Abstract
Androgen receptor (AR) transcriptional activity depends on interactions between the AR NH2-terminal region and transcriptional coregulators. A yeast two-hybrid screen of a human testis library using predicted α-helical NH2-terminal fragment AR-(370–420) as bait identified suppressor of variegation 3–9 homolog 2 (SUV39H2) histone methyltransferase as an AR interacting protein. SUV39H2 interaction with AR and the AR coregulator, melanoma antigen-A11 (MAGE-A11), was verified in two-hybrid, in vitro glutathione S-transferase affinity matrix and coimmunoprecipitation assays. Fluorescent immunocytochemistry colocalized SUV39H2 and AR in the cytoplasm without androgen, in the nucleus with androgen, and with MAGE-A11 in the nucleus independent of androgen. Chromatin immunoprecipitation using antibodies raised against SUV39H2 demonstrated androgen-dependent recruitment of AR and SUV39H2 to the androgen-responsive upstream enhancer of the prostate-specific antigen gene. SUV39H2 functioned cooperatively with MAGE-A11 to increase androgen-dependent AR transcriptional activity. SUV39H2 histone methyltransferase is an AR coactivator that increases androgen-dependent transcriptional activity through interactions with AR and MAGE-A11.
Keywords: androgen receptor, SUV39H2, histone methyltransferase, melanoma antigen-A11, MAGE-A11
INTRODUCTION
Androgen receptor (AR) transcriptional activity depends on high affinity binding of testosterone or dihydrotestosterone (DHT). The cascade of molecular events includes release of heat shock proteins, formation of the AR NH2- and carboxyl-terminal (N/C) interaction, AR dimerization and nuclear translocation, interaction with coregulatory proteins and binding to androgen response element DNA. The predominant androgen-dependent AR transactivation domains are located in the NH2-terminal region and carboxyl-terminal ligand binding domain (Simental et al., 1991; He et al., 1999). Human AR NH2-terminal amino acid residues 142–337 are considered the dominant transactivation domain in mammalian cells (Simental et al., 1991).
AR transcriptional activity is regulated through post-translational modification by enzymes that result in AR acetylation (Fu et al., 2000; Gaughan et al., 2002), methylation (Gaughan et al., 2011; Ko et al., 2011; Coffey et al., 2013), ubiquitination (Gaughan et al., 2005), deubiquitination (Burska et al., 2013) and sumoylation (Poukka et al., 2000). AR coactivators with acetyltransferase activity include p300 and the p160 steroid receptor coactivator family (Heemers and Tindall, 2007). Other AR coregulators include CARM-1 (coactivator-associated arginine methyltransferase-1) with methyltransferase activity (Chen et al., 1999) and LSD1 (lysine-specific demethylase 1) with demethylase activity (Metzger et al., 2005). The sites of AR modification by most of these enzymes are within the AR NH2-terminal region.
The intrinsically disordered AR NH2-terminal region is interrupted by short predicted α-helical regions that serve as interaction sites for coregulatory proteins (He et al., 2004b). Notable among these is the AR NH2-terminal FXXLF motif sequence 23FQNLF27 that is highly conserved across species (He et al., 2000, 2002a). In response to high affinity androgen binding, the AR NH2-terminal FXXLF motif interacts with the activation function 2 surface of the AR carboxyl-terminal ligand binding domain to mediate the androgen-dependent AR N/C interaction required for optimal gene activation (He et al., 2000). The AR NH2-terminal FXXLF motif also serves as the interaction site for melanoma antigen-A11 (MAGE-A11), a transcriptional coregulator that increases AR transcriptional activity by competing with the AR N/C interaction and exposing activation function 2 for p160 coactivator recruitment (Bai et al., 2005). Two features of AR interaction with MAGE-A11 are specific to human and nonhuman primates. First, sequence flanking the AR FXXLF motif evolved during the early mammalian transition to primates (He et al., 2002a). Hydrophobic residue valine-33 that flanks the human AR NH2-terminal FXXLF motif is alanine in AR of lower mammals. Human AR valine-33 is required for AR to interact with MAGE-A11, but valine-33 is not required for the androgen-dependent AR N/C interaction (Liu et al., 2011). Second, MAGE-A11 itself is a member of the MAGE gene family that evolved in primates (Brasseur et al., 1995; Scanlan et al., 2002; Dhodapkar et al., 2003). MAGE-A11 increases human AR transcriptional capacity through mechanisms that include stabilization of transcriptionally active AR dimers (Minges et al., 2013) and recruitment of p160 and p300 coactivators (Askew et al., 2009, 2010).
Other short predicted α-helical regions in the human AR NH2-terminal region involved in AR transcriptional activity include the WXXLF motif sequence 433WHTLF437 and the highly conserved AR-(234–247) region. The human AR WXXLF motif is evolutionarily less well conserved than the AR FXXLF motif (He et al., 2002a) and provides only a minor contribution to the androgen-dependent AR N/C interaction (He et al., 2000, 2002a). The AR WXXLF motif is required to mediate the AR transcriptional response to MAGE-A11 and p300 (Lagarde et al., 2012; Dehm et al., 2007). The importance of MAGE-A11 and AR WXXLF motif region in enhancing AR transcriptional activity was demonstrated by a naturally occurring human AR-R405S mutation near the WXXLF motif that caused partial androgen insensitivity in a newborn by interfering with the AR transcriptional response to MAGE-A11 (Lagarde et al., 2012). The highly conserved AR-(234–247) region predicted to form an α-helix is the interaction site for CHIP (carboxyl-terminus of the Hsp70-intereacting protein), an E3 ubiquitin ligase that directs the ubiquitination of Hsp70 and Hsp90 chaperone complexes for degradation by the proteasome (Jiang et al., 2001; Demand et al., 2001; He et al., 2004a). CHIP is a negative AR coregulator that promotes AR degradation in the absence or presence of androgen (He et al., 2004a).
The present study extends the use of comparative sequence alignment and α-helical structure predictions within the intrinsically disordered AR NH2-terminal region. A yeast two-hybrid screen of a human testis library was performed using the predicted AR-(370–420) NH2-terminal α-helical region as bait. The studies identify and characterize SUV39H2 (human suppressor of variegation 3–8 homolog 2), a histone methyltransferase that directs trimethylation of lysine 9 on histone H3, as an AR interacting protein recruited to a classical androgen response gene.
MATERIALS AND METHODS
Yeast two-hybrid screen
An amplified human testis two-hybrid library was screened using YEASTMAKER Transformation System 2 (Clontech) (He et al., 2004a). The yeast pBD-GAL4-Cam-AR-(370–420) bait vector was created by PCR amplification of pCMV-AR-(370–420) and cloning the fragment into EcoRI and SalI sites of pBD-GAL4-Cam (Agilent Technologies). Yeast strain HF7c was transformed with pBD-GAL4-Cam-AR-(370–420) coding for human AR NH2-terminal amino acid residues 370–420 and plated on synthetic medium without Trp and His using the pBD-GAL4 Cam Phagemid Vector Kit (Agilent Technologies). A yeast clone expressing pBD-GAL4-Cam-AR-(370–420) was transformed with 100 µg of amplified human testis Matchmaker cDNA library (BD Clontech) and plated in synthetic medium without Leu, Trp and His. Colonies were scored after several days and transferred to plates containing synthetic medium lacking Leu, Trp and His with the addition of 5 mM 3-amino-1,2,4-triazole. Approximately 140 colonies were retested in a two-hybrid β-galactosidase filter assay (Clontech). Freshly prepared Z buffer containing 60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 50 mM 2-mercaptoethanol, pH 7.0, and 5-bromo-4-chloro-3-indolyl-β-D-galactoside was added to the filters and incubated at room temperature to score blue colonies after 2 h or overnight. Plasmids from 17 positive yeast colonies in the β-galactosidase filter assay were rescued. Colonies that grew in synthetic medium lacking Leu gradually lost the bait vector but not the library plasmids. The library vector was purified using Yeastmaker plasmid isolation kit K1611-1 (Clontech) and sequenced.
Expression, immunoblot and transcription assays
Immunoblot and immunoprecipitation experiments were performed using monkey kidney COS1 cells (2 × 106 cells/10 cm dish) and DEAE dextran DNA transfection (He et al., 2002b; Askew et al., 2007; Minges et al., 2015). For siRNA expression, COS1 cells (4 × 105/well) plated in 6-well plates were transfected the next day with 0.5 µg FLAG-SUV39H2-L/well using Lipofectamine 2000 (Thermofisher). Antibodies used to probe the immunoblots included rabbit HA antibody ab9110 (Abcam, dilution 1:1000–2000), mouse FLAG-M2 antibody F3165 (Sigma-Aldrich, dilution 1:500–2000), rabbit anti-AR peptide antibodies AR32 (1 µg/ml) and AR52 (10 µg/ml) (Lubahn et al., 1988; Quarmby et al., 1990), rabbit MAGE1 and MAGE2 antibodies against baculovirus expressed human FLAG-MAGE-A11 (0.2–5 µg/ml) (Su et al., 2013), and mouse β-actin antibody ab-6276 (Abcam, 1:5000 dilution). Immunoblots were calibrated using Precision Plus Dual Stain molecular weight markers (Bio-Rad). Chemiluminescence was detected using SuperSignal West Dura Extended Duration Substrate (Pierce).
AR transcriptional activity was measured by transfection of CV1 cells using calcium phosphate and prostate-specific antigen (PSA) enhancer reporter PSA-Enh-Luc with pCMV-AR (Lubahn et al., 1988), pCMV-AR-(1–660) (Simental et al., 1991), pSG5-MAGE (Bai et al., 2005) and/or pSG5-SUV39H2. Cells were incubated in serum-free medium with or without 10 nM DHT for 24 h before harvest. Two-hybrid interaction assays were performed using Effectene transfection reagent (Qiagen) in HepG2 human hepatocellular carcinoma cells (2 × 105 cells/well) in 12-well plates with GAL-SUV39H2, pVP16-AR-(1–660), pVP16-AR-(370–420), VP-MAGE-A11 and 5XGAL4Luc3 reporter gene (He et al., 2001; He and Wilson, 2003; Bai et al., 2005). CWR-R1 cells (2 × 105/well) plated in 12-well plates were transfected the next day using Effectine transfection regent (Qiagen) with 0.25 µg of the androgen responsive mouse mammary tumor virus luciferase reporter vector (MMTV-Luc) with or without 20 nM SUV39H2 siRNA (Dharmacon), nonspecific siRNA or AR siRNA that decreased AR levels (Ponguta et al. 2008; Askew et al., 2009; Minges et al., 2013). The next day CWR-R1 cells were transferred to serum-free medium and 24 h later treated with or without 0.1 nM DHT in serum-free medium. Luciferase activity was measured using an automated Lumistar Galaxy luminometer (BMG Labtech) with the mean (S.D.) representative of at least three independent experiments.
In vitro interaction assay
GST in vitro interaction assays were performed using bacterial expressed GST fusion proteins of MAGE-A11 and AR by incubation with in vitro translated 35S-methionine-labeled SUV39H2-S. pGEX-3X-1-AR-(1–566) and pGEX-5X-1-AR-(1–660) were described (He et al., 1999, 2000). pGEX-4T-1-MAGE-(2–429) was created by PCR amplification of full-length MAGE-A11 with EcoRI and XhoI ends from pSG5-MAGE and ligation into the same sites of pGEX-4T-1. GST fusion proteins were expressed in log phase XL1-Blue Escherichia coli treated with 0.5 mM isopropyl-1-thio-β-D-galactopyranoside for 3 h (He et al., 1999; 2000). Cell extracts were prepared by sonication in 0.5% Nonidet P-40, 0.1 M NaCl, 1 mM EDTA and 20 mM Tris, pH 8.0 and incubated for 1 h at 4°C with glutathione-agarose beads (Amersham). Washed beads were incubated for 2 h at 4°C with 25 µCi of 35S-methionine (PerkinElmer Life Sciences)-labeled SUV39H2-S using the TNT T7 Quick-coupled Transcription/Translation System (Promega). Washed beads were eluted at high temperature in an SDS containing buffer and analyzed on a 12% acrylamide gel containing SDS.
Immunocytochemistry
pCMV-AR, pCMV-FLAG-SUV39H2 and/or pSG5-MAGE (0.1 µg) were expressed in COS1 cells (1.5 × 105 cells/well) plated in 12-well plates with glass coverslips using Effectene transfection reagent (Qiagen) (He et al., 2004a; Bai et al, 2005). Cells were fixed using 3% paraformaldehyde, permeabilized using 0.5% Trion X-100, blocked with 0.5% bovine serum albumin, and incubated with AR primary rabbit antibody ab-3510 (Abcam, 1:250 dilution), FLAG-M2 mouse monoclonal antibody F3165 (Sigma, 1:1000 dilution) and/or MAGE-A11-(94–108) rabbit polyclonal antipeptide antibody (10 µg/ml) (Bai et al., 2008). Secondary antibodies were rhodamine (tetramethyl rhodamine isocyanate)-conjugated AffiniPure donkey anti-mouse IgG (Jackson ImmunoResearch Laboratories, 1:75 dilution) and fluorescein isothiocyanate-conjugated AffiniPure donkey anti-rabbit IgG (Jackson ImmunoResearch Laboratories, 1:50 dilution). Slides were viewed using a Zeiss LSM 210 confocal microscope.
SUV39H2 antibody
Rabbit polyclonal antibodies were prepared against the SUV39H2-L peptide C142DELNRRKNHKGMIFVEN158, which is equivalent to SUV39H2-S-(82–98). A cysteine residue was included at the NH2-terminus for conjugation to Keyhole limpet hemocyanin (Pocono Rabbit Farm & Laboratory, Canadensis, PA). This region of SUV29H2 was predicted to be immunogenic based on the Princeton Biomolecules algorithm. Immunoreactivity was indicated on immunoblots by expression of full-length SUV39H2-L and partial SUV39H2-S-(67–350) identified in the yeast two-hybrid screen. Antibody purification was performed using antigen affinity chromatography of activated immunoaffinity Affi-Gel 10 gel (BioRad) coupled to the peptide antigen in 0.2 M ethanolamine, pH 8.0. Antiserum was incubated for 2 h at 4°C, eluted with 0.1 M glycine, pH 3.0 in 0.1 volume 1 M Tris-HCl, pH 8.0 and neutralized.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed by plating 8 × 106 LAPC-4 cells/10 cm dish in RPMI medium containing 10% charcoal-stripped fetal bovine serum (Atlantic Biologicals) (Askew et al., 2010). After 3 days cells were placed in fresh medium with or without 10 nM methyltrienolone (R1881, PerkinElmer), a synthetic androgen agonist resistant to metabolism, and incubated for 24 h. Cells were crosslinked using 1% formaldehyde for 10 min, treated with 0.125 M glycine and extracted (Askew et al., 2010). Cell extracts were precleared using protein-A agarose and immunoprecipitated using 10 µg of normal rabbit IgG sc-2027 (Santa Cruz Biotechnology), rabbit anti-AR H-280 sc-13062 (Santa Cruz Biotechnology) or peptide-purified rabbit anti-SUV39H2 antibody. PCR was performed using Taq polymerase (Qiagen) and PSA upstream enhancer primers 5'-GGGACAACTTGCAAACCTG-3' and 5'-GTATCTGTGTGTCTTCTGAGC-3' to amplify a 285-bp fragment.
RESULTS
Identification of SUV39H2 as AR NH2-terminal interacting protein
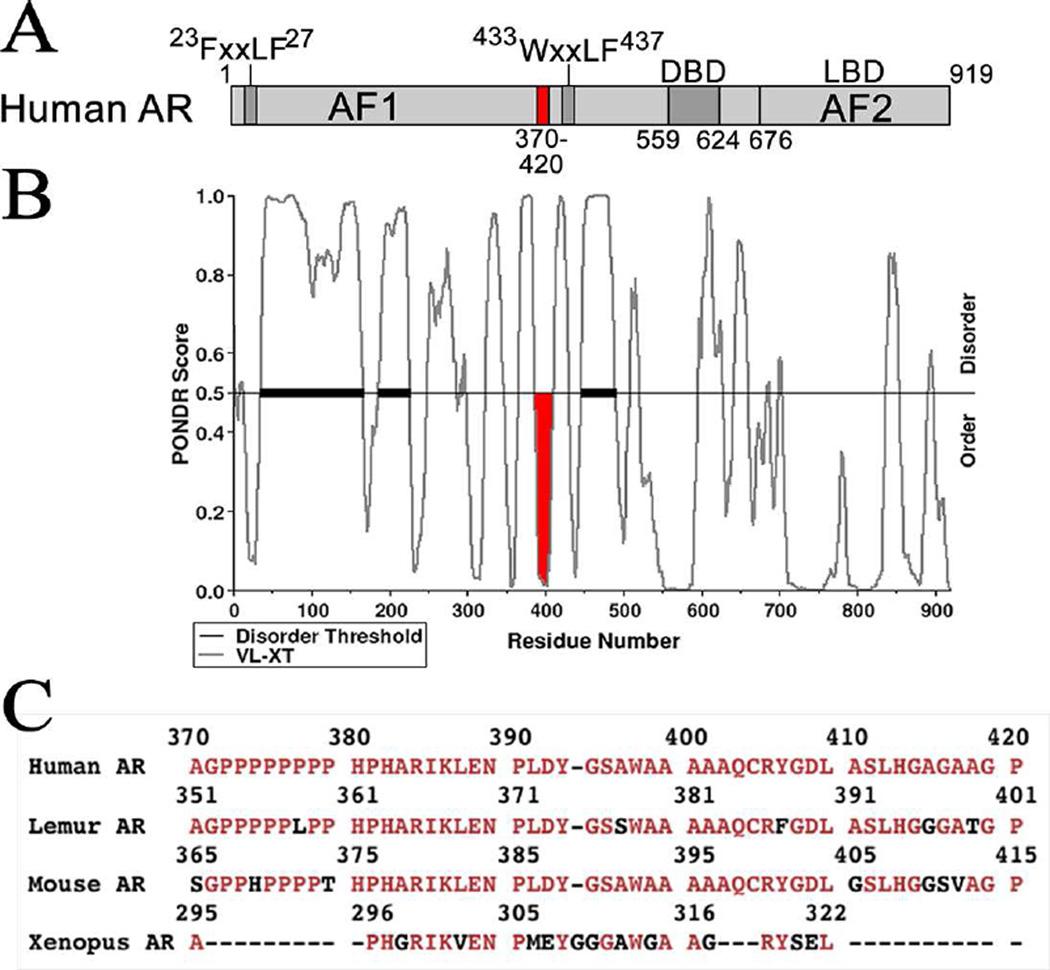
Human AR NH2-terminal amino acid residues 370–420 (Fig. 1A, shown in red) are adjacent to the WXXLF motif sequence 433WHTLF437 involved in the AR transcriptional response to p300 and MAGE-A11 (Dehm et al., 2007; Lagarde et al., 2012). AR-(370–420) is predicted to form an ordered structure based on the PONDR algorithm of ordered and disordered regions deduced from the linear sequence (Fig. 1B). Comparative sequence alignment of human AR-(370–420) in the less well conserved AR NH2-terminal region compared to the AR DNA and ligand binding domains (Choong et al., 1998) showed sequence conservation with corresponding regions of AR from Eulemur collaris (lemur) and Mus musculus (mouse) and lack of similarity with Xenopus laevis (frog) (Fig. 1C) or fish species (not shown). AR-(370–420) is central to a Tau-5 region (human AR amino acid residues 361–537) that contains a partially conserved 372–379 polyproline tract, a more highly conserved 398–402 polyalanine tract, and WXXLF motif sequence 433WHTLF437 involved in AR transcriptional activation (Lubahn et al., 1988; Jenster et al., 1995; He et al., 2000; Dehm et al., 2007; Lagarde et al., 2012; De Mol et al., 2016). A high α-helical propensity was identified for AR NH2-terminal amino acid residues 390–410 (De Mol et al., 2016).
Figure 1. Human AR domains and sequence.
[A] Human AR functional domains include the FXXLF motif sequence 23FQNLF27 that mediates the androgen-dependent AR N/C interaction (He et al., 2000), WXXLF motif sequence 433WHTLF437 involved in AR activation by MAGE-A11 and p300 (Dehm et al., 2007; Lagarde et al., 2012), activation function 1 (AF1) NH2-terminal amino acid residues 142–337 (Simental et al., 1991), DNA binding domain (DBD) amino acid residues 559–624, and ligand binding domain (LBD) amino acid residues 676–919 (Lubahn et al., 1988). The human AR-(370–420) region is shown in red. Human AR amino acid numbering is as initially reported (Lubahn et al., 1988). [B] PONDR structure prediction of human AR. Downward peaks represent predicted ordered regions and upward peaks represent predicted disordered regions. Human AR-(370–420) in red is a predicted α-helix. [C] Human AR NH2-terminal 370–420 amino acid sequence (NCBI M20132, J30180) is compared to corresponding regions of Eulemur collaris collared brown lemur (NCBI O97776), Mus musculus mouse (NCBI P19091) and Xenopus laevis frog AR (NCBI AAC97386) that precedes the human AR-(433–437) WXXLF motif shown in (A) (He et al., 2002a). Pairwise protein sequence alignment was performed using EMBOSS Needle (http://www.ebi.ac.uk/Tools/psa/emboss_needle/).
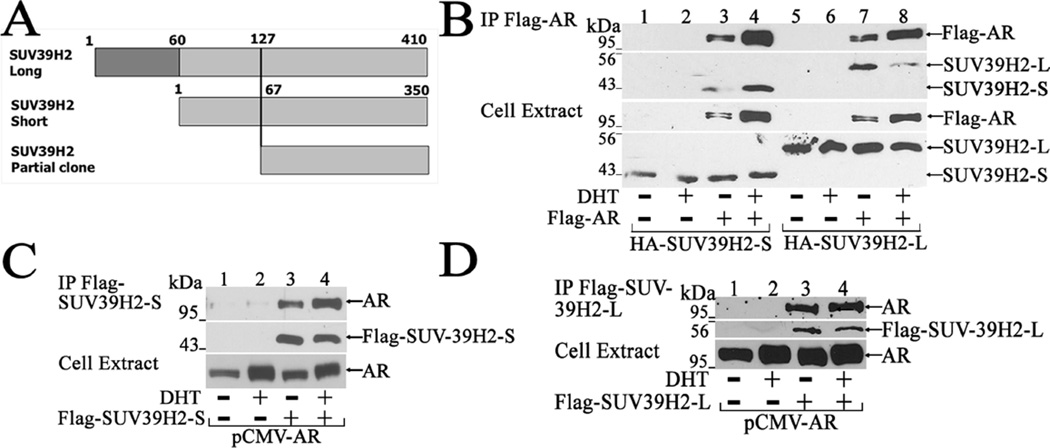
To determine whether human AR-(370–420) is an interaction site for an AR coregulator, a yeast two-hybrid screen of a human testis library was performed using pBD-GAL4-Cam-AR-(370–420) as bait. From 140 colonies selected for further testing, 17 were positive in a β-galactosidase assay. From these a partial sequence was identified that corresponded to the carboxyl-terminal region of the histone-lysine N-methyltransferase SUV39H2 (Fig. 2A). Human SUV39H2 occurs in a canonical SUV39H2-L long form (isoform-3) of 410 amino acid and the shorter SUV39H2-S (isoform-1). SUV39H2-S is identical to SUV39H2-L except that the short form lacks the first 60 NH2-terminal amino acids of SUV39H2-L (Fig. 2A) (UniProt.org). Both SUV39H2-L and SUV39H2-S have the evolutionarily conserved 124 amino acid SET (Su(var)3–9, Enhancer-of-zeste, Trithorax) domain (SUV39H2-L-(250–373)) required for histone H3 methylation (Rea et al., 2000).
Figure 2. AR interacts with SUV39H2.
[A] Schematic diagram of 410 amino acid SUV39H2-L long isoform and 350 amino acid SUV39H2-S short isoform histone 3 methyltransferase identified in a human testis two-hybrid library as SUV39H2-L-(127–410) carboxyl-terminal fragment equivalent to SUV39H2-S-(67–350) selected using pBD-GAL4-Cam-AR-(370–420) as bait. SUV39H2-L and SUV39H2-S are identical except SUV39H2-S lacks 60 NH2-terminal amino acids in SUV39H2-L. [B] Coimmunoprecipitation of AR and SUV39H2 was performed by expressing 2 µg pCMV-FLAG (−) or 2 µg pCMV-FLAG-AR in COS1 cells with 2 µg pSG5-HA-SUV39H2-S or 2 µg pSG5-HA-SUV39H2-L. The day after transfection and 24 h before harvest, cells were incubated in serum-free medium with or without 10 nM DHT. Equivalent amounts of cell extract protein (40 µg/lane) and immunoprecipitates were probed on immunoblots using HA and FLAG-M2 antibodies. [C] pCMV-FLAG (5 µg) (−) or 5 µg pCMV-FLAG-SUV39H2-S was expressed in COS1 cells with 2 µg pCMV-AR. The day after transfection and 24 h before harvest, cells were incubated in serum-free medium with 0.1 µg/ml epidermal growth factor (EGF) with or without 10 nM DHT. Equivalent amounts of cell extract protein (40 µg/lane) and immunoprecipitates were probed on immunoblots using AR52, AR32, HA and FLAG-M2 antibodies. [D] pCMV-FLAG (5 µg) (−) or 5 µg pCMV-FLAG-SUV39H2-L was expressed in COS1 cells with 2 µg pCMV-AR and analyzed as in (B).
SUV39H2 interacts with AR NH2-terminal region
Coimmunoprecipitation studies were performed using SUV39H2-S or SUV39H2-L with FLAG-tagged full-length AR to verify the histone methyltransferase interaction with AR. SUV39H2-S (Fig. 2B, lanes 1–4) and SUV39H2-L (Fig. 2B, lanes 5–8) coimmunoprecipitated with FLAG-AR in the absence or presence of DHT when assayed using equivalent amounts of protein on immunoblots. AR interaction with SUV39H2-S increased in the presence of 10 nM DHT and decreased with SUV39H2-L, and there was no interaction with the FLAG vector control (Fig. 2B). AR interaction with SUV39H2 was also seen in reciprocal experiments where full-length AR immunoprecipitated with FLAG-SUV39H2-S or FLAG-SUV39H2-L in the absence or presence of 10 nM DHT as shown on immunoblots using equivalent amounts of protein (Fig. 2C and D).
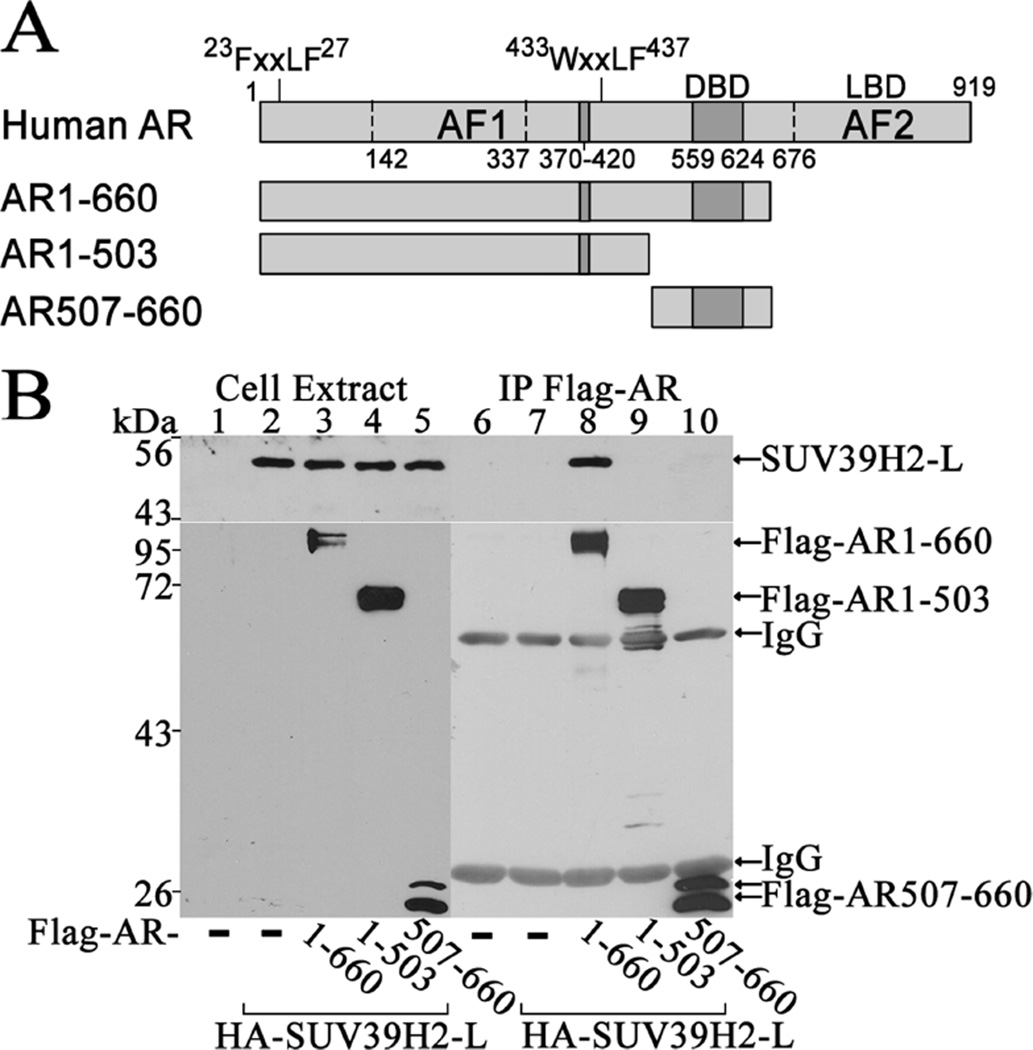
Coimmunoprecipitation studies were also performed using FLAG-AR fragments to determine whether AR interaction with SUV39H2 was mediated solely by the AR NH2-terminal domain (Fig. 3A). FLAG-AR-(1–660) contains the AR NH2-terminal region and DNA binding domain and interacted with SUV39H2-L (Fig. 3B, lane 8). However, neither FLAG-AR-(1–503) AR NH2-terminal fragment that lacks the DNA binding domain or FLAG-AR-(507–660) DNA binding domain interacted with SUV39H2-L (Fig. 3B, lanes 9 and 10).
Figure 3. SUV39H2-L interacts with AR NH2-terminal region.
[A] Schematic diagram of full-length human AR that contains the NH2-terminal FXXLF motif sequence 23FQNLF27, WXXLF motif sequence 433WHTLF437, NH2-terminal activation function 1 (AF1), DNA binding domain (DBD) and activation function 2 (AF2) in the ligand binding domain (LBD). AR NH2-terminal and DNA binding domain fragment AR-(1–660), AR NH2-terminal fragment AR-(1–503) and AR DNA binding domain fragment AR-(507–660) were expressed as FLAG-tagged fusion proteins. [B] Coimmunoprecipitation was performed by expressing 3 µg pCMV-FLAG (−), 3 µg pCMV-FLAG-AR-(1–660), 3 µg pCMV-FLAG-AR-(1–503) or 6 µg pCMV-FLAG-AR-(507–919) with or without 4 µg pSG5-HA-SUV39H2-L in COS1 cells. The day before harvest cells were incubated in serum-free medium containing 0.1 µg/ml EGF. Equivalent amounts of cell extract protein (50 µg/lane) and immunoprecipitates were probed on immunoblots using FLAG-M2 and HA antibodies.
The results suggest that short and long forms of SUV39H2 interact with AR and that the AR DNA binding domain, which itself did not interact with SUV39H2, facilitates AR interaction of SUV39H2-L in the context of the AR NH2-terminal region in coimmunoprecipitation assays. The findings suggest that the DNA binding domain may impose structural constraints on the NH2-terminal region to facilitate SUV39H2 binding, consistent with the effects of the progesterone receptor DNA binding domain on the progesterone receptor NH2-terminal region (Bain et al., 2000).
SUV39H2 interacts with AR and MAGE-A11
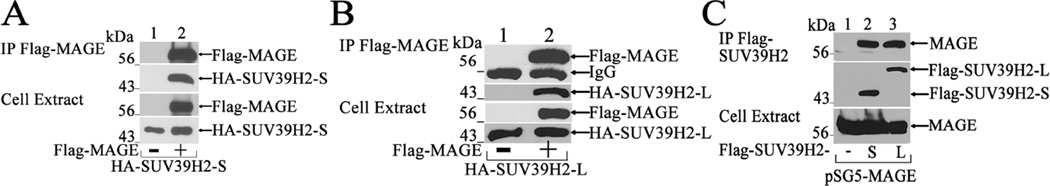
The NH2-terminal AR-(370–420) interaction site for SUV39H2 lies adjacent to the 433WXXLF437 motif involved in the AR transcriptional response to MAGE-A11 (Fig. 1A) (Lagarde et al., 2012). This raised the possibility that SUV39H2 also interacts with MAGE-A11. To test this, studies were performed using FLAG-MAGE-A11 and SUV39H2 assayed on immunoblots using equivalent amounts of protein. SUV39H2-S and SUV39H2-L each coimmunoprecipitated with FLAG-MAGE (Fig. 4A and B). Similarly, MAGE-A11 coimmunoprecipitated with FLAG-SUV39H2-S or FLAG-SUV39H2-L (Fig. 4C, lanes 2 and 3).
Figure 4. SUV39H2 interacts with MAGE-A11.
[A, B] Coimmunoprecipitation was performed in COS1 cells by expressing 4 µg pCMV-FLAG (−) or 4 µg pCMV-FLAG-MAGE with 2 µg pSG5-HA-SUV39H2-S (A) or 2 µg pSG5-HA-SUV39H2-L (B). The day after transfection and 24 h before harvest, cells were incubated in serum-free medium containing 0.1 µg/ml EGF. Equivalent amounts of cell extract protein (40 µg/lane) were probed on immunoblots using HA and FLAG-M2 antibodies. [C] pCMV-FLAG (6 µg) (−), 6 µg pCMV-FLAG-SUV39H2-S or 6 µg pCMV-FLAG-SUV39H2-L was expressed with 2 µg pSG5-MAGE in COS1 cells. The day after transfection and 24 h before harvest, cells were incubated in serum-free medium containing 0.1 µg/ml EGF and 1 µM MG132 proteasome inhibitor. Equivalent amounts of cell extract protein (0.1 mg/lane) and immunoprecipitates pooled from three 10 cm dishes were probed on immunoblots using FLAG-M2 and MAGE1 antibodies.
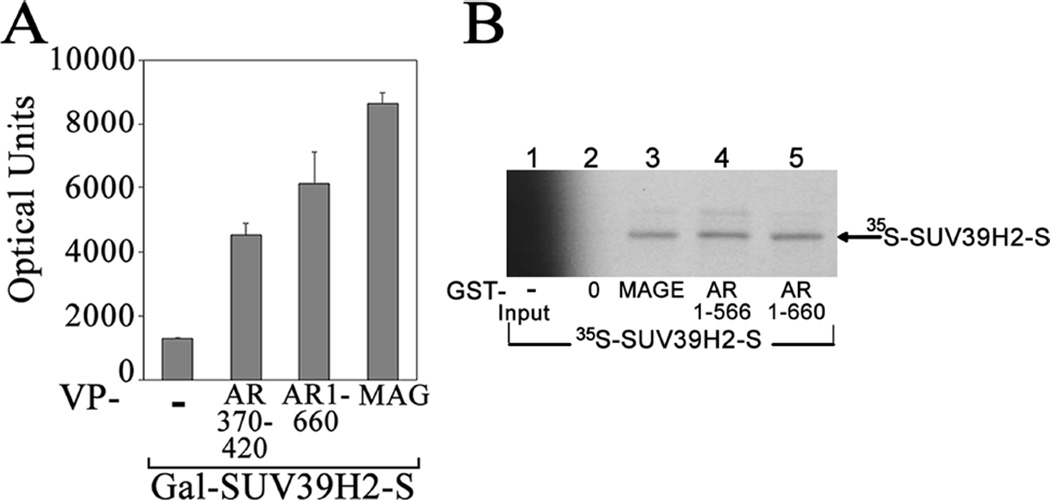
Mammalian two-hybrid assays were performed to obtain additional evidence that SUV39H2 interacts with AR and MAGE-A11. The GAL4 DNA binding domain-SUV39H2-S fusion protein GAL-SUV39H2-S increased transcriptional activation of a GAL4-responsive luciferase reporter by 5 to 9-fold in the presence of VP-AR-(370–420), VP-AR-(1–660) or VP-MAGE-A11 compared to the VP16 vector control (Fig. 5A). The results provided additional evidence that AR-(370–420) is the principal interaction site for SUV39H2 and that SUV39H2 also interacts with MAGE-A11.
Figure 5. SUV39H2 interacts with AR and MAGE-A11.
[A] Mammalian two-hybrid interaction assay was performed in HepG2 cells using 50 ng GAL-SUV39H2-S expressed with 50 ng pVP16 (−), VP-AR-(370–420), VP-AR-(1–660) or VP-MAGE-A11 (VP-MAG) and 0.1 µg 5XGAL4Luc3 reporter gene using Effectene. Cells were incubated in serum-free medium for 24 h before harvest and measurement of luciferase activity. Shown is the mean ± S.D. representative of 3 independent experiments. [B] GST affinity matrix binding assays were performed using partially purified GST-0, GST-MAGE-A11, GST-AR-(1–566) or GST-AR-(1–660) expressed in E. coli and incubated with in vitro translated 35S-methionine-labeled SUV39H2-S as described in Methods. The input lane 1 contained 7% of the reaction.
SUV39H2 interaction with the AR NH2-terminal region and MAGE-A11 was also investigated in in vitro glutathione S-transferase (GST) affinity matrix assays. In vitro translated 35S-methionine-labeled SUV39H2-S was adsorbed to affinity matrix containing GST-MAGE-A11, GST-AR-(1–566) or GST-AR-(1–660) but not to the GST control (Fig. 5B). Interaction of SUV39H2 with the GST-AR-(1–566) NH2-terminal fragment that lacks the DNA binding domain suggested that the AR DNA binding domain was not required for SUV39H2 to interact with the AR NH2-terminal region using in vitro protein expression. This result was consistent with the AR NH2-terminal domain serving as the principal interaction site for SUV39H2.
SUV39H2 colocalizes with AR and MAGE-A11
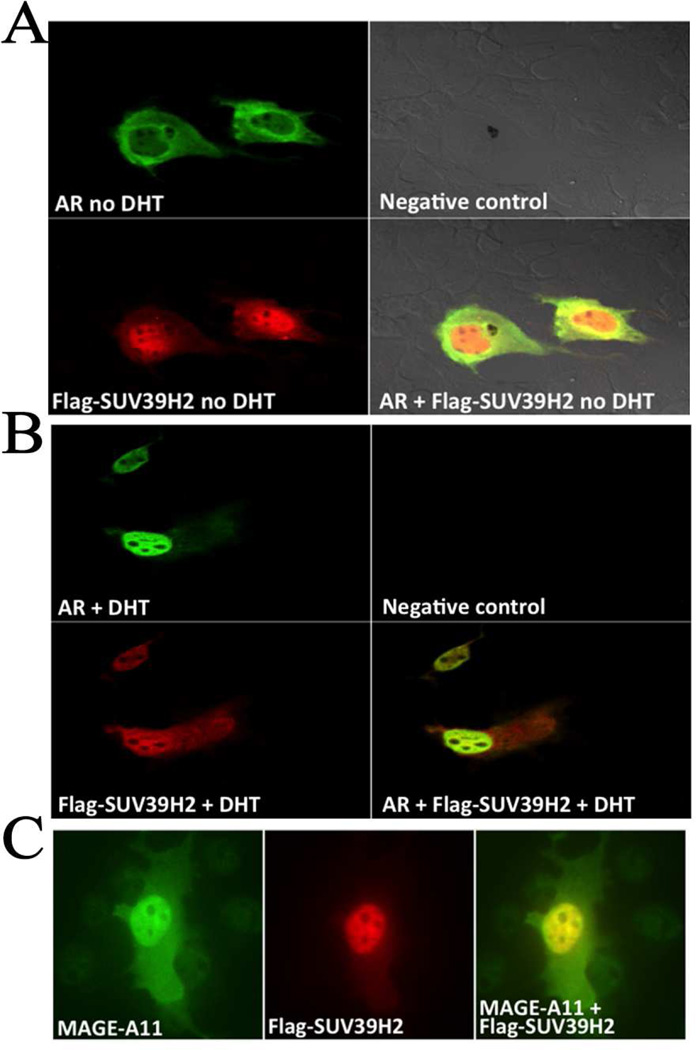
The subcellular relationship of SUV39H2, AR and MAGE-A11 was investigated using immunocytochemistry. In the absence of androgen, fluorescein isothiocyanate-linked AR localized in the cytoplasm (Fig. 6A, green immunostaining), whereas rhodamine-linked SUV39H2 was predominantly nuclear (Fig. 6A, red immunostaining)). A merge of rhodamine and fluorescein fluorescence (yellow) showed colocalization of AR and SUV39H2 in the cytoplasm in the absence of androgen, with SUV39H2 also detected in the nucleus without AR (Fig. 6A). In the presence of 10 nM DHT, AR colocalized with SUV39H2 in the nucleus, with persistent residual SUV39H2 in the cytoplasm without AR (Fig. 6B). The immunocytochemical results were consistent with the coimmunoprecipitation data in Fig. 2B that AR interacts with SUV39H2 in the absence or presence of androgen.
Figure 6. SUV39H2 colocalizes with AR and MAGE-A11.
[A, B] pCMV-AR (0.1 µg) and 0.1 µg pCMV-FLAG-SUV39H2-L were expressed in COS1 cells incubated in the absence (A) or presence of 10 nM DHT (B) for 24 h before fixation and immunostaining as described in Methods. Fixed and permeabilized cells were immunostained using AR ab-3510 and FLAG-M2 antibodies. Green fluorescence represents AR, red fluorescence represents FLAG-SUV39H2-L and yellow is the merged data. No primary antibody addition served as the negative control. [C] pSG5-MAGE (0.1 µg) and 0.1 µg pCMV-FLAG-SUV39H2-S were expressed in COS1 cells. Fixed and permeabilized cells were immunostained using MAGE-(94–108) and FLAG-M2 antibodies. Green fluorescence represents MAGE-A11, red fluorescence represents FLAG-SUV39H2-S and yellow is the merged data. Original magnification ×63.
Application of the same methodology showed that fluorescein isothiocyanate-linked MAGE-A11 (green immunostaining) was predominantly nuclear as reported previously (Bai et al., 2005) and colocalized in the nucleus with SUV39H2, with residual coimmunostaining in the cytoplasm (Fig. 6C). The results suggest that SUV39H2 is predominantly a nuclear protein that associates with AR in the nucleus in the presence of androgen. SUV39H2 colocalizes with MAGE-A11 in the cytoplasm and nucleus independent of androgen.
SUV39H2 enhances AR transcriptional activity
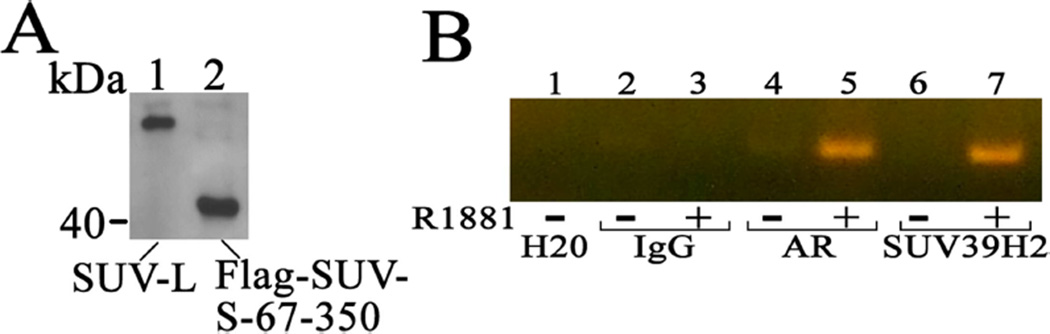
To determine whether SUV39H2 influences androgen-dependent gene regulation, chromatin immunoprecipitation was performed using LAPC-4 prostate cancer cells and primers that amplify the upstream androgen-responsive enhancer region of the PSA gene (Askew et al., 2010). For this purpose polyclonal antibodies were raised in rabbits against SUV39H2-L-(142–158) peptide C142DELNRRKNHKGMIFVEN158, which is equivalent to SUV39H2-S-(82–98). Immunoreactivity of the peptide-purified antibody for SUV39H2 was demonstrated on immunoblots using SUV39H2-L and FLAG-SUV39H2-S-(67–350) (Fig. 7A) that contained the carboxyl-terminal fragment initially identified in the yeast two-hybrid screen (Fig. 2A). Amplification of the upstream enhancer of the PSA gene in LAPC-4 cells treated for 24 h with or without 10 nM R1881, a synthetic androgen agonist, provided evidence that AR and SUV39H2 were recruited to the androgen-responsive enhancer region of the PSA gene in the presence of androgen (Fig. 7B).
Figure 7. Chromatin immunoprecipitation of AR and SUV39H2.
[A] Preparation of a rabbit SUV39H2 polyclonal antibody was verified on immunoblots by expressing in COS1 cells 5 µg pSG5-SUV39H2-L (lane 1) or 5 µg pCMV-FLAG-SUV39H2-S-(67–350) (lane 2) identified as the original isolate in the yeast two-hybrid library screen. Equivalent amounts of cell extract protein (0.1 mg/lane) were probed on the immunoblot using SUV39H2 antibody (1:1000 dilution). [B] Chromatin immunoprecipitation analysis of AR and SUV39H2 binding to the androgen-responsive upstream enhancer region of the PSA gene was performed using LAPC-4 cells incubated in the absence or presence of 10 nM R1881 as described in Methods. Equivalent amounts of cell extract were immunoprecipitated using 10 µg normal rabbit IgG (lanes 2 and 3), AR H-280 sc-13062 antibody (lanes 4 and 5) and SUV39H2 antibody (lanes 6 and 7) with the PCR negative H2O control in lane 1.
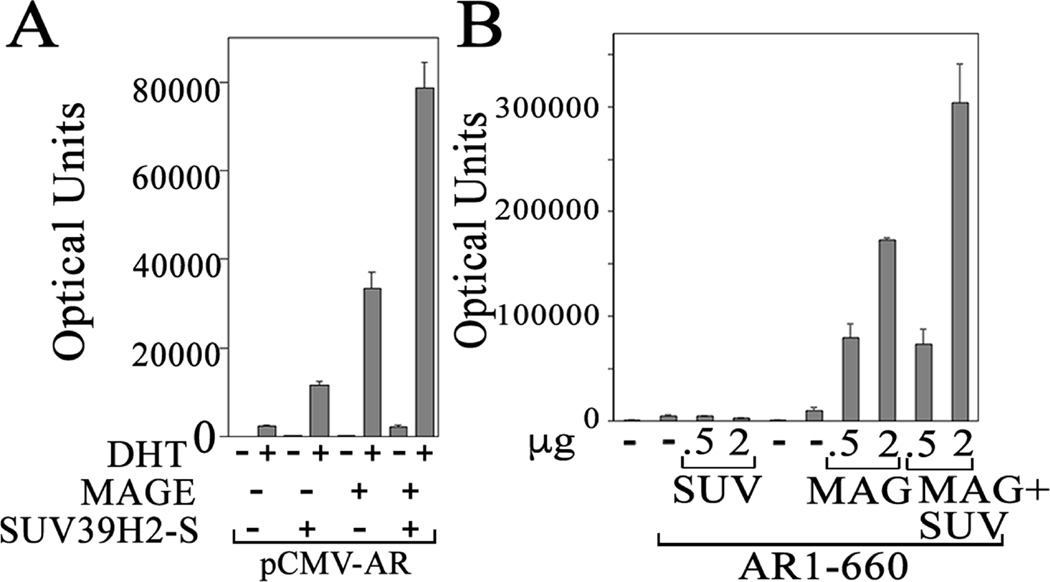
The ability of SUV39H2 to increase AR transcriptional activity was tested in androgen-dependent reporter gene assays. SUV39H2-S or MAGE-A11 increased full-length AR transcriptional activity in the presence of DHT (Fig. 8A). These results were consistent with previous evidence that MAGE-A11 increases AR transcriptional activity (Bai et al., 2005). SUV39H2-S and MAGE-A11 functioned cooperatively to increase androgen-dependent AR transcriptional activity (Fig. 8A).
Figure 8. Cooperative effects of MAGE-A11 and SUV39H2 on AR transcriptional activity.
[A] pCMV-AR (0.1 µg) was expressed with 2 µg pSG5 (−), 2 µg pSG5-SUV39H2-S and/or 0.5 µg pSG5-MAGE and 2.5 µg PSA-Enh-Luc/6 cm dish of CV1 cells transfected using calcium phosphate. The day after transfection and 24 h before harvest, cells were incubated in serum-free medium with or without 1 nM DHT and luciferase activity was measured. [B] pCMV5 (0.1 µg) (−) or 0.05 µg pCMV-AR-(1–660) was expressed with 0.5 or 2 µg pSG5-SUV39H2-S (SUV) and/or 0.5 or 2 µg pSG5-MAGE (MAG) and 5 µg PSA-Enh-Luc reporter gene using calcium phosphate. Cells were incubated for 24 h in serum-free medium the day before harvest and assayed for luciferase activity. Shown is the mean ± S.D. representative of 3 independent experiments.
The transcriptional effects of SUV39H2-S and MAGE-A11 on constitutive activity of the AR-(1–660) NH2-terminal and DNA binding domain fragment were assayed based on evidence that SUV39H2 and MAGE-A11 interact with the AR NH2-terminal region (Bai et al., 2005). The results show that SUV39H2 alone did not increase constitutive activity of AR-(1–660) (Fig. 8B) in contrast to MAGE-A11, which increased AR-(1–660) activity as reported previously (Bai et al., 2005). However, there was evidence that SUV39H2 and MAGE-A11 function cooperatively to increase AR-(1–660) constitutive activity (Fig. 8B).
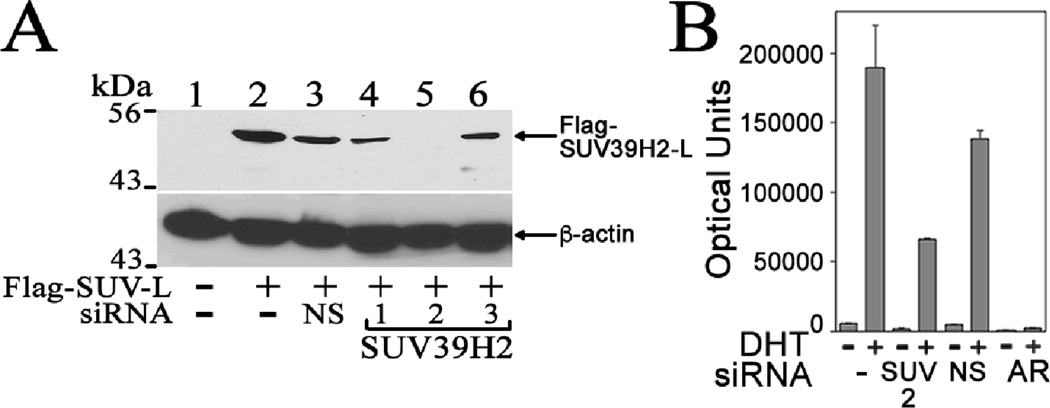
The potentiating effect of SUV39H2 on AR activity was investigated further by determining the effect of siRNA knockdown of SUV39H2 or AR on endogenous AR transcriptional activity in CWR-R1 prostate cancer cells. Of the three siRNAs tested, SUV39H2 siRNA-2 lowered FLAG-SUV39H2-L levels in COS1 cells (Fig. 9A, lane 5). SUV39H2 siRNA-2 expressed in CWR-R1 prostate cancer cells with the MMTV-Luc reporter gene decreased AR mediated transcriptional activity compared to no siRNA or nonspecific siRNA, but was less effective than knockdown of AR in decreasing AR activity (Fig. 9B).
Figure. 9. Inhibition of endogenous AR transcriptional activity in CWR-R1 cells after knockdown of SUV39H2 or AR.
[A] COS1 cells were plated without penicillin or streptomycin in 1 ml medium and transfected with 0.5 µg pCMV-FLAG (−) or 0.5 µg pCMV-FLAG-SUV39H2-L (Flag-SUV-L) with or without 10 nM nonspecific (NS) siRNA or 10 nM SUV39H2 siRNA-1, 2 or 3 (Dharmacon) using Lipofectamine 2000. The immunoblot with equivalent amounts of COS1 protein extract (80 µg/lane) was probed using FLAG-M2 and β-actin antibodies. [B] CWR-R1 prostate cancer cells in 12-well plates were transfected using Effectine with 0.25 µg MMTV-Luc without siRNA (−) or with 20 nM SUV39H2-2 siRNA, nonspecific siRNA (NS) or AR pool siRNA (Dharmacon) that decreased AR levels (Ponguta et al. 2008; Askew et al., 2009; Minges et al., 2013). Cells were treated for 24 h in serum-free medium with or without 0.1 nM DHT. Shown is the mean ± S.D. representative of 3 independent experiments.
The results suggest that SUV39H2 is an AR coactivator that increases androgen-dependent AR transcriptional activity through interactions with AR and MAGE-A11.
DISCUSSION
Histone methyltransferases, AR and MAGE-A11
The histone code hypothesis states that covalent modification of histones by acetylation and methylation determines the functional state of a gene (Strahl and Allis, 2000). Nuclear receptors activate target genes by altering chromatin structure through interactions with histone modifying enzymes such as histone tail lysine acetylation by p300 acetyltransferase during gene transcription, and lysine and arginine methylation during gene activation or repression (Strahl and Allis, 2000; Lachner et al., 2003; Leader et al., 2006). p300 increases AR transactivation through interactions with MAGE-A11 (Askew et al., 2010), but evidence was lacking whether SUV39H histone methyltransferase regulates AR transcriptional activity.
In this report we show that interactions of AR and MAGE-A11 with SUV39H2 methyltransferase increases AR transcriptional activity. The principal AR interaction site for SUV39H2 is a short AR-(370–420) predicted α-helical region in the AR NH2-termimal domain. However, interaction of SUV39H2-L with the AR NH2-termimal region in coimmunoprecipitation studies was stabilized by the presence of the AR DNA binding domain. This suggested that the AR-(507–660) DNA binding and hinge region, which alone did not interact with SUV39H2-L, may function cooperatively with AR-(370–420) in the interaction with SUV39H2. However, GST in vitro binding assays showed that AR-(1–566), which also lacks the DNA binding domain, was sufficient to interact with SUV39H2-S. This was consistent with AR-(370–420) serving as the principal interaction site for the partial SUV39H2-(67–350) fragment identified in the yeast two-hybrid screen of a human testis library. The results raise the possibility that AR preferentially interacts with SUV39H2-S compared to SUV39H2-L. This conclusion was supported by the increase in SUV39H2-S coimmunoprecipitation with FLAG-AR in the presence of androgen that was not seen with SUV39H2-L. However, the potential significance of these findings is not clear since no compelling distinguishing features have been reported for SUV39H2-S and SUV39H2-L.
Coimmunoprecipitation and subcellular localization studies suggest that SUV39H2 interacts with AR in the absence or presence of androgen and that SUV39H2 increases AR transcriptional activity in the presence of androgen. This was supported by the androgen-dependent recruitment of AR and SUV39H2 to the androgen-responsive upstream enhancer region of the PSA gene, and by the increase in androgen-dependent AR transcriptional activity in the presence of SUV39H2 and MAGE-A11. Although ChIPseq was not performed, it might be expected that a histone methyltransferase such as SUV39H2 is recruited to many other genes including other androgen-regulated genes. The findings suggest an AR transcription complex in which SUV39H2 and MAGE-A11 up-regulate androgen-dependent gene expression (Fig. 10). The ability of the experimental drug EPI-001 proposed for treatment of castration-recurrent prostate cancer (Andersen et al., 2010) to interact with human AR NH2-terminal amino acid residues 354–448 (De Mol et al, 2016) suggests that inhibition of AR interaction with SUV39H2 may be the molecular mechanism for inhibition of AR transcriptional activity by EPI-001. While our molecular studies that demonstrate recruitment of AR and SUV39H2 to the androgen response region of the PSA gene were limited to LAPC-4 prostate cancer cells, the findings suggest that SUV39H2 contributes to prostate cancer development and progression by increasing AR transcriptional activity. This was supported by the inhibition of endogenous AR transcriptional activity in CWR-R1 prostate cancer cells associated with knockdown of SUV39H2.
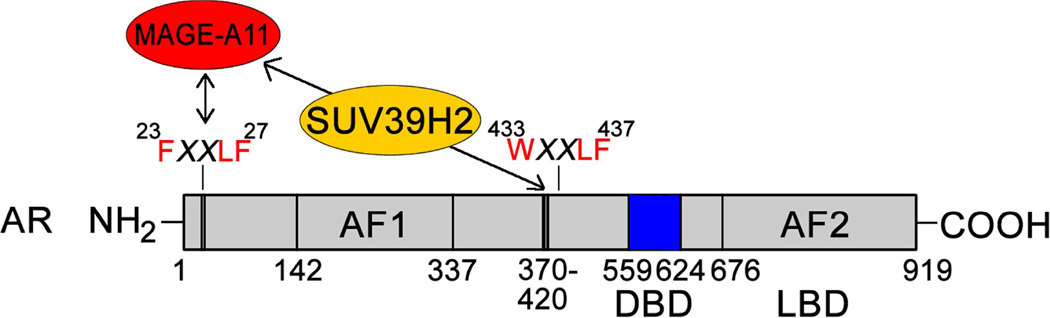
Figure 10. Model of SUV39H2 interaction with AR and MAGE-A11.
SUV39H2 histone methyltransferase interacts with AR-(370–420), a predicted α-helical region in the AR NH2-terminal domain adjacent to AR WXXLF motif sequence 433WHTLF437 required to increase AR activation in response to MAGE-A11. SUV39H2 also interacts with MAGE-A11, an AR coregulator that interacts with the human AR NH2-terminal FXXLF motif sequence 23FQNLF27 (Bai et al., 2005). SUV39H2 and MAGE-A11 function cooperatively to increase AR transcriptional activity.
SUV39H2 histone methyltransferase has not been previously linked to gene activation by steroid receptors. SUV39H1 and SUV39H2 isoforms catalyze trimethylation of histone 3 on lysine 9 (Rea et al., 2000; Schuhmacher et al., 2015), a conserved epigenetic mark that recruits heterochromatin protein 1 gene repressor protein (Nielsen et al., 2001; Du et al., 2015). Trimethylation of histone H3 at lysine 9 by SUV39H1 and SUV39H2 was also linked to active gene transcription involved in cell cycle-dependent processes (Vakoc et al., 2005; Aagaard et al., 2000). Dynamic changes in histone methylation may be required for androgen-dependent up-regulation of genes by AR and MAGE-A11.
Several methyltransferases catalyze mono, di and trimethylation of lysine residues in histones H3 and H4 during gene regulation (Santos-Rosa et al., 2002; Gerber and Shilatifard, 2003; Fritsch et al., 2010). SUV39H1 (412 amino acids) and SUV39H2 (410 amino acids, SUV39H2-L) in the G9a subgroup are similar in length but have only 58% sequence identity (Rea et al., 2000; Lee et al., 2006; Fritsch et al., 2010). Notable differences between mouse Suv39h1 and Suv39h2 include an 82 NH2-terminal basic amino acid-enriched extension in Suv39h2 not present in Suv39h1 (O’Carroll et al., 2000). Mouse Suv39h1 and Suv39h2 have 48% sequence identity. Predominant sites of histone 3 methylation by mouse Suv39h are endogenous retrovirus regions and retrotransposons involved in gene evolution inside and outside heterochromatin (Bulut-Karslioglu et al., 2014).
The two isoforms of human SUV39H2 studied in this report, SUV39H2-L (410 amino acids) and SUV39H2-S (350 amino acids, GenBank CAG33653.1) are identical except SUV39H2-S lacks 60 NH2-terminal amino acids in SUV39H2-L. SUV39H2-L and SUV39H2-S contain the ~130 amino acid SET domain (SUV39H2-L-(250–373)) in the catalytic site and chromodomain (SUV39H2-L-(47–105)) that interacts with trimethylated histone 3 on lysine 9 (Rea et al., 2000; Firestein et al., 2000). Although initially reported as testis specific, SUV39H2 is ubiquitously expressed in human tissues (O'Carroll et al., 2000; Mauger et al., 2015). Human SUV39H2 has a preformed docking platform for the histone 3 tail similar to other histone H3 lysine 9 methyltransferases (Wu et al., 2010). Although short and long forms of human SUV39H2 are thought to have histone methyltransferase activity, differences in nuclear localization were suggested by diffuse nuclear immunostaining of SUV39H2-L compared to SUV39H2-S localized in nuclear foci (Mauger et al., 2015). Data presented here suggest that AR and MAGE-A11 interact with SUV39H2-L and SUV39H2-S.
AR, MAGE-A11 and SUV39H2 in cancer
DNA and histone methyltransferases have been implicated in human health and disease (Grewal and Moazedm, 2003; Kouzarides, 2007). SUV39H2 was considered an oncogene up-regulated in residual breast cancer compared to primary breast tumors (Franci et al., 2013). SUV39H2 was increased in clinically localized prostate cancer compared to normal prostate (Vieira et al., 2013), and SUV39H1 was increased in colorectal cancer (Ozdağ et al., 2006). Mutations in SUV39H1 and SUV39H2 were identified in ovarian and breast cancer (Ozdağ et al., 2006). Single nucleotide polymorphisms in SUV39H2 were linked to greater susceptibility to lung cancer (Yoon et al., 2006). In this report we provide evidence that SUV39H2 increases AR transcriptional activity in CWR-R1 prostate cancer cells. In contrast, loss of mouse Suv39h correlated with chromosomal instability, impaired cell viability and increased tumor risk (Peters et al., 2001), and human SUV39H2 was considered a tumor suppressor in B-cell lymphoma (Cloos et al., 2008; Patani et al., 2011).
A link between SUV39H and cancer was also suggested by SUV39H1 interaction with retinoblastoma protein (pRb) or Rb-related protein p107, which are regulators of E2F transcriptional activity (Nicolas et al., 2003). This is relevant because MAGE-A11 interacts with p107 and increases E2F1 transcriptional activity (Su et al., 2013). pRb mutants in cancer can lose their ability to bind SUV39H2 (Nielsen et al., 2001). pRb interaction with E2F1 transcriptionally silences E2F1 by blocking recruitment of coactivators and more directly by recruiting histone deacetylase transcriptional repressors that remove acetyl groups from histone 3 lysine 9 (Flemington et al., 1993; Helin et al., 1993; Nielsen et al., 2001). SUV39H1 subsequently methylates the same histone 3 lysine 9 in E2F1 target genes (Nielsen et al., 2001; Vandel et al., 2001), and when complexed with pRb-E2F creates a binding site for heterochromatin protein 1 (Trimarchi and Lees, 2002). Accumulation of SUV39H, pRb and heterochromatin protein 1 on E2F-responsive proliferative leads to gene repression and induction of cell senescence (Trimarchi and Lees, 2002).
SUV39H2 also forms a complex with Rb-related pocket proteins that promote silencing of E2F-responsive genes, exit from the cell cycle and onset of cell differentiation (Ait-Si-Ali et al., 2004; Cobrinik, 2005). However, trimethylation of histone H3 on lysine 9 of E2F-responsive genes by SUV39H2 is a reversible process that contributes to cell cycle regulation (Aagaard et al., 2000; Berger, 2007). Although relatively little is known about SUV39H2 and steroid receptor signaling, our studies suggest that AR and MAGE-A11 interact with SUV39H2 to increase androgen-dependent transcriptional activity and may contribute to the regulation of E2F1.
HIGHLIGHTS.
A yeast two-hybrid screen of a human testis library used AR-(370–420) as bait.
Suppressor of variegation 3–9 homolog 2 (SUV39H2) methyltransferase was identified.
SUV39H2 interacts with androgen receptor (AR) and melanoma antigen-A11 (MAGE-A11).
Fluorescent immunohistochemistry colocalized SUV39H2, AR and MAGE-A11.
SUV39H2 was recruited to the prostate-specific antigen enhancer with androgen.
Acknowledgments
The work was supported by United States Public Health Service National Cancer Institute, National Institutes of Health (Grant P01-CA77739). We thank Frank S. French for reviewing the manuscript.
Abbreviations
- AR
androgen receptor
- SUV39H2
suppressor of variegation 3–9 homolog 2 histone H3 methyltransferase
- MAGE-A11
melanoma antigen-A11
- DHT
dihydrotestosterone
- N/C
NH2- and carboxyl-terminal
- PSA
prostate-specific antigen
- GST
glutathione S-transferase
- pRb
retinoblastoma protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors have no conflict of interest with the contents of this article.
REFERENCES
- Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J. Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- Ait-Si-Ali S, Guasconi V, Fritsch L, Yahi H, Sekhri R, Naguibneva I, Robin P, Cabon F, Polesskaya A, Harel-Bellan A. A Suv39h-dependent mechanism for silencing S-phase genes in differentiating but not in cycling cells. EMBO J. 2004;23:605–615. doi: 10.1038/sj.emboj.7600074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen RJ, Mawji NR, Wang J, Wang G, Haile S, Myung JK, Watt K, Tam T, Yang YC, Bañuelos CA, Williams DE, McEwan IJ, Wang Y, Sadar MD. Regression of castrate-recurrent prostate cancer by a small-molecule inhibitor of the amino-terminus domain of the androgen receptor. Cancer Cell. 2010;17:535–546. doi: 10.1016/j.ccr.2010.04.027. [DOI] [PubMed] [Google Scholar]
- Askew EB, Bai S, Blackwelder AJ, Wilson EM. Transcriptional synergy between melanoma antigen gene protein-A11 (MAGE-11) and p300 in androgen receptor signaling. J. Biol. Chem. 2010;285:21824–21836. doi: 10.1074/jbc.M110.120600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew EB, Bai S, Hnat AT, Minges JT, Wilson EM. Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators. J. Biol. Chem. 2009;284:34793–34808. doi: 10.1074/jbc.M109.065979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Askew EB, Gampe RT, Stanley TB, Faggart JL, Wilson EM. Modulation of androgen receptor activation function 2 by testosterone and dihydrotestosterone. J. Biol. Chem. 2007;282:25801–25816. doi: 10.1074/jbc.M703268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, Grossman G, Yuan L, Lessey BA, French FS, Young SL, Wilson EM. Hormone control and expression of androgen receptor coregulator MAGE-11 in human endometrium during the window of receptivity to embryo implantation. Mol. Hum. Reprod. 2008;14:107–116. doi: 10.1093/molehr/gam080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai S, He B, Wilson EM. Melanoma antigen gene protein MAGE-11 regulates androgen receptor function by modulating the interdomain interaction. Mol. Cell. Biol. 2005;25:1238–1257. doi: 10.1128/MCB.25.4.1238-1257.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bain DL, Franden MA, McManaman JL, Takimoto GS, Horwitz KB. The N-terminal region of the human progesterone A-receptor. Structural analysis and the influence of the DNA binding domain. J. Biol. Chem. 2000;275:7313–7320. doi: 10.1074/jbc.275.10.7313. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Brasseur F, Rimoldi D, Liénard D, Lethé B, Carrel S, et al. Expression of MAGE genes in primary and metastatic cutaneous melanoma. Int. J. Cancer. 1995;63:375–380. doi: 10.1002/ijc.2910630313. [DOI] [PubMed] [Google Scholar]
- Bulut-Karslioglu A, De La Rosa-Velázquez IA, Ramirez F, Barenboim M, Onishi-Seebacher M, Arand J, Galán C, Winter GE, Engist B, Gerle B, O'Sullivan RJ, Martens JH, Walter J, Manke T, Lachner M, Jenuwein T. Suv39h-dependent H3K9me3 marks intact retrotransposons and silences LINE elements in mouse embryonic stem cells. Mol. Cell. 2014;55:277–290. doi: 10.1016/j.molcel.2014.05.029. [DOI] [PubMed] [Google Scholar]
- Burska UL, Harle VJ, Coffey K, Darby S, Ramsey H, O'Neill D, Logan IR, Gaughan L, Robson CN. Deubiquitinating enzyme Usp12 is a novel co-activator of the androgen receptor. J. Biol. Chem. 2013;288:32641–32650. doi: 10.1074/jbc.M113.485912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- Choong CS, Kemppainen JA, Wilson EM. Evolution of the primate androgen receptor: a structural basis for disease. J. Mol. Evol. 1998;47:334–342. doi: 10.1007/pl00006391. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- Coffey K, Rogerson L, Ryan-Munden C, Alkharaif D, Stockley J, Heer R, Sahadevan K, O'Neill D, Jones D, Darby S, Staller P, Mantilla A, Gaughan L, Robson CN. The lysine demethylase, KDM4B, is a key molecule in androgen receptor signalling and turnover. Nucleic Acids Res. 2013;41:4433–4446. doi: 10.1093/nar/gkt106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Mol E, Fenwick RB, Phang CT, Buzón V, Szulc E, de la Fuente A, Escobedo A, García J, Bertoncini CW, Estébanez-Perpiñá E, McEwan IJ, Riera A, Salvatella X. EPI-001, a compound active against castration-resistant prostate cancer, targets transactivation unit 5 of the androgen receptor. ACS Chem. Biol. 2016;11:2499–2505. doi: 10.1021/acschembio.6b00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehm SM, Regan KM, Schmidt LJ, Tindall DJ. Selective role of an NH2-terminal WXXLF motif for aberrant androgen receptor activation in androgen depletion independent prostate cancer cells. Cancer Res. 2007;67:10067–10077. doi: 10.1158/0008-5472.CAN-07-1267. [DOI] [PubMed] [Google Scholar]
- Demand J, Alberti S, Patterson C, Höhfeld J. Cooperation of a ubiquitin domain protein and an E3 ubiquitin ligase during chaperone/proteasome coupling. Curr. Biol. 2001;11:1569–1577. doi: 10.1016/s0960-9822(01)00487-0. [DOI] [PubMed] [Google Scholar]
- Dhodapkar MV, Osman K, Teruya-Feldstein J, Filippa D, Hedvat CV, Iversen K, Kolb D, Geller MD, Hassoun H, Kewalramani T, Comenzo RL, Coplan K, Chen YT, Jungbluth AA. Expression of cancer/testis (CT) antigens MAGE-A1, MAGE-A3, MAGE-A4, CT-7, and NY-ESO-1 in malignant gammopathies is heterogeneous and correlates with site, stage and risk status of disease. Cancer Immun. 2003;3:9. [PubMed] [Google Scholar]
- Du J, Johnson LM, Jacobsen SE, Patel DJ. DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 2015;16:519–532. doi: 10.1038/nrm4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestein R, Cui X, Huie P, Cleary ML. Set domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3–9. Mol. Cell. Biol. 2000;20:4900–4909. doi: 10.1128/mcb.20.13.4900-4909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemington EK, Speck SH, Kaelin WG. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. USA. 1993;90:6914–6918. doi: 10.1073/pnas.90.15.6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franci C, Zhou J, Jiang Z, Modrusan Z, Good Z, Jackson E, Kouros-Mehr H. Biomarkers of residual disease, disseminated tumor cells, and metastases in the MMTV-PyMT breast cancer model. PLoS One. 2013;8:e58183. doi: 10.1371/journal.pone.0058183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol. Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, Ogryzko V, Avantaggiati ML, Pestell RG. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J. Biol. Chem. 2000;275:20853–20860. doi: 10.1074/jbc.M000660200. [DOI] [PubMed] [Google Scholar]
- Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- Gaughan L, Logan IR, Neal DE, Robson CN. Regulation of androgen receptor and histone deacetylase 1 by Mdm2-mediated ubiquitylation. Nucleic Acids Res. 2005;33:13–26. doi: 10.1093/nar/gki141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaughan L, Stockley J, Wang N, McCracken SR, Treumann A, Armstrong K, Shaheen F, Watt K, McEwan IJ, Wang C, Pestell RG, Robson CN. Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res. 2011;39:1266–1279. doi: 10.1093/nar/gkq861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber M, Shilatifard A. Transcriptional elongation by RNA polymerase II and histone methylation. J. Biol. Chem. 2003;278:26303–26306. doi: 10.1074/jbc.R300014200. [DOI] [PubMed] [Google Scholar]
- Grewal SI, Moazedm D. Heterochromatin and epigenetic control of gene expression. Science. 2003;301:798–802. doi: 10.1126/science.1086887. [DOI] [PubMed] [Google Scholar]
- He B, Bai S, Hnat AT, Kalman RI, Minges JT, Patterson C, Wilson EM. An androgen receptor NH2-terminal conserved motif interacts with the COOH terminus of the Hsp70-interacting protein (CHIP) J. Biol. Chem. 2004a;279:30643–30653. doi: 10.1074/jbc.M403117200. [DOI] [PubMed] [Google Scholar]
- He B, Bowen NT, Minges JT, Wilson EM. Androgen-induced NH2- and COOH-terminal interaction inhibits p160 coactivator recruitment by activation function 2. J. Biol. Chem. 2001;276:42293–42301. doi: 10.1074/jbc.M107492200. [DOI] [PubMed] [Google Scholar]
- He B, Gampe RT, Kole AJ, Hnat AT, Stanley TB, An G, Stewart EL, Kalman RI, Minges JT, Wilson EM. Structural basis for androgen receptor interdomain and coactivator interactions suggests a transition in nuclear receptor activation function dominance. Mol. Cell. 2004b;16:425–438. doi: 10.1016/j.molcel.2004.09.036. [DOI] [PubMed] [Google Scholar]
- He B, Kemppainen JA, Voegel JJ, Gronemeyer H, Wilson EM. Activation function 2 in the human androgen receptor ligand binding domain mediates interdomain communication with the NH2- terminal domain. J. Biol. Chem. 1999;274:37219–37225. doi: 10.1074/jbc.274.52.37219. [DOI] [PubMed] [Google Scholar]
- He B, Kemppainen JA, Wilson EM. FXXLF and WXXLF sequences mediate the NH2-terminal interaction with the ligand binding domain of the androgen receptor. J. Biol. Chem. 2000;275:22986–22994. doi: 10.1074/jbc.M002807200. [DOI] [PubMed] [Google Scholar]
- He B, Lee LW, Minges JT, Wilson EM. Dependence of selective gene activation on the androgen receptor NH2- and COOH-terminal interaction. J. Biol. Chem. 2002a;277:25631–25639. doi: 10.1074/jbc.M202809200. [DOI] [PubMed] [Google Scholar]
- He B, Minges JT, Lee LW, Wilson EM. The FXXLF motif mediates androgen receptor-specific interactions with coregulators. J. Biol. Chem. 2002b;277:10226–10235. doi: 10.1074/jbc.M111975200. [DOI] [PubMed] [Google Scholar]
- He B, Wilson EM. Electrostatic modulation in steroid receptor recruitment of LXXLL and FXXLF motifs. Mol. Cell. Biol. 2003;23:2135–2150. doi: 10.1128/MCB.23.6.2135-2150.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helin K, Harlow E, Fattaey A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell. Biol. 1993;13:6501–6508. doi: 10.1128/mcb.13.10.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heemers HV, Tindall DJ. Androgen receptor (AR) coregulators: a diversity of functions converging on and regulating the AR transcriptional complex. Endocr. Rev. 2007;28:778–808. doi: 10.1210/er.2007-0019. [DOI] [PubMed] [Google Scholar]
- Jenster G, van der Korput HA, Trapman J, Brinkmann AO. Identification of two transcription activation units in the N-terminal domain of the human androgen receptor. J. Biol. Chem. 1995;270:7341–7346. doi: 10.1074/jbc.270.13.7341. [DOI] [PubMed] [Google Scholar]
- Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Höhfeld J, Patterson C. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 2001;276:42938–42944. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Ko S, Ahn J, Song CS, Kim S, Knapczyk-Stwora K, Chatterjee B. Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol. Endocrinol. 2011;25:433–444. doi: 10.1210/me.2010-0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachner M, O'Sullivan RJ, Jenuwein T. An epigenetic road map for histone lysine methylation. J. Cell Sci. 2003;116:2117–2124. doi: 10.1242/jcs.00493. [DOI] [PubMed] [Google Scholar]
- Lagarde WH, Blackwelder AJ, Minges JT, Hnat AT, French FS, Wilson EM. Androgen receptor exon 1 mutation causes androgen insensitivity by creating phosphorylation site and inhibiting melanoma antigen-A11 activation of NH2- and carboxyl-terminal interaction-dependent transactivation. J. Biol. Chem. 2012;287:10905–10915. doi: 10.1074/jbc.M111.336081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leader JE, Wang C, Fu M, Pestell RG. Epigenetic regulation of nuclear steroid receptors. Biochem. Pharmacol. 2006;72:1589–1596. doi: 10.1016/j.bcp.2006.05.024. [DOI] [PubMed] [Google Scholar]
- Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Su S, Blackwelder AJ, Minges JT, Wilson EM. Gain in transcriptional activity by primate-specific coevolution of melanoma antigen-A11 and its interaction site in androgen receptor. J. Biol. Chem. 2011;286:29951–29963. doi: 10.1074/jbc.M111.244715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubahn DB, Joseph DR, Sar M, Tan J, Higgs HN, Larson RE, French FS, Wilson EM. The human androgen receptor: complementary deoxyribonucleic acid cloning, sequence analysis and gene expression in prostate. Mol. Endocrinol. 1988;2:1265–1275. doi: 10.1210/mend-2-12-1265. [DOI] [PubMed] [Google Scholar]
- Mauger O, Klinck R, Chabot B, Muchardt C, Allemand E, Batsché E. Alternative splicing regulates the expression of G9A and SUV39H2 methyltransferases, and dramatically changes SUV39H2 functions. Nucleic Acids Res. 2015;43:1869–1882. doi: 10.1093/nar/gkv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger E, Wissmann M, Yin N, Müller JM, Schneider R, Peters AH, Günther T, Buettner R, Schüle R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- Minges JT, Grossman G, Zhang P, Kafri T, Wilson EM. Post-translational down-regulation of melanoma antigen-A11 (MAGE-A11) by human p14-ARF tumor suppressor. J. Biol. Chem. 2015;290:25174–25187. doi: 10.1074/jbc.M115.663641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minges JT, Su S, Grossman G, Blackwelder AJ, Pop EA, Mohler JL, Wilson EM. Melanoma antigen-A11 (MAGE-A11) enhances transcriptional activity by linking androgen receptor dimers. J. Biol. Chem. 2013;288:1939–1952. doi: 10.1074/jbc.M112.428409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas E, Roumillac C, Trouche D. Balance between acetylation and methylation of histone H3 lysine 9 on the E2F-responsive dihydrofolate reductase promoter. Mol. Cell. Biol. 2003;23:1614–1622. doi: 10.1128/MCB.23.5.1614-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- O'Carroll D, Scherthan H, Peters AH, Opravil S, Haynes AR, Laible G, Rea S, Schmid M, Lebersorger A, Jerratsch M, Sattler L, Mattei MG, Denny P, Brown SD, Schweizer D, Jenuwein T. Isolation and characterization of Suv39h2, a second histone H3 methyltransferase gene that displays testis-specific expression. Mol. Cell. Biol. 2000;20:9423–9433. doi: 10.1128/mcb.20.24.9423-9433.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozdağ H, Teschendorff AE, Ahmed AA, Hyland SJ, Blenkiron C, Bobrow L, Veerakumarasivam A, Burtt G, Subkhankulova T, Arends MJ, Collins VP, Bowtell D, Kouzarides T, Brenton JD, Caldas C. Differential expression of selected histone modifier genes in human solid cancers. BMC Genomics. 2006;7:90. doi: 10.1186/1471-2164-7-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patani N, Jiang WG, Newbold RF, Mokbel K. Histone-modifier gene expression profiles are associated with pathological and clinical outcomes in human breast cancer. Anticancer Res. 2011;31:4115–4125. [PubMed] [Google Scholar]
- Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- Ponguta LA, Gregory CW, French FS, Wilson EM. Site-specific androgen receptor serine phosphorylation linked to epidermal growth factor-dependent growth of castration-recurrent prostate cancer. J. Biol. Chem. 2008;283:20989–21001. doi: 10.1074/jbc.M802392200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poukka H, Karvonen U, Janne OA, Palvimo JJ. Covalent modification of the androgen receptor by small ubiquitin-like modifier 1 (SUMO-1) Proc. Natl. Acad. Sci. USA. 2000;97:14145–14150. doi: 10.1073/pnas.97.26.14145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quarmby VE, Kemppainen JA, Sar M, Lubahn DB, French FS, Wilson EM. Expression of recombinant androgen receptor in cultured mammalian cells. Mol. Endocrinol. 1990;4:1399–1407. doi: 10.1210/mend-4-9-1399. [DOI] [PubMed] [Google Scholar]
- Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol. Rev. 2002;188:22–32. doi: 10.1034/j.1600-065x.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- Simental JA, Sar M, Lane MV, French FS, Wilson EM. Transcriptional activation and nuclear targeting signals of the human androgen receptor. J. Biol. Chem. 1991;266:510–518. [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Su S, Minges JT, Grossman G, Blackwelder AJ, Wilson EM. Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins. J. Biol. Chem. 2013;288:24809–24824. doi: 10.1074/jbc.M113.468579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2002;3:11–20. doi: 10.1038/nrm714. [DOI] [PubMed] [Google Scholar]
- Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira FQ, Costa-Pinheiro P, Ramalho-Carvalho J, Pereira A, Menezes FD, Antunes L, Carneiro I, Oliveira J, Henrique R, Jerónimo C. Deregulated expression of selected histone methylases and demethylases in prostate carcinoma. Endocr. Relat. Cancer. 2013;21:51–61. doi: 10.1530/ERC-13-0375. [DOI] [PubMed] [Google Scholar]
- Wu H, Min J, Lunin VV, Antoshenko T, Dombrovski L, Zeng H, Allali-Hassani A, Campagna-Slater V, Vedadi M, Arrowsmith CH, Plotnikov AN, Schapira M. Structural biology of human H3K9 methyltransferases. PLoS One. 2010;5:e8570. doi: 10.1371/journal.pone.0008570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KA, Hwangbo B, Kim IJ, Park S, Kim HS, Kee HJ, Lee JE, Jang YK, Park JG, Lee JS. Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer. Carcinogenesis. 2006;27:2217–2222. doi: 10.1093/carcin/bgl084. [DOI] [PubMed] [Google Scholar]