Abstract
Sequences that influence nucleosome positioning in promoter regions, and their relation to gene regulation, have been the topic of much research over the last decade. In yeast, significant nucleosome-depleted regions are found, which facilitate transcription. With the arrival of nucleosome positioning maps for the human genome, it was discovered that in our genome, unlike in that of yeast, promoters encode for high nucleosome occupancy. In this work, we look at the genomes of a range of different organisms, to provide a catalog of nucleosome positioning signals in promoters across the tree of life. We utilize a computational model of the nucleosome, based on crystallographic analyses of the structure and elasticity of the nucleosome, to predict the nucleosome positioning signals in promoter regions. To be able to apply our model to large genomic datasets, we introduce an approximative scheme that makes use of the limited range of correlations in nucleosomal sequence preferences to create a computationally efficient approximation of the full biophysical model. Our predictions show that a clear distinction between unicellular and multicellular life is visible in the intrinsically encoded nucleosome affinity. Furthermore, the strength of the nucleosome positioning signals correlates with the complexity of the organism. We conclude that encoding for high nucleosome occupancy, as in the human genome, is in fact a universal feature of multicellular life.
Introduction
Nucleosomes are the fundamental packaging units of DNA that eukaryotic organisms employ to render their genomes compact enough to fit inside a cell, consisting of ∼147 basepairs worth of DNA wrapped around a histone core. This packaging also restricts access to the genome: DNA bound to histones is unavailable for coupling to many other DNA-binding complexes, such as the transcriptional machinery. Therefore, the positioning of nucleosomes along the genome interacts with gene expression, as was already realized some three decades ago (1).
This interplay suggests that nucleosomes may play a role in gene regulation, and nucleosomes are in fact actively displaced to regulate gene expression (2, 3). Genomic sequences may also have evolved to position nucleosomes in specific, beneficial locations. This possibility is suggested both by the fact that the degeneracy of the genetic code, in principle, allows for multiplexing of such positioning signals with genetic information (4), and by the observation that the mutation patterns of DNA bound to histones differ from those of linker DNA (5).
Research into such nucleosome positioning signals, hardcoded into eukaryotic genomes, has veritably exploded over the last decade, primarily due to the development of experimental methods that allow for efficient genomewide nucleosome mapping (6). This research has provided insight into the importance of nucleosomal sequence preferences for chromatin organization (7), and has allowed for the creation, refinement, and testing of many models for predicting nucleosome positioning along genomes (8, 9). The intrinsic nucleosome-DNA affinity of genomic sequences appears to play a significant role in vivo in positioning nucleosomes in certain regions of the genome, such as transcription start sites (TSSs) and origins of replication (7), alongside other effects like the presence of proteins that compete for the same DNA stretch or the action of chromatin remodelers (10, 11).
Around the TSS of Saccharomyces cerevisiae (baker’s yeast), nucleosomes have been found to be depleted on average, both in vitro and in vivo (12, 13, 14, 15, 16, 17, 18). The persistence of this depletion in vitro, in the absence of active remodeling, identifies the sequence preferences of nucleosomes as the dominant cause. Those preferences have been measured and utilized in various models to explain the observed nucleosome depletion (15, 16, 18, 19, 20). These nucleosome-depleted regions (NDRs) in gene promoters are thought to be encoded into the genomic sequence to allow RNA polymerases more ready access to the TSS, thereby facilitating transcription (13).
Since the earliest studies on baker’s yeast, inquiries into nucleosome positioning have been extended to the genomes of many other organisms, such as Schizosaccharomyces pombe (21) and various other species of yeast (22), Caenorhabditis elegans (23, 24), Plasmodium falciparum (25), flies (26), zebrafish (27), Arabidopsis thaliana (28), mice (29, 30), and humans (30, 31, 32, 33, 34, 35). Most of these studies were conducted in vivo, and therefore do not allow for isolation of effects encoded into the genomic sequences. This body of research shows, however, that sequence effects alone are not generally sufficient to explain in vivo observations (11). An important role is also played by the active regulation of transcription. In yeast, the promoters of actively transcribed genes show much more pronounced nucleosome depletion than those of inactive genes (21).
In human cells, as in yeast, NDRs were found in vivo only for actively expressed genes (31). However, in vitro nucleosome mapping reveals that the human genome does not share yeast’s strategy of depletion-by-default. Instead, it was found that promoter regions in the human genome showed enhanced nucleosome occupancy. One interpretation is that this reflects the differentiated nature of human cells: it may be more beneficial to keep genes relatively inaccessible by default, and to actively open the promoter region only when needed (33, 34). This idea seems to be countered by newer results, however, which find stronger intrinsic nucleosome-attracting regions (NARs) for housekeeping genes than for tissue-specific genes, directly opposite of what one would expect (36). Those results indicate that the function of the NARs in the human genome may be to retain nucleosomes in sperm cells (in which most nucleosomes are removed from the chromatin) and so pass on epigenetic information to the next generation.
Whichever is the case, these ideas raise the question whether the presence of an NDR in yeast versus that of an NAR in humans might be a general distinguishing feature between unicellular and multicellular life. To answer this question, we utilize a purely mechanics-based model for the sequence-dependent DNA-nucleosome affinity to predict in vitro nucleosome positioning signals, and compare the signals encoded into the promoter regions of a wide range of genomes.
Materials and Methods
Data acquisition
All genomic sequences and gene (cDNA) data were downloaded from ensemblgenomes.org, release 31 (37). The in vitro nucleosome map produced by Kaplan et al. (18) was retrieved from GEO accession number GEO: GSE13622. The map from Valouev et al. (34) was downloaded from ccg.vital-it.ch/mga/hg18/valouev11/valouev11.html. The map from Locke et al. (38) was downloaded from http://nucleosome.rutgers.edu/nucenergen/celegansnuc/. The data from Ercan et al. (24) was taken directly from Fig. 1 C in that reference. TSS locations in S. cerevisiae were derived from David et al. (39) in the manner described in Vaillant et al. (40).
Model
We employed a statistical model inspired by that of Segal et al. (13), Field et al. (16), and Kaplan et al. (18). However, whereas their models are trained on experimental data, we employed this type of model to create a computationally inexpensive approximation to the theoretical nucleosome model recently published in Eslami-Mossallam et al. (4). The predictiveness of the Eslami-Mossallam nucleosome model has been examined in Eslami-Mossallam et al. (4), where it was found to outperform the experimentally informed models mentioned above, and in de Bruin et al. (41), where it is shown to be applicable not only to predictions for nucleosome positioning along a genome, but also the sequence-dependent response of nucleosomes to external forces.
We employed an extended version of the model presented in Segal et al. (13), which is informed by trinucleotide distributions, rather than dinucleotide distributions, because we found that this trinucleotide model leads to a more accurate approximation (see the Supporting Material and Fig. S1 for more information).
The model of Segal et al. (13) requires as input position-dependent (di)nucleotide probabilities for the nucleosome. These can be derived from suitable sequence ensembles, as done in their article and its followup work. Such ensembles can also be generated in silico using the mutation Monte Carlo method of Eslami-Mossallam et al. (4) We applied the mutation Monte Carlo method to generate an ensemble of 107 high-affinity nucleosome sequences, from which we calculated the necessary di- and trinucleotide probability distributions. We found that the bioinformatical model approximated the full biophysical model with a root mean square deviation of 0.85 kT.
For this work, the parameterization of the nucleosome model was changed from the hybrid parameterization described in Eslami-Mossallam et al. (4), to a parameterization informed solely by crystallography data. We found that this improves its applicability to long-range effects. See the Supporting Material for more information.
Sequence analysis
For every genome analyzed, we calculated the averaged signal as follows. For every annotated gene, we looked up the location of the TSS, and extracted the 1146 bp before and after. For each of the resulting sequences, we calculated a probability landscape for nucleosome positioning using the trinucleotide model mentioned above. We would like to calculate occupancies from these landscapes and average over all genes. Unfortunately, because the probabilities vary over several orders of magnitude, the number of genes is generally not large enough to provide a meaningful average; it tends to be dominated by the highest probabilities. Therefore, we instead consider the average energy landscape for a given organism.
From the predicted probabilities, an energy landscape can be calculated up to a constant shift, because such a probability is the normalized Boltzmann weight of a state. We took the average of the energy landscapes of all the sequences as a representative energy landscape for a given organism. For each basepair (−1000 to +1000), we then calculated the nucleosome occupancy by summing the Boltzmann probabilities of all 147 nucleosome positions that lead to that basepair being covered by the nucleosome. This gives us a prediction of the intrinsic nucleosome affinity encoded in the genomic sequences.
Results and Discussion
Opposing nucleosome occupancy signals in yeast and human genomes
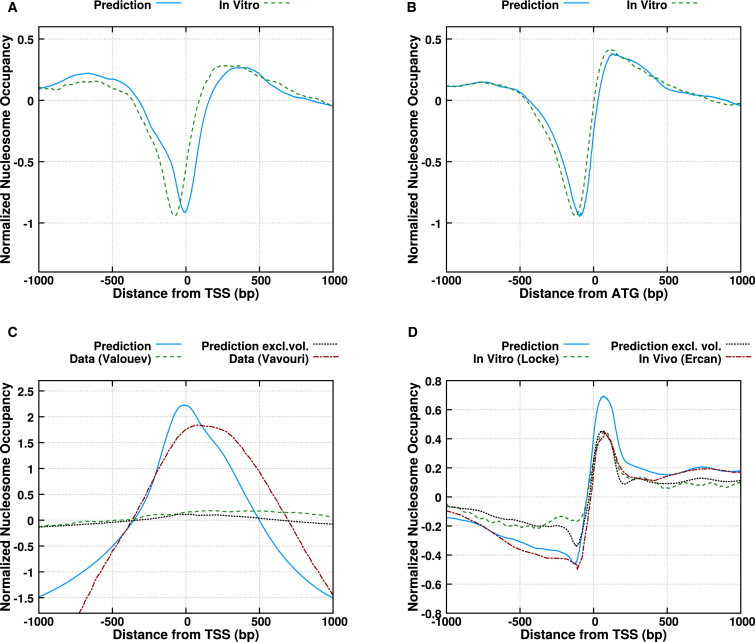
The high-coverage S. cerevisiae nucleosome maps provide the standard testing ground for any model designed to predict nucleosome occupancy. Applying our nucleosome affinity model (see Materials and Methods), we find we can correctly predict NDRs in the promoter regions of S. cerevisiae. The comparisons, for regions centered on the TSSs and on the start codons, are shown in Fig. 1, A and B, respectively.
Figure 1.
Comparison of predicted and measured intrinsic nucleosome positioning signals in promoter regions. The quantities plotted are the natural logarithms of the occupancies and the signals have been normalized such that they average to zero. (Solid blue curves) Our predictions in the limit of low nucleosome density, which give an account of the strength of the signals intrinsically encoded; (dashed green curves) in vitro measurements; (dotted black curves) predictions taking into account the steric interactions. Using the same treatment as in Chevereau et al. (44), these curves have a free parameter , i.e., the difference between the chemical potential and the average energy of the landscape, which we determined to be −8.5 kT for yeast (curves not shown due to similarity with the low-density limit), −5.7 kT for C. elegans, and −1.38 kT for humans. (A and B) S. cerevisiae, average nucleosome occupancy centered on the TSS and start codons, respectively. Data from Kaplan et al. (18). (C) Like (A), for Homo sapiens. The in vitro data is from Valouev et al. (34). Additionally shown is the nucleosome retention signal from Vavouri and Lehner (36). (D) Like (A), for C. elegans. The in vivo data is from Ercan et al. (24); the in vitro data is from Locke et al. (38). To see this figure in color, go online.
For the human genome, a map of in vitro nucleosome occupancy has been published by Valouev et al. (34), and, as predicted by Tillo et al. (33), it reveals occupancy signals opposite to that of yeast: human promoters seem to encode for high, rather than low, nucleosome occupancy. Vavouri and Lehner (36) similarly find an increased retention of nucleosomes when nucleosomes are depleted in human sperm cells. Correspondingly, when applying our model to the promoter regions of the human genome, we find a very strong NAR around the TSS, as can be seen in Fig. 1 C.
Initially surprisingly, the signal found by Valouev et al. (34) is an order-of-magnitude smaller than that predicted by our model and that found by Vavouri and Lehner (36). This discrepancy can be explained when we consider that the nucleosome density cannot exceed 1 per 147 bp due to excluded volume. The experiment attempts to measure enrichment of nucleosomes in the promoter regions relative to the average density of nucleosomes. Unlike in experiments that look at nucleosome depletion or retention, the excluded volume between nucleosomes puts a limit on how strong the enrichment can be in practice.
This is the reason for the discrepancy between the in vitro results of Valouev et al. (34) and ours and those of Vavouri and Lehner (36). To approximate the effects of steric interactions, we applied Percus’ equation (42) to our average energy landscapes, and solved it as described in Vanderlick et al. (43). The solution depends on the chemical potential of the nucleosomes binding to the DNA (see also Chevereau et al. (44)), which we adjust to get a good fit with the in vitro data. We see that steric interactions can indeed explain the very weak signal for humans (dotted black curve in Fig. 1 C) as well as the apparent overshoot of our prediction for C. elegans (same in Fig. 1 D).
This means that at physiological conditions, the nucleosome density will be saturated at much smaller values due to steric interactions. However, we stress that independent of this saturation effect, a nucleosome at the peak of the nucleosome occupancy signal will be strongly energetically bound, and so hinder transcription if it is not actively removed, as well as be more stable under a nucleosome-depleting force.
The results of Vavouri and Lehner (36) when examining where nucleosomes are retained when they are depleted from chromatin in human sperm are more in line with our predictions, as can also be seen in Fig. 1 C. When depleting nucleosomes, excluded-volume interactions are not a constraint and our predictions can be probed. Although these authors studied a special in vivo situation, the nucleosome retention signals were found to correlate strongly with DNA sequence. Because the depletion of nucleosomes in sperm is an out-of-equilibrium process, and our model therefore does not make direct numerical predictions for this situation, we note the similarity between our predictions and the in vivo nucleosome retention signal.
We thus have interesting observations and predictions on two ends of a spectrum. A very simple, unicellular eukaryote shows nucleosome depletion as its most prominent, intrinsically encoded nucleosome positioning feature. A complex multicellular one shows high nucleosome occupancy instead. What happens in between these two extremes?
In Fig. 1 D we present a comparison between our predicted signal for C. elegans and the signals found in vitro by Locke et al. (38) and in vivo by Ercan et al. (24). We find remarkable agreement in the shape of the signal, indicating that the data is indeed indicative of intrinsically encoded nucleosome positioning. Somewhat surprisingly, the in vitro and in vivo signals are similar to each other, which is not as strongly the case for yeast, and even less so for humans (see e.g., Fig. 3 in Vavouri and Lehner (36)). It has been noted that an in vivo nucleosome occupancy map of the nematode C. elegans lacks many of the features that distinguish in vivo maps from in vitro maps of yeast, such as strongly phased nucleosomes. Valouev et al. (23) find much flexibility in nucleosome positions in C. elegans. Such variability may average out some of the effects of active remodeling, rendering the two maps similar.
C. elegans seems to show a nucleosome positioning signal that is a hybrid of the signals found in the yeast and human genomes. It has an NDR upstream of the TSS, like yeast, but it also shows a significant NAR just after the TSS.
Intrinsic nucleosome positioning signals are indicative of multicellularity
The hybrid behavior in C. elegans may be hypothetically explained. As suggested by Tillo et al. (33), organisms may wish to tune their genomic sequences to intrinsically deactivate genes that are active only in some cell types, while intrinsically activating those that are common to all of its cells. In unicellular life, most genes will not be permanently silenced, leading to an overall average depletion signal. In complex multicellular life, the signal may be dominated by the many genes that are intrinsically deactivated, leading to an overall attractive signal. C. elegans may then represent a range of organisms where the two contributions are more equal, leading to both a depleted region just before the start codon (where it is also observed in yeast) and an attractive region just after (the peak in occupancy in the human genome is also skewed toward the right).
The results of Vavouri and Lehner (36), however, suggest that, at least in the human genome, the hypothesis of Tillo et al. (33) does not hold, and the function of the NARs is to retain nucleosomes in sperm cells. The hybrid signal we find in C. elegans may in this case similarly play a dual role of facilitating initiation of transcription, but at the same time assist in nucleosome retention.
We can extend our observation of these signals to other genomes using our model. We mapped the nucleosome positioning signals for promoters in genomes across the tree of life and discovered organisms that have intrinsically encoded NDRs and NARs, as well as many that fall into the hybrid category.
Most archaea (14 genomes analyzed) show a signal similar to that of yeast, in that a nucleosome-depleted region is the most prominent feature (Fig. S2). Archaea are unicellular organisms that do not have histone octamers, but employ only tetramers of (archaeal) histones to compactify their DNA. We expect these tetramers to obey positioning rules similar enough to nucleosomes that our model is predictive of their occupancy. We therefore analyzed the octamer affinity landscapes, for the sake of comparison to eukaryotes, even though archaea do not possess them. The signals show that these simple unicellular organisms almost all fall into the depletion-by-default category.
Fungi (seven genomes analyzed) show somewhat more diverse signals, Fig. S3. While S. cerevisiae has a prominent NDR, many of the other fungi analyzed lack both a localized depleted region and a localized attractive region, but retain a step-function signal centered on the TSS. Fungal cells are not highly differentiated, but some fungi are dimorphic (they switch between unicellular and filamentous states), possibly causing these more hybridlike signals.
Plants (four genomes analyzed) come in many forms, from unicellular algae to complex multicellular life. As expected, we see various signals (Fig. S4). The genome of Chlamydomonas reinhardtii, a unicellular alga, shows an NDR. Among the multicellular plants, we see two signals with a strong NAR, and one with hybrid behavior.
Among animals (24 genomes analyzed) we also find various signals. In worms, like C. elegans, we find both hybrid signals and more NAR-like signals (Fig. S5). Drosophila melanogaster and other members of its genus show strong hybrid signals, with a swift rise in nucleosome occupancy at the TSS (Fig. S6). Finally, the zebrafish genome and all mammalian genomes analyzed (human, chimpanzee, and mouse) have strong NARs (Fig. S7).
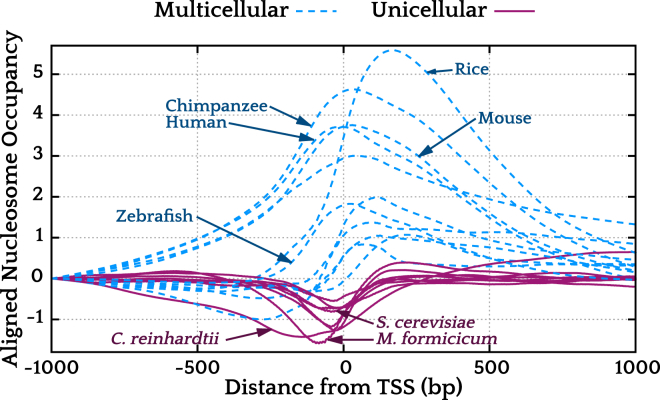
We see a clear separation between unicellular and multicellular organisms. Although some signals from unicellular lifeforms show some hybrid characteristics, the dominant feature is generally an NDR. All multicellular genomes, on the other hand, either encode for high nucleosome occupancy in the promoter region, or show hybrid signals. This distinction persists across the eukaryotic phylogenetic tree and is clearly visible in Fig. 2, where we have plotted a representative set of signals, divided into unicellular and multicellular classes. We finally note that these signals qualitatively correlate well with GC content (Fig. S8), suggesting that GC content is a prominent factor in shaping mechanical signals in promoter regions.
Figure 2.
A representative selection of nucleosome positioning signals from various genomes. As a visual aid, the signals have been shifted vertically such that the logarithmic nucleosome occupancy at position −1000 is 0. The signals clearly fall into two distinct classes, based on whether the organism is unicellular or multicellular. To see this figure in color, go online.
Intrinsic nucleosome positioning signals correlate with complexity
One proposed measure for organism complexity is the number of different cell types an organism possesses (45), and the ideas presented here clearly have a link to this measure. Unfortunately, numerical data describing the numbers of cell types does not appear to be readily available in the literature, so we were unable to define a numerical measure of complexity. Therefore, we have restricted ourselves to ordering the organisms, by making assumptions about the cell type numbers. From simple to complex, we list: archaea, unicellular eukaryotes, filamentous and dimorphic fungi, multicellular plants, nematodes, Drosophila flies, zebrafish, and mammals.
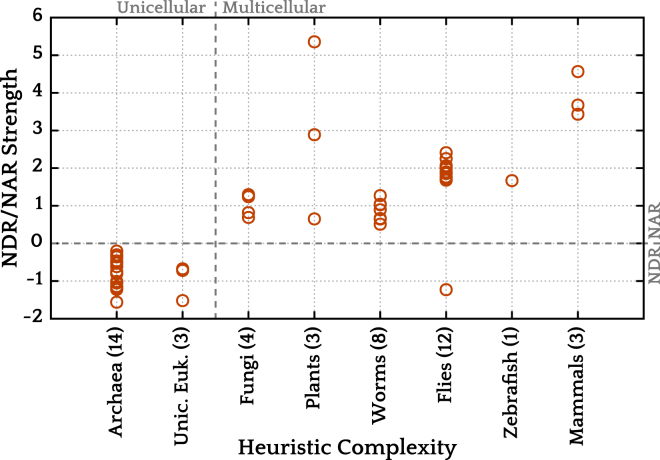
We then considered the strength and direction of the NDR/NAR signals. To quantify this, we calculated the maximum and minimum of the signal and took the difference with the signal value at position −1000 relative to the start codon. We then took the largest of these two values (in the absolute sense) and designated this value as the signal’s strength (not in the absolute sense; a dominant NDR gives a negative signal strength).
The signal strength as thus defined clearly distinguishes unicellular and multicellular lifeforms (Welch’s t (39.051) = 10.5512, p-value 5.4 × 10−13) and the signals for multicellular organisms show correlation with our complexity ordering (Spearman rs = 0.52, p-value 82.3 × 10−3), as shown in Fig. 3. The ordering of the organisms is almost certainly imperfect, for example because all multicellular plants have been lumped together; without more accurate knowledge of the cell type numbers, there is no way to place them more realistically. However, the NDR/NAR strengths show a tentative trend. All unicellular eukaryotes have a negative signal strength, indicating an NDR, as noted in the previous section. All multicellular eukaryotes (with one exception, D. melanogaster) have a stronger NAR than NDR, and the strength of this NAR roughly increases with complexity. This observation concurs with the hypothesis of Tillo et al. (33). Our expectation based on that hypothesis would be that a more differentiated organism will have more genes that are nucleosome-occupied by default, leading to a higher NAR signal. It is not clear what purpose this correlation might serve in the context of nucleosome retention in the germline.
Figure 3.
Promoter nucleosome positioning signal strength grouped by a heuristic measure of complexity of the organisms. The numbers in parentheses indicate how many genomes fall in each category. To see this figure in color, go online.
Conclusions
We found that the recently discovered fact that the human genome, unlike the yeast genome, encodes (on average) for an NAR rather than an NDR in the promoter region, is in fact a universal feature of multicellular life. The hypothesis put forth by Tillo et al. (33) is that this NAR suppresses gene transcription and that this suppression helps an organism with differentiated cell types manage its gene expression. Genes that are not needed in every cell type are suppressed by default, and only activated in those cells where they are necessary. In unicellular lifeforms, however, most genes will be in constant use, and keeping those genes easily accessible is more favorable.
On the other hand, Vavouri and Lehner (36) have found that the NARs found in humans in fact serve a different purpose, namely the retention of certain nucleosomes in sperm cells, and their study of the signals found for housekeeping genes versus tissue-specific genes directly contradicts the hypothesis of Tillo et al. (33). The NARs we find in multicellular life may therefore instead be indicative of the need to retain nucleosomes in the germ cells of multicellular organisms.
NARs are common to complex multicellular lifeforms, while almost all unicellular lifeforms we analyzed have NDRs. In-between there is a range of organisms with hybrid positioning signals. In almost all of these signals, however, the NAR is a more prominent feature than the NDR. This leads to a clear distinction between uni- and multicellular life based on the type of nucleosome positioning signals found in the promoter regions.
Furthermore, the strength of the NAR appears to increase with organism complexity. This fits the hypothesis of Tillo et al. (33), because organisms with more cell differentiation will have more genes suppressed by an NAR (and possibly by stronger ones). If the purpose of the NARs is solely to retain nucleosomes in the germline, it seems that more complex life cares more strongly about retaining its nucleosomes and passing on epigenetic information. More research will be needed to explore this idea.
Given the presence of hybrid signals, we speculate that the encoding of NARs versus NDRs in promoter regions is not an all-or-nothing choice for organisms. Whether the NARs serve to close off genes by default, or to retain nucleosomes in the germline, they compete with an apparent need to create an NDR to facilitate the initiation of transcription. The organisms showing hybrid signals seem to strike a balance between the two.
Outlook
We hope that our results will motivate the experimental community to expand the available catalog of in vitro nucleosome maps to a greater number and variation of organisms. This will help not only verify our findings but also be of great service to any followup inquiries into the deeper nature and meaning of the signals we have found. We also suggest that nucleosome maps be generated at lower nucleosome densities, because steric hindrance will hide strong enrichment signals.
We also hope to encourage further examination of housekeeping versus tissue-specific genes in other organisms to further test the hypothesis of Tillo et al. (33), and an expansion of the results of Vavouri and Lehner (36) to other organisms, to test whether nucleosome retention in the germline is a goal served by the mechanical signals we find in the genomes of other complex organisms. If so, our results raise an intriguing question: why do more complex organisms tend to favor stronger nucleosome retention?
Author Contributions
H.S. and C.V. designed the study; M.T. devised and built the model; M.T. and C.V. performed the analyses; and M.T., C.V., and H.S. contributed to the article.
Acknowledgments
We thank Alain Arneodo, Benjamin Audit, Remus Dame, and Bram Henneman for discussions.
This work was supported by the Netherlands Organisation for Scientific Research (NWO/OCW), as part of the Frontiers of Nanoscience program.
Editor: Tamar Schlick.
Footnotes
Supporting Materials and Methods, Supporting Results, and eight figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(17)30035-8.
Supporting Citations
References (46, 47, 48) appear in the Supporting Material.
Supporting Material
References
- 1.Han M., Grunstein M. Nucleosome loss activates yeast downstream promoters in vivo. Cell. 1988;55:1137–1145. doi: 10.1016/0092-8674(88)90258-9. [DOI] [PubMed] [Google Scholar]
- 2.Becker P.B., Workman J.L. Nucleosome remodeling and epigenetics. Cold Spring Harb. Perspect. Biol. 2013;5:a017905. doi: 10.1101/cshperspect.a017905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lorch Y., Kornberg R.D. Chromatin-remodeling and the initiation of transcription. Q. Rev. Biophys. 2015;48:465–470. doi: 10.1017/S0033583515000116. [DOI] [PubMed] [Google Scholar]
- 4.Eslami-Mossallam B., Schram R.D., Schiessel H. Multiplexing genetic and nucleosome positioning codes: a computational approach. PLoS One. 2016;11:e0156905. doi: 10.1371/journal.pone.0156905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Makova K.D., Hardison R.C. The effects of chromatin organization on variation in mutation rates in the genome. Nat. Rev. Genet. 2015;16:213–223. doi: 10.1038/nrg3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tolkunov D., Morozov A.V. Genomic studies and computational predictions of nucleosome positions and formation energies. Adv. Protein Chem. Struct. Biol. 2010;79:1–57. doi: 10.1016/S1876-1623(10)79001-5. [DOI] [PubMed] [Google Scholar]
- 7.Iyer V.R. Nucleosome positioning: bringing order to the eukaryotic genome. Trends Cell Biol. 2012;22:250–256. doi: 10.1016/j.tcb.2012.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Teif V.B. Nucleosome positioning: resources and tools online. Brief. Bioinform. 2015;17:745–757. doi: 10.1093/bib/bbv086. [DOI] [PubMed] [Google Scholar]
- 9.Liu H., Zhang R., Zhou S. A comparative evaluation on prediction methods of nucleosome positioning. Brief. Bioinform. 2014;15:1014–1027. doi: 10.1093/bib/bbt062. [DOI] [PubMed] [Google Scholar]
- 10.Struhl K., Segal E. Determinants of nucleosome positioning. Nat. Struct. Mol. Biol. 2013;20:267–273. doi: 10.1038/nsmb.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Z., Wippo C.J., Pugh B.F. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science. 2011;332:977–980. doi: 10.1126/science.1200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan G.-C., Liu Y.-J., Rando O.J. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 13.Segal E., Fondufe-Mittendorf Y., Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Albert I., Mavrich T.N., Pugh B.F. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 15.Lee W., Tillo D., Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 16.Field Y., Kaplan N., Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shivaswamy S., Bhinge A., Iyer V.R. Dynamic remodeling of individual nucleosomes across a eukaryotic genome in response to transcriptional perturbation. PLoS Biol. 2008;6:e65. doi: 10.1371/journal.pbio.0060065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan N., Moore I.K., Segal E. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ioshikhes I.P., Albert I., Pugh B.F. Nucleosome positions predicted through comparative genomics. Nat. Genet. 2006;38:1210–1215. doi: 10.1038/ng1878. [DOI] [PubMed] [Google Scholar]
- 20.Yuan G.C., Liu J.S. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lantermann A.B., Straub T., Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 22.Tsankov A.M., Thompson D.A., Rando O.J. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valouev A., Ichikawa J., Johnson S.M. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ercan S., Lubling Y., Lieb J.D. High nucleosome occupancy is encoded at X-linked gene promoters in C. elegans. Genome Res. 2011;21:237–244. doi: 10.1101/gr.115931.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bunnik E.M., Polishko A., Le Roch K.G. DNA-encoded nucleosome occupancy is associated with transcription levels in the human malaria parasite Plasmodium falciparum. BMC Genomics. 2014;15:347. doi: 10.1186/1471-2164-15-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavrich T.N., Jiang C., Pugh B.F. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Vastenhouw N.L., Liu X.S. Canonical nucleosome organization at promoters forms during genome activation. Genome Res. 2014;24:260–266. doi: 10.1101/gr.157750.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M., Seddon A.E., Shiu S. Determinants of nucleosome positioning and their influence on plant gene expression. Genome Res. 2015;25:1182–1195. doi: 10.1101/gr.188680.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teif V.B., Vainshtein Y., Rippe K. Genome-wide nucleosome positioning during embryonic stem cell development. Nat. Struct. Mol. Biol. 2012;19:1185–1192. doi: 10.1038/nsmb.2419. [DOI] [PubMed] [Google Scholar]
- 30.Fenouil R., Cauchy P., Andrau J.C. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res. 2012;22:2399–2408. doi: 10.1101/gr.138776.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ozsolak F., Song J.S., Fisher D.E. High-throughput mapping of the chromatin structure of human promoters. Nat. Biotechnol. 2007;25:244–248. doi: 10.1038/nbt1279. [DOI] [PubMed] [Google Scholar]
- 32.Schones D.E., Cui K., Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tillo D., Kaplan N., Hughes T.R. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One. 2010;5:e9129. doi: 10.1371/journal.pone.0009129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Valouev A., Johnson S.M., Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–520. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gaffney D.J., McVicker G., Pritchard J.K. Controls of nucleosome positioning in the human genome. PLoS Genet. 2012;8:e1003036. doi: 10.1371/journal.pgen.1003036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vavouri T., Lehner B. Chromatin organization in sperm may be the major functional consequence of base composition variation in the human genome. PLoS Genet. 2011;7:e1002036. doi: 10.1371/journal.pgen.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kersey P.J., Allen J.E., Staines D.M. EnsemblGenomes 2016: more genomes, more complexity. Nucleic Acids Res. 2016;44(D1):D574–D580. doi: 10.1093/nar/gkv1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Locke G., Haberman D., Morozov A.V. Global remodeling of nucleosome positions in C. elegans. BMC Genomics. 2013;14:284. doi: 10.1186/1471-2164-14-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.David L., Huber W., Steinmetz L.M. A high-resolution map of transcription in the yeast genome. Proc. Natl. Acad. Sci. USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vaillant C., Palmeira L., Arneodo A. A novel strategy of transcription regulation by intragenic nucleosome ordering. Genome Res. 2010;20:59–67. doi: 10.1101/gr.096644.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de Bruin L., Tompitak M., Schiessel H. Why do nucleosomes unwrap asymmetrically? J. Phys. Chem. B. 2016;120:5855–5863. doi: 10.1021/acs.jpcb.6b00391. [DOI] [PubMed] [Google Scholar]
- 42.Percus J.K. Equilibrium state of a classical fluid of hard rods in an external field. J. Stat. Phys. 1976;15:505–511. [Google Scholar]
- 43.Vanderlick T.K., Scriven L.E., Davis H.T. Solution of Percus’s equation for the density of hard rods in an external field. Phys. Rev. A Gen. Phys. 1986;34:5130–5131. doi: 10.1103/physreva.34.5130. [DOI] [PubMed] [Google Scholar]
- 44.Chevereau G., Palmeira L., Vaillant C. Thermodynamics of intragenic nucleosome ordering. Phys. Rev. Lett. 2009;103:188103. doi: 10.1103/PhysRevLett.103.188103. [DOI] [PubMed] [Google Scholar]
- 45.Valentine J.W., Collins A.G., Meyer C.P. Morphological complexity increase in metazoans. Paleobiology. 1994;20:131–142. [Google Scholar]
- 46.Olson W.K., Gorin A.A., Zhurkin V.B. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl. Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Calladine C.R., Drew H.R. A base-centred explanation of the B-to-A transition in DNA. J. Mol. Biol. 1984;178:773–782. doi: 10.1016/0022-2836(84)90251-1. [DOI] [PubMed] [Google Scholar]
- 48.Becker N.B., Wolff L., Everaers R. Indirect readout: detection of optimized subsequences and calculation of relative binding affinities using different DNA elastic potentials. Nucleic Acids Res. 2006;34:5638–5649. doi: 10.1093/nar/gkl683. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.