Recording only the statistics of driven, asynchronous dynamics reveals a network’s interaction structure.
Keywords: nonlinear dynamics, network dynamics, network reconstruction, physical connectivity, structural connectivity, gene networks
Abstract
Revealing physical interactions in complex systems from observed collective dynamics constitutes a fundamental inverse problem in science. Current reconstruction methods require access to a system’s model or dynamical data at a level of detail often not available. We exploit changes in invariant measures, in particular distributions of sampled states of the system in response to driving signals, and use compressed sensing to reveal physical interaction networks. Dynamical observations following driving suffice to infer physical connectivity even if they are temporally disordered, are acquired at large sampling intervals, and stem from different experiments. Testing various nonlinear dynamic processes emerging on artificial and real network topologies indicates high reconstruction quality for existence as well as type of interactions. These results advance our ability to reveal physical interaction networks in complex synthetic and natural systems.
INTRODUCTION
Many complex systems in physics and biology constitute networks of dynamically interacting units (1). Examples range from gene regulatory networks in the cell (2–5) and neural circuits in the brain (6–8) to food webs in ecosystems (9) and power grids (10–13), as well as other supply systems of engineering (14, 15). These systems’ interaction networks fundamentally underlie their collective dynamics and function, thus rendering the knowledge of their interaction topology essential. For instance, identifying new pathways in gene regulatory networks and understanding long-range feedback in engineering systems require exact knowledge of their physical interaction networks.
A fundamental question about both natural and artificial networks is thus which units directly act upon which other units. For instance, for networks of interacting units i ∈ {1, …, N} described by ordinary nonlinear differential equations dxi/dt = Fi(x), this question mathematically becomes which of the variables xj among (x1, …, xN) =: x explicitly appear in Fi(x). In many settings, this physical connectivity is not directly accessible. Revealing these interaction networks then poses a high-dimensional nonlinear reconstruction problem. Evaluating the dynamics of the system exploiting heuristic approximations, such as thresholding correlations or other measures of statistical dependency between units’ dynamics, is simple, efficient, and generally feasible, yet often an unreliable predictor for physical connectivity (16). A conceptual reason is that more than one statistical dependency network may emerge for the same physical network, for example, due to multistability (17–19). In turn, methods aiming at directly identifying physical interactions generally require either a priori knowledge of a detailed model of the system, rely on the system being in simple states (such as close to fixed points), or need high-resolution, synchronized, temporally ordered observations for all units with connections of interest (20). For example, transfer entropy (21) and cross-embedding (22) require temporally ordered measurements; a direct method for inferring structural connectivity described by Shandilya and Timme (23) requires synchronous, temporally ordered, high-resolution measurements and prior knowledge of the system’s model; and the system identification method described by Gardner et al. (5) requires the system to be at steady state and the dynamics to be essentially linear in the activity of the nodes of the network. However, real systems, such as genetic regulatory networks, other biological circuits, and even some human-made systems (24), often do not fulfill these requirements. To the best of our knowledge, a method capable of inferring physical interaction networks without requiring at least one of these constraints does not exist to date.
Here, we propose a generic strategy to reveal physical interactions from responses of invariant measures (that is, distributions of points sampled in state space) to small driving signals. The strategy does not rely on any of the requirements above. Via compressed sensing, the resulting equations obtained from driving-response experiments yield the network structure even if the number of available experiments is small compared to the network size. Because only statistics of recorded system states are evaluated to reveal the physical connectivity, measurements of dynamic states can be temporally disordered, be acquired at large sampling intervals, come from different experiments, and be collected at different times for different units. In addition, no detailed prior knowledge for the model of the system is required.
THEORY BASED ON TIME INVARIANTS
To introduce the basic strategy of reconstructing networks from time invariants (Fig. 1), we consider networks of units i ∈ {1, …, N} represented by state variables evolving in time t according to
| (1) |
Fig. 1. Strategy of network reconstruction from responses of time invariants.
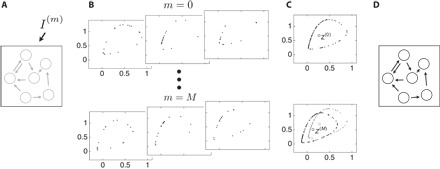
(A) A networked dynamical system with unknown topology (gray) is perturbed by external driving signals Im, m ∈ {1, …, M}. (B) Potentially noisy, disordered, low-resolution data are collected from several different experiments. (C) The centers of mass z(m) of each of these distributions of points sampled in state space are calculated. (D) The changes in response to driving signals I(m) yield the network topology.
Here, is the state vector of the entire network, denotes the temporal derivative of the variable xi(t), and ξi(t) represents noise with zero average. Systems of higher dimensional units, , d > 1, are discussed further below and in note S1.
Driving the system (Eq. 1) with signals
| (2) |
modifies its dynamics. For temporally constant or otherwise stationary driving signals, the temporal trajectories of the system after potential transients may exhibit collective dynamics that generate a defined statistics of points in state space. These states are recorded under M different driving conditions m ∈ {1, …, M} with signals and, for each experiment, generate an invariant density ρ(m) characteristic of the dynamics defined by Eq. 2. One simple quantity induced by such a density is given by its center of mass . If these data sample the invariant measure well (25), then the average may be approximated by the sample mean z(m) = < x(m)(t) > t∈T = |T|−1∑t∈Tx(m)(t) (see note S1 for a more detailed discussion). Here, T is the set of time points at which the data are recorded.
How can we reconstruct interaction networks from these data? Approximating Eq. 2 up to first order around z(0) yields
| (3) |
The exact conditions under which this approximation is justified are elaborated in note S1. By averaging over the recorded sample points T, we get
| (4) |
where are temporal averages of the stationary driving signals, and we set for all i. Last, substituting the expression for the undriven dynamics Fi(z(0)) by setting m = 0 in Eq. 4, we obtain (see note S1 for a detailed derivation)
| (5) |
where are the elements of the Jacobian . We take , because the centers of mass do not change in time if the recorded points sample the invariant density well (see note S2 for error estimates for sampling). This yields
| (6) |
where is the vector of averaged driving signals , is the matrix of response differences of the centers of mass, and is the respective row of the Jacobian matrix.
Evidently, the differences in the invariant density’s centers of mass are jointly determined by the driving vector and the network topology. We remark that the reconstruction problem decomposes over nodes in the network such that the set of incoming interactions to each node can be reconstructed independently. A sufficient number of driving-response experiments thus yield a set of linear equations (Eq. 6) for each node i, restricting the potential interaction networks estimated by J. Our goal of identifying which variables xj appear in Fi(x) is thus equivalent to finding those pairs (i, j) where Jij ≠ 0 such that unit j directly acts on i (and thus also those where no such direct interactions exist, Jij = 0). Notice that, because the Jacobian is evaluated at the center of mass of the unperturbed invariant density, the reconstruction approach is expected to recover the correct interactions if they consistently exist across the relevant fractions of state space, which include the observed driven dynamics and the unperturbed centers of mass. Here, we consider constant driving signals, , complemented by additive noise. If the number M of experiments is larger than the network size N, then reconstructing a given network becomes possible via a least squares solution to Eq. 6. However, in many experimental settings, the number of available experiments is relatively small. To overcome this limitation, we exploit compressed sensing theory by determining an L1-norm minimum solution Ĵi to Eq. 6 such that the number of experiments M can be much smaller than the network size N [see Methods; (26–29)]. Last, we rank the resulting absolute values and vary a threshold Jc to distinguish between existing and absent interactions. Hence, evaluation of inference performance is done in a parameter-free manner (note S3).
PERFORMANCE ON ARTIFICIAL AND REAL NETWORK TOPOLOGIES
Our strategy is successful in reconstructing the topology of physical interaction networks across a range of systems and collective dynamics (Figs. 2 to 5). To evaluate the quality of reconstruction, we initially consider a class of random networks where each unit is a Goodwin oscillator (30), a prototypical biological oscillator that characterizes various biological processes from circadian clocks to somitogenesis (see Methods for model description) (31–33). Under mild constraints, the strategy readily generalizes to systems of other, also higher-dimensional units (see below) and more complex dynamics (see note S1 for a complete derivation).
Fig. 2. Evaluation scheme illustrating robust reconstruction.
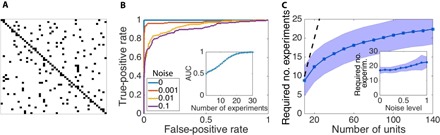
(A) Representative adjacency matrix indicating network connectivity as defined by present (black) and absent (white) links. (B) ROC curve obtained by varying a threshold Jc separating links classified as existing from those classified as absent (see note S3). The AUC increases with decreasing noise level, with perfect ranking of reconstructed links in the limit of noiseless dynamics. Inset: The quality of network reconstruction, as specified by the AUC, increases with the number of driving-response experiments. (C) The number of experiments required for high-quality reconstruction (here, AUC > 0.95) increases sublinearly (compared to the dotted line) with network size and (inset) changes only weakly with the noise level. Data are shown for random networks of (default size) N = 50 Goodwin oscillators with a regular incoming degree of 4, a default noise level of 0.5, a default number of experiments of 25, and a number of sampled time points of 100; shading indicates SD across ensembles of network realizations.
Fig. 5. Reconstruction of the circadian clock network in Drosophila.
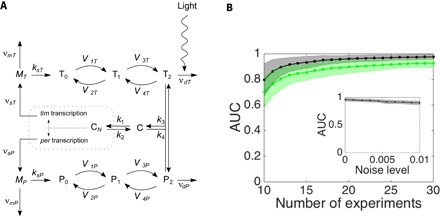
The quality of reconstruction (AUC) of the circadian clock (A) increases with the number of driving-response experiments, for both noiseless (gray curve) and noisy (green curve) dynamics (B), and changes only weakly with noise level (inset). Number of sampled time points, 300; default number of experiments, 20; noise level for noisy case, 0.01; shading indicates SD across ensembles of network realizations. (A) Modified with permission from Leloup and Goldbeter (39) (Fig. 1).
We first analyze one network realization (Fig. 2A) and collect individual results to network ensembles to obtain a robust quality analysis. Reconstruction quality is quantified by the area under the receiver operating characteristic (ROC) curve (AUC), a score typically between one-half (chance level) and one (perfect ranking of the reconstructed interactions) (see note S3). As expected, the reconstruction quality improves with the number of driving-response experiments (Fig. 2B). With compressed sensing methods [see (34–38) and Methods], the number of experiments required to obtain a given quality increases sublinearly with network size and continuously changes with increasing noise (Fig. 2C). Furthermore, the sparser a given network is, the lower the number of required experiments for robust reconstruction (see note S4). The invariants-based reconstruction strategy works analogously for different network topologies and different nodal coupling strengths, both homogeneous and heterogeneous (note S5) and for various dynamic processes; beyond networks of noisy oscillators, robust reconstruction is achieved for networks exhibiting qualitatively distinct dynamics, such as a biological network of transcription factor regulators close to a fixed point and a network of Rössler oscillators exhibiting chaotic dynamics (notes S6 and S7). Finally, the invariants-based strategy outperforms available standard reconstruction baselines, including measures of mutual information and correlation between the activity patterns of every two nodes in the network, partial correlation between the pairwise activity patterns (given the activity patterns of the other nodes in the network), and transfer entropy (note S8 and figs. S7 and S8).
In addition to disentangling existing and missing interactions within a network, the strategy can be extended to reveal finer topological features and to require only partial, lower dimensional dynamical information. First, separately ranking the reconstructed Jij values as deduced from Eq. 6 to find activating interactions above a certain threshold and negative ones below a second threshold yields two sets, one for consistently activating and one for consistently inhibiting interactions (Fig. 3). The separate identification of different types of interactions not only provides more information about their nature but also enhances the quality of reconstruction (Fig. 3 and note S9). Second, partial dynamical information, such as experimental data for networks of high-dimensional units limited to only one out of several dynamical variables for each unit, may suffice to reconstruct complete interaction networks (Fig. 4). See note S10 for a systematic evaluation of the effect of missing dynamical information on the quality of reconstruction. These results establish that reconstruction of network interactions is possible in a generic class of dynamical systems under a broad range of conditions.
Fig. 3. Revealing interaction types.
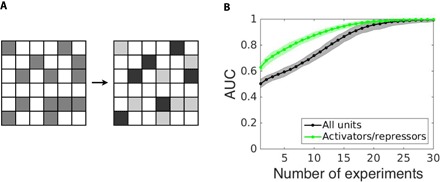
(A) Beyond distinguishing existing from missing interactions (schematically represented by the medium gray and white adjacency matrix), activating and inhibiting interactions may be separately detected (dark gray, light gray, and white matrix). (B) The reconstruction quality (AUC) benefits from the separate reconstruction of different types of interactions (green curve) compared to joint reconstruction of existing and missing interactions (gray curve), and increases with the number of driving-response experiments. Data are shown for random networks of N = 50 Goodwin oscillators with a regular incoming degree of 4 and a number of sampled time points of 100; shading indicates SD across ensembles of network realizations.
Fig. 4. Robust reconstruction from one-dimensional sampling of multidimensional unit dynamics.
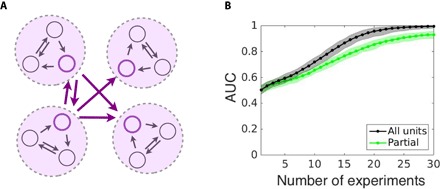
(A) Scheme illustrates three-dimensional units (encircled), coupled through one observed variable (colored), whereas the other two variables are unobserved (gray). (B) The reconstruction quality (AUC) stays robust and reduces only slightly for reconstruction based on partial, one-dimensional measurements (green curve) relative to reconstruction based on three-dimensional measurements (gray curve), and increases with the number of driving-response experiments. Data are shown for random networks of N = 50 Goodwin oscillators with a regular incoming degree of 4 and a number of sampled time points of 100; shading indicates SD across ensembles of network realizations.
Beyond generic model systems, the reconstruction strategy also yields promising results for specific biological settings. We demonstrate this on a genetic regulatory network characterizing the circadian clock in Drosophila (see note S11 for model description) (39). This clock coordinates the biological response to the day-night cycle. The quality of reconstruction of the circadian clock benefits from increasing the number of driving-response experiments and is robust to noisy signals (Fig. 5).
DISCUSSION
The presented theory relating responses of invariant measures and resulting observables to small driving signals enables us to reveal physical interaction networks from statistics of essentially arbitrary dynamical data that are sufficiently broadly sampled. The theory relies on the system being stationary or otherwise exhibiting an invariant measure that is sampled well by the data. It also relies on the option to drive the system with signals that are sufficiently small to still yield linear responses and, at the same time, sufficiently large to outweigh the noise and finite sampling influence. However, no fine-tuning of the driving signals is needed (see note S12). In addition, the method relies on changes in the stationary dynamics of the system following perturbations. Hence, it suits systems that do not exhibit an extreme form of perfect saturation or systems that adapt on fast time scales in response to external signals. If adaptation time scales are slow relative to the period during which the dynamic sampling is performed, then the method we suggest naturally becomes applicable again, because the system does not fully adapt and is effectively stationary during the measurement time. Thus, shifts in averages can be naturally observed following the same framework. The strategy is successful for generic mathematical model schemes and specific biological settings. Extended options include distinguishing between activating and inhibiting interactions as well as revealing the existence of interactions even if access is limited to only one out of several dynamical variables for individual units.
Because the strategy relies solely on the invariant density, state space points recorded may be used without known temporal order, may have been recorded at varying sampling intervals, and may even come from several experiments (performed under the same conditions). Furthermore, different units of the same system may, in principle, be recorded separately and at different times. All these options emerge because only statistical information (a rough estimate of an observable, such as center of mass) of the dynamical data is used. This strategy for revealing physical interactions from statistics thus constitutes a previously unknown intermediate approach between purely statistical methods for inferring effective connectivity (for example, correlations) and approaches inferring physical connectivity from high-resolution, time-ordered recordings of the full dynamics (5, 20, 27, 40–42).
The full range of features may be useful under various experimental conditions. For instance, many high-quality measurements of single-cell gene expression levels obtained simultaneously are available either at a system-wide level for a single time point [for example, Taniguchi et al. (43)] or for a few genes for several time points (44), yet there is no restriction in principle to measuring all genes in a sequence of different experiments. Together, these results offer a novel perspective on inferring physical and not only correlative (effective) connectivity of networked systems from statistically sampling their dynamics.
METHODS
Goodwin oscillators
To evaluate the quality of the reconstruction approach in a controlled setting, we considered networks of N prototypical Goodwin oscillators i ∈ {1, …, N} (30), each with three variables—xi, yi, and zi—evolving in time according to , , and . In addition, f(zi) = vo/[1 + (zi/Km)n] constitutes a local nonlinearity and g(yi, yj) = yi − yj constitutes the diffusive interactions. In direct numerical simulations, the parameters are ai = bi = ci = 0.4, vo = Km = 1, and n = 17.
Network topologies
For generic evaluations (Figs. 2 to 4), we used random networks of N = 50 Goodwin oscillators with a regular incoming degree of 4. We fixed the degree for Figs. 2 to 4 and varied it to compare networks of different degrees in fig. S2. In addition, we used an Erdős Rényi random network of N = 50 genetic regulators, with edge probability p = 0.01, where a genetic regulator is chosen to be either an activator or a repressor with equal probability (fig. S3). For the topology of a real biological system, we considered the circadian clock network of Drosophila [(39, 45); see note S11] (Fig. 5).
Compressed sensing
The framework of compressed sensing (34–38) enables us to reconstruct a high-dimensional sparse signal based on linear measurements, where the number of measurements is small relative to the dimension of the signal. In our context, the goal was to reconstruct the network physical connections for a given unit i, by solving the linear set of equations (Eq. 6) for averaged driving signals , driving-response matrix , and M ≪ N. Given that Ji is sufficiently sparse and Δz fulfills certain conditions, as elaborated in note S13, this problem can be posed as an L1-norm convex optimization problem with guarantees for a robust and stable solution, and solved using standard software such as CVX, a MATLAB package for specifying and solving convex problems (46, 47).
Supplementary Material
Acknowledgments
We thank I. Kanter, C. Kirst, B. Lünsmann, and M. Peer for valuable discussions and L. J. Deutsch for help with figure preparation. M.N. is grateful to the Azrieli Foundation for the award of an Azrieli Fellowship. Funding: This study was supported by the Federal Ministry of Education and Research (BMBF; grant 03SF0472E) and the Max Planck Society (to M.T.). Author contributions: All authors conceived the research and contributed materials and analysis tools. M.N. and M.T. designed the research. All authors worked out the theory. M.N. and J.C. developed the algorithms and carried out the numerical experiments. All authors analyzed the data, interpreted the results, and wrote the manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/2/e1600396/DC1
note S1. Details of the derivation of invariant-based reconstruction.
note S2. Error estimates for observables from sampled invariant density.
note S3. Reconstruction evaluation.
note S4. Moderate influence of link density.
note S5. Reconstructing homogeneous and heterogeneous networks.
note S6. Reconstruction of systems near fixed points.
note S7. Reconstruction of chaotic systems.
note S8. Performance compared with available standard baselines.
note S9. Distinguishing activating from inhibiting interactions.
note S10. The effect of missing information.
note S11. Model descriptions.
note S12. The effect of various driving conditions on reconstruction quality.
note S13. Compressed sensing.
fig. S1. Approximating the center of mass of invariant densities by the sample mean.
fig. S2. Sparser networks require fewer experiments for robust reconstruction.
fig. S3. Reconstruction is robust across network topologies.
fig. S4. The quality of reconstruction increases with the number of experiments for a network of genetic regulators.
fig. S5. Reconstruction of a network of Rössler oscillators exhibiting chaotic dynamics.
fig. S6. Comparison of reconstruction quality across different approaches.
fig. S7. Comparison of reconstruction quality against transfer entropy.
fig. S8. Separate reconstruction of activating and inhibiting interactions enhances the quality of reconstruction.
fig. S9. Quality of reconstruction (AUC score) decreases gradually with the fraction of hidden units in the network.
fig. S10. Quality of reconstruction increases as driving signals overcome noise and finite sampling effects.
REFERENCES AND NOTES
- 1.Strogatz S. H., Exploring complex networks. Nature 410, 268–276 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Karlebach G., Shamir R., Modelling and analysis of gene regulatory networks. Nat. Rev. Mol. Cell Biol. 9, 770–780 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Erwin D. H., Davidson E. H., The evolution of hierarchical gene regulatory networks. Nat. Rev. Genet. 10, 141–148 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Davidson E. H., Emerging properties of animal gene regulatory networks. Nature 468, 911–920 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gardner T. S., di Bernardo D., Lorenz D., Collins J. J., Inferring genetic networks and identifying compound mode of action via expression profiling. Science 301, 102 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Takemura S.-y., Bharioke A., Lu Z., Nern A., Vitaladevuni S., Rivlin P. K., Katz W. T., Olbris D. J., Plaza S. M., Winston P., Zhao T., Horne J. A., Fetter R. D., Takemura S., Blazek K., Chang L.-A., Ogundeyi O., Saunders M. A., Shapiro V., Sigmund C., Rubin G. M., Scheffer L. K., Meinertzhagen I. A., Chklovskii D. B., A visual motion detection circuit suggested by Drosophila connectomics. Nature 500, 175–181 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sporns O., Tononi G., Kötter R., The human connectome: A structural description of the human brain. PLOS Comput. Biol. 1, e42 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schneidman E., Berry M. J., Segev R., Bialek W., Weak pairwise correlations imply strongly correlated network states in a neural population. Nature 440, 1007–1012 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.B. Drossel, A. J. McKane, Modelling food webs, in Handbook of Graphs and Networks: From the Genome to the Internet, S. Bornholdt, H. G. Schuster, Eds. (Wiley-VCH, 2003). [Google Scholar]
- 10.Filatrella G., Nielsen A. H., Pedersen N. F., Analysis of a power grid using a Kuramoto-like model. Eur. Phys. J. B 61, 485–491 (2008). [Google Scholar]
- 11.Rohden M., Sorge A., Timme M., Witthaut D., Self-organized synchronization in decentralized power grids. Phys. Rev. Lett. 109, 064101 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Witthaut D., Timme M., Braess’s paradox in oscillator networks, desynchronization and power outage. New J. Phys. 14, 083036 (2012). [Google Scholar]
- 13.Menck P. J., Heitzig J., Kurths J., Schellnhuber H. J., How dead ends undermine power grid stability. Nat. Commun. 5, 3969 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Carvalho R., Buzna L., Just W., Helbing D., Arrowsmith D. K., Fair sharing of resources in a supply network with constraints. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 85, 046101 (2012). [DOI] [PubMed] [Google Scholar]
- 15.Rubido N., Grebogi C., Baptista M. S., Resiliently evolving supply-demand networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 89, 012801 (2014). [DOI] [PubMed] [Google Scholar]
- 16.B. Lünsmann, “Reconstruction of physical interactions in stationary stochastic network dynamics,” thesis, University of Göttingen, Germany (2015). [Google Scholar]
- 17.Prinz A. A., Bucher D., Marder E., Similar network activity from disparate circuit parameters. Nat. Neurosci. 7, 1345–1352 (2004). [DOI] [PubMed] [Google Scholar]
- 18.Kirst C., Timme M., Battaglia D., Dynamic information routing in complex networks. Nat. Commun. 7, 11061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Timme M., Wolf F., Geisel T., Coexistence of regular and irregular dynamics in complex networks of pulse-coupled oscillators. Phys. Rev. Lett. 89, 258701 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Timme M., Casadiego J., Revealing networks from dynamics: An introduction. J. Phys. A Math. Theor. 47, 343001 (2014). [Google Scholar]
- 21.Schreiber T., Measuring information transfer. Phys. Rev. Lett. 85, 461 (2000). [DOI] [PubMed] [Google Scholar]
- 22.Tajima S., Yanagawa T., Fujii N., Toyoizumi T., Untangling brain-wide dynamics in consciousness by cross-embedding. PLOS Comput. Biol. 11, e1004537 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shandilya S. G., Timme M., Inferring network topology from complex dynamics. New J. Phys. 13, 013004 (2011). [Google Scholar]
- 24.I. Dzafic, P. Mohapatra, Impedance based fault location for weakly meshed distribution networks, in 2011 IEEE PES Innovative Smart Grid Technologies (IEEE, 2011), January 2011, pp. 1–6.
- 25.E. Ott, Chaos in Dynamical Systems (Cambridge Univ. Press, 2002), 478 pp. [Google Scholar]
- 26.S. Boyd, L. Vandenberghe, Convex Optimization (Cambridge Univ. Press, 2004). [Google Scholar]
- 27.Timme M., Revealing network connectivity from response dynamics. Phys. Rev. Lett. 98, 224101 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Napoletani D., Sauer T. D., Reconstructing the topology of sparsely connected dynamical networks. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 77, 026103 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Wang W.-X., Yang R., Lai Y.-C., Kovanis V., Grebogi C., Predicting catastrophes in nonlinear dynamical systems by compressive sensing. Phys. Rev. Lett. 106, 154101 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goodwin B. C., Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 3, 425–438 (1965). [DOI] [PubMed] [Google Scholar]
- 31.Ruoff P., Vinsjevik M., Monnerjahn C., Rensing L., The Goodwin oscillator: On the importance of degradation reactions in the circadian clock. J. Biol. Rhythms 14, 469–479 (1999). [DOI] [PubMed] [Google Scholar]
- 32.François P., Despierre N., Siggia E. D., Adaptive temperature compensation in circadian oscillations. PLOS Comput. Biol. 8, e1002585 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeiser S., Müller J., Liebscher V., Modeling the Hes1 oscillator. J. Comput. Biol. 14, 984–1000 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Candès E. J., Wakin M. B., An introduction to compressive sampling. IEEE Signal Process. Mag. 25, 21–30 (2008). [Google Scholar]
- 35.Candes E. J., Tao T., Near-optimal signal recovery from random projections: Universal encoding strategies? IEEE Trans. Inf. Theory 52, 5406–5425 (2006). [Google Scholar]
- 36.Candès E. J., Romberg J., Tao T., Robust uncertainty principles: Exact signal reconstruction from highly incomplete frequency information. IEEE Trans. Inf. Theory 52, 489–509 (2006). [Google Scholar]
- 37.Candes E. J., Romberg J. K., Tao T., Stable signal recovery from incomplete and inaccurate measurements. Commun. Pure Appl. Math. 59, 1207–1223 (2006). [Google Scholar]
- 38.Donoho D. L., Compressed sensing. IEEE Trans. Inf. Theory 52, 1289–1306 (2006). [Google Scholar]
- 39.Leloup J.-C., Goldbeter A., A model for circadian rhythms in Drosophila incorporating the formation of a complex between the PER and TIM proteins. J. Biol. Rhythms 13, 70–87 (1998). [DOI] [PubMed] [Google Scholar]
- 40.Yu D., Parlitz U., Driving a network to steady states reveals its cooperative architecture. Europhys. Lett. 81, 48007 (2008). [Google Scholar]
- 41.Yu D., Righero M., Kocarev L., Estimating topology of networks. Phys. Rev. Lett. 97, 188701 (2006). [DOI] [PubMed] [Google Scholar]
- 42.Tomovski I., Kocarev L., Network topology inference from infection statistics. Phys. A 436, 272–285 (2015). [Google Scholar]
- 43.Taniguchi Y., Choi P. J., Li G.-W., Chen H., Babu M., Hearn J., Emili A., Xie X. S., Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science 329, 533–538 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Longo D., Hasty J., Dynamics of single-cell gene expression. Mol. Syst. Biol. 2, 64 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leloup J.-C., Goldbeter A., Chaos and birhythmicity in a model for circadian oscillations of the PER and TIM proteins in Drosophila. J. Theor. Biol. 198, 445–459 (1999). [DOI] [PubMed] [Google Scholar]
- 46.M. Grant, S. Boyd, CVX: Matlab Software for Disciplined Convex Programming, Version 2.1 (CVX Research, 2014); http://cvxr.com/cvx.
- 47.M. Grant, S. Boyd, Graph implementations for non-smooth convex programs, Recent Advances in Learning and Control, Lecture Notes in Control and Information Sciences, V. Blondel, S. Boyd, H. Kimura, Eds. (Springer-Verlag Limited, 2008), pp. 95–110. [Google Scholar]
- 48.Rössler O. E., An equation for continuous chaos. Phys. Lett. A 57, 397–398 (1976). [Google Scholar]
- 49.Natarajan B. K., Sparse approximate solutions to linear systems. SIAM J. Comput. 24, 227–234 (1995). [Google Scholar]
- 50.Cai T. T., Wang L., Orthogonal matching pursuit for sparse signal recovery with noise. IEEE Trans. Inf. Theory 57, 4680–4688 (2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/3/2/e1600396/DC1
note S1. Details of the derivation of invariant-based reconstruction.
note S2. Error estimates for observables from sampled invariant density.
note S3. Reconstruction evaluation.
note S4. Moderate influence of link density.
note S5. Reconstructing homogeneous and heterogeneous networks.
note S6. Reconstruction of systems near fixed points.
note S7. Reconstruction of chaotic systems.
note S8. Performance compared with available standard baselines.
note S9. Distinguishing activating from inhibiting interactions.
note S10. The effect of missing information.
note S11. Model descriptions.
note S12. The effect of various driving conditions on reconstruction quality.
note S13. Compressed sensing.
fig. S1. Approximating the center of mass of invariant densities by the sample mean.
fig. S2. Sparser networks require fewer experiments for robust reconstruction.
fig. S3. Reconstruction is robust across network topologies.
fig. S4. The quality of reconstruction increases with the number of experiments for a network of genetic regulators.
fig. S5. Reconstruction of a network of Rössler oscillators exhibiting chaotic dynamics.
fig. S6. Comparison of reconstruction quality across different approaches.
fig. S7. Comparison of reconstruction quality against transfer entropy.
fig. S8. Separate reconstruction of activating and inhibiting interactions enhances the quality of reconstruction.
fig. S9. Quality of reconstruction (AUC score) decreases gradually with the fraction of hidden units in the network.
fig. S10. Quality of reconstruction increases as driving signals overcome noise and finite sampling effects.


