Abstract
Other than an association with HPV infection, little is known about the genetic alterations determining the development of penile cancer. Although penile cancer is rare in the developed world, it presents a significant burden in developing countries. Here, we report the findings of whole-exome sequencing (WES) to determine the somatic mutational landscape of penile cancer. WES was performed on penile cancer and matched germline DNA from 27 patients undergoing surgical resection. Targeted resequencing of candidate genes was performed in an independent 70 patient cohort. Mutation data were also integrated with DNA methylation and copy-number information from the same patients. We identified an HPV-associated APOBEC mutation signature and an NpCpG signature in HPV-negative disease. We also identified recurrent mutations in the novel penile cancer tumor suppressor genes CSN1(GPS1) and FAT1. Expression of CSN1 mutants in cells resulted in colocalization with AGO2 in cytoplasmic P-bodies, ultimately leading to the loss of miRNA-mediated gene silencing, which may contribute to disease etiology. Our findings represent the first comprehensive analysis of somatic alterations in penile cancer, highlighting the complex landscape of alterations in this malignancy.
Introduction
The etiology of penile cancer is multifactorial with smoking, phimosis, poor personal hygiene, and low socioeconomic status all being risk factors for tumor development (1). However, other than the oncogenic impact of high-risk human papillomavirus (HPV) infection, which is responsible for approximately 30% of penile cancers, little is known about the molecular alterations involved in the development of this disease.
Penile cancer is relatively rare in the developed world, but represents a global health problem, with high prevalence and significant associated morbidity and mortality in developing countries (1, 2). The age standardized incidence of penile cancer is 0.3 to 1.0 per 100,000 men in European countries and the United States, equating to approximately 1,600 new cases per annum in the United States (3). In contrast, the incidence in developing nations varies from 3 to 8.3 per 100,000 (3). Five-year survival is between 50% and 80% dependent on tumor stage and age at presentation, and over 40% of patients present with lymph node metastases (2).
We have previously reported genome-wide epigenetic events involved in an aggressive penile cancer phenotype and identified potential epigenetic drivers of penile cancer (4). In this study, we sought to determine the genetic component involved in penile cancer development, by performing the first whole-exome sequencing (WES) analysis of penile cancer to identify the somatic alterations.
Materials and Methods
Sample cohort
Samples were collected from penile cancer patients at UCLH NHS Trust, UK. Informed consent was obtained from all subjects and ethical approval for this study was granted by the University College London (UCL)/University College London Hospital (UCLH) BioBank for Health and Human Disease (NC06.11). All samples were reviewed by a consultant histopathologist and confirmed to be ≥80% tumor content. The samples have previously been reported in other studies (4). The histologic characteristics of our discovery and validation cohorts are shown in Supplementary Table S1.
Whole-exome sequencing
Exome capture was performed using 50 ng of tumor or matched germline DNA. Library preparation was performed using the Illumina TrueSeq or Nextrea Exome Capture Kit according to the manufacturers guidelines (Ilumina). Samples underwent paired end sequencing on a Hi-Seq2000 platform with 100 bp read length to a mean coverage of ×60. Validation samples were subjected to targeted sequencing candidate genes (CSN1/GPS1, FAT1, and TP53) using Fluidigm custom amplification. A mean coverage of ×156 was achieved across the validation cohort. Detailed methods for sequencing analysis can be found in the Supplementary Methods.
Sequencing data have been submitted to European Genomephenome Archive (EGA; Accession: EGAD00001001785). Access requests are through the UCL Translational Uro-Oncology Data Access Committee (DAC).
Variant detection
The Varscan (5) algorithm (v2.2.10, varscan.sourceforge. net) was used to call both single-nucleotide alterations and indels. Alterations were annotated to the Human reference genome, version 38, using Annovar (v.517, http://www.openbioinformatics.org/annovar/; ref. 6). SNPs were annotated using Oncotator from the Broad Institute (http://www.broadinstitute.org/oncotator/). Common germline SNPs as recorded in either dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), 1000 genomes (http://www.1000genomes.org) were excluded from further analysis. Germline SNP analysis was used to confirm sample identify between tumor and normal pairs. Significantly mutated genes were defined as genes with recurrent mutations that showed a functional mutation bias q value of 0.1 or less following analysis by the IntOgen mutation analysis platform (7). Mutations in potential candidate drivers were visualized after filtering for genes not in the MutSig5000 set of pan-tissue cancer–associated genes (8).
Annotation and visualization of mutations
SNPs were annotated using Oncotator from the Broad Institute http://www.broadinstitute.org/oncotator/. Significantly mutated genes were defined as genes with recurrent mutations that showed a functional mutation bias q value of 0.1 or less following analysis by the IntOgen mutation analysis platform (7). Mutations in potential candidate drivers were visualized after filtering for genes not in the MutSig5000 set of pan-tissue cancer–associated genes (8).
Pathway analysis
Pathway analysis was performed using IntOgen pathway analysis (9). Significantly, mutated KEGG pathways were identified using IntOgen pathway analysis based on functional mutation bias with a q value of less than 0.05. Selected coding mutations were visualized in the context of cancer associated pathways.
Supervised analysis of potential deamination signatures
Overall mutational analysis was carried out by visualizing the breakdown of mutations into 12 categories of base changes. A high proportion of mutations were C>T or G>A, which were candidate cytosine deamination events. To examine the role of cytosine deamination mediated by the APOBEC family of cytosine deaminases and the spontaneous deamination of methylcytosines in contributing to the overall mutational burden of the tumor samples, supervised analysis was carried out to model mutagenesis as the fraction of all mutations in the tumor. Candidate APOBEC-induced mutations were defined as C→K changes in the TCW context on either strand, where K = T/G, W = A/T whereas CpG deamination events were defined as C→T changes in the context of ACG/CCG/GCG (8). TCG mutations were not considered for analysis because of potential confounding between APOBEC-induced and CpG-deamination induced C→T mutations (7). To test for associations between fractions of deamination mutations were carried out for CpG deamination mutations and TCW→TKW mutations by HPV status we performed a multivariate binomial regression of APOBEC and CpG deamination mutations. We regressed the fraction of TCW→TKW mutations and the A/C/G CG → A/C/G TG mutations using a binomial glm with HPV status by viral load, age, stage, and grade as potential explanatory variables.
Unsupervised extraction of mutational signatures
Mutations were parsed into a mutation motif matrix using the SomaticSignatures R package. 100 runs of NMF using the brunet algorithm in the NMF R package were used to decompose the motif matrix into signatures and sample-exposures (basis and coefficient respectively) for 2 to 5 clusters. This was followed by selecting the number of signatures on the basis of maximum consensus silhouette and the cophenetic correlation coefficient. Two signatures were chosen based on a high cophenetic correlation and maximum consensus silhouette width (Supplementary Fig. S6).
Analysis of enrichment for CpG deamination events among TP53 mutations
Enrichment for CpG mutations in TP53 was carried out using binomial testing against a background probability distribution. The background distribution (under the null hypothesis of no enrichment) was defined to factor in the number of CpG and non-CpG sites in the longest TP53 transcript according to the IARC P53 database (123 and 3001) and the average CpG mutation fraction in the four TP53 mutant tumors normalized to the exome wide frequency of CpG sites.
Viral detection
HPV viral load was assessed in penile cancer DNA using qPCR for low risk HPV 6 and 11 and high risk HPV 16, 18 and 31 by qPCR. Primers and probes are listed in Supplementary Table S3. The reference genes GAPDH and ACTB were used to normalize all HPV Q-PCR reactions. High and low viral load cases were defined as >1 HPV copy/cell and <1 HPV copy per cell, respectively. VirusSeq (odin.mdacc.tmc.edu/~xsu1/VirusSeq.html; ref. 10) and ViralFusionSeq (v. vfs-2012-12-07, http://sourceforge.net/projects/viralfusionseq/; ref. 11) were used to identify HPV integration from exome sequencing data.
DNA methylation analysis
DNA methylation and copy-number data were analyzed as previously described (4).
CSN1 mutations
Constructs expressing Flag-tagged wild-type GPS1/CSN1 were a kind gift from Ning Wei (Yale University). Mutations in GPS1/CSN1 were made using the Q5 Site-Directed Mutagenesis Kit (NEB) and confirmed by sequencing.
miRNA reporter assays
HeLa cells were transfected with wild-type or the indicated mutant CSN1 with Renilla reporter constructs using Fugene 6 transfection reagent (Promega). For miR-100 assays, the reporter construct contained miR-100–binding sites in the 3′ untranslated region (UTR) with seed sequence matches or mismatches. The Let-7 reporter construct contained seven partially complementary let-7a sites. The activity of Renilla luciferase was assayed and normalized to firefly luciferase activity using the Dual-Luciferase Reporter Assay system (Promega). The activity of the miR-100 reporter containing seed matches was compared with that of the reporter containing mismatches. For the let-7 reporter, the activity was compared with a reporter construct with no miRNA-binding sites in the Renilla 3′UTR.
Immunofluorescence
HeLa cells were plated on to 13 mm glass coverslips (VWR) and transfected with the indicated GPS1/CSN1–expressing constructs using Jetprime (Polyplus Transfection). Cells were fixed in 4% PFA for five minutes at 37°C and post-fix permeabilized for 90 seconds using 0.25% Triton X-100/PBS. Cells were stained with anti-Flag M2 (Sigma) and Alexa-Flour–conjugated secondary (Life Technologies).
Results
Mutational spectrum of penile cancer
Analysis revealed 810 genes containing somatic mutations among the 27 tumors (24 tumor germline pairs, 3 single tumors), with a mean somatic mutation rate of 30 per tumor. This represents 1.78 non-silent mutations per megabase (range, 0.72–7.5), relatively low when compared with other adult tumors that have a similar overall Ti/Tv (Transition/Transversion) ratio (12). There was no significant association between mutational burden and tumor stage, grade, age, or overall HPV status; however, when stratified for HPV viral load, high viral load tumors had a significantly lower mutational load (P < 0.05) when compared with HPV negative (Fig. 1).
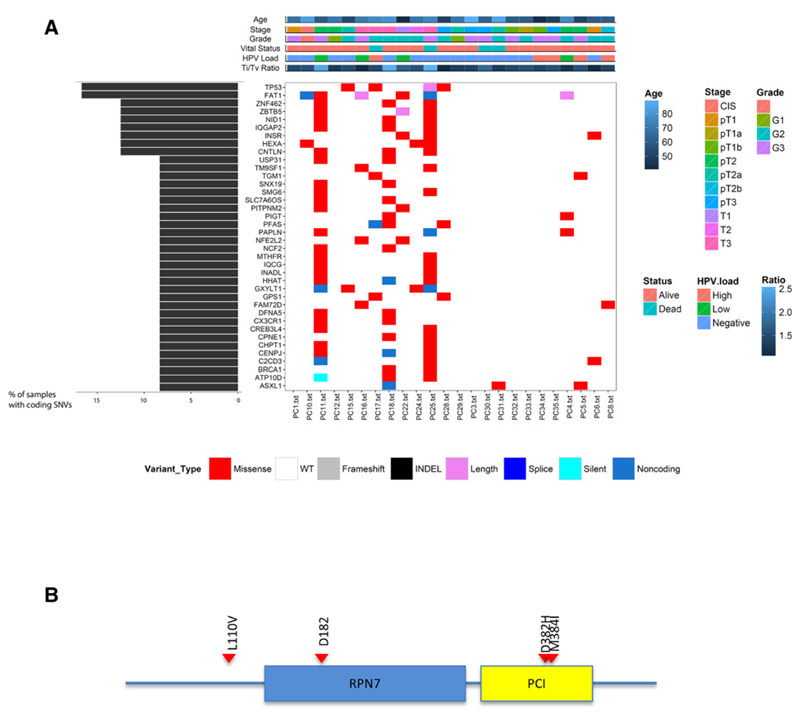
Figure 1. Somatic variants in penile cancer.
A, recurrently mutated genes in penile cancer. The central heatmap shows the mutation status of the recurrently mutated genes for each tumor. Somatic mutations are colored according to functional class (lower). Left, mutation count for each individual gene. Top, patient phenotype data for age, stage grade, Ti/Tv ratio and HPV viral status. B, schematic representation of the CSN1 protein. The conserved RPN7 and PCI domains are shown in blue and yellow, respectively. CSN1 alterations, position, and aa substitution are highlighted. Red, missense mutations.
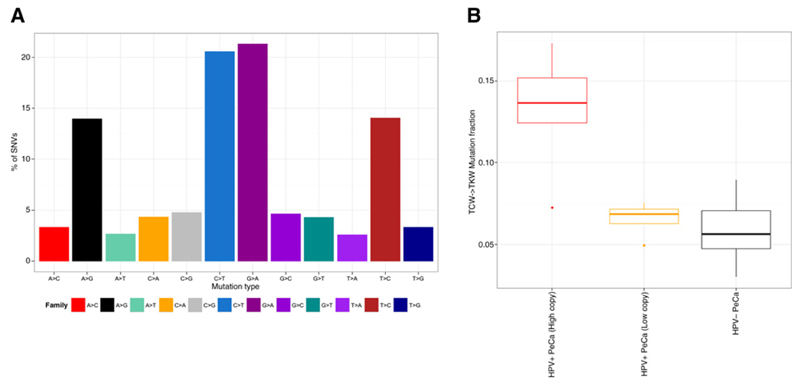
As mutational patterns can be indicative of specific mutagenic mechanisms driving tumorigenesis, we assessed whether any mutational signatures exist within penile cancer. We found C>T alterations to be the most frequent substitution type. C>T mutations at CpG sites are ubiquitously present in cancers across multiple tissue types (13) and have been linked to the spontaneous deamination of methylated cytosine (mCpG; ref. 13). Active cytosine deamination mediated by one or more 'TC-specific" apolipoprotein B mRNA-editing enzyme, catalytic polypeptide-like (APOBEC3) has recently been implicated as a significant driver of C>T and C>G mutations in HPV-associated cancers (14, 15). To distinguish between mutations caused by methyl-cytosine deamination and APOBEC-catalyzed cytosine deamination, we examined the trinucleotide sequence contexts within which these mutations occur. We considered C>T mutations occurring at VCG sites (where V represents A, C or G) as mCpG deamination events and C>K (where K represents T or G) mutations in the TCW context as APOBEC-mediated (14, 15). Multivariate modeling implicated APOBEC-induced mutagenesis to be significantly different between HPV status and did not implicate clinical covariates and age as confounders (P < 2.2e−16). CpG mutations, were also associated with HPV status (HPV negative disease P = 4.10e−07 and low-copy HPV+ P = 1.55e−07; Fig. 2 and Supplementary Fig. S1), but not with age, stage or grade.
Figure 2. Mutational signatures in penile cancer.
A, bar plot showing the breakdown of mutations by type of substitution reveals a high frequency of C>T and G>A mutations. B, boxplot of proportion of TCW alterations with patients stratified by HPV viral status (HPV high, HPV low, and HPV negative).
The potential association of HPV viral load and the APOBEC3 (A3)-associated signature may help clarify the association of APOBEC-induced mutagenesis. Previously, a lack of correlation between expression of A3 family transcripts and the A3 signature was noted (15). Similarly, E6/E7 oncogene expression is correlated with neither the A3 signature nor with A3 expression in HPV-associated tumors (14). These data further highlight the role of HPV integration in the etiology of a subset of penile cancer, and although we cannot rule out the potential oncogenic effect of a cleared HPV infection, the lack of A3 signature mutations in HPV low/negative samples suggests mechanisms other than high-risk HPV infection are responsible for the development of penile cancer in a significant number of cases.
A significant enrichment of C>T alterations at [A/C/G]CG dinucleotides was demonstrated in both HPV low and negative disease compared with HPV high. Besides deamination of methylated cytosine resulting in somatic mutation, it can also affect DNA methylation levels. We observe a significant (P < 0.001) decrease in DNA methylation in HPV low and negative samples compared with high viral load samples and matched normal both globally and over gene bodies (Supplementary Fig. S2). There is no difference in methylation (global, gene body or CpG island) between HPV high samples and matched normal tissue (Supplementary Fig. S2), and the effect persists when comparing aged matched cases. CG deamination has been suggested to be a non-random enzyme-catalyzed event rather than a spontaneous occurrence (16). Although the catalytic drivers of this non-random CG deamination have yet to be identified, both A3A and A3B have been shown to deaminate cytosines in a CpG context (17). Besides potentially affecting epigenetic regulation, this CG deamination process may generate mutations integral to the carcinogenic process (14), for example, in TP53, where three quarters of somatic alterations in penile cancer are CG deamination events (P < 0.005).
Somatic alterations in penile squamous cell carcinoma
Recurrent somatic mutations in penile cancer were catalogued to identify putative pathogenic drivers. Of the 810 mutated genes, only 137 (17%) were recurrent alterations; 756 (93%) have previously been identified as mutated in other tumor types. The lack of significant overlap in recurrently mutated genes in our cohort suggests that many are likely passenger mutations (12).
Of the private alterations (637/810) several have been identified as putative drivers in the pan-cancer analysis of 4742 cancers (8), and include tumor suppressors CDKN2A and NF1, oncogenes such as HRAS and potential therapeutic targets such as kinases FLT1 and TGFBR2. Because of the low rate of recurrent alterations, enrichment of somatic mutations in canonical pathways was assessed (7). The p53 signaling pathway was identified as the most significantly mutated pathway (P 5.37 × 10−5).
To identify putative cancer drivers, mutations that were recurrent and showed significant functional mutation bias were characterized using IntOgen (Fig. 1 and Supplementary Fig. S3; ref. 7). Recurrent mutations were identified in the tumor suppressor genes TP53 (4/27), FAT1 (4/27), and the G-protein pathway suppressor CSN1(GPS1; 3/27; Fig. 1 and Supplementary Fig. S3). Orthogonal validation by sanger sequencing confirmed 100% of TP53, FAT1 and CSN1 mutations in the original cohort. TP53, CNS1, and FAT1 were also analyzed in independent cohort of 70 penile cancer (50 FFPE, 20 fresh) and found to be mutated in 19%, 17%, and 14%, respectively. As matched germline DNA was only available for 20 of 70 of the validation cohort, the possibility that some of these potential inactivating mutations may result from germline alterations cannot be excluded; therefore, only mutations that were predicted to be functionally deleterious were included. As we only observe mutation in a total of 18 of 97 cases, we also assessed whether TP53 under goes significant disruption through changes in DNA methylation or genomic copy number. Only 3 cases exhibited genomic loss of TP53, none of which overlapped with those harboring mutation (although copy-number neutral LOH cannot be ruled out) and none showed significant changes in promoter methylation. This low TP53 mutation rate suggests that disruption of p53 is a key feature in only a subset of penile cancer (18).
Somatic CSN1 mutations result in aberrant miRNA processing
Of the genes containing novel mutations, CSN1 is mutated in 11% of cases (Fig. 1), with a single case harboring two CSN1 alterations. CSN1 is a suppressor of G-proteins and mitogen-activated signal transduction, and is an essential part of the COP9 signalosome complex (CSN), involved in the regulation of stem cell self-renewal and differentiation (19). Sequencing of an independent penile cancer cohort demonstrated a 14% (10/70) frequency of mutation. CSN1 is also reported to be mutated at low frequency in several other tumor types (9), and although little is known about its role in cancer development, it may be involved in the suppression of the AP-1 transcription factor pathway (20) and inhibit JUN dependent transcription activity (21).
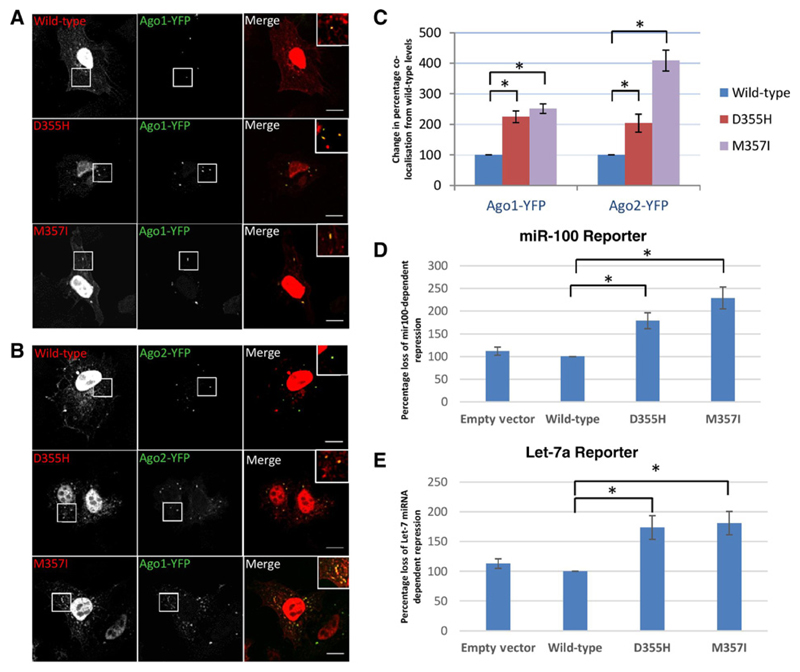
As none of the alterations associated with CSN1 are predicted to be truncating, to assess their role we recapitulated the 4 mutations identified in the initial discovery cohort in vitro. Recapitulation of the CSN1 mutations resulted in the translocation of CSN1 to the cytoplasm and colocalization with Argonaute1– and Argonaute2–positive P-bodies (Fig. 3). As the Argonaute (AGO) family of proteins are key components of the RNA-induced silencing complex (RISC) that mediates microRNA-dependent repression, we sought to assess whether CSN1 mutation effected miRNA-mediated gene repression. Expression of CSN1 point mutants resulted in a significant (P < 0.005) inhibition of miRNA-mediated gene repression compared with wild-type (Fig. 3). The greatest effect was seen with mutations (D382H and M384I) in the PCI (Protease Component) domain of CSN1 (Figs. 2 and 3); five of the 10 alterations identified in the validation cohort were also located within the PCI domain.
Figure 3. Mutations in CSN1 increase its localization to Ago1- and 2-positive P-bodies and leads to inhibition of miR-100 and Let-7–dependent repression of gene expression.
A, the D355H and M357I mutations on CSN1 1 localize to both the nucleus of HeLa cells and to Ago1-YFP–positive foci in the cytoplasm. Wild-type CSN1 localizes predominantly to the nucleus with occasional foci being formed. B, CSN1 mutants also localize to Ago2-YFP–positive foci in cytoplasm. C, quantification of Ago-positive foci colocalizing with CSN1 reveals the percentage of Ago-positive foci colocalizing with CSN1 is significantly increased with both mutants. Results are presented as the percentage of change in colocalization seen with wild-type. D and E, miR-100 and let-7a reporter assays in HeLa cells show that overexpression of both D355H and M357I CSN1 mutants inhibit miRNA-dependent repression of a Renilla reporter gene containing miR-100 or let-7a–binding sites in the 3′UTR. Results are presented as a percentage change from repression in cells transfected with wild-type CSN1. Wild-type CSN1 shows no significant difference from repression seen with empty vector. The activity of Renilla luciferase was assayed and normalized to firefly luciferase activity. Data are expressed relative to the activity of the reporter containing nontargeted miRNA sites. *, P < 0.05 according to Student t test.
Additional candidate penile cancer drivers
Mutation of FAT1 has been reported in other cancer types, including head and neck squamous cell carcinoma, colon cancer, and glioblastoma (22). FAT1 mutation promotes tumorigenicity through deregulation of the Wnt signaling cascade promoting cell proliferation and tumor growth (22). Analysis of the validation cohort identified FAT1 mutations in a further 17% of samples, in which stopgain mutations represent the most frequent alteration. Deletion of 4q35, the locus harboring FAT1, has also been frequently identified in human cancers (22). Integrated analysis of genomic and epigenomic data was, therefore, performed to assess whether FAT1 is disrupted by changes in genomic copy number or DNA methylation. No changes in copy number were observed; however, increased FAT1 promoter methylation (compared with normal penile squamous epithelium) was observed in a further 20% of cases lacking mutation (Supplementary Fig. S4). These data indicate that FAT1 is potentially deregulated in up to 37% of penile cancer, and suggest that FAT1 to be a key tumor suppressor in penile cancer development.
Discussion
The genetic and epigenetic mechanisms driving the development of penile cancer are poorly understood. Through WES, we have performed the most comprehensive analysis to date of the genetic alterations in penile cancer and implicated novel somatic mutations in penile cancer pathogenesis.
Penile cancer appear genetically quiet with strikingly few recurrent somatic mutations identified when compared with other adult tumors. However, with the caveat that, as with many other exome sequencing projects, we are underpowered to detect low frequency (<10%) alterations. The concept that penile cancer are relatively genomically quiet, is further highlighted by the low number of copy-number alterations seen in penile cancer, with approximately 50% of penile cancer showing no significant CNAs and only 13 regions of significant recurrent CNA (Supplementary Fig. S5). Although analysis of the mutational spectrum suggests a “APOBEC” mutational phenotype in HPV-driven disease, as in other HPV-driven tumors. The relative paucity of somatic alterations suggests that penile cancer development is driven by a complex interaction of molecular aberrations. The potential CpG deamination signature and concomitant reduction in DNA methylation in HPV-negative tumors, suggest that changes to the epigenome may represent a major pathogenic mechanism in penile cancer development (4).
Analysis of recurrent somatic mutations identified novel candidate drivers in penile cancer. These include the G Protein suppressor and member of the COP-9 signalosome complex, CSN1. The COP-9 complex and in particularly CSN1 has been implicated in the phosphorylation and activation of p53 (23). Previous screening studies in Drosophila identified other members of the COP-9 signalosome being involved in the miRNA pathway. Depletion of CSN3 and CSN7 by RNAi in S2 cells led to increases in siRNA and miRNA-dependent silencing of reporter genes (24). Dysregulation of the miRNA pathway is frequently observed in many cancers and further work is required to determine the mechanism by which singlepoint mutations of CSN1 can disrupt miRNA-mediated silencing and define its role in the development of human malignancies. However, data presented here suggest CSN1 to be a novel tumor suppressor, which when mutated disrupts miRNA-mediated gene silencing, and thus contributes to the development of penile cancer. The identification of CSN1 mutation, and its role in aberrant miRNA processing, and that APOBEC family members also interact with AGO2 to inhibit miRNA-dependent silencing (25), suggest that miRNA dysregulation may be an important driver of penile cancer pathogenesis.
We also observe the frequent mutation of the FAT1 tumor suppressor gene. FAT1 is somatically altered (by mutation or deletion) in multiple tumor types. Here, we show for the first time that in penile cancer FAT1 is not only inactivated through somatic mutation but also through promoter hypermethylation, resulting in FAT1 deregulation in over a third of penile cancer. This further highlights the potential important role of aberrant epigenetic changes in penile cancer development.
Our results, the first WES of penile cancer, provides novel insight into the relationship between somatic alterations and penile cancer development. These data significantly enhance our current understanding of genetic alterations driving penile cancer by showing that whilst recurrent somatic alterations can explain a proportion of penile cancer, epigenetic mechanisms play a significant role in penile cancer development. The discovery that mutations in the novel tumor suppressor gene CSN1 results in aberrant miRNA processing, and the high rate of CpG deamination suggests that deregulated gene silencing and changes in epigenetic regulation play an important role in penile cancer and particularly in non–HPV-associated tumors.
Supplementary Material
Acknowledgments
We would like to thank Debbie Hughes and Dr. Alan Pitman at the Institute of Neurology, UCL, London for their help running the exomes and also Dr. Chaz Mein at the Barts Cancer Institute for his help with the validation.
Grant Support
A. Feber and J.D. Kelly are supported by the UCLH/UCL Comprehensive Biomedical Research Program and the MRC (MR/M025411/1). Research involved in this project was funded by Orchid (The Male Cancer Charity). D.C. Worth, M. Arya, T. Powles, and J.D. Kelly are also supported by Orchid. Research in the S. Beck laboratory was supported by the Welcome Trust (WT084071, WT093855), Royal Society Wolfson Research Merit Award (WM100023), MRC (G100041), IMI-JU OncoTrack (115234), and EU-FP7 projects EPIGENESYS (257082), IDEAL (259679), and BLUEPRINT (282510). T.V. Sharp is supported by a BBRSC Project Grant (BB/I007571/2). T. Fenton is supported by the Rosetrees Trust. A. Feber also received support from the Rosetrees Trust. A. Chakravarthy is funded through a UCL Postgraduate Research Scholarship.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Authors' Contributions
Conception and design: A. Feber, M. Arya, M. Saqib, A. Freeman, T. Powles, S. Beck, J.D. Kelly
Development of methodology: A. Feber, D.C. Worth, M. Arya, T. Powles, T.V. Sharp
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A. Feber, D.C. Worth, P. de Winter, K. Shah, M. Arya, M. Saqib, R. Nigam, P.R. Malone, W.S. Tan, C. Jameson, T. Powles, A. Muneer, J.D. Kelly
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A. Feber, D.C. Worth, A. Chakravarthy, S. Rodney, G.A. Wilson, T. Fenton, T.V. Sharp, J.D. Kelly
Writing, review, and/or revision of the manuscript: A. Feber, A. Chakravarthy, P. de Winter, M. Arya, S. Rodney, A. Freeman, T. Fenton, T.V. Sharp, A. Muneer, J.D. Kelly
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): A. Feber, P. de Winter, M. Arya, A. Freeman, S. Beck, J.D. Kelly
Study supervision: A. Muneer, J.D. Kelly
References
- 1.Dillner J, von Krogh G, Horenblas S, Meijer CJ. Etiology of squamous cell carcinoma of the penis. Scand J Urol Nephrol Suppl. 2000;205:194–200. doi: 10.1080/00365590050509913. [DOI] [PubMed] [Google Scholar]
- 2.Arya M, Li R, Pegler K, Sangar V, Kelly JD, Minhas S, et al. Long-term trends in incidence, survival and mortality of primary penile cancer in England. Cancer Causes Control. 2013;24:2169–76. doi: 10.1007/s10552-013-0293-y. [DOI] [PubMed] [Google Scholar]
- 3.Hernandez BY, Barnholtz-Sloan J, German RR, Giuliano A, Goodman MT, King JB, et al. Burden of invasive squamous cell carcinoma of the penis in the United States, 1998–2003. Cancer. 2008;113:2883–91. doi: 10.1002/cncr.23743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feber A, Arya M, de Winter P, Saqib M, Nigam R, Malone PR, et al. Epigenetics markers of metastasis and HPV-induced tumorigenesis in penile cancer. Clin Cancer Res. 2015;21:1196–206. doi: 10.1158/1078-0432.CCR-14-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, et al. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics. 2009;25:2283–5. doi: 10.1093/bioinformatics/btp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods. 2013;10:1081–2. doi: 10.1038/nmeth.2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y, Yao H, Thompson EJ, Tannir NM, Weinstein JN, Su X, et al. VirusSeq: software to identify viruses and their integration sites using next-generation sequencing of human cancer tissue. Bioinformatics. 2013;29:266–7. doi: 10.1093/bioinformatics/bts665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li JW, Wan R, Yu CS, Co NN, Wong N, Chan TF, et al. ViralFusionSeq: accurately discover viral integration events and reconstruct fusion transcripts at single-base resolution. Bioinformatics. 2013;29:649–51. doi: 10.1093/bioinformatics/btt011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis JM, Kiezun A, Ramos AH, Serra S, Pedamallu CS, Qian ZR, et al. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nat Genet. 2013;45:1483–6. doi: 10.1038/ng.2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature. 2013;500:415–21. doi: 10.1038/nature12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson S, Chakravarthy A, Su X, Boshoff C, Fenton TR. APOBEC-mediated cytosine deamination links PIK3CA helical domain mutations to human papillomavirus-driven tumor development. Cell Rep. 2014;7:1833–41. doi: 10.1016/j.celrep.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 15.Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat Genet. 2013;45:970–6. doi: 10.1038/ng.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen JC, Rideout WM, III, Jones PA. The rate of hydrolytic deamination of 5-methylcytosine in double-stranded DNA. Nucleic Acids Res. 1994;22:972–6. doi: 10.1093/nar/22.6.972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suspene R, Aynaud MM, Vartanian JP, Wain-Hobson S. Efficient deamination of 5-methylcytidine and 5-substituted cytidine residues in DNA by human APOBEC3A cytidine deaminase. PLoS ONE. 2013;8:e63461. doi: 10.1371/journal.pone.0063461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Castren K, Vahakangas K, Heikkinen E, Ranki A. Absence of p53 mutations in benign and pre-malignant male genital lesions with over-expressed p53 protein. Int J Cancer. 1998;77:674–8. doi: 10.1002/(sici)1097-0215(19980831)77:5<674::aid-ijc2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 19.Pan L, Wang S, Lu T, Weng C, Song X, Park JK, et al. Protein competition switches the function of COP9 from self-renewal to differentiation. Nature. 2014;514:233–6. doi: 10.1038/nature13562. [DOI] [PubMed] [Google Scholar]
- 20.Tsuge T, Matsui M, Wei N. The subunit 1 of the COP9 signalosome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J Mol Biol. 2001;305:1–9. doi: 10.1006/jmbi.2000.4288. [DOI] [PubMed] [Google Scholar]
- 21.Tsuge T, Menon S, Tong Y, Wei N. CSN1 inhibits c-Jun phosphorylation and down-regulates ectopic expression of JNK1. Protein Cell. 2011;2:423–32. doi: 10.1007/s13238-011-1043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morris LG, Kaufman AM, Gong Y, Ramaswami D, Walsh LA, Turcan S, et al. Recurrent somatic mutation of FAT1 in multiple human cancers leads to aberrant Wnt activation. Nat Genet. 2013;45:253–61. doi: 10.1038/ng.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bech-Otschir D, Kraft R, Huang X, Henklein P, Kapelari B, Pollmann C, et al. COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 2001;20:1630–9. doi: 10.1093/emboj/20.7.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou R, Hotta I, Denli AM, Hong P, Perrimon N, Hannon GJ. Comparative analysis of argonaute-dependent small RNA pathways in Drosophila. Mol Cell. 2008;32:592–9. doi: 10.1016/j.molcel.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang J, Liang Z, Yang B, Tian H, Ma J, Zhang H. Derepression of microRNA-mediated protein translation inhibition by apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G (APOBEC3G) and its family members. J Biol Chem. 2007;282:33632–40. doi: 10.1074/jbc.M705116200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.