Abstract
Helix–junction–helix (HJH) motifs are flexible building blocks of RNA architecture that help define the orientation and dynamics of helical domains. They are also frequently involved in adaptive recognition of proteins and small molecules and in the formation of tertiary contacts. Here, we use a battery of nuclear magnetic resonance techniques to examine how deleting a single bulge residue (C24) from the human immunodeficiency virus type 1 (HIV-1) transactivation response element (TAR) trinucleotide bulge (U23-C24-U25) affects dynamics over a broad range of time scales. Shortening the bulge has an effect on picosecond-to-nanosecond interhelical and local bulge dynamics similar to that casued by increasing the Mg2+ and Na+ concentration, whereby a preexisting two-state equilibrium in TAR is shifted away from a bent flexible conformation toward a coaxial conformation, in which all three bulge residues are flipped out and flexible. Surprisingly, the point deletion minimally affects microsecond-to-millisecond conformational exchange directed toward two low-populated and short-lived excited conformational states that form through reshuffling of bases pairs throughout TAR. The mutant does however, adopt a slightly different excited conformational state on the millisecond time scale, in which U23 is intrahelical, mimicking the expected conformation of residue C24 in the excited conformational state of wild-type TAR. Thus, minor changes in HJH topology preserve motional modes in RNA occurring over the picosecond-to-millisecond time scales but alter the relative populations of the sampled states or cause subtle changes in their conformational features.
Graphical abstract
Many regulatory RNAs undergo large changes in conformation when performing biological functions. Such conformational changes can allow adaptive interactions with protein and ligand binding partners,1–3 ensure that ribonucleoprotein complexes assemble in a directional and hierarchical manner,4,5 allow riboswitches to regulate gene expression in response to an array of cellular signals,6–8 and allow catalytic ribozymes to adopt the multitude of conformations required to complete multistep catalytic cycles.9–11 The importance of dynamics to RNA folding and function has motivated studies that seek a detailed quantitative description of RNA flexibility with the goal of elucidating dynamic properties important for folding and function.12–15 Studying a broader RNA dynamic landscape is also important for rational structure-based design of RNA-targeting therapeutics.16–18
Helix–junction–helix (HJH) motifs, such as bulges and internal loops, are flexible building blocks of RNA architecture that adjoin helical domains.19–21 Because local flexibility within HJH motifs can allow helical domains to adopt different orientations, HJH motifs play an essential role in defining the global structure and dynamics of RNA. Additionally, HJH motifs frequently undergo conformational adaptation upon binding to proteins,1,22 ligands,23 small molecule therapeutics,24,25 and catalytically essential metals26,27 and upon formation of tertiary contacts.19–21 Dynamic studies indicate that HJH motifs undergo motions over a broad range of time scales. These include rigid body interhelical motions and local fluctuations of junction residues on picosecond-to-microsecond time scales28–34 and conformational exchange directed toward low-populated (typically <5%) and short-lived (lifetime typically <2 ms) “excited conformational states” (ESs) that feature reshuffling of base pairs in and around the HJH motif on slower microsecond-to-millisecond time scales.35–41
Here, we use a battery of NMR techniques to examine how shortening a bulge HJH motif from three to two nucleotides affects motional modes occurring over picosecond-to-millisecond time scales. We focus on the transactivation response element (TAR) from human immunodeficiency virus type 1 (HIV-1) (Figure 1A), which has served as a model system for studying HJH dynamics.28,42 Prior studies employing NMR,28,35,37,42–51 gel mobility,52 transient electric birefringence,53 fluorescence,30,54 X-ray crystallography,55 electron paramagnetic resonance (EPR),32–34 molecular dynamics (MD) simulations,56 and combinations of NMR and molecular dynamics29 have shown that TAR undergoes complex dynamics over time scales spanning 12 orders of magnitude. These dynamics are proposed to play important roles in the adaptive recognition of TAR by cognate proteins and small molecule ligands designed to inhibit TAR–protein interactions in the development of anti-HIV therapeutics54,55,57–59 and may additionally play a crucial role in the dimerization of the HIV genome.60
Figure 1.
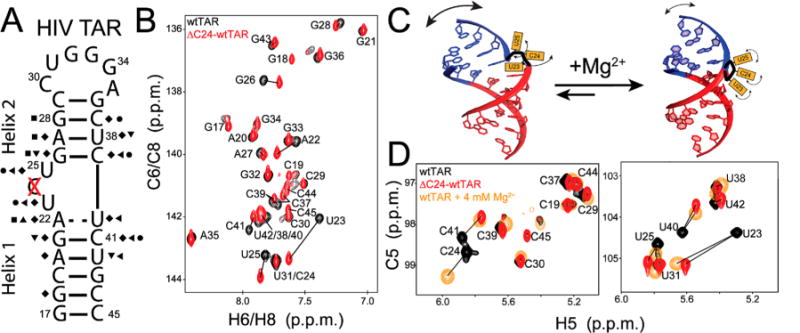
Chemical shift comparison of wtTAR and ΔC24-wtTAR. (A) Secondary structure of HIV-1 TAR and ΔC24-wtTAR where the deleted bulge residue is highlighted with a red X. Symbols on the secondary structure indicate sites with significant chemical shift perturbations [ > 0.25 ppm (see Methods)]: C6 H6 (circles), C8 H8 (squares), C2 H2 (triangle), C5 H5 (left triangles), C1′ H1′ (diamonds), and N1 H1/N3 H3 (inverse triangles). (B) Aromatic two-dimensional (2D) HSQC overlaid spectra for wtTAR (black) and ΔC24-wtTAR (red), with lines highlighting resonances showing significant chemical shift perturbations. (C) Two-state TAR equilibrium. (D) Overlay of 2D HSQC spectra of wtTAR (black), ΔC24-wtTAR (red), and wtTAR in the presence of 4 mM Mg2+ (orange),91 with lines highlighting resonances with significant chemical shift perturbations relative to those of wtTAR under low-salt conditions.
In this study, we examined the consequence of deleting bulge residue C24 on HIV-1 TAR dynamics (Figure 1A). This TAR variant (hereafter termed ΔC24-TAR) is also of biological interest because it is a rare but naturally occurring variant of TAR in HIV-1mal and HIV-1U455 isolates.61 We previously used NMR residual dipolar couplings (RDCs)62,63 to characterize interhelical dynamics in a ΔC24-TAR variant in which the wild-type apical loop, which is involved in microsecond-to-millisecond conformational exchange, was replaced with a UUCG tetraloop (ΔC24-TARuucg).42 These studies showed that shortening the bulge leads to a significant restriction of interhelical dynamics.42,64 In this study, we used a broader set of NMR techniques to more comprehensively characterize motions in ΔC24-TAR containing the wild-type apical loop (ΔC24-wtTAR) over a wide range of time scales. Our results show that ΔC24-wtTAR retains the basic motional modes observed in HIV-1 TAR and that shortening the bulge causes subtle changes in the relative populations and structure of the sampled conformational states.
METHODS
Sample Preparation
In Vitro Transcription
HIV-1 wild-type TAR (wtTAR) and ΔC24-wtTAR samples were synthesized by in vitro transcription using T7 RNA polymerase (Fischer Scientific), uniformly 13C- and 15N-labeled nucleotides (Cambridge Isotope Laboratories, Inc.), and synthetic DNA templates (Integrated DNA Technologies) containing the T7 promoter sequence and RNA sequence. The reaction mixture was filtered and concentrated to 1 mL using a centrifugal concentrator (3 kDa molecular weight cutoff, EMD Millipore), mixed with a formamide denaturing RNA loading dye, and fully denatured by being heated at 95 °C for 5 min. The mixture was loaded onto a 13 cm × 102 cm, 20% (w/v) polyacrylamide gel in 8 M urea and 1× Tris/borate/EDTA and run for 12 h. The target RNA was excised from the gel by briefly shadowing at 365 nm with a UV hand lamp, followed by electro-elution (Whatmann, GE Healthcare) in 1× Tris/acetic acid/EDTA, and ethanol precipitation. The precipitate was dissolved in water, annealed by being heated at 95 °C for 5 min followed by rapid cooling on ice, and then buffer exchanged using a centrifugal concentrator (EMD Milipore) into NMR buffer [15 mM sodium phosphate, 25 mM sodium chloride, and 0.1 mM EDTA (pH 6.4)]; 10% D2O was added to the sample before NMR data were collected. This buffer was used for all experiments in this study unless stated otherwise.
Solid-Phase Oligonucleotide Synthesis
ES2uucg-TAR and G28U-TAR were synthesized using the MerMade 6 DNA/RNA synthesizer (BioAutomation) with standard phosphoramidite RNA chemistry, and deprotection protocols for bases and the 2′-hydroxyl. Samples were purified using Glen-Pak RNA purification cartridges, following the product protocol, which can be found online (www.glenresearch.com). Once purified, the RNA was precipitated with ethanol, desalted using buffer exchange, and annealed as described earlier.
NMR Experiments
Resonance Assignments
Chemical shift assignment experiments were conducted on an 800 MHz Agilent DirectDrive2 NMR spectrometer equipped with a triple-resonance HCN cold probe. NMR assignments for wtTAR, TARuucg, and ΔC24-wtTAR were obtained from prior studies42,44,65 (see Figure 1 and Figure S1). Resonance assignments for exchangeable and nonexchangeable protons in G28U-TAR-ΔC24-wtTAR, and ΔC24-wtTAR-ES2uucg were obtained using two-dimensional (2D) 1H–1H nuclear Overhauser effect spectroscopy (NOESY) experiments with mixing times of 250 and 150 ms, respectively,66 along with conventional 2D HSQC of aliphatic [13C,1H] resonances and band-selective optimized flip angle short transient (SOFAST) 2D HMQC67 spectra of aromatic [13C,1H], and imino [15N,1H] resonances at 25 and 10 °C. wtTAR-ES2uucg assignment experiments were conducted on an 800 MHz Varian Inova spectrometer equipped with a HCN cyrogenic probe using 2D 1H–1H NOESY with a mixing time of 150 ms, with aliphatic HSQC and aromatic and imino SOFAST-HMQC spectra at 25 and 10 °C to obtain assignments. All G28U and ES2uucg resonances were assigned as described previously,68 and all data were processed and analyzed using NMRPipe69 and SPARKY.70
Chemical Shift Perturbations
Chemical shift differences were computed using the equation
where Δδ is the change in chemical shift, X is carbon or nitrogen, and α = γH/γX, where γ is the gyromagnetic ratio. A chemical shift change was considered significant when Δδ ≥ 0.25 ppm.
Estimating the Population of the Coaxial State Using Chemical Shifts
Populations of the bent (pbent) and coaxial (pcoaxial) states at various Mg2+ concentrations in TARuucg were reported previously.46 Briefly, the populations for ΔC24-wtTAR were estimated from the observed chemical shifts (δobs) using the equation δobs = pbent(δbent) + pcoaxial(δcoaxial), where pbent + pcoaxial = 1 and δbent and δcoaxial are the chemical shifts of the bent and coaxial states, respectively. Here, we assume that the bent and coaxial states are in fast exchange on the NMR time scale, such that the observed chemical shift is a population-weighted average and that the chemical shift of the bent state is the same for all TAR variants. The chemical shift of U23 H6 was used to report on the relative populations of the bent and coaxial states of various TAR constructs. This resonance experiences linear and significant perturbations with Mg2+ and Na+ consistent with two-state exchange providing a sensitive probe of the bent–coaxial transition. δbent was estimated to be the observed U23 H6 chemical shift of TARuucg in the absence of Mg2+ at 25 mM NaCl, and δcoaxial was estimated to be the observed U23 H6 chemical shift at 4 mM Mg2+ for ΔC24-wtTAR. Similar results were obtained when using the chemical shifts of U23 C6, A22 C8, and A22 H8 (data not shown).
Measurement and Analysis of 13C Spin Relaxation
13C longitudinal (R1) and rotating frame (R1ρ) spin relaxation rates for aromatic (C8, C6, and C2) and sugar (C1′) resonances were measured on a 600 MHz Bruker NMR spectrometer equipped with an HCN cryogenic probe as previously described.71 Spectra were recorded in a pseudo-three-dimensional manner using a transverse relaxation-optimized spectroscopy (TROSY)72 detected experiment and five unique delay times (Table S1). Peaks in each spectrum were fit to a Gaussian shape using a NMRPipe autofit script, and R1 and R1ρ rates were determined by fitting resonance intensities at each delay to a monoexponential function using in-house python scripts. Standard jackknife resampling was used to estimate error. Transverse relaxation rates (R2) were derived from longitudinal and rotating frame relaxation rates by eq 1
| (1) |
where θ = arctan(ωSL/ΔΩ), ΔΩ = Ω − ωrf, Ω is the offset value in hertz, ωrf is the reference frequency in hertz, and ωSL is the spin-lock power set to 3.5 kHz to suppress chemical exchange during the experiment. R1 and R2 values for wtTAR and ΔC24-wtTAR are provided in Tables S2 and S3 of the Supporting Information.
A relative order parameter (Srel2) for each residue was calculated as described previously.44 Briefly, 2R2 –R1 values were computed for each site, and the values were normalized relative to the largest value measured in the A-form helix for each given spin type (C8, C2, C6, and C1′). Because of incomplete suppression of chemical exchange previously characterized35 at G34 C8, C30 C1′, U31 C1′, G34 C1′, A27 C1′, and A35 C1′, these sites have Srel2 values of >1 and were not considered further.
Measurement of RDCs
One-bond C–H (1DCH) and N–H (1DNH) RDCs were measured at 800 MHz using 2D TROSY72 for C2 H2, C8 H8, C1′ H1′, and C6 H6, in which splittings are encoded along the 1H or 13C dimension and IPAP-SOFAST-HMQC73 for N1 H1 and N3 H3 in which splittings are encoded along the 1H dimension. RDCs were calculated as the difference in splittings measured in the absence (J) and presence (J+D) of an alignment medium. Errors in RDCs were estimated on the basis of the rmsd of values measured along the 1H and 13C dimensions as described previously62,63[RDC rmsd = 3.7 Hz (Figure S5A)]. The final RDCs reported (Table S4) are the average of the 1H and 13C RDCs obtained in the two dimensions. The aligned sample was prepared by adding 25 mg/mL Pf1 phage (Asla biotech, Ltd.) in NMR buffer to a 1 mM sample of uniformly 13C- and 15N-labeled ΔC24-wtTAR in the same NMR buffer, resulting in a D2O splitting of 28 Hz. Careful inspection of HSQC overlays shows little to no change in the chemical shifts of ΔC24-wtTAR in the absence and presence of Pf1 phage, consistent with previous studies that show that the addition of phage does not affect TAR RNA structure and/or dynamics42,44 (Figure S5B).
Order Tensor Analysis of RDC
RDCs measured in Watson–Crick base pairs in helices 1 and 2 of ΔC24-wtTAR were subjected to an order tensor analysis using idealized A-form helices generated by 3DNA,74 as described previously.75–77 RDCs measured in flexible residues in helix 1 (G17, C45, A22, and U40) and helix 2 (C29 and G36) were omitted from the analysis.78 The best-fit order tensor for each helix was determined by singular-value decomposition (SVD)75 as implemented in RAMAH.79 The uncertainty in order tensor elements due to RDC uncertainty and A-form structural noise was estimated using AFORM-RDC.78 The average interhelical orientation was determined by rotating each input idealized A-form helix into the principal axis system (PAS) of the corresponding best-fit order tensor frame (Sxx Syy Szz) using an in-house C program based on Euler-RNA.77 All structures were visualized using PyMol.80 Degeneracies in possible helical orientations due to 180° rotation around the Sxx, Syy, and Szz axes of the PAS75 resulted in four degenerate solutions, three of which could be excluded as described previously.77 Specifically, 180° rotations about Sxx and Syy resulted in significant interhelical steric clashes, while 180° rotation about Szz resulted in a A22 (O3′)–G26 (P) distance that cannot be satisfactorily linked by two bulge residues (4.9 Å per nucleotide). RDCs measured in wtTAR and TARuucg were normalized to that of ΔC24-wtTAR to account for differences in the degree of alignment by multiplication of a scaling factor given by the ratio in the generalized degree of order (ϑ)76 measured in helix 2, which dominates overall molecular alignment.43 Scaling factors of 1.6 and 2.0 were applied to wtTAR and TARuucg RDCs, respectively.
13C and 15N R1ρ Relaxation Dispersion
15N and 13C R1ρ relaxation dispersion (RD) experiments were conducted on a 700 MHz Bruker Avance III spectrometer equipped with an HCN resonance cryogenic probe and a 600 MHz Bruker Avance III spectrometer equipped with an HCN resonance cryogenic probe.81–83 All data were measured on a 1 mM uniformly 13C- and 15N-labeled sample at 25 °C in NMR buffer unless otherwise noted. On- and off-resonance profiles were measured using various spin lock offsets (Ω) and spin lock powers (ωSL) for each spin of interest and 2–12 unique delay times. Data were processed using NMRPipe69 to determine peak intensities at each delay time, which were then fit to a monoexponential function using in-house python scripts to determine the rotating frame relaxation rate (R1ρ). Standard Monte Carlo simulations (500 iterations) were used to estimate error. Data points that meet the 13C–13C Hartmann–Hahn matching conditions were omitted from the analysis, as previously described.81 On- and off-resonance data were individually and globally fit to the two-state Laguerre equation82 by eq 2 using Origin 2015 (OriginLab)
| (2) |
or by a three-state Laguerre equation35 with no minor exchange for a linear topology by eq 3 using an in-house python code
| (3) |
where R1 and R2 are the longitudinal and transverse relaxation rates (in inverse seconds), respectively, ω/2π is the strength of the spin lock in hertz, Ω/2π is an offset from a reference frequency (ωrf) in hertz, and kex is the exchange rate between the ground state (GS) and the excited state (ES) in inverse seconds. The tilt angle in the rotating frame θ = arctan(ωSL/ΔΩ), where ωSL is the strength of the spin lock power for the carrier in hertz and is the difference in the average spin lock offset and the reference frequency (ωrf) in hertz. In addition, , where pGS and pES are the populations of the ground state and excited state, respectively, and ΩGS and ΩES are the resonance offsets from the spin lock carrier for the respective states in hertz. Finally, the spin lock strengths at the GS (ωGS) and ES (ωES) are defined as ωGS2 =(ΩGS + ωrf)2 + ωSL2 and ωES2 = (ΩES + ωrf)2 + ωSL2, respectively; the effective spin lock field strength is ωeff2 = ΔΩ2 + ωSL2, and the difference in the offset between the ES and the GS is ΔωES = ΩES − ΩGS. To clearly show exchange parameters, relaxation dispersion profiles are plotted as R2 + Rex to remove the contributions of R1 to R1ρ, which do not provide any information about chemical exchange.84
MC-Fold Predictions of RNA Secondary Structure
RNA secondary structure prediction was preformed using MC-Fold using standard input options returning the 20 best, 15% suboptimal structures predicted.85
RESULTS AND DISCUSSION
Chemical Shift Mapping
To analyze the impact of deleting the C24 bulge residue, we initially compared NMR 2D HSQC spectra of uniformly 13C- and 15N-labeled ΔC24-wtTAR with those of wtTAR. The excellent overlay of 2D NH HSQC spectra indicates that ΔC24-wtTAR adopts the same secondary structure as wtTAR (Figure 1A). In both cases, we did not observe the imino resonance of U40, indicating that the A22-U40 base pair does not form a stable Watson–Crick (WC) base pair in either ΔC24-wtTAR or wtTAR. In contrast, comparison of 2D CH HSQC spectra of base and sugar moieties reveals large changes in chemical shifts, particularly for bulge residues (U23 and U25) and surrounding WC base pairs A22-U40 and G26-C39 (Figure 1B and Figure S1A), which indicates a conformational change at these sites. As expected, insignificant perturbations are observed for apical loop residues, which are separated from the mutation site by a 4 bp helix.
Interestingly, the changes in chemical shift arising from shortening the bulge are similar to those reported previously when adding Mg2+ or Na+ to HIV-1 TAR, using a slightly modified TAR construct that is capped by a more stable UUCG apical loop46 (TARuucg). These studies proposed that TAR exists in a rapid two-state equilibrium between a globally flexible, bent state and a globally rigid, coaxially stacked state (Figure 1C). In the bent state, bulge residues C24 and U25 are flipped out and highly flexible while U23 is intrahelically stacked on A22. In the linear state, all three bulge residues are flipped out and flexible, allowing the helices to coaxially stack46,48 (Figure 1C). The addition of Mg2+ or Na+ shifts this preexisting equilibrium toward the coaxial conformation.46
Therefore, chemical shift data indicate that shortening the TAR bulge shifts this equilibrium in favor of the coaxial state (Figure 1D and Figure S2). On the basis of the Mg2+−dependent chemical shift changes, a prior study46 estimated the population of the coaxial state of TAR in the absence of Mg2+ and 25 mM NaCl to be ≈10%. Using a similar approach (see Methods), we estimated the coaxial state of ΔC24-wtTAR in the absence of Mg2+ and 25 mM NaCl to be >60%.
To examine whether Mg2+ can also shift the equilibrium toward the coaxial state, we titrated Mg2+ into ΔC24-wtTAR. As expected, the addition of Mg2+ to ΔC24-wtTAR results in similar perturbations as observed for wtTAR but required a lower Mg2+ concentration to reach saturation (≈2 and 4 mM for wtTAR and ΔC24-wtTAR, respectively) (Figure S3A). Additionally, spectra began to converge upon comparison of ΔC24-wtTAR and wtTAR in the presence of 4 mM Mg2+, indicating more similar populations of the bent and coaxial states in the presence of magnesium (Figure S3B). On the basis of the chemical shifts, we estimated that in the presence of 4 mM Mg2+, the coaxial state is >99 and 83% populated in ΔC24-wtTAR and wtTAR, respectively46 (see Methods).
Comparison of Picosecond-to-Nanosecond Dynamics Using Spin Relaxation
To further characterize the differences in dynamics between wtTAR and ΔC24-wtTAR in the absence of Mg2+, we compared 13C longitudinal (R1) and transverse (R2) spin relaxation rate constants for base (C8, C6, and C2) and sugar (C1′) nuclei in wtTAR and ΔC24-wtTAR at 25 °C (Figure S4 and Tables S2 and S3). Spin relaxation data provide information about both overall and internal motions occurring on picosecond-to-nanosecond time scales. As expected, because of their similar molecular size, similar R2/R1 ratios are measured for wtTAR and ΔC24-wtTAR (Figure 2A). To gain insights into differences in internal motions, we calculated the relative order parameter, Srel2, for each site (see Methods), which ranges from 1 to 0 for minimal to maximal amplitude internal motions, respectively.44,86 Indeed, lower Srel2 values were observed for bulge residues U23 and U25 in ΔC24-wtTAR as compared to wtTAR, consistent with increased picosecond-to-nanosecond dynamics and a flipped out flexible conformation for bulge residues (Figure 2B). Therefore, these data also support the idea that shortening of the TAR bulge favors a coaxial conformation with flipped out bulge residues.
Figure 2.
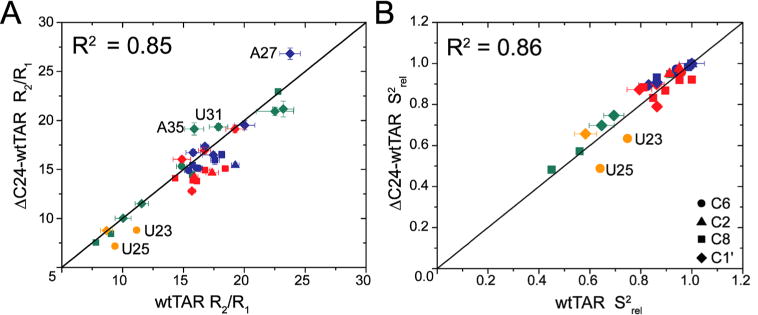
Characterization of picosecond-to-nanoseconds motions using spin relaxation. Shown are comparisons of (A) 13C R2/R1 ratios and (B) the relative order parameter, Srel2, measured in wtTAR and ΔC24-wtTAR. Data are colored according to helix 1 (red), helix 2 (blue), bulge (orange), and apical loop residues (green). Symbols represent spin type, C6 (circles), C2 (triangles), C8 (squares), and C1′ (diamonds).
Structural and Dynamic Comparison Utilizing Residual Dipolar Couplings
To further characterize differences in interhelical structure and dynamics between wtTAR and ΔC24-wtTAR, we measured NMR residual dipolar couplings (RDCs)62,63 in the absence of Mg2+. RDCs provide information about the orientation and dynamics of bond vectors relative to a common alignment frame over time scales ranging from picoseconds to milliseconds.63 We measured 45 one-bond CH and NH RDCs in uniformly 13C- and 15N-labeled ΔC24-wtTAR (1 mM) at 25 °C using the bacteriophage (Pf1) alignment medium75,76 (Table S4). A comparison of RDCs measured in wtTAR44 and ΔC24-wtTAR (Figure 3A) revealed significant differences even after accounting for differences in the degree of overall alignment (Figure 3B). Once again, the largest differences are observed for bulge residues U23 and U25 where RDCs are near zero in ΔC24-wtTAR (Figure 3A), consistent with motional averaging and a flexible, extrahelical conformation of bulge residues. Strikingly, RDCs measured in ΔC24-wtTAR show improved agreement with those previously measured in TARuucg in the presence of 4 mM Mg2+, consistent with a larger population of the coaxial conformation for ΔC24-wtTAR in the absence of Mg2+ (Figure 3C).46
Figure 3.
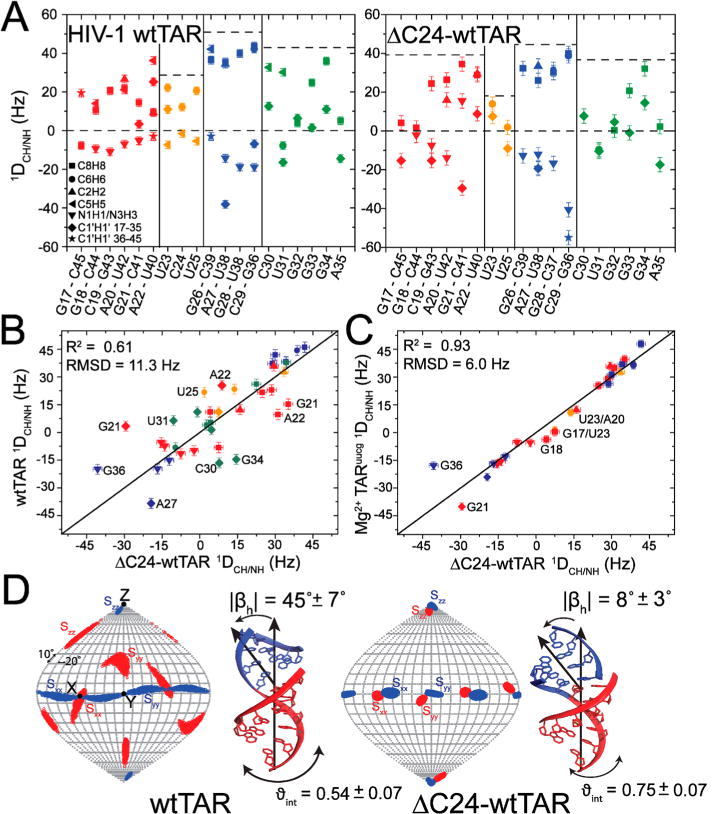
Characterizing interhelical dynamics using residual dipolar couplings. (A) RDCs measured in wtTAR44 and ΔC24-wtTAR, where wtTAR RDCs have been scaled by 1.5 to account for differences in the degree of alignment (see Methods). Dotted lines represent the largest positive RDC measured in each region of the structure (helix 1, helix 2, bulge, and apical loop) to highlight variation in the degree of alignment and dynamics. Comparison of RDCs measured in ΔC24-wtTAR with values measured in (B) wtTAR in the absence of Mg2+44 and (C) TARuucg in the presence of 4 mM Mg2+.46 RDCs are normalized to the degree of alignment (ϑ) measured in helix 2 of ΔC24-wtTAR. Error bars represent the experimental error (one standard deviation) in RDC measurements (see Methods). The notable outlier, G36, likely reflects differences in the apical loop. RDCs are colored according to helix 1 (red), helix 2 (blue), bulge (orange), and apical loop (green) residues. See the inset of panel A for the legend. (D) Order tensor frame (Sxx, Syy, Szz) determined for helix 1 (red) and helix 2 (blue) shown on a Sanson–Flamsteed map projection obtained from the order tensor analysis of RDCs using RAMAH.79 Additionally shown are the average interhelical orientations of wtTAR (left) and ΔC24-wtTAR [right (see Methods)] obtained by superimposing the order tensor frames for each helix. The errors in ϑint and |βh| were estimated using AFORM-RDC.78
To characterize the average interhelical orientation and amplitude of interhelical dynamics, RDCs measured in A-form helices were subjected to an order tensor analysis as described previously (see Methods and Table 1).76–78,87,88 Excellent agreement between the measured and back-calculated RDCs using the best-fit order tensor indicates that like those in wtTAR, the helices in ΔC24-wtTAR adopt an idealized A-form geometry (Figure S5C). The average interhelical structure obtained by superimposing order tensor frames [Sxx, Syy, Szz (see Methods)] shows a significant reduction in the average interhelical bend angle (βh) for ΔC24-wtTAR (|βh| = 8 ± 3°) as compared to that of wtTAR (|βh| = 45 ± 7°). This is accompanied by a reduction in the amplitude of interhelical dynamics as judged by an increase in the internal generalized degree of order (ϑint = ϑi/ϑii;ϑi < ϑiv, where ϑint varies from 0 to 1 for maximal to minimal amplitude interhelical motions, respectively) from wtTAR (ϑint = 0.54 ± 0.07) to ΔC24-wtTAR (ϑint = 0.75 ± 0.06) (Figure 3D). The ϑint value measured for ΔC24-wtTAR agrees well with the previously reported value for domain-elongated ΔC24-TARuucg (ϑint = 0.77 ± 0.04).42 These results provide further support that shortening the TAR bulge preserves interhelical motions but biases the ensemble of conformations toward the coaxially stacked state.
Table 1.
Order Tensor Analysis of wtTAR and ΔC24-wtTARa
| helix | N | rmsd (Hz) | CN | R | η | ϑ (×10−3) | ϑint | βh (deg) | ξ (deg) | |
|---|---|---|---|---|---|---|---|---|---|---|
| wtTAR | 1 | 14 | 1.2 | 5.7 | 0.99 | 0.36 ± 0.10 | 0.65 ± 0.08 | 0.54 ± 0.07 | 45 ± 7 | −41 ± 50 |
| 2 | 13 | 1.7 | 3.0 | 0.99 | 0.10 ± 0.05 | 1.19 ± 0.05 | ||||
| AC24-wtTAR | 1 | 11 | 2.5 | 4.4 | 0.99 | 0.59 ± 0.15 | 1.4 ± 0.1 | 0.75 ± 0.07 | −8 ± 3 | 23 ± 50 |
| 2 | 11 | 1.5 | 5.9 | 0.99 | 0.14 ± 0.09 | 1.9 ± 0.1 |
Statistics for the order tensor analysis of RDCs measured in wtTAR44 and ΔC24-wtTAR. Shown are the number of RDCs used (N), the condition number (CN),76 the root-mean-square deviation (rmsd) and Pearson’s correlation coefficient (R) between measured RDCs and back-calculated RDCs, order tensor asymmetry (η = |Syy −Sxx|/Szz),76 the generalized degree of order ,76 the internal generalized degree of order (ϑint = ϑi/ϑii;ϑi < ϑii),76 the interhelical bend angle (βh), and the interhelical twist (ξ). Angles were calculated using an in-house program based on Euler-RNA,77 and errors inr β, ξ, η, ϑ, and ϑint were estimated using AFORM-RDC.78
Impact of Mutation on Microsecond-to-Millisecond Conformational Exchange
Next, we used carbon and nitrogen spin relaxation dispersion (RD) in the rotating frame (R1ρ) to examine how deleting bulge residue C24 affects slower microsecond-to-millisecond conformational exchange directed toward two ESs35,37 [ES1 and ES2 (Figure 4A,B)]. To remain consistent with prior studies of wtTAR,35,37 RD data were measured for ΔC24-wtTAR in the absence of Mg2+. In the RD experiment, the contribution to the transverse relaxation rate (R2) due to chemical exchange (Rex) is measured as a function of the power and offset frequency of a radiofrequency spin lock, which is applied during the relaxation period. The resulting dependence of R1ρ on spin lock power and offset can be fitted using algebraic expressions82 to extract exchange parameters of interest, including population of the ES, forward and backward exchange rates, and the difference in chemical shift between ES and GS (Δω = ωES − ωGS). Note that the RD experiment is not sensitive to interhelical motions, which have been shown to occur on the nanosecond-to-microsecond time scales and fall outside the detection limits of this class of NMR experiments.42,71
Figure 4.
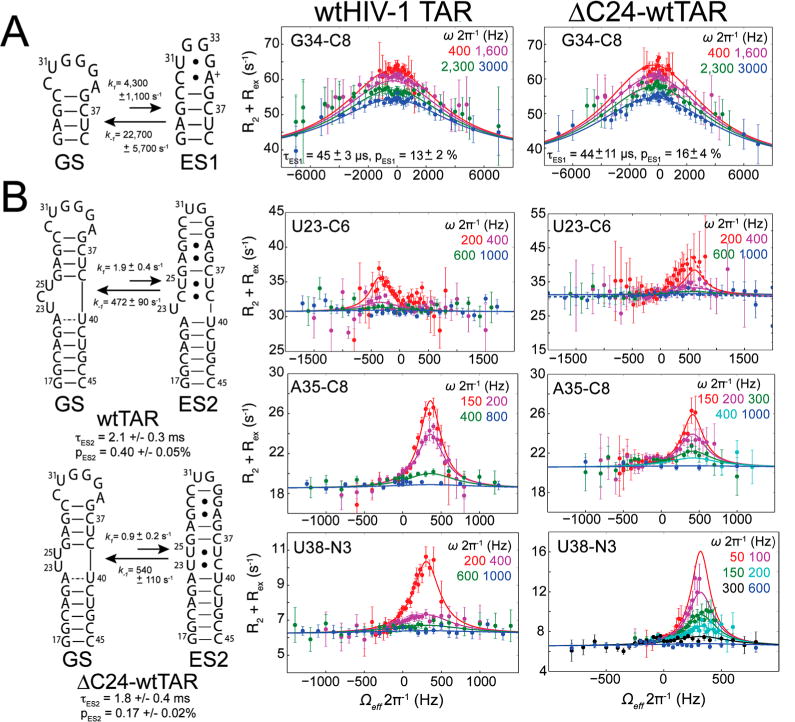
Measuring microsecond-to-millisecond motions using relaxation dispersion in the rotating frame. Secondary structure (left) and representative RD profiles for (A) ES1 and (B) ES2 in wtTAR35,37 (middle) and ΔC24-wtTAR (right). RD profiles show the dependence of R2 + Rex on spin lock power (ωeff/2π) and offset (Ω/2π) with global fits (—) and individual fits (–) to the two-state Laguerre equation (eq 2).82 Error bars represent experimental uncertainty [one standard deviation (see Methods)].
The deletion of bulge nucleotide C24 was not expected to affect ES1 conformational exchange, which involves local reshuffling of base pairs within the wild-type apical loop (Figure 4A). Indeed, all apical loop sites that showed ES1 RD in wtTAR (C30 C1′, U31 C1′, U31 C6, G34 C8, G34 C1′, and A35 C1′) showed similar RD in ΔC24-wtTAR (Figure 4A and Figure S6A). A global two-state fit of the RD data yielded an ES1 population (pES1 = 16 ± 4%) and exchange rate (kex = 27000 ± 1400 s−1) similar to those reported previously for wtTAR (pES1 = 13 ± 2%, and kex = 26000 ± 700 s−1).35
ES2 features a larger rearrangement of secondary structure, which reshuffles nucleotides in the bulge, upper helix, and apical loop (Figure 4B). In ES2 of wtTAR, C24 forms a weak C24-C39 mispair, while U23 is bulged out.37 Therefore, deletion of C24 was expected to impact ES2 exchange and structure. Surprisingly, RD data similar to those of ES2 in wtTAR were obtained for ΔC24-wtTAR for several sites in the bulge, upper helix, and apical loop (Figure 4B and Figure S6A). A global two-state fit of the RD data yielded exchange parameters similar to those obtained for ES2 in wtTAR. The ES2 ΔC24-wtTAR population is reduced by 2-fold (pES2 of 0.17 ± 0.02% compared to a value of 0.40 ± 0.05% in wtTAR), whereas the exchange rate remains the same within experimental error (kex of 541 ± 93 s−1 compared to a kex value of 474 ± 69 s−1 in wtTAR). Additionally, sites that do not show RD in wtTAR also lack RD in ΔC24-wtTAR (Figure S6A). Therefore, deleting the bulge minimally affects the thermodynamics and kinetics of an exchange process that is experienced by residues in the bulge, upper helix, and apical loop. This suggests that for wtTAR, flipping out U23 contributes little to the overall thermodynamics and kinetics of the GS–ES2 transition. This is possible because any loss of stacking interactions between U23 and A22 in the GS is partly compensated for by a favorable increase in entropy due to a flexible flipped out conformation in ES2.
Similar RD profiles were also observed for wtTAR and ΔC24-wtTAR in the presence of 5 mM Mg2+. However, the addition of Mg2+ altered the populations, exchange rates, and possibly chemical shift differences between the GS and ESs (Figure S6B and Table S6). The effects of Mg2+ on exchange will be characterized more fully in future studies. It should be noted that during the course of these studies, we also obtained evidence of a third distinct ES (ES3) in the absence of Mg2+, for both wtTAR and ΔC24-wtTAR at residues G26 C8 and A27 C1′ with a population of ≈0.2% and a kex of ≈5000 s−1 in ΔC24-wtTAR at 25 °C and a population of ≈0.2% and a kex of ≈2300 s−1 in wtTAR at 10 °C (Figure S6C and Table S6). Furthermore, A27 C1′ has RD consistent with ES2 in wtTAR at 25 °C; thus, it is likely that this resonance experiences three-state exchange involving the GS, ES2, and ES3 at 25 °C in ΔC24-wtTAR. Although the A27 C1′ RD profile is dominated by exchange with ES3, fitting to a three-state linear model without minor exchange (eq 3) yields exchange parameters consistent with those obtained for ES2 and ES3 (Table S7 and Methods). This new ES3 will be more fully characterized in future studies.
ΔC24-wtTAR Adopts an ES2 Conformation Different from That of wtTAR
The RD analysis yields the difference in chemical shift between ES2 and the GS (Δω = ωES −ωGS). With the exception of U23 C6 and A22 C1′, similar Δω values were determined for various ES2 sites in wtTAR and ΔC24-wtTAR, indicating that they adopt a similar ES2 conformation (Table S6). However, the Δω values for U23 C6 and A22 C1′ in ΔC24-wtTAR were opposite in sign to those measured in wtTAR. In wtTAR, Δω(U23 C6) = +2.3 ppm and was attributed to loss of U23-A22 stacking in the GS to form a bulged out conformation in ES2. In contrast, in ΔC24-wtTAR, Δω(U23 C6) = −3.4 ppm, suggesting an opposite transition from a flipped out conformation in the GS to a flipped in conformation in ES2. Similarly, in wtTAR, a Δω(A22 C1′) of −2.2 ppm suggests sugar repuckering farther from C3′-endo in the GS toward C2′-endo in ES2, possibly due to the bulging out of U23, whereas in ΔC24-wtTAR, a Δω(A22 C1′) of +2.0 ppm indicates a shift toward a more helical C3′-endo conformation in ES2, consistent with a flipped in helical U23 conformation. A simple explanation for these data is that, in ΔC24-TAR, U23 replaces C24 to form an U23-C39 mispair (Figure 4B and Figure S7).
To test the proposed ΔC24-wtTAR ES2 secondary structure, we used a G28U point mutation, which was previously used to trap ES2 secondary structure in wtTAR.37 As expected, the G28U-ΔC24-wtTAR mutant adopts the proposed ES2 secondary structure based on imino 2D SOFAST-HMQC spectra and NOE connectivities (Figure S8). The differences in chemical shifts between G28U-ΔC24-wtTAR and the ground state ΔC24-wtTAR are in excellent agreement with the differences in chemical shifts between the ES and GS measured by RD in ΔC24-wtTAR (Figure 5A). In addition, comparing spectra of ΔC24-wtTAR and wtTAR with those of their G28U mutant counterpart reveals that the G28U mutation recapitulates the opposite changes in the chemical shifts observed for U23 C6 and A22 C1′ (Figure 5A). The NOE connectivity between A22 H1′ and U23 H6, which is not observed in G28U-wtTAR, also helps to establish that in G28U-ΔC24-wtTAR U23 is not flipped out but rather intrahelical, consistent with formation of an U23-C39 base pair (Figure 5B).
Figure 5.
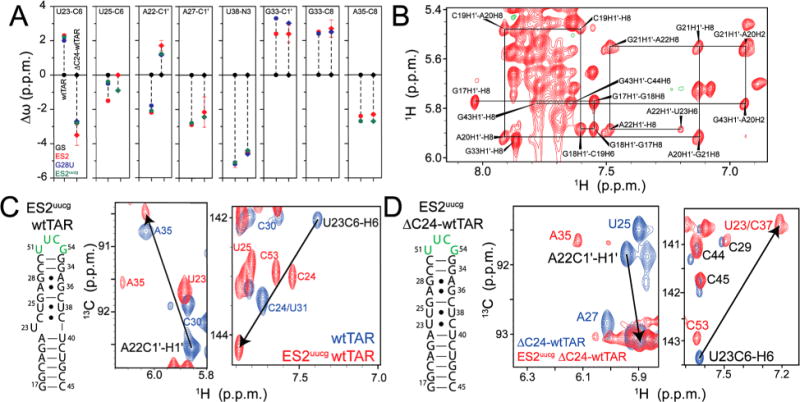
Trapping ES2 in ΔC24-wtTAR using mutations. (A) Comparison of Δω between ES2 and the GS of wtTAR (circles) and ΔC24-wtTAR (diamonds) as determined by RD (red), and the G28U (blue) and ES2uucg (green) ES2 trapped mutants. Errors in RD chemical shifts were determined from the standard error from the global fit. (B) Nonexchangeable region of the 1H–1H NOESY spectrum for G28U-ΔC24-wtTAR, highlighting the sequential assignments from G17 to U23 (solid black lines). (C and D) Secondary structure of ES2uucg for (C) wtTAR and (D) ΔC24-wtTAR, highlighting the mutated apical loop residues in green, with 2D HSQC overlays of the wtTAR and ΔC24-wtTAR (blue) and their respective ES2uucg trap (red). Black arrows highlight the opposite sign of Δω between GS and ES2 for A22 C1′ and U23 C6 in wtTAR and ΔC24-wtTAR.
Stabilizing ES2 in wtTAR and ΔC24-wtTAR Using a UUCG Loop Mutation
As noted previously,37 nonexchangeable NOE connectivities were difficult to resolve for all residues in the G28U-wtTAR mutant, leading to ambiguous assignments for several resonances in base and sugar 2D HSQC spectra. To resolve these ambiguities and further confirm the observed differences in the ES2 secondary structure between wtTAR and ΔC24-wtTAR, we designed a second ES2 mutant in which we replaced the ES2 apical loop with a stable UUCG tetraloop [ES2uucg (Figure 5C,D)]. We used this mutation to trap ES2 in both wtTAR (wtTAR-ES2uucg) and ΔC24-wtTAR (ΔC24-wtTAR-ES2uucg). Both mutants adopted the expected ES2 secondary structure with an UUCG apical loop as judged on the basis of imino 2D SOFAST-HMQC spectra and NOE connectivities (Figure S9).
The new mutants allowed us to resolve many ambiguous assignments and to correct a subset of assignments in G28U-wtTAR.37 Updated assignments do not affect any previous conclusions reached about wtTAR-ES2 (Supporting Discussion 1); in fact, better agreement is observed between the ES2 chemical shifts measured in wtTAR using RD and those observed in the mutant G28U-wtTAR (Figure 5A) when using updated assignments. Comparison of wtTAR and ΔC24-wtTAR spectra with those of their ES2uucg trapped counterparts revealed opposite changes in the chemical shifts for U23 C6 and A22 C1′ (Figure 5C,D). We also observe excellent agreement between the ES2 and G28U-wtTAR chemical shifts for seven new resonances whose assignments could be resolved (Figure 5A). These data support our previously proposed ES2 structure of HIV-1 TAR and also confirm that ΔC24-wtTAR-ES2uucg adopts a slightly different conformation with flipped in U23 (Figure 4B).
Chemical exchange to ES3 required the use of a three-state exchange equation for A27 C1′ to obtain Δω for both ES2 and ES3 (see Methods). The ES2 Δω(A27 C1′) value of −2.2 ppm obtained from three-state fitting of the A27 C1′ RD data (Table S7) is in excellent agreement with the Δω based on ES2 mutants [Δω(A27 C1′) = −2.4 ppm (Figure 5A)]. Additionally, no RD was observed for U25 C6 (Figure S8), which is consistent with the relatively small chemical shift perturbations predicted by the ES2uucg mutant [Δω(U25 C6) = −0.9 ppm] and smaller population of ES2 in ΔC24-wtTAR compared to that in wtTAR.37
Impact of Mutation on the Dynamic RNA Energy Landscape
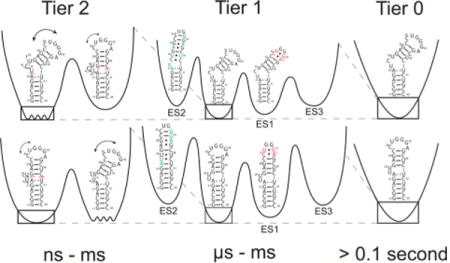
The RNA free energy landscape can be hierarchically organized into local energetic minima containing conformational states (CSs) separated by large kinetic barriers; each minimum is then, in turn, subdivided into a greater number of local energetic minima of conformational substates separated by lower kinetic barriers (Figure 6). These hierarchically organized energetic layers can be classified into different tiers of RNA dynamics (tier 0, tier 1, and tier 2) that are organized in terms of transitions between CSs within each tier.89 Such a framework was first introduced by Frauenfelder et al.90 to describe protein dynamics. In RNA, tier 0 motions represent large-scale changes in secondary structure that can remodel entire hairpins occurring on the second time scale, tier 1 represents localized changes in base pairing occurring on the microsecond-to-millisecond time scale, and tier 2 represents librational and interhelical motions occurring on time scales faster than microseconds.89
Figure 6.
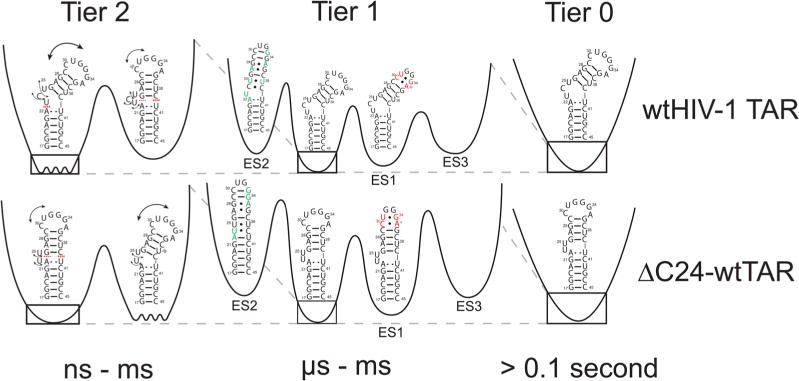
Hierarchical energy landscape of wtTAR and ΔC24-wtTAR in the absence of Mg2+. Shown are the tiers of the energy landscape for wtTAR (top) and ΔC24-wtTAR (bottom). The dashed red lines denote stacking and black arrows flexibility. Residues involved in ES1 and ES2 exchange are colored red and green, respectively. Barrier heights were estimated as the activation energy calculated from the Arrhenius equation at 25 °C.
In this study, we set out to characterize how deleting a single bulge residue in HIV-1 TAR affects motions on each tier and how each tier is then further affected by the addition of Mg2+. The results show that shortening the TAR bulge does not substantially reorganize the motional modes or alter sampled conformational substates. For example, in tier 2, we still observe a two-state equilibrium between a flexible bent state and coaxial state with extrahelical bulge residues. Shortening the bulge primarily shifts the two-state equilibrium toward the coaxial state. These differences in dynamics are diminished at high Mg2+ concentrations, which favors the coaxial state in both wtTAR and ΔC24-wtTAR. Shortening the bulge most likely favors the coaxial state due to both a reduction in the entropy of the bent state (and therefore reduction of the entropic penalty accompanying coaxial stacking) and a reduction in the degree of phosphate–backbone electrostatic charge repulsion due to loss of a bulge nucleotide. A dissection of these energetic contributions will require further studies.
Interestingly, the point deletion mutant had minimal effects on slower tier 1 microsecond-to-millisecond exchange directed toward two ESs even in the absence of Mg2+. This was the case even though ΔC24-wtTAR adopts a slightly different ES2 secondary structure. The addition of Mg2+ also affects microsecond-to-millisecond exchange, but the effects were similar for ΔC24-wtTAR and wtTAR (Figure S6B). The preservation of exchange directed to ES1 and ES2 in two distinct but naturally occurring TAR molecules provides additional support for the potential functional importance of these excited conformational states. One potential functional role for the TAR ES2 conformational state is in HIV genome dimerization. Prior studies have shown that deleting the TAR UCU bulge significantly impairs genomic dimerization, which is essential for viral infectivity.60 Deletion of the UCU bulge has also been shown to inhibit formation of ES2.37 Moreover, our results indicate that the G28U point mutant, which traps the ES2 state, has a strong propensity to form duplex or kissing dimers (see Supporting Discussion 1). Future studies will examine more deeply how Mg2+ and other physiologically important metals impact the tier 1 conformational landscape of wtTAR, how other mutations that have not been selected by the HIV virus affect these dynamics, and how the ES2 conformation could potentially play a role in HIV genomic dimerization.
Supplementary Material
Acknowledgments
We thank Bharathwaj Sathyamoorthy, Zachary W. Stein, and Huiqing Zhou for their input and assistance, Daniel Herschlag for critical input and suggestions, and the Duke Magnetic Resonance Spectroscopy Center for their resources and technical assistance.
Funding
This work was supported by the National Institutes of Health (NIH) (Grant PO1 GM0066275 awarded to H.M.A.-H.), the NIH Structural Biology and Biophysics Training Program (5T32GM008487 awarded to D.K.M.), and a Duke University Joe Taylor Adams Endowed Fellowship awarded to D.K.M.
ABBREVIATIONS
- HIV
human immunodeficiency virus
- TAR
transactivation response element
- HJH
helix–junction–helix
- NMR
nuclear magnetic resonance
- RD
relaxation dispersion
- RDC
residual dipolar coupling
- NOE
nuclear Overhauser effect
- HSQC
heteronuclear single-quantum coherence
- HMQC
heteronuclear multiple-quantum coherence
- TROSY
transverse relaxation-optimized spectroscopy
- WC
Watson–Crick
- ES
excited conformational state
- CS
conformational state
- rmsd
root-mean-square deviation
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.bio-chem.6b00285.
Supporting discussion, additional data, and figures (PDF)
Notes
The authors declare the following competing financial interest(s): H.M.A. is an advisor to and holds an ownership interest in Nymirum, an RNA-based drug discovery company.
References
- 1.Leulliot N, Varani G. Current Topics in RNA–Protein Recognition: Control of Specificity and Biological Function through Induced Fit and Conformational Capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 2.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 3.Kligun E, Mandel-Gutfreund Y. Conformational readout of RNA by small ligands. RNA Biol. 2013;10:981–989. doi: 10.4161/rna.24682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Woodson SA. RNA folding pathways and the self-assembly of ribosomes. Acc Chem Res. 2011;44:1312–1319. doi: 10.1021/ar2000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shajani Z, Sykes MT, Williamson JR. Assembly of Bacterial Ribosomes. Annu Rev Biochem. 2011;80:501–526. doi: 10.1146/annurev-biochem-062608-160432. [DOI] [PubMed] [Google Scholar]
- 6.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 7.Serganov A, Nudler E. A Decade of Riboswitches. Cell. 2013;152:17–24. doi: 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furtig B, Nozinovic S, Reining A, Schwalbe H. Multiple conformational states of riboswitches fine-tune gene regulation. Curr Opin Struct Biol. 2015;30:112–124. doi: 10.1016/j.sbi.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 9.Solomatin SV, Greenfeld M, Chu S, Herschlag D. Multiple native states reveal persistent ruggedness of an RNA folding landscape. Nature. 2010;463:681–U117. doi: 10.1038/nature08717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 11.Zhuang X, Kim H, Pereira MJ, Babcock HP, Walter NG, Chu S. Correlating structural dynamics and function in single ribozyme molecules. Science. 2002;296:1473–1476. doi: 10.1126/science.1069013. [DOI] [PubMed] [Google Scholar]
- 12.Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. Functional complexity and regulation through RNA dynamics. Nature. 2012;482:322–330. doi: 10.1038/nature10885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brion P, Westhof E. Hierarchy and Dynamics of RNA Folding. Annu Rev Biophys Biomol Struct. 1997;26:113–137. doi: 10.1146/annurev.biophys.26.1.113. [DOI] [PubMed] [Google Scholar]
- 14.Halder S, Bhattacharyya D. RNA structure and dynamics: a base pairing perspective. Prog Biophys Mol Biol. 2013;113:264–283. doi: 10.1016/j.pbiomolbio.2013.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Korostelev A, Ermolenko DN, Noller HF. Structural dynamics of the ribosome. Curr Opin Chem Biol. 2008;12:674–683. doi: 10.1016/j.cbpa.2008.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermann T. Rational ligand design for RNA: the role of static structure and conformational flexibility in target recognition. Biochimie. 2002;84:869–875. doi: 10.1016/s0300-9084(02)01460-8. [DOI] [PubMed] [Google Scholar]
- 17.Stelzer AC, Frank AT, Kratz JD, Swanson MD, Gonzalez-Hernandez MJ, Lee J, Andricioaei I, Markovitz DM, Al-Hashimi HM. Discovery of selective bioactive small molecules by targeting an RNA dynamic ensemble. Nat Chem Biol. 2011;7:553–559. doi: 10.1038/nchembio.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gomez Pinto I, Guilbert C, Ulyanov NB, Stearns J, James TL. Discovery of ligands for a novel target, the human telomerase RNA, based on flexible-target virtual screening and NMR. J Med Chem. 2008;51:7205–7215. doi: 10.1021/jm800825n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann T, Patel DJ. Stitching Together RNA Tertiary Architectures. J Mol Biol. 1999;294:829–849. doi: 10.1006/jmbi.1999.3312. [DOI] [PubMed] [Google Scholar]
- 20.Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure. 2000;8:R47–R54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- 21.Batey RT, Rambo RP, Doudna JA. Tertiary Motifs in RNA Structure and Folding. Angew Chem, Int Ed. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 22.Durney MA, D’Souza VM. Preformed protein-binding motifs in 7SK snRNA: structural and thermodynamic comparisons with retroviral TAR. J Mol Biol. 2010;404:555–567. doi: 10.1016/j.jmb.2010.08.042. [DOI] [PubMed] [Google Scholar]
- 23.Chow CS, Bogdan FM. A structural basis for RNA-ligand interactions. Chem Rev. 1997;97:1489–1513. doi: 10.1021/cr960415w. [DOI] [PubMed] [Google Scholar]
- 24.Meyer ST, Hergenrother PJ. Small molecule ligands for bulged RNA secondary structures. Org Lett. 2009;11:4052–4055. doi: 10.1021/ol901478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hermann T, Westhof E. Rational drug design and high-throughput techniques for RNA targets. Comb Chem High Throughput Screening. 2000;3:219–234. doi: 10.2174/1386207003331652. [DOI] [PubMed] [Google Scholar]
- 26.Pyle AM. Metal ions in the structure and function of RNA. JBIC, J Biol Inorg Chem. 2002;7:679–690. doi: 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 27.Huppler A, Nikstad LJ, Allmann AM, Brow DA, Butcher SE. Metal binding and base ionization in the U6 RNA intramolecular stem-loop structure. Nat Struct Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Q, Sun X, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 29.Salmon L, Bascom G, Andricioaei I, Al-Hashimi HM. A General Method for Constructing Atomic-Resolution RNA Ensembles using NMR Residual Dipolar Couplings: The Basis for Interhelical Motions Revealed. J Am Chem Soc. 2013;135:5457–5466. doi: 10.1021/ja400920w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu J, Kadakkuzha BM, Zhao L, Fan M, Qi X, Xia T. Dynamic Ensemble View of the Conformational Landscape of HIV-1 TAR RNA and Allosteric Recognition. Biochemistry. 2011;50:5042–5057. doi: 10.1021/bi200495d. [DOI] [PubMed] [Google Scholar]
- 31.Grant GP, Boyd N, Herschlag D, Qin PZ. Motions of the substrate recognition duplex in a group I intron assessed by site-directed spin labeling. J Am Chem Soc. 2009;131:3136–3137. doi: 10.1021/ja808217s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim NK, Murali A, DeRose VJ. A distance ruler for RNA using EPR and site-directed spin labeling. Chem Biol. 2004;11:939–948. doi: 10.1016/j.chembiol.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 33.Edwards TE, Okonogi TM, Sigurdsson ST. Investigation of RNA-Protein and RNA-Metal Ion Interactions by Electron Paramagnetic Resonance Spectroscopy: The HIV TAR-Tat Motif. Chem Biol. 2002;9:699–706. doi: 10.1016/s1074-5521(02)00150-3. [DOI] [PubMed] [Google Scholar]
- 34.Edwards TE, Sigurdsson ST. Electron paramagnetic resonance dynamic signatures of TAR RNA-small molecule complexes provide insight into RNA structure and recognition. Biochemistry. 2002;41:14843–14847. doi: 10.1021/bi026299a. [DOI] [PubMed] [Google Scholar]
- 35.Dethoff EA, Petzold K, Chugh J, Casiano-Negroni A, Al-Hashimi HM. Visualizing transient low-populated structures of RNA. Nature. 2013;491:724–728. doi: 10.1038/nature11498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Zhang Q. Characterizing excited conformational states of RNA by NMR spectroscopy. Curr Opin Struct Biol. 2015;30:134–146. doi: 10.1016/j.sbi.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee J, Dethoff EA, Al-Hashimi HM. Invisible RNA state dynamically couples distant motifs. Proc Natl Acad Sci U S A. 2014;111:9485–9490. doi: 10.1073/pnas.1407969111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Houck-Loomis B, Durney MA, Salguero C, Shankar N, Nagle JM, Goff SP, D’Souza VM. An equilibrium-dependent retroviral mRNA switch regulates translational recoding. Nature. 2011;480:561–564. doi: 10.1038/nature10657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blad H, Reiter NJ, Abildgaard F, Markley JL, Butcher SE. Dynamics and Metal Ion Binding in the U6 RNA Intramolecular Stem–Loop as Analyzed by NMR. J Mol Biol. 2005;353:540–555. doi: 10.1016/j.jmb.2005.08.030. [DOI] [PubMed] [Google Scholar]
- 40.Cordero P, Das R. Rich structure landscapes in both natural and artificial RNAs revealed by mutate-and-map analysis. PLoS Comput Biol. 2015;11:e1004473. doi: 10.1371/journal.pcbi.1004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kim HD, Nienhaus GU, Ha T, Orr JW, Williamson JR, Chu S. Mg2+-dependent conformational change of RNA studied by fluorescence correlation and FRET on immobilized single molecules. Proc Natl Acad Sci U S A. 2002;99:4284–4289. doi: 10.1073/pnas.032077799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Q, Stelzer AC, Fisher CK, Al-Hashimi HM. Visualizing spatially correlated dynamics that directs RNA conformational transitions. Nature. 2007;450:1263–1267. doi: 10.1038/nature06389. [DOI] [PubMed] [Google Scholar]
- 43.Al-Hashimi HM, Gosser Y, Gorin A, Hu W, Majumdar A, Patel DJ. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315:95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 44.Dethoff EA, Hansen AL, Musselman C, Watt ED, Andricioaei I, Al-Hashimi HM. Characterizing Complex Dynamics in the Transactivation Response Element Apical Loop and Motional Correlations with the Bulge by NMR, Molecular Dynamics, and Mutagenesis. Biophys J. 2008;95:3906–3915. doi: 10.1529/biophysj.108.140285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fonseca R, Pachov DV, Bernauer J, van den Bedem H. Characterizing RNA ensembles from NMR data with kinematic models. Nucleic Acids Res. 2014;42:9562–9572. doi: 10.1093/nar/gku707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Casiano-Negroni A, Sun X, Al-Hashimi HM. Probing Na + -Induced Changes in the HIV-1 TAR Conformational Dynamics Using NMR Residual Dipolar Couplings: New Insights into the Role of Counterions and Electrostatic Interactions in Adaptive Recognition. Biochemistry. 2007;46:6525–6535. doi: 10.1021/bi700335n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dayie KT, Brodsky AS, Williamson JR. Base flexibility in HIV-2 TAR RNA mapped by solution 15N, 13C NMR relaxation. J Mol Biol. 2002;317:263–278. doi: 10.1006/jmbi.2001.5424. [DOI] [PubMed] [Google Scholar]
- 48.Al-Hashimi HM, Pitt SW, Majumdar A, Xu W, Patel DJ. Mg2+-induced Variations in the Conformation and Dynamics of HIV-1 TAR RNA Probed Using NMR Residual Dipolar Couplings. J Mol Biol. 2003;329:867–873. doi: 10.1016/s0022-2836(03)00517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brodsky AS, Williamson JR. Solution Structure of the HIV-2 TAR- Argininamide Complex. J Mol Biol. 1997;267:624–639. doi: 10.1006/jmbi.1996.0879. [DOI] [PubMed] [Google Scholar]
- 50.Huang W, Varani G, Drobny GP. 13C/15N-19F intermolecular REDOR NMR study of the interaction of TAR RNA with Tat peptides. J Am Chem Soc. 2010;132:17643–17645. doi: 10.1021/ja1051439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Olsen GL, Edwards TE, Deka P, Varani G, Sigurdsson ST, Drobny GP. Monitoring tat peptide binding to TAR RNA by solid-state 31P-19F REDOR NMR. Nucleic Acids Res. 2005;33:3447–3454. doi: 10.1093/nar/gki626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riordan FA, Bhattacharyya A, McAteer S, Lilley DMJ. Kinking of RNA Helices by Bulged Bases, and the Structure of the Human Immunodeficiency Virus TransactivatorResponse Element. J Mol Biol. 1992;226:305–310. doi: 10.1016/0022-2836(92)90947-i. [DOI] [PubMed] [Google Scholar]
- 53.Zacharias M, Hagerman PJ. The bend in RNA created by the trans-activation responce element bulge of human immunodeficiency virus is straightended by arginine and Tat-derived peptide. Proc Natl Acad Sci U S A. 1995;92:6052–6056. doi: 10.1073/pnas.92.13.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murchie AIH, Davis B, Isel C, Afshar M, Drysdale MJ, Bower J, Potter AJ, Starkey ID, Swarbrick TM, Mirza S, Prescott CD, Vaglio P, Aboul-ela F, Karn J. Structure-based Drug Design Targeting an Inactive RNA Conformation: Exploiting the Flexibility of HIV-1 TAR RNA. J Mol Biol. 2004;336:625–638. doi: 10.1016/j.jmb.2003.12.028. [DOI] [PubMed] [Google Scholar]
- 55.Ippolito JA, Steitz T. A 1.3-Å resolution crystal structure of the HIV-1 trans-activation response region RNA stem reveals a metal ion-dependentbulge conformation. Proc Natl Acad Sci U S A. 1998;95:9819–9824. doi: 10.1073/pnas.95.17.9819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mustoe AM, Al-Hashimi HM, Brooks CL., III Coarse Grained Models Reveal Essential Contributions of Topological Constraints to the Conformational Free Energy of RNA Bulges. J Phys Chem B. 2014;118:2615–2627. doi: 10.1021/jp411478x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faber C, Sticht H, Schweimer K, Rosch P. Structural rearrangements of HIV-1 Tat-responsive RNA upon binding of neomycin B. J Biol Chem. 2000;275:20660–20666. doi: 10.1074/jbc.M000920200. [DOI] [PubMed] [Google Scholar]
- 58.Du Z, Lind KE, James TL. Structure of TAR RNA complexed with a Tat-TAR interaction nanomolar inhibitor that was identified by computational screening. Chem Biol. 2002;9:707–712. doi: 10.1016/s1074-5521(02)00151-5. [DOI] [PubMed] [Google Scholar]
- 59.Davis B, Afshar M, Varani G, Murchie AI, Karn J, Lentzen G, Drysdale M, Bower J, Potter AJ, Starkey ID, Swarbrick T, Aboul-ela F. Rational design of inhibitors of HIV-1 TAR RNA through the stabilisation of electrostatic ″hot spots″. J Mol Biol. 2004;336:343–356. doi: 10.1016/j.jmb.2003.12.046. [DOI] [PubMed] [Google Scholar]
- 60.Jalalirad M, Saadatmand J, Laughrea M. Dominant role of the 5′ TAR bulge in dimerization of HIV-1 genomic RNA, but no evidence of TAR-TAR kissing during in vivo virus assembly. Biochemistry. 2012;51:3744–3758. doi: 10.1021/bi300111p. [DOI] [PubMed] [Google Scholar]
- 61.Berkhout B. Structural Features in TAR RNA of Human and Simian Immunodeficiency Viruses: A Phylogentic Analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tjandra N, Bax A. Direct measurement of distances and angles in biomolecules by NMR in a dilute liquid crystalline medium. Science. 1997;278:1111–1114. doi: 10.1126/science.278.5340.1111. [DOI] [PubMed] [Google Scholar]
- 63.Tolman JR, Flanagan JM, Kennedy MA, Prestegard JH. NMR evidence for slow collective motions in cyanometmyoglobin. Nat Struct Biol. 1997;4:292–297. doi: 10.1038/nsb0497-292. [DOI] [PubMed] [Google Scholar]
- 64.Frank AT, Stelzer AC, Al-Hashimi HM, Andricioaei I. Constructing RNA dynamical ensembles by combining MD and motionally decoupled NMR RDCs: new insights into RNA dynamics and adaptive ligand recognition. Nucleic Acids Res. 2009;37:3670–3679. doi: 10.1093/nar/gkp156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Carlomagno T, Amata I, Williamson JR, Hennig M. NMR assignments of HIV-2 TAR RNA. Biomol NMR Assignments. 2008;2:167–169. doi: 10.1007/s12104-008-9112-x. [DOI] [PubMed] [Google Scholar]
- 66.Hwang TL, Shaka AJ. Water Suppresion That Works. Exciatation Sculpting Using Arbitary Waveforms and Pulsed Field Gradients. J Magn Reson, Ser A. 1995;112:275–279. [Google Scholar]
- 67.Sathyamoorthy B, Lee J, Kimsey I, Ganser LR, Al-Hashimi H. Development and application of aromatic [13C, 1H] SOFAST-HMQC NMR experiment for nucleic acids. J Biomol NMR. 2014;60:77–83. doi: 10.1007/s10858-014-9856-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fürtig B, Richter C, Wöhnert J, Schwalbe H. NMR Spectroscopy of RNA. ChemBioChem. 2003;4:936–962. doi: 10.1002/cbic.200300700. [DOI] [PubMed] [Google Scholar]
- 69.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 70.Goddard TD, Kneller DG. SPARKY 3. University of California; San Franciso: 2008. [Google Scholar]
- 71.Hansen AL, Al-Hashimi HM. Dynamics of Large Elongated RNA by NMR Carbon Relaxation. J Am Chem Soc. 2007;129:16072–16082. doi: 10.1021/ja0757982. [DOI] [PubMed] [Google Scholar]
- 72.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci U S A. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schanda P, Kupče Ē, Brutscher B. SOFAST-HMQC Experiments for Recording Two-dimensional Deteronuclear Correlation Spectra of Proteins within a Few Seconds. J Biomol NMR. 2005;33:199–211. doi: 10.1007/s10858-005-4425-x. [DOI] [PubMed] [Google Scholar]
- 74.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Prestegard JH, Al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Q Rev Biophys. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- 76.Tolman JR, Al-Hashimi HM, Kay LE, Prestegard JH. Structural and Dynamic Analysis of Residual Dipolar Coupling Data for Proteins. J Am Chem Soc. 2001;123:1416–1424. doi: 10.1021/ja002500y. [DOI] [PubMed] [Google Scholar]
- 77.Bailor MH, Musselman C, Hansen AL, Gulati K, Patel DJ, Al-Hashimi HM. Characterizing the relative orientation and dynamics of RNA A-form helices using NMR residual dipolar couplings. Nat Protoc. 2007;2:1536–1546. doi: 10.1038/nprot.2007.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Musselman C, Pitt SW, Gulati K, Foster LL, Andricioaei I, Al-Hashimi HM. Impact of static and dynamic A-form heterogeneity on the determination of RNA global structural dynamics using NMR residual dipolar couplings. J Biomol NMR. 2006;36:235–249. doi: 10.1007/s10858-006-9087-9. [DOI] [PubMed] [Google Scholar]
- 79.Hansen AL, Al-Hashimi HM. Insight into the CSA tensors of nucleobase carbons in RNA polynucleotides from solution measurements of residual CSA: Towards new long-range orientational constraints. J Magn Reson. 2006;179:299–307. doi: 10.1016/j.jmr.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 80.The PyMOL Molecular Graphics System. Enhanced for Mac OS X. Schrodinger, LLC; Portland, OR: 2010. version 1.7.0.3. [Google Scholar]
- 81.Hansen AL, Nikolova EN, Casiano-Negroni A, Al-Hashimi HM. Extending the Range of Microsecond-to-Millisecond Chemical Exchange Detected in Labeled and Unlabeled Nucleic Acids by Selective Carbon R 1ρNMR Spectroscopy. J Am Chem Soc. 2009;131:3818–3819. doi: 10.1021/ja8091399. [DOI] [PubMed] [Google Scholar]
- 82.Massi F, Johnson E, Wang C, Rance M, Palmer AG. NMR R1ρRotating-Frame Relaxation with Weak Radio Frequency Fields. J Am Chem Soc. 2004;126:2247–2256. doi: 10.1021/ja038721w. [DOI] [PubMed] [Google Scholar]
- 83.Korzhnev DM, Orekhov VY, Kay LE. Off-Resonance R1ρ NMR Studies of Exchange Dynamics in Proteins with Low Spin-Lock Fields: An Application to a Fyn SH3 Domain. J Am Chem Soc. 2005;127:713–721. doi: 10.1021/ja0446855. [DOI] [PubMed] [Google Scholar]
- 84.Xue Y, Kellogg D, Kimsey IJ, Sathyamoorthy B, Stein ZW, McBrairty M, Al-Hashimi HM. Characterizing RNA Excited States Using NMR Relaxation Dispersion. 1st. Elsevier Inc; Amsterdam: 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Parisien M, Major F. The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature. 2008;452:51–55. doi: 10.1038/nature06684. [DOI] [PubMed] [Google Scholar]
- 86.Fushman D, Tjandra N, Cowburn D. An approach to direct determination of protein dynamics from N-15 NMR relaxation at multiple fields, independent of variable N-15 chemical shift anisotropy and chemical exchange contributions. J Am Chem Soc. 1999;121:8577–8582. [Google Scholar]
- 87.Losonczi JA, Andrec M, Fischer MWF, Prestegard JH. Order Matrix Analysis of Residual Dipolar Couplings Using Singular Value Decomposition. J Magn Reson. 1999;138:334–342. doi: 10.1006/jmre.1999.1754. [DOI] [PubMed] [Google Scholar]
- 88.Mollova ET, Hansen MR, Pardi A. Global Structure of RNA Determined with Residual Dipolar Couplings. J Am Chem Soc. 2000;122:11561–11562. [Google Scholar]
- 89.Mustoe AM, Brooks CL, Al-Hashimi HM. Hierarchy of RNA Functional Dynamics. Annu Rev Biochem. 2014;83:441–466. doi: 10.1146/annurev-biochem-060713-035524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frauenfelder H, Sligar SG, Wolynes PG. The energy landscapes and motions of proteins. Science. 1991;254:1598–1603. doi: 10.1126/science.1749933. [DOI] [PubMed] [Google Scholar]
- 91.Lee J, Vogt CE, McBrairty M, Al-Hashimi HM. Influence of Dimethylsulfoxide on RNA Structure and Ligand Binding. Anal Chem. 2013;85:9692–9698. doi: 10.1021/ac402038t. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


