Highlight
Here we cloned and intensively characterized the first natural embryo-specific bidirectional promoter which could facilitate multi-gene expression in maize, and proposed a model of regulation of bidirectional transcription initiation.
Key words: Bidirectional promoter, cis-elements, embryo specificity, immature embryo transient expression system, transgenic maize, truncation analysis.
Abstract
Bidirectional promoters are identified in diverse organisms with widely varied genome sizes, including bacteria, yeast, mammals, and plants. However, little research has been done on any individual endogenous bidirectional promoter from plants. Here, we describe a promoter positioned in the intergenic region of two defensin-like protein genes, Def1 and Def2 in maize (Zea mays). We examined the expression profiles of Def1 and Def2 in 14 maize tissues by qRT-PCR, and the results showed that this gene pair was expressed abundantly and specifically in seeds. When fused to either green fluorescent protein (GFP) or β-glucuronidase (GUS) reporter genes, P ZmBD1, P ZmDef1, and P ZmDef2 were active and reproduced the expression patterns of both Def1 and Def2 genes in transformed immature maize embryos, as well as in developing seeds of transgenic maize. Comparative analysis revealed that PZmBD1 shared most of the expression characteristics of the two polar promoters, but displayed more stringent embryo specificity, delayed expression initiation, and asymmetric promoter activity. Moreover, a truncated promoter study revealed that the core promoters only exhibit basic bidirectional activity, while interacting with necessary cis-elements, which leads to polarity and different strengths. The sophisticated interaction or counteraction between the core promoter and cis-elements may potentially regulate bidirectional promoters.
Introduction
Promoters in the intergenic regions of divergent genes are arranged in one of three ways: head-to-head, tail-to-tail, or overlapping (Beck and Warren, 1988). Studies of overlapping promoters, defined as bidirectional promoters, in various species have increased in number over the past few decades. Adachi and Lieber (2002) presented an overview of bidirectional promoters in the human genome. A study reported that 10% of genes in the human genome with high GC content and less than 1kb in length shared bidirectional promoters (Trinklein et al., 2004). Bidirectional promoters are widespread in yeast genomes and generate pervasive transcription (Neil et al., 2009; Xu et al., 2009). Krom and Ramakrishna (2008) performed a comparative analysis of the expression patterns of gene pairs whose expression was directed by a shared bidirectional promoter in rice, Arabidopsis, and Populus and showed that they were mostly co-expressed and were usually involved in the same pathway. Using a bidirectional promoter search model created by Trinklein et al. (2004), ~2471 bidirectional promoters were identified in Arabidopsis (Wang et al., 2009). Given differences in the gene organization of various species, diverse threshold values for bidirectional promoter length have been used in other plant genome-wide analyses. A more stringent criterion (<0.25kb) was used to identify 212, 462, and 141 bidirectional promoters from rice, Arabidopsis, and Populus (Dhadi et al., 2009). Kourmpetli et al. (2013) coupled two criteria – a bidirectional promoter length between 0.1 and 0.6kb and the presence of at least two G-box sequences (CACGTG) within the intergenic region – to identify 70 bidirectional promoters involved in seed development in Arabidopsis. Our previous work showed that more bidirectional promoters can be identified in rice, Arabidopsis, Populus, soybean, and maize by using the transcript physical location instead of the gene physical location as coordinates to create a more effective search model (Liu et al., 2014b ).
Studies of bidirectional promoters on a genome-wide scale in plants are not so extensive as in animals, and fewer studies focus on individual bidirectional promoters in plants. Keddie et al. (1994) indicated the presence of the possible bidirectional promoter only based on gene expression in Brassica napus. Xie et al. (2001) created the first artificial plant bidirectional promoter and showed that any polar promoter could be bidirectionalized by fusing a minimal promoter in a head-to-head orientation. Subsequently, based on the same strategy artificial constitutive (Zhang et al., 2008) and vascular-specific (Lv et al., 2009) bidirectional promoters were generated in tobacco. A 955-bp intergenic region between the CaTin1 and CaTin2 genes, which directed the expression of the two genes in response to tobacco mosaic virus (TMV), was the first natural bidirectional promoter to be reported (Shin et al., 2003). Two rice chymotrypsin protease inhibitor genes, OCPI1 and OCPI2, share a 1126-bp natural bidirectional promoter that drove the simultaneous expression of reporter genes encoding β-glucuronidase (GUS) and green fluorescent protein (GFP) in transiently transformed onion epidermal cells (Singh et al., 2009). Most studies of natural bidirectional promoters have focused on the model plant Arabidopsis. Examples of bidirectional promoters identified in Arabidopsis include two gene pairs, cab1/cab2 and At5g06290/At5g06280, involved in photosynthesis which have 2177- and 351-bp bidirectional promoters, respectively (Bondino and Valle, 2009; Mitra et al., 2009); 668- and 461-bp intergenic regions in the At3g17140/At3g17150 and At4g10596/At4g110600 gene pairs identified by characterization of two T-DNA promoter trap lines (Pratibha et al., 2013; Sharma et al., 2014); an asymmetric bidirectional promoter in the At1g71850/At1g71860 gene pair (Liu et al., 2015); and a heat-inducible bidirectional promoter in the AtClpB-C/AtCK2 gene pair (Mishra and Grover, 2014).
Here, we show that two defensin-like genes are driven by a 635-bp bidirectional promoter. Def1 and Def2 encoded two peptides consisting of 79 and 82/121 amino acids, respectively. Fusing the GUS and GFP genes to the ends of PZmBD1 and expressing the construct in transient and stable transformation systems showed that it could drive expression of the two reporter genes specifically and strongly in transgenic maize embryos. Truncation analysis of the PZmBD1 region revealed that the interaction between core promoter and cis-elements responsible for bidirectional transcription strength and polarity.
Materials and methods
RNA preparation and transcript analysis
Experimental tissues, including 10 DAP (10 days after pollination) whole kernels, 15, 20, 25, and 30 DAP embryos and endosperms; and ligules, sheaths, leaves, stems, and roots of large trumpet-stage inbred line B73 maize plants were ground into powder in liquid nitrogen using a mortar and pestle. TRIzol reagent (TransGen, Beijing, China) was used to extract total RNA from the powdered tissues. Genomic DNA removal and first strand cDNA synthesis were carried out simultaneously in the same reaction system using cDNA Synthesis SuperMix (TransGen, Beijing, China). The cDNA quality was tested using maize actin gene primers. The cDNA was then used as a template for quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) using SYBR Green reagent according to the manufacturer’s instructions (TaKaRa, Dalian, China). Primers used in this work are listed in Supplementary Table S1 at JXB online.
Polar and bidirectional promoter cloning and expression vector construction
Genomic sequences extending ~2.0kb upstream from the translation start sites (ATG) of the Def1 and Def2 genes were cloned using the high-fidelity KOD polymerase (TOYOBO, Japan). The isolated sequences were cloned into the pEASY-Blunt vector (TransGen, Beijing, China) and confirmed by sequencing. The primers used are listed in Supplementary Table S1. The 635-bp intergenic region located between the two ATG codons containing a putative bidirectional promoter was cloned and the sequence was confirmed as above. The two 2.0-kb genomic sequences and the 635-bp intergenic region were substituted for the 35S promoter located upstream of the β-glucuronidase gene (GUS) in the pCAMBIA3301 vector to form new expression vectors. An EGFP-polyA fragment from the pRTL2GFP vector was inserted into pCAMBIA3301 upstream of the 35S promoter in the opposite orientation to that of the GUS gene and the 635-bp putative bidirectional promoter and all of the truncation fragments replaced the 35S promoter to create a dual reporter gene expression vector. Promoter sequences and cloning primers are listed in Supplementary Table S1.
Immature embryo transient expression system
Hi-II maize seeds were collected at 20 DAP for isolation of immature embryos. The peeled embryos were transferred to liquid Murashige and Skoog (MS) medium in an ultraclean hood and washed twice. The embryos were placed on solid hypertonic medium (six or nine embryos per dish) for 4h and then bombarded with particles coated with DNA constructs using a particle delivery system (Bio-Rad, Hercules, CA). The bombarded embryos were transferred to solid MS medium and incubated at 28°C for 24h in the dark, followed by detection of reporter gene expression.
Generation of transgenic maize plants
The five expression constructs described above were used to generate transgenic maize plants by Agrobacterium tumefaciens-mediated transformation of immature Hi-II maize embryos (Frame et al., 2002). Immature embryos 1.0–2.0mm in diameter were collected from 10–12 DAP ears, treated with 5% sodium hypochlorite solution for 30min, washed twice with sterile water, and transferred to infection medium. A. tumefaciens strain EHA105 harboring the expression vector was grown for 3 d on solid YEB medium containing rifampicin and kanamycin, scraped from the medium, transferred into the liquid infection medium containing 1% acetosyringone (AS), and cultured at 28°C with agitation at 160rpm until an OD550 of 0.3–0.4 was reached. Embryos were then infected with the prepared A. tumefaciens for 5min and transferred to callus induction medium followed by selection of calli resistant to 3mg l−1 bialaphos. Transgenic T0 plants were identified by PCR analysis and herbicide leaf painting.
Detection of reporter gene activity
Histochemical staining was used for qualitative analysis of GUS activity in transiently transformed embryos and in tissues of promoter-GUS transgenic maize plants as described previously (Jefferson et al., 1987). Tissues from bidirectional promoter-GUS/GFP transgenic maize plants were analyzed for GFP signals using a Leica fluorescence stereoscope equipped with a GFP filter followed by histochemical staining for GUS activity detection as described above. Duplicate samples of all the tissues analyzed above were collected and ground into powder in liquid nitrogen using a mortar and pestle. Total protein was isolated from the powdered tissues in extraction buffer (50mM sodium phosphate buffer, 10mM Na2EDTA, 1% sodium N-lauroylsarcosine, 1% Triton X-100, 0.1% beta mercaptoethanol). The GUS substrate 4-methylumbelliferyl β-D-glucuronide (4-MUG) was used for quantitative analysis. Each biological sample had three replicates.
Results
Structure and expression characteristics of the Def1 and Def2 genes in maize
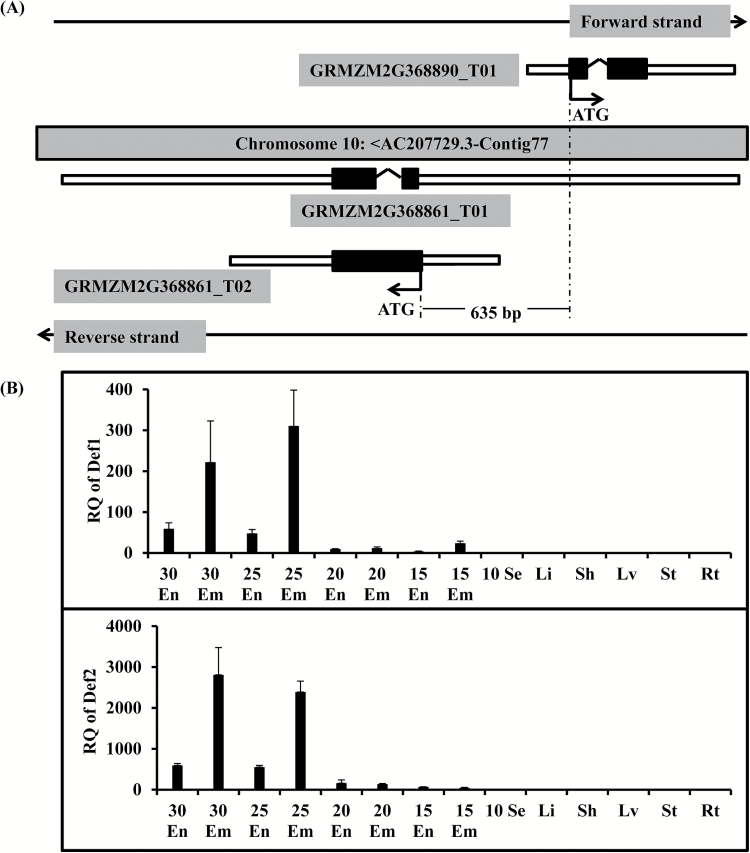
The genomic structure of the bidirectional gene pair is shown in Fig. 1A. The gene pair is located in contig 77 on chromosome 10. The Def1 gene (GRMZM2G368890) transcribes one transcript from the forward strand, while the Def2 gene (GRMZM2G368861) produces two transcripts from the reverse strand. This gene pair would be excluded by Trinklen’s model because the 5ʹ-UTR of the GRMZM2G368861_T01 transcript sequence completely overlaps the physical location of the GRMZM2G368890_T01 transcript sequence. However, under our modified model (Liu et al., 2014b ), it could be included based on the presence of the 635-bp intergenic region between the translation start sites of the GRMZM2G368861_T02 and GRMZM2G368890_T01 transcripts (Fig. 1A). According to gene annotation data of the National Center for Biotechnology Information (NCBI) database, the GRMZM2G368890 gene encodes a 79-amino acid protein named defensin-like protein 1/low-molecular-weight cysteine-rich protein (LCR68) belonging to the Knot1 super family with a putative receptor-binding site. The GRMZM2G368861 gene encodes an 82/121-amino acid protein named flower-specific gamma-thionin precursor/hypothetical protein that also belongs to the Knot1 super family. According to the gene annotation data of the Gramene and UniProt databases, GRMZM2G368890 and GRMZM2G368861 encode defensin-like protein 1/gamma-zeathionin-1 and defensin-like protein 2/gamma-zeathionin-2, respectively. We chose the names defensin-like protein 1 (DEF1) and defensin-like protein 2 (DEF2) as the protein names corresponding to the GRMZM2G368890 and GRMZM2G368861 genes for the convenience of understanding of their functions.
Fig. 1.
Physical location and expression characteristics of the two defensin-like protein genes Def1 and Def2 in maize. (A) Schematic representation of the genomic organization of the Def1 and Def2 genes on chromosome 10 in maize. The Def1 gene produces only one transcript (GRMZM2G368890_T01) while the Def2 gene produces two transcripts (GRMZM2G368861_T01 and GRMZM2G368861_T02). (B) qPCR data showing the relative transcript levels of both genes during different developmental stages of endosperm and embryos and in various vegetative tissues of large trumpet-stage inbred line B73 maize plants. En, Em, Se, Li, Sh, Lv, St, and Rt represent endosperm, embryo, seed, ligule, sheath, leaf, stem, and root tissues, respectively, of large trumpet-stage inbred line B73 maize plants; ATG, translation start site; RQ, relative expression quantity (2–ΔΔCt), with all the relative expression levels relative to that in root.
To confirm the expression characteristics of Def1 and Def2 in maize described in our previous study (Liu et al., 2014a ), we examined their expression profiles in 14 maize tissues using qRT-PCR analysis (Fig. 1B). The Def1 and Def2 expression profiles determined by qRT-PCR coincided with the results of our previous study (Liu et al., 2014a ). These results indicated that Def1 and Def2 were expressed specifically in the maize embryo and aluerone layer, and that the Def2 expression level was higher than that of Def1. Moreover, the Def1 and Def2 expression levels were considerably higher in embryos than in endosperm.
Functional testing of polar and bidirectional promoter activity using a maize immature embryo transient expression system
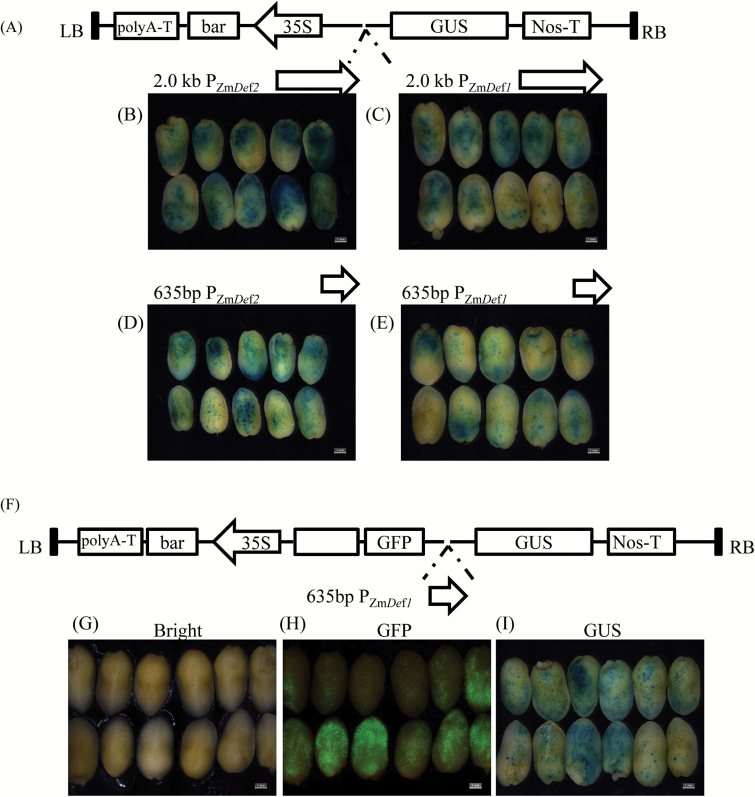
To verify the activities of the Def1 and Def2 polar promoters and to determine whether the intergenic region between the two genes functions as a bidirectional promoter, upstream sequences extending 2.0kb 5ʹ of the Def1 and Def2 translation start sites and the 635-bp intergenic region between the two translation start sites were amplified using PCR and inserted upstream of the GUS reporter gene in the pCAMBIA3301 vector to create the 635-bp PZmDef1::GUS, 635-bp PZmDef2::GUS, 2.0-kp PZmDef1::GUS, and 2.0-kb PZmDef2::GUS constructs (Fig. 2A). In addition, the 635-bp PZmDef2 was inserted between the GUS and GFP genes contained in a dual reporter gene GUS/GFP vector derived from the pCAMBIA3301 and pRTL2GFP plasmids (Zhou et al., 2013) (Fig. 2F). Twenty days after pollination, immature embryos of Hi-II maize plants were bombarded with the DNA constructs to test promoter activity using GUS staining and GFP fluorescence visualization. As shown in Fig. 2B–E, the promoters of 2.0-kb PZmDef2 (Fig. 2B), 2.0-kb PZmDef1 (Fig. 2C), 635-bp PZmDef2 (Fig. 2D), and 635-bp PZmDef1 (Fig. 2E) drove GUS gene expression in the bombarded maize embryos. As shown in Fig. 2G–I, the 635-bp PZmDef2 drove simultaneous expression of the GUS and GFP reporter genes in transiently transformed maize embryos. These results indicated that the cloned 5ʹ flanking regions functioned as promoters and that the 635-bp intergenic region functioned as a bidirectional promoter.
Fig. 2.
Functional analysis of various putative promoters using a maize immature embryo transient expression system. (A) Four GUS constructs that were placed under the regulation of the putative polar or bidirectional promoter of Def1 and Def2, respectively. GUS staining of immature embryos bombarded with (B) the putative promoter of Def2, 2-kb PzmDef2::GUS; (C) the putative promoter of Def1, 2-kb PzmDef1::GUS; (D) 635-bp PzmDef2::GUS; and (E) 635-bp PzmDef1::GUS. (F) Diagrammatic representation of the construct containing the dual reporter genes GFP/GUS, driven by a 635-bp putative bidirectional promoter. Bombarded immature embryos imaged in (G) bright field mode; (H) dark field model using a GFP filter set; and in (I) bright field mode after histochemical staining. LB, left border; RB, right border; T, terminator. Bars, 0.5mm.
Comparative analysis of tissue specificity and expression levels of the polar and bidirectional promoters in transgenic maize plants
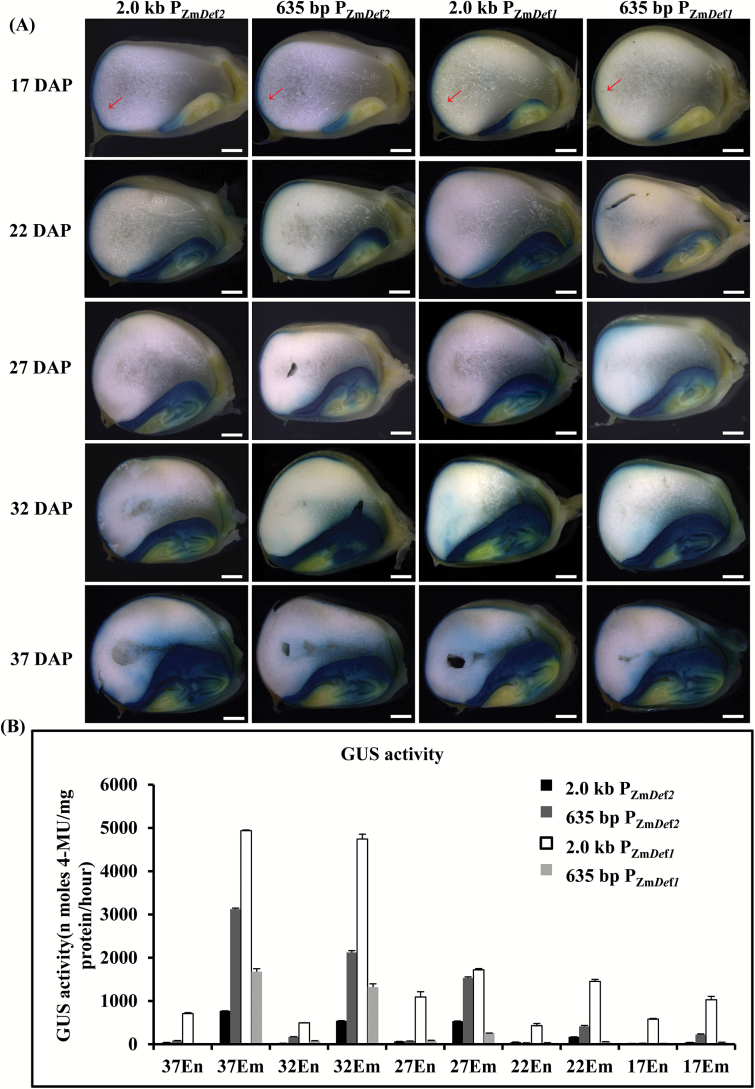
To compare the tissue specificity and promoter strengths of the promoters, GUS or GFP was placed under the regulation of the 635-bp intergenic region and the 2.0-kb PZmDef1 and PZmDef2 polar promoters. GUS staining in diverse tissues at various developmental stages provided qualitative analysis of promoter tissue specificity and GUS activity was used to quantify promoter strength. As shown in Fig. 3A, all four promoters displayed strong expression activity in embryos at different kernel developmental stages, consistent with the results of transient expression assay shown in Fig. 2B–E. In addition, the aleurone layer showed GUS staining between 17 and 37 DAP. Moreover, the promoter strengths of the four promoters increased with seed development. The 635-bp and 2.0-kb PZmDef2 promoters did not produce detectable GUS activity in vegetative tissues at the trefoil-stage (Supplementary Fig. S1) or in large trumpet-stage tassels, silk, or pollen of the transgenic maize plants (Supplementary Fig. S2). Like the PZmDef1 promoters, the 635-bp PZmDef1 also exhibited stringent embryo specificity share aluerone-specific characters (Supplementary Figs S1, S2). The 2.0-kb PZmDef1 drove GUS expression that was induced in leaves by mechanical injury with GUS activity detected only in the knife-cut edges of the leaves, as indicated by red arrows in Supplementary Figs S1 and S2. Quantitative analysis of the GUS activities of the four promoters showed that the GUS expression levels increased during seed development (Fig. 3B). Interestingly, the strength of the 635-bp PZmDef2 was considerably stronger than that of the 2.0-kb PZmDef2 during all stages of embryo development, howerver the relative strengths of the 635-bp PZmDef1 was weaker than 2.0-kb PZmDef1 promoters. The 635-bp PZmDef2 was stronger than the 635-bp PZmDef1, but the 2.0-kb PZmDef2 was markedly weaker than the 2.0-kb PZmDef1.
Fig. 3.
Comparative analysis of the tissue specificity and expression strength of the polar and bidirectional promoters in transgenic maize plants. (A) Analysis of promoter tissue specificity by GUS histochemical staining in transgenic maize plants. (B) GUS activities during various developmental stages of seeds from transgenic maize plants. DAP, days after pollination; En, endosperm; Em, embryo. Red arrow indicates GUS staining sites in the aleurone layer. Bars, 1mm.
To further analyze the differences between the 2.0-kb and 635-bp PZmDef1 and PZmDef2 promoters, we used GUS staining to analyze their expression characteristics during the early stages of seed development. As shown in Supplementary Fig. S3, initial expression of the 635-bp and 2.0-kb PZmDef2 and the 635-bp PZmDef1 started in the endosperm aleurone layer, while the initial expression of the 2.0-kb PZmDef1 started in the embryo. Expression of both the 635-bp and 2.0-kb PZmDef2 promoters in embryos began at 16 DAP with different expression levels, whereas weak expression of the 2.0-kb PZmDef1 began at 12 DAP and expression of the 635-bp PZmDef1 began at 17 DAP.
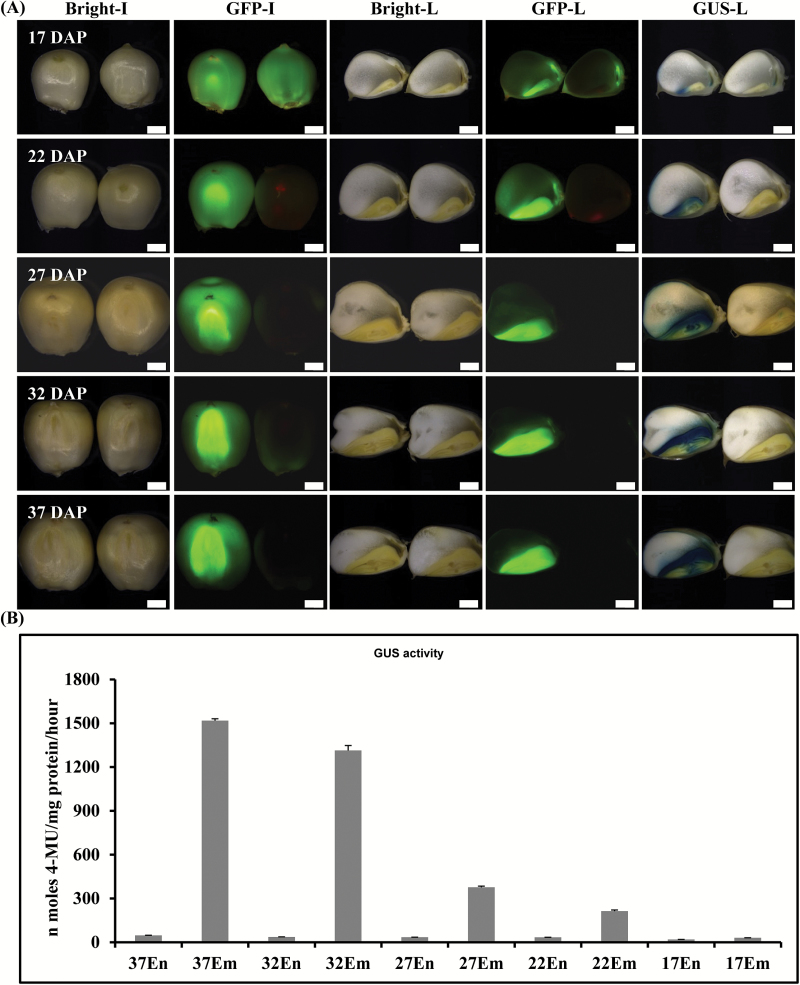
The expression characteristics of the 635-bp PZmDef1 and PZmDef2 promoters were confirmed by analyzing the expression of the dual reporter genes in the GFP::PZm Def1:: GUS construct (Fig. 2F) in transgenic maize plants (Fig. 4). GUS staining and quantitative analysis of GUS activity in kernels were performed as described above accompanied by GFP fluorescence visualization. As shown in Fig. 4A, PZmdef1 drove GUS and GFP reporter gene expression in embryos and in the endosperm aleurone layer from 17 to 37 DAP and quantitative analysis of GUS activity in seeds showed that the expression level increased with seed development (Fig. 4B). GUS activity was not detected in non-seed tissues (Supplementary Fig. S4). The results generated by the 635-bp PzmDef2 with dual reporter genes in transgenic maize plant (Supplementary Fig. S5) also reproduced the expression pattern of Def1 and Def2 as PzmDef1 did. These results were consistent with the results obtained from transgenic maize plants expressing constructs in which the GUS reporter gene was fused to either end of the bidirectional promoter (Fig. 3), and the intergenic region (635-bp PZmdef1 or 635-bp PZmdef2) between the ZmDef1 and ZmDef2 genes functions as an embryo-specific and asymmetric bidirectional promoter: PZmBD1.
Fig. 4.
Characterization of the tissue specificity and expression strength of the PZmdef1 in transgenic maize plants. (A) Qualitative analysis of bidirectional expression activities of PZmdef1 in transgenic maize seeds. Bright-I and GFP-I, intact seeds imaged in bright field mode and using a GFP filter set, respectively; Bright-L, GFP-L, and GUS-L, longitudinal sections imaged in bright field mode, using a GFP filter set, and in bright field mode after histochemical staining, respectively. Control tissues corresponding to the various developmental stages are shown on the right side of each photograph. Bars, 1mm. (B) Quantification of GUS activities level during various developmental stages of seeds from transgenic maize plants. En, endosperm; Em, embryo.
Taken together, these results indicated that the 2.0-kb PZmDef2 and PZmDef1 were stringent embryo-specific promoters that share alerone-specific characters and the 635-bp intergenic region functions as an embryo-specific and asymmetric bidirectional promoter (PZmBD1). Moreover, PZmBD1 had different expression characteristics compared to the 2.0-kb polar promoters of the two genes. We designated this 635-bp bidirectional promoter PZmBD1 to reflect that it is the first natural bidirectional promoter to be cloned from maize and characterized intensively.
Truncation analysis of the cis-elements involved in regulating bidirectional transcription
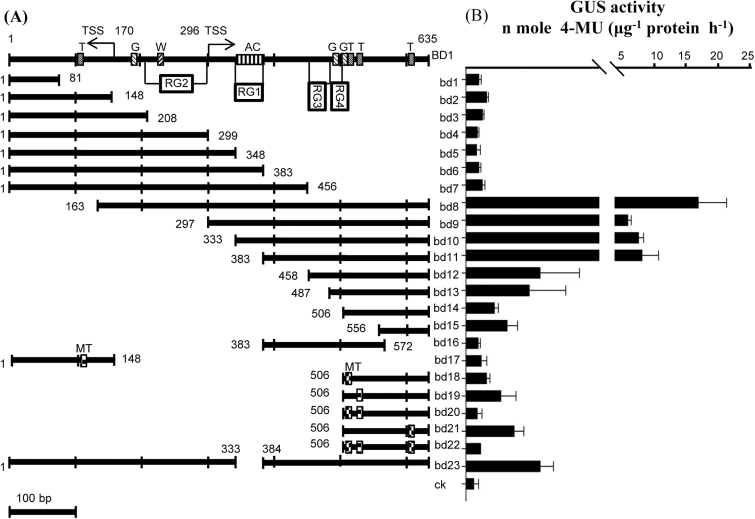
To investigate the potential mechanism regulating a bidirectional promoter, a dual reporter gene transient system was used to test the transcriptional activity of a series of truncation, internal deletion, and point mutation constructs (Fig. 5A) of PZmBD1. As shown in Figs 5 and 6, we found that some elements control the promoter activity and direction of transcription initiation. The two longer fragments, bd6 (positions 1 to 383) and bd7 (positions 1 to 456), produced a stronger GFP signal (Figs 5B, 6F, 6G) than the two shorter fragments, bd4 (positions 1 to 299) and bd5 (positions 1 to 348), (Fig. 5B, 6D, 6E). While the two even shorter fragments, bd2 (positions 1 to 148) and bd3 (positions 1 to 208), recovered the strong GFP-expression activity and showed GUS-expression activity (Figs 5B, 6B, 6C), but the 81bp fragment (bd1) lost the activity in both directions (Figs 5B, 6A). These results indicated that the common region (positions 348 to 383, RG1) shared by bd6 and bd7 functions as an enhancer in the GFP-expression direction, the common region (positions 208 to 299, RG2) shared by bd4 and bd5 served as a repressor regulating bidirectional expression, and RG1 has dominant epistasis to RG2. Sequence analysis showed that RG1 is an AC-I/AC-II fused element (Fig. 5A), which could interact with the G (positions 196 to 202) box to enhance gene expression (Hatton et al., 1995). RG2 contained a W box (positions 229 to 235) (Fig. 5A). The W box was reported to interact with transcription factor (TF) OsWRKY71, which represses GA induction of the Amy32b promoter in aleurone cells (Zhang et al., 2004). Furthermore, the region between bd2 and bd1 suggested that a TATA box (positions 100 to 103, TATA) plays a key role in regulating bidirectional expression (Figs 5B, 6A, 6B). This speculation was verified by the TATA box mutant (CACA) construct in which the bidirectional expression was dramatically decreased (Figs 5B, 6V).
Fig. 5.
Truncation analysis of cis-elements of PZmBD1. (A) Schematic representation of the GFP::mPZmBD1::GUS constructs used for maize immature embryo transient transformation. TSS, transcription start site; T, TATA box; G, G box; W, W box; AC, AC-I/AC-II fused cis-elements; MT, mutated TATA box; RG, region; BD1, the intact bidirectional promoter PZmBD1; bd1-bd23, the series of mutated bidirectional promoters of PZmBD1; ck, maize immature embryos bombarded with empty vector. All mutated bidirectional promoters (mPZmBD1) used GUS and GFP as dual reporter genes in one construct. (B) Quantification for GUS activity of mPZmBD1 in immature embryos. Each bar represents data from between three representative independent bombardments, each bombardment contains nine immature maize embryos transformed with 1 μg plasmid of each constuct.
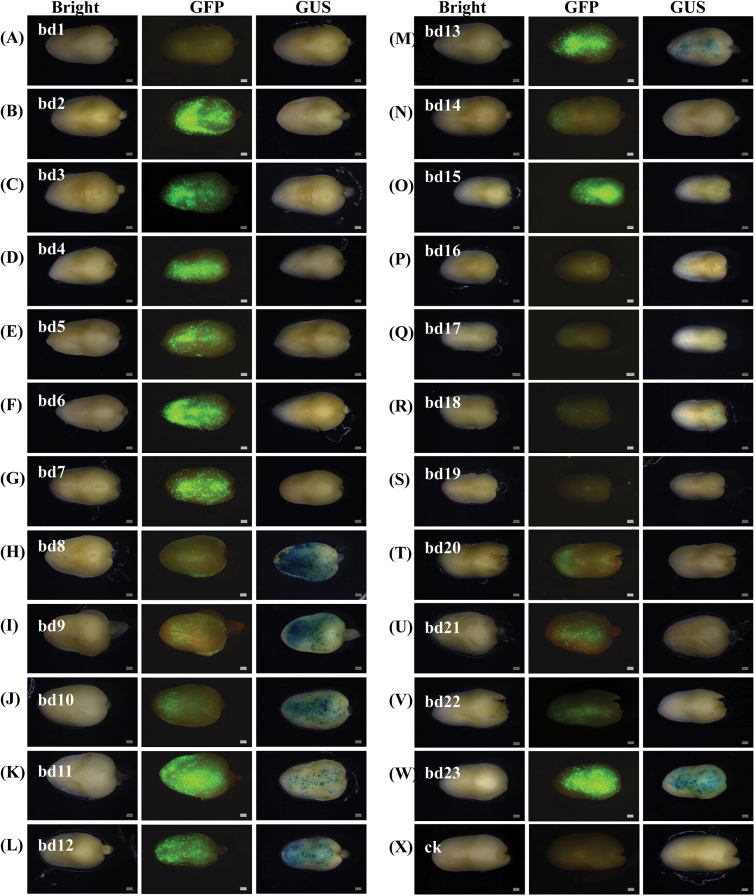
Fig. 6.
The activity of the different GFP::mPZmBD1::GUS constructs assayed using maize immature embryo transient transformation. (A) to (W) represent the 23 constructs pbd1GG3 to pbd23GG3, respectively. Bright, pictures of the embryos were taken 24h after bombardment under a bright field using a Leica fluorescence stereoscope; GFP, pictures taken under dark field with ultraviolet light and a GFP filter following the bright field pictures; GUS, pictures taken after histochemical staining following the dark field pictures. Each construct was subject to two individual bombardments with six immature embryos; this figure shows one embryo. More information is shown in Supplementary Fig. S6. BD1, full length of the bidirectional promoter, and truncated from (A) 81 to 635, (B) 148 to 635, (C) 208 to 635, (D) 299 to 635, (E) 348 to 635, (F) 383 to 635, (G) 456 to 635, (H) 1 to 163, (I) 1 to 297, (J) 1 to 333, (K) 1 to 383, (L) 1 to 458, (M) 1 to 487, (N) 1 to 506, (O) 1 to 556, (P) 1 to 383 and 572 to 635, (Q) 148 to 635 and a point mutation in the TATA box, (R) 1 to 506 and a point mutation in TATA box 1, (S) 1 to 506 and a point mutation in TATA box 2, (T) 1 to 506 and point mutations in TATA boxes 1 and 2, (U) 1 to 506 and point mutations in TATA box 3, (V) 1 to 506 and point mutations in TATA boxes 1–x3, (W) 333 to 384, and (X) empty vector. Bars, 0.5mm.
Three truncated fragments, bd8 (positions 163 to 635), bd9 (positions 297 to 635), and bd10 (positions 333 to 635), displayed weak GFP-expression activity and strong GUS-expression activity (Figs 5B, 6H–J), the opposite expression activity to that of the shorter truncation fragments bd11 (positions 383 to 635) and bd12 (positions 458 to 635) (Figs 5B, 6K, 6L). bd13 maintained the promoter strength in the GFP-expression direction as in bd11 and bd12, while the activity in the GUS-expression direction decreased (Figs 5B, 6M). However, the bidirectional transcription activity decreased to a lower expression level when the truncated fragments were 129bp (bd14) long (Figs 5B, 6N). These results indicate that the common region (positions 348 to 383, RG1) shared by bd8, bd9, and bd10 functions as an enhancer in the GUS-expression direction, the common region shared by bd11, bd12, and bd13 (positions 458 to 487, RG3) functions as another enhancer in the GUS-expression direction, and the fourth region (positions 487 to 506, RG4) functions as another enhancer in the GFP-expression direction. Sequence analysis showed that the two fused elements ABRE/IRO2OS (positions 484 to 491 and 499 to 506) overlap two G boxes (positions 487 to 492 and 502 to 507) in RG3+RG4 that could interact with the AC-I/AC-II fused element in RG1 enhancement of transcription activity (Figs 5B, 6A). bd12, which contained two fused ABRE/IRO2OS elements, has strong bidirectional promoter activity (Figs 5B, 6L), while the activity decreased dramatically in one direction when one ABRE/IRO2OS (bd13) was broken (Figs 5B, 6M), and vice versa (bd14) (Figs 5B, 6N). Therefore, AC-I/AC-II and ABRE/IRO2OS are involved in regulating bidirectional activity.
Sequence analysis also identified three TATA boxes (box 1, 508 to 514; box 2, 529 to 534; box 3, 603 to 608) in bd14 (129bp) (Fig. 5A). The mutated box 1 led to the loss of GUS expression, but significantly increased the GFP signal (Figs 5B, 6O), while the mutated box 2 showed the opposite results (Figs 5B, 6P). The results with the mutated box 3 were similar to those of box 1 (Figs 5B, 6R). The fragments lost promoter activity when boxes 1 and 2 were mutated simultaneously or all three boxes were mutated (Figs 5B, 6Q, 6S). However, fragment bd15 (positions 556 to 635), which contains only box 3, had bidirectional promoter activity (Figs 5B, 6T) and fragment bd16 (positions 383 to 572), containing boxes 1 and 2, and lacking the bd15 region, just maintained GFP expression (Figs 5B, 6U). These results indicate that TATA box 1 is responsible for transcription initiation in one direction and antagonizes TATA box 2, which regulates transcription initiation in the other direction, and they are both are involved in coordinated bidirectional expression.
Discussion
Maize is a critical food and feed crop that is also used as an efficient and safe plant bioreactor to produce industrial enzymes, antibodies, vaccines, and pharmaceutical compounds (Ramessar et al., 2008). Moreover, metabolic engineering and trait stacking have been used in maize to modify complex metabolic pathways and to improve crop quality (Naqvi et al., 2010). Embryo-specific promoters and efficient strategies are required to achieve these goals. In this study, we report the identification of a defensin-like protein bidirectional gene pair, Def1 and Def2, which are expressed at high levels in maize seeds. The intergenic region of the gene pair functions as an embryo-specific bidirectional promoter, and the PZmDef1 and PZmDef2 function as embryo-specific promoters. Transient and stable transformation assays revealed that the polar promoters of the Def1 and Def2 genes function as strong and stringent embryo-specific promoters (Figs 3, 4, Supplementary Figs S1, S2). The 2.0-kb PZmDef1 exhibited trace expression in leaves (Supplementary Figs S1, S2) and the 635-bp intergenic region of Def1 and Def2 functioned as a strong and stringent embryo-specific asymmetric bidirectional promoter sharing aluerone-specific characters, which we designated PZmBD1 (Fig. 4, Supplementary Fig. S4). The PZmBD1 bidirectional promoter could serve as promoter resources for maize and monocot biotechnology, which could be used for gene stacking and metabolic pathway engineering in maize and other monocot plant species.
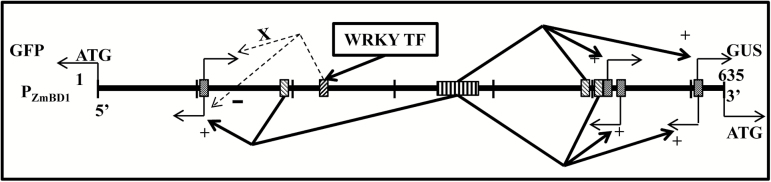
In vertebrates, studies of promoter sequences and TF binding sites showed that a small set of TFs interact with cis-regulatory sequences in bidirectional promoters and may constitute a simple mechanism for the regulation of bidirectional transcription (Li et al., 2006; Lin et al., 2007; Trinklein et al., 2004). In plants, 1, 16, and 39 cis-regulatory sequences were shown to be overrepresented in bidirectional promoters of Populus, rice, and Arabidopsis, respectively (Dhadi et al., 2009), but solid, direct evidence that would establish a clear model of bidirectional promoter regulation was not presented. Recent studies have revealed that bidirectionality is an inherent feature of promoters (Jacquier, 2009; Carninci, 2010; Wei et al., 2011). Nascent transcripts in both directions at the transcription start site of a promoter were detected in almost equal amounts (Core et al., 2008); however, most promoters produce final transcripts in just one direction. A potential model for bidirectional regulation of a promoter via the initiation, elongation, and termination steps of the transcription process may provide an explanation (Wei et al., 2011). An assumed 5ʹ checkpoint in this model may control the transcription process between pause, elongation, and termination. With transcription from a promoter initiated equally in both directions, the RNA polymerase would pause at the 5ʹ checkpoint and epigenetic modification associated with the 5ʹ checkpoint would determine whether subsequent elongation or termination would occur. Transcript stability would then control the amount of transcript accumulation. Polar promoters display orientation preference, whereas bidirectional promoters produce stable transcripts in both directions; however, the mechanism that regulates bidirectional promoters remains unclear. Our study suggests that the mechanism of bidirectional transcription initiation of a bidirectional promoter involves one core promoter, located at both ends with relatively low bidirectional transcription initiation activity and cis-elements with various combinations of antagonistic or synergistic effects to negatively or positively regulate the level of transcription (Fig. 7). The two core promoters bd2 and bd15 in our model are additional evidence that bidirectionality is an inherent feature of promoters (Jacquier, 2009; Carninci, 2010; Wei et al., 2011), and the W box interaction with WRKY TFs may serve as a 5ʹ checkpoint (Wei et al., 2011). There may be other 5ʹ checkpoints near the other core promoter. However, the activity of all of the 5ʹ checkpoints can be eliminated by enhancer-liked activity so that a promoter develops a new characteristic: bidirectionality.
Fig. 7.
Model showing the potential mechanism regulating a maize bidirectional promoter. –, negative regulation of the core promoter activity; +, positive regulation of the core promoter activity; x, eliminates the activity of the bidirectional core promoter in one direction. WRKY TF, WRKY transcription factors. The core promoters contain a TATA box at the proximal end to ATG with low transcription activity in one or more directions forming the backbone of a bidirectional promoter. An enhancer-like AC-I/AC-II fused cis-element interacts with some cis-elements that positively enhance the bidirectional promoter activity, while WRKY TF binding to the W box down-regulates or eliminates the activity of one core promoter. Due to the number of core promoters and because the regulating effects at both ends are asymmetric, there has some differences in the promoter activity of PZmBD1 in the two directions.
In this study, we demonstrated that two genes with similar functions were arranged in a head-to-head fashion with the 635-bp intergenic region serving as an embryo-specific, bidirectional promoter. The relative expression levels of two reporter genes driven simultaneously by the bidirectional promoter showed that the promoter strength was asymmetric. This gives rise to questions as to what the relationship between the two genes in the defense pathway is and whether the asymmetry is caused by differences in the responses of the two genes to signals during various stages of development, as part of a defense pathway, or in response to other factors. We also constructed a potential model in which core promoters with low bidirectional expression activity interacted with cis-elements, which conferred bidirectionality to the promoter, making it a so-called bidirectional promoter. A more detailed study of the role of the cis-elements is needed and is a subject of future research.
Supplementary data
Supplementary data is available at JXB online.
Fig. S1. Comparative analysis of the tissue specificity of the polar and bidirectional promoters in transgenic maize plants.
Fig. S2. Comparative analysis of the tissue specificity of the polar and bidirectional promoters in transgenic maize plants.
Fig. S3. Comparative analysis of the expression patterns of the polar and bidirectional promoters during early developmental stages of transgenic maize plants.
Fig. S4. Analysis of bidirectional promoter tissue specificity in transgenic maize.
Fig. S5. Characterization of the tissue specificity and expression strength of the PZmdef2 in transgenic maize plants.
Fig. S6. The activity of different GFP::mPZmBD1::GUS constructs assayed using maize immature embryo transient transformation.
Table S1. Promoter sequences and cloning primers used in the study.
Acknowledgments
We thank professor Jun Zhao, Lei Wang and Yubin Li for providing the constructive suggestions. This work was supported by the National Natural Science Foundation of China (grant number 31500301), the National Special Program for GMO Development of China (grant number 2014ZX08003-002), and by the Trans Research Dream Fund (Trans-RasDF-013). The authors declare no conflict of interest.
Glossary
Abbreviations:
- Bd
bidirectional fragment
- DAP
days after pollination
- GFP
green fluorescent protein
- GUS
β-glucuronidase
- P
promoter
- RG
region.
References
- Adachi N, Lieber MR. 2002. Bidirectional gene organization: a common architectural feature of the human genome. Cell 109, 807–809. [DOI] [PubMed] [Google Scholar]
- Beck CF, Warren RA. 1988. Divergent promoters, a common form of gene organization. Microbiological Reviews 52, 318–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondino HG, Valle EM. 2009. A small intergenic region drives exclusive tissue-specific expression of the adjacent genes in Arabidopsis thaliana . BMC Molecular Biology 10, 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P. 2010. RNA dust: where are the genes? DNA Research 17, 51–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Core LJ, Waterfall JJ, Lis JT. 2008. Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322, 1845–1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhadi SR, Krom N, Ramakrishna W. 2009. Genome-wide comparative analysis of putative bidirectional promoters from rice, Arabidopsis and Populus. Gene 429, 65–73. [DOI] [PubMed] [Google Scholar]
- Frame BR, Shou H, Chikwamba RK, et al. 2002. Agrobacterium tumefaciens-mediated transformation of maize embryos using a standard binary vector system. Plant Physiology 129, 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatton D, Sablowski R, Yung MH, Smith C, Schuch W, Bevan M. 1995. Two classes of cis sequences contribute to tissue-specific expression of a PAL2 promoter in transgenic tobacco. The Plant Journal 7, 859–876. [DOI] [PubMed] [Google Scholar]
- Jacquier A. 2009. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nature Reviews Genetics 10, 833–844. [DOI] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. 1987. GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. The EMBO Journal 6, 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddie JS, Tsiantis M, Piffanelli P, Cella R, Hatzopoulos P, Murphy DJ. 1994. A seed-specific Brassica napus oleosin promoter interacts with a G-box-specific protein and may be bi-directional. Plant Molecular Biology 24, 327–340. [DOI] [PubMed] [Google Scholar]
- Kourmpetli S, Lee K, Hemsley R, Rossignol P, Papageorgiou T, Drea S. 2013. Bidirectional promoters in seed development and related hormone/stress responses. BMC Plant Biology 13, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krom N, Ramakrishna W. 2008. Comparative analysis of divergent and convergent gene pairs and their expression patterns in rice, Arabidopsis, and Populus. Plant Physiology 147, 1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YY, Yu H, Guo ZM, Guo TQ, Tu K, Li YX. 2006. Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Computational Biology 2, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JM, Collins PJ, Trinklein ND, Fu Y, Xi H, Myers RM, Weng Z. 2007. Transcription factor binding and modified histones in human bidirectional promoters. Genome Research 17, 818–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SJ, Yue QJ, Zhang W. 2015. Structural and functional analysis of an asymmetric bidirectional promoter in Arabidopsis thaliana . Journal of Integrative Plant Biology 57, 162–170. [DOI] [PubMed] [Google Scholar]
- Liu X, Tian J, Zhou X, Chen R, Wang L, Zhang C, Zhao J, Fan Y. 2014. a Identification and characterization of promoters specifically and strongly expressed in maize embryos. Plant Biotechnology Journal 12, 1268–1296. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhou X, Li Y, Tian J, Zhang Q, Li S, Wang L, Zhao J, Chen R, Fan Y. 2014. b Identification and functional characterization of bidirectional gene pairs and their intergenic regions in maize. BMC Genomics 15, 338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv X, Song X, Rao G, Pan X, Guan L, Jiang X, Lu H. 2009. Construction vascular-specific expression bi-directional promoters in plants. Journal of Biotechnology 141, 104–108. [DOI] [PubMed] [Google Scholar]
- Mishra RC, Grover A. 2014. Intergenic sequence between Arabidopsis caseinolytic protease B-cytoplasmic/heat shock protein100 and choline kinase genes functions as a heat-inducible bidirectional promoter. Plant Physiology 166, 1646–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A, Han J, Zhang ZJ. 2009. The intergenic region of Arabidopsis thaliana cab1 and cab2 divergent genes functions as a bidirectional promoter. Planta 229, 1015–1022. [DOI] [PubMed] [Google Scholar]
- Naqvi S, Farre G, Sanahuja G, Capell T, Zhu C, Christou P. 2010. When more is better: multigene engineering in plants. Trends in Plant Science 15, 48–56. [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d’Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457, 1038–1042. [DOI] [PubMed] [Google Scholar]
- Pratibha P, Singh SK, Sharma I, Kumar R, Srinivasan R, Bhat SR, Ahuja PS, Sreenivasulu Y. 2013. Characterization of a T-DNA promoter trap line of Arabidopsis thaliana uncovers a cryptic bi-directional promoter. Gene 524, 22–27. [DOI] [PubMed] [Google Scholar]
- Ramessar K, Sabalza M, Capell T, Christou P. 2008. Maize plants: An ideal production platform for effective and safe molecular pharming. Plant Science 174, 409–419. [Google Scholar]
- Sharma I, Srinivasan R, Ahuja P, Bhat S, Sreenivasulu Y. 2014. Identification and characterization of a T-DNA promoter trap line of Arabidopsis thaliana uncovers an embryo sac-specific bi-directional promoter. Plant Molecular Biology Reporter, 1–9. [Google Scholar]
- Shin R, Kim MJ, Paek KH. 2003. The CaTin1 (Capsicum annuum TMV-induced clone 1) and CaTin1-2 genes are linked head-to-head and share a bidirectional promoter. Plant and Cell Physiology 44, 549–554. [DOI] [PubMed] [Google Scholar]
- Singh A, Sahi C, Grover A. 2009. Chymotrypsin protease inhibitor gene family in rice: Genomic organization and evidence for the presence of a bidirectional promoter shared between two chymotrypsin protease inhibitor genes. Gene 428, 9–19. [DOI] [PubMed] [Google Scholar]
- Trinklein ND, Aldred SF, Hartman SJ, Schroeder DI, Otillar RP, Myers RM. 2004. An abundance of bidirectional promoters in the human genome. Genome Research 14, 62–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wan L, Li D, Zhu L, Qian M, Deng M. 2009. Searching for bidirectional promoters in Arabidopsis thaliana . BMC Bioinformatics 10 Suppl 1, S29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W, Pelechano V, Jarvelin AI, Steinmetz LM. 2011. Functional consequences of bidirectional promoters. Trends in Genetics 27, 267–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie M, He Y, Gan S. 2001. Bidirectionalization of polar promoters in plants. Nature Biotechnology 19, 677–679. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457, 1033–1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Gai Y, Wang W, Zhu Y, Chen X, Jiang X. 2008. Construction and analysis of a plant transformation binary vector pBDGG harboring a bi-directional promoter fusing dual visible reporter genes. Journal of Genetics and Genomics 35, 245–249. [DOI] [PubMed] [Google Scholar]
- Zhang ZL, Xie Z, Zou X, Casaretto J, Ho TH, Shen QJ. 2004. A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells. Plant Physiology 134, 1500–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Li S, Zhao Q, Liu X, Zhang S, Sun C, Fan Y, Zhang C, Chen R. 2013. Genome-wide identification, classification and expression profiling of nicotianamine synthase (NAS) gene family in maize. BMC Genomics 14, 238. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.