Abstract
A combination of silver nanoparticles (AgNPs) and an antibiotic can synergistically inhibit bacterial growth, especially against the drug-resistant bacteria Salmonella typhimurium. However, the mechanism for the synergistic activity is not known. This study chooses four classes of antibiotics, β-lactam (ampicillin and penicillin), quinolone (enoxacin), aminoglycoside (kanamycin and neomycin), and polykeptide (tetracycline) to explore their synergistic mechanism when combined with AgNPs against the multidrug-resistant bacterium Salmonella typhimurium DT 104. Enoxacin, kanamycin, neomycin, and tetracycline show synergistic growth inhibition against the Salmonella bacteria when combined with AgNPs, while ampicillin and penicillin do not. UV–vis and Raman spectroscopy studies reveal that all these four synergistic antibiotics can form complexes with AgNPs, while ampicillin and penicillin do not. The presence of tetracycline enhances the binding of Ag to Salmonella by 21% and Ag+ release by 26% in comparison to that without tetracycline, while the presence of penicillin does not enhance the binding of Ag or Ag+ release. This means that AgNPs first form a complex with tetracycline. The tetracycline–AgNPs complex interacts more strongly with the Salmonella cells and causes more Ag+ release, thus creating a temporal high concentration of Ag+ near the bacteria cell wall that leads to growth inhibition of the bacteria. These findings agree with the recent findings that Ag+ release from AgNPs is the agent causing toxicity.
Graphical abstract
INTRODUCTION
Antibacterial resistance has become a global issue. According to the 2014 Antimicrobial Resistance Global Report, some bacteria have begun to develop resistance even to the third generation of antibiotics.1 Since the 1980s, no new antibacterial drugs have been developed, making the situation even more urgent.
Bulk silver has a long history of being used as an antimicrobial.2,3 In the past decade, combining silver nanoparticles (AgNPs) with antibiotics has been considered a potential method to overcome bacterial drug resistance.4 AgNPs used for this purpose have been synthesized using different methods and modified with many different antibacterial agents to achieve high antimicrobial activity.5–12 AgNPs are also effective against multidrug-resistant bacteria.13,14 Recently, some publications presented evidence for the synergistic antibacterial activity of AgNPs combined with conventional antibiotics, especially against multidrug-resistant bacteria including Staphylococcus aureus and Escherichia coli.15–17 The activity of selected antibiotics was enhanced to different degrees in the presence of AgNPs, regardless of the synthetic method.18 Most researchers studying antibacterial activity used the disk diffusion method to determine MIC/IC50/IC80 values or zones of inhibition. Hwang et al.16 found that combination of AgNPs with ampicillin, chloramphenicol, and kanamycin against various pathogenic bacteria inhibits the formation of biofilm that is associated with the resistance to antimicrobial agents and chronic bacterial infections. Other mechanistic studies19–21 pointed out that AgNPs generate hydroxyl radicals to enhance the bactericidal effect. Li et al.22 hypothesized that AgNPs and amoxicillin either kill the bacteria with a different mechanism individually or they form a complex in that amoxicillin molecules surround the AgNPs core. While AgNPs’ ability as a drug carrier may play a role for the synergistic effect, the release of Ag+ is also considered to enhance the antibacterial activity. Chelation of Ag+ is believed to prohibit DNA unwinding,23 leading to bacterial cell damage, but a satisfactory synergistic mechanism is lacking.
In this study, we examined the synergistic antibacterial mechanism of four different classes of conventional antibiotics in combination with AgNPs, 1) β-lactam (ampicillin and penicillin), 2) quinolone (enoxacin), 3) aminoglycoside (kanamycin A and neomycin), and 4) polykeptide (tetracycline), against the multidrug-resistant bacterium Salmonella typhimurium DT 104 (abbreviated as Salmonella).24 We hypothesize that the synergistic antibacterial activity correlates with the complex formation between AgNPs and the antibiotic. Since Ag+ is a known antibacterial,25,26 the release of Ag+ from AgNPs into solution was also examined under various experimental conditions.
EXPERIMENTAL SECTION
Materials
Multidrug-resistant Salmonella (ATCC 700408) was obtained from the American Type Culture Collection (Manassas, VA). Silver nitrate, sodium borohydride, sodium citrate, NaOH, tetracycline hydrochloride (TET), neomycin sulfate (NEO), kanamycin sulfate (KAN), tryptic soy broth (TSB), and tryptic soy agar (TSA) were purchased from Sigma-Aldrich (St. Louis, MO). Penicillin G sodium salt (PEN) and nitric acid (trace metal grade, 67%–70%) were from Fisher Scientific (Houston, TX). Ampicillin sodium salt (AMP) was from Research Products International Corp (Prospect, IL), and enoxacin hydrochloride (ENO) was from MP Biomedicals LLC (Solon, OH). The structures of the antibiotics are shown in Figure SI 1.
AgNPs Synthesis and Interaction with Antibiotics
Spherical AgNPs of 29.8 ± 6.4 nm in diameter were prepared according to the citrate reduction method as described in our previous reports.27–29 Sodium citrate was used as both the reducing agent and the stabilizer for AgNPs. The synthesized AgNPs were purified by three centrifugations at 5000 rpm for 1 h each to remove residual Ag+ and then resuspended in nanopure water. The thus-prepared AgNPs remain stable without aggregation for 3 months, but they are freshly prepared and used for experiments since Ag+ release from AgNPs occurs during long-term storage.30 The size of the purified AgNPs was determined to be 29.8 ± 6.4 nm by TEM (JEOL 2100). To determine the concentration of the AgNPs, an AgNPs solution was added with nitric acid to oxidize Ag atoms to Ag+ for determination of total concentration of Ag by ICP-MS (Varian 820-MS).
The interaction of AgNPs with the antibiotics was investigated by addition of each antibiotic separately to an AgNPs solution (50 μM). The antibiotic/AgNPs ratio varied from 0.1 to 100 or with the concentration of the antibiotic of 5, 50, 500, and 5000 μM. The spectra were recorded by UV–vis and Raman spectroscopy after mixing for 2 h. Raman spectra were recorded using 670 nm as excitation wavelength via a miniaturized QE65000 Scientific-grade Spectrometer (Ocean Optics) equipped with TE cooled 2048 pixel CCD as detector and interfaced to a computer via a USB port. The spectral response range was 220–3600 cm−1. The UV–vis absorption spectra were recorded by a UV-2600 spectrophotometer (Shimadzu, Japan) with 0.9 mL semimicro cuvette.
Bacterial Culture and Treatment with Antibiotics and in Combination with AgNPs
Growth of Salmonella was carried out as described in our previous report.24 Briefly, 50 μL of the bacterium was removed from a frozen (‒80 °C) sample and added to 12 mL of TSB, followed by incubation in a C25 class incubator with continuous agitation at 220 rpm for 10 h at 37 °C. Inoculums of about 1 × 108 colony forming units per mL (CFU/mL) were achieved.31 The bacterial solution was further diluted to different concentrations for treatments.
Different concentrations of an antibiotic of 0.5, 2, 8, and 16 μM, in the presence or absence of 50 μM AgNPs (in Ag atoms), were tested to evaluate the possible synergistic effect. The Salmonella (1 × 105 CFU/mL) in PBS was treated with a desired concentration of AgNPs, AgNO3, an individual antibiotic, or the combination of the antibiotic with AgNPs for 2 h. Then, 100 μL of the treated bacterial solution was taken to be plated, followed by incubation for 24 h and colony counting as described before.24 As a control, tetracycline was chosen as a representative to confirm whether Ag+ has a synergistic antibacterial effect when combined with antibiotics. Combinations of 5 μM Ag+ with 0, 0.5, 2, 8, and 16 μM of tetracycline were tested for inhibition of the 1 × 105 CFU/mL bacteria cells. This indicated that all groups inhibited 42.7%–46.8% of bacteria growth with no significant differences (Figure SI 2), demonstrating there is no synergistic effect between Ag+ and tetracycline.
Binding of AgNPs to Salmonella in the Presence or Absence of an Antibiotic
AgNPs (1 μM) in suspension were mixed with 0, 1, and 10 μM of each antibiotic for at least 30 min. Then, a Salmonella solution was added and shaken in an incubator shaker at 37 °C for 2 h. The final Salmonella concentration was 1 × 107 CFU/mL unless otherwise stated. To separate free AgNPs from those tightly interacted with or internalized in the Salmonella cells, the solutions were centrifuged at 2000 rpm for 15 min.32 It is confirmed that 94 ± 5% of the bacteria were brought to pellet under this centrifugation condition while 99 ± 4% of AgNPs (in case of 1 μM, examined by ICP-MS) remained in the supernatant (Table SI 1). The pellets were collected and went through nitric acid treatment. The resultant solution was diluted until the estimated Ag+ concentration ranged from 20 to 100 ppb. The diluted solution was centrifuged at 5000 rpm for 30 min to remove the bacterial residue before ICP-MS analysis. All the Ag binding experiments were repeated at least seven times until consistent results were achieved. The average binding percent of Ag was obtained.
Release of Ag+ from AgNPs in the Presence or Absence of Antibiotic and/or Salmonella
The release of Ag+ from AgNPs into solution was examined by ICP-MS to further investigate the role that Ag+ plays for the synergistic effect. In the absence of Salmonella, 10 μM of each of the six antibiotics were mixed with 1 or 50 μM of AgNPs and allowed to interact for 2 h. In the presence of Salmonella (1 × 107 CFU/mL), 1 or 10 μM tetracycline was mixed with 1 or 50 μM AgNPs, respectively, while AgNPs alone were used as a control. After interaction for 2 h, the mixtures were centrifuged at 12,000 rpm for 2 h so that the nanoparticles were removed as pellets while Ag+ remained in the supernatant. The supernatant solution was then analyzed by ICP-MS to determine the concentration of Ag+.
Statistical Analysis
To confirm statistical significance between tetracycline, penicillin, or the negative control without any added antibiotic, one-way ANOVA in SPSS software was used. A p < 0.05 is used for significant responses.
RESULTS AND DISCUSSION
Synergistic Antibacterial Activity of the Selected Six Antibiotics in Combination with AgNPs
The chosen Salmonella is known to resist antibiotics, such as penicillin and tetracycline.33–35 As shown in Figure 1, all six antibiotics do not effectively inhibit the growth of Salmonella when used alone in the chosen concentration range, confirming the resistance of the Salmonella to these antibiotics. Controls show that AgNPs alone inhibits 10% of the Salmonella growth. When combined with a β-lactam antibiotic, penicillin or ampicillin, the growth inhibition at the highest concentration (16 μM) of any of these two antibiotics is less than 20% when compared with the control. Therefore, there is no synergistic effect of ampicillin or penicillin when combined with AgNPs against Salmonella.
Figure 1.
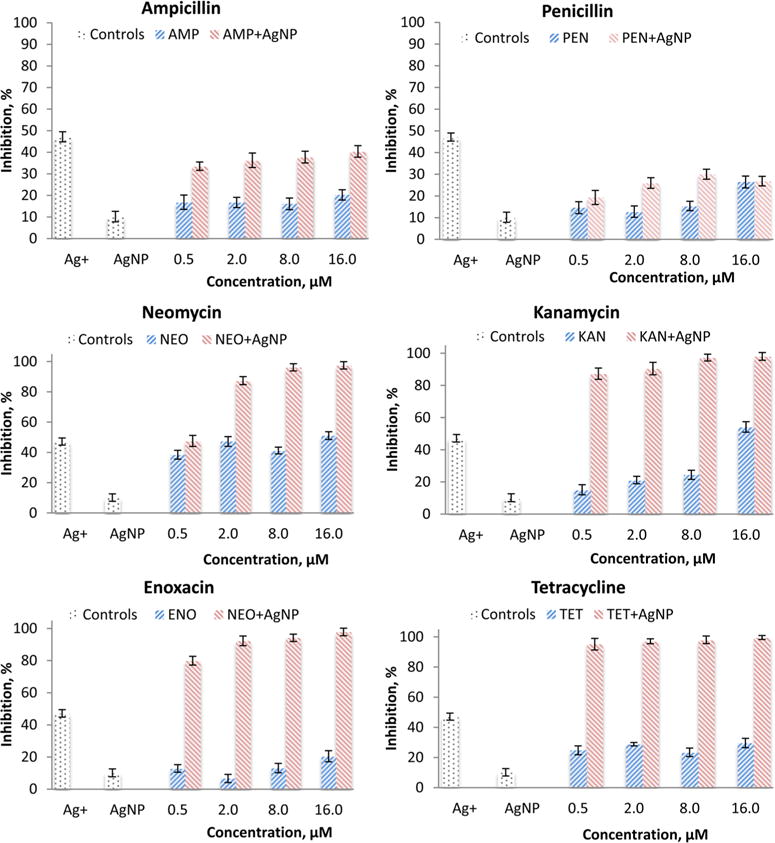
Growth inhibition of Salmonella typhimurium DT104 after exposure to an antibiotic at 0, 0.5, 2, 8, 16 μM in combination with AgNPs (50 μM) for 2 h. Ag+ is the positive control for AgNO3 at 5 μM.
However, Salmonella growth is greatly inhibited by the combination of AgNPs with tetracycline, enoxacin, neomycin, or kanamycin. Even at the lowest tested concentration of 0.5 μM of any antibiotics (except neomycin) in combination with AgNPs, the inhibition is much greater than AgNPs alone or the antibiotic alone. At higher neomycin concentrations (1.0 to 10 μM), the combination of AgNPs with neomycin also inhibits much more than AgNPs alone or neomycin alone. The strongest inhibition is observed with tetracycline–AgNPs. The inhibition is nearly 100% at the antibiotic concentration of 8 or 16 μM. Therefore, all these four antibiotics have synergistic effects when combined with AgNPs against Salmonella in a concentration-dependent manner. The IC50 values were determined to be 0.15 and 0.72 μM for tetracycline and neomycin, respectively, when combined with AgNPs.
Interaction between AgNPs and Antibiotics
UV–vis absorption spectra of AgNPs alone, each antibiotic alone, and AgNPs–antibiotic combinations are shown in Figure 2. AgNPs have a characteristic extinction at around 392 nm. Kanamycin, ampicillin, and neomycin have no absorption in the range of 300–700 nm. Enoxacin and penicillin have no absorption above 380 nm, which is distinguishable from the peak of AgNPs at 392 nm. Tetracycline has an absorption extending to 500 nm and interferes with the absorption peak of AgNPs at high concentrations.
Figure 2.
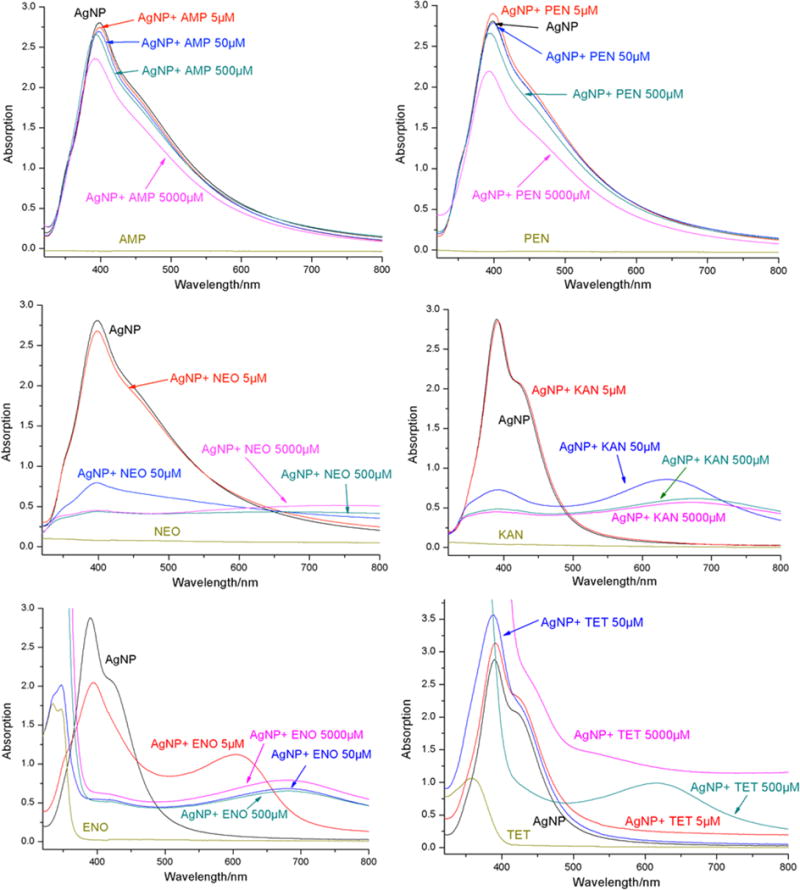
UV–vis spectra of AgNPs (50 μM) in the absence and presence of ampicillin (AMP), penicillin (PEN), neomycin (NEO), kanamycin (KAN), enoxacin (ENO), and tetracycline (TET) at 5, 50, 500, and 5000 μM.
Upon addition of ampicillin or penicillin, the extinction at 392 nm of AgNPs slightly decreases in the entire concentration range from 5 to 5000 μM but does not lead to any significant aggregation of AgNPs. Adding 5 μM enoxacin to an AgNPs solution causes a decrease in AgNPs’ extinction at 392 nm, while another broad band at 615 nm appears, which is attributed to the aggregated AgNPs.36 When the enoxacin concentration increases to 50 μM and higher, the AgNPs’ extinction at 392 nm falls sharply and red-shifts and finally forms a very broad band around 700 nm. This demonstrates that enoxacin forms a complex with AgNPs leading to aggregation of AgNPs. Similar phenomena are observed when neomycin or kanamycin is added to AgNPs solutions. Although introduction of 5 μM of either antibiotic has no effect on the extinction of AgNPs, an increase in the antibiotic concentration to 50 μM abruptly causes aggregation of AgNPs. Judging from the speedy color change of AgNPs solutions upon addition of the two antibiotics, addition of neomycin leads to quicker aggregation than kanamycin.
For tetracycline, no significant change of the AgNPs extinction is observed for concentrations below 50 μM. Addition of 500 μM tetracycline causes a broad band at 625 nm due to aggregation of AgNPs. Simultaneously, the characteristic peak of AgNPs at 392 nm greatly decreases, although the tetracycline absorption below 500 nm offsets the extinction level at this wavelength range. The broadband dominates the spectrum when tetracycline concentration further increases. The spectrum of 50 μM AgNPs mixed with 500 μM tetracycline signifies the beginning of aggregation of AgNPs. This means that all these four antibiotics form complexes with AgNPs that cause aggregation of the AgNPs.
Raman spectroscopy was used to further verify the interaction between AgNPs and individual antibiotic molecules. As shown in Figure 3, there is no significant Raman activity from AgNPs or from any of the individual antibiotics, except for tetracycline, which exhibits weak Raman signals. No Raman enhancement is observed when AgNPs are combined with ampicillin and penicillin at any test concentrations. This implies that the antibiotics do not interact with AgNPs strongly to replace the stabilizer molecules (e.g., citrate) on the surface of AgNPs. However, no surface resonance enhancement was observed in the case of neomycin even when its concentration is 5000 μM. The reason may be that neomycin has a very weak Raman scattering, especially when excited with a 760 nm laser light source. Therefore, the enhancement is not detectable under current experimental conditions.
Figure 3.
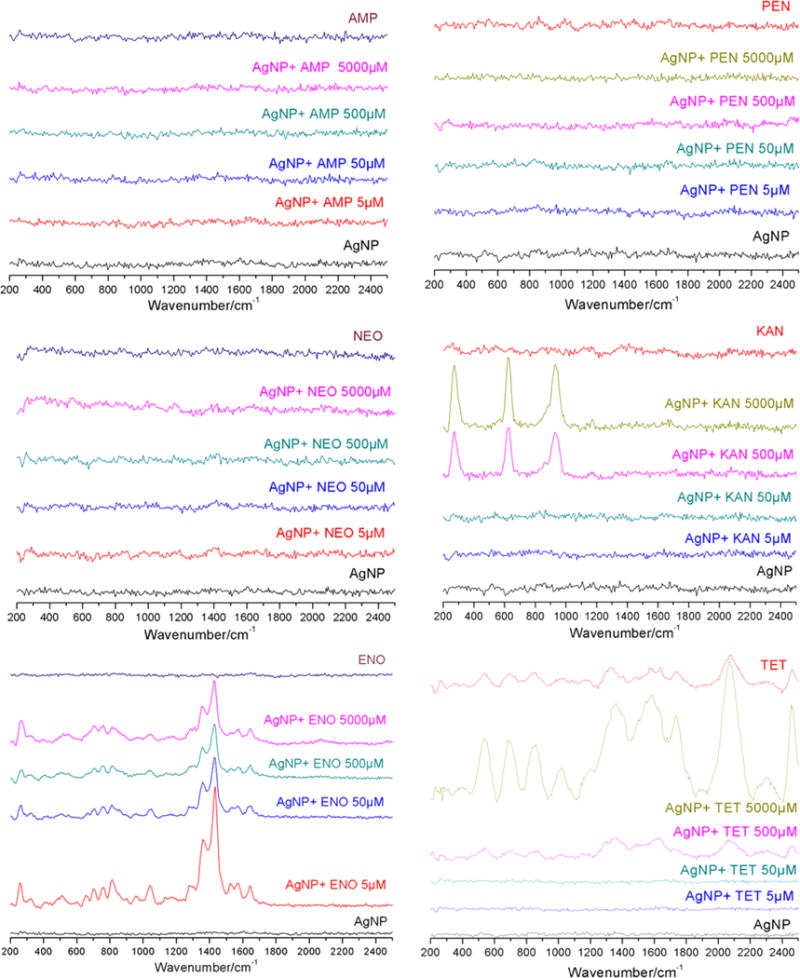
Raman spectra of AgNPs (50 μM) in the presence of ampicillin (AMP), penicillin (PEN), neomycin (NEO), kanamycin (KAN), enoxacin (ENO), and tetracycline (TET) at concentrations of 5, 50, 500, and 5000 μM.
In the presence of AgNPs, multiple Raman bands of enoxacin are observed around 1400 cm−1 that can be assigned to stretching of the pyridine ring.37 The Raman signals for kanamycin at 270, 620, and 890 cm−1 are attributed to skeletal deformation of the tetrahydropyran rings, O–Ag stretching, and skeletal stretching of tetrahydropyran rings, respectively.37–39 For tetracycline, the band near 1600 cm−1 is the typical C═O stretching vibration. The signals between 1230 and 1360 cm−1 are located in the region of C–C and C–N stretching vibrations.40 The signals at 500 to 870 cm−1 regions are characteristic for cyclohexene ring vibration and breathing.37 The band at 1000 cm−1 is assigned to a phenyl ring breathing vibration. The intense band at 2010 cm−1 could be from the pseudoantisymmetric C═O stretching when oxygen interacts with the Ag atoms of AgNPs.39
The spectral information from Raman and UV–vis spectroscopy clearly demonstrate that enoxacin, kanamycin, neomycin, and tetracycline interact with AgNPs strongly, replacing the surface citrate molecules and forming antibiotic-AgNPs complex(es). Enoxacin, kanamycin, and neomycin readily cause aggregation of AgNPs. Aggregation occurs at higher concentrations for tetracycline, while no such effects were observed for ampicillin and penicillin. It should be noted that enoxacin, kanamycin, neomycin, and tetracycline are also the ones which have synergistic antibacterial activity when combined with AgNPs. These results clearly point out that the synergistic effect of AgNPs–antibiotic combination against Salmonella is mediated by complexes that are produced from the interaction between AgNPs and the antibiotics, instead of individual or additive action of AgNPs or antibiotic. In other words, the bacterial cells are killed more effectively by “AgNPs-antibiotic complexes”. However, the nature of the interaction between the AgNPs and the antibiotics needs further investigation.
AgNPs Binding to Salmonella in the Presence or Absence of Antibiotics
Among the six antibiotics examined, synergistic effects were observed for enoxacin, kanamycin, neomycin, and tetracycline that were also found to strongly bind to AgNPs. Enoxacin, kanamycin, and neomycin induced aggregation of AgNPs at low antibiotic/AgNPs molar ratios. In the case of tetracycline, aggregation of AgNPs occurred only at high concentrations of tetracycline; therefore, tetracycline of low concentration (up to 10 μM) was chosen as a representative to quantitatively study the binding of AgNPs to Salmonella cells. Control titration of tetracycline into an AgNPs solution is carried out to confirm that tetracycline solutions of tested concentrations are safe for AgNPs aggregation (Figure SI 3). Because of the substantial aggregation, it was not feasible to separate AgNPs attached to bacteria cells from those unattached AgNPs aggregates. Also, aggregation will influence the Ag+ release kinetics due to the changes in the available surface areas of AgNPs.41 Penicillin did not cause any aggregation of AgNPs nor did it show any synergistic inhibition of the Salmonella growth and thus was chosen as a negative control.
To better understand the binging of Ag (either in form of AgNPs or Ag+), the ratio of AgNPs to Salmonella cells was estimated in different concentrations. The number of Ag atoms per AgNP was calculated by the equation,
where ρ is the density of Ag (10.49 g/cm3), M is the atomic mass of Ag (107.87 g/mol), and d is the average diameter of AgNPs (30 nm).42,43 This yields 8.28 × 105 Ag atoms per nanoparticle. Thus, the number of Ag nanoparticles per milliliter of solution (1 μM AgNPs in Ag atoms) is 1 × 10−6 mole Ag L−1 × 6.023 × 1023 Ag atoms mol−1 ÷ 8.28 × 105 Ag atoms AgNP−1 × 1 × 10−3 mL/L = 7.3 × 108 AgNP/mL. Thus, the ratio of AgNPs to Salmonella cells is about 73:1 in a medium with 1 μM AgNPs and 1 × 107 CFU/mL Salmonella cells.
AgNPs (1 μM) were tested for binding to Salmonella cells (1 × 107 CFU/mL) when combining with 0, 1, and 10 μM of tetracycline or penicillin. Percent Ag bindings is shown in Figure 4. The binding of Ag in the presence of 1 μM tetracycline is 42.2%, and it is 42.4% for the same concentration of penicillin. These values are not significantly different from that in the absence of the antibiotics (41.5%) (p > 0.05). In the presence of 10 μM of tetracycline, the percent binding significantly increases to 50.3% (p < 0.05) but that with penicillin decreases to 38.3%. This indicates that the presence of 10 μM of tetracycline facilitates the interaction between AgNPs and Salmonella cells resulting in more Ag to be attached to the cells, which refers to the total Ag attached to the cell surface and internalized as AgNPs or Ag+.
Figure 4.
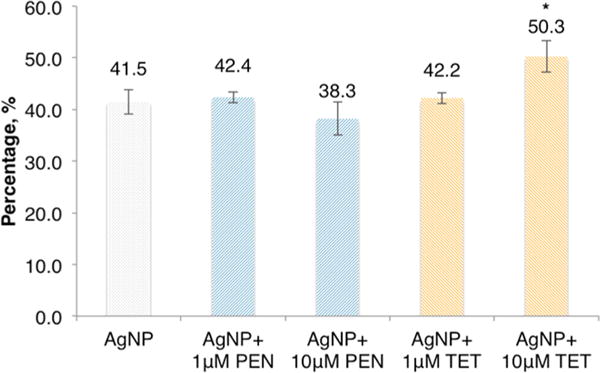
Percent binding of Ag to Salmonella (1 × 107 CFU/mL) in the absence and presence of tetracycline (TET) and penicillin (PEN). Concentration of AgNPs is 1 μM (73 AgNPs/cell). Values are mean ± SD from seven experiments (n = 7). * Significance is confirmed by one-way ANOVA, p < 0.05.
Different concentrations of Salmonella were used to further confirm the effect of the concentrations of the bacterial cells on the Ag binding. As shown in Figure 5, the Ag binding increases as the concentration of the Salmonella increases from 0.25 to 4 × 107 CFU/mL. However, the increase is not linear. It is also confirmed that the combination of tetracycline can enhance the binding of Ag, and the enhancement varies at different Salmonella concentrations. The presence of 1 μM of tetracycline can increase the binding by 0.6%–7.6%, while the presence of 10 μM of tetracycline can lead to a 3.6%–13.8% increase when compared to AgNPs alone. The AgNP binding percentage also changes when the concentration of Salmonella cells varies, confirming that the change of binding amount is actually caused by the cells instead of by the centrifugation.
Figure 5.
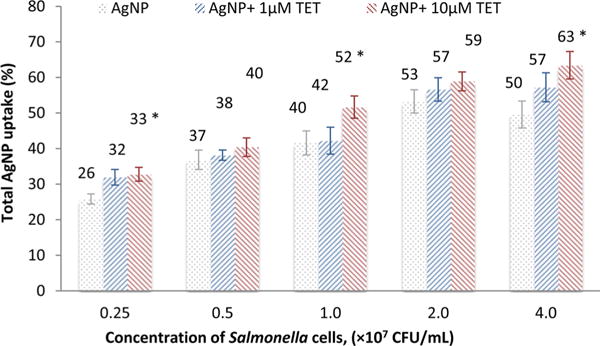
Effect of Salmonella concentrations on the binding of Ag in the presence of tetracycline (TET). * Significance is confirmed by ANOVA (p < 0.05) compared to control group with AgNPs alone.
These results confirm that the 73:1 of AgNPs to Salmonella cells is an ideal ratio for quantitative study of Ag binding since a relatively small change of AgNPs may be detected. To illustrate, 41.5% of Ag binding by Salmonella cells (without added tetracycline) in the control group represents 30 AgNPs per cell, while 51.7% of Ag binding (with 10 μM tetracycline) counts 37 AgNPs per cell. The 7 AgNPs increase represents a 23% increase. However, this 7 AgNPs difference would not be detectable if the AgNPs/Salmonella cells ratio were 7300. This would be the case if we used AgNPs at 100 μM (about 7300 AgNPs/cell).
Combining 50 μM AgNPs with 1 μM tetracycline kills 90% of the bacteria, while the same combination with 1 μM penicillin only kills 15% (Figure 1). The percent binding of Ag in these two cases is essentially the same at ~42%, indicating that the Ag binding may not be the only factor causing the growth inhibition of Salmonella cells. At a higher concentration of the antibiotics of 10 μM, the Ag binding percentages are 50% and 38% for tetracycline and penicillin, respectively (Figure 4), but bacterial growth inhibitions are 95% and 15%, respectively. Although it has been proposed that the antibacterial mechanism of AgNPs is due to the binding of AgNPs to the bacterial cell wall, thus impairing the membrane structure and enzyme activity,10,11,44 the results here show that bacterial binding of AgNPs is not the only reason that AgNPs cause bacterial growth inhibition.
Ag+ release
It has been reported that Ag+ is the culprit for the antibacterial activity of AgNPs,20,45 while intact AgNPs may or may not directly induce toxicity.25,26 It has also been reported that binding of AgNPs to the bacterial cells led to collapse of cell wall and bacterial death.10,11,44 In this study, we examined Ag+ release from AgNPs in the presence of the antibiotics and the Salmonella in cell culture medium to determine the effect of Ag+ release on the inhibition of bacterial growth. Ultracentrifugation (12,000 rpm) for 1 h was used to separate intact AgNPs from the solution, leaving the Ag+ in the supernatant solution. AgNPs within a range of 23 to 37 nm were effectively settled by centrifugation at the bottom of the centrifuge tube. The extinction of AgNPs at 400 nm in the supernatant solution was not significant after centrifugations, indicating successful removal of the AgNPs from solution. The released Ag+ concentration and the corresponding percentage of the total AgNP are listed in Table 1 (see Tables SI 2 and 3 for detailed Ag+ release from 1 and 50 μM AgNPs solutions). The concentration of Ag+ release ranges from 97 to 154 nM (9.7% to 15.4%). Ampicillin and penicillin have no significant effect on the release of Ag+, whereas the presence of enoxacin, kanamycin, neomycin, or tetracycline increases Ag+ release from 9.7% to 13.4%–15.4%.
Table 1.
Concentration (nM) and Percentage (%) of Ag+ Release from AgNPs (1 μM, 73 AgNPs/cell) upon Interaction with Antibiotics (10 μM) and/or Salmonella (1 × 107 CFU/mL)
| Ag+ (nM) | Ag released (%) | |
|---|---|---|
| AgNPs | 97 ± 20 | 9.7 ± 2.0 |
| AgNPs + AMP | 103 ± 27 | 10.3 ± 2.7 |
| AgNPs + PEN | 97 ± 44 | 9.7 ± 4.4 |
| AgNPs + Salmonella | 121 ± 34 | 12.1 ± 3.4 |
| AgNPs + PEN + Salmonella | 123 ± 26 | 12.3 ± 2.6 |
| AgNPs + ENO | 134 ± 35 | 13.4 ± 3.5 |
| AgNPs + KAN | 135 ± 36 | 13.5 ± 3.6 |
| AgNP + NEO | 154 ± 28 | 15.4 ± 2.8 |
| AgNP + TETa | 145 ± 21 | 14.5 ± 2.1 |
| AgNP + TET + Salmonellaa | 182 ± 26 | 18.2 ± 2.6 |
Significance is confirmed by ANOVA, p < 0.05.
The amount of Ag+ released increases from 9.7% to 12.1% in the presence of Salmonella and further increases to 18.2% with 10 μM tetracycline. The percent of Ag+ release by the presence of Salmonella only is 12.1%, which is 25% more than that in the absence of Salmonella. The Ag+ release in the presence of both penicillin and Salmonella is essentially the same as that in the presence of Salmonella alone (12.1% versus 12.3%). However, Ag+ release in the presence of both tetracycline and Salmonella is 26% more than that in the presence of Salmonella alone (18.2% versus 14.5%) (p < 0.05). This demonstrates that the presence of tetracycline facilitates Ag+ release. Ag+ has a strong binding affinity to sulfur-containing proteins and can be easily taken up by bacterial cells.44,46 Therefore, the Ag+ release in the presence of bacterial cells is higher due to cellular binding. The further increase in Ag+ release in the presence of tetracycline signifies that the tetracycline–AgNPs complex produces more Ag+ and may cause more growth inhibition of the bacterium. Assuming that 37 AgNPs are bound on each Salmonella cell when tetracycline is present versus 30 when tetracycline is not, the estimated “effective” Ag+ concentration would be 100 mM in the vicinity of the Salmonella cell, 20,000 times of the Ag+ control concentration of 5 μM, which killed nearly 50% of the bacteria (see detailed calculation in Table SI 4 and discussions associated with the table).
Mechanism for Synergistic Antibacterial Activity of Combined AgNPs and Antibiotics
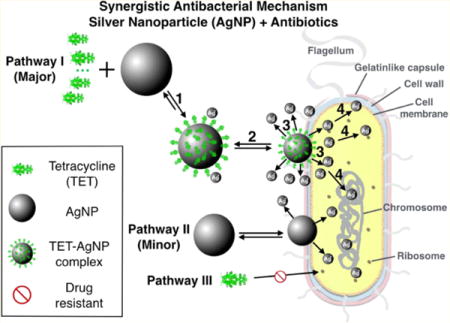
Enoxacin, kanamycin, neomycin, and tetracycline are found to exhibit a synergistic antibacterial effect against Salmonella when combined with AgNPs. These antibiotic molecules are also found to bind AgNPs in solution to form antibiotic–AgNPs complexes. We propose a four-step pathway leading to the synergistic activity observed (Figure 6): (1) Antibiotic molecules, using tetracycline as an example, form complexes with AgNPs (tetracyclilne–AgNPs). (2) The tetracycline–AgNPs complexes bind to a bacterium. (3) The bacterium-attached tetracycline– AgNPs complexes release Ag+, more than AgNPs alone would release under the same conditions. Thus, it creates a temporary and local high Ag+ concentration near the surface of the bacterium. (4) Ag+ acts as the agent to cause bacterium toxicity by binding to the proteins and DNA molecules of the cell walls as well as those inside the cells, disabling the bacterial functions leading to bacterial death. Although AgNPs can do the same thing as shown in Pathway II, it is believed that when such an antibiotic is present, the primary pathway is through the antibiotic–AgNPs complex. Pathway III is not effective due to resistance to the antibiotics the bacterium developed.
Figure 6.
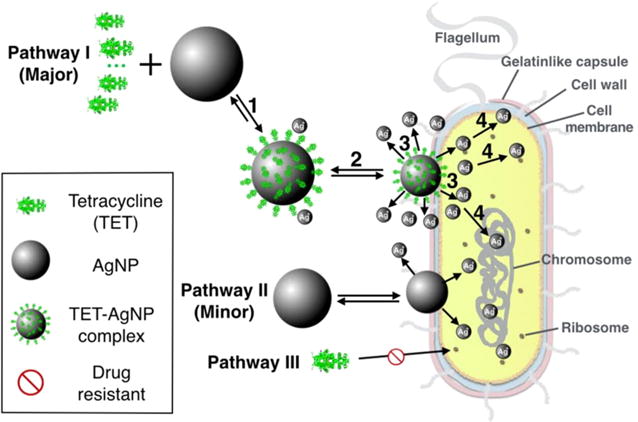
Schematic drawing of the synergistic antibacterial pathway of AgNPs with tetracycline against multidrug-resistant Salmonella. A four-step procedure is proposed as the major pathway leading to cell death. Pathway II is a minor pathway, and Pathway III is not effective due to antibacterial resistance by Salmonella.
Previous studies demonstrate that AgNPs have many different pathways of bactericidal activity, as discussed in several review papers.2,3,22,47 They include the role of surface coating agents,29,48,49 generation of reactive oxygen species,50 free radicals derived from the surface of AgNPs,2 and released Ag+51 and silver ion stress,52 as well as other fine interactions with bacterial cells53,54 that could further trigger downstream damages to the bacterial cell wall via linkages with the respiratory enzymes of bacterial cells10,11 or by depleting the levels of intracellular ATP.44 The exact mode of action for the bactericidal effect by AgNPs remains debatable, whether AgNPs or the released Ag+ are the actual active agent.55 Most of the recent studies show that Ag+ is more likely the agent causing cell death.26,56,57 Our result here also demonstrates that Ag+, not AgNPs, ultimately causes cell death.
Supplementary Material
Acknowledgments
This research was supported by the National Science Foundation JSU-UCSB Partnership for Research and Education in Materials (Grant DMR-0611539) and Department of Education P031B090210-11 through Title III-HBGI. Core research facilities were supported by grants from the National Institutes of Health (NCRR 2G12RR013459-11).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.est.6b00998.
Information is mentioned in the text. (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Antimicrobial Resistance: Global Report on Surveillance. World Health Organization; 2014. [Google Scholar]
- 2.Kim JS, Kuk E, Yu KN, Kim J-H, Park SJ, Lee HJ, Kim SH, Park YK, Park YH, Hwang C-Y, et al. Antimicrobial effects of silver nanoparticles. Nanomedicine. 2007;3(1):95–101. doi: 10.1016/j.nano.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Hajipour MJ, Fromm KM, Ashkarran AA, de Aberasturi DJ, de Larramendi IR, Rojo T, Serpooshan V, Parak WJ, Mahmoudi M. Antibacterial properties of nanoparticles. Trends Biotechnol. 2012;30(10):499–511. doi: 10.1016/j.tibtech.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Allahverdiyev AM, Kon KV, Abamor ES, Bagirova M, Rafailovich M. Coping with antibiotic resistance: combining nanoparticles with antibiotics and other antimicrobial agents. Expert Rev Anti-Infect Ther. 2011;9(11) doi: 10.1586/eri.11.121. 1035–1–52. [DOI] [PubMed] [Google Scholar]
- 5.Feng J, Shi Q, Li W, Shu X, Chen A, Xie X, Huang X. Antimicrobial activity of silver nanoparticles in situ growth on TEMPO-mediated oxidized bacterial cellulose. Cellulose. 2014;21(6):4557–4567. [Google Scholar]
- 6.Xiao W, Xu J, Liu X, Hu Q, Huang J. Antibacterial hybrid materials fabricated by nanocoating of microfibril bundles of cellulose substance with titania/chitosan/silver-nanoparticle composite films. J Mater Chem B. 2013;1(28):3477–3485. doi: 10.1039/c3tb20303d. [DOI] [PubMed] [Google Scholar]
- 7.Cheng F, Betts JW, Kelly SM, Schaller J, Heinze T. Synthesis and antibacterial effects of aqueous colloidal solutions of silver nanoparticles using aminocellulose as a combined reducing and capping reagent. Green Chem. 2013;15(4):989–998. [Google Scholar]
- 8.Yang G, Xie J, Hong F, Cao Z, Yang X. Antimicrobial activity of silver nanoparticle impregnated bacterial cellulose membrane: effect of fermentation carbon sources of bacterial cellulose. Carbohydr Polym. 2012;87(1):839–845. doi: 10.1016/j.carbpol.2011.08.079. [DOI] [PubMed] [Google Scholar]
- 9.Drogat N, Granet R, Sol V, Memmi A, Saad N, Koerkamp CK, Bressollier P, Krausz P. Antimicrobial silver nanoparticles generated on cellulose nanocrystals. J Nanopart Res. 2011;13(4):1557–1562. [Google Scholar]
- 10.Li W-R, Xie X-B, Shi Q-S, Zeng H-Y, Ou-Yang O-S, Chen Y-B. Antibacterial activity and mechanism of silver nanoparticles on Escherichia coli. Appl Microbiol Biotechnol. 2010;85(4):1115–1122. doi: 10.1007/s00253-009-2159-5. [DOI] [PubMed] [Google Scholar]
- 11.Rai M, Yadav A, Gade A. Silver nanoparticles as a new generation of antimicrobials. Biotechnol Adv. 2009;27(1):76–83. doi: 10.1016/j.biotechadv.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 12.Pinto RJ, Marques PA, Neto CP, Trindade T, Daina S, Sadocco P. Antibacterial activity of nanocomposites of silver and bacterial or vegetable cellulosic fibers. Acta Biomater. 2009;5(6):2279–2289. doi: 10.1016/j.actbio.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 13.Lara HH, Ayala-Núnez NV, Turrent LDCI, Padilla CR. Bactericidal effect of silver nanoparticles against multidrug-resistant bacteria. World J Microbiol Biotechnol. 2010;26(4):615–621. [Google Scholar]
- 14.Rai M, Deshmukh S, Ingle A, Gade A. Silver nanoparticles: the powerful nanoweapon against multidrug-resistant bacteria. J Appl Microbiol. 2012;112(5):841–852. doi: 10.1111/j.1365-2672.2012.05253.x. [DOI] [PubMed] [Google Scholar]
- 15.Shahverdi AR, Fakhimi A, Shahverdi HR, Minaian S. Synthesis and effect of silver nanoparticles on the antibacterial activity of different antibiotics against Staphylococcus aureus and Escherichia coli. Nanomedicine. 2007;3(2):168–171. doi: 10.1016/j.nano.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 16.Hwang I-S, Hwang JH, Choi H, Kim K-J, Lee DG. Synergistic effects between silver nanoparticles and antibiotics and the mechanisms involved. J Med Microbiol. 2012;61(Pt 12):1719–1726. doi: 10.1099/jmm.0.047100-0. [DOI] [PubMed] [Google Scholar]
- 17.Kora AJ, Rastogi L. Enhancement of Antibacterial Activity of Capped Silver Nanoparticles in Combination with Antibiotics, on Model Gram-Negative and Gram-Positive Bacteria. Bioinorg Chem Appl. 2013;2013:1. doi: 10.1155/2013/871097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dasari TPS, Deng H, McShan D, Yu H. Nanosilver-Based Antibacterial Agents for Food Safety. In: Ray PC, editor. Food Poisoning: Outbreaks, Bacterial Sources and Adverse Health Effects. NOVA Science Publishers; 2014. pp. 35–62. [Google Scholar]
- 19.Fabrega J, Fawcett SR, Renshaw JC, Lead JR. Silver nanoparticle impact on bacterial growth: effect of pH, concentration, and organic matter. Environ Sci Technol. 2009;43(19):7285–7290. doi: 10.1021/es803259g. [DOI] [PubMed] [Google Scholar]
- 20.Sotiriou GA, Pratsinis SE. Antibacterial activity of nanosilver ions and particles. Environ Sci Technol. 2010;44(14):5649–5654. doi: 10.1021/es101072s. [DOI] [PubMed] [Google Scholar]
- 21.Tejamaya M, Römer I, Merrifield RC, Lead JR. Stability of citrate, PVP, and PEG coated silver nanoparticles in ecotoxicology media. Environ SciTechnol. 2012;46(13):7011–7017. doi: 10.1021/es2038596. [DOI] [PubMed] [Google Scholar]
- 22.Li P, Li J, Wu C, Wu Q, Li J. Synergistic antibacterial effects of β-lactam antibiotic combined with silver nanoparticles. Nanotechnology. 2005;16(9):1912. [Google Scholar]
- 23.Batarseh KI. Anomaly and correlation of killing in the therapeutic properties of silver (I) chelation with glutamic and tartaric acids. J Antimicrob Chemother. 2004;54(2):546–548. doi: 10.1093/jac/dkh349. [DOI] [PubMed] [Google Scholar]
- 24.McShan D, Zhang Y, Deng H, Ray PC, Yu H. Synergistic Antibacterial Effect of Silver Nanoparticles Combined with Ineffective Antibiotics on Drug Resistant Salmonella typhimurium DT104. Journal of Environmental Science and Health, Part C. 2015;33(3):369–384. doi: 10.1080/10590501.2015.1055165. [DOI] [PubMed] [Google Scholar]
- 25.Yang X, Gondikas AP, Marinakos SM, Auffan M, Liu J, Hsu-Kim H, Meyer JN. Mechanism of silver nanoparticle toxicity is dependent on dissolved silver and surface coating in Caenorhabditis elegans. Environ Sci Technol. 2012;46(2):1119–1127. doi: 10.1021/es202417t. [DOI] [PubMed] [Google Scholar]
- 26.Xiu Z-M, Zhang Q-B, Puppala HL, Colvin VL, Alvarez PJ. Negligible particle-specific antibacterial activity of silver nanoparticles. Nano Lett. 2012;12(8):4271–4275. doi: 10.1021/nl301934w. [DOI] [PubMed] [Google Scholar]
- 27.Lee P, Meisel D. Adsorption and surface-enhanced Raman of dyes on silver and gold sols. J Phys Chem. 1982;86(17):3391–3395. [Google Scholar]
- 28.Tiwari VS, Oleg T, Darbha GK, Hardy W, Singh J, Ray PC. Non-resonance SERS effects of silver colloids with different shapes. Chem Phys Lett. 2007;446(1):77–82. [Google Scholar]
- 29.Lu W, Senapati D, Wang S, Tovmachenko O, Singh AK, Yu H, Ray PC. Effect of surface coating on the toxicity of silver nanomaterials on human skin keratinocytes. Chem Phys Lett. 2010;487(1):92–96. doi: 10.1016/j.cplett.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kittler S, Greulich C, Diendorf J, Koller M, Epple M. Toxicity of silver nanoparticles increases during storage because of slow dissolution under release of silver ions. Chem Mater. 2010;22(16):4548–4554. [Google Scholar]
- 31.Allen CA, Fedorka-Cray PJ, Vazquez-Torres A, Suyemoto M, Altier C, Ryder LR, Fang FC, Libby SJ. In vitro and in vivo assessment of Salmonella enterica serovar Typhimurium DT104 virulence. Infection and immunity. 2001;69(7):4673–4677. doi: 10.1128/IAI.69.7.4673-4677.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Daima HK, Selvakannan P, Shukla R, Bhargava SK, Bansal V. Fine-tuning the antimicrobial profile of biocompatible gold nanoparticles by sequential surface functionalization using polyoxometalates and lysine. PLoS One. 2013;8(10):e79676. doi: 10.1371/journal.pone.0079676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Threlfall EJ. Epidemic Salmonella typhimurium DT 104—a truly international multiresistant clone. J Antimicrob Chemother. 2000;46(1):7–10. doi: 10.1093/jac/46.1.7. [DOI] [PubMed] [Google Scholar]
- 34.Poppe C, Smart N, Khakhria R, Johnson W, Spika J, Prescott J. Salmonella typhimurium DT104: a virulent and drug-resistant pathogen. Canadian Veterinary Journal. 1998;39(9):559. [PMC free article] [PubMed] [Google Scholar]
- 35.Mather A, Reid S, Maskell D, Parkhill J, Fookes M, Harris S, Brown D, Coia J, Mulvey M, Gilmour M, et al. Distinguishable epidemics of multidrug-resistant Salmonella Typhimurium DT104 in different hosts. Science. 2013;341(6153):1514–1517. doi: 10.1126/science.1240578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Canamares M, Garcia-Ramos J, Gomez-Varga J, Domingo C, Sanchez-Cortes S. Comparative study of the morphology, aggregation, adherence to glass, and surface-enhanced Raman scattering activity of silver nanoparticles prepared by chemical reduction of Ag+ using citrate and hydroxylamine. Langmuir. 2005;21(18):8546–8553. doi: 10.1021/la050030l. [DOI] [PubMed] [Google Scholar]
- 37.Dollish FR, Fateley WG, Bentley FF. Characteristic Raman Frequencies of Organic Compounds. Wiley; 1974. [Google Scholar]
- 38.Zengin A, Tamer U, Caykara T. Extremely sensitive sandwich assay of kanamycin using surface-enhanced Raman scattering of 2-mercaptobenzothiazole labeled gold@ silver nanoparticles. Anal Chim Acta. 2014;817:33–41. doi: 10.1016/j.aca.2014.01.042. [DOI] [PubMed] [Google Scholar]
- 39.Nakamoto K. Infrared and Raman Spectra of Inorganic and Coordination ompounds. Wiley Online Library; 1986. [Google Scholar]
- 40.Krafft C, Hinrichs W, Orth P, Saenger W, Welfle H. Interaction of Tet repressor with operator DNA and with tetracycline studied by infrared and Raman spectroscopy. Biophys J. 1998;74(1):63–71. doi: 10.1016/S0006-3495(98)77767-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Yao Y, Sullivan N, Chen Y. Modeling the primary size effects of citrate-coated silver nanoparticles on their ion release kinetics. Environ Sci Technol. 2011;45(10):4422–4428. doi: 10.1021/es104205a. [DOI] [PubMed] [Google Scholar]
- 42.Henglein A, Giersig M. Formation of colloidal silver nanoparticles: capping action of citrate. J Phys Chem B. 1999;103(44):9533–9539. [Google Scholar]
- 43.Liu X, Atwater M, Wang J, Huo Q. Extinction coefficient of gold nanoparticles with different sizes and different capping ligands. Colloids Surf B. 2007;58(1):3–7. doi: 10.1016/j.colsurfb.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 44.Lok C-N, Ho C-M, Chen R, He Q-Y, Yu W-Y, Sun H, Tam PK-H, Chiu J-F, Che C-M. Proteomic analysis of the mode of antibacterial action of silver nanoparticles. J Proteome Res. 2006;5(4):916–924. doi: 10.1021/pr0504079. [DOI] [PubMed] [Google Scholar]
- 45.Liu J, Hurt RH. Ion release kinetics and particle persistence in aqueous nano-silver colloids. Environ Sci Technol. 2010;44(6):2169–2175. doi: 10.1021/es9035557. [DOI] [PubMed] [Google Scholar]
- 46.Deng H, Yu H. A Mini Review on Controlling the Size of Ag Nanoclusters by Changing the Stabilizer to Ag Ratio and by Changing DNA Sequence. Advances in Natural Science. 2015;8(2):1–9. [Google Scholar]
- 47.Maillard JY, Hartemann P. Silver as an antimicrobial: facts and gaps in knowledge. Crit Rev Microbiol. 2013;39(4):373–383. doi: 10.3109/1040841X.2012.713323. [DOI] [PubMed] [Google Scholar]
- 48.Fortunati E, Latterini L, Rinaldi S, Kenny JM, Armentano I. PLGA/Ag nanocomposites: In vitro degradation study and silver ion release. J Mater Sci: Mater Med. 2011;22(12):2735–2744. doi: 10.1007/s10856-011-4450-0. [DOI] [PubMed] [Google Scholar]
- 49.Zhao CM, Wang WX. Importance of surface coatings and soluble silver in silver nanoparticles toxicity to Daphnia magna. Nanotoxicology. 2012;6(4):361–370. doi: 10.3109/17435390.2011.579632. [DOI] [PubMed] [Google Scholar]
- 50.Xu H, Qu F, Xu H, Lai W, Wang YA, Aguilar ZP, Wei H. Role of reactive oxygen species in the antibacterial mechanism of silver nanoparticles on Escherichia coli O157:H7. BioMetals. 2012;25(1):45–53. doi: 10.1007/s10534-011-9482-x. [DOI] [PubMed] [Google Scholar]
- 51.Le Ouay B, Stellacci F. Antibacterial activity of silver nanoparticles: A surface science insight. Nano Today. 2015;10(3):339–354. [Google Scholar]
- 52.McQuillan JS, Groenaga Infante H, Stokes E, Shaw AM. Silver nanoparticle enhanced silver ion stress response in Escherichia coli K12. Nanotoxicology. 2012;6(8):857–866. doi: 10.3109/17435390.2011.626532. [DOI] [PubMed] [Google Scholar]
- 53.Grigor’Eva A, Saranina I, Tikunova N, Safonov A, Timoshenko N, Rebrov A, Ryabchikova E. Fine mechanisms of the interaction of silver nanoparticles with the cells of Salmonella typhimurium and Staphylococcus aureus. BioMetals. 2013;26(3):479–488. doi: 10.1007/s10534-013-9633-3. [DOI] [PubMed] [Google Scholar]
- 54.Radzig MA, Nadtochenko VA, Koksharova OA, Kiwi J, Lipasova VA, Khmel IA. Antibacterial effects of silver nanoparticles on gram-negative bacteria: Influence on the growth and biofilms formation, mechanisms of action. Colloids Surf B. 2013;102:300–306. doi: 10.1016/j.colsurfb.2012.07.039. [DOI] [PubMed] [Google Scholar]
- 55.Dorobantu LS, Fallone C, Noble AJ, Veinot J, Ma G, Goss GG, Burrell RE. Toxicity of silver nanoparticles against bacteria, yeast, and algae. J Nanopart Res. 2015;17(4):172. [Google Scholar]
- 56.Xiu ZM, Ma J, Alvarez PJJ. Differential effect of common ligands and molecular oxygen on antimicrobial activity of silver nanoparticles versus silver ions. Environ Sci Technol. 2011;45(20):9003–9008. doi: 10.1021/es201918f. [DOI] [PubMed] [Google Scholar]
- 57.Kang F, Alvarez PJ, Zhu D. Microbial extracellular polymeric substances reduce Ag+ to silver nanoparticles and antagonize bactericidal activity. Environ Sci Technol. 2014;48(1):316–322. doi: 10.1021/es403796x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


