Abstract
The breast cancer resistance protein (BCRP) is an ABC transporter playing a crucial role in the pharmacokinetics of drugs. The early identification of substrates and inhibitors of this efflux transporter can help to prevent or foresee drug-drug interactions. In this work, we built a ligand-based in silico classification model to predict the inhibitory potential of drugs toward BCRP. The model was applied as a virtual screening technique to identify potential inhibitors among the small-molecules subset of DrugBank. Ten compounds were selected and tested for their capacity to inhibit mitoxantrone efflux in BCRP-expressing PLB985 cells. Results identified cisapride (IC50 = 0.4 µM) and roflumilast (IC50 = 0.9 µM) as two new BCRP inhibitors. The in silico strategy proved useful to prefilter potential drug-drug interaction perpetrators among a database of small molecules and can reduce the amount of compounds to test.
Keywords: BCRP inhibition, virtual screening, logistic regression, cisapride, roflumilast
Introduction
The breast cancer resistance protein (BCRP, gene: ABCG2) is a transmembrane protein that belongs to the superfamily of the ABC transporters. In humans, these proteins use the energy of adenosine triphosphate (ATP) hydrolysis to actively transport their substrates out of the cells. BCRP in particular is expressed at crucial barriers and organs such as the canalicular membrane of the hepatocytes, the blood-brain barrier, the gastrointestinal tract, and the placenta.1 BCRP therefore restricts access to crucial organs and increases elimination of its substrates.
Inhibition or dysfunction of BCRP has been shown to produce gout and drug-drug interactions,2,3 but it can also be used as a strategy to increase the bioavailability of drugs.4 Furthermore, BCRP is involved in multidrug resistance (MDR), a phenomenon by which cancerous cells overexpress efflux transporters, which pump out cytotoxic drugs and therefore resist to the treatment. Developing BCRP inhibitors to increase the concentration of anticancer drugs into resistant cancer cells has been seen as a potential strategy to overcome MDR.5
Because of its involvement in drug-drug interactions, the Food and Drug Administration (FDA) recommends testing compounds in development for their capacity to inhibit BCRP.6 In this context, implementing a fast and inexpensive in silico method to identify potential BCRP inhibitors would allow prioritizing compounds in the early drug discovery phase.
Many in silico studies around specific chemical families such as flavonoids or tariquidar analogues have led to local quantitative structure-activity relationship models.7,8 While these kinds of models are useful for gaining understanding of the chemical features that drive activity within a family, they are of little use for other chemical families. Thus,also several global machine learning-based classification models have been proposed to predict BCRP inhibition. Erić et al.9 collected and merged literature data on BCRP inhibition and built neural network and support vector machine models on 96 compounds. They report test set accuracies over 82%, sensitivities over 83%, and specificities over 80%. Their models were also used on a completely different data set (extracted from another publication) of 147 compounds and yielded accuracies between 67% and 70%. Matsson and colleagues10 used a diverse training set of 80 compounds and the descriptors logD and polarizability to distinguish between BCRP inhibitors and noninhibitors. They obtained a sensitivity of 83% and a specificity of 76% on a test set of 43 compounds. Pan and colleagues11 trained a Bayesian categorization model and a set of pharmacophores on 203 compounds. Their models were used to prospectively screen the Collaborative Drug Discovery (CDD) database, and 33 compounds were selected and tested. Among them, two (flunarizine and pimozide) showed significant BCRP inhibition activity at 10 µM. All these models share the fact that they were built on rather small data sets, not making use of all the data available at the respective times of their studies. Also, only one of them challenged their model in a prospective study where compounds were actually bought and tested.
In this study, we are making use of a 978-compound BCRP inhibition data set previously collected and curated12 to build a global binary classification model for prediction of BCRP inhibition. We used the model in a prospective manner to screen DrugBank and to identify potentially new BCRP inhibitors. Ten compounds were selected and tested in BCRP-expressing PLB985 cells, and two of them (cisapride and roflumilast) revealed considerable inhibition at 10 µM.
Materials and Methods
BCRP Inhibition Data Set
Data for building the BCRP inhibition prediction model were taken as is from Montanari and Ecker.12 The data set contains 433 inhibitors and 545 noninhibitors, collected from 47 publications.
Descriptor Sets
Four descriptor sets were used to build the models. MACCS fingerprints were computed using RDKit.13 Morgan fingerprints were computed using RDKit,13 with a bit vector length of 1024 and a radius of 4. These fingerprints are equivalent to extended-connectivity fingerprints of diameter 8 (ECFP8). VolSurf descriptors were computed using the Molecular Operating Environment (MOE) implementation.14 Two-dimensional physicochemical descriptors were computed with CDKdescUI.15
Machine-Learning Models
Four different learning algorithms were used to train the models. Logistic regression, naive Bayes, support vector machine (SVM), and random forest were built in python with the scikit-learn library.16 For logistic regression, the regularization parameters (penalty and C parameters) were optimized with GridSearch with an internal cross-validation. For naive Bayes and random forest, the default parameters were kept. For SVM, an RBF kernel was chosen, where the parameters C and gamma were optimized via GridSearch in a 5-fold internal cross-validation. The code for building and cross-validating the final model (logistic regression based on Morgan fingerprints) and predicting new compounds is available as a python file in the supplementary information. Along with the training set, it allows interested users to quickly build the same model and use it for their own purposes. The required libraries are NumPy,17 scikit-learn,16 and RDKit.13
Evaluation Method
The models were evaluated by external 10-fold cross-validation (i.e., independently from parameter selection). To evaluate the performance in a more demanding way, sets of publications that make up the BCRP inhibition data set were randomly drawn and kept as external validation sets. Each time, the publications left formed the training set, on which the modeling process previously described was applied. The validation set was then passed through the trained model. This “leave-sources-out” evaluation was repeated 166 times to rule out any chance effect. In both validation schemes, the confusion matrix, Matthews’s correlation coefficient (MCC), and the area under the receiver operating characteristic curve (ROC AUC) are reported as measures of predictive power of the model. For the leave-sources-out evaluation, an average of these metrics over the 166 splits is given.
Chemical Similarity
Chemical similarity between the tested compounds and the training set was measured using RDKit fingerprints and the Tanimoto similarity metrics. Candidate compounds with similarity over 0.75 to any of the compounds in the training set were considered too close to the training set.
Virtual Screening
After training on the complete training set, the model was used to screen the DrugBank database. In total, 1780 compounds were retrieved from the DrugBank database (www.drugbank.ca, as of October 2013) from the small-molecules section, containing all the approved drugs and a few experimental compounds. The data were curated using the following protocol: (1) inorganic compounds (not containing carbon atoms) were removed; (2) mixtures were separated and salts were removed; (3) organometallic compounds were removed; (4) compounds containing rare atoms (selenium, silicon, gold, platinum) were removed; (5) compounds were standardized using the following options: clean 2D, aromatize, mesomerize, neutralize, tautomerize, and all the transform options in ChemAxon’s Standardizer; (6) nonunique structures were removed; and (7) compounds permanently charged were removed.
The software used for these different tasks was ChemAxon’s Instant JChem v.5.3 and Standardizer v.5.11.3 (http://www.chemaxon.com) and MOE 2011.10.14
After curation, the screening database contained 1702 compounds. These compounds were passed through the model and ranked according to their predicted inhibitor score, which roughly corresponds to their probability of inhibiting BCRP according to the model.
In Vitro Testing
Materials
RPMI 1640 for cell culture was purchased from LifeTechnologies (Rockville, MD). Supplements for cell culture, including fetal bovine serum (FBS), and the antibiotics penicillin and streptomycin were purchased from Sigma (St. Louis, MO). The fluorescent substrate mitoxantrone as well as Ko143, DMSO, phosphate-buffered saline (PBS), all compounds used for HPMI preparation, and the 10 test compounds were also purchased from Sigma.
Cell culture
The human myeloid leukemia PLB985 parental and stably expressing BCRP18 cell lines were kindly provided by B. Sarkadi (Institute of Enzymology, Research Centre for Natural Sciences, Budapest, Hungary) and K. Nemet (Creative Cell Ltd., Budapest, Hungary). Both cell lines were cultured in RPMI 1640 supplemented with 10% FBS and 100 IU/mL penicillin and streptomycin. Cell lines were maintained at 37 °C in an atmosphere containing 5% CO2 with 95% relative humidity.
Steady-state mitoxantrone accumulation experiments
For BCRP inhibition studies, the steady-state accumulation of mitoxantrone (7 µM) was performed as previously described.19
Briefly, cells were harvested, pelleted (300 g, 5 min, 4 °C), diluted to a concentration of 12 × 106 cells/mL in HPMI (10 mM HEPES, 120 mM NaCl, 5 mM KCl, 0.4 mM MgCl2, 0.04 mM CaCl2, 10 mM glucose, 10 mM NaHCO3, 5 mM Na2HPO4, pH 7.4 with NaOH), and kept on ice for further processing. Every compound tested was dissolved in DMSO and diluted to a concentration of 40 µM with HPMI, resulting in a concentration of 2% DMSO in the 40-µM stocks. For each data point, 25 µL of cell suspension was preincubated for 5 min at 37 °C with 25 µL of the 40-µM stock of the compound to test. HPMI containing 2% DMSO alone was included as DMSO control. Thereafter, cells were preloaded with 50 µL of a 14-µM mitoxantrone stock in DMSO for 20 min at 37 °C. Consequently, the final concentrations for mitoxantrone, the compounds to test, and DMSO in the final assay volume of 100 µL were 7 µM, 10 µM, and 1.5%, respectively. Preloading was stopped by chilling cells on ice for 5 min followed by the addition of 400 µL ice-cold PBS. Cells were pelleted (300 g, 5 min, 4 °C), supernatant was discarded, and cell pellet was resuspended in 150 µL ice-cold PBS. Until measurement, cells were kept on ice and in the dark. Immediately before the measurement of the fluorescence intensity in cells by flow cytometry (MACSQuant; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), 50 µL DAPI solution (4 µg/mL in PBS) was added to gate out dead cells immediately before FACS measurement. Ko143 was used as a positive control for BCRP inhibition at a final concentration of 1 µM. The background fluorescence of the cell suspension was measured in the presence of 1.5% DMSO.
Data and statistical analysis
The mitoxantrone fluorescence intensity in every sample was corrected by the background fluorescence of the cells. Data were then normalized to the positive control Ko143, whose fluorescence intensity was set as 100%. Statistical analysis were performed for normalized data by using an unpaired, parametric t test with GraphPad Software (San Diego, CA) Prism version 6.05 to compare each single compound with the DMSO control.
IC50 value measurements and calculations
For cisapride and roflumilast, IC50apparent values were estimated measuring steady-state accumulation of 7 µM mitoxantrone as described above in the presence of 10 or 12 different compound concentrations ranging from 0.001 to 100 µM. Ko143 at a final concentration of 1 µM was included as a positive control for BCRP inhibition. After correcting for background fluorescence of unstained cells, IC50apparent values were calculated by performing nonlinear regression analyses (GraphPad Prism 6, “log(Agonist) vs. response – Variable slope”), using the following equation:
where X is the log of compound concentration; Y is the response in fluorescent intensity units; “Bottom” and “Top” are the lower and the higher plateaus of the nonlinear fit curve, respectively; IC50apparent refers to the IC50apparent value; and Hill slope is a factor that describes the steepness of the curve.
To correct for the expression-level dependency of IC50apparent values and the pump-leak kinetics as reviewed in Stein,20 IC50 values were calculated using the following equation:
where the fluorescence intensity at zero inhibitor concentration [I]0 is according to the “Bottom” level and the fluorescence intensity at infinite inhibitor concentration [I]∞ is according to the “Top” level given by nonlinear regression analysis.21 The mean IC50 values ± SDs given were calculated from three or five independent experiments for cisapride and roflumilast, respectively.
Results
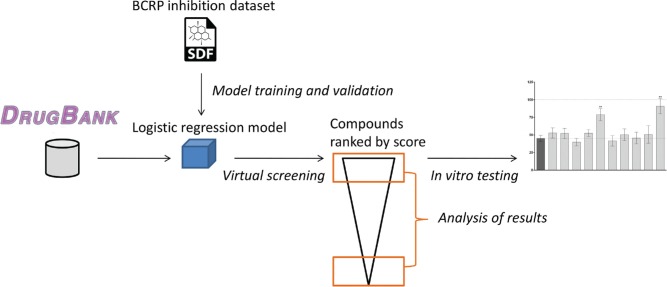
The overall workflow followed in this study is depicted in Figure 1 . Briefly, the data set containing 978 compounds annotated for their inhibition of BCRP was used to train a logistic regression model. The model was used to predict the BCRP inhibition capability of compounds belonging to DrugBank. The score given by the model was used to rank compounds, and top- and low-scoring compounds were checked against the literature. For those top-scoring compounds for which no information was available, we selected 10 that were structurally diverse and also not similar to the training set. These 10 compounds were bought and tested in vitro in a cell-based mitoxantrone accumulation assay, which revealed two new inhibitors of BCRP. The results of the different steps are detailed in the following section.
Figure 1.
General workflow of the steps followed throughout the study.
Classification Model
The training set used for building the in silico predictive model is, to our knowledge, the largest curated pharmacological data set for BCRP inhibition. The compounds were collected from open sources such as ChEMBL,22 PubChem,23 and Medline. The whole process of merging together compounds measured in different assays has been previously described.12 Compounds are annotated as inhibitor or noninhibitor for BCRP, and this categorization is what the model is expected to provide. Thus, the model will not be able to predict inhibition potency.
Among the 16 different models tried (four descriptor sets times four learning algorithms; see Materials and Methods section), random forest, logistic regression, and SVM performed consistently well in simple cross-validation, except with VolSurf descriptors (see Suppl. Table S1 for details).
Models were validated by two methods: 10-fold stratified cross-validation and repeated sampling of data sources (individual publications or database entries, leave-sources-out method). The first method is the most commonly employed to evaluate the capacity of the models to predict compounds from the same distribution as the training set. The second method is more demanding since excluding entire sources from the training set mimics a prospective validation. Both validation results are reported in Table 1 for the final model in terms of confusion matrix, MCC, and area under the ROC curve. Logistic regression and SVM built on ECFP-like fingerprints gave the highest area under the ROC curve (0.90 in cross-validation for both models). The model based on logistic regression was selected as the final one, because its training time was much shorter than the one for the SVM. Based on the final model settings, a sensitivity analysis was performed to assess the potential influence of the thresholds applied when assigning the class labels to the training set. The results demonstrate the robustness of the obtained model when used for ranking the compounds (for further details, see supplementary information).
Table 1.
Confusion Matrix, Matthew’s Correlation Coefficient, and Area under the ROC Curve of the Logistic Regression Model in Cross-Validation and Prospective Evaluation.
| TN (n) | FN (n) | TP (n) | FP (n) | MCC | AUC ROC | |
|---|---|---|---|---|---|---|
| 10-fold CV | 478 | 101 | 332 | 67 | 0.65 | 0.90 |
| Leave-sources-outa | 71 | 35 | 49 | 34 | 0.26 | 0.71 |
AUC, area under the curve; CV, cross-validation; FN, false negatives; FP, false positives; MCC, Matthews’s correlation coefficient; ROC, receiver operating characteristic; TN, true negatives; TP, true positives.
Average of 166 models built on a randomly chosen subset of data sources (i.e., publications) and validated by the left-out sources.
Virtual Screening
The 1702 compounds from the DrugBank small-molecules set were prepared as described in the Materials and Methods section. The same fingerprints as for the training set were computed and sent to the model for prediction. In total, 129 compounds received a score higher than 0.5 and thus were predicted as BCRP inhibitors. Among them, 32 were present in the training set and already annotated as BCRP inhibitors. Another three were not in the training set and reported in DrugBank as BCRP inhibitors. Finally, seven were found inactive in a PubChem Bioassay (AID1325). These seven compounds represent therefore false positives.
The 100 compounds with the lowest score (between 0 and 0.03) were also individually examined. We found eight compounds from the training set annotated as noninhibitors and another 18 that were found inactive in the PubChem Bioassay AID1325. No false negative could be found among the low-scored compounds that were examined.
A list of 56 compounds with unknown BCRP activity from the highest scored ones served as a basis to select compounds for experimental testing. From this initial list of candidates, xhanthophyll was removed because it is a lipid. We also checked for intralist similarity and removed the compound having the lowest predicted score from each pair having a Tanimoto similarity over 0.75. The refined list of candidates for experimental validation therefore contained 43 compounds. Finally, we checked the similarity between our candidates and the training set and removed all compounds with a Tanimoto similarity to the closest neighbor in the training set higher than 0.75. This left us with 29 compounds, from which 10 could be bought according to vendor availabilities ( Table 2 ).
Table 2.
Probability Score Given by the Model and Tanimoto Similarity to the Closest Neighbor in the Training Set for the 10 Compounds Selected for In Vitro Testing.
| DrugBank ID | Compound Name | Score of the Model | Highest Tanimoto Similarity |
|---|---|---|---|
| DB01395 | Drospirenone | 0.85 | 0.74 |
| DB01228 | Encainide | 0.67 | 0.63 |
| DB01218 | Halofantrine | 0.67 | 0.51 |
| DB00276 | Amsacrine | 0.64 | 0.52 |
| DB01656 | Roflumilast | 0.60 | 0.52 |
| DB00514 | Dextromethorphan | 0.59 | 0.60 |
| DB00857 | Terbinafine | 0.55 | 0.53 |
| DB00169 | Cholecalciferol | 0.55 | 0.54 |
| DB01091 | Butenafine | 0.54 | 0.51 |
| DB00604 | Cisapride | 0.54 | 0.72 |
In Vitro Testing
To evaluate BCRP inhibition by the 10 selected compounds, the intracellular accumulation of the fluorescent BCRP substrate mitoxantrone was measured in BCRP-overexpressing PLB985 cells by flow cytometry. Protein expression and functionality of BCRP in overexpressing PLB985 cells were verified as given in the supplementary information.
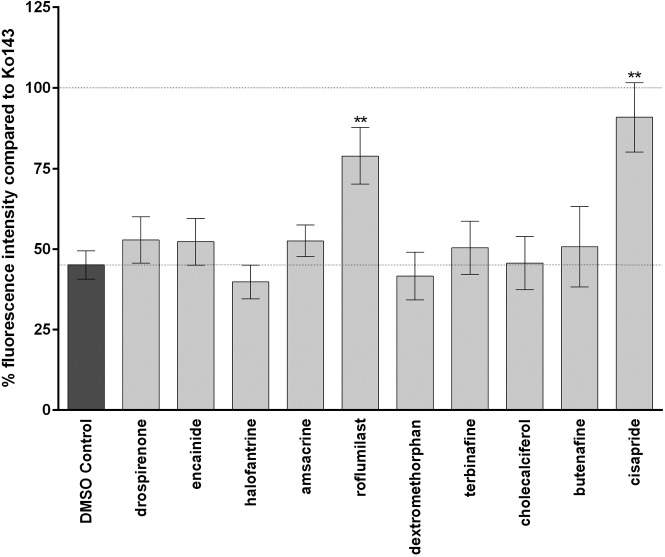
The inhibitory potency of the compounds on BCRP function was tested at a concentration of 10 µM. As shown in Figure 2 , two compounds—namely, roflumilast and cisapride (for chemical structures see Fig. 3 )—showed a significant inhibitory activity on BCRP compared with the negative control. Compared with the positive control Ko143 (set to 100%), roflumilast showed an inhibitory effect of 62% ± 16%. Cisapride was even more effective with an inhibitory activity of 84% ± 20%. For all other compounds, only a marginal inhibitory activity (<15% compared with the positive control Ko143) was obtained.
Figure 2.
Effect of different compounds on breast cancer resistance protein (BCRP) transport function. Intracellular accumulation of the fluorescent BCRP substrate mitoxantrone (7 µM) was measured in the absence and presence of the test compounds as described in the Materials and Methods section. Data are given as the mean percentage of fluorescence intensity ± SD of three independent experiments normalized to the positive control Ko143, which was set as 100%. Each individual experiment was performed in duplicates. Compounds showing significant different fluorescence intensity compared with the DMSO control are marked with asterisks (**p < 0.01).
Figure 3.
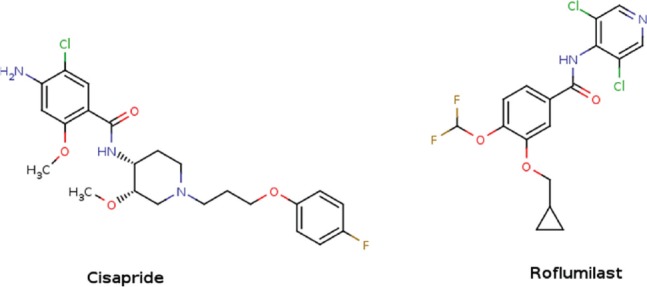
Structure of the two compounds (cisapride and roflumilast) that inhibit breast cancer resistance protein (BCRP) transport function in the in vitro assay.
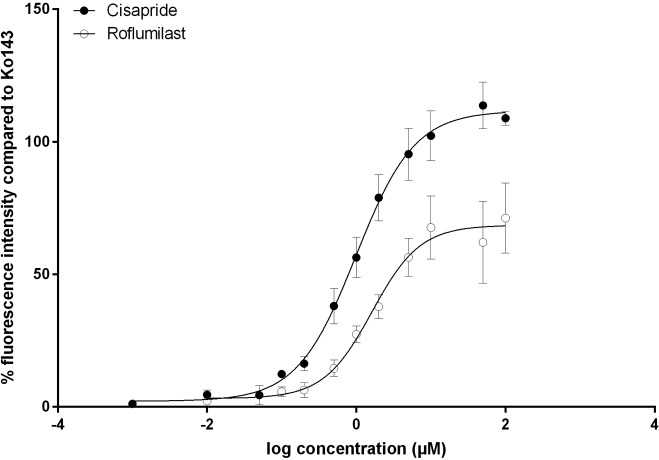
Based on their inhibitory activity on BCRP shown at 10 µM, IC50 values were determined for roflumilast and cisapride. The inhibitory activity of both compounds indeed could be confirmed by IC50 values of 0.9 ± 0.2 µM and 0.4 ± 0.1 µM, respectively, with cisapride being 2.25-fold more active than roflumilast ( Fig. 4 ).
Figure 4.
IC50apparent measurement of cisapride and roflumilast. Steady-state accumulation of mitoxantrone (7 µM) in the absence and presence of 10 or 12 different concentrations ranging from 0.001 to 100 µM of roflumilast and cisapride, respectively, was measured as described in the Materials and Methods section. Data given here show the mean percentage fluorescence intensity after subtracting the background fluorescence of unstained cells and the fluorescence of the DMSO control and subsequent normalization to the positive control Ko143, which was set as 100%, ± SD of three and five independent experiments for cisapride and roflumilast, respectively. Each experiment was performed at least in technical duplicates.
Discussion
Pharmacokinetics problems are a major cause of failures in late stages of drug development. While for a long time, most of the focus was on cytochromes and metabolism, the importance of transporters at the different steps of drug absorption, distribution, and elimination starts to be recognized. BCRP, because of its diverse substrate profile and its expression at crucial tissues, actively participates in the absorption and elimination of drugs.24 It is also involved in drug-drug interactions.25 For this reason, being able to predict whether a drug will be a substrate or an inhibitor of BCRP is of great interest. Until recently, researchers viewed BCRP as a drug target to reduce multidrug resistance (MDR), therefore trying to develop potent and selective inhibitors. Although no such candidates passed clinical trials, this trend allowed growing the pharmacological knowledge behind BCRP inhibition. These data are crucial to build statistical models that can predict inhibition of BCRP. As previously mentioned, most existing models are either restricted to a structural family of compounds or to a very small data set. Here, we were able to build a predictive model on 978 compounds, up to now the largest training set ever used for BCRP inhibition prediction. The model gave very good cross-validation results, similar to recently published models.9 The second validation, which consists of randomly selecting a set of publications from the data set to play the role of the external test set (leave-sources-out approach), led to results similar to what was observed by Erić and colleagues9 on their external set. The difference to using a test set is that we are repeating the splits many times, building and validating the model for each new subset of the data, and averaging the results. Thus, the results are statistically representative of an average prospective study. While there is quite a drop between the cross-validation results and the repeated external set results (AUC of 0.90 vs. 0.71), this is a frequent phenomenon when working with medicinal chemistry data sets. The model, implemented in a python script to allow quick reproduction of the results and easy utilization to predict new compounds, is very fast. In 4.5 s, the 1700 compounds of our DrugBank set were screened and the predictions written to a file for further use. The script is available for the interested reader, who can train and cross-validate the model and then use it to predict BCRP inhibition on new data (given to the model as an sd file). More information on how to install and run the script is also available as supplementary information.
Prospective validation of the model was performed by in silico screening of DrugBank and testing a set of top-ranked compounds. This identified two new potent BCRP inhibitors among known drugs: cisapride (84% inhibition at 10 µM, IC50 of 0.40 µM) and roflumilast (62% inhibition at 10 µM, IC50 of 0.9 µM). Roflumilast was approved by the Food and Drug Administration in 2011 for the treatment of chronic obstructive pulmonary disease (COPD) exacerbations in patients for whom previous treatments failed.26 The usual dose is 500 µg/d, taken once. The maximum plasma concentration in Caucasian men is observed after 1 h at 2 µg/L, which corresponds to a concentration of roflumilast in blood of 5 nM,27 well below the observed IC50 for BCRP inhibition. This may explain why no drug-drug interactions involving roflumilast as the perpetrator have been reported so far.
Cisapride is a serotonin 5-HT4 agonist, indicated for treatment of gastroesophageal reflux in adults. It was approved in 1988 by the European Medicines Agency. Since 2000, cisapride has been partially withdrawn for cardiotoxicity since it has been found to inhibit the hERG potassium channel,28 leading to prolonged QT and cardiac arrhythmia.
Taken together, the results of the DrugBank screening are the following: five true positive (three compounds identified in the literature and two in the in vitro testing), 15 false positive, 18 true negative, and no false negative. Many of the false positives came from the results of the PubChem Assay AID1325, which is a primary multiplex screening for both BCRP and P-gp inhibition. This assay tested 194,480 compounds, and only 200 were annotated as active at BCRP. The threshold used for classifying a compound as active in the assay is 80% inhibition, which is quite high and could explain the surprisingly low hit rate for that particular screen (0.1%), especially given the promiscuous behavior of BCRP. While the amount of false positives obtained with our in silico model may seem high, one has to bear in mind that for detecting drug-drug interactions, models with low false-negative rates are desirable. Given the speed and ease of use of the in silico model, it could be useful as an early screening tool to detect potential drug-drug interaction perpetrators before confirming them by an in vitro assay.
Beyond in silico screening, the model was recently also used to support the design of BCRP-selective compounds.29 There, the predictive model assisted in prioritizing the compounds to be synthesized. In that sense, the method can be viewed as a help for BCRP inhibitor design.
In conclusion, in the present work, an in silico classification model based on a manually curated set of compounds has been developed to identify potential BCRP inhibitors. It showed excellent predictivity in cross-validation (ROC AUC of 0.9) and good predictivity in prospective validation (ROC AUC of 0.7). Subsequent virtual screening of DrugBank small molecules, followed by experimental testing of top-ranked hits, allowed the discovery of roflumilast and cisapride as previously unknown potent BCRP inhibitors, with IC50 values of 0.9 µM and 0.4 µM, respectively. The model can be used for quick prefiltering of compounds that may be drug-drug interaction perpetrators and therefore reduce the necessary amount of in vitro tests.
Supplementary Material
Acknowledgments
We thank the software company ChemAxon for providing us kindly with their software, B. Sarkadi and K. Nemet for providing PLB985 parental and PLB985BCRP cells, and K. Kuchler (Medical University of Vienna) for providing place and resources for the in vitro experiments described in the supplementary information.
Footnotes
Supplementary material for this article is available on the Journal of Biomolecular Screening Web site at http://jbx.sagepub.com/supplemental.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreements 115002 (eTOX), resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007-2013) and EFPIA Companies’ in kind contribution. We acknowledge additional financial support by the Austrian Science Fund (FWF), grant F03502.
References
- 1. Maliepaard M., Scheffer G. L., Faneyte I. F., et al. Subcellular Localization and Distribution of the Breast Cancer Resistance Protein Transporter in Normal Human Tissues. Cancer Res. 2001, 61, 3458–3464. [PubMed] [Google Scholar]
- 2. Matsuo H., Takada T., Ichida K., et al. ABCG2/BCRP Dysfunction as a Major Cause of Gout. Nucleosides Nucleotides Nucleic Acids 2011, 30, 1117–1128. [DOI] [PubMed] [Google Scholar]
- 3. Lee C. A., O’Connor M. A., Ritchie T. K., et al. Breast Cancer Resistance Protein (ABCG2) in Clinical Pharmacokinetics and Drug Interactions: Practical Recommendations for Clinical Victim and Perpetrator Drug-Drug Interaction Study Design. Drug Metab. Dispos. Biol. Fate Chem. 2015, 43, 490–509. [DOI] [PubMed] [Google Scholar]
- 4. Breedveld P., Beijnen J. H., Schellens J. H. M. Use of P-Glycoprotein and BCRP Inhibitors to Improve Oral Bioavailability and CNS Penetration of Anticancer Drugs. Trends Pharmacol. Sci. 2006, 27, 17–24. [DOI] [PubMed] [Google Scholar]
- 5. Nakagawa H., Saito H., Ikegami Y., et al. Molecular Modeling of New Camptothecin Analogues to Circumvent ABCG2-Mediated Drug Resistance in Cancer. Cancer Lett. 2006, 234, 81–89. [DOI] [PubMed] [Google Scholar]
- 6. Food and Drug Administration. Guidance for Industry Drug Interaction Studies—Study Design, Data Analysis, Implications for Dosing, and Labeling Recommendations; Draft Guidance; U.S. Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER): Rockville, MD, 2012. [Google Scholar]
- 7. Cramer J., Kopp S., Bates S. E., et al. Multispecificity of Drug Transporters: Probing Inhibitor Selectivity for the Human Drug Efflux Transporters ABCB1 and ABCG2. ChemMedChem 2007, 2, 1783–1788. [DOI] [PubMed] [Google Scholar]
- 8. Zhang S., Yang X., Coburn R. A., et al. Structure Activity Relationships and Quantitative Structure Activity Relationships for the Flavonoid-Mediated Inhibition of Breast Cancer Resistance Protein. Biochem. Pharmacol. 2005, 70, 627–639. [DOI] [PubMed] [Google Scholar]
- 9. Erić S., Kalinić M., Ilić K., et al. Computational Classification Models for Predicting the Interaction of Drugs with P-Glycoprotein and Breast Cancer Resistance Protein. SAR QSAR Environ. Res. 2014, 25, 939–966. [DOI] [PubMed] [Google Scholar]
- 10. Matsson P., Englund G., Ahlin G., et al. A Global Drug Inhibition Pattern for the Human ATP-Binding Cassette Transporter Breast Cancer Resistance Protein (ABCG2). J. Pharmacol. Exp. Ther. 2007, 323, 19–30. [DOI] [PubMed] [Google Scholar]
- 11. Pan Y., Chothe P. P., Swaan P. W. Identification of Novel Breast Cancer Resistance Protein (BCRP) Inhibitors by Virtual Screening. Mol. Pharm. 2013, 10, 1236–1248. [DOI] [PubMed] [Google Scholar]
- 12. Montanari F., Ecker G. F. BCRP Inhibition: From Data Collection to Ligand-Based Modeling. Mol. Inform. 2014, 33, 322–331. [DOI] [PubMed] [Google Scholar]
- 13. RDKit. http://www.rdkit.org/ (accessed Jun 16, 2015).
- 14. Molecular Operating Environment (MOE), 201308; Chemical Computing Group: Montreal, Canada. [Google Scholar]
- 15. Guha R. CDK Descriptor UI. https://github.com/rajarshi/cdkdescui (accessed Jan 7, 2016).
- 16. Pedregosa F., Varoquaux G., Gramfort A., et al. Scikit-Learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- 17. van der Walt S., Colbert S. C., Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput. Sci. Eng. 2011, 13, 22–30. [Google Scholar]
- 18. Ozvegy-Laczka C., Várady G., Köblös G., et al. Function-Dependent Conformational Changes of the ABCG2 Multidrug Transporter Modify Its Interaction with a Monoclonal Antibody on the Cell Surface. J. Biol. Chem. 2005, 280, 4219–4227. [DOI] [PubMed] [Google Scholar]
- 19. Homolya L., Orbán T. I., Csanády L., et al. Mitoxantrone Is Expelled by the ABCG2 Multidrug Transporter Directly from the Plasma Membrane. Biochim. Biophys. Acta BBA Biomembr. 2011, 1808, 154–163. [DOI] [PubMed] [Google Scholar]
- 20. Stein W. D. Kinetics of the Multidrug Transporter (P-Glycoprotein) and Its Reversal. Physiol. Rev. 1997, 77, 545–590. [DOI] [PubMed] [Google Scholar]
- 21. Litman T., Skovsgaard T., Stein W. D. Pumping of Drugs by P-Glycoprotein: A Two-Step Process? J. Pharmacol. Exp. Ther. 2003, 307, 846–853. [DOI] [PubMed] [Google Scholar]
- 22. Bento A. P., Gaulton A., Hersey A., et al. The ChEMBL Bioactivity Database: An Update. Nucleic Acids Res. 2014, 42, D1083–D1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y., Xiao J., Suzek T. O., et al. PubChem’s BioAssay Database. Nucleic Acids Res. 2012, 40, D400–D412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jonker J. W., Smit J. W., Brinkhuis R. F., et al. Role of Breast Cancer Resistance Protein in the Bioavailability and Fetal Penetration of Topotecan. J. Natl. Cancer Inst. 2000, 92, 1651–1656. [DOI] [PubMed] [Google Scholar]
- 25. Xia C. Q., Yang J. J., Gan L.-S. Breast Cancer Resistance Protein in Pharmacokinetics and Drug-Drug Interactions. Expert Opin. Drug Metab. Toxicol. 2005, 1, 595–611. [DOI] [PubMed] [Google Scholar]
- 26. Baye J. Roflumilast (Daliresp). Pharm. Ther. 2012, 37, 149–161. [PMC free article] [PubMed] [Google Scholar]
- 27. Nassr N., Huennemeyer A., Herzog R., et al. Effects of Rifampicin on the Pharmacokinetics of Roflumilast and Roflumilast N-Oxide in Healthy Subjects. Br. J. Clin. Pharmacol. 2009, 68, 580–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rampe D., Roy M.-L., Dennis A., et al. A Mechanism for the Proarrhythmic Effects of Cisapride (Propulsid): High Affinity Blockade of the Human Cardiac Potassium Channel HERG. FEBS Lett. 1997, 417, 28–32. [DOI] [PubMed] [Google Scholar]
- 29. Schwarz T., Montanari F., Cseke A., et al. Subtle Structural Differences Trigger Inhibitory Activity of Propafenone Analogues at the Two Polyspecific ABC Transporters: P-Glycoprotein (P-Gp) and Breast Cancer Resistance Protein (BCRP). ChemMedChem 2016, 11, 1380–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.