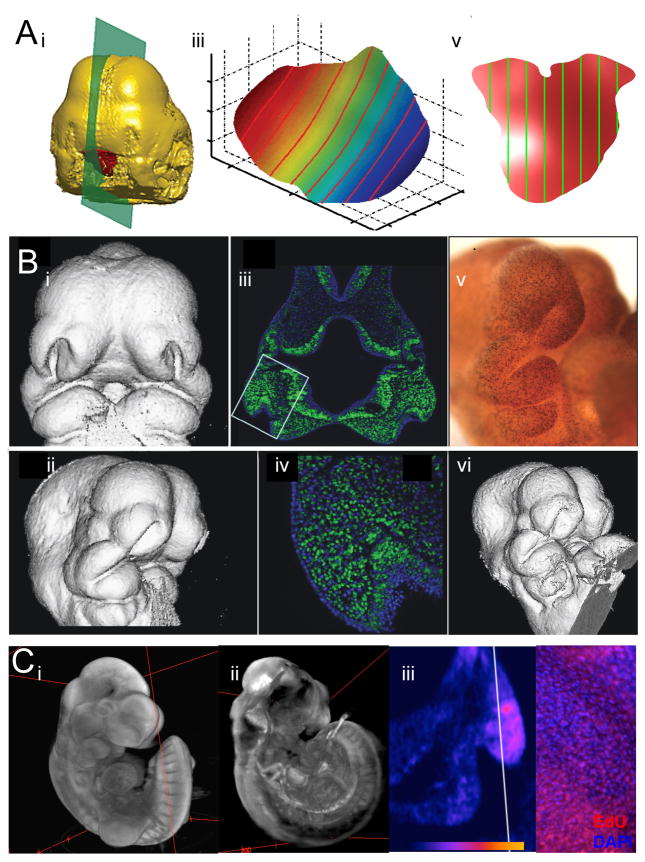
The questions of developmental biology are multilayered and increasingly complex. The broader opportunity for the combination of imaging and morphometrics lies in the challenge of quantitative integration across the genotype-phenotype map; that is, the development of techniques and statistical methods that quantify morphology in combination with molecular and cellular-level data. We are now in the very early stages of realizing this goal using a combination of innovative imaging technology, molecular methods and quantitative methods. OPT imaging allows us to step toward this goal. Using OPT we have been able to, for example, relate variation in the 3D shape of a gene expression domain to morphogenesis (Hu et al. 2015c; Mio et al. 2015). OPT allowed the simultaneous imaging of a 3D gene expression pattern and external embryonic morphology. Quantifying the shape of the gene expression pattern required a novel morphometric method for the analysis of fuzzy shapes. In collaboration with Washington Mio, we solved this problem by applying spherical harmonics analysis to the Shh gene expression domains in chick embryos (Figure 13a). OPT is currently limited in that it can only image expression for 3 genes in the same embryos.
Figure 13.
A) Analysis of Shape of Gene Expression. i. OPT scan of whole mount in situ showing Shh expression (red). The green plane indicates the slices in i and ii. ii. Section of FEZ parallel to sagittal plane. iii. Surface spline extracted from raw FEZ. B) MicroCT renderings and cell proliferation data from the same specimens. i and ii) 3D reconstruction of μCT taken after processing but before sectioning. iii and iv) Hoescht 33342 staining to visualize cell nuclei (blue) with cells in S phase visualized using EdU + Alexa Fluor® 488 labeling (green) at 5X and 200X. v) Specimen processed wholemount for anti-PHH3 primary antibody to identify M-phase cells. vi) MicroCT rendering of the same specimen after treatment. C) Images derived from OPT imaging of EdU stained embryo counterstained with Sytox Green. B) Volume view of Sytox Green channel showing regional difference in cell density. C) Volume view of EdU channel showing regional difference in cell proliferation. D) Heat map of virtual section of lateral nasal prominence of (C) showing local proliferation differences. E) EdU incorporation as viewed in a traditional confocal section EdU stain (red), DAPI (blue).
Combining data on cellular dynamics and morphology offers tremendous potential to study mechanisms of morphogenesis and their relation to structural birth defects. This is also a difficult problem. MicroCT and OPT are both potential avenues towards this goal. It is possible to successfully perform whole mount immunohistochemistry on mouse embryos to obtain both scans and sections (Figure 13b). This is very labour intensive, however, and the partial digestion of the embryo that is required for the whole mount immunohistochemistry step greatly complicates the μCT scanning. OPT offers the ability to obtain such data simultaneously from the same embryo. The challenge here is spatial resolution which at a nominal level of 5 microns is currently insufficient to make out signals in individual cells. However, spatial variation in staining intensity may be provide useful cellular level data (Figure 13c).
At a methodological level, developmental biology presents morphometrics with a massive challenge. 3D imaging modalities offer rich datasets and we lack methods to make effective use of the information contained in these images. Conducting statistical tests on quantifications derived from the full complexity of such image sets will likely run into computational power issues. Integrating this information across modalities and across levels poses an additional challenge in terms of both methods and computational power. These challenges are well worth investing in. Integrating 3D imaging based on a firm foundation of morphometric theory will be key if developmental biology is to move towards quantitative frameworks that allow for predictive modeling of morphogenesis (Boehm et al. 2010). Personalized or precision medicine is unlikely to make significant inroads for structural birth defects based on genomics alone (Hallgrimsson et al. 2014). Developmental processes are simply too complex for prediction to bypass quantitative understanding the role of development in the genotype-phenotype map. That can only be realized through significant investment in imaging and morphometrics as applied to development.
Acknowledgments
This work is supported by NSERC grant #238992-12 to BH, NIH-NIDCR grants 1R01DE021708 and 1U01DE024440 to BH and RM and 1R01DE021708 to Rich Spritz and BH as well as the Canadian Foundation for Innovation, Alberta Innovates Health Solutions and the University of Calgary.
Contributor Information
Benedikt Hallgrimsson, Dept. of Cell Biology and Anatomy, Alberta Children’s Hospital, Research Institute, and McCaig Bone and Joint Institute, University of Calgary, Alberta.
Christopher J. Percival, Dept. of Cell Biology and Anatomy, Alberta Children’s Hospital, Research Institute, and McCaig Bone and Joint Institute, University of Calgary, Alberta
Rebecca Green, Dept. of Cell Biology and Anatomy and the Alberta Children’s Hospital, Research Institute, and McCaig Bone and Joint Institute, University of Calgary, Alberta.
Nathan M. Young, Department of Orthopaedic Surgery, San Francisco General Hospital, The University of California San Francisco, California
Washington Mio, Florida State University, Tallahassee, Florida.
Ralph Marcucio, Department of Orthopaedic Surgery, San Francisco General Hospital, The University of California San Francisco, California.
References Cited
- Alberch P. Developmental constraints in evolutionary processes. In: Bonner JT, editor. Development in Evolution. Berlin and New York: Springer-Verlag; 1982. pp. 313–332. [Google Scholar]
- Baghdadi L, Zamyadi M, Sled JG, Schneider JE, Bhattacharya S, Henkelman RM, Lerch JP. Semi-automatic segmentation of multiple mouse embryos in MR images. BMC Bioinformatics. 2011;12:237. doi: 10.1186/1471-2105-12-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastir M, Rosas A. Hierarchical nature of morphological integration and modularity in the human posterior face. Am J Phys Anthropol. 2005;128(1):26–34. doi: 10.1002/ajpa.20191. [DOI] [PubMed] [Google Scholar]
- Billington CJ, Jr, Schmidt B, Marcucio RS, Hallgrimsson B, Gopalakrishnan R, Petryk A. Impact of retinoic acid exposure on midfacial shape variation and manifestation of holoprosencephaly in Twsg1 mutant mice. Dis Model Mech. 2015;8(2):139–146. doi: 10.1242/dmm.018275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm B, Westerberg H, Lesnicar-Pucko G, Raja S, Rautschka M, Cotterell J, Swoger J, Sharpe J. The role of spatially controlled cell proliferation in limb bud morphogenesis. PLoS Biol. 2010;8(7):e1000420. doi: 10.1371/journal.pbio.1000420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bookstein B, Chernoff B, Elder R, Humphries J, Smith G, Strauss R. Morphometrics in Evolutionary Biology. Philadelphia: The Academy of Natural Sciences of Philadelphia; 1985. [Google Scholar]
- Bookstein FL. Size and Shape Spaces for Landmark Data in Two Dimensions. Statistical Science. 1986;1(2):181–222. [Google Scholar]
- Bookstein FL. Principal Warps: Thin-Plate Splines and the Decomposition of Deformations. IEEE Transactions on Patterns Analysis and Machine Intelligence. 1989;11(6):567–585. [Google Scholar]
- Bookstein FL. Morphometric Tools for Landmark Data. Cambridge: Cambridge University Press; 1991. [Google Scholar]
- Bookstein FL. “Voxel-based morphometry” should not be used with imperfectly registered images. Neuroimage. 2001;14(6):1454–1462. doi: 10.1006/nimg.2001.0770. [DOI] [PubMed] [Google Scholar]
- Bookstein FL, Gunz P, Mitteroecker P, Prossinger H, Schaefer K, Seidler H. Cranial integration in Homo: singular warps analysis of the midsagittal plane in ontogeny and evolution. J Hum Evol. 2003;44(2):167–187. doi: 10.1016/s0047-2484(02)00201-4. [DOI] [PubMed] [Google Scholar]
- Boughner JC, Wat S, Diewert VM, Young NM, Browder LW, Hallgrimsson B. Short-faced mice and developmental interactions between the brain and the face. J Anat. 2008;213(6):646–662. doi: 10.1111/j.1469-7580.2008.00999.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett AD, Taylor CJ. Information Processing in Medical Imaging. Springer; 1999. A framework for automated landmark generation for automated 3D statistical model construction; pp. 376–381. [Google Scholar]
- Burian RM. The epistemology of development, evolution, and genetics. Cambridge: Cambridge University Press; 2005. [Google Scholar]
- Chakravarty MM, Aleong R, Leonard G, Perron M, Pike GB, Richer L, Veillette S, Pausova Z, Paus T. Automated analysis of craniofacial morphology using magnetic resonance images. 2011 doi: 10.1371/journal.pone.0020241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheverud J. Quantitative genetic analysis of cranial morphology in the cotton-top (Saguinus oedipus) and saddle-back (S. fuscicollis) tamarins. Journal of Evolutionary Biology. 1996a;9(1):5–42. [Google Scholar]
- Cheverud JM. Developmental integration and the evolution of pleiotropy. Am Zool. 1996b;36:44–50. [Google Scholar]
- Chinthapalli K, Bartolini E, Novy J, Suttie M, Marini C, Falchi M, Fox Z, Clayton LM, Sander JW, Guerrini R, et al. Atypical face shape and genomic structural variants in epilepsy. Brain. 2012;135(Pt 10):3101–3114. doi: 10.1093/brain/aws232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong HJ, Young NM, Hu D, Jeong J, McMahon AP, Hallgrimsson B, Marcucio RS. Signaling by SHH rescues facial defects following blockade in the brain. Dev Dyn. 2011 doi: 10.1002/dvdy.23726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WJ, Albertson RC. Quantification and variation in experimental studies of morphogenesis. Dev Biol. 2008;321(2):295–302. doi: 10.1016/j.ydbio.2008.06.025. [DOI] [PubMed] [Google Scholar]
- Douglas TS. Image processing for craniofacial landmark identification and measurement: a review of photogrammetry and cephalometry. Comput Med Imaging Graph. 2004;28(7):401–409. doi: 10.1016/j.compmedimag.2004.06.002. [DOI] [PubMed] [Google Scholar]
- Dryden IL, Mardia KV. Statistical Shape Analysis. Chichester: John Wiley & Sons; 1998. [Google Scholar]
- Frost SR, Marcus LF, Bookstein FL, Reddy DP, Delson E. Cranial allometry, phylogeography, and systematics of large-bodied papionins (primates: Cercopithecinae) inferred from geometric morphometric analysis of landmark data. Anat Rec. 2003;275A(2):1048–1072. doi: 10.1002/ar.a.10112. [DOI] [PubMed] [Google Scholar]
- Gonzalez PN, Lotto FP, Hallgrimsson B. Canalization and developmental instability of the fetal skull in a mouse model of maternal nutritional stress. Am J Phys Anthropol. 2014;154(4):544–553. doi: 10.1002/ajpa.22545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. Allometry and size in ontogeny and phylogeny. Biol Rev. 1966;41(4):587–640. doi: 10.1111/j.1469-185x.1966.tb01624.x. [DOI] [PubMed] [Google Scholar]
- Gower JC. Generalized Procrustes Analysis. Psychometrika. 1975;40(1):33–51. [Google Scholar]
- Green RM, Feng W, Phang T, Fish JL, Li H, Spritz RA, Marcucio RS, Hooper J, Jamniczky H, Hallgrimsson B, et al. Tfap2a-dependent changes in mouse facial morphology result in clefting that can be ameliorated by a reduction in Fgf8 gene dosage. Dis Model Mech. 2015;8(1):31–43. doi: 10.1242/dmm.017616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunz P, Mitteroecker P. Semilandmarks: a method for quantifying curves and surfaces. Hystrix, the Italian Journal of Mammalogy. 2013;24(1):103–109. [Google Scholar]
- Gunz P, Mitteroecker P, Bookstein FL. Modern morphometrics in physical anthropology. Springer; 2005. Semilandmarks in three dimensions; pp. 73–98. [Google Scholar]
- Hall BK. Homoplasy and homology: Dichotomy or continuum? J Hum Evol. 2007;52(5):473–479. doi: 10.1016/j.jhevol.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Hallgrímsson B, JCB, Turinsky A, Logan C, Sensen CW. Geometric Morphometrics and the Study of Development. In: Hallgrimsson CWSaB., editor. Advanced Imaging in Biology and Medicine: Technology, Software Environments, Applications. Springer; Verlag: 2009. [Google Scholar]
- Hallgrimsson B, Jamniczky H, Young NM, Rolian C, Parsons TE, Boughner JC, Marcucio RS. Deciphering the Palimpsest: Studying the Relationship Between Morphological Integration and Phenotypic Covariation. Evolutionary Biology. 2009;36(4):355–376. doi: 10.1007/s11692-009-9076-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallgrimsson B, Mio W, Marcucio RS, Spritz R. Let’s face it--complex traits are just not that simple. PLoS Genet. 2014;10(11):e1004724. doi: 10.1371/journal.pgen.1004724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann T, Meinzer H-P. Statistical shape models for 3D medical image segmentation: a review. Med Image Anal. 2009;13(4):543–563. doi: 10.1016/j.media.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Heuze Y, Martinez-Abadias N, Stella JM, Arnaud E, Collet C, Garcia Fructuoso G, Alamar M, Lo LJ, Boyadjiev SA, Di Rocco F, et al. Quantification of facial skeletal shape variation in fibroblast growth factor receptor-related craniosynostosis syndromes. Birth Defects Res A Clin Mol Teratol. 2014;100(4):250–259. doi: 10.1002/bdra.23228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CA, Martinez-Abadias N, Motch SM, Austin JR, Wang Y, Jabs EW, Richtsmeier JT, Aldridge K. Postnatal brain and skull growth in an Apert syndrome mouse model. Am J Med Genet A. 2013;161A(4):745–757. doi: 10.1002/ajmg.a.35805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopman SM, Merks JH, Suttie M, Hennekam RC, Hammond P. Face shape differs in phylogenetically related populations. Eur J Hum Genet. 2014;22(11):1268–1271. doi: 10.1038/ejhg.2013.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D. Colloquium papers: Numbering the hairs on our heads: the shared challenge and promise of phenomics. Proc Natl Acad Sci U S A. 2010;107(Suppl 1):1793–1799. doi: 10.1073/pnas.0906195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle D, Govindaraju DR, Omholt S. Phenomics: the next challenge. Nat Rev Genet. 2010;11(12):855–866. doi: 10.1038/nrg2897. [DOI] [PubMed] [Google Scholar]
- Hu D, Young N, Xu Q, Jamniczky H, Green R, Mio W, MarcuciO R, Hallgrimsson S. Signals from the brain induce variation in avian facial shape. Dev Dyn. 2015a doi: 10.1002/dvdy.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Young NM, Li X, Xu Y, Hallgrimsson B, Marcucio RS. A dynamic Shh expression pattern, regulated by SHH and BMP signaling, coordinates fusion of primordia in the amniote face. Development. 2015b;142(3):567–574. doi: 10.1242/dev.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, Young NM, Xu Q, Jamniczky H, Green RM, Mio W, Marcucio RS, Hallgrimsson B. Signals from the brain induce variation in avian facial shape. Dev Dyn. 2015c doi: 10.1002/dvdy.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamniczky H. Phenotypic Integration Patterns Support an Account of Homology as a Manifestation of Evolvability. Evolutionary Biology. 2008;35(4):312–316. [Google Scholar]
- Jamniczky HA, Hallgrimsson B. A comparison of covariance structure in wild and laboratory muroid crania. Evolution. 2009;63(6):1540–1556. doi: 10.1111/j.1558-5646.2009.00651.x. [DOI] [PubMed] [Google Scholar]
- Joshi SH, Srivastava A, Mio W. Elastic shape models for interpolations of curves in image sequences. Inf Process Med Imaging. 2005;19:541–552. doi: 10.1007/11505730_45. [DOI] [PubMed] [Google Scholar]
- Kendall DG. Shape-manifolds, procrustean metrics and complex projective spaces. Bulletin of the London Mathematical Society. 1984;16:81–121. [Google Scholar]
- Klingenberg CP. Heterochrony and allometry: the analysis of evolutionary change in ontogeny. Biol Rev Camb Philos Soc. 1998;73(1):79–123. doi: 10.1017/s000632319800512x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP. Novelty and “Homology-free” morphometrics: What’s in a name? Evolutionary Biology. 2008;35(3):186–190. [Google Scholar]
- Klingenberg CP. Visualizations in geometric morphometrics: how to read and how to make graphs showing shape changes. Hystrix-Italian Journal of Mammalogy. 2013;24(1):15–24. [Google Scholar]
- Klingenberg CP, Badyaev AV, Sowry SM, Beckwith NJ. Inferring developmental modularity from morphological integration: analysis of individual variation and asymmetry in bumblebee wings. Am Nat. 2001;157(1):11–23. doi: 10.1086/317002. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, McIntyre GS. Geometric morphometrics of developmental instability: Analyzing patterns of fluctuating asymmetry with Procrustes methods. Evolution. 1998;52(5):1363–1375. doi: 10.1111/j.1558-5646.1998.tb02018.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Mebus K, Auffray JC. Developmental integration in a complex morphological structure: how distinct are the modules in the mouse mandible? Evol Dev. 2003;5(5):522–531. doi: 10.1046/j.1525-142x.2003.03057.x. [DOI] [PubMed] [Google Scholar]
- Klingenberg CP, Zimmermann M. Static, ontogenetic, and evolutionary allometry: a multivariate comparison in nine species of water striders. Am Nat. 1992:601–620. [Google Scholar]
- Kristensen E, Parsons TE, Hallgrimsson B, Boyd SK. A novel 3-D image-based morphological method for phenotypic analysis. IEEE Trans Biomed Eng. 2008;55(12):2826–2831. doi: 10.1109/TBME.2008.923106. [DOI] [PubMed] [Google Scholar]
- Lande R. Quantitative genetic analysis of multivariate evolution: applied to brain:body size allometry. Evolution. 1979;33:203–215. doi: 10.1111/j.1558-5646.1979.tb04694.x. [DOI] [PubMed] [Google Scholar]
- Lele S, Cole TM., III A new test for shape differences when variance-covariance matrices are unequal. J Hum Evol. 1996;31(3):193–212. [Google Scholar]
- Lele S, Richtsmeier JT. Euclidean distance matrix analysis: a coordinate-free approach for comparing biological shapes using landmark data. Am J Phys Anthropol. 1991;86(3):415–427. doi: 10.1002/ajpa.1330860307. [DOI] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT. Euclidean distance matrix analysis: confidence intervals for form and growth differences. Am J Phys Anthropol. 1995;98(1):73–86. doi: 10.1002/ajpa.1330980107. [DOI] [PubMed] [Google Scholar]
- Lele S, Richtsmeier JT. An Invariant Approach to the Statistical Analysis of Shapes. Boca Raton: Chapman & Hall; 2001. [Google Scholar]
- Love AC. The erotetic organization of developmental biology. Towards a Theory of Development. 2014:33. [Google Scholar]
- Marcus LF, Corti M, Loy A, Naylor GJ, Slice DE. Advances in morphometrics. Springer Science & Business Media; 2013. [Google Scholar]
- Marquez EJ, Cabeen R, Woods RP, Houle D. The Measurement of Local Variation in Shape. Evol Biol. 2012;39(3):419–439. doi: 10.1007/s11692-012-9159-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Burian R, Kauffman S, Alberch P, Campbell J, Goodwin B, Lande R, Raup D, Wolpert L. Developmental constraints and evolution. Quart Rev Biol. 1985;60(3):265–287. [Google Scholar]
- Metscher BD. MicroCT for developmental biology: A versatile tool for high-contrast 3D imaging at histological resolutions. Dev Dyn. 2009;238(3):632–640. doi: 10.1002/dvdy.21857. [DOI] [PubMed] [Google Scholar]
- Mio W, Xu Q, Jamniczky H, Hu D, Green RM, Marcucio RS, Hallgrimsson B. Correlations between the morphology of Sonic hedgehog expression domains and embryonic craniofacial shape. Evolutionary Biology. 2015;(3) doi: 10.1007/s11692-015-9321-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitteroecker P. The developmental basis of variational modularity: Insights from quantitative genetics, morphometrics, and developmental biology. Evolutionary Biology. 2009;36(4):377–385. [Google Scholar]
- Mitteroecker P, Bookstein F. The conceptual and statistical relationship between modularity and morphological integration. Syst Biol. 2007;56(5):818–836. doi: 10.1080/10635150701648029. [DOI] [PubMed] [Google Scholar]
- Mitteroecker P, Bookstein F. The Ontogenetic Trajectory of the Phenotypic Covariance Matrix, with Examples from Craniofacial Shape in Rats and Humans. Evolution. 2008 doi: 10.1111/j.1558-5646.2008.00587.x. [DOI] [PubMed] [Google Scholar]
- Mitteroecker P, Gunz P. Advances in Geometric Morphometrics. Evolutionary Biology. 2009;36(2):235–247. [Google Scholar]
- Mitteroecker P, Gunz P, Neubauer S, Muller G. How to Explore Morphological Integration in Human Evolution and Development? Evolutionary Biology. 2012;39(4):536–553. [Google Scholar]
- Mitteroecker P, Huttegger SM. The concept of morphospaces in evolutionary and developmental biology: Mathematics and metaphors. Biological Theory. 2009;4(1):54–67. [Google Scholar]
- Monteiro LR. Multivariate regression models and geometric morphometrics: the search for causal factors in the analysis of shape. Systematic Biology. 1999:192–199. doi: 10.1080/106351599260526. [DOI] [PubMed] [Google Scholar]
- Motch Perrine SM, Cole TM, 3rd, Martinez-Abadias N, Aldridge K, Jabs EW, Richtsmeier JT. Craniofacial divergence by distinct prenatal growth patterns in Fgfr2 mutant mice. BMC Dev Biol. 2014;14:8. doi: 10.1186/1471-213X-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieman BJ, Wong MD, Henkelman RM. Genes into geometry: imaging for mouse development in 3D. Curr Opin Genet Dev. 2011;21(5):638–646. doi: 10.1016/j.gde.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Norris FC, Wong MD, Greene ND, Scambler PJ, Weaver T, Weninger WJ, Mohun TJ, Henkelman RM, Lythgoe MF. A coming of age: advanced imaging technologies for characterising the developing mouse. Trends Genet. 2013;29(12):700–711. doi: 10.1016/j.tig.2013.08.004. [DOI] [PubMed] [Google Scholar]
- Olson EC, Miller RA. Morphological Integration. Chicago: University of Chicago Press; 1958. [Google Scholar]
- Oxnard C, O’Higgins P. Biology clearly needs morphometrics. Does morphometrics need biology? Biological Theory. 2009;4(1):84–97. [Google Scholar]
- Parsons TE, Kristensen E, Hornung L, Diewert VM, Boyd SK, German RZ, Hallgrimsson B. Phenotypic variability and craniofacial dysmorphology: increased shape variance in a mouse model for cleft lip. J Anat. 2008;212(2):135–143. doi: 10.1111/j.1469-7580.2007.00845.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons TE, Schmidt EJ, Boughner JC, Jamniczky HA, Marcucio RS, Hallgrimsson B. Epigenetic integration of the developing brain and face. Dev Dyn. 2011;240(10):2233–2244. doi: 10.1002/dvdy.22729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlicev M, Cheverud J, Wagner GP. Measuring Morphological Integration Using Eigenvalue Variance. Evolutionary Biology. 2009;36:157–170. [Google Scholar]
- Percival C, Tapaltsyan V, Klein O, Hallgrimsson B. Inbred background effects on the effect of Sprouty gene deletions on craniofacial morphology In Prep. [Google Scholar]
- Percival CJ, Green R, Marcucio R, Hallgrimsson B. Surface landmark quantification of embryonic mouse craniofacial morphogenesis. BMC Dev Biol. 2014;14:31. doi: 10.1186/1471-213X-14-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiet AE, Kaufman MH, Goddeeris MM, Brandenburg J, Elmore SA, Johnson GA. High-resolution magnetic resonance histology of the embryonic and neonatal mouse: a 4D atlas and morphologic database. Proceedings of the National Academy of Sciences. 2008;105(34):12331–12336. doi: 10.1073/pnas.0805747105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell K, Wilson D. 3-dimensional imaging modalities for phenotyping genetically engineered mice. Veterinary Pathology Online. 2012;49(1):106–115. doi: 10.1177/0300985811429814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raup DM. Geometric analysis of shell coiling. J Paleont. 1966;40:1178–1190. [Google Scholar]
- Richtsmeier JT. An Invariant Approach to the Study of Fluctuating Asymmetry. In: Slice DE, editor. Modern Morphometrics in Physical Anthropology. New York: Kluwer/Plenum Publishers; 2005. pp. 187–212. [Google Scholar]
- Richtsmeier JT, Cheverud J, Danahey S, Corner B, Lele S. Sexual dimorphism of ontogeny in the crab-eating macaque (Macaca fascicularis) J Hum Evol. 1993a;25(1):1–30. [Google Scholar]
- Richtsmeier JT, Corner BD, Grausz HM, Cheverud JM, Danahey SE. The role of postnatal growth pattern in the production of facial morphology. Systematic Biology. 1993b:307–330. [Google Scholar]
- Rohlf FJ. Shape Statistics: Procrustes Superimposition and Tangent Spaces. Journal of Classification. 1999;16:197–223. [Google Scholar]
- Rohlf FJ. Statistical power comparisons among alternative morphometric methods. Am J Phys Anthropol. 2000;111(4):463–478. doi: 10.1002/(SICI)1096-8644(200004)111:4<463::AID-AJPA3>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ, Bookstein FL. Proceedings of the Michigan Morphometrics Workshop. Ann Arbor, Mich: University of Michigan Museum of Zoology; 1990. p. vii.p. 380. [Google Scholar]
- Rohlf FJ, Corti M. Use of two-block partial least-squares to study covariation in shape. Systematic Biology. 2000;49(4):740–753. doi: 10.1080/106351500750049806. [DOI] [PubMed] [Google Scholar]
- Rohlf FJ, Slice DE. Extensions of the Procrustes method for the optical superimposition of landmarks. Syst Zool. 1990;39(1):40–59. [Google Scholar]
- Roth VL. On homology. Biological Journal of the Linnean Society. 1984;22:13–29. [Google Scholar]
- Roth VL. Homology and hierarchies: Problems solved and unresolved. J Evol Biol. 1991;4:167–194. [Google Scholar]
- Schmidt EJ, Parsons TE, Jamniczky HA, Gitelman J, Trpkov C, Boughner JC, Logan CC, Sensen CW, Hallgrimsson B. Micro-computed tomography-based phenotypic approaches in embryology: procedural artifacts in assessments of embryonic craniofacial growth and development. BMC Dev Biol. 2010a;10(1):18. doi: 10.1186/1471-213X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt EJ, Parsons TE, Jamniczky HA, Gitelman J, Trpkov C, Boughner JC, Logan CC, Sensen CW, Hallgrimsson B. Micro-computed tomography-based phenotypic approaches in embryology: procedural artifacts on assessments of embryonic craniofacial growth and development. BMC Dev Biol. 2010b;10:18. doi: 10.1186/1471-213X-10-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpe J. Advanced Imaging in Biology and Medicine. Springer; 2009. Optical Projection Tomography; pp. 199–224. [Google Scholar]
- Sharpe J, Ahlgren U, Perry P, Hill B, Ross A, Hecksher-Sorensen J, Baldock R, Davidson D. Optical projection tomography as a tool for 3D microscopy and gene expression studies. Science. 2002;296(5567):541–545. doi: 10.1126/science.1068206. [DOI] [PubMed] [Google Scholar]
- Smith BR. Visualizing human embryos. Sci Am. 1999;280(3):76–81. doi: 10.1038/scientificamerican0399-76. [DOI] [PubMed] [Google Scholar]
- Smith BR, Effmann EL, Johnson GA. MR microscopy of chick embryo vasculature. J Magn Reson Imaging. 1992;2(2):237–240. doi: 10.1002/jmri.1880020220. [DOI] [PubMed] [Google Scholar]
- Smith BR, Johnson GA, Groman EV, Linney E. Magnetic resonance microscopy of mouse embryos. Proc Natl Acad Sci U S A. 1994;91(9):3530–3533. doi: 10.1073/pnas.91.9.3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BR, Linney E, Huff DS, Johnson GA. Magnetic resonance microscopy of embryos. Comput Med Imaging Graph. 1996;20(6):483–490. doi: 10.1016/s0895-6111(96)00046-8. [DOI] [PubMed] [Google Scholar]
- Smith FJ, Percival CJ, Young NM, Hu D, Schneider RA, Marcucio RS, Hallgrimsson B. Divergence of craniofacial developmental trajectories among avian embryos. Dev Dyn. 2015 doi: 10.1002/dvdy.24262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneath PH, Sokal RR. The principles and practice of numerical classification. 1973. Numerical taxonomy. [Google Scholar]
- Sober E. Conceptual issues in evolutionary biology. Mit Press; 1994. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry. New York: W.H. Freeman and Company; 1995a. [Google Scholar]
- Sokal RR, Rohlf FJ. Biometry: the principles andpractice of statistics in biological research. WH Freeman & Co; San Francisco: 1995b. SokalBiometry: the principles and practice of statistics in biological research 1995 [Google Scholar]
- Srivastava A, Joshi SH, Mio W, Liu X. Statistical shape analysis: clustering, learning, and testing. IEEE Trans Pattern Anal Mach Intell. 2005;27(4):590–602. doi: 10.1109/TPAMI.2005.86. [DOI] [PubMed] [Google Scholar]
- Steppan SJ, Phillips PC, Houle D. Comparative quantitative genetics: evolution of the G matrix. Trends in Ecology & Evolution. 2002;17(7):320–327. [Google Scholar]
- Subburaj K, Ravi B, Agarwal M. Automated identification of anatomical landmarks on 3D bone models reconstructed from CT scan images. Comput Med Imaging Graph. 2009;33(5):359–368. doi: 10.1016/j.compmedimag.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Thompson A. On Growth and Form. Camrbidge: Cambridge University Press; 1942 (1961). [Google Scholar]
- Thompson DAW. On growth and form. Cambridge [Eng.]: University press; 1917. p. xv.p. 793. [Google Scholar]
- Van Valen LM. Homology and causes. J Morphol. 1982;173(3):305–312. doi: 10.1002/jmor.1051730307. [DOI] [PubMed] [Google Scholar]
- Wagner GP. A comparative study of morphological integration in Apis mellifera (Insecta, Hymenoptera) Z zool Syst Evolut - Forsch. 1990;28:48–61. [Google Scholar]
- Wagner GP. The developmental genetics of homology. Nat Rev Genet. 2007;8(6):473–479. doi: 10.1038/nrg2099. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Booth G, Bagheri-Chaichian H. A population genetic theory of canalization. Evolution. 1997;51(2):329–347. doi: 10.1111/j.1558-5646.1997.tb02420.x. [DOI] [PubMed] [Google Scholar]
- Wong MD, Maezawa Y, Lerch JP, Henkelman RM. Automated pipeline for anatomical phenotyping of mouse embryos using micro-CT. Development. 2014;141(12):2533–2541. doi: 10.1242/dev.107722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Chong HJ, Hu D, Hallgrimsson B, Marcucio RS. Quantitative analyses link modulation of sonic hedgehog signaling to continuous variation in facial growth and shape. Development. 2010a;137(20):3405–3409. doi: 10.1242/dev.052340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Hu D, Lainoff AJ, Smith FJ, Diaz R, Tucker AS, Trainor PA, Schneider RA, Hallgrimsson B, Marcucio RS. Embryonic bauplans and the developmental origins of facial diversity and constraint. Development. 2014;141(5):1059–1063. doi: 10.1242/dev.099994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NM, Wagner GP, Hallgrimsson B. Development and the evolvability of human limbs. Proc Natl Acad Sci U S A. 2010b;107(8):3400–3405. doi: 10.1073/pnas.0911856107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelditch ML. Geometric morphometrics for biologists: a primer. Boston, MA: Elsevier Academic Press; 2004. [Google Scholar]
- Zelditch ML, Lundrigan BL, Garland T. Developmental regulation of skull morphology. I. Ontogenetic dynamics of variance. Evol Dev. 2004;6(3):194–206. doi: 10.1111/j.1525-142X.2004.04025.x. [DOI] [PubMed] [Google Scholar]