Summary
Imprinted genes are regulated according to parental origin and can influence embryonic growth and metabolism and confer disease susceptibility. Here, we designed sensitive allele-specific reporters to non-invasively monitor imprinted Cdkn1c expression in mice and showed that expression was modulated by environmental factors encountered in utero. Acute exposure to chromatin-modifying drugs resulted in de-repression of paternally inherited (silent) Cdkn1c alleles in embryos that was temporary and resolved after birth. In contrast, deprivation of maternal dietary protein in utero provoked permanent de-repression of imprinted Cdkn1c expression that was sustained into adulthood and occurred through a folate-dependent mechanism of DNA methylation loss. Given the function of imprinted genes in regulating behavior and metabolic processes in adults, these results establish imprinting deregulation as a credible mechanism linking early-life adversity to later-life outcomes. Furthermore, Cdkn1c-luciferase mice offer non-invasive tools to identify factors that disrupt epigenetic processes and strategies to limit their long-term impact.
Keywords: imprinting, Cdkn1c, environmental stress, bioluminescence, luciferase reporter mice
Graphical Abstract

Highlights
-
•
Allele-specific expression of imprinted Cdkn1c imaged in vivo using bioluminescence
-
•
Chromatin-modifying drugs applied in utero transiently de-repress Cdkn1c imprinting
-
•
In utero exposure to low-protein diet permanently disrupts the Cdkn1c imprint
-
•
Folate supplements during gestation protect against loss of Cdkn1c imprinting
Van de Pette et al. use sensitive allele-specific reporters to longitudinally image imprinted Cdkn1c expression in mice and show that expression is modulated by environmental factors encountered in utero. These results establish imprinting deregulation as a mechanism linking early-life adversity to later-life outcomes and provide tools to detect imprinting changes in vivo.
Introduction
Epigenetics is the study of heritable changes in gene expression that arise from non-genetic influences. Genomic imprinting is an epigenetic process found in eutherian and metatherian mammals that results in parent-of-origin-specific allelic expression (John and Surani, 2000). A relatively small subset of genes within the mammalian genome (0.4%) is imprinted (Surani et al., 1984, McGrath and Solter, 1984), and these show mono-allelic expression either universally or in specific tissues that favors the maternal (e.g., Cdkn1c and Ube3a) or the paternal allele (e.g., Dlk1 and Nnat; Monk et al., 2006). Imprinted expression is initially determined by differential DNA methylation that is established in the germline (Surani, 1998). Although the rationale for genomic imprinting remains uncertain, the critical role of imprinted genes in embryonic growth, placental development, and neurogenesis (Cleaton et al., 2014) suggests that imprinting may serve to balance the selective pressures between parental genomes and control in utero offspring demand (Wolf and Hager, 2006, Day and Bonduriansky, 2004, Haig, 2004). Imprinted genes encode proteins that have a wide range of roles in nutrient transport, signaling, cell-cycle control, protein synthesis and degradation, and ion channel function. Their impact extends into postnatal life with key roles in the regulation of both metabolic and neuronal processes. Alterations at imprinted gene loci in humans are associated with rare disorders, such as Beckwith-Wiedemann syndrome (Lam et al., 1999), and also more common pathological conditions, including mental disability, impaired neuro-behavioral function, diabetes, obesity, muscle hypertrophy, and also with cancer (Radford et al., 2011).
Despite their importance, imprinted genes are particularly challenging to study. This in part reflects experimental difficulties that are common to mono-allelic genes, in that gene deletion experiments show all or nothing effects, whereas alterations in gene dosage can result in complex phenotypes in which isolating genetic and epigenetic traits is problematic (Cleaton et al., 2014, John, 2010). In addition, imprinted genes are often clustered within genomic domains in which regulation is achieved through multiple levels of epigenetic control, including DNA methylation, non-coding RNAs, and modified histones (Bartolomei and Ferguson-Smith, 2011). Finally, studies to assess the impact of chromatin-modifying drugs or environmental stress on imprinted gene expression require the maternal and paternal alleles to be discriminated on the basis of heterozygous SNPs or have used LacZ-based targeting of endogenous alleles (John, 2010). Whereas such approaches provide valuable generic tools to examine imprint dynamics through development and in response to stress, the invasive nature of the allelic readout means that it is not feasible to longitudinally monitor imprinted gene expression in different individuals throughout their life course. Models using fluorescence-based reporters have provided non-invasive readouts at whole-body and single-cell resolution (Jones et al., 2011, Swanzey and Stadtfeld, 2016), but tissue depth and sensitivity constraints may limit their general applicability. To provide new tools for investigating the consequences of early-life adversity, we generated a series of knockin embryonic stem cells (ESCs) and mouse lines in which firefly luciferase reports endogenous imprinted gene expression, and non-invasive bioluminescent imaging provides a means of monitoring expression longitudinally in vivo.
Cdkn1c is a maternally expressed imprinted gene that lies within the imprinting cluster 2 (IC2) on mouse chromosome 7 and is imprinted in both mice and humans (Hatada and Mukai, 1995, Hatada et al., 1996). The gene encodes a cyclin-dependent kinase inhibitor that is transiently expressed during embryogenesis in cells exiting proliferation (Lee et al., 1995, Matsuoka et al., 1995) and is particularly abundant within neural and skeleto-muscular tissue around mid-gestation (Westbury et al., 2001). Cdkn1c has an important role in regulating fetal growth and placental development (Andrews et al., 2007, Takahashi et al., 2000, Tunster et al., 2011) as well as lineage-specific roles, including in brown adipose tissue (Van De Pette et al., 2016), skeletal muscle (Osborn et al., 2011), and in adult quiescent stem cells (Zacharek et al., 2011, Matsumoto et al., 2011, Joseph et al., 2009). Cdkn1c lies within a complex imprinted domain regulated by an imprinting center that acquires DNA methylation in the maternal germline (gametic DMR; KvDMR1; John and Lefebvre, 2011, Hatada and Mukai, 1995, Hatada et al., 1996, Mohammad et al., 2012). This differentially methylated region spans the promoter of the paternally expressed long non-coding RNA Kcnq1ot1 required for continuous domain-wide imprinting. The Cdkn1c promoter and gene body are also directly DNA methylated on the paternal allele post-fertilization, after allelic silencing has been established (somatic DMR; Cdkn1c-sDMR [somatic differentially methylated region]; Bhogal et al., 2004). Given the profound effect of modest dosage alteration of this gene on post-natal metabolic and behavioral processes (Andrews et al., 2007, Van De Pette et al., 2016, McNamara et al., 2016), it provides an ideal candidate to study using sensitive allele-specific reporters.
Results
Generating Luciferase-Based Allelic Reporters for Mouse Cdkn1c
Mouse ESC lines were generated in which firefly luciferase (FLuc) alone, or in combination with β-galactosidase (FLucLacZ), was knocked into the endogenous Cdkn1c locus (Figures S1A and 1A, respectively). In some of the resulting targeted clones, low-level bioluminescence was detected after adding the luciferase substrate D-luciferin, consistent with insertion of luciferase into the maternal allele in selected clones (Figure 1B, blue). Upon differentiation, we observed increased expression of Cdkn1c (Figures 1C, left, and S1B, left), as anticipated from previous studies (Wood et al., 2010). In clones with a presumed maternal insertion, increased Cdkn1c expression was coupled to a corresponding increase in luciferase expression (Figures 1C and S1B). In clones with a presumed paternal insertion (KIpat), increased levels of Cdkn1c expression were not accompanied by luciferase expression (Figures 1C and S1B), consistent with maintenance of the silent imprint.
Figure 1.
Visualizing Cdkn1c Gene Expression In Vivo Using Bioluminescence
(A) Scheme of alternative knockin (KI) strategies used to generate Cdkn1c-FLucLacZ and Cdkn1c-FLuc embryonic stem cells (ESCs) and reporter mouse lines, in which sequences coding for the T2A peptide, the open reading frame of FLuc, a second T2A peptide, and the open reading frame of LacZ were inserted between the last amino acid and the translation termination codon in exon 3 (…KRLREGRG…; Cdkn1c-FLucLacZ) or IRES elements and the open reading frame of FLuc was inserted into a unique HindIII in the 3′ UTR (Cdkn1c-FLuc).
(B) Low-level bioluminescence (blue-green) in Cdkn1c-FLucLacZ and Cdkn1c-FLuc ESCs was detected in clones with a presumed maternal insertion (KImat), but not in clones with a paternal insertion (KIpat) or in wild-type ESCs (wt) (scale bar represents levels of bioluminescence).
(C) Total Cdkn1c expression (left), determined by RT-PCR, was increased in ESC clones with either a KImat (dashed line) or a KIpat (solid line) insertion over 21 days of embryoid body differentiation. Luciferase expression (right), determined by RT-PCR, was detected uniquely in KImat clones. Samples were normalized to β-actin and expressed as the mean ± SE.
(D) Bioluminescent imaging of representative P28 female Cdkn1c-FLucLacZ mice. Luciferase activity was observed in Cdkn1c-FLucLacZ KImat, with very low/negligible signals detectable upon paternal inheritance (KIpat) or in wild-type mice (wt). Strongest signal was evident in the skin, with low level signal detected in the internal organs.
(E) Bioluminescence detected in pregnancies with maternal inheritance of Cdkn1c-FLucLacZ (KImat, left) in utero, but not paternal inheritance (KIpat, right; less than twice background). Lower panels show bioluminescence imaging of dissected E11.5 embryos, where luciferase activity was seen in head and back of KImat embryos and placental tissue and quantified (flux). All Cdkn1c-FLucLacZ embryos imaged showed predicted parent-of-origin-specific bioluminescent activity.
(F) Total Cdkn1c gene expression in embryos (E11.5) was determined by RT-PCR, and levels were similar in samples from wild-type and where Cdkn1c-FLucLacZ was transmitted maternally (KImat) or paternally (KIpat; left). Luciferase (black) and Cdkn1c-Luciferase (gray) transcripts were detected uniquely from KImat. Samples were normalized to β-actin and expressed as the mean ± SE.
(G) Scheme of the mouse IC2 imprinting domain, showing the two DMRs that regulate Cdkn1c imprinted expression (KvDMR1 and Cdkn1c sDMR) and the position of bi-allelic (white), maternally expressed (dark gray), and paternally expressed (light gray) genes. Bisulfite analysis showing DNA methylation at KvDMR1 and Cdkn1c sDMR is similar in KImat and wt embryos at E11.5 (closed circles, methylated; open circles, un-methylated; where number indicates fully un-methylated strands).
Mice were generated from targeted ESCs to test whether bioluminescence was observed in offspring (Figures 1D and S1C) and to verify that this activity was transmitted in the appropriate parent-of-origin manner. Maternal transmission of the FLucLacZ transgene resulted in bioluminescent signal in the skin and internal organs of transgenic offspring (blue signal; KImat) at 4 weeks of age, with no signal evident in offspring after paternal inheritance (KIpat) or in non-transgenic (wild-type [WT]) controls (Figure 1D). Strikingly, pregnant females carrying embryonic day 11.5 (E11.5) Cdkn1c-FLucLacZ KImat embryos (14/14), but not KIpat embryos (0/10), showed a strong bioluminescent signal in the abdominal region (Figure 1E, upper). On dissection, transgenic embryos and placenta carrying the maternal targeted allele appropriately expressed luciferase, whereas those carrying the paternal targeted allele show no bioluminescence (Figure 1E, lower). Similar results were obtained with Cdkn1c-FLuc mice (Figure S1C).
Staining of E11.5 Cdkn1c-FLucLacZ KImat embryos for LacZ (Figure S1D) confirmed spatially appropriate expression in the hindbrain, spine, and developing cartilage, consistent with the published distribution of Cdkn1c (Westbury et al., 2001). This was further verified by 3D imaging using optical projection tomography (OPT) of cleared embryos (Figure S1D, lower; Movie S1), combined with photoacoustic tomography (Figure S1E). Importantly, no staining was detected in KIpat embryos by this sensitive approach, confirming global repression of the paternal allele. Consistent with this, luciferase mRNA was only detectable after maternal inheritance (Figure 1F) alongside wild-type levels of the Cdkn1c transcript. Amplification with a primer set that spanned Cdkn1c exon 3 and luciferase exon 1 confirmed linked expression of luciferase and endogenous Cdkn1c transcripts (Figure 1F). Bisulfite analysis of the two differentially methylated regions associated with Cdkn1c imprinting (Bhogal et al., 2004, Mancini-Dinardo et al., 2006) showed normal DNA methylation patterns in heads of Cdkn1c-FLucLacZ KImat embryos (Figure 1G). Collectively, these data indicate that luciferase accurately reports Cdkn1c expression without impairing the methylation or regulation of the endogenous locus.
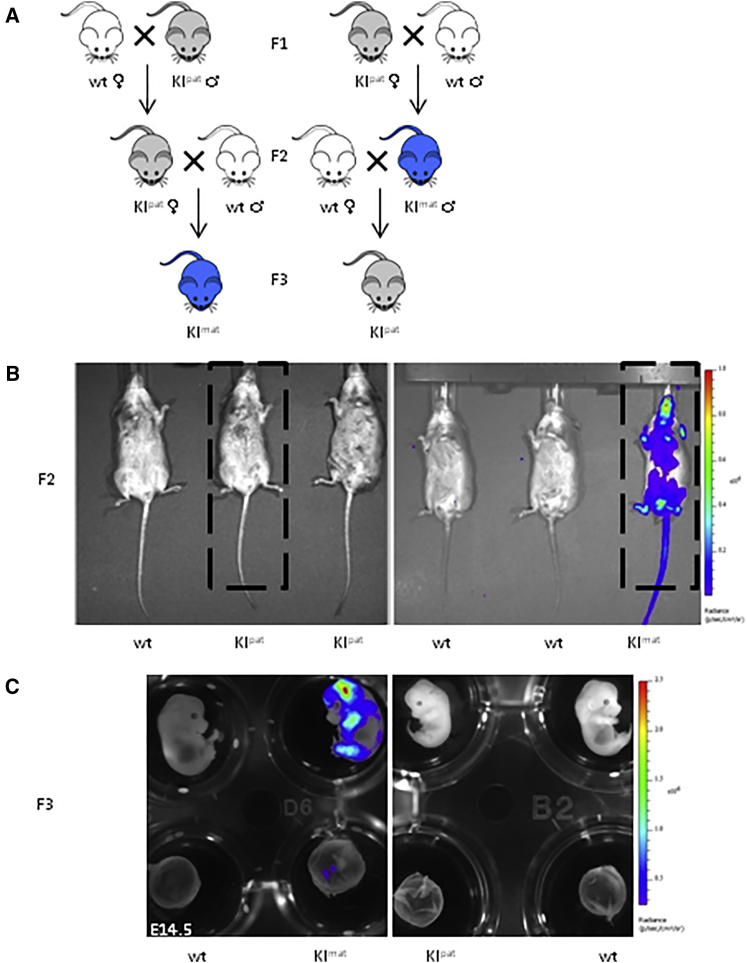
Imprinted Cdkn1c-FlucLacZ Expression Is Appropriately Reset through the Germline
Epigenetic marks that establish and maintain imprinting are normally erased and reset in the germline so that allelic expression is correctly maintained in subsequent generations (Bartolomei and Ferguson-Smith, 2011). To check whether erasure and resetting of imprints occurred normally in the luciferase-targeted mice, we tracked bioluminescence (blue) among reciprocal genetic crosses of Cdkn1c-FLucLacZ (gray) and wild-type mice (white) across generations (Figure 2A; F1, F2, and F3). Tracing bioluminescence activity across three generations revealed epigenetic inheritance as predicted (Figures 2B and 2C), in which allelic silencing of Cdkn1c-FLucLacZ was reversed through maternal transmission and re-established through paternal transmission. The ability to image Cdkn1c expression longitudinally in vivo through successive generations suggested that these reporter mice might be useful and robust models to screen for factors and environmental stresses that could interfere with imprinting. Importantly, as female mice inheriting Cdkn1c-FLucLacZ paternally (KIpat; left box, Figure 2B) were devoid of luciferase signal, these animals offered an optimal setting (minimal background) to detect bioluminescence signals in utero from KImat embryos and placental tissue (Figures 1E, left, and 2C).
Figure 2.
Correct Imprint Resetting of Cdkn1c-FLucLacZ across Generations
(A) Diagram showing predicted expression and inheritance of a maternally expressed imprinted gene (such as Cdkn1c) or transgene (Cdkn1c-FLucLacZ) in reciprocal crosses across three generations. Wild-type mice are shown in white, expression through maternal inheritance is shown in blue, and inheritance of a silent imprint (Cdkn1c-FLucLacZ; KIpat) is indicated in gray.
(B) Experimental evidence of imprint resetting; bioluminescent imaging of adult F2 mice, Cdkn1c-FLucLacZ KIpat females (left box), and KImat males (right box), showing predicted parent-of-origin-specific luciferase activity (blue). Highlighted animals were then used to generate F3 (as outlined in A).
(C) Bioluminescent image of E14.5 embryos, generated from the indicated transgenic parents; signal was detected upon maternal inheritance of luciferase in embryos (upper panel) and in placental tissue (lower panel; left), which had been silent in the previous generation. Conversely, paternal inheritance of the previously active luciferase was sufficient to silence the previously active allele (right).
Chromatin-Modifying Drugs Transiently Disrupt Paternal Silencing of Cdkn1c In Utero
5′azacytidine (5′Aza) disrupts DNA methylation in cells by inhibiting DNMT1 activity, thereby preventing the incorporation of 5-methylcytosine into hemi-methylated DNA strands at S phase. In dividing cells in culture, 5′Aza treatment has been shown to reduce DNA methylation at the Cdkn1c promoter (Flotho et al., 2009). Trichostatin A (TSA) inhibits histone deacetylase activity and has been shown to deplete repressive histone marks at the Cdkn1c promoter (Yang et al., 2009). We reasoned that drugs that alter chromatin, such as 5′Aza and TSA, might be effective at disrupting Cdkn1c expression when epigenetic marks are consolidated (Bhogal et al., 2004, Umlauf et al., 2004). To examine this possibility, wild-type female mice were crossed with homozygous Cdkn1c-FLucLacZ (KI/KI) males to produce heterozygous offspring in which the Cdkn1c-FLucLacZ imprint was repressed. The pregnant females were then treated with drugs at E12.5–E13.5, and bioluminescence was evaluated at E14.5, at birth (P1), and at 4 weeks of age (postnatal day 28 [P28]; Figure 3A). Bioluminescence was detected in utero at E14.5 with strongest signal seen following combined drug treatment (Combi) (Figure 3B). Control vehicle-treated KIpat embryos were consistently negative throughout these studies. For 5′Aza- (3/9) and TSA-treated (4/9) pregnancies, bioluminescence was not detected in all the transgenic embryos, whereas all the transgenic embryos (7/7) displayed increased luciferase activity upon combination treatment (Figure 3C). These animals showed a corresponding decrease in DNA methylation across the Cdkn1c somatic DMR at E14.5 as compared with controls (Figure 3D). We noticed that the levels of bioluminescence were generally lower than in age-matched Cdkn1c-FLucLacZ KImat embryos, consistent with partial de-repression of the paternal allele. Furthermore, de-repression appeared transient and was variable among combination drug-treated animals, as shown in pups imaged at birth (P1; Figure 3E). Four weeks after birth (P28), bioluminescence signal was no longer evident in drug-treated KIpat animals (Figure 3F), and DNA methylation in the brain was similar in vehicle- and Combi-treated Cdkn1c-FLucLacZ KIpat mice (Figure 3G). Taken together, these data show that conventional chromatin-modifying drugs alone or in combination are capable of relieving imprinted Cdkn1c-FLucLacZ repression in developing embryos.
Figure 3.
Silencing of Paternal Cdkn1c-FLucLacZ Is Transiently Released by In Utero Exposure to Epigenetic Drugs
(A) Embryos carrying silent (paternally inherited) Cdkn1c-FLucLacZ were generated by mating wild-type (wt) females with homozygous Cdkn1c-FLucLacZ males. Pregnant females were treated with trichostatin A (TSA) or 5′ azacytidine (5′Aza) alone or together at the times indicated. Offspring were imaged at E14.5, at birth (P1), and at 4 weeks of age (P28).
(B) Low-level bioluminescence was occasionally detected in 5′Aza- and TSA-alone treated pregnancies, whereas stronger and consistent signal (blue) was detected in combination-treated (Combi) embryos in utero (left) or individually dissected embryos (right) in the head and back.
(C) Cdkn1c-FLucLacZ-derived bioluminescent activity was consistently elevated in E14.5 embryos exposed to combined drug treatment.
(D) Bisulfite analysis of DNA methylation at the KvDMR1 and Cdkn1c sDMR in the brain of E14.5 Cdkn1c-FLucLacZ KIpat embryos shows reduced methylation in embryos exposed to combination drug treatment versus controls (closed circles, methylated; open circles, un-methylated).
(E) Variable increases in luciferase activity (blue, flux) characterize combination-drug-treated Cdkn1c-FLucLacZ KIpat animals at P1 (right), with no signal detected in vehicle-treated controls (left).
(F) Luciferase activity was no longer detected in Cdkn1c-FLucLacZ KIpat mice at P28 that had been exposed to combination drug treatment in utero.
(G) Bisulfite analysis of DNA methylation at the KvDMR1 and Cdkn1c sDMR in the brain of P28 Cdkn1c-FLucLacZ KIpat mice shows that previously ablated methylation is restored by adulthood to normal levels (closed circles, methylated; open circles, un-methylated).
Dietary Protein Restriction In Utero Provokes De-repression of Paternal Cdkn1c into Adulthood
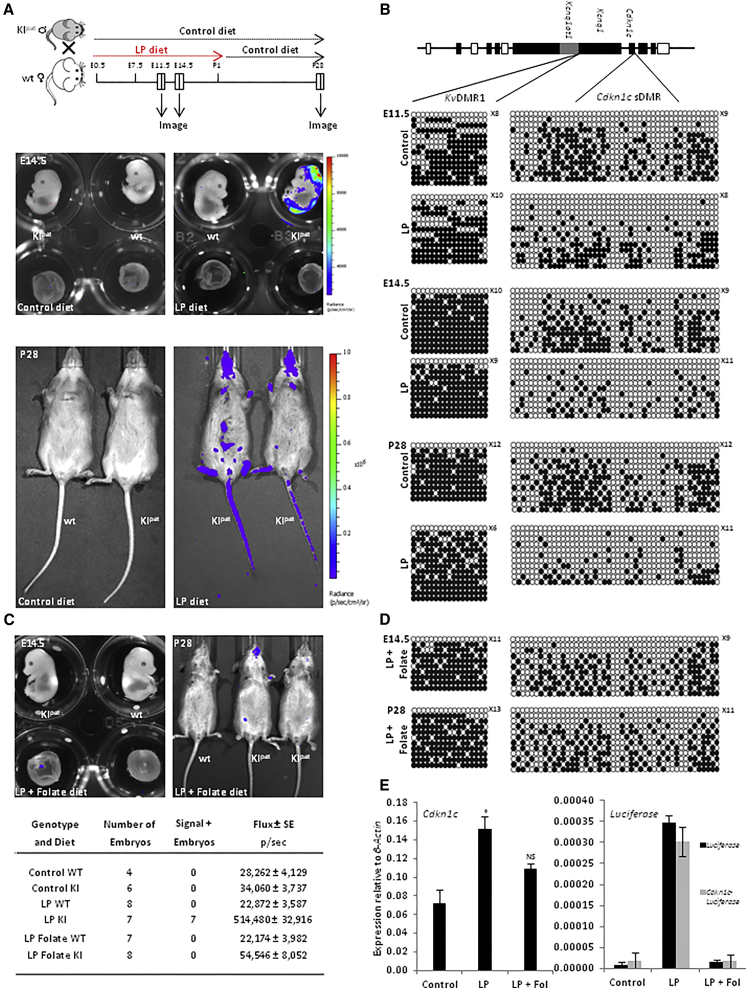
Cdkn1c has previously been proposed to be sensitive to in utero dietary protein restriction (Vucetic et al., 2010). In particular, mice that were fed a low-protein diet through pregnancy (as a surrogate for early-life adversity) produced offspring with elevated levels of Cdkn1c in the midbrain associated with DNA hypo-methylation at the promoter. To examine whether exposure to low-protein diet in utero provokes de-repression of the silent paternal Cdkn1c-FLucLacZ, we crossed wild-type female mice with heterozygous Cdkn1c-FLucLacZ (WT/KI) males (Figure 4A, schematic). Pregnant mice were fed calorie-balanced, low-protein diet (LP) from the detection of vaginal plugs until birth. All newborn offspring were maintained thereafter on a normal (unrestricted) diet. This window of exposure ensures that the influence of LP diet is restricted to a specific period of development. Although bioluminescence signal was not detected at E11.5 (Figure S2), by E14.5, all Cdkn1c-FLucLacZ KIpat embryos expressed luciferase following maternal exposure to low-protein diet (exemplified in Figure 4A, middle right). Signal was most pronounced in the head, and luciferase re-expression among KIpat embryos was prominent in the midbrain region (Figure S3). De-repression was sustained in mice imaged subsequently at 4 weeks of age (Figure 4A, lower right) and throughout adulthood, despite no longer being exposed to a restricted diet. These data establish that in utero exposure to a low-protein diet results in permanent de-repression of the normally silent paternal allele.
Figure 4.
Stable Silencing of Paternally Inherited Cdkn1c-FLucLacZ during Life Course Depends upon the Availability of Methyl Donors In Utero
(A) Offspring with a silent (paternally inherited) Cdkn1c-FLucLacZ were generated by mating wild-type (wt) females with heterozygous Cdkn1c-FLucLacZ males. Upon detection of a vaginal plug, a group of pregnant females were switched to a calorie-matched but low-protein (LP) diet for the duration of their pregnancy, with mothers and litters returning to a normal diet after birth. Pregnancies were imaged/examined at the times indicated (E11.5, E14.5, and P28). No mis-expression of luciferase was observed at day E11.5 (Figure S2), irrespective of diet; however, by E14.5, luciferase activity was detected selectively in embryos of mothers fed LP diet (upper right) and expression continued as these matured into adults (lower right). No signal was detected in animals fed normal (control) diet at any time.
(B) Comparative bisulfite analysis of DNA methylation at the Cdkn1c locus in the brain of Cdkn1c-FLucLacZ KIpat animals (E11.5, E14.5, and P28) born to mothers fed control versus LP diet during pregnancy. Cdkn1c sDMR becomes hypo-methylated in LP conditions in utero, and methylation is not restored subsequently (closed circles, methylated; open circles, un-methylated). Methylation at the KvDMR1 is unaltered.
(C) Pregnant females as in (A) were fed LP diet supplemented with increased folate. Bioluminescent imaging of embryos (E14.5) from mothers fed LP + folate showed reduced mis-expression of Cdkn1c-FLucLacZ KIpat as compared with those fed LP alone (A), with luciferase activity remaining low or negligible as they matured into adults (P28; upper right; image scales same as A).
(D) Bisulfite analysis showing DNA methylation at KvDMR1 and Cdkn1c sDMR in E14.5 (upper) and P28 (lower) animals born to mothers fed LP + folate diet. Progressive hypo-methylation of the Cdkn1c sDMR was buffered against by the increased dietary folate.
(E) Total Cdkn1c gene expression in E14.5 brain was determined by RT-PCR, and levels were elevated in samples from LP-exposed litters (p < 0.033), compared to control and LP + folate litters (p < 0.38). Luciferase (black) and Cdkn1c-Luciferase (gray) transcripts were detected using RT-PCR uniquely in LP brain samples, demonstrating loss of imprinting. Samples were normalized to β-actin and expressed as the mean ± SE.
To further explore the mechanism underlying Cdkn1c re-expression, we compared DNA methylation in the brain at E11.5, E14.5, and in adults at 4 weeks of age (Figure 4B). Appropriate DNA methylation at the somatic DMR was evident at E11.5, when no luciferase activity was detected (Figure S2), but this was progressively eroded in animals exposed to LP diet during gestation (Figure 4B). These data show that, under these conditions, the somatic DMR is established correctly, but not maintained, suggesting that dietary protein may be required to sustain DNA methylation at the paternal allele. In contrast, DNA methylation at KvDMR1 was unaffected by LP diet (Figure 4B), consistent with previous reports (Ivanova et al., 2012).
Rescue of Dietary-Induced Loss of Paternal Cdkn1c Silencing by Folate Supplementation
As dietary protein is known to be an important source of methyl donors required for DNA methylation, we hypothesized that a paucity of methyl donors might contribute to the failure to sustain repression of Cdkn1cpat alleles in vivo. To test this, we repeated the dietary experiments using the low-protein diet with increased folate supplementation as a source of methyl donors. This had a dramatic effect, reducing paternal Cdkn1c-FLucLacZ bioluminescence to background levels in embryos (Figure 4C, left) and in resulting adults (Figure 4C, right). We also found that, following folate supplementation, methylation of Cdkn1c somatic DMR was indistinguishable from normal controls (Figures 4B and 4D). De-repression and restoration of Cdkn1c-FlucLacZ silencing in response to LP and LP + folate diet, respectively, was directly validated by allele-specific transcript analysis (Figure 4E). Thus, although previous studies have shown Cdkn1c upregulation in response to LP diet (Vucetic et al., 2010), our data now establish that dietary restriction can cause loss of imprinting.
Discussion
In utero development is critically dependent on imprinted gene dose (Radford et al., 2011). This necessary control has been shown to extend into the programming of adult metabolism (Da Rocha et al., 2009; Charalambous et al., 2012). Here, we show that maternal dietary restriction has a profound impact on Cdkn1c expression in the embryo, provoking a partial loss of imprinting that persists through adult life, even when a normal diet is resumed. Prolonged exposure to low-protein diet during gestation erodes DNA methylation at the Cdkn1c somatic DMR and results in re-expression of the paternal allele. Because deregulation is rescued by elevated folate supplementation, methyl donor deprivation appears to be the most likely cause of imprint erosion. Although we do not yet know whether this reflects a specific window of vulnerability in embryonic development or simply an increased demand engendered by proliferating cells in the embryo, the observation that the gametic KvDMR1 DMR resists DNA de-methylation supports previous findings that gametic differentially methylated regions (gDMRs) are relatively stable (Ivanova et al., 2012). Mechanistically, gametic and somatic DMRs both require the maintenance of DNA methylation by DNMT1 (Caspary et al., 1998, Bhogal et al., 2004). However, whereas zygotic deficiency of the de novo methylases Dnmt3a or 3b has no effect on gDMR methylation, loss of Dnmt3b results in de-methylation of Cdkn1c-sDMR independent of KvDMR1 status (Auclair et al., 2014). These data provide a precedent for the differential sensitivities of the gametic and somatic DMRs and implicate Dnmt3b as a candidate in preventing hypo-methylation at the Cdkn1c sDMR.
Imprinted genes are pivotal for regulating growth and metabolism, and yet the intricacies of imprinting have remained challenging to study. This reflects the intrinsic complexity of imprinting control regions (ICRs) but also a paucity of markers needed to reliably distinguish maternal from paternal alleles. Here, we describe two independent mouse lines in which luciferase-based bioluminescence reports allelic Cdkn1c expression, without disruption of endogenous gene output. The value of using this non-invasive approach is that it allows allelic expression to be imaged in individuals throughout life course so that epigenetic changes and their consequences can be evaluated directly. The close correspondence of luciferase expression in Cdkn1c-FLuc and Cdkn1c-FLucLacZ mouse lines suggests similar approaches might also be useful in studying allelic expression from other imprinted loci. Consistent with this idea, we have generated a series of ESC lines that report maternally expressed (Ube3a) or paternally expressed imprinted genes (Dlk1, Nnat, and Igf2) and are characterizing luciferase expression in mouse lines derived from such (Table S1). These lines, together with the Cdkn1c-FLucLacZ and Cdkn1c-FLuc lines described herein, provide novel genetic tools to interrogate the epigenetic mechanisms that establish, maintain, and reprogram imprinted gene expression in the female and the male germlines.
The observation that the Cdkn1c imprint is permanently disrupted by altered maternal diet provides a clear link between early-life adversity and the subsequent epigenetic mis-regulation in adult life. Our results suggest that a deficiency in methyl donor supply in utero is the most likely cause of imprint disruption, whereas limited exposure to well-characterized chromatin-modifying drugs in utero transiently deregulates imprint silencing. The basis of these different epigenetic outcomes is interesting and could reflect differences in the timing or length of exposure, inherent differences in cell proliferation, or the susceptibility of developing tissue to certain agents. Although future studies will be required to discriminate these possibilities, our ability to detect transient and permanent changes in imprint silencing in vivo offers an exciting new opportunity to explore the plasticity of epigenetic processes and their phenotypic outcome. More broadly, these luciferase-based imaging models will facilitate the rapid screening of epigenetic drugs and environmental stresses relevant for drug discovery programs and for understanding how epigenome deregulation in early life impacts upon longer-term health.
Experimental Procedures
Animal Maintenance
Mice were handled and all in vivo studies were performed in accordance with the United Kingdom Animals (Scientific Procedures) Act (1986), were approved by the Imperial College AWERB committee, and performed under a UK Home Office project license.
Epidrug Injections
5′Aza (Sigma-Aldrich) and TSA (Sigma-Aldrich) were dissolved as 0.75 μg/μL and 0.3 μg/μL stocks in PBS and 30% ethanol, respectively. Wild-type 129S2/SvHsd dams were set up with Cdkn1c-FLucLacZ males, and upon vaginal plug discovery, matings were separated. For 5′Aza administration, pregnant dams were injected with 5 μg/g body weight at E12.5 intraperitoneally (i.p.). For TSA administration, pregnant dams were injected i.p. with 1 μg/g body weight at E12.5 and E13.5. Vehicle injections were performed with 30% ethanol at the same time points as TSA injections. Pregnant dams and embryos were imaged at E14.5; offspring were imaged at P1 and P28.
Low Protein Study
Wild-type 129S2/SvHsd dams were set up with Cdkn1c-FLucLacZ males, and upon vaginal plug discovery, matings were separated. Females were fed either a low-protein chow (5769; TestDiet), a calorie-matched control chow (5755; TestDiet), or a low-protein chow with elevated folate supplement (5769 with 20 PPM Folate; TestDiet) until E11.5 or E14.5 for embryonic studies or birth for adult studies. Pregnant dams and embryos were imaged at E14.5; offspring were imaged at P28.
Bioluminescent Imaging
D-Luciferin (PerkinElmer) was dissolved in H20 at 30 mg/mL. For in vitro studies, cells were grown to 90% confluence, 150 μg/mL was added to the medium, and plates were imaged after 2 min. For in vivo studies, mice were weighed and injected i.p. with 0.15 mg/g body weight before being anesthetized with isoflurane. Mice were imaged 10 min post-injection in an IVIS Spectrum (PerkinElmer) under anesthesia. Images of cell plates, adult mice, and pregnant dams were taken at field of view (FOV) C, with binning 4 and 180 s exposure. For imaging of embryos, pregnant females were injected with D-Luciferin at least 10 min prior to imaging. Embryos were dissected into 24-well dishes containing PBS and placed in the IVIS Spectrum. Images of embryos were taken at FOV A, with binning 1, focus 1 cm, and 180 s exposure. For epidrug and low protein imaging, settings were the same, with the exception of binning 4 in embryos. No additional D-Luciferin was added, and imaging continued for up to 35 min post-injection. Analysis of images was performed on Living Image software (Caliper Life Sciences). For quantification of bioluminescent signal, regions of interest were drawn around embryos and signal flux within the region was calculated.
Author Contributions
M.V.d.P., R.M.J., and A.G.F. conceived of and wrote the manuscript. R.M.J., A.C.F.-S., A.U., D.J.W., and M.M. were instrumental in designing the vectors used to generate and characterize the mice. A.S., J.M., and P.M.W.F. helped with the development of imaging protocols. A.F., L.B., W.K.T., A.D., G.M., A.A., E.J.P., M.A.U., and Z.W. contributed to the experiments described.
Acknowledgments
The work was funded by the MRC, the European Research Council, and institutional support to Imperial College from the Wellcome Trust, NIHR Imperial BRC (to A.A.). We thank FUJIFILM VisualSonics for their help in the development and production of photoacoustic imaging. We thank Taconic Biosciences for embryonic stem cell and animal services. We are indebted to John Savill for his advice and suggestion to develop methods allowing epigenetic change to be imaged in vivo. R.M.J.’s research is funded by MRC grant MR/M013960/1 and BBSRC grant BB/J015156. A.G.F. is funded by ERC Advanced Grant 294627.
Published: January 31, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, one table, and one movie and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.01.010.
Supplemental Information
Absorbance (green) was measured in the developing cartilage, spine, hindbrain and liver in KImat embryos. Weaker absorbance was also detectable in liver. Very low absorbance was observed in KIpat and wt embryos.
References
- Andrews S.C., Wood M.D., Tunster S.J., Barton S.C., Surani M.A., John R.M. Cdkn1c (p57Kip2) is the major regulator of embryonic growth within its imprinted domain on mouse distal chromosome 7. BMC Dev. Biol. 2007;7:53. doi: 10.1186/1471-213X-7-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auclair G., Guibert S., Bender A., Weber M. Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse. Genome Biol. 2014;15:545. doi: 10.1186/s13059-014-0545-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M.S., Ferguson-Smith A.C. Mammalian genomic imprinting. Cold Spring Harb. Perspect. Biol. 2011;3:a002592. doi: 10.1101/cshperspect.a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhogal B., Arnaudo A., Dymkowski A., Best A., Davis T.L. Methylation at mouse Cdkn1c is acquired during postimplantation development and functions to maintain imprinted expression. Genomics. 2004;84:961–970. doi: 10.1016/j.ygeno.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Caspary T., Cleary M.A., Baker C.C., Guan X.J., Tilghman S.M. Multiple mechanisms regulate imprinting of the mouse distal chromosome 7 gene cluster. Mol. Cell. Biol. 1998;18:3466–3474. doi: 10.1128/mcb.18.6.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charalambous M., Ferron S.R., da Rocha S.T., Murray A.J., Rowland T., Ito M., Schuster-Gossler K., Hernandez A., Ferguson-Smith A.C. Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 2012;15:209–221. doi: 10.1016/j.cmet.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaton M.A.M., Edwards C.A., Ferguson-Smith A.C. Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes. Annu. Rev. Genomics Hum. Genet. 2014;15:93–126. doi: 10.1146/annurev-genom-091212-153441. [DOI] [PubMed] [Google Scholar]
- da Rocha S.T., Charalambous M., Lin S.P., Gutteridge I., Ito Y., Gray D., Dean W., Ferguson-Smith A.C. Gene dosage effects of the imprinted delta-like homologue 1 (dlk1/pref1) in development: implications for the evolution of the imprinting. Plos Genetics. 2009;5:e1000392. doi: 10.1371/journal.pgen.1000392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day T., Bonduriansky R. Intralocus sexual conflict can drive the evolution of genomic imprinting. Genetics. 2004;167:1537–1546. doi: 10.1534/genetics.103.026211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho C., Claus R., Batz C., Schneider M., Sandrock I., Ihde S., Plass C., Niemeyer C.M., Lübbert M. The DNA methyltransferase inhibitors azacitidine, decitabine and zebularine exert differential effects on cancer gene expression in acute myeloid leukemia cells. Leukemia. 2009;23:1019–1028. doi: 10.1038/leu.2008.397. [DOI] [PubMed] [Google Scholar]
- Haig D. Genomic imprinting and kinship: how good is the evidence? Annu. Rev. Genet. 2004;38:553–585. doi: 10.1146/annurev.genet.37.110801.142741. [DOI] [PubMed] [Google Scholar]
- Hatada I., Mukai T. Genomic imprinting of p57KIP2, a cyclin-dependent kinase inhibitor, in mouse. Nat. Genet. 1995;11:204–206. doi: 10.1038/ng1095-204. [DOI] [PubMed] [Google Scholar]
- Hatada I., Inazawa J., Abe T., Nakayama M., Kaneko Y., Jinno Y., Niikawa N., Ohashi H., Fukushima Y., Iida K. Genomic imprinting of human p57KIP2 and its reduced expression in Wilms’ tumors. Hum. Mol. Genet. 1996;5:783–788. doi: 10.1093/hmg/5.6.783. [DOI] [PubMed] [Google Scholar]
- Ivanova E., Chen J.H., Segonds-Pichon A., Ozanne S.E., Kelsey G. DNA methylation at differentially methylated regions of imprinted genes is resistant to developmental programming by maternal nutrition. Epigenetics. 2012;7:1200–1210. doi: 10.4161/epi.22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John R.M. Engineering mouse models to investigate the function of imprinting. Brief. Funct. Genomics. 2010;9:294–303. doi: 10.1093/bfgp/elq010. [DOI] [PubMed] [Google Scholar]
- John R.M., Lefebvre L. Developmental regulation of somatic imprints. Differentiation. 2011;81:270–280. doi: 10.1016/j.diff.2011.01.007. [DOI] [PubMed] [Google Scholar]
- John R.M., Surani M.A. Genomic imprinting, mammalian evolution, and the mystery of egg-laying mammals. Cell. 2000;101:585–588. doi: 10.1016/s0092-8674(00)80870-3. [DOI] [PubMed] [Google Scholar]
- Jones M.J., Bogutz A.B., Lefebvre L. An extended domain of Kcnq1ot1 silencing revealed by an imprinted fluorescent reporter. Mol. Cell. Biol. 2011;31:2827–2837. doi: 10.1128/MCB.01435-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Andersson E.R., Vlachos P., Södersten E., Liu L., Teixeira A.I., Hermanson O. p57Kip2 is a repressor of Mash1 activity and neuronal differentiation in neural stem cells. Cell Death Differ. 2009;16:1256–1265. doi: 10.1038/cdd.2009.72. [DOI] [PubMed] [Google Scholar]
- Lam W.W.K., Hatada I., Ohishi S., Mukai T., Joyce J.A., Cole T.R.P., Donnai D., Reik W., Schofield P.N., Maher E.R. Analysis of germline CDKN1C (p57KIP2) mutations in familial and sporadic Beckwith-Wiedemann syndrome (BWS) provides a novel genotype-phenotype correlation. J. Med. Genet. 1999;36:518–523. [PMC free article] [PubMed] [Google Scholar]
- Lee M.H., Reynisdóttir I., Massagué J. Cloning of p57KIP2, a cyclin-dependent kinase inhibitor with unique domain structure and tissue distribution. Genes Dev. 1995;9:639–649. doi: 10.1101/gad.9.6.639. [DOI] [PubMed] [Google Scholar]
- Mancini-Dinardo D., Steele S.J.S., Levorse J.M., Ingram R.S., Tilghman S.M. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A., Takeishi S., Kanie T., Susaki E., Onoyama I., Tateishi Y., Nakayama K., Nakayama K.I. p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell. 2011;9:262–271. doi: 10.1016/j.stem.2011.06.014. [DOI] [PubMed] [Google Scholar]
- Matsuoka S., Edwards M.C., Bai C., Parker S., Zhang P., Baldini A., Harper J.W., Elledge S.J. p57KIP2, a structurally distinct member of the p21CIP1 Cdk inhibitor family, is a candidate tumor suppressor gene. Genes Dev. 1995;9:650–662. doi: 10.1101/gad.9.6.650. [DOI] [PubMed] [Google Scholar]
- McGrath J., Solter D. Completion of mouse embryogenesis requires both the maternal and paternal genomes. Cell. 1984;37:179–183. doi: 10.1016/0092-8674(84)90313-1. [DOI] [PubMed] [Google Scholar]
- McNamara G.I., Davis B.A., Dwyer D.M., John R.M., Isles A.R. Behavioural abnormalities in a novel mouse model for Silver Russell syndrome. Hum. Mol. Genet. 2016 doi: 10.1093/hmg/ddw357. Published online October 24, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad F., Pandey G.K., Mondal T., Enroth S., Redrup L., Gyllensten U., Kanduri C. Long noncoding RNA-mediated maintenance of DNA methylation and transcriptional gene silencing. Development. 2012;139:2792–2803. doi: 10.1242/dev.079566. [DOI] [PubMed] [Google Scholar]
- Monk D., Arnaud P., Apostolidou S., Hills F.A., Kelsey G., Stanier P., Feil R., Moore G.E. Limited evolutionary conservation of imprinting in the human placenta. Proc. Natl. Acad. Sci. USA. 2006;103:6623–6628. doi: 10.1073/pnas.0511031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn D.P.S., Li K., Hinits Y., Hughes S.M. Cdkn1c drives muscle differentiation through a positive feedback loop with Myod. Dev. Biol. 2011;350:464–475. doi: 10.1016/j.ydbio.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford E.J., Ferrón S.R., Ferguson-Smith A.C. Genomic imprinting as an adaptative model of developmental plasticity. FEBS Lett. 2011;585:2059–2066. doi: 10.1016/j.febslet.2011.05.063. [DOI] [PubMed] [Google Scholar]
- Surani M.A. Imprinting and the initiation of gene silencing in the germ line. Cell. 1998;93:309–312. doi: 10.1016/s0092-8674(00)81156-3. [DOI] [PubMed] [Google Scholar]
- Surani M.A.H., Barton S.C., Norris M.L. Development of reconstituted mouse eggs suggests imprinting of the genome during gametogenesis. Nature. 1984;308:548–550. doi: 10.1038/308548a0. [DOI] [PubMed] [Google Scholar]
- Swanzey E., Stadtfeld M. A reporter model to visualize imprinting stability at the Dlk1 locus during mouse development and in pluripotent cells. Development. 2016;143:4161–4166. doi: 10.1242/dev.138255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Nakayama K., Nakayama K. Mice lacking a CDK inhibitor, p57Kip2, exhibit skeletal abnormalities and growth retardation. J. Biochem. 2000;127:73–83. doi: 10.1093/oxfordjournals.jbchem.a022586. [DOI] [PubMed] [Google Scholar]
- Tunster S.J., Van de Pette M., John R.M. Fetal overgrowth in the Cdkn1c mouse model of Beckwith-Wiedemann syndrome. Dis. Model. Mech. 2011;4:814–821. doi: 10.1242/dmm.007328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D., Goto Y., Cao R., Cerqueira F., Wagschal A., Zhang Y., Feil R. Imprinting along the Kcnq1 domain on mouse chromosome 7 involves repressive histone methylation and recruitment of Polycomb group complexes. Nat. Genet. 2004;36:1296–1300. doi: 10.1038/ng1467. [DOI] [PubMed] [Google Scholar]
- Van De Pette M., Tunster S.J., McNamara G.I., Shelkovnikova T., Millership S., Benson L., Peirson S., Christian M., Vidal-Puig A., John R.M. Cdkn1c boosts the development of brown adipose tissue in a murine model of Silver Russell syndrome. PLoS Genet. 2016;12:e1005916. doi: 10.1371/journal.pgen.1005916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucetic Z., Totoki K., Schoch H., Whitaker K.W., Hill-Smith T., Lucki I., Reyes T.M. Early life protein restriction alters dopamine circuitry. Neuroscience. 2010;168:359–370. doi: 10.1016/j.neuroscience.2010.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westbury J., Watkins M., Ferguson-Smith A.C., Smith J. Dynamic temporal and spatial regulation of the cdk inhibitor p57(kip2) during embryo morphogenesis. Mech. Dev. 2001;109:83–89. doi: 10.1016/s0925-4773(01)00512-3. [DOI] [PubMed] [Google Scholar]
- Wolf J.B., Hager R. A maternal-offspring coadaptation theory for the evolution of genomic imprinting. PLoS Biol. 2006;4:e380. doi: 10.1371/journal.pbio.0040380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood M.D., Hiura H., Tunster S.J., Arima T., Shin J.Y., Higgins M.J., John R.M. Autonomous silencing of the imprinted Cdkn1c gene in stem cells. Epigenetics. 2010;5:214–221. doi: 10.4161/epi.5.3.11275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X., Murthy Karuturi R.K., Sun F., Aau M., Yu K., Shao R., Miller L.D., Boon Ooi Tan P., Yu Q. CDKN1C (p57KIP2) is a direct target of EZH2 and suppressed by multiple epigenetic mechanisms in breast cancer cells. PLoS One. 2009;4:e5011. doi: 10.1371/journal.pone.0005011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharek S.J., Fillmore C.M., Lau A.N., Gludish D.W., Chou A., Ho J.W., Zamponi R., Gazit R., Bock C., Jäger N. Lung stem cell self-renewal relies on BMI1-dependent control of expression at imprinted loci. Cell Stem Cell. 2011;9:272–281. doi: 10.1016/j.stem.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Absorbance (green) was measured in the developing cartilage, spine, hindbrain and liver in KImat embryos. Weaker absorbance was also detectable in liver. Very low absorbance was observed in KIpat and wt embryos.