Abstract
Environmental enteric dysfunction (EED) is often measured with a dual sugar absorption test and implicated as a causative factor in childhood stunting. Disturbances in the gut microbiota are hypothesized to be a mechanism by which EED is exacerbated, although this supposition lacks support. We performed 16S ribosomal RNA gene sequencing of fecal samples from 81 rural Malawian children with varying degrees of EED to determine which bacterial taxa were associated with EED. At the phyla level, Proteobacteria abundance is reduced with severe EED. Among bacterial genera, Megasphaera, Mitsuokella, and Sutterella were higher in EED and Succinivibrio, Klebsiella, and Clostridium_XI were lower in EED. Bacterial diversity did not vary with the extent of EED. Though EED is a condition that is typically believed to affect the proximal small bowel, and our focus was on stool, our data do suggest that there are intraluminal microbial differences that reflect, or plausibly lead to, EED.
Environmental enteric dysfunction (EED), characterized by small bowel villous atrophy and inflammation, often develops early in childhood and is believed to impact the physical, immunological, and cognitive development of children.1 EED is associated with unsanitary living conditions, asymptomatic presence of intestinal pathogens, and malnutrition in developing countries.2 At present the dual sugar absorption test, one form of which is the lactulose:mannitol (L:M) test, is the most widely used method to detect EED.3
Disturbances in the gut microbiota may cause or be a critical cofactor in the pathogenesis of EED, but published data have not yet supported this notion. Here, we used fecal 16S rRNA gene sequencing to identify phylogenetic groupings of bacteria to detect dysbiosis associated with EED in rural African children.
Eighty-one 12- to 23-month-old healthy Malawian children (47 boys and 34 girls) selected from the rural areas of Machinga and Nsanje districts enrolled in a legume intervention trial participated in the study (Table 1).4 All samples were collected before distribution of any legume supplements. No child at entry was reported to have had diarrhea in the previous 7 days. The children lived in subsistence farming families in unelectrified mud huts often without access to clean water. The children consumed a plant-based diet, predominantly composed of maize with animal source foods no more frequent that twice per month. The study was approved by the Institutional Review Board from Washington University School of Medicine in St. Louis, and the University of Malawi College of Medicine Research and Ethics Committee.
Table 1.
Characteristics of the Malawian children studied*
| Characteristic | No EED L:M < 0.15 (N = 8) | Moderate EED 0.15 ≤ L:M ≤ 0.45 (N = 49) | Severe EED L:M > 0.45 (N = 24) | P value† |
|---|---|---|---|---|
| Age, months | 20.5 ± 3.7 | 20.9 ± 2.9 | 18.7 ± 4.6 | 0.050‡ |
| Female, n (%) | 3 (38) | 21 (43) | 10 (42) | 0.960 |
| Mid-upper arm circumference, cm | 14.1 ± 1.4 | 14.9 ± 0.9 | 14.6 ± 1.0 | 0.082 |
| Height-for-age z score | −1.2 ± 1.4 | −1.1 ± 1.2 | −1.6 ± 0.9 | 0.217 |
| Stunted,§ n (%) | 2 (25) | 13 (27) | 7 (29) | 0.962 |
| Caregiver is mother, n (%) | 8 (100) | 46 (94) | 24 (100) | 0.361 |
| Father is alive, n (%) | 8 (100) | 49 (100) | 23 (96) | 0.300 |
| Siblings | 2.6 ± 2.1 | 2.7 ± 1.8 | 2.8 ± 1.8 | 0.958 |
| Individuals that sleep in the same room as child | 3.5 ± 0.5 | 3.3 ± 0.6 | 3.3 ± 0.6 | 0.664 |
| Home with a metal roof, n (%) | 1 (13) | 12 (24) | 3 (13) | 0.416 |
| Family owns bicycle, n (%) | 4 (50) | 32 (65) | 11 (46) | 0.254 |
| Animals sleep in house, n (%) | 3 (38) | 14 (29) | 6 (25) | 0.793 |
| Water from a clean source, n (%) | 5 (63) | 30 (61) | 20 (83) | 0.155 |
| Uses a pit latrine, n (%) | 3 (38) | 6 (12) | 2 (8) | 0.103 |
EED = environmental enteric dysfunction; L:M = lactulose:mannitol.
Data are expressed as: means ± standard deviations for continuous measures or counts (percentages) for categorical measures.
For continuous characteristics, P value is calculated with one-way analysis of variance with Tukey's correction.
Main effects and interactions were considered significant at P values < 0.05.
Height-for-Age z score < −2.
An L:M test was carefully conducted and stools during L:M test were collected from each child.5 Each stool sample was snap frozen in liquid nitrogen within minutes of collection and frozen (−80°C) until processing. L:M was calculated as the ratio of the lactulose to mannitol concentrations in the urine by high-performance liquid chromatography at Baylor College of Medicine, Texas. Three categories of EED were designated: no EED (L:M < 0.15), moderate EED (0.15 ≤ L:M ≤ 0.45), and severe EED (L:M > 0.45), as previously described.6
Stool processing was performed as previously described.7 DNA was extracted by using the NucliSENS easyMAG “SpecificA” (BioMerieux, Durham, NC) following manufacturer's instructions. The V1–V2 hypervariable regions of the 16S rRNA gene were amplified by polymerase chain reaction (PCR) using the primers V1_V2_F “AGAGTTTGATCMTGGCTCAG” and V1_V2_R “CTGCTGCCTYCCGTA.” The PCR products were purified and sequenced using the MiSeq Genome Sequencer (Illumina, San Diego, CA).
Sample sequences were binned based on Illumina index sequences. Paired-end fastq files were generated, and the primers were removed from the 3′ end of the sequence using Trimmomatic and Flexbar allowing one mismatch in addition to primer degeneracies. Low-quality bases were removed using Mothur software with the parameter trim.seqs(qaverage = 35).8 Paired reads were assembled using FLASH and then assembled sequences less than 200 bases were removed and taxonomic calls were generated for each assembly using the Ribosomal Database Project (RDP) Naive Bayesian Classifier version 2.5 with training set 9.9,10 Chimeric sequences were identified and removed using ChimeraSlayer with default parameters.9
To analyze the diversity at various taxonomic levels, RDP-generated taxonomic calls were analyzed using in-house Perl script to generate sample versus taxonomy matrices, where a 0.5 confidence level was required to accept a call at each taxonomic level and reads with < 0.5 confidence at a level, for example, genus, were considered unclassified at the family level. Because different samples yielded different sequence depth, read subsampling, or rarefaction, at a depth of 33,000 reads was done using the Vegan package in R.11 The V1–V2 reads were assigned to nine phyla and 272 genera. Fold difference in expression between classifications of EED were determined using reads per kilobase of exon model per million mapped reads (Partek Genomics Suite; Partek, St. Louis, MO).
Bacterial genera were considered to be associated with EED if the following four criteria were met: 1) Spearman's correlation coefficient between genera abundance and L:M was significant at a level of P < 0.05; 2) the abundance showed a decrease or increase with increasing EED severity using a one-way, linear trend analysis of variance (ANOVA) test; 3) there was a 5-fold difference in abundance between children with no EED and severe EED; and 4) linear discriminate analyses effect size (LefSe) showed a significant difference (Supplemental Table 1).
Among the 81 children studied, eight (10%), 49 (60%), and 24 (30%) had no, moderate, and severe EED, respectively (Table 1). The baseline categorization of EED was associated with change in height-for-age z score in the subsequent 3 months (P = 0.01, Tukey–Kramer multiple comparison test).
At the phyla level, there was a significant reduction in Proteobacteria abundance: 9.19 ± 5.61, 4.22 ± 5.50, and 4.20 ± 4.59 between no EED and moderate EED, and severe EED, respectively (ANOVA P value = 0.045); no other significant differences were found (Supplemental Figure 1).
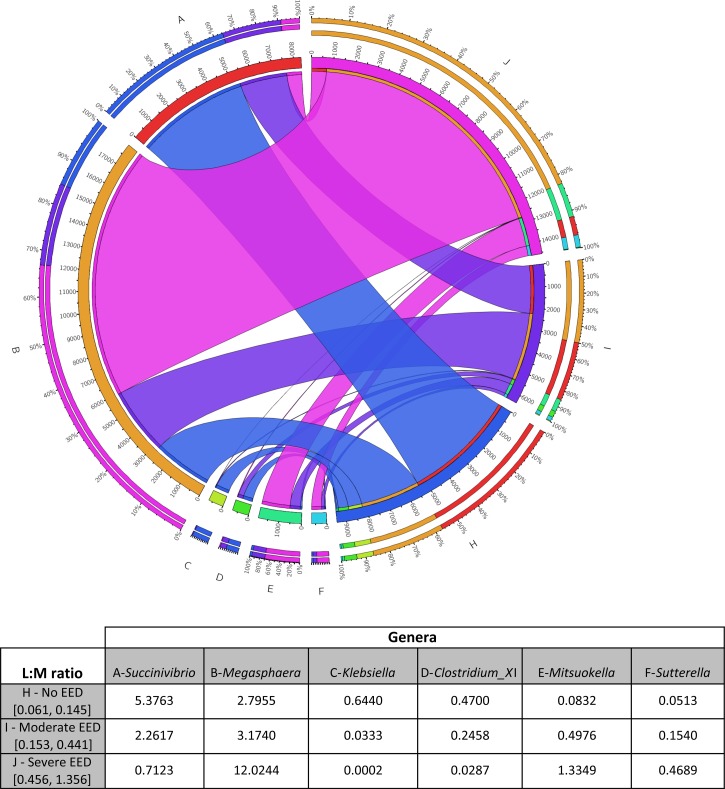
Megasphaera, Mitsuokella, and Sutterella were more prevalent in EED versus no EED, and Succinivibrio, Klebsiella, and Clostridium_XI were less prevalent in EED versus no EED (Figure 1 ).
Figure 1.
Circos plot and table showing associations between six genera abundance (% of total bacteria) and each category of lactulose:mannitol (L:M). The circus plot is a circular representation of the association of the different genera labeled A–G and the environmental enteric dysfunction (EED) status (no EED, moderate EED, and severe EED), labeled H–J. The inner circle shows the percentage of genera abundance multiplied by 1,000, the outer circle the relative abundance of the genera among the six genera. The lines indicate the association of each genera with the EED status: no EED is blue, moderate EED is purple, and severe EED is pink.
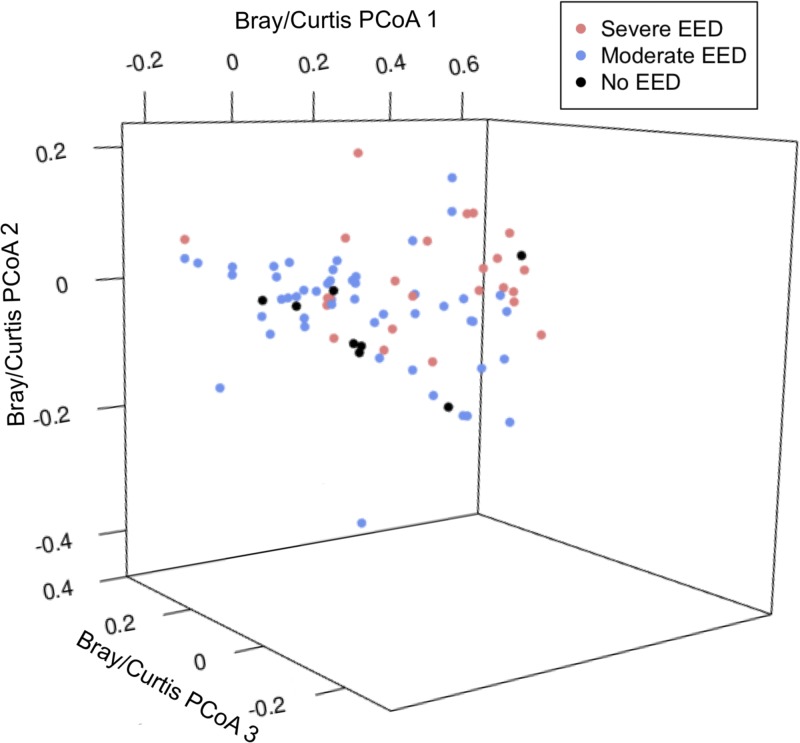
The microbiota diversity at the phyla and genera levels as assessed by Bray–Curtis dissimilarity analyses shows that the fecal microbiota communities between EED groups were significantly different based on multivariate permutation testing using Adonis from the R library “Vegan” (100 permutations, P < 0.001) (Figure 2 ).11 The Shannon H diversity among no, moderate, and severe EED at the genera level were 1.60 ± 0.44, 1.64 ± 0.48, and 1.62 ± 0.40 (as mean ± standard deviation), respectively (ANOVA P value = 0.9655).
Figure 2.
Microbial community variation represented by principal coordinates plot (PCoA, genus-level Brady–Curtis dissimilarity). There are no significant differences in the community structures of children based on their environmental enteric dysfunction (EED) status (P > 0.05).
At the genus level, two genera were present only in no EED individuals, 51 genera only in moderate EED, and 28 in severe EED; 138 genera were present in all individuals in different proportions depending on the individual EED status (Supplemental Figure 2).
This study was limited by the use of fecal samples for 16S rRNA gene sequencing, which largely reflect the bacteria in the colon, whereas EED is a disease of the proximal bowel, where bacteria concentrations are estimated to be 109-fold less abundant (duodenum, 103; jejunum, 104; ileum, 107; and colon, 1012 bacteria/g).12 Bacteria of potential interest in EED may well be present in small quantities, but difficult to detect. We attempted to control for this limitation by looking for differences in bacterial relative abundance on the basis of the L:M test. A much more informative and direct way to look for dysbiosis of the small intestine would require direct sampling using endoscopy, which is not justifiable in this population of asymptomatic children. Because of this limitation, the species that we identified cannot be considered to be causal.
Considering the three genera that were decreased with EED, Succinivibrio is a fiber-degrading bacterium, typical in rural African populations with diets high in fiber and complex carbohydrates.13 Succinivibrio has been associated with altered carbohydrate metabolism and niche disruptions in mucosal interfaces induced by parasitic infections. Klebsiella is normally found in the human intestine and feces and usually does not cause enteric disease. Clostridium_XI deconjugates primary bile acids in the distal ileum.14 Recent work demonstrated different amounts and conjugation patterns of serum bile acids in EED.15 This observation might be related to changes in Clostridium_XI.
Among the three genera that were higher in EED, Megasphaera ferments carbohydrates and produces fatty acids. This organism is involved in lactic acid catabolism and amino acid deamination.16 Adults with celiac disease, another inflammatory condition of the upper small bowel, have higher proportions of Megasphaera in their fecal microbiota.17 Individuals with asymptomatic human immunodeficiency virus enteritis also carry larger fractions of Megasphaera in their feces.18 Mitsuokella is a strict anaerobic bacterium that inhabits the intestinal tract. Mitsuokella metabolites produce butyrate, which can have an anti-inflammatory effect and also inhibit intestinal epithelial proliferation. Mitsuokella also inhibits Salmonella growth.19 The increase in abundance of Mitsuokella in EED might limit the inflammation of EED, but the operative mechanisms could be complex. Sutterella has associated with Crohn's disease, and with abnormal intestinal permeability.20 It is interesting that both Megasphaera and Sutterella have been associated with other clinical conditions in which small bowel inflammation is prominent, so perhaps these bacteria, overall, evoke an inflammatory response.
EED is not associated with a profound fecal dysbiosis, but six genera were identified as having significantly different abundances in EED. The significance of this may be elucidated in the future by investigation of the duodenal and jejunal microbiota isolated from endoscopy samples.
Supplementary Material
Supplemental figures and tables.
Footnotes
Financial support: This study was supported by the Feed the Future Program, USAID, and the Children's Discovery Institute of Washington University and St. Louis Children's Hospital.
Conflict of interest: Phillip I. Tarr is on the Scientific Advisory Board of MediBeacon, and is the coinventor of a novel technology to measure gut permeability.
Authors' addresses: M. Isabel Ordiz, Kevin Stephenson, Sophia Agapova, Kristine M. Wylie, Indi Trehan, Phillip I. Tarr, and Mark J. Manary, Department of Pediatrics, Washington University School of Medicine, St. Louis, MO, E-mails: ordiz_i@wustl.edu, kbstephe@gmail.com, shtepaz@gmail.com, kwylie@wustl.edu, itrehan@wustl.edu, tarr@wustl.edu, and manary@wustl.edu. John Martin, The McDonnell Genome Institute, St. Louis, MO, E-mail: jmartin@wustl.edu. Ken Maleta, Department of Community Health, College of Medicine, University of Malawi, Malawi, Africa, E-mail: ken.maleta@gmail.com.
References
- 1.Keusch GT, Denno DM, Black RE, Duggan C, Guerrant RL, Lavery JV, Nataro JP, Rosenberg IH, Ryan ET, Tarr PI, Ward H, Bhutta ZA, Coovadia H, Lima A, Ramakrishna B, Zaidi AK, Hay Burgess DC, Brewer T. Environmental enteric dysfunction: pathogenesis, diagnosis, and clinical consequences. Clin Infect Dis. 2014;59((Suppl 4)):S207–S212. doi: 10.1093/cid/ciu485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prendergast AJ, Kelly P. Interactions between intestinal pathogens, enteropathy and malnutrition in developing countries. Curr Opin Infect Dis. 2016;29:229–236. doi: 10.1097/QCO.0000000000000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Denno DM, VanBuskirk K, Nelson ZC, Musser CA, Hay Burgess DC, Tarr PI. Use of the lactulose to mannitol ratio to evaluate childhood environmental enteric dysfunction: a systematic review. Clin Infect Dis. 2014;59((Suppl 4)):S213–S219. doi: 10.1093/cid/ciu541. [DOI] [PubMed] [Google Scholar]
- 4.Trehan I, Benzoni NS, Wang AZ, Bollinger LB, Ngoma TN, Chimimba UK, Stephenson KB, Agapova SE, Maleta KM, Manary MJ. Common beans and cowpeas as complementary foods to reduce environmental enteric dysfunction and stunting in Malawian children: study protocol for two randomized controlled trials. Trials. 2015;16:520. doi: 10.1186/s13063-015-1027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stauber J, Shaikh N, Ordiz MI, Tarr PI, Manary MJ. Droplet digital PCR quantifies host inflammatory transcripts in feces reliably and reproducibly. Cell Immunol. 2016;303:43–49. doi: 10.1016/j.cellimm.2016.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ordiz MI, Shaikh N, Trehan I, Maleta K, Stauber J, Shulman R, Devaraj S, Tarr PI, Manary MJ. Environmental enteric dysfunction is associated with poor linear growth and can be identified by host fecal mRNAs. J Pediatr Gastroenterol Nutr. 2016;63:453–459. doi: 10.1097/MPG.0000000000001315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ordiz MI, May TD, Mihindukulasuriya K, Martin J, Crowley J, Tarr PI, Ryan K, Mortimer E, Gopalsamy G, Maleta K, Mitreva M, Young G, Manary MJ. The effect of dietary resistant starch type 2 on the microbiota and markers of gut inflammation in rural Malawi children. Microbiome. 2015;3:37. doi: 10.1186/s40168-015-0102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schloss PD. A high-throughput DNA sequence aligner for microbial ecology studies. PLoS One. 2009;4:e8230. doi: 10.1371/journal.pone.0008230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G, Ciulla D, Tabbaa D, Highlander SK, Sodergren E, Methé B, DeSantis TZ, Human Microbiome Consortium. Petrosino JF, Knight R, Birren BW. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011;21:494–504. doi: 10.1101/gr.112730.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM. The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oksanen J, Blanchet FG, Kindt R, Legendre P, O'Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Wagner H. Vegan: Community Ecology Package 2016. 2016. https://CRAN.R-project.org/package=vegan Available at.
- 12.Verdu EF, Galipeau HJ, Jabri B. Novel players in coeliac disease pathogenesis: role of the gut microbiota. Nat Rev Gastroenterol Hepatol. 2015;12:497–506. doi: 10.1038/nrgastro.2015.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graf D, Di Cagno R, Fåk F, Flint HJ, Nyman M, Saarela M, Watzl B. Contribution of diet to the composition of the human gut microbiota. Microb Ecol Health Dis. 2015;26:26164. doi: 10.3402/mehd.v26.26164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 15.Semba RD, Gonzalez-Freire M, Moaddel R, Trehan I, Maleta K, Khadeer M, Ordiz MI, Ferrucci L, Manary MJ. J Pediatr Gastroenterol Nutr. 2016. Environmental enteric dysfunction is associated with altered bile acid metabolism. doi:10.1097/MPG.0000000000001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marounek M, Fliegrova K, Bartos S. Metabolism and some characteristics of ruminal strains of Megasphaera elsdenii. Appl Environ Microbiol. 1989;55:1570–1573. doi: 10.1128/aem.55.6.1570-1573.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacklin P, Kaukinen K, Tuovinen E, Collin P, Lindfors K, Partanen J, Mäki M, Mättö J. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm Bowel Dis. 2013;19:934–941. doi: 10.1097/MIB.0b013e31828029a9. [DOI] [PubMed] [Google Scholar]
- 18.Vázquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrús ML, Madrid N, Vallejo A, Sainz T, Martínez-Botas J, Ferrando-Martínez S, Vera M, Dronda F, Leal M, Del Romero J, Moreno S, Estrada V, Gosalbes MJ, Moya A. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal Immunol. 2015;8:760–772. doi: 10.1038/mi.2014.107. [DOI] [PubMed] [Google Scholar]
- 19.Levine UY, Bearson SMD, Stanton TB. Mitsuokella jalaludinii inhibits growth of Salmonella enterica serovar Typhimurium. Veterinarian Microbiology. 2012;159:115–122. doi: 10.1016/j.vetmic.2012.03.027. [DOI] [PubMed] [Google Scholar]
- 20.Mangin I, Bonnet R, Seksik P, Rigottier-Gois L, Sutren M, Bouhnik Y, Neut C, Collins MD, Colombel JF, Marteau P, Doré J. Molecular inventory of faecal microflora in patients with Crohn's disease. FEMS Microbiol Ecol. 2004;50:25–36. doi: 10.1016/j.femsec.2004.05.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental figures and tables.