Abstract
Burning mouth syndrome (BMS) is a chronic pain disorder characterized by severe burning sensation in normal looking oral mucosa. Diagnosis of BMS remains to be a challenge to oral healthcare professionals because the method for definite diagnosis is still uncertain. In this study, a quantitative saliva proteomic analysis was performed in order to identify target proteins in BMS patients’ saliva that may be used as biomarkers for simple, non-invasive detection of the disease. By using isobaric tags for relative and absolute quantitation labeling and liquid chromatography-tandem mass spectrometry to quantify 1130 saliva proteins between BMS patients and healthy control subjects, we found that 50 proteins were significantly changed in the BMS patients when compared to the healthy control subjects (p ≤ 0.05, 39 up-regulated and 11 down-regulated). Four candidates, alpha-enolase, interleukin-18 (IL-18), kallikrein-13 (KLK13), and cathepsin G, were selected for further validation. Based on enzyme-linked immunosorbent assay measurements, three potential biomarkers, alpha-enolase, IL-18, and KLK13, were successfully validated. The fold changes for alpha-enolase, IL-18, and KLK13 were determined as 3.6, 2.9, and 2.2 (burning mouth syndrome vs. control), and corresponding receiver operating characteristic values were determined as 0.78, 0.83, and 0.68, respectively. Our findings indicate that testing of the identified protein biomarkers in saliva might be a valuable clinical tool for BMS detection. Further validation studies of the identified biomarkers or additional candidate biomarkers are needed to achieve a multi-marker prediction model for improved detection of BMS with high sensitivity and specificity.
Keywords: Burning mouth syndrome, orofacial pain, quantitative proteomics, biomarkers, isobaric tags for relative and absolute quantitation, liquid chromatography-tandem mass spectrometry
Introduction
Burning mouth syndrome (BMS) is a chronic pain disorder characterized by severe burning sensation in normal looking oral mucosa. The International Association for the Study of Pain defines BMS as “chronic oral mucosal pain or discomfort that has no identifiable causative lesions and is not caused by any other condition or disease.” The overall prevalence of BMS in the general population is roughly 4%.1 However, women are three to seven times more likely than men of a similar age to experience BMS symptoms,1,2 especially after the menopause, when its prevalence may be 18% to 33%.3 BMS is rarely observed in patients younger than 30 years of age and prevalence may increase from 3- to 12-fold with increasing age.1 The disease often affects the tongue (particularly the tip and lateral borders), lips, and palate. In addition to a burning sensation, the patients may present unremitting oral mucosal pain, dysgeusia, and xerostomia.2
BMS was initially considered as a psychogenic illness. However, over the years, researchers have discovered objectively measured abnormalities in physiologic responses of the trigeminal nerves and histopathologic changes in nociceptive fibers of BMS patients,4–6 suggesting that BMS might be a neuropathic pain disorder. In fact, studies focusing on trigeminal nerve alterations have shown both hypersensitivity and hyposensitivity as well as large- and small-fiber neuropathy, which indicate that BMS may have multiple etiologies producing similar symptoms.5–8 Recently, investigators have examined BMS patients for central neural changes, specifically on dopamine receptors in the basal ganglia, and found that BMS is associated with a decline in endogenous dopamine levels in the putamen, which results in altered central nociceptive signal processing.9,10 These findings suggest that brain function alters along with peripheral nerve changes and support the notion that central modulation of sensory signal occurs in BMS patients. Nevertheless, the etiology and pathophysiology of BMS are not well understood, and the exact cause of neuropathy in BMS remains unknown. Consequently, there has been no effective treatment for the patients with BMS. A thorough understanding of the pathogenesis and etiology of this disease, combined with novel diagnostics and therapeutic interventions, is required for an improved management of BMS patients.
Quantitative proteomics is a powerful methodology for biomedical applications because it allows global profiling of proteomic samples from disease patients to reveal significantly altered proteins that may serve as targets for disease diagnosis, prognosis, and/or therapeutics.11–13 In order to profile proteins present in the saliva samples from BMS patients, we used a quantitative proteomics approach based on isobaric tags for relative and absolute quantitation (iTRAQ) and liquid chromatography-tandem mass spectrometry (LC-MS/MS). iTRAQ is a stable isotope labeling method which allows for concurrent identification and quantification of proteins from up to eight samples within a same experiment.14 Upon labeling by iTRAQ 8-plex isobaric tags (reporter ions, m/z 113, 114, 115, 116, 117, 118, 119, and 121), peptide samples derived from eight protein samples can be combined and analyzed with a single LC-MS/MS run. The isobaric tags are cleaved during higher energy collision dissociation (HCD) to yield an isotope series (reporter ions) representing the quantity of a single peptide of known mass from eight different samples. Since the peptide remains attached to the isobaric tags until HCD is conducted, the peptide is simultaneously fragmented for sequence identification. Therefore, the relative intensities of the reporter ions (ratios) represent the relative abundance of each peptide among the eight samples, and the relative abundance of each protein among the eight samples is computed from the ratios of each unique peptides derived from the protein.
The purpose of the present study is to identify target proteins present in BMS patients’ saliva that may be used as biomarkers for simple, non-invasive detection of BMS. First, a quantitative proteomics approach based on LC-MS/MS and iTRAQ was used to relatively quantify all saliva proteins between BMS and healthy control groups. Second, statistical analysis was used to identify proteins at significantly altered levels between BMS and healthy control groups, and a panel of candidate biomarkers were further validated with enzyme-linked immunosorbent assay (ELISA). Lastly, statistical analysis was performed to evaluate the performance of the validated potential biomarkers for BMS detection.
Materials and methods
Sample collection
This study was approved by the institutional review board committee at the University of California Los Angeles, and all BMS and healthy control subjects signed the consent form prior to sample collection. In total, 38 subjects were recruited for this study, including 19 BMS patients and 19 healthy control subjects. Both BMS patients and healthy control subjects do not have intraoral inflammation such as gingivitis and periodontal disease to avoid other putative biomarkers, pro-inflammatory mediators, being detected in proteomic profile data. Originally, there were more than 19 BMS patients; however, since they were either taking other medication for other disorders/diseases or had visual analog scale (VAS) scale less than 3, they were excluded for this project to reduce accounting various variables. All BMS patients were recently diagnosed and had not received any prior treatment before sample collection, and all control subjects had never experienced any oral burning. These all BMS patients claimed having burning sensation inside of their mouth, dried mouth, and idiopathic pain inside their mouth. Whole saliva samples were collected using a well-defined and standardized protocol.15–18 The whole saliva samples were unstimulated, and each individual does not have any other inflammation in his or her mouth. In order to prevent any contamination from other sources, we asked patients to rinse their mouth thoroughly before collecting saliva. Once collected, the samples were centrifuged at 10,000 × g for 10 min at 4℃ to separate supernatant from cell pellets and debris. The supernatant was then aliquoted and stored at −80℃. The samples were thawed out only once for measurement in order to prevent protein degradation. Total protein concentration of saliva samples was measured with the 2-D Quant Kit (GE Healthcare, Pittsburgh, PA, USA).
Gel electrophoresis
Saliva samples with a total protein amount of 60 µg were separated with the NUPAGE Novex 4%–12% Bis-Tris gels (Invitrogen, Carlsbad, CA, USA), and the gels were fixed for 30 min with 50% methanol and 10% acetic acid. (Note: In order to normalize the total protein amount for all samples and visualize fine protein bands, we selected the 60 µg as minimum quantity for each sample.) After fixation, the gels were stained with the SimplyBlue SafeStain (Invitrogen) for 40 min at room temperature to visualize the protein bands. Gel lanes were excised for subsequent in-gel tryptic digestion and iTRAQ labeling of the resulting peptides.
Quantitative proteomic analysis
Each lane of protein gel pieces were reduced using tris (2-carboxyethyl) phosphine (TCEP) and alkylated with methyl methanethiosulfonate (MMTS) followed by trypsin digestion in 50 mM TEAB buffer at 37℃ for 16 h. After proteolytic cleavage, the resulting peptides from each sample were labeled with iTRAQ labeling reagents (8-plex kit; AB SCIEX, Framingham, MA, USA). The BMS samples were labeled with iTRAQ reagents 113 to 116 and the control subjects were labeled with iTRAQ 117 to 121. The resulting peptides were combined and separated by SCX using a polySULFOETHYL A column (4.6 mm × 200 mm, 5 µm, 300 Å; Poly LC Inc., Columbia, MD, USA). Total 20 fractions were collected. Each fraction was desalted using the PepClean C18 spin columns (Pierce, Rockford, IL, USA) and further analyzed by RPLC-MS/MS on the Orbitrap Velos instrument. The peptide samples were loaded onto a C18 trap column at a flow rate of 30 µl/min. Then, the peptides were separated on a C18 nano column (75 µm × 150 mm, 3 µm, 100 Å, C18; Dionex, Sunnyvale, CA, USA) at a flow rate of 250 nl/min on an UltiMate 3000 LC system (Dionex) using 3 h gradient. The MS/MS spectra were acquired on the Orbitrap Velos mass spectrometer using a data-dependent analysis mode, which the top 10 most abundant ions in each MS scan (m/z 350–2000) were selected for MS/MS analysis. The capillary temperature was set to 275℃ and the spray voltage set to 2 kV. The resolution for MS scan is 60,000 full width at half maximum (FWHM) and 7500 FWHM for MS/MS scan. The lock mass was used for accurate mass measurement. The MS/MS spectra were searched against the UniRef100 human database (120,982 entries) using the Mascot Daemon (Version 2.3). The mass tolerance was 10 ppm for MS and 0.1 Da for MS/MS. The variable modifications include methionine oxidation and tyrosine iTRAQ labeling. The fixed modifications include N-terminus and Lysine side chain 8-plex iTRAQ labeling and cysteine MMTS alkylation. All the protein and peptides were identified with a false discovery rate less than 1%.
The relative quantitation was calculated as the average fold changes of identified proteins in BMS patients as compared to the average fold changes of the proteins in control subjects. The median ratio was used for normalization.
Enzyme-linked immunosorbent assay
ELISA was used to quantify the levels of four candidate biomarkers in BMS patients (n = 15) and healthy control subjects (n = 15). The following commercially available ELISA kits were used: alpha-enolase (Abcam Inc., USA, #ab181417); interleukin-18 ((IL-18)) (R&D Systems Inc., USA, #7620); kallikrein-13 (KLK13) (Sino Biological Inc., China, #SEK10199-5); and cathepsin G (Abnova Corporation., USA, #KA1265).
Statistical analysis
Statistical analysis of the proteomics data was performed with simple Student t-test. Statistical analysis of the validation data was performed with the MedCalc statistical software (MedCalc Software Co., Ostend, Belgium), and the receiver operating characteristic (ROC) analysis was used to estimate the performance of each potential biomarkers.
Results
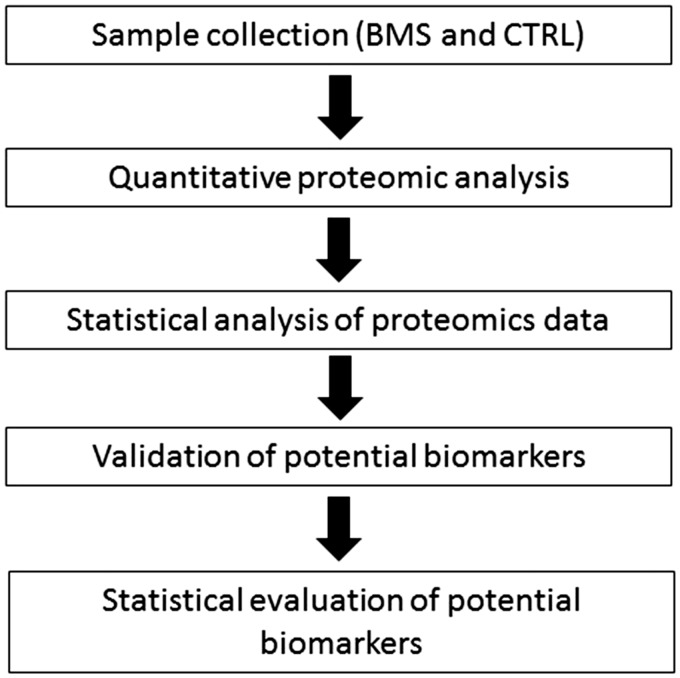
The purpose of this study is to identify target proteins present in BMS patients’ saliva that may be used for non-invasive detection of the disease. The study design is very straightforward, as shown in Figure 1. First, a quantitative proteomics approach was used to relatively quantify global saliva protein levels between BMS patients and healthy control subjects. Second, statistical analysis was used to identify differentially expressed proteins between BMS and healthy control subjects. Next, a panel of candidate biomarkers were chosen for ELISA validation, and statistical analysis was performed to evaluate the performance of the potential biomarkers for BMS detection.
Figure 1.
The workflow for discovery of potential saliva protein biomarkers of BMS.
Quantitative proteomic analysis
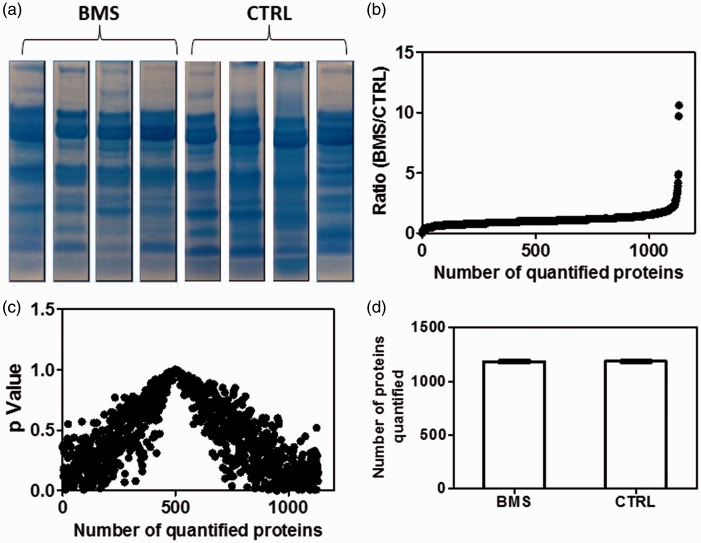
Since the 8-plex iTRAQ kit allows to label eight protein samples, we chose four BMS and four control samples for a two-group comparative proteomic analysis. These BMS patients and control subjects were well matched in terms of gender, age, and ethnicity to minimize potential bias from these factors during the discovery phase. Prior to iTRAQ labeling, total protein concentrations of the eight samples were measured so that equal amount of proteins (60 µg in total) from each samples was used for iTRAQ labeling. Afterwards, SDS-PAGE was used to separate the proteins from the eight samples (Figure 2(a)), and individual gel lanes were excised for in-gel tryptic digestion and iTRAQ labeling of the resulting peptides.
Figure 2.
Relative quantitation of salivary proteins between BMS patents (n = 4) and healthy control subjects (n = 4). Quantitative proteomic analysis was based on SDS-PAGE separation of saliva proteins, tryptic digestion of gel-separation proteins, iTRAQ labeling of extracted peptides, and LC-MS/MS analysis of iTRAQ-labeled peptides. (a) SDS-PAGE separation of saliva proteins of BMS and control subjects. (b) Relative ratios of iTRAQ reporter ions which represent the relative levels of 1130 saliva proteins between BMS and healthy control subjects. (c) p values for 1130 quantified proteins between BMS and healthy control groups. (d) Average number of proteins quantified from the BMS or control groups.
After 8-plex iTRAQ labeling, the eight labeled peptide samples (four BMS and four controls) were combined and analyzed with LC-MS/MS. In total, we quantified more than 1100 proteins with more than 7500 identified peptides from each BMS or control samples (Figure 2(c)). The relative quantitation was computed as the average fold change of the identified protein in four BMS samples as compared to the average fold change of the protein from four control subjects, and the median ratio was used for normalization. Figure 2(b) illustrates the iTRAQ reporter ion ratios for 1130 proteins, which show the relative levels of these proteins between the BMS and healthy control groups. The complete list of quantified proteins from all the eight samples (four BMS and four healthy controls) is shown in Supplemental Table 1.
Statistical analysis of the quantitative proteomics data revealed that 50 proteins were significantly (p ≤ 0.05) changed in the BMS patients when compared to the healthy control subjects. Among this group of proteins, 39 ones showed significantly increased levels whereas the remaining 11 ones showed significantly decreased levels in BMS patients versus healthy control subjects (Supplemental Tables 2 and 3).
ELISA for validation
Among the 39 proteins that showed significant increased levels in BMS patients, we ranked them by fold changes. Based on the fold change as well as previous studies in the literature about their function, we selected four candidates, KLK13, cathepsin G, alpha-enolase, and IL-18 for further validation.
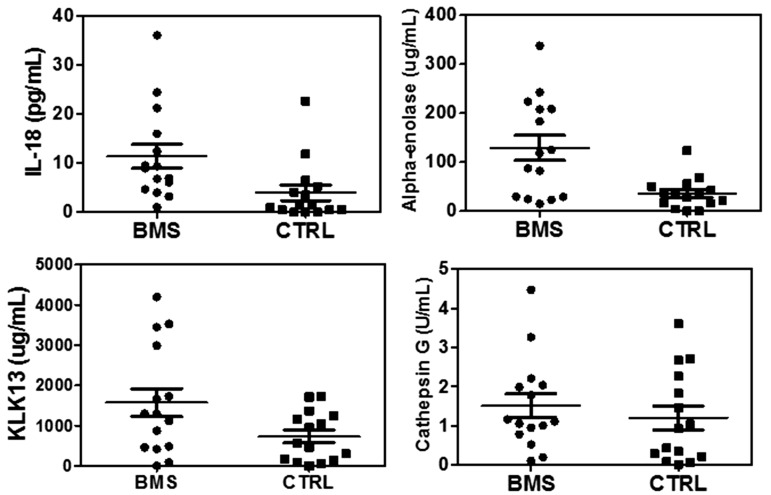
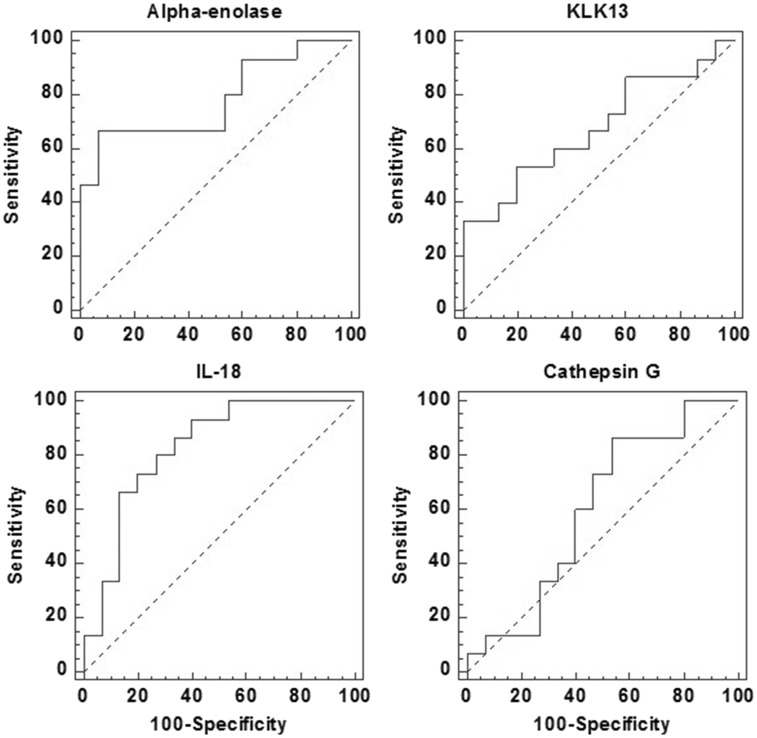
The validation results for KLK13, cathepsin G, alpha-enolase, and IL-18 are shown in Figure 3. Based on ELISA measurement, the levels of alpha-enolase, IL-18, and KLK13 were found to be significantly elevated in BMS versus control group (n = 15 per group). The fold changes (BMS vs. control) for alpha-enolase, KLK13, and IL-18 were determined as 3.6, 2.2, and 2.9, with p values of 0.002, 0.04, and 0.02, respectively (Table 1). However, ELISA measurement suggests that cathepsin G levels were not significantly different between BMS patients and control subjects. ROC analysis was performed to estimate the performance of these candidate biomarkers (Figure 4) and the results are shown in Table 1.
Figure 3.
ELISA validation of four candidates, IL-18, alpha-enolase, KLK13, and cathepsin G, between BMS and healthy control subjects (n = 15 per group).
Table 1.
Performance of the potential biomarkers based on ELISA validation.
| Protein biomarker | Fold change | p | Sensitivity (%) | Specificity (%) | ROC value |
|---|---|---|---|---|---|
| Alpha-enolase | 3.6 | 0.002 | 66.7 | 93.3 | 0.78 |
| Kallikrein-13 | 2.2 | 0.04 | 53.3 | 80.0 | 0.68 |
| Interleukin 18 | 2.9 | 0.02 | 93.3 | 60.0 | 0.83 |
| Cathepsin G | 1.3 | 0.47 | 86.7 | 46.7 | 0.60 |
ROC: receiver operating characteristic.
Figure 4.
ROC analysis of the four potential biomarkers based on ELSIA validation data. The performance of these candidate biomarkers is shown in Table 1.
Discussion
BMS is a chronic pain disorder characterized by severe burning and stinging of the oral cavity in the absence of any organic disease. It may last at least four months to a number of years and significantly deteriorates patients’ quality of life. Diagnosis of BMS remains to be a challenge to oral healthcare professionals due to the discrepancy between the severity, extensive objective pain felt by the patient, and the presence of limited clinical changes of the oral mucosa. Despite a number of clinical tests that have been proposed for BMS patients, the methods of definite diagnosis for BMS are still uncertain. In addition, the symptoms and clinical tests of BMS are not well-known to general population, which causes significant delay in diagnosis of the disease.
Currently, a number of tests/examinations have been recommended for helping diagnose BMS in oral medicine clinics, such as measuring salivary flow rate, testing hematology (complete blood count, glucose levels, and nutritional factors), examining oral cavities, testing pain intensity using a VAS, culturing fungus from oral mucosa, and testing allergy from medication or foods by eliminating one at a time.19–21 Due to various and unknown causes of BMS, clinicians even perform a number of different tests to exam BMS patients, so that they can propose suitable treatment for individual BMS patient in respect to their clinical histories. Although there are classification approaches for BMS patients based on their conditions, background, or symptoms, clinicians cannot explicitly tell the patients they have definite diagnosis of BMS and what causes them to have BMS during their first clinic visits because there is no standardized test available yet. BMS may share similar symptoms to Sjogren’s syndrome which is an autoimmune disease that causes patients to have clinically dry mouth and eyes symptoms.16,17,22 A common practice in oral medicine clinics is that, when BMS patients claim dry mouth symptoms, clinicians would refer the patients to test first for Sjögren’s syndrome. In this regard, a reliable clinical test for BMS would be very valuable to differentiate BMS from Sjögren’s syndrome patients.
One effective way to contribute to finding a clinical test for BMS is to discover multiple biomarkers that can be combined to achieve a sensitive and specific detection of the disease. Saliva is a body fluid that can be easily and repeatedly collected from patients to help diagnose and monitor diseases. Therefore, in this study, we aim to conduct a quantitative proteomic analysis of saliva samples from BMS patients and healthy control subjects in order to identify differentially expressed proteins that may serve as biomarkers for BMS detection.
Our quantitative proteomic analysis based on iTRAQ labeling and LC-MS/MS indicated that the saliva proteomes of BMS patients are distinct from those of healthy control subjects. The proteomic alterations may result from the peripheral nerve changes in oral cavity, particularly in the tongue and salivary gland tissues. Meanwhile, these proteomic changes, particularly the decreased expression of important functional saliva proteins, might contribute to the dysgeusia and xerostomia symptoms felted by certain BMS patients. Our studies suggested that there are similarities and dissimilarities between the saliva proteomes of BMS patients and those of Sjögren’s syndrome patients. For instance, carbonic anhydrase 6 and β2-microglobulin were significantly up-regulated in patients with Sjögren’s syndrome but barely changed in those with BMS. Prolactin-inducible protein was down-regulated in Sjögren’s syndrome patients but up-regulated in BMS patients. However, alpha-enolase and carbonic anhydrase 2 were up-regulated in both BMS and Sjögren’s syndrome whereas lysozyme C was down-regulated in both BMS and Sjögren’s syndrome.22
Among the 39 proteins that showed significantly increased levels in BMS patients, four candidates, IL-18, alpha-enolase, KLK13, and cathepsin G, were chosen for further validation. Three potential biomarkers, IL-18, alpha-enolase, and KLK13, were successfully validated with ELISA, which showed consistently elevated expression levels in BMS patients as revealed by the proteomic analysis. IL-18 is a pro-inflammatory cytokine which acts as an inflammatory mediator in body fluids.23 In fact, IL-18 is considered as a potential factor in immune response that induces chronic inflammation in different diseases such as rheumatoid arthritis, psoriatic arthritis, Crohn’s disease, and Sjogren’s syndrome.23,24 Human kallikreins are proteolytic enzymes known to be overexpressed in many types of carcinomas to enhance invasiveness and metastatic characteristics of tumors.25 They are also involved in inflammatory response by activating kinins (polypeptides formed by kallikrein) through cleaving the high-molecular-weight kininogen.26 These kinins play important roles in regulating inflammatory process and pain responses because activated kinins enhance the vascular permeability. A previous study actually predicted that kallikreins are elevated in BMS patients and increased activity of kallikrein promotes the inflammation process in oral mucosa of BMS patients.26 Alpha-enolase, a glycolytic enzyme, also plays a role in immune response as a cell surface receptor for plasminogen on pathogens, such as streptococci, and activated immune cells, leading to systemic infection or tissue invasion.27 Anti-alpha enolase is often found in different types of autoimmune and inflammatory diseases and cancers where alpha-enolase is upregulated compared to healthy controls.28 Our previous study also demonstrated that alpha-enolase was at elevated levels in saliva from Sjögren’s syndrome patients.22
As shown in Figure 3, elevated levels of these proteins were only observed in a partial group of BMS patients, and there were overlaps for their expression between BMS and control group. This might reduce the power of the analysis if these markers are used in clinic. Therefore, we need further estimate the performance of these potential biomarkers with ROC analysis. As shown in Figure 4, according to the ROC analysis, alpha-enolase and IL-18 appeared to be more valuable biomarkers than KLK13 and cathepsin G because they showed higher ROC values and their expression levels were significantly higher in BMS patients than the control subjects. In addition, further studies are warranted to confirm the clinical utility of these potential biomarkers. IL-18 and KLK13 seem to be unique biomarkers to BMS as they were not significantly changed in Sjögren’s syndrome patients.22 However, saliva alpha-enolase levels were significantly elevated in both BMS and Sjögren’s syndrome patients. Nevertheless, we need further test these potential biomarkers in new BMS patient population and also Sjögren’s syndrome patients. With additional studies to further validate their clinical value among larger patient cohorts, we may be able combine these potential biomarkers or with other clinical indicators to accomplish a significantly improved detection of BMS.
Conclusion
In summary, our quantitative proteomic analysis indicated that there are significant changes to the saliva proteomes of BMS patients compared to healthy control subjects. By using ELISA for validation, we found that alpha-enolase, IL-18, and KLK13 are potentially valuable biomarkers for BMS. Since these proteins are over-expressed in saliva from BMS patients, they are possibly associated with peripheral nerve damage or increased inflammatory response in the oral cavity of the patients. To the best of our knowledge, this is the first quantitative proteomics study of BMS. Our findings have shown that testing of the identified protein biomarkers in saliva might be a valuable tool for BMS detection. In the future, a multi-center clinical trial is needed to further validate the identified biomarkers or additional candidate biomarkers so that a multi-marker prediction model can be achieved for an improved detection of BMS with high sensitivity and specificity.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was partially supported by the faculty research grant from the UCLA School of Dentistry (DM, SH) and the American Academy of Orofacial Pain (RM).
Supplemental Material
Supplementary material for this paper can be found at http://journals.sagepub.com/doi/suppl/10.1177/1744806916686796.
References
- 1.Bergdahl M, Bergdahl J. Burning mouth syndrome: prevalence and associated factors. J Oral Pathol Med 1999; 28: 350–354. [DOI] [PubMed] [Google Scholar]
- 2.Scala A, Checchi L, Montevecchi M, et al. Update on burning mouth syndrome: overview and patient management. Crit Rev Oral Biol Med 2003; 14: 275–291. [DOI] [PubMed] [Google Scholar]
- 3.Wardrop R, Hailes J, Burger H, et al. Oral discomfort at menopause. Oral Surg Oral Med Oral Pathol 1989; 67: 535–540. [DOI] [PubMed] [Google Scholar]
- 4.Beneng K, Yilmaz Z, Yiangou Y, et al. Sensory purinergic receptor p2x3 is elevated in burning mouth syndrome. Int J Oral Maxillofac Surg 2010; 39: 815–819. [DOI] [PubMed] [Google Scholar]
- 5.Forssell H, Jääskeläinen S, Tenovuo O, et al. Sensory dysfunction in burning mouth syndrome. Pain 2002; 99: 41–47. [DOI] [PubMed] [Google Scholar]
- 6.Grushka M, Sessle B, Howley T. Psychophysical assessment of tactile, pain and thermal sensory functions in burning mouth syndrome. Pain 1987; 28: 169–184. [DOI] [PubMed] [Google Scholar]
- 7.Lauria G, Majorana A, Borgna M, et al. Trigeminal small-fiber sensory neuropathy causes burning mouth syndrome. Pain 2005; 115: 332–337. [DOI] [PubMed] [Google Scholar]
- 8.Yilmaz Z, Renton T, Yiangou Y, et al. Burning mouth syndrome as a trigeminal small fibre neuropathy: increased heat and capsaicin receptor trpv1 in nerve fibres correlates with pain score. J Clin Neurosci 2007; 14: 864–871. [DOI] [PubMed] [Google Scholar]
- 9.Jääskeläinen SK. Pathophysiology of primary burning mouth syndrome. Clin Neurophysiol 2012; 123: 71–77. [DOI] [PubMed] [Google Scholar]
- 10.Maltsman-Tseikhin A, Moricca P, Niv D. Burning mouth syndrome: will better understanding yield better management? Pain Practice 2007; 7: 151–162. [DOI] [PubMed] [Google Scholar]
- 11.Hu S, Loo JA, Wong DT. Human body fluid proteome analysis. Proteomics 2006; 6: 6326–6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Misuno K, Liu X, Feng S, et al. Quantitative proteomic analysis of sphere-forming stem-like oral cancer cells. Stem Cell Res Ther 2013; 4: 156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Misuno K, Tran SD, Khalili S, et al. Quantitative analysis of protein and gene expression in salivary glands of Sjogren’s-like disease nod mice treated by bone marrow soup. PLoS One 2014; 9: e87158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chahrour O, Cobice D, Malone J. Stable isotope labelling methods in mass spectrometry-based quantitative proteomics. J Pharm Biomed Anal 2015; 113: 2–20. [DOI] [PubMed] [Google Scholar]
- 15.Hu S, Arellano M, Boontheung P, et al. Salivary proteomics for oral cancer biomarker discovery. Clin Cancer Res 2008; 14: 6246–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu S, Gao K, Pollard R, et al. Preclinical validation of salivary biomarkers for primary sjogren’s syndrome. Arthritis Care Res (Hoboken) 2010; 62: 1633–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu S, Vissink A, Arellano M, et al. Identification of autoantibody biomarkers for primary sjogren’s syndrome using protein microarrays. Proteomics 2011; 11: 1499–1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu S, Xie Y, Ramachandran P, et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics 2005; 5: 1714–1728. [DOI] [PubMed] [Google Scholar]
- 19.Aggarwal A, Panat SR. Burning mouth syndrome: a diagnostic and therapeutic dilemma. J Clin Exp Dent 2012; 4: e180–e185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Coculescu EC, Radu A, Coculescu BI. Burning mouth syndrome: a review on diagnosis and treatment. J Med Life 2014; 7: 512–515. [PMC free article] [PubMed] [Google Scholar]
- 21.Dahiya P, Kamal R, Kumar M, et al. Burning mouth syndrome and menopause. Int J Prev Med 2013; 4: 15–20. [PMC free article] [PubMed] [Google Scholar]
- 22.Hu S, Wang J, Meijer J, et al. Salivary proteomic and genomic biomarkers for primary sjogren’s syndrome. Arthritis Rheum 2007; 56: 3588–3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banu S, Jabir NR, Mohan R, et al. Correlation of toll-like receptor 4, interleukin-18, transaminases, and uric acid in patients with chronic periodontitis and healthy adults. J Periodontol 2015; 86: 431–439. [DOI] [PubMed] [Google Scholar]
- 24.Colafrancesco S, Priori R, Alessandri C, et al. Il-18 serum level in adult onset still’s disease: a marker of disease activity. Int J Inflam 2012; 2012: 156890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hashem N, Mara T, Mohamed M, et al. Human kallikrein 14 (klk14) expression in salivary gland tumors. Int J Biol Markers 2010; 25: 32–37. [DOI] [PubMed] [Google Scholar]
- 26.Loeb LM, Naffah-Mazzacoratti MG, Porcionatto MA, et al. Chondroitin sulfate and kallikrein in saliva: markers for glossodynia. Int Immunopharmacol 2008; 8: 1056–1058. [DOI] [PubMed] [Google Scholar]
- 27.Pancholi V. Multifunctional alpha-enolase: its role in diseases. Cell Mol Life Sci 2001; 58: 902–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Terrier B, Degand N, Guilpain P, et al. Alpha-enolase: a target of antibodies in infectious and autoimmune diseases. Autoimmun Rev 2007; 6: 176–182. [DOI] [PubMed] [Google Scholar]