Abstract
Somatic mutations in CREBBP occur frequently in B-cell lymphoma. Here, we show that loss of CREBBP facilitates development of germinal center derived lymphomas in mice. In both human and murine lymphomas CREBBP loss of function resulted in focal depletion of enhancer H3K27 acetylation and aberrant transcriptional silencing of genes that regulate B-cell signaling and immune responses including class II MHC. Mechanistically, CREBBP regulated enhancers are counter-regulated by the BCL6 transcriptional repressor in a complex with SMRT and HDAC3, which we find bind extensively to MHC class II loci. HDAC3 loss of function rescued repression of these enhancers and corresponding genes including MHC class II, and more profoundly suppress CREBPP mutant lymphomas in vitro and in vivo. Hence CREBBP loss of function contributes to lymphomagenesis by enabling unopposed suppression of enhancers by BCL6/SMRT/HDAC3 complexes, suggesting HDAC3 targeted therapy as a precision approach for CREBBP mutant lymphomas.
Keywords: Lymphoma, CREBBP, H3K27ac, Enhancer, MHC II
Introduction
Follicular lymphoma (FL) and diffuse large B-cell lymphoma (DLBCL) are the two most common subtypes of non-Hodgkin lymphoma (NHL). Although initially exhibiting an indolent phenotype, FLs are nevertheless mostly incurable, and 40–50% eventually transform into an aggressive and lethal form of DLBCL (1). DLBCLs manifest an aggressive and fast growing phenotype, but can be eradicated by chemo-immunotherapy (R-CHOP) in approximately 60% of cases. Both FL and DLBCLs arise from B-cells undergoing the germinal center (GC) reaction, a stage of the humoral immune response where B-cells undergo immunoglobulin affinity maturation. GCs B-cells form transiently in response to T-cell dependent antigen stimulation and are characterized by their unique ability to simultaneously proliferate and tolerate genomic instability. These features make these cells inherently prone to malignant transformation.
Genome sequencing studies have shown that recurrent mutation of histone modifying enzymes is a genetic hallmark of FL and DLBCL. Some of the most highly recurrent disease alleles in FL and DLBCL correspond to somatic mutation affecting two closely related histone-acetyltransferase (HAT) encoding genes: CREBBP and EP300. Between these two loci, CREBBP somatic mutations are more frequent, reported in 6.4–22.3% of patients with DLBCL and 30.8–68% of FLs (2–7). These mutations are frequently monoallelic and tend to occur within the HAT enzymatic domain. HAT domain missense mutants may loose their ability to acetylate protein substrates such as histones or transcription factors due to reduced binding of acetyl-CoA (2). Mutations in EP300 occur in 5.4–9.7% of DLBCL and 8.7–23.1% of FL (2, 3, 6, 7), and are often mutually exclusive with those of CREBBP suggesting that loss of either of these two genes might affect a common downstream pathway. It is not yet known how these mutations might contribute to malignant transformation. Notably, CREBBP mutations tend to be early events in lymphomagenesis, and for example in FL, likely occur in B-cells that already harbor the t(14;18) translocation that drives constitutive expression of BCL2 (7).
CREBBP mediates acetylation of certain histone lysine residues and among these the best characterized from the functional standpoint is H3K27 acetylation, which is required for the activation of enhancers. Enhancers are largely responsible for cell-context specific transcriptional programming, and are located at sites distal to gene coding regions or within introns. For a genomic region to manifest enhancer activity it must acquire H3K4 mono/dimethylation (H3K4me1/2) (8). However for enhancers to become fully active they require the further addition of H3K27ac mark by HATs such as CREBBP and EP300 (9). Therefore, it is possible that loss of CREBBP and EP300 HAT activity may disrupt activity of enhancers in B-cells, and perhaps through this mechanism contributes to lymphomagenesis. Such an effect has been suggested by several recent reviews (10, 11). In this study we explore the role and epigenetic mechanism of action of CREBBP in lymphomagenesis.
RESULTS
Crebbp deficiency promotes lymphomagenesis in vivo
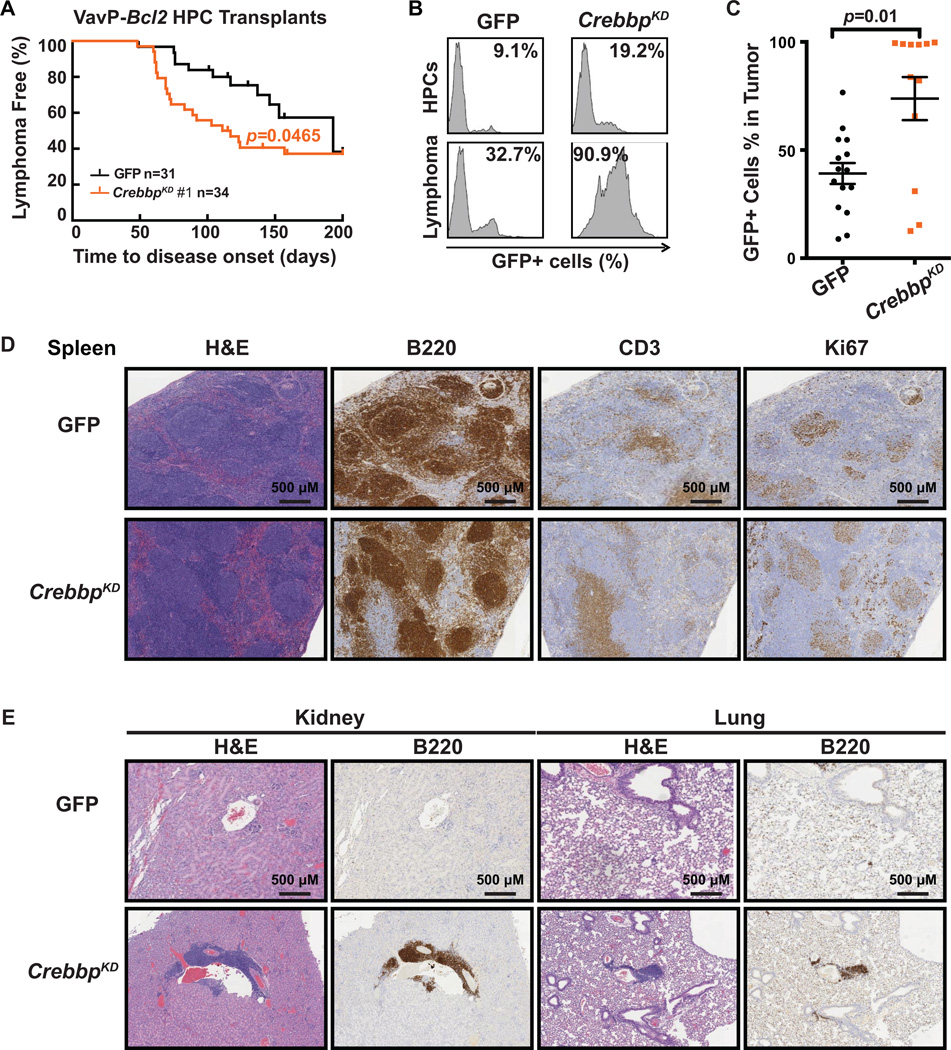
Although CREBBP mutations are among the most frequent genetic lesions in NHL, their functional significance is unknown. We modeled CREBBP deficiency in vavBcl2 transgenic mice, which closely recapitulate the genetics and pathology of human FL (12–14). VavBcl2 hematopoietic progenitor cells (HPCs) were transduced with GFP encoding retroviruses expressing shRNAs against Crebbp (VavP-Bcl2/CrebbpKD) or control (VavP-Bcl2/GFP), and transplanted into lethally irradiated wild-type recipients (Supplementary Fig. S1A). Crebbp shRNAs induced robust knockdown of CREBBP expression measured by RT-qPCR in murine FL5–12 cells (Supplementary Fig. S1B). We observed significant acceleration of lymphoma onset in VavP-Bcl2/CrebbpKD (shRNA #1, n=34, p=0.0465) as compared to VavP-Bcl2/GFP group (n=31, Fig. 1A). This was reproduced with a second shRNA (shRNA #3, Supplementary Fig. S1C). The contribution of CREBBP was evident in that only 13 to 19% of HPCs expressed GFP whereas 90% of lymphoma cells were GFP+ in the VavP-Bcl2/CrebbpKD (Fig. 1B). Overall VavP-Bcl2/CrebbpKD tumors contained a significantly higher ratio of transduced cells than VavP-Bcl2/GFP tumors (p=0.01, Mann-Whitney U test, Fig. 1C). We purified B220+ lymphoma cells from spleen and examined the global H3K27ac level by Western blotting. We observed that acetylation of H3K27 was reduced in lymphoma cells transduced with Crebbp shRNA, confirming the functional impact of Crebbp knockdown (Supplementary Fig. S1D). The lymphoma cells from VavP-Bcl2/CrebbpKD were B220+, CD19+, IgM+, and CD3- (Fig. 1D and Supplementary Fig. S1E), confirming their B-cell identity. VavP-Bcl2/CrebbpKD lymphomas were more aggressive and widely disseminated than VavP-Bcl2 control tumors as shown by scoring tissue invasion and disease morphology by histopathology of spleen and other organs (Fig. 1D and 1E, and Supplementary Fig. S1F). PCR analysis of the murine IgL Vλ-Jλ locus indicated that these lymphomas were clonal (Supplementary Fig. S1G). Somatic hypermutation is a hallmark of GC B-cells and GC derived lymphomas (15). Indeed sequencing the VDJH4 locus of both VavP-Bcl2/CrebbpKD and VavP-Bcl2 control lymphomas revealed the presence of somatic hypermutation, confirming the GC origin of these tumors (Supplementary Fig. S1H). A similar general phenotype was observed using shRNA against Ep300 in vavP-Bcl2 transgenic mice including the significant acceleration of lymphoma onset (n=37, p=0.0143, log-rank test), more aggressive histology, B-cell immunophenotype, clonality and GC origin (Supplementary Fig. S2). Hence, Crebbp and Ep300 deficiency can promote GC lymphomagenesis and function as tumor suppressor genes.
Figure 1. Crebbp deficiency accelerates B-cell lymphoma development in mice.
A, Kaplan-Meier curve of C57BL/6 mice transplanted with VavP-Bcl2 HPCs transduced with MSCV-GFP retroviral vector alone (GFP, black, n=31), or containing shRNAs against Crebbp (red, n=34). Statistical significance of survival difference was determined by the log-rank test between shCrebbp and vector alone. B, Representative flow cytometry histograms showing the GFP positive cell percentage of the pre-injection HPCs and the splenic murine lymphoma cells that derived from the same HPC. C, Dot plot representing the GFP positive cell percentage in the splenic murine lymphoma cells in individual recipient animal. The mean and standard error of the mean (S.E.M.) were represented for each transplant group. Statistical significance was determined by Mann-Whitney test. D, H&E, B220, CD3, and Ki67 staining of spleen tissues extracted from recipient mice upon sacrifice. Scale bars, 500 µm. E, H&E and B220 staining of kidney and lung tissues extracted from recipient mice upon sacrifice. Scale bars, 500 µm.
Crebbp deficiency leads to preferential loss of H3K27acetylation at enhancers
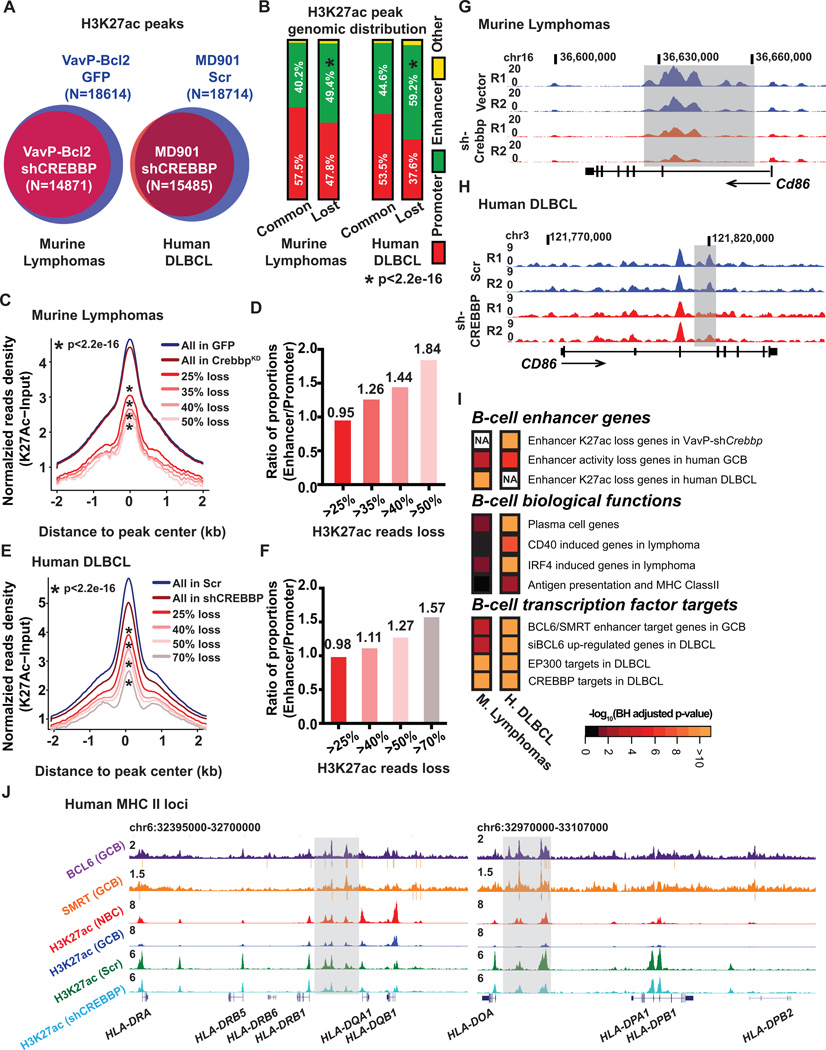
For downstream mechanistic studies exploring how loss of histone acetyl transferases could accelerate lymphomagenesis we focused primarily on Crebbp since i) Crebbp KD and Ep300 KD resulted in similar phenotype, ii) CREBBP mutations are more frequent in humans, and iii) are mutually exclusive with EP300 suggesting overlapping mechanisms of action. To determine the impact of CREBBP loss of function on H3K27 acetylation patterning we performed ChIP-seq for H3K27ac in B220+ lymphoma cells from VavP-Bcl2/GFP mice (n=4), as well as in VavP-Bcl2/CrebbpKD (n=6) mice. We identified 18,614 H3K27ac “peaks” in VavP-Bcl2/GFP lymphomas (Fig. 2A). Interesting, there was a net loss of 3,660 (19.7%) peaks and practically zero gain of new peaks in VavP-Bcl2/CrebbpKD lymphomas (Fig. 2A). A similar effect was observed in human GC derived lymphoma B-cells where CREBBP was depleted using two independent shRNAs, with focal loss of 17.3% of H3K27ac peaks after CREBBP knockdown (Fig. 2A and Supplementary Fig. S3A). As in the mice, depletion of CREBBP in the human cells also resulted in global reduction of H3K27ac (Supplementary Fig. S3B).
Figure 2. Crebbp deficiency results in focal H3K27ac loss in mouse and human lymphoma.
A, Venn diagrams showing the overlap between the H3K27ac peaks in B220+ cells from VavP-Bcl2/GFP tumors (n=4) or from VavP-Bcl2/shCrebbp tumors (n=6) (left panel), or the overlap between the H3K27ac peaks in MD901 cells transduced with either control scramble shRNA (n=3) or shRNAs against CREBBP (n=6) (right panel). B, Stacked bar plot representing the genomic distribution of common and lost H3K27ac peaks between VavP-Bcl2/GFP and VavP-Bcl2/shCrebbp tumor cells (left panel), or between MD901 cells transduced with either control scramble or CREBBP shRNAs (n=6). C, Normalized average H3K27ac read density plot at loci identified as H3K27ac peaks in MACS-purified B220+ B cells from VavP-Bcl2/GFP tumors. The black line represents the average values in VavP-Bcl2/EV tumors (n=4), and the dark red line represents the values in VavP-Bcl2/shCrebbp tumors (n=6). Average values of peaks that exhibited more than 25%, 35%, 40%, and 50% reads loss in VavP-Bcl2/shCrebbp tumors as compared to VavP-Bcl2/GFP tumors are shown as lines in different shades of red. * representing statistical significant loss of normalized H3K27ac read density as determined by Kolmogorov-Smirnov test. D, Bar plot representing ratio of the proportions of enhancer peaks or promoter peaks that exhibited more than 25%, 35%, 40%, and 50% reads loss in VavP-Bcl2/shCrebbp tumors as compared to VavP-Bcl2/GFP tumors. E, Normalized average H3K27ac read density plot at loci identified as H3K27ac peaks in scramble shRNA transduced MD901 cells. The black line represents the average values in scramble shRNA transduced MD901 cells (n=3), and the dark red line represents the values in CREBBP shRNAs transduced MD901 cells (n=6). Average values of peaks that exhibited more than 25%, 40%, 50%, and 70% reads loss in CREBBP KD cells as compared to control scramble cells are shown as lines in different shades of red. * representing statistical significant loss of normalized H3K27ac read density as determined by Kolmogorov-Smirnov test. F, Bar plot representing ratio of the proportions of enhancer peaks or promoter peaks that exhibited more than 25%, 40%, 50%, and 70% reads loss in CREBBP KD cells as compared to control scramble MD901 cells. G, UCSC read-density tracks of normalized H3K27ac ChIP-seq reads at murine Cd86 locus in two representative VavP-Bcl2/GFP tumors and two representative VavP-Bcl2/shCrebbp tumors. Shaded areas highlight the regions showed loss of H3K27ac in VavP-Bcl2/shCrebbp tumors. H, UCSC read-density tracks of normalized H3K27ac ChIP-seq reads at human CD86 locus in two representative biological replicates of control scramble (Scr) MD901 cells and two representative biological replicates of CREBBP KD (shCREBBP) MD901 cells. Shaded areas highlight the regions showed loss of H3K27ac in KD cells. I, Pathways analysis of genes (n=1147) with > 25% reduction of H3K27ac reads at enhancers in murine VavP-Bcl2/shCrebbp tumors, or genes (n=2928) with > 25% reduction of H3K27ac reads at enhancers in CREBBP knockdown cells. Heatmap represents the BH-adjusted p value of each geneset tested. J, UCSC read-density tracks of normalized BCL6 (purple) and SMRT (orange) ChIP-seq reads in human tonsilar GCBs, H3K27ac ChIP-seq reads in human tonsilar NBCs (red) and GCBs (blue), and H3K27ac ChIP-seq in control scramble (Scr, green) and CREBBP KD (shCREBBP, turquoise) MD901 cells at the human MHC II loci. BCL6 and SMRT peaks determined by MACS2 are indicated by grey bars under the read density track. Shaded areas highlight the enhancers that were bound by BCL6 and SMRT, and showed loss of H3K27ac in CREBBP KD cells.
We focused on H3K27ac sites disproportionately affected by CREBBP loss of function since these would be most strongly linked to functional impact. In human lymphoma cells we observed that 76.9% of CREBBP binding sites are located at putative enhancers, including introns, and distal regions (Supplementary Fig. S3C). Accordingly, we observed that H3K27ac peaks lost in VavP-Bcl2/CrebbpKD lymphomas were disproportionately located at intergenic or intronic locations corresponding to putative enhancers (p<2.2e-16, Fig. 2B), whereas unaffected peaks were preferentially located at promoters. Similarly, ChIP-seq studies in human cells indicated preferential loss of H3K27ac at putative enhancers (59.2% vs. 37.6% at promoters, p<2.2e-16, Fig. 2B). Confirming this assessment, quantitative assessment of H3K27ac ChIP-seq profiles revealed increasing skewing towards enhancers among sites with progressively greater loss of H3K27ac after CREBBP loss of function in murine and human lymphoma cells (p<2.2e-16, Fig. 2C to 2F, examples shown in Fig. 2G and 2H), and expression of genes nearby these enhancers was reduced in CREBBP deficient cells, as determined by RNA-seq performed in both the murine and human CREBBP knockdown systems (Supplementary Fig. S4A and S4B). We did not observe significant correlation between gene expression reduction and promoter H3K27ac loss in mice, although there was correlation in humans (data not shown), so that enhancer effects were the most consistent effects of CREBBP loss of function. Finally, there was significant enrichment of the murine CREBBP enhancer H3K27ac loss gene signature in human lymphoma cell enhancer H3K27ac loss genes with CREBBP knockdown (p=6.88e-23, Fig. 2I), supporting the human relevance of the murine model. Hence CREBBP loss of function preferentially affects H3K27 acetylation at enhancers and affects similar genes in humans and mice.
CREBBP loss of function represses enhancers that are poised in GC B-cells
To determine more precisely how CREBBP loss of function perturbs enhancer function we next mapped the location of enhancers in primary human GC B-cells, which are the cell of origin of DLBCL and FL. To identify the enhancers relevant and specific to this cell type we performed replicate ChIP-seq for H3K4me2, H3K4me3 and H3K27ac in purified primary human naïve B cells (NBs) and GC B-cells. We identified 7577 active enhancer H3K4me2+H3K4me3−H3K27ac+ peaks in NBC and 5219 in GC B-cells (Supplementary Fig. S5A), the great majority (two thirds) of which were unique and specific to each cell type. Changes in enhancer activation state were accompanied by corresponding changes in gene expression (Supplementary Fig. S5B and S5C, examples are shown in Supplementary Fig. S5D). Notably, among sites that lose H3K27 acetylation after CREBBP knockdown in mouse and human lymphomas, there was significant enrichment for genes that also lose enhancer activity in GC B-cells (Fig. 2I). Hence CREBBP loss of function strengthens repression of B-cell enhancers that are deactivated in GCs. This could contribute to lymphomagenesis by aberrantly maintaining aspects of the GC phenotype. Indeed, further analysis of CREBBP ChIP-seq and RNA-seq signatures in mice and humans revealed significant enrichment for genes linked to termination of the GC reaction such as those induced by CD40, IRF4 signaling and plasma cell differentiation (Fig. 2I, Supplementary Fig. S6, for a complete list of enriched pathways please check the Supplementary Tables S1 and S2). In the human cells we also observed enrichment for antigen presentation and MHC Class II genes. This result is consistent with reported down regulation of these genes in CREBBP mutant FLs (7). CREBBP loss of function thus results in failure to activate genes involved in GC exit and immune recognition.
To probe CREBBP function we compared CREBBP knockdown RNA-seq and H3K27ac ChIP-seq profiles in mice and human cells to a database of B-cell ChIP-seq and transcriptional profiles. This approach revealed a striking enrichment for genes whose enhancers are bound by the BCL6/SMRT transcriptional repressor complex, and accordingly, genes found to be derepressed upon BCL6 knockdown in GC derived DLBCL cells (Fig. 2I and Supplementary Fig. S6). CREBBP and EP300 target genes in DLBCL were similarly enriched (Fig. 2I and Supplementary Fig. S6), consistent with EP300 and CREBBP having similar functions in these cells. By contrast, plasma cell genes and IRF4 signaling genes were not significantly enriched in the genes whose promoters lost H3K27ac enrichment in murine VavP-Bcl2/CrebbpKD lymphoma cells (Supplementary Fig. S7). Taken together, these data indicate that CREBBP/EP300 and BCL6-SMRT complexes have opposing effects on enhancers that are silenced in GC B-cells and are activated upon GC exit and antigen presentation.
BCL6 has not been previously implicated in antigen and MHC class II presentation, therefore we analyzed BCL6 and SMRT ChIP-seq profiles performed in primary human Naïve and GC B-cells for MHC II enhancer binding. We readily identified multiple binding sites in enhancers for MHC class II (Fig. 2J) and related genes such as CD74 and CIITA (Supplementary Fig. S8). Notably the H3K27ac level at these enhancers were reduced in GC B-cells as compared to that in NBCs, suggesting that BCL6 normally attenuates the MHC II locus during the GC reaction (Fig. 2J). H3K27ac at these enhancers was also reduced by CREBBP KD (Fig. 2J, Supplementary Fig. S8). These results indicate novel and opposing functions for BCL6/SMRT complexes and CREBBP in immune recognition.
CREBBP mutant lymphomas exhibit a GC/BCL6 target enhancer repression signature
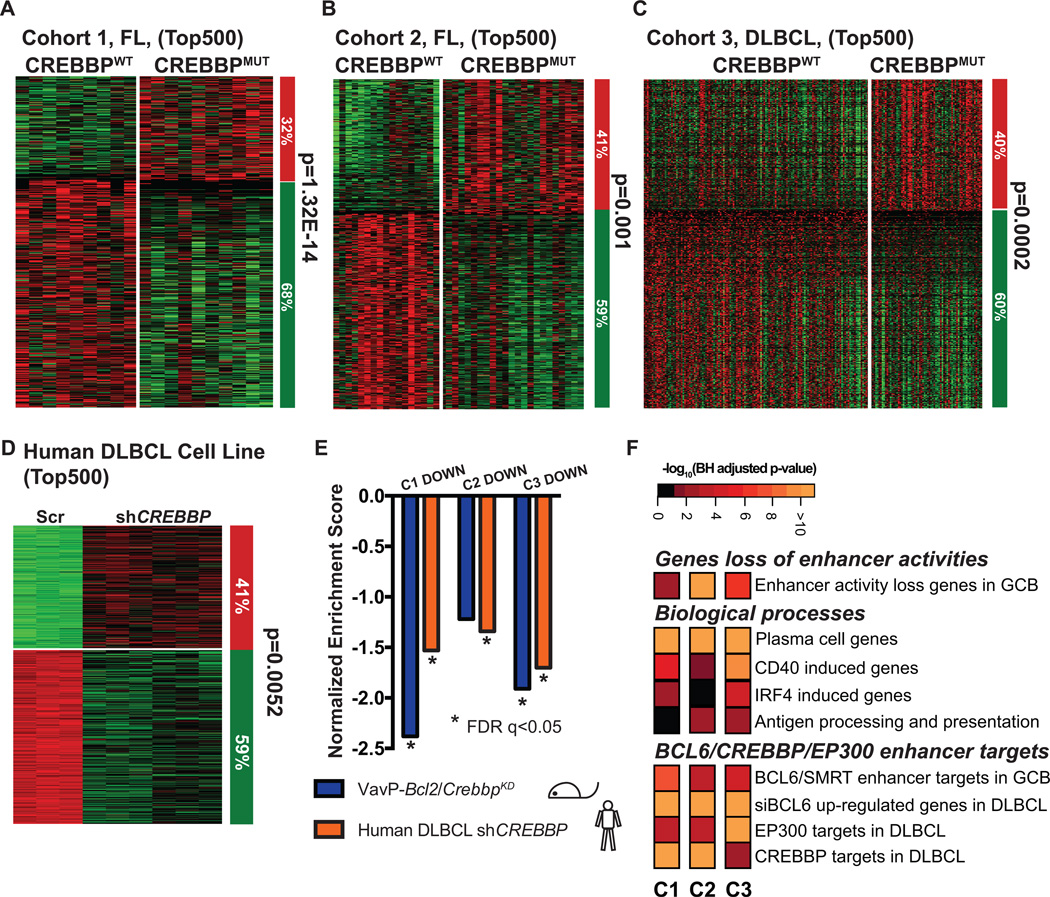
To determine the relevance of these experimental CREBBP signatures to those occurring in the human disease, we next performed RNAseq and mutation profiling in DNA of two cohorts of FL patients (Supplementary Table S3). Non-synonymous CREBBP cSNVs were detected in 38.5% of cohort 1 and 46.1% in cohort 2 mostly clustering to four hotspots in the HAT domain (Supplementary Fig. S9A and S9B, and Supplementary Table S4). Somatic mutations were validated by Sanger Sequencing in a random subset of tumors and their germline controls (Supplementary Fig. S9C). RNAseq was also performed in 347 DLBCL patient samples, of which 20.5% (n=71) carried CREBBP mutations that were determined by targeted sequencing of tumor DNA (Supplementary Fig. S9A. CREBBP mutations were significantly more frequent in GCB-DLBCL than ABC-DLBCL (p=1.42E-5, cell of origin classification determined by Nanostring Lymph2Cx (16) Supplementary Fig. S9D) and again preferentially involved the HAT domain (Supplementary Table S5). We examined differential gene expression between CREBBP mutant and CREBBP WT samples lacking other epigenetic mutations that might introduce confounding effects. In all three cohorts there was significant skewing towards genes being more repressed than activated in CREBBP mutant patients when we examined the top differentially expressed genes (Fig. 3A to 3C, and Supplementary Fig. S10A). Comparison of the signatures of the three cohorts indicated that they were extremely similar (GSEA FDR q<0.05) confirming that similar genes were affected across CREBBP mutant FL and DLBCL patients (Supplementary Fig. S10B). The same repressive pattern was induced by CREBBP knockdown in human GC derived DLBCL cell lines (Fig. 3D). Indeed GSEA also revealed that the FL and DLBCL CREBBP mutant repressed signatures were also highly similar to the murine VavP-Bcl2/CrebbpKD lymphomas and human CREBBP knockdown cell lines (FDR q<0.05, Fig. 3E), demonstrating that these models properly reflect the biology of the human disease, and are furthermore consistent with the fact that HAT domain mutations of CREBBP in human patients are known to result in enzymatic loss of function (2). In accordance with this notion, genes downregulated in human CREBBP mutant lymphomas were highly significantly enriched for genes that normally lose enhancer H3K27ac in GC B-cells, GC exit genes including CD40 induced, IRF4 induced, plasma cell differentiation, and MHC class II antigen processing and presentation (Fig. 3F, for a complete list of enriched pathways please check Supplementary Table S2). Again, there was significant enrichment for genes with enhancers regulated by BCL6/SMRT complexes, genes induced by BCL6 siRNA and genes that are direct targets of EP300 and CREBBP in DLBCL cells (Fig. 3F). These results identify a conserved signature of CREBBP and BCL6-SMRT regulated genes that control GC exit and immune recognition.
Figure 3. CREBBP loss of function results in gene expression repression signature.
A–D, Supervised analysis of the top 500 most differentially expression genes between CREBBP WT (CREBBPWT) and mutant (CREBBPMUT) FL (A, B), DLBCL patients (C), or between scramble and CREBBP knockdown MD901 cells (D). Columns represent individual samples, rows correspond to the genes. Heatmap represents the z-scores of the expression value (RPKM) characterized by RNA-seq. The column on the right represents the proportion of the genes that were repressed (green) or upregulated (red) CREBBPMUT patient samples as compared to CREBBPWT patient samples (A–C), or in CREBBP knockdown cells as compared to scramble control samples of the respective cohorts (D). Statistical significance was determined by Fisher Exact test. E, Summary of the GSEA of the down-regulated genes in the top 500 most differentially expressed genes of respective cohorts as compared to ranked gene expression changes between either murine VavP-Bcl2/CrebbpKD and VavP-Bcl2/EV murine tumors (purple bars) or between CREBBP KD and Scr MD901 cells (orange bars). * indicates statistical significant enrichment (FDR q<0.05). F, Pathway analysis of the down-regulated genes within the top 500 most differentially expressed genes of respective cohorts. Heatmap represents the –log10 BH-adjusted p-value of each geneset tested.
CREBBP mutant lymphoma cells are preferentially dependent on HDAC3
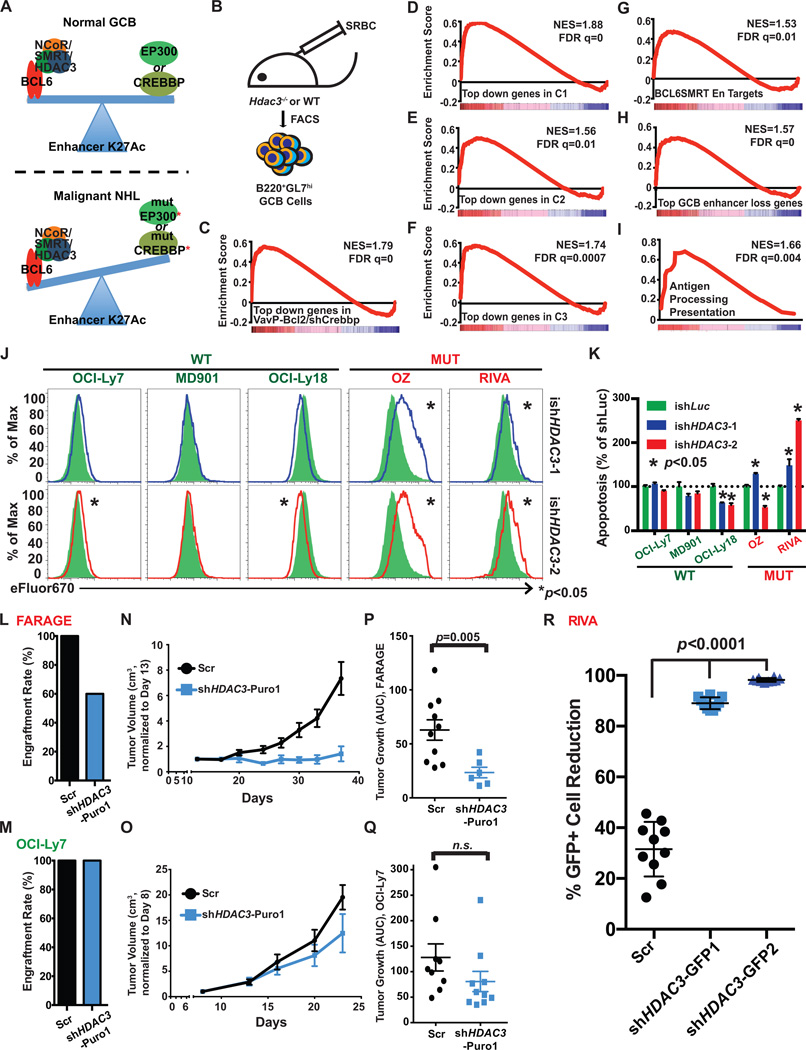
In normal GC B-cells, induction of BCL6 results in recruitment of the SMRT corepressor and its close homolog NCOR to enhancers, both of which in turn recruit HDAC3. The histone deacetylase HDAC3 is a component of SMRT/NCOR complexes and mediates H3K27 deacetylation of enhancers by BCL6 (17). Thus, BCL6-SMRT/NCOR-HDAC3 complexes “toggle” enhancers that are active in mature B-cells likely due to EP300 and CREBBP HAT activity (based on the above data), to a poised H3K27 deacetylated configuration resulting in reduced gene expression (17). This enhancer repressive effect is reversed when B-cells exit the GC reaction through post-translational modification of SMRT and NCOR and downregulation of BCL6, which results in re-acquisition of histone acetylation (18–20). Hence we hypothesized that a major mechanism through which CREBBP mutation/loss of function leads to lymphomagenesis is by partially disabling dynamic reversal of enhancer repression mediated by BCL6-SMRT/NCOR-HDAC3 complexes (Fig. 4A).
Figure 4. Loss of HDAC3 inhibits CREBBP mutant lymphoma growth in vitro and in vivo.
A, A schematic model showing the opposing effects on enhancer H3K27ac regulation between CREBBP and EP300 and BCL6/SMRT/HDAC3 corepressor complex in normal GCB cells and in malignant lymphoma cells. B, A cartoon outlining the generation and collection of GC B-cells in Hdac3−/− and Hdac3fl/fl mice for RNAseq. C–I, GSEA enrichment plots showing correlation of different genesets with ranked expression change between murine Hdac3−/− and Hdac3fl/fl germinal centre B-cells, n=2 from each group. NES, normalized enrichment score. J, Representative flow cytometry histograms demonstrating proliferation of CREBBP WT lymphoma cell lines (OCI-Ly7, MD901 and OCI-Ly18) and mutant lymphoma cell lines (OZ and RIVA). Each cell line was transduced with inducible shRNAs against either control Luciferase gene (ishLuc, green shaded area) or HDAC3 (ishHDAC3-1, blue, top panel, and ishHDAC3-2, red, bottom panel). Transduced cells were labeled with cell proliferation dye eFluor 670 and cultured for 5 days with the presence of 0.2 µg/ml Doxycycline. * represents statistical difference between ishLuc (n=3) and respective ishHDAC3 (n=3), determined by T-test. K, Bar plot showing the relative apoptotic cells in CREBBP mutant or WT cells with ishHDAC3-1 (blue), and ishHDAC3-2 (red) as compared to ishLuc (green), measured by Annexin V staining. The numbers represent mean and S.E.M. of the percentage of Annexin V+/DAPI- cells of three replicates normalized to the average in the shLuc samples. L,M, Engraftment rate of FARAGE (L, CREBBP mutant) or OCI-Ly7 (M, CREBBP WT) in SCID mice. SCID mice were s.c. injected with 1E7 FARAGE or OCI-Ly7 cells either transduced with a scramble shRNA (black) or a shRNA against HDAC3 (blue), 10 animals per group. N,O, Tumor growth plots of FARAGE (N) and OCI-Ly7 (O) xenografted mice. P,Q. Dot plots showing the growth of each tumor measured as area under the curve. Average tumor growth (mean ± S.E.M.) is represented on the y-axis, which represents tumor volume (cm3)/time (days). Statistical significance was determined by Mann-Whitney U test. R, Dot plots showing the percentages of GFP positive tumor cell population reduction in RIVA xenograft model with either HDAC3 shRNAs (shHDAC3-GFP1, shHDAC3-GFP2) or scramble shRNA (Scr). For each group, mean ± S.D. was presented. Statistical significance was calculated by Mann-Whitney test.
To first assess the requirement for HDAC3 in mediating BCL6-SMRT/NCOR effects in GC B-cells we performed two experiments. In the first we obtained mice genetically engineered to simultaneously express mutant forms of NCOR and SMRT that are unable to associate with HDAC3 in a stable manner (21). These mice were immunized with a T-cell dependent antigen and splenic tissue assessed after 10 days for GC formation. In these animals we observed an ~50% reduction in the abundance of GC B-cells as determined by flow cytometry (Supplementary Fig. S11). Since this model is constitutive to rule out that this effect could be due to non-B-cell autonomous effects we next performed a similar experiment in mice engineered for conditional deletion of HDAC3, driven by Cd19Cre. These mice revealed the same phenotype, with ~50% reduction in the abundance of GC B-cells after T-cell dependent antigen stimulation (S.W. Hiebert, data not shown, Fig. 4B). Taken together, these animal models confirm that HDAC3 is required by BCL6/SMRT complex to fully establish the GC reaction.
Next, to determine whether the opposing effects between CREBBP and BCL6-SMRT complexes on gene expression is driven by HDAC3, we analyzed RNA-seq data obtained from purified FACS sorted GC B-cells from Cd19Cre/Hdac3−/− mice. Genes induced in Hdac3 knockout GC B-cells were strikingly and significantly enriched for genes that are repressed in VavP-Bcl2/CrebbpKD lymphomas and in human CREBBP mutant FL and DLBCLs (Fig. 4C to 4F). Accordingly we also observed enrichment for the BCL6-SMRT enhancer target genes, enhancers that are repressed in GC B-cells, and the class II MHC and antigen presentation gene set among genes induced in Cd19Cre/Hdac3−/− GC B-cells (Fig. 4G to 4I). Hence, HDAC3 antagonizes CREBBP function such that CREBBP mutation leads to unopposed HDAC3 activity in GC B cells and lymphomas.
It is well established that BCL6 maintains the growth of lymphoma cells partly through its recruitment of SMRT/HDAC3 complexes (17). Given that our data suggests that CREBBP loss of function particularly strengthens this aspect of BCL6 function we postulated that CREBBP mutant lymphoma cells would be especially sensitive (to a greater degree than CREBBP WT cells) to HDAC3 loss of function. We therefore transduced CREBBP mutant and wild-type DLBCL cells with inducible lentiviral shRNAs against HDAC3 that exhibited robust knockdown of HDAC3 expression (Supplementary Fig. S12A). HDAC3 knockdown significantly impaired proliferation of CREBBP mutant (OZ and RIVA) but not CREBBP WT DLBCL cells (Fig. 4J and Supplementary Fig. S12B). HDAC3 knockdown also caused more apoptosis in CREBBP mutant RIVA cells but not WT cell lines (Fig. 4K and Supplementary Fig. S12C). We next evaluated the effect of HDAC3 knockdown in CREBBP mutant and wild-type DLBCL in vivo. We performed this experiment in two ways. First, we enriched CREBBP mutant (FARAGE) and WT (OCI-Ly7) cells transduced with control shRNA or shHDAC3 with a puromycin selection mark, and implanted these cells into SCID mice. We observed that knockdown of HDAC3 impaired engraftment of CREBBP mutant but not CREBBP WT DLBCL cell lines, as compared to control (Fig. 4L and 4M). Among engrafted mice HDAC3 knockdown resulted in much more significant retardation of tumor growth in the CREBBP mutant (p=0.005) than wild type DLBCL (Fig. 4N to 4Q). Immunohistochemistry analysis showed a trend towards increased necrotic areas in the CREBBP mutant mice with HDAC3 knockdown, but no difference in in the CREBBP wild type lymphomas (Supplementary Fig. S12D). Second, we implanted NSG mice with CREBBP mutant RIVA cells containing a mixed population of GFP+shHDAC3 or control shRNA transduced with non-transduced cells. Five weeks after implantation, the mice were sacrificed and lymphomas evaluated to ascertain the percentage of GFP positive cells. HDAC3 knockdown lymphomas featured almost complete loss of GFP representing a highly significant depletion of GFP+ cells as opposed to control shRNA transduced cells (p<0.0001, Fig. 4R). Hence, CREBBP mutant DLBCLs are addicted to HDAC3 for their proliferation and survival.
Repression of antigen processing and presentation genes due to CREBBP loss can be rescued by HDAC3 inhibition
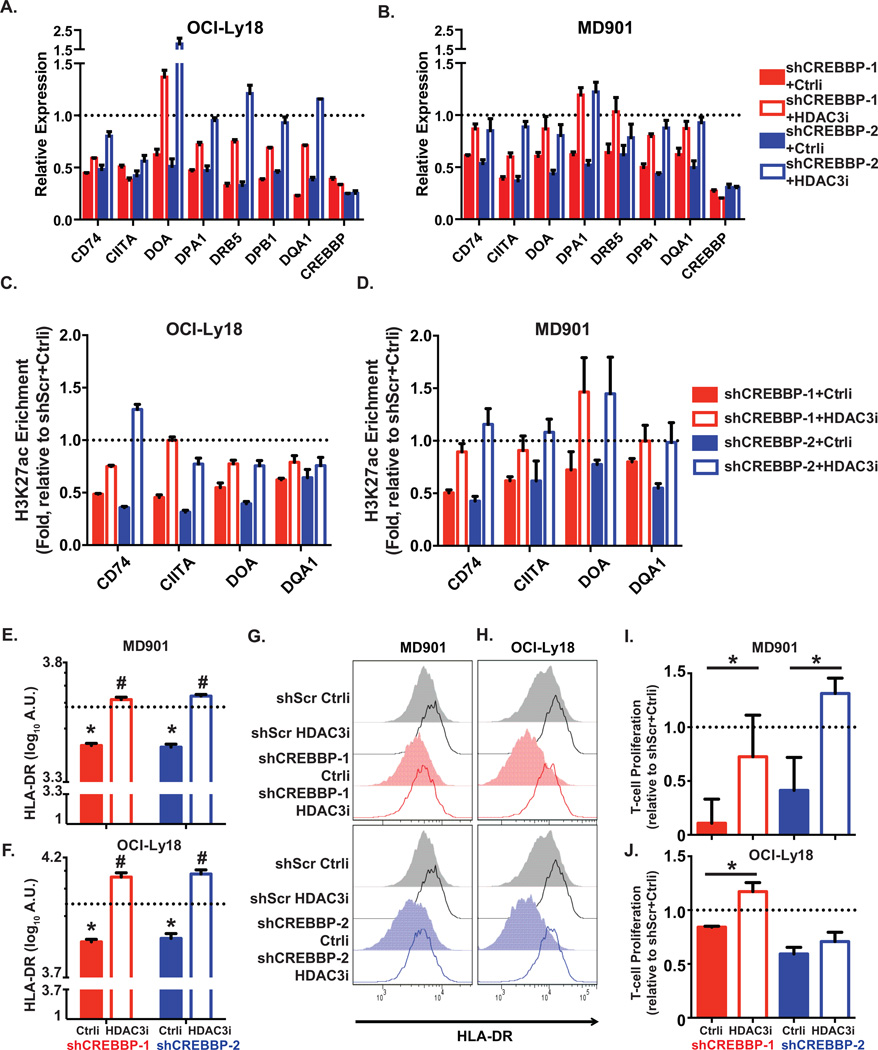
Finally, to confirm the opposing actions and potential novel biological effects of CREBBP and HDAC3 we examined in greater detail their counter-regulatory effects at the MHC class II loci, which as shown above are counter-regulated by CREBBP and BCL6-SMRT-HDAC3 complexes. First, to confirm that recruitment of HDAC3 to these loci was dependent on BCL6, we performed HDAC3 ChIP-qPCR in DLBCL cells after transduction with siBCL6 or control siRNA. BCL6 siRNA but not control induced the expected reduction in BCL6, as well as loss of enrichment of HDAC3 at MHC class II related loci (Supplementary Figure S13). Next, we observed that transduction of CREBBP WT DLBCL cells (OCI-Ly18 and MD901) with either of the two CREBBP shRNAs induced significant reduction in the transcript abundance of MHC class II genes in these cells (Fig. 5A and 5B, primer sequences see Supplementary Table S6). Furthermore, this gene expression reduction was accompanied by loss of H3K27ac at enhancers of the MHC class II genes (Fig. 5C and 5D, primer sequences see Supplementary Table S6). CREBBP knockdown likewise accordingly induced significant reduction of cell surface HLA-DR as detected by flow cytometry in two independent experiments (Figure 5E to 5H, and Supplementary Fig. S14A and 14B). Notably, treatment of CREBBP knockdown cells with a selective HDAC3 inhibitor (22) rescued the expression of most of these MHC class II genes (Figure 5A and 5B), H3K27ac enrichment at their enhancers (Figure 5C and 5D), and restored expression of cell surface HLA-DR (Figure 5E to 5H, and Supplementary Fig. S14A and 14B). Consistent with results from Green et al. showing that primary CREBBP mutant DLBCLs less efficiently induce allogeneic T-cell activation (7), we showed that CREBBP knockdown cells failed to stimulate human T-cell proliferation to the same extend as control knockdown cells (Figure 5I and 5J, and Supplementary Fig. S14C). This loss of the T-cell stimulation ability was at least partially rescued by treatment with the selective HDAC3 inhibitor in both cell lines (Figure 5I and 5J, and Supplementary Fig. S14C). Taken together, our data suggest that in GC B-cells MHC class II gene expression is controlled by enhancer toggling through BCL6/SMRT/HDAC3 complexes in contraposition with CREBBP. CREBBP mutation or loss of function allows greater BCL6/SMRT/HDAC3 mediated repression resulting in the reduction of surface expression of these genes thus potentially impairing the ability of T-cells to suppress the outgrowth of malignant B-cells.
Figure 5. CREBBP regulates antigen processing and presentation gene enhancers.
A,B, Bar plots representing the relative expression of antigen presentation and MHC II genes in OCI-Ly18 (A) and MD901 (B) cells upon CREBBP depletion and treated with either a selective HDAC3 inhibitor (HDAC3i) or a control compound (Ctrli), as compared to scramble shRNA induced cells (set as 1, dotted lines). Bar graph represents mean and S.E.M. from three replicates. C,D, Bar plots representing the relative H3K27ac enrichment at enhancers of MHC II genes in OCI-Ly18 (C) and MD901 (D) cells upon CREBBP depletion and treated with either a selective HDAC3 inhibitor (HDAC3i) or a control compound (Ctrli), as compared to scramble shRNA induced cells (set as 1, dotted lines). Bar graph represents mean and S.E.M. from three replicates. E,F, Quantification of HLA-DR measured by flow cytometry in shCREBBP or scramble transduced lymphoma MD901 (E) and OCI-Ly18 (F) cells treated with either a selective HDAC3 inhibitor (HDAC3i) or a control compound (Ctrli). Cells were transduced with shRNAs for 3 days and then treated with compounds for 96hr. The data was represented as mean ± S.D. Statistical significance was determined by Student’s T-test. * indicates significant difference between control compound-treated scamble or shCREBBP transduced cells (p<0.05). # indicates significant difference between control compound and selective HDAC3 inhibitor treated cells (p<0.05). G,H, Representative flow cytometry histograms showing cell surface level of MHC II molecule HLA-DR that were quantified in E and F. I,J, Bar plots showing the relative proliferation of T-cells stimulated by shCREBBP or scramble transduced lymphoma MD901 (I) or OCI-Ly18 (J) cells treated with either a selective HDAC3 inhibitor (HDAC3i) or a control compound (Ctrli). The data indicate relative folds of T cell proliferation, represented as ratio of fluorescence (560nm/590nm) from each treatments to that from scramble shRNA transduced cells treated with control compound (set as 1, dotted lines). Statistical significance was determined by Student’s T-test.
DISCUSSION
CREBBP is one the most frequently mutated genes in human lymphomas and our study demonstrates its tumor suppressor action and provides insight into its mechanism. We find that CREBBP loss of function disrupts a finely tuned mechanism of enhancer toggling that controls gene expression during the GC reaction. Specifically, CREBBP and likely EP300 maintain H3K27 acetylation of specific enhancers required for terminal differentiation and immune signaling in mature B-cells. This enables B-cells to undergo plasma cell differentiation upon immunization. However a subset of B-cells follows an alternative fate and instead enters the proliferative and mutagenic GC reaction. In GC B-cells these enhancers are transiently repressed by BCL6/SMRT/HDAC3 complexes through H3K27 deacetylation. This effect is terminated through CD40 signaling that disrupts BCL6/SMRT repression complexes upon GC exit, upon which enhancers recover their H3K27actylated active state (18–20).
However, CREBBP mutation results in a failure to reactivate these enhancers, which impairs GC exit and immune recognition programs, and ultimately promotes malignant transformation. Because GC enhancer deacetylation is driven by HDAC3, CREBBP mutant lymphomas become especially dependent on this protein as observed in our functional assays in vitro and in vivo. Hence HDAC3 dependency is an exploitable target in lymphoma patients with CREBBP somatic mutations. Because FLs and DLBCLs are dependent on BCL6 regardless of CREBBP mutation status it is expected that CREBBP WT lymphomas would manifest some degree of HDAC3 dependency as well. Accordingly, our data show that HDAC3 knockdown still has mild deleterious effects in CREBBP wild type DLBCL cell lines in vitro and in vivo.
Mutant CREBBP induced loss of enhancer function is reminiscent to recent findings indicating that somatic mutations of the histone methyltransferase KMT2D in B-cell lymphomas induce malignant transformation by disrupting enhancer H3K4 mono and dimethylation (14, 23). In the case of KMT2D the impact of loss of function alleles is to attenuate enhancer response to CD40 and other signaling pathways. Hence induction of gene expression linked to GC exit signals is dampened and there is aberrant persistence of GCs beyond their usual transient time frame, and affected B-cells manifest a defect in class switch recombination (14). Even though KMT2D and CREBBP are both enhancer regulators, they seem to have at least partially distinct functions given that: i) somatic mutation of these two genes often occur together in DLBCL and FL and ii) their target genes and downstream biological effects are not identical (e.g. such as enhanced proliferation in KMT2D (14, 23) or reduced MHC class II expression in CREBBP loss of function).
It is important to underline that the transcriptional and epigenetic signatures induced by CREBBP knockdown in mice and human cells was highly similar to that of human patients with CREBBP mutations. Moreover the critical mechanistic point of HDAC3 dependency was observed in both CREBBP mutant DLBCL cells as well as CREBBP loss of function experiments. Finally, the loss of MHC class II gene expression and loss of the ability of lymphomas to stimulate a T-cell response was similar in CREBBP knockdown as well as CREBBP mutant primary patients compared to respectively controls (7). These three points confirm the relevance of our findings to the natural process of CREBBP somatic mutation. We underline that MHC class II loss of function is an important event in lymphomas, associated with inferior clinical outcome (24). Along these lines it is notable that the BCL6/SMRT/HDAC3/CREBBP mechanism regulates not only enhancers within the MHC class II loci but also genes that themselves play important roles in supporting MHC class II expression such as CIITA, or that enable activity of MHC class II surface proteins such as CD74. In addition to being a target gene of BCL6 and CREBBP, CIITA itself is mutated in approximately 10% of DLBCLs based on publicly available TCGA data (data not shown), and even more frequently in primary mediastinal large B cell lymphoma (PMBL) (25). The occurrence of somatic mutations of CREBBP and its downstream target CIITA in DLBCLs support the notion that loss of function of antigen presentation functions is an important aspect of lymphomagenesis. Yet we do not rule out that CREBBP mutation might have additional functions. For example it was proposed that CREBBP mutation could impair TP53 function and enhance BCL6 through reduced acetylation (2). Although the most pronounced loss of H3K27 acetylation occurred at enhancers, we point out that at least part of the function of CREBBP may still be related to effects on promoter function as shown in Supplementary Table S2. Future studies should help to determine if any other putative effects of CREBBP are relevant to lymphomagenesis beyond those described in this manuscript.
Finally, it is intriguing that the CREBBP-HDAC3 enhancer toggling mechanism controls expression of antigen processing and presentation genes, which could potentially disrupt immunosurveillance by T-cells. As noted, CREBBP mutant lymphomas feature less T-cell infiltration and impaired ability to activate T-cells ex vivo (7). We show that CREBBP loss of function indeed directly mediates loss of MHC class II expression and the ability of T-cells to recognize B-cells. Most strikingly we show that this effect is rescued by targeting HDAC3. This finding is consistent with and refines the notion that targeting pan-HDAC inhibitors could induce MHC class II expression in lymphoma cells (26). Hence our findings have direct ramifications for lymphoma therapy since CREBBP mutant lymphomas may exhibit continued requirement for HDAC3 activity to hide from T-cells through epigenetic silencing of MHC class II. Synthetic HDAC3 dependence in CREBBP mutant tumors may thus have dual actions on both immune-surveillance and other growth promoting functions of BCL6-SMRT complexes. These mechanisms indicate an opportunity for HDAC3 specific inhibitors as a precision medicine approach for CREBBP mutant lymphomas.
METHODS
Characterization of human NHL samples
Characterization of NHL samples, including two cohorts of follicular lymphoma samples and one cohort of newly diagnosed DLBCL samples, was previously described in Ortega-Molina et al (14). These studies were approved by either the Institutional Review Board (IRB) at Weill Cornell Medical College (IRB#0107004999), or the Research Ethics Board at the University of British Columbia, British Columbia Cancer Agency (REB#H13-01478). Informed consent was obtained from all subjects.
Tonsilar B-cell isolation
Primary cells were isolated from normal fresh de-identified human tonsillectomy specimens (IRB#0805009767). Naïve B cells were labeled with anti-IgD-FITC followed by anti-FITC Microbeads (MACS). GC B-cells were first labeled with anti-CD77 (rat IgM), then mouse anti-rat IgM, IgG1 isotype, and followed with rat anti-mouse IgG1 Microbeads. Beads labeled cells were then enriched by passing through an autoMACS instrument using the PosselD protocol. The purity of the isolated cells is normally greater than 90%.
Generation of mice
Animal studies have been approved by Institutional Animal Care and Use Committees at MSKCC (protocol #: 07-01–002) and WCMC (protocol #: 2011-0034). The vavP-Bcl2 mouse model of FL (27) was adapted to the adoptive transfer approach using retrovirally transduced HPCs (28). Briefly, vavPBcl2 transgenic hematopoietic progenitor cells (HPCs) from fetal livers (E14.5) were transduced with retroviruses expressing short hairpin RNAs against Crebbp or empty vector and monitored the recipients for 200 days. 8–10 week old C57BL/6 females were used for all transplantation experiments.
The retroviral vectors are based on MSCV (29).
Mouse shRNAs were design using Designer of Small Interfering RNA.
shCrebbp #1: ATGCATCAGATTTGTGTTCTA
shCrebbp #2: CAGGACCTACGGAGTCATCTA
shCrebbp #3 CTGGTTGCCTATGCTAAGAAA
shEp300 #1: CAGGTACAAGCAAAGAATCAA
Mouse B220+ tumor sample preparation
B220+ cells were purified from mouse lymphomas by immunomagnetic enrichment with CD45R microbeads (Miltenyi Biotech).
Histology
Tissues were fixed overnight in formalin, embedded in paraffin blocks and sectioned. Tissue sections were stained with hematoxylin/eosin, B220, CD3, Ki67, and TUNEL following standard procedures.
Histologic score system
Semi-quantitative scale of organ infiltration was used based on the size of the infiltrate of atypical lymphocytes, corresponding to 1+ (size of the infiltrate <50% of 40X field); 2+ (size of the infiltrate >50% of 40X and < 100% of 40X field), 3+ (size of the infiltrate > one 40X field) for all organs, except spleen. Splenic infiltrate was evaluated based on the size of the white pulp and atypical lymphocytes outside of the white pulp, as follows: 1+ atypical lymphocytes occupying <60% of spleen surface; 2+ atypical lymphocytes occupying between 60% and 80% of spleen surface; 3+ atypical lymphocytes occupying >80% of spleen surface.
Flow cytometric analysis
Mouse tumor cell suspensions were stained as described (28). The antibodies used were B220/CD45R (BD Biosciences, 553092) or IgG1 (BD Biosciences, 560089) conjugated with APC, CD19 (BD Biosciences, 557399), IgM (BD Biosciences, 553409), IgD (BD Biosciences, 558597), GL7 (BD Biosciences, 561530) conjugated with phycoerythrin, and analyzed with BD LSRFortessa™ cell analyzer. Analysis was performed with FlowJo software (Tree Star).
IgVH rearrangement analysis
PCR to evaluate IgVH rearrangements was performed on cDNA of lymphoma cells with a set of forward primers that anneal to the framework region of the most abundantly used IgVH gene families and reverse primers located in the JH1–4 gene segments (30).
Somatic hypermutation
The genomic sequences from VH to the intron downstream of JH4 were PCR-amplified from cDNA of B220+ cells using degenerate forward primers for the different VH families (31) and a reverse primer (5'-AGGCTCTGAGATCCCTAGACAG-3' (32)) downstream JH4. Proofreading polymerase (Phusion High Fidelity, NEB) was used for amplification with PCR conditions previously published (31). Amplification products were isolated from agarose gel, cloned into pGEMT and single colonies were submitted to Sanger sequencing. Sequences were compared with IgBLAST (33).
Cell culture
Human DLBCL cell lines were grown in medium either containing 90% Iscove’s (OCI-Ly1, OCI-Ly7) or 90% RPMI1640 (FARAGE, MD901, OCI-Ly18, RIVA, OZ), 10% fetal bovine serum, 1% HEPES, 1% Glutamine, and 1% penicillin/streptomycin. OCI-Ly1, OCI-Ly7, and OCI-Ly18 were obtained from Ontario Cancer Institute in June 2011. FARAGE was obtained from ATCC in May 2011. MD901 was provided by Jose Angel Martinez-Climent, Centre for Applied Medical Research (CIMA), Pamplona, Spain in June 2011. OCI-Ly1, OCI-Ly7, OCI-Ly18, FARAGE, MD901 were recently authenticated by Biosynthesis (Lewisville, TX) using their STR Profiling and Comparison Analysis Service in February 2015. RIVA and OZ were authenticated by immunophenotyping in October 2016. These cell lines have also been routinely tested for mycoplasma contamination in the laboratory.
Immunoblot analysis
The protein level of CREBBP and HDAC3 in whole cell extracts were detected by western blotting using rabbit anti-CREBBP (Santa Cruz Biotechnology, sc-369) and rabbit anti-HDAC3 (Santa Cruz Biotechnology, sc-11417) respective. Levels of H3K27ac were detected by western blotting using histone extracts collected using the acid extraction method and blotted with Rabbit anti-H3K27ac (abcam, ab4729) and Rabbit anti-total H3 (abcam, ab1791).
Knocking down of human CREBBP and HDAC3
The shRNAs sequences used to knockdown CREBBP or HDAC3 expression in human cell lines are as following:
-
shRNAs against human CREBBP
shCREBBP-1: GTAACTCTGGCCATAGCTTAA
shCREBBP-2: GATGCATCAGATTTGCGTTCT
-
shRNA against human HDAC3 used in xenograft experiment
shHDAC3-GFP1 or Puro1 (same vector backbone but with different selection markers):
CCGGCAAGAGTCTTAATGCCTTCAACTCGAGTTGAAGGCATTAAGACTCTTGTTTTTTG
shHDAC3-GFP2:
CCGGGCCTGACAATGGTACCTATTACTCGAGTAATAGGTACCATTGTCAGGCTTTTTTG
-
Inducible shRNAs against human HDAC3 used in cell proliferation and apoptosis experiments:
ishHDAC3-1: TGCTGTTGACAGTGAGCGCCGGGGTTCAAGAAGCTTTCTATAGTGAAGCCACAGATGTATAGAAAGCTTCTTGAACCCCGTTGCCTACTGCCTCGGA
ishHDAC3-2: TGCTGTTGACAGTGAGCGAACCATGACAATGACAAGGAAATAGTGAAGCCACAGATGTATTTCCTTGTCATTGTCATGGTCTGCCTACTGCCTCGGA
mRNA-seq library preparation and sequencing analysis
Total RNA was extract from patient samples using Trizol (LifeTechnologies). RNA concentration was determined using Qubit (LifeTechnologies) and integrity was verified using Agilent 2100 Bioanalyzer (Agilent Technologies). 100 – 200 ng high quality total RNA from each sample was subjected to sequencing library preparation following the protocols of either Illumina TruSeq RNA kit (human cohort 1) or TruSeq Stranded Total RNA with Ribo-Zero kit (human cohort 2, mouse tumors, and MD901 CREBBP KD). Libraries were multiplexed (6 samples per lane) and sequenced on Illumina HiSeq 2500 as either 50-bp single-read runs (TruSeq RNA) or 50-bp paired-end runs (Stranded Total RNA).
Reads were mapped to the reference human (hg19) or mouse (mm10) genome sequence, using Tophat2 aligner (34) with the default parameters. The mRNA expression level for each gene was represented as FPKM, called by Cufflinks (35). For downstream RNAseq analysis, the comparison of gene expression between two groups was determined by the Student t test and the Benjamini–Hochberg adjusted P values.
ChIP and ChIP-seq library preparation and sequencing analysis
Histone ChIPs, including H3K4me1/2, H3K4me3 and H3K27ac ChIPs in human tonsilar Naïve and GC B-cells, human DLBCL cell lines and mouse splenic tumor cells, were performed as previously described (14). BCL6 and HDAC3 ChIP-qPCR as well as BCL6 and SMRT ChIPseq data was adapted from Hatzi et al (17). Briefly, 1–5 × 106 cells were fixed with 1% formaldehyde for 10 min at room temperature, followed by nuclei preparation and sonication (Branson Sonicator, Branson or Covaris E220, Covaris) to achieve enrichment of short fragment chromatin (~200 bp). 1–2 ug of antibody (H3K4me1/2, abcam, ab32356; H3K4me3, abcam, ab8580; H3K27ac, abcam, ab4729, all tested using histone-peptide array (Active Motif 13001) for specificity) was added to precleared chromatin lysate and incubated overnight at 4 °C. Enriched chromatin was collected by using protein-A beads (Roche) pull-down and subsequent reverse cross-linking and purification. ChIPseq libraries were prepared from 5–10 ng ChIP DNA following instructions of Illumina TruSeq ChIP Sample Prep Kit. Pooled libraries (6 samples per lane) were then sequenced on Illumina HiSeq 2500 as 50-bp single-end runs.
CREBBP ChIPseq in DLBCL cell line OCI-Ly7 was performed as described (17) with modifications. Briefly, 2 × 107 cells were fixed firstly with 2 mM of disuccinimidyl glutarate (DSG, ProteoChem, C1104) for 45 min at room temperature, followed by 1% formaldehyde for 10 min at room temperature. Fragmentation of fixed chromatin was performed by sonication of nuclei (Branson Sonicator, Branson) to achieve enrichment of short fragment chromatin (100 – 500 bp). 5 ug of antibody (CREBBP, Santa Cruz Biotechnology, sc-369; P300, Santa Cruz Biotechnology, sc-584) conjugated with Dynabeads protein A (Thermo Fisher Scientific) was added to precleared chromatin lysate and incubated overnight at 4 °C. Enriched chromatin was isolated through extensive wash steps and subsequent reverse cross-linking and purification. ChIPseq libraries were prepared from 5–10 ng ChIP DNA following instructions of Illumina TruSeq ChIP Sample Prep Kit. Libraries were then sequenced on Illumina HiSeq 2500 as 50-bp single-end runs.
Single-end reads were mapped to the reference human (hg18) or mouse (mm10) genome, using BWA aligner (36) with the default parameters. Only reads mapping uniquely to the genome with not more than 2 mismatches were retained for downstream analysis. Peak detection in each sample was performed with the ChIPseeqer program (37) and annotated to genes and/or promoters based on hg18 or mm10 refseq genes.
The consensus peaks from mouse H3K27Ac ChIP-seq for the normal and tumor samples were defined by the overlapping peaks which appear in >=3 out of the 4 normal samples, and >=4 out of the 6 tumors samples (shCrebbp) respectively. The consensus peaks from MD901 H3K27Ac ChIP-seq for the scramble shRNA (control) and shCREBBP samples were defined by the overlapping peaks which appear in >=2 out of the 3 samples for the control, and 4 out of 6 samples for shCREBBP (i.e., 2 out of 3 samples for both shRNA, or 3 out 3 samples for one shRNA and 1 out of 3 samples for the other shRNA), respectively.
Pathway analysis
The pathway analysis was performed as previously published (38). Briefly, the gene sets for canonical pathways and Gene Ontology (GO) terms were downloaded from the Molecular Signatures Database (MSigDB) (39) using C5 collection. The B-cell and lymphoid specific signatures were curated by Staudt laboratory (downloaded from (40)) or Melnick laboratory from previous publications. This database contains ~250 gene sets/signatures associated with normal lymphoid biology and lymphoid neoplasms. For the complete list of pathways please check Extended Data Table 1. Fisher’s Exact test was used to calculate enrichment p values for each of those gene sets and the BH method (41) was used for False Discovery Rate (FDR) control.
Gene set enrichment (GSEA) analysis
All the GSEA analysis results in this manuscript were generated from GSEA preranked mode (39). The input files are all the genes and their gene expression level logFC (log2 fold change) from each RNA-seq dataset, including (i) the three FL or DLBCL patient cohorts (CREBBP mutation vs WT), and (ii) the shRNA data for the mouse or human MD901 cell line (shCrebbp vs scramble shRNA). All the gene sets used in GSEA were curated by Staudt laboratory (40) or Melnick laboratory from previous publications.
Mutation discovery
Exome sequencing of cohort 1 using Aglient SureSelectXTHuman All Exon 50 MB Target Enrichment System for Illumina Pair-End Sequencing Library kit (Agilent Technologies, Santa Clara, CA), Raindance targeted re-sequencing of cohort 2 (RainDance Technologies, Billerica, MA), targeted re-sequencing of cohort 3 (347 tumor samples and 67 matched normal samples), Cell-of-origin calling of cohort3 by Lymph2Cx 20-gene GEP assay on the NanoString platform (NanoString Technologies, WA), as well as mutational calling algorithms were described previously in Ortega-Molina et al (14). Aglient Exome and Raindance platforms were cross-validated using DNA from DLBCL cell lines. Greater than 95% concordance was observed. Cohort 3 DLBCL targeted re-sequencing results were further validated by the Fluidigm Access Array system (Fluidigm Corporation, CA).
NS-DAD animal and experiment
NS-DAD mice were a generous gift from Dr. Mitchell Lazar (21). Age- and sex-matched mice were immunized intraperitoneally with 0.5 ml of a 2% sheep red blood cell (SRBC) suspension in PBS (Cocalico Biologicals), and sacrificed after 10 days. For staining of GC B cell populations, single-cell suspensions from mouse spleens were stained using the following fluorescent-labeled anti-mouse antibodies: PE-Cy7 conjugated anti-B220, APC conjugated anti-B220, PE conjugated anti-FAS, APC conjugated anti-CD38. DAPI was used for the exclusion of dead cells.
Cell proliferation assay
After DLBCL cell lines transduced with inducible shRNAs against Luciferase gene (Luc, control) and human HDAC3 (ishHDAC3-1, ishHDAC3-2) were treated with 0.2 µg/ml doxycycline (DOX) for 24 hours, cells were labeled with 2.5 µM Cell Proliferation Dye eFluor 670 (eBioscience, 65–0840), and cultured for additional 5 days. The fluorescence intensity of the dye (APC channel) in the GFP+ (FITC channel) cells was measured by flow cytometry right after labeling and every 24hr subsequently until the end of the experiment.
DLBCL xenotransplant models
Ten millions of puromycin selected, scramble or HDAC3 shRNA transduced human DLBCL cells (FARAGE or OCI-Ly7) were subcutaneously injected in the left flank of 8-week-old SCID mice. Tumor volume was monitored every 3–4 days using electronic digital callipers (Fisher Scientific, Pittsburgh, PA) in 2 dimensions when they became palpable. Mice were sacrificed by cervical dislocation under anaesthesia at the end of the experiment when tumors in the scramble shRNA group reached maximum size allowed by the animal protocol.
Fifteen millions of either scramble or HDAC3 shRNA transduced human DLBCL RIVA cells were subcutaneously injected in the flank of 8-week-old NSG mice. The shRNA vector in this experiment contains a GFP marker. The GFP positive cell percentage before injection was measured by flow cytometry. Tumor volume was monitored every 3–4 days using electronic digital callipers in 2 dimensions when they became palpable. Mice were sacrificed by cervical dislocation under anaesthesia at the end of the experiment when tumors in the scramble shRNA group reached maximum size allowed by the animal protocol. Single cell suspension was prepared from individual tumor, and was stained with anti-human CD19-PE (BD Biosciences, 555413) antibody and DAPI to determine the percentage of GFP positive cells in in the CD19 positive human B-cell population by flow cytometry.
siRNA Transfections
OCI-Ly1 cells transfected using Nucleofector 96-well Shuttle system (Lonza) with 1uM siRNA against BCL6 (HSS100968) or non-targeted siRNA (46–2001) (Stealth RNAi, Invitrogen).
siBCL6: 5′-CCAUUGUGAGAAGUGUAACCUGCAU-3′
Mixed lymphocyte reaction
MD901 and OCI-Ly18 B-lymphoblastoid cell lines (BLCL) were transduced with scramble or CREBBP shRNAs for 3 days and then treated with a selective HDAC3 inhibitor or a control compound for 96hr. BLCLs were irradiated (9000r 131Cs) added to 1.5×105 CD3+ T cells at a ratio of 10:1 T:BLCL in triplicate flat-bottom microwells. Cells were incubated for 5d at 37°C 5%CO2, after which T cell proliferation was assessed by Cell Titer Blue assay (Promega G8081).
Statistical methods
Sample sizes for comparisons between cell types or between mouse genotypes followed Mead's recommendations (42). Samples were allocated to their experimental groups according to their predetermined type (i.e., mouse genotype) and, therefore, there was no randomization. Investigators were not blinded to the experimental groups unless indicated. In the case in Fig. 1A, only mice that developed lymphomas were considered; mice that didn't develop lymphomas were censored and indicated with ticks in the Kaplan-Meier curves. Survival in mouse experiments was represented with Kaplan-Meier curves, and significance was estimated with the log-rank test. For contingency analysis we used the chi-squared exact test.
Data Deposition
The RNA-seq and ChIP-seq data are available at GEO database (SuperSeries: GSE79686; MD901 H3K27Ac ChIP-seq: GSE79639; MD901 RNA-seq: GSE79684; mouse H3K27Ac ChIP-seq: GSE79640; mouse RNA-seq: GSE79685).
Supplementary Material
Statement of Significance.
Our findings establish the tumor suppressor function of CREBBP in GC lymphomas in which CREBBP mutations disable acetylation and result in unopposed deacetylation by BCL6/SMRT/HDAC3 complexes at enhancers of B-cell signaling and immune response genes. Hence, inhibition of HDAC3 can restore the enhancer histone acetylation and may serve as a targeted therapy for CREBBP mutant lymphomas.
Acknowledgments
We thank the members in the Melnick and the Wendel groups for the thoughtful discussions related to this study. We thank the Weill Cornell Medical College Epigenomics core for performing the NGS library preparation and sequencing for this study. We thank the Rockefeller University Genomics Resource Center for performing NGS sequencing of CREBBP/P300 ChIP for this study. We thank the members of the MSK Antitumor assessment core, the MSK Laboratory of Comparative Pathology, the Weill Cornell Cell Sorting core, the Weill Cornell Leukemia Biorepository and Personalized Medicine Centre, the MSK Flow Cytometry core, and the MSK Molecular Cytology core. We thank Dr. Mitch Lazar from University of Pennsylvania for providing the NSDAD mouse model.
Financial support:
Y.J. was a Scholar of the American Society of Hematology. A.O.-M is supported by funding from The Leukemia & Lymphoma Society. H.G. is supported by a Junior Investigator award from the UCSF Academic Senate. H.Y. is supported by the 2016 AACR-Takeda Oncology Fellowship in Lymphoma Research, Grant Number 16-40-38-YING, and was supported by Ministry of Science and Technology, ROC. S.W. is supported by Postdoctoral fellowship from Swedish Research Council (VR) and by the 2015 AACR-Millennium Fellowship in Lymphoma Research, Grant Number 15-40-38-WANG. D.E., D.W.S, and R.D.G. are supported by a Terry Fox Program Project grant # 1023. D.W.S. is also funded by the BC Cancer Foundation. S. S. and J. W. Y. are supported by P01 CA23766 from the National Cancer Institute, NIH and Swim Across America. C.C. was supported by a fellowship from Ministry of Science and Technology, ROC. R.G.R. was supported by NIH grant R01CA178765. O.E. is supported by NSF CAREER and R01 CA194547. S.W.H. is supported by NCI R01-CA164605. H.G.W. is supported by the American Cancer Society grant RSG-13-048-01-LIB, the Lymphoma Research Foundation, Cycle for Survival, Functional Genomics Initiative, W.H. Goodwin and A. Goodwin and the Commonwealth Foundation for Cancer Research, Steven Greenberg, the Center for Experimental Therapeutics at Memorial Sloan Kettering Cancer Center, US National Institutes of Health (NIH) grants RO1CA183876-01 and 1R01CA19038-01 and Core Grant P30 CA008748 and P50 CA192937- 01A1. H.G.W. is a Scholar of the Leukemia and Lymphoma Society and is supported by an LLS SCOR. AM is supported by NIH R01 CA187109, LLS TRP 6457-15, LLS SCOR#7012-16, and the Chemotherapy Foundation.
Footnotes
Disclosure of Potential Conflicts of Interest:
E. Holson is chief scientific officer of KDAc Therapeutics, Inc. No potential conflicts of interest were disclosed by other authors.
Authors’ Contributions
Conception and design: Y. Jiang, A. Ortega-Molina, H. Geng, H. Wendel, A.M. Melnick
Acquisition of data: Y. Jiang, A. Ortega-Molina, H. Ying, D. McNally, L. Wang, X.A. Ena, D. Poloway, S. Parsa, S. Wang, K.R. Stengel, D. Ennishi, K. Hatzi, C. Chu, S. Sharma
Analysis and interpretation of data: Y. Jiang, A. Ortega-Molina, H. Geng, H. Ying, A.S. Doane, M. Teater, C. Meydan, D. Ennishi, D.W. Scott, J.W. Young, K. Hatzi, C. Chu, R.G. Roeder, R. Shaknovich, S.W. Hiebert, R.D. Gascoyne, W. Tam, O. Elemento, H. Wendel, A.M. Melnick
Writing, review, and/or revision of the manuscript: Y. Jiang, A. Ortega-Molina, H. Geng, H. Ying, S.W. Hiebert, O. Elemento, H. Wendel, A.M. Melnick
Administrative, technical, or material support: H. Geng, D. McNally, Z. Li, J.E. Kranz, E. Holson, W. Tam
Study supervision: Y. Jiang, O. Elemento, H. Wendel, A.M. Melnick
References
- 1.Al-Tourah AJ, Gill KK, Chhanabhai M, Hoskins PJ, Klasa RJ, Savage KJ, et al. Population-based analysis of incidence and outcome of transformed non-Hodgkin's lymphoma. J Clin Oncol. 2008;26:5165–5169. doi: 10.1200/JCO.2008.16.0283. [DOI] [PubMed] [Google Scholar]
- 2.Pasqualucci L, Dominguez-Sola D, Chiarenza A, Fabbri G, Grunn A, Trifonov V, et al. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 2011;471:189–195. doi: 10.1038/nature09730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat Genet. 2011;43:830–837. doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lohr JG, Stojanov P, Lawrence MS, Auclair D, Chapuy B, Sougnez C, et al. Discovery and prioritization of somatic mutations in diffuse large B-cell lymphoma (DLBCL) by whole-exome sequencing. Proc Natl Acad Sci U S A. 2012;109:3879–3884. doi: 10.1073/pnas.1121343109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Grubor V, Love CL, Banerjee A, Richards KL, Mieczkowski PA, et al. Genetic heterogeneity of diffuse large B-cell lymphoma. Proc Natl Acad Sci U S A. 2013;110:1398–1403. doi: 10.1073/pnas.1205299110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, et al. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature. 2011;476:298–303. doi: 10.1038/nature10351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A. 2015;112:E1116–E1125. doi: 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lunning MA, Green MR. Mutation of chromatin modifiers; an emerging hallmark of germinal center B-cell lymphomas. Blood Cancer J. 2015;5:e361. doi: 10.1038/bcj.2015.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loeffler M, Kreuz M, Haake A, Hasenclever D, Trautmann H, Arnold C, et al. Genomic and epigenomic co-evolution in follicular lymphomas. Leukemia. 2015;29:456–463. doi: 10.1038/leu.2014.209. [DOI] [PubMed] [Google Scholar]
- 12.Oricchio E, Nanjangud G, Wolfe AL, Schatz JH, Mavrakis KJ, Jiang M, et al. The Eph-receptor A7 is a soluble tumor suppressor for follicular lymphoma. Cell. 2011;147:554–564. doi: 10.1016/j.cell.2011.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell. 2013;23:677–692. doi: 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med. 2015;21:1199–1208. doi: 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basso K, Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol. 2015;15:172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- 16.Scott DW, Wright GW, Williams PM, Lih CJ, Walsh W, Jaffe ES, et al. Determining cell-of-origin subtypes of diffuse large B-cell lymphoma using gene expression in formalin-fixed paraffin-embedded tissue. Blood. 2014;123:1214–1217. doi: 10.1182/blood-2013-11-536433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, et al. A Hybrid Mechanism of Action for BCL6 in B Cells Defined by Formation of Functionally Distinct Complexes at Enhancers and Promoters. Cell Rep. 2013;4:578–588. doi: 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polo JM, Ci W, Licht JD, Melnick A. Reversible disruption of BCL6 repression complexes by CD40 signaling in normal and malignant B cells. Blood. 2008;112:644–651. doi: 10.1182/blood-2008-01-131813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell. 2007;12:280–292. doi: 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 20.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol. 2007;8:705–714. doi: 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 21.You SH, Lim HW, Sun Z, Broache M, Won KJ, Lazar MA. Nuclear receptor co-repressors are required for the histone-deacetylase activity of HDAC3 in vivo. Nat Struct Mol Biol. 2013;20:182–187. doi: 10.1038/nsmb.2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wagner FF, Lundh M, Kaya T, McCarren P, Zhang YL, Chattopadhyay S, et al. An Isochemogenic Set of Inhibitors To Define the Therapeutic Potential of Histone Deacetylases in beta-Cell Protection. ACS Chem Biol. 2016;11:363–374. doi: 10.1021/acschembio.5b00640. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med. 2015;21:1190–1198. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rimsza LM, Roberts RA, Miller TP, Unger JM, LeBlanc M, Braziel RM, et al. Loss of MHC class II gene and protein expression in diffuse large B-cell lymphoma is related to decreased tumor immunosurveillance and poor patient survival regardless of other prognostic factors: a follow-up study from the Leukemia and Lymphoma Molecular Profiling Project. Blood. 2004;103:4251–4258. doi: 10.1182/blood-2003-07-2365. [DOI] [PubMed] [Google Scholar]
- 25.Mottok A, Woolcock B, Chan FC, Tong KM, Chong L, Farinha P, et al. Genomic Alterations in CIITA Are Frequent in Primary Mediastinal Large B Cell Lymphoma and Are Associated with Diminished MHC Class II Expression. Cell Rep. 2015;13:1418–1431. doi: 10.1016/j.celrep.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 26.Cycon KA, Mulvaney K, Rimsza LM, Persky D, Murphy SP. Histone deacetylase inhibitors activate CIITA and MHC class II antigen expression in diffuse large B-cell lymphoma. Immunology. 2013;140:259–272. doi: 10.1111/imm.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egle A, Harris AW, Bath ML, O'Reilly L, Cory S. VavP-Bcl2 transgenic mice develop follicular lymphoma preceded by germinal center hyperplasia. Blood. 2004;103:2276–2283. doi: 10.1182/blood-2003-07-2469. [DOI] [PubMed] [Google Scholar]
- 28.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, et al. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–337. doi: 10.1038/nature02369. [DOI] [PubMed] [Google Scholar]
- 29.Dickins RA, Hemann MT, Zilfou JT, Simpson DR, Ibarra I, Hannon GJ, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
- 30.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehlich A, Martin V, Muller W, Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr Biol. 1994;4:573–583. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- 32.Gostissa M, Bianco JM, Malkin DJ, Kutok JL, Rodig SJ, Morse HC, 3rd, et al. Conditional inactivation of p53 in mature B cells promotes generation of nongerminal center-derived B-cell lymphomas. Proc Natl Acad Sci U S A. 2013;110:2934–2939. doi: 10.1073/pnas.1222570110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ye J, Ma N, Madden TL, Ostell JM. IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 2013;41:W34–W40. doi: 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giannopoulou EG, Elemento O. An integrated ChIP-seq analysis platform with customizable workflows. BMC bioinformatics. 2011;12:277. doi: 10.1186/1471-2105-12-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geng H, Brennan S, Milne TA, Chen WY, Li Y, Hurtz C, et al. Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia. Cancer Discov. 2012;2:1004–1023. doi: 10.1158/2159-8290.CD-12-0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaffer AL, Wright G, Yang L, Powell J, Ngo V, Lamy L, et al. A library of gene expression signatures to illuminate normal and pathological lymphoid biology. Immunol Rev. 2006;210:67–85. doi: 10.1111/j.0105-2896.2006.00373.x. [DOI] [PubMed] [Google Scholar]
- 41.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;57:289–300. [Google Scholar]
- 42.Festing MFW. Laboratory Animals Ltd., Royal Society of Medicine (Great Britain). The design of animal experiments : reducing the use of animals in research through better experimental design. London: Royal Society of Medicine; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.