Abstract
The γ-secretase complex represents an evolutionarily conserved family of transmembrane aspartyl proteases that cleave numerous type-I membrane proteins, including the β-amyloid precursor protein (APP) and the receptor Notch. All known rare mutations in APP and the γ-secretase catalytic component, presenilin, which lead to increased amyloid βpeptide production, are responsible for early-onset familial Alzheimer’s disease. β-amyloid protein precursor-like (APPL) is the Drosophila ortholog of human APP. Here, we created Notch- and APPL-based Drosophila reporter systems for in vivo monitoring of γ-secretase activity. Ectopic expression of the Notch- and APPL-based chimeric reporters in wings results in vein truncation phenotypes. Reporter-mediated vein truncation phenotypes are enhanced by the Notch gain-of-function allele and suppressed by RNAi-mediated knockdown of presenilin. Furthermore, we find that apoptosis partly contributes to the vein truncation phenotypes of the APPL-based reporter, but not to the vein truncation phenotypes of the Notch-based reporter. Taken together, these results suggest that both in vivo reporter systems provide a powerful genetic tool to identify genes that modulate γ-secretase activity and/or APPL metabolism.
Keywords: γ-secretase, Alzheimer’s disease, APPL, Notch, presenilin
INTRODUCTION
The γ-secretase complex, which consists of at least four proteins-Presenilin (PS), Nicastrin, Aph-1, and Pen-2, serves as a transmembrane aspartyl protease that plays a critical role in Alzheimer’s disease (AD) and Notch signaling pathway (Bai et al., 2015; De Strooper, 2003; Fortini, 2009). Amyloid βpeptides (Aβs), which are the main constituents of senile plaques present in the brain affected with AD, are generated by sequential cleavages of the β-amyloid precursor protein (APP) by β-secretase (BACE) and γ-secretase (Esler and Wolfe, 2001; Goedert, 2015). Another well-known substrate for γ-secretase is Notch, whose signaling controls a large number of cell fate decisions during development (Fortini, 2009; Louvi and Artavanis-Tsakonas, 2006). The cleavage of both APP and Notch within their single-pass transmembrane domains by γ-secretase releases cytosolic fragments and allows them to enter the nucleus, thereby regulating gene transcription (Brown et al., 2000; Cao and Südhof, 2001; Louvi and Artavanis-Tsakonas, 2006; Wang et al., 2014; Figs. 1A and 1B). This γ-secretase-dependent regulated intramembrane proteolysis (RIP) is tightly controlled (Brown et al., 2000), and thus alterations in the RIP of APP and Notch result in developmental defects and diseases (Esler and Wolfe, 2001; Louvi and Artavanis-Tsakonas, 2006).
Fig. 1. Generation of the Notch- and APPL-based Fusion Reporters.
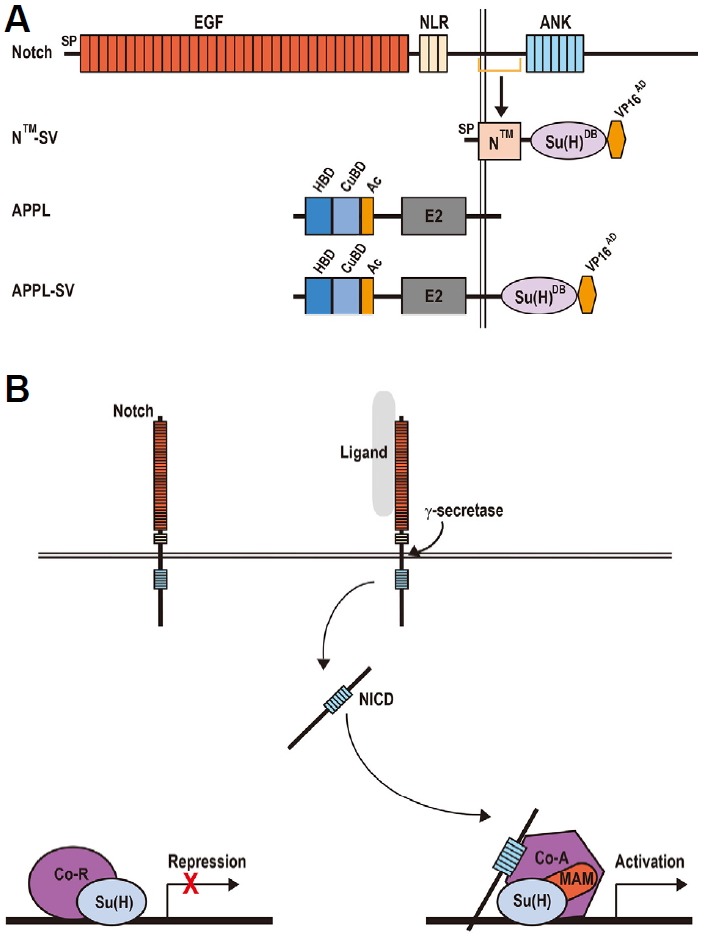
(A) Schematic diagram of the Notch- and APPL-based reporter transgenes. SP, signal peptide; EGF, epidermal growth factor-like; LNR, Lin12-Notch repeats; ANK, seven ankyrin repeat domains; NICD, Notch intracellular domain; Su(H)DB, the DNA-binding domain of Su(H); VP16AD, the transcriptional activation domain of the virus VP16; HBD, heparin-binding domain; CuBD, copper-binding domain; Ac, acidic domain; E2, APP extracellular carbohydrate domain. (B) Transcriptional switch in the Drosophila canonical Notch signaling pathway. In the absence of Notch signaling, Su(H) protein functions as a transcriptional repressor through recruiting co-repressors (Co-R) including Hairless, Groucho and CtBP. Notch activation is triggered by ligand binding and sequential cleavages of ADAM family metalloproteases and γ-secretase. The second cleavage liberates the NICD, which then enters the nucleus and associates with Su(H) and Mastermind (MAM) to drive the transcription of Notch target genes. Co-A, co-activators.
AD is the most common form of senile dementia characterized by the deposition of aggregation-prone Aβs and neurofibrillary tangles in the brain (Selkoe, 1998). Interestingly, early-onset familial Alzheimer’s disease (EOAD) is caused by rare mutations in APP, PS1, and PS2 (Tanzi, 2012). This is consistent with the fact that PS is the catalytic subunit of the γ-secretase complex whereas APP is a substrate for γ-secretase (De Strooper, 2003). In addition, γ-secretase activity and Aβ production were not detected in PS1/PS2 double-knockout cells (Herreman et al., 2000; Zhang et al., 2000). Therefore, γ-secretase serves as a therapeutic target to reduce Aβ production and development of γ-secretase inhibitors can provide an effective therapy for AD. However, there is still one major concern that γ-secretase inhibitors might cause deleterious side effects because they also affect the RIP of numerous other substrates including Notch (Esler and Wolfe, 2001; McCarthy et al., 2009). One possible approach to overcome this problem is to identify new therapeutic target genes that specifically affect the γ-secretase-dependent processing of APP rather than other substrates. To this end, we built a Drosophila reporter system for in vivo detection of γ-secretase activity by taking advantage of the Notch signaling pathway in the wing.
Canonical Notch signaling, which is highly conserved in the animal kingdom, is initiated by the binding of ligands including Delta, Serrate, and LAG-2 (Fortini, 2009; Louvi and Artavanis-Tsakonas, 2006). Ligand binding induces sequential cleavages of the Notch receptor by ADAM family metalloproteases and γ-secretase, releasing the Notch intracellular domain (NICD) (Fortini, 2009; Louvi and Artavanis-Tsakonas, 2006). NICD then enters the nucleus and forms an active transcriptional complex with the DNA-binding protein Suppressor of Hairless (Su(H)) and Mastermind (MAM), triggering expression of Notch target genes (Fortini, 2009; Louvi and Artavanis-Tsakonas, 2006; Fig. 1B). In the absence of Notch signaling, Su(H) proteins associate with various core-pressors to actively repress the transcription of Notch target genes (Fortini, 2009; Louvi and Artavanis-Tsakonas, 2006; Fig. 1B). These findings demonstrate that Su(H) is the key effector of Notch signaling.
The Drosophila homolog of human APP is known as β-amyloid protein precursor-like (APPL) (Martin-Morris and White, 1990). Flies homozygous for Appl deletion allele showed a fast phototaxis defect that can be restored by human APP as well as fly APPL, revealing their functional conservation (Luo et al., 1992). Furthermore, APPL is proteolytically cleaved by a BACE-like secretase (dBACE) and γ-secretase, producing a neurotoxic Aβ-like fragment in Drosophila (Carmine-Simmen et al., 2009). However, dBACE does not appear to recognize and cleave human APP at the β-secretase (BACE) cleavage site, suggesting the lack of a clear homolog of human BACE in Drosophila (Carmine-Simmen et al., 2009; Greeve et al., 2004). In contrast, each component and the activity of the γ-secretase complex appear to be evolutionarily conserved across invertebrates and vertebrates (De Strooper, 2003; Selkoe and Wolfe, 2007). In support of this claim, Drosophila γ-secretase can cleave human APP and thus produce Aβ peptides in combination with human BACE (Greeve et al., 2004).
Here, we have created a Drosophila reporter system for in vivo monitoring of γ-secretase activity and it provides a genetic tool to discover novel genes that modulate γ-secretase-mediated RIP of Notch and APP-like (APPL), and APPL metabolism.
MATERIALS AND METHODS
Drosophila strains
The w1118 Drosophila strain was used as a wild-type control. The following flies were obtained from the Bloomington Stock Center: UAS-psn RNAi (#27681), UAS-p35 (#5073), UAS-psn (#8310), e16E-GAL4 (#30557), and NAx-E2 (#51660). The e16E-GAL4 enhancer trap line was shown to drive expression of a reporter gene in the striped pattern of engrailed during embryonic development (Weiss et al., 2001) and also in the posterior compartment of the wing imaginal disc during third instar larval stage (Johnson et al., 1995).
Expression constructs
The UAS-N™-SV and UAS-APPL-SV constructs were generated by PCR and/or restriction enzyme-based strategies and subcloned into the pUAST vector (Brand and Perrimon, 1993). For the Litmus 28-Su(H)-VP16 (SV), a NcoI-PstI fragment from pGEX-Su(H) (Bailey and Posakony, 1995), which contains the DNA-binding domain (amino acids 109–457; GenBank: AAF53434.1) of Su(H), and a PstI-XbaI fragment from pAct-GAL4-VP16 (Han and Manley, 1993), which contains the transcriptional activation domain (amino acids 2–79; GenBank: AIZ65950.1) of VP16, were subcloned into the Litmus 28 vector (New England Biolabs, Inc.). For the Litmus 28-N™-SV, a SalI-NcoI fragment from a Notch minigene clone (Wharton et al., 1985), which contains the N™ (amino acids 1710–1891; GenBank: AAF45848.2), and a BglII-SalI fragment from Litmus 28-NΔECN (Ju et al., 2000), which contains the signal peptide region (amino acids 1–60) of Notch, were subcloned into the Litmus 28-SV. For the UAS-N™-SV, a BglII-XbaI fragment from the Litmus 28-N™-SV was subcloned into the pUAST vector. For the UAS-APPL-SV construct, a EcoRI-NcoI fragment from GH04413, which encodes the full-length APPL (amino acids 1–887; GenBank: AAF45520), and a NcoI-XbaI fragment from the Limus 28-SV, were subcloned into the pUAST vector. These construct sequences were confirmed by DNA sequencing. Transgenic flies carrying these constructs were created according to standard D. melanogaster transformation procedures (KAIST transgenic fly service). Multiple independent lines for the same transgenic construct are indicated by superscripts. For example, N™-SV61f and N™-SV42m are independent insertion lines for the construct UAS-N™-SV.
Immunohistochemistry
Expression patterns of transgenes were visualized using anti-VP16 monoclonal antibody (14-5, Santa Cruz Biotech.) and 3,3′-diaminobenzidine reaction, as described previously (Jeong et al., 2012; Kim et al., 1995).
Preparation of Drosophila wings and phenotypic characterization
Using two pairs of forceps, the wings of female and male adult flies (younger than 3 days) were carefully cut off and subsequently were arranged in the same orientation on a glass slide. A coverslip was applied and each corner of the coverslip was sealed with a regular nail polish. In wild-type wing, the average L4 to L3 vein length ratio was 0.97 and the average L5 to L3 vein length ratio was 0.58 (see Supplementary Fig. S1A). The average percentages of L4 and L5 vein truncations were calculated by the following equations (see Supplementary Fig. S1B):
Statistical analysis
All quantitative data are presented as mean ± standard error of the mean (S.E.M.). Statistical significance was determined using unpaired t-test (*p < 0.05, **p < 0.01, and ***p < 0.001).
RESULTS
Generation of transgenic reporters for in vivo detection of the processing of Notch and APPL
We decided to take advantage of the Notch signaling pathway to generate transgenic reporters for in vivo detection of the activity of γ-secretase, which acts as a key player in Alzheimer’s disease (AD) (Esler and Wolfe, 2001; Selkoe, 1998). Notch is cleaved within its single-pass transmembrane domain by γ-secretase (Fortini, 2009; Louvi and Artavanis-Tsakonas, 2006). The cleavage of the Notch receptor releases its intracellular domain (NICD) and translocates it into the nucleus, thereby driving the expression of Notch target genes through the assembly of an active transcriptional complex with Su(H) and Mastermind (Fortini, 2009; Louvi and Artavanis-Tsakonas, 2006; Figs. 1A and 1B). Vein pattern formation in the Drosophila wing is controlled by Notch signaling (de Cellis, 1998). Reduction in Notch signaling activity leads to the formation of thicker veins, whereas increased Notch activity results in loss of veins (de Cellis and Garcia-Bellido, 1994a). These observations prompted us to examine whether Notch activity is correlated with vein phenotypes in a dose-dependent manner. Normal vein patterning was observed in wild-type females (Fig. 2A), whereas in females heterozygous for NAx-E2, a dominant gain-of-function (GOF) Abruptex (Ax) allele (de Cellis and Garcia-Bellido, 1994b), vein L5 consistently failed to reach the wing margin (5.4% L5 vein truncation; Figs. 2B and 2I). This Ax phenotype of vein L5 was significantly increased up to 26.1% vein truncation in females homozygous for NAx-E2 (p = 0.002, t-test; Figs. 2C and 2I). In addition, these homozygous mutants displayed a moderately truncated L4 vein (4.8% vein truncation; Figs. 2C and 2I). These findings suggest that the severity of Ax vein phenotype is proportional to Notch signaling activity. To further address whether γ-secretase activity is required for patterning of the Drosophila wing veins, we took advantage of the RNA interference (RNAi) technique. When the Drosophila presenilin (psn) gene was knocked down by overexpression of psn RNAi transgene using e16E-GAL4, which is expressed in the posterior compartment of the wing (Johnson et al., 1995), wing patterns showed robust vein thickening but no vein truncation phenotypes (Figs. 2D and 2I). This vein thickening phenotype, which is reminiscent of the phenotype found in Notch loss-of-function alleles (de Cellis and Garcia-Bellido, 1994a), was completely restored by coexpression of wild-type Psn (Fig. 2F). Overexpression of wild-type Psn alone hardly affected vein patterning (Figs. 2E and 2I). These results indicate target gene specificity of the psn RNAi transgene. Furthermore, knockdown of psn in the posterior compartment using e16E-GAL4 completely suppressed the vein truncation phenotype observed in males hemizygous for NAx-E2 (Figs. 2G and 2H). Taken together, these findings strongly suggest that γ-secretase activity contributes to vein pattern formation in the Drosophila wing through the Notch signaling pathway.
Fig. 2. Wing vein phenotypes of Notch gain-of-function and presenilin loss-of-function alleles.
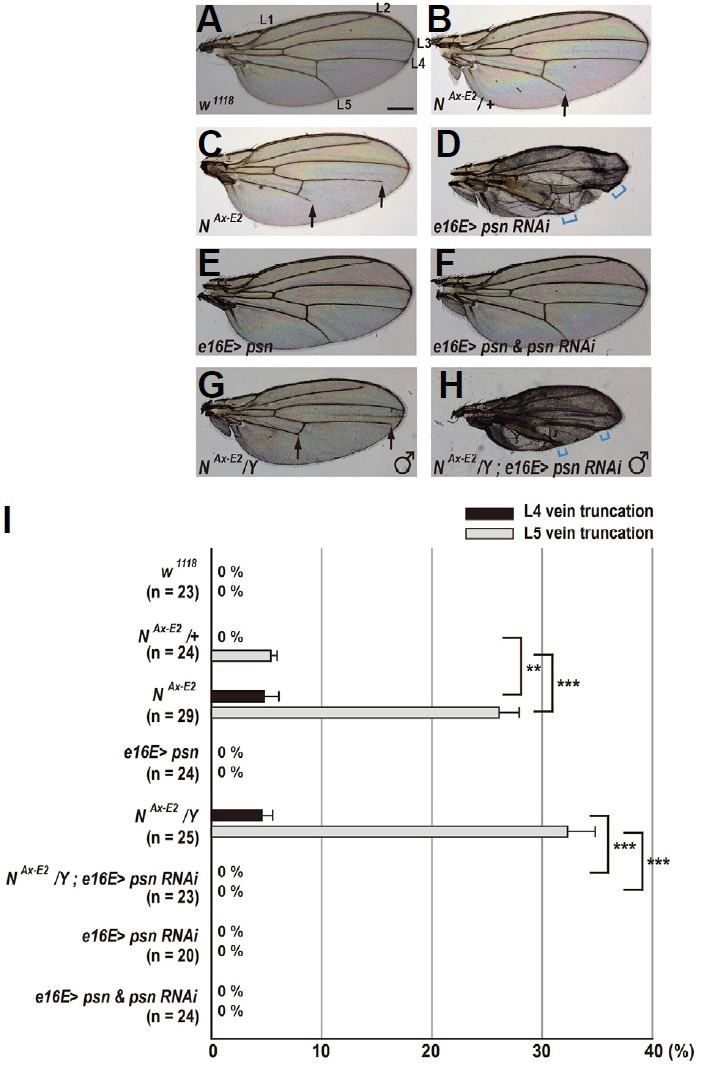
(A) Wild-type wing shows a normal pattern of five longitudinal veins (L1–L5). Scale bar indicates 50 μm. (B) In females heterozygous for NAx-E2, the wing displays mild truncation only in L5 vein (arrow). (C) In females homozygous for NAx-E2, the wing displays mild truncation in L4 vein and moderate truncation in L5 vein (arrows). (D) Overexpression of psn RNAi in the posterior compartment results in vein thickening (square brackets in blue). (E) Female wing overexpressing wild-type Psn shows a normal vein pattern. (F) Overexpression of wild-type Psn completely rescues the psn RNAi-induced vein thickening phenotype. (G) In males hemizygous for NAx-E2, the wing exhibits mild truncation in L4 vein and moderate truncation in L5 vein (arrows). (H) Vein truncation phenotype observed in NAx-E2 hemizygotes is suppressed by RNAi-mediated knockdown of psn. Square brackets indicate vein thickening phenotype.(I) Percentages of wing vein truncations in females and males with indicated genotypes (**p < 0.01 and ***p < 0.001, t-test). Error bars indicate S.E.M. by t-test. n = number of wings scored for each genotype.
To monitor the proteolytic cleavage of Notch and APPL by γ-secretase within their transmembrane domains, we explored a sensitive reporter system, in which the chimeric transcriptional activator Su(H)-VP16 (SV) is fused to either a subfragment of Notch (N™) or full-length APPL (UAS-N™-SV and UAS-APPL-SV in Fig. 1A). The N™ subfragment possesses a transmembrane domain and the N-terminal region of NICD (Fig. 1A). The SV was created by fusion of the DNA-binding domain of Su(H) transcription factor (Bailey and Posakony, 1995) and the transcriptional activation domain of the virus VP16 (Sadowski et al., 1988) (Fig. 1A). Therefore, in case of nuclear translocation of the SV proteins, they are supposed to activate some of the Notch target genes, mimicking increased Notch pathway activity. Increased Notch activity in the wing can be visualized by the severity of the resulting vein phenotypes. Based on the fact that the N™ subfragment and APPL contain a γ-secretase-mediated cleavage site, we reasoned that the expression of both reporter transgenes under the control of e16E-GAL4 provides a sensitive assay for γ-secretase-dependent processing of Notch and APPL.
Expression of the notch- and APPL-based chimeric reporters in wings caused loss of wing veins
To determine whether both the N™-SV and APPL-SV chimeric reporter proteins transduce Notch signaling activity, we ectopically overexpressed these transgenes using e16E-GAL4 (Figs. 3E and 3H). Ectopic expression of N™-SV61f in the posterior compartment of the wing resulted in moderate truncations of L4 and L5 veins in females (Fig. 3D). These vein truncation phenotypes were not observed in wild-type, e16E-GAL4/+, and N™-SV61f/+ female wings (Figs. 3A–3C). Interestingly, in females expressing APPL-SV85m, we observed severe truncations of L4 and L5 veins (Fig. 3G). However, vein pattern formation in females heterozygous for the APPL-SV85m transgene was normal (Fig. 3F). These findings might suggest that both the N™-SV and APPL-SV reporter proteins were processed at least by γ-secretase, and they further demonstrate that the γ-secretase-cleaved C-terminal fragments of both reporters including the SV can transduce Notch activity in the wing.
Fig. 3. Ectopic expression of Notch- and APPL-based reporters in wings results in vein truncations.
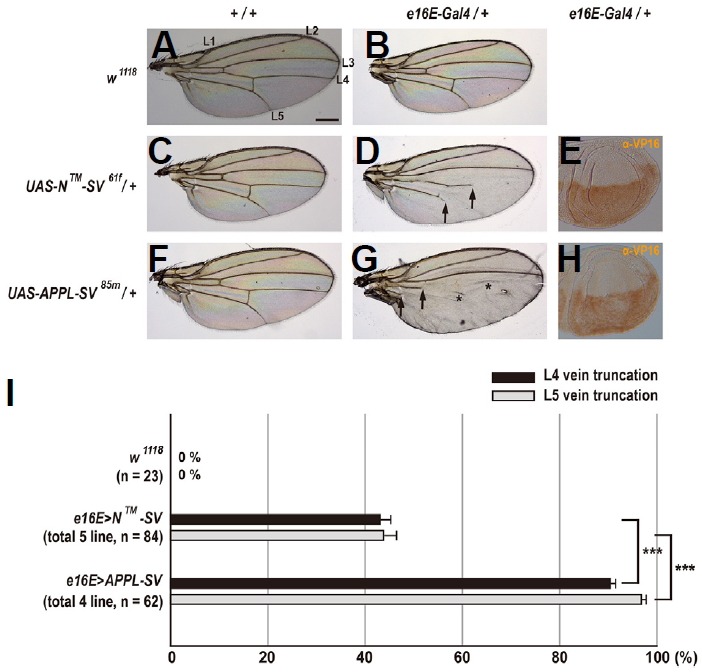
(A) Wild-type wing shows a normal pattern of five longitudinal veins (L1–L5). Scale bar indicates 50 μm. (B) Female heterozygous for e16E-GAL4 displays a normal vein pattern. (C) Female heterozygous for UAS-N™-SV61f shows a normal vein pattern. (D) Overexpression of N™-SV61f in the posterior compartment leads to moderate vein truncation (arrows). (E) The N™-SV proteins are expressed homogeneously in the posterior compartment of the wing imaginal disc. Anterior is up. (F) Female heterozygous for UAS-APPL-SV85m shows a normal vein pattern. (G) Overexpression of APPL-SV85m in the posterior compartment causes severe vein truncation (arrows). Asterisks indicate a false vein. (H) Expression pattern of the APPL-SV proteins is relatively sparse in the posterior compartment of the wing imaginal disc. Anterior is up. (I) Percentages of wing vein truncations in females with indicated genotypes (***p < 0.001, t-test). Error bars indicate S.E.M. by t-test. n = number of wings scored for each genotype.
To further address the difference in phenotypic severity between N™-SV and APPL-SV (Figs. 3D and 3G), we obtained a total of 5 independent transgenic lines for N™-SV and a total of 4 lines for APPL-SV. On average, ectopic expression in 5 different lines for N™-SV caused 43.1% L4 vein truncations and 43.8% L5 vein truncations (n = 84), while expression in 4 different lines for APPL-SV resulted in 90.4% L4 vein truncations and 96.8% L5 vein truncations (n = 62) (Fig. 3I). An almost 2-fold difference in the percentage of both vein truncations between N™-SV and APPL-SV (p < 0.001, t-test; Fig. 3I) strongly suggests that the N™-SV and APPL-SV proteins are differentially processed by γ-secretase and/or the γ-secretase-cleaved C-terminal portions of both reporter proteins generate different levels of Notch transducing activity. Another possibility is that the APPL-SV proteins are more abundantly expressed than the N™-SV proteins.
Reporter-mediated wing vein phenotypes are enhanced by Notch and suppressed by presenilin
Based on wing vein phenotypes, we deduced that the N™-SV and APPL-SV reporter proteins transduce Notch activity (Fig. 3). To further address this issue, we investigated genetic interactions among N™-SV, Notch, and psn. Ectopic expression of N™-SV61f in the posterior compartment resulted in 38.3% L4 vein truncations and 33.6% L5 vein truncations (Figs. 4B and 4G). These N™-SV61f vein phenotypes were significantly enhanced by one copy of NAx-E2 allele: from 38.3% to 69.2% for L4 vein truncations (p < 0.001, t-test) and from 33.6% to 41.0% for L5 vein truncations (p = 0.001, t-test) (Figs. 4C, 4D, and 4G). Similar patterns of genetic interactions were also observed with a different insertion line, N™-SV42m (p < 0.001, t-test for both veins; Supplementary Figs. S2C, S2D, and S2G). Overexpression of N™-SV42m in the wing led to 57.0% L4 vein truncations and 57.7% L5 vein truncations, which are greater than those obtained after ectopic expression of N™-SV61f (Supplementary Figs. S2B and S2G). These findings support the claim that the N™-SV proteins transduce Notch activity in the wing. In contrast, the N™-SV61f and N™-SV42m vein truncation phenotypes were completely suppressed by RNAi-mediated knockdown of psn (Figs. 4E–4G and Supplementary Figs. S2E–S2G), indicating that Psn is required for the gain of N™-SV function in the wing. Interestingly, the psn RNAi-mediated vein thickening phenotype was also suppressed, but it was not completely suppressed by overexpression in either N™-SV transgenic line (Figs. 4E, 4F, Supplementary Figs. S2E and S2F). Therefore, knockdown of psn is epistatic to N™-SV overexpression with respect to vein pattern formation in the wing.
Fig. 4. Genetic interactions among N™-SV61f, Notch, and presenilin alleles.
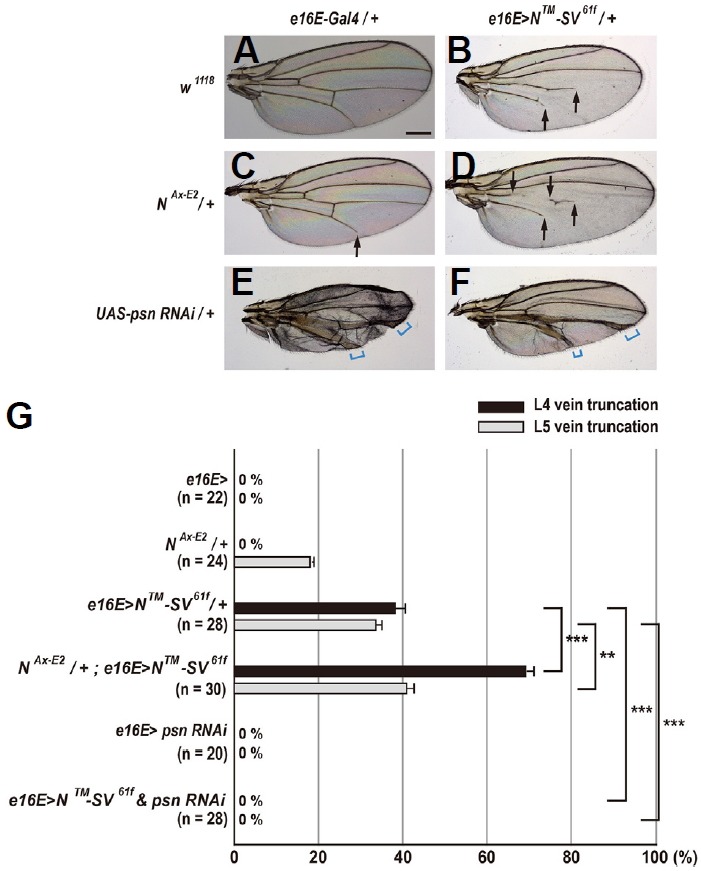
(A) Females heterozygous for e16E-Gal4 show a normal pattern of five longitudinal veins. Scale bar indicates 50 μm. (B) Overexpression of N™-SV61f in the posterior compartment results in moderate vein truncation (arrows). (C) In females heterozygous for NAx-E2, the wing shows mild truncation only in L5 vein (arrow). (D) One copy of NAx-E2mutation slightly enhances the gain-of-function (GOF) phenotype of N™-SV61f (arrows). (E) Overexpression of psn RNAi in the posterior compartment results in vein thickening (square brackets). (F) Overexpression of psn RNAi completely suppresses the GOF phenotype of N™-SV61f. Square brackets indicate vein thickening phenotype. (G) Percentages of wing vein truncations in females with indicated genotypes (**p < 0.01 and ***p < 0.001, t-test). Error bars indicate S.E.M. by t-test. n = number of wings scored for each genotype.
We next analyzed genetic interactions among APPL-SV, Notch, and psn. Females overexpressing APPL-SV85m in the posterior compartment showed highly penetrant vein truncation defects: 84.8% L4 vein truncations and 90.0% L5 vein truncations (Figs. 5B and 5G). These APPL-SV85m vein phenotypes were enhanced by one copy of NAx-E2 allele: from 84.8% to 94.7% for L4 vein truncations (p = 0.001, t-test) and from 90.0% to 98.2% for L5 vein truncations (p = 0.003, t-test), suggesting that the APPL-SV proteins can act as a Notch signal transducer in the wing (Figs. 5C, 5D, and 5G). Similar patterns of genetic interactions for both L4 and L5 vein phenotypes were not observed with the APPL-SV82m transgene, a different insertion line (p > 0.3, t-test; Supplementary Figs. S3C, S3D, and S3G), probably due to high penetrance of vein truncation defects in females overexpressing APPL-SV82m alone (92.8% for L4 veins and 100% for L5 veins; Supplementary Fig. S3G). In addition, we found that the APPL-SV85m vein truncation phenotypes were robustly suppressed by RNAi-mediated knockdown of psn: from 84.8% to 51.2% for L4 vein truncations (p<0.001, t-test) and from 90.0% to 1.6% for L5 vein truncations (p < 0.001, t-test) (Figs. 5E–5G). Partial suppression especially in L4 veins, which is different from that observed in females coexpressing psn RNAi and N™-SV (Figs. 4F and 4G), might indicate that APPL-SV-induced Notch activity antagonizes the knockdown effects of psn to a greater extent than N™-SV. Furthermore, similar patterns of suppressive genetic interactions were also observed with the APPL-SV82m transgene (p < 0.001, t-test for both L4 and L5 vein truncations; Supplementary Figs. S3E–S3G), demonstrating the requirement of Psn for the gain of APPL-SV function in the wing. Females coexpressing psn RNAi and APPL-SV exhibited large reduction in vein thickening phenotype compared to females overexpressing psn RNAi alone (Figs. 5E and 5F, Supplementary S3E and S3F), which is indicative of the antagonistic relationships between knockdown of psn and APPL-SV overexpression in patterning of the wing veins.
Fig. 5. Genetic interactions among APPL-SV85m, Notch, and presenilin alleles.
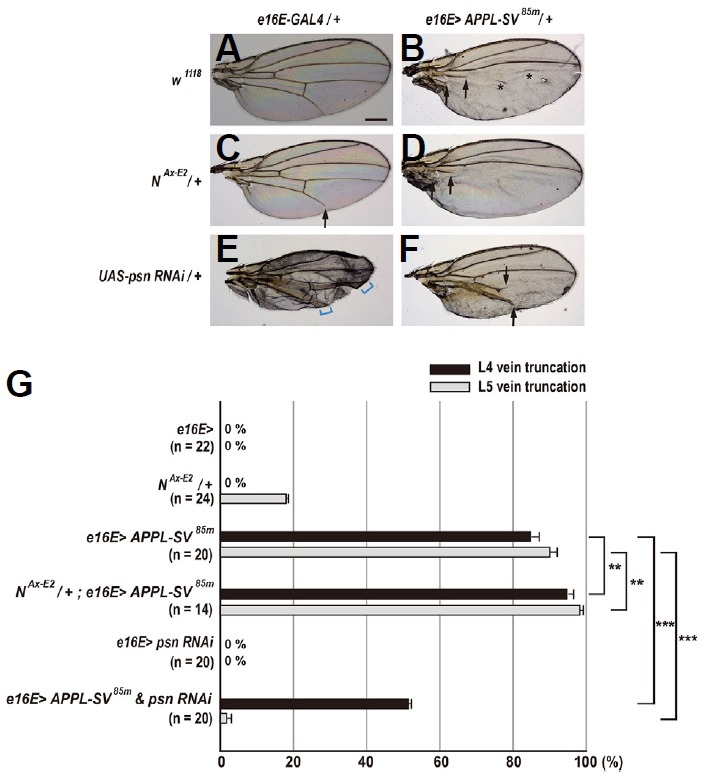
(A) Females heterozygous for e16E-Gal4 show a normal pattern of five longitudinal veins. Scale bar indicates 50 μm. (B) Overexpression of APPL-SV85m in the posterior compartment results in severe vein truncation (arrows). Asterisks indicate a false vein. (C) In females heterozygous for NAx-E2, the wing shows mild truncation only in L5 vein (arrow). (D) One copy of NAx-E2mutation slightly enhances the GOF phenotype of APPL-SV85m (arrows). (E) Overexpression of psn RNAi in the posterior compartment results in vein thickening (square brackets). (F) Overexpression of psn RNAi partly suppresses the GOF vein phenotype of N™-SV61f. (G) Percentages of wing vein truncations in females with indicated genotypes (**p < 0.01 and ***p < 0.001, t-test). Error bars indicate S.E.M. by t-test. n = number of wings scored for each genotype.
The role of apoptosis in reporter-induced wing vein truncations
Since Notch plays a role in apoptosis during Drosophila visual system development (Bertet et al., 2014; Brachmann and Cagan, 2003), we investigated whether Notch-mediated regulation of apoptosis is involved in patterning of the wing veins. Interestingly, females overexpressing the baculovirus pan-caspase inhibitor p35, which has been shown to prevent apoptotic death in Drosophila (Hay et al., 1994), exhibited a mild phenotype only in L5 vein, indicating a positive, but a minor role of apoptosis in vein pattern formation (3.5% vein truncation; Figs. 6D and 6G). In females expressing both p35 and N™-SV61f in the posterior compartment, we observed a small but significant increase in vein phenotypes compared to females overexpressing N™-SV61f: from 38.3% to 51.1% for L4 vein truncations (p < 0.001, t-test) and from 33.6% to 46.1% for L5 vein truncations (p < 0.001, t-test) (Figs. 6B, 6E and 6G). Similar enhancement in vein phenotype was also found in females coexpressing p35 and N™-SV42m in the posterior compartment: from 57.0% to 60.5% for L4 vein truncations (p = 0.03, t-test) and from 57.7% to 76.6% for L5 vein truncations (p < 0.001, t-test) (Supplementary Figs. S4B, S4E, and S4G). Taken together, this seemingly additive genetic interaction between p35 and N™-SV suggests that N™-SV-induced vein truncation is not due to apoptosis.
Fig. 6. Apoptosis partly contributes to vein truncations induced by the APPL-based reporter.
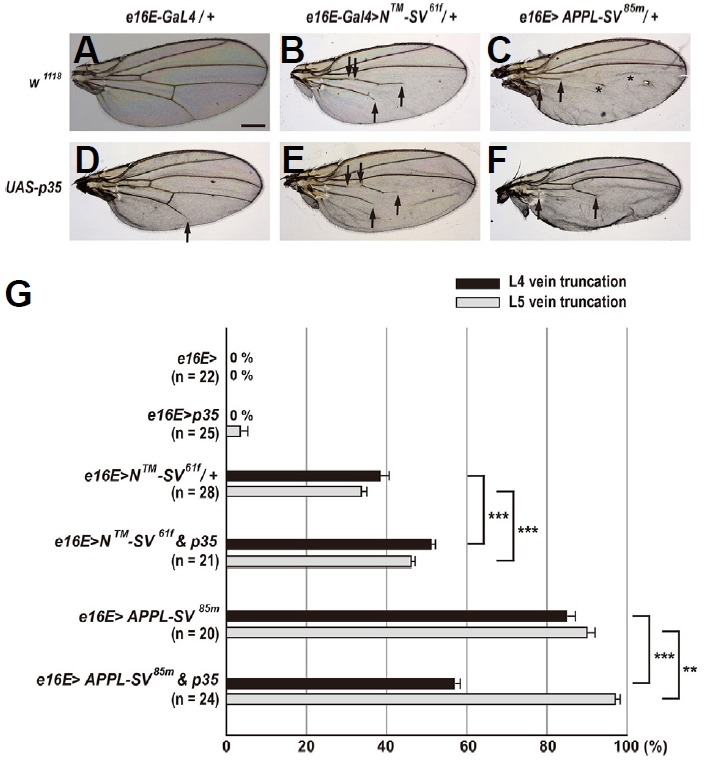
(A) Females heterozygous for e16E-Gal4 show a normal pattern of five longitudinal veins. Scale bar indicates 50 μm. (B) Overexpression of N™-SV61f in the posterior compartment results in moderate vein truncation (arrows). (C) Overexpression of APPL-SV85m in the posterior compartment leads to severe vein truncation (arrows). Asterisks indicate a false vein. (D) Overexpression of p35 in the posterior compartment results in slight vein truncation (arrow). (E) Coexpression of p35in the posterior compartment slightly increases the GOF vein phenotype of N™-SV61f (arrows). (F) Coexpression of p35in the posterior compartment differentially affects the GOF vein phenotype of APPL-SV85m (arrows). (G) Percentages of wing vein truncations in females with indicated genotypes (**p < 0.01 and ***p < 0.001, t-test). Error bars indicate S.E.M. by t-test. n = number of wings scored for each genotype.
To examine whether APPL-SV-induced vein phenotypes result from apoptosis, we coexpressed p35 and APPL-SV85m in the posterior compartment of the wing. Unexpectedly, we observed that coexpression of p35 and APPL-SV85m robustly suppressed L4 vein truncation phenotypes of APPL-SV85m from 84.8% to 56.8% (p < 0.001, t-test), whereas coexpression of p35 and APPL-SV85m enhanced L5 vein truncation phenotypes from 90.0% to 97.0% (p = 0.003, t-test) (Figs. 6C, 6F, and 6G). Suppressive genetic interactions were also observed in L4 veins, but not in L5 veins with the APPL-SV82m transgene: from 92.8% to 54.8% for L4 vein truncations (p < 0.001, t-test) and no change was observed in L5 vein truncations (Supplementary Figs. S4C, S4F, and S4G). Strong suppressive interactions between p35 and APPL-SV, especially in L4 veins suggest that overexpression of APPL-SV leads to cell death, and they further demonstrate that APPL-SV-induced cell death partly contributes to L4 vein truncation phenotypes. However, phenotypic enhancement observed in L5 veins, which is similar to that observed in females coexpressing p35 and N™-SV (Fig. 6G), suggests that apoptosis is not attributable to L5 vein truncation phenotypes of APPL-SV.
DISCUSSION
Drosophila melanogaster provides a powerful genetic tool to analyze gene function associated with human diseases including Alzheimer’s disease (AD) and evolutionarily conserved signaling pathways (Bier, 2005; Lu and Vogel, 2009). Indeed, many different Drosophila models of AD, which showed progressive degeneration through elevated production of amyloid β-peptides (Aβs), were developed (Fernandez-Funez et al., 2015). Some of these AD models are based on overexpression of wild-type human APP, EOAD-associated mutant APP, or a secreted form of human Aβ42 peptide (Fernandez-Funez et al., 2015). These findings demonstrate highly conserved mechanisms underlying Aβ-mediated neurotoxicity and neurodegeneration between Drosophila and humans. This is further supported by the fact that the fly genome contains functional homologs for most of the EOAD-associated genes including APP, PS1, and PS2 (Martin-Morris and White, 1990; Stempfle et al., 2010). In combination with human BACE, Drosophila γ-secretase is able to cleave human APP to generate Aβ peptides, indicating its conserved intramembrane proteolytic activity (Greeve et al., 2004). Although the Aβ sequence is not well conserved among APP homologs from different species, a growing body of evidence suggests that all APP homologs play an important and conserved role in neural development including neurite growth, axon guidance, and synaptogenesis (Nicolas and Hassan, 2014). However, little is known about how γ-secretase-mediated proteolytic processing of APP is regulated at the molecular and genetic levels.
One of the hallmarks of AD is senile plaques largely consisting of Aβs, which are produced by sequential cleavages of APP by β-secretase (BACE) and γ-secretase (Esler and Wolfe, 2001; Goedert, 2015). Given that all known mutations responsible for early-onset familial Alzheimer’s disease (EOAD) are localized in APP, PS1, and PS2 (Tanzi, 2012), γ-secretase represents a good therapeutic target for developing Aβ-lowering drugs. Therefore, brain-penetrant inhibitors of γ-secretase are emerging as one of the most effective therapies for AD (De Strooper et al., 2010). However, toxicity of γ-secretase inhibitors was found to be a major hurdle in preclinical studies since γ-secretase is also required for the RIP of numerous other substrates including Notch (De Strooper et al., 2010; Esler and Wolfe, 2001; McCarthy et al., 2009). Here, as an alternative approach to overcome this target-based toxicity, we report the development of a Drosophila reporter system for in vivo detection of γ-secretase activity. Based on several observations, we expect that this in vivo reporter system will provide a genetic tool to discover novel genes, which specifically modulate γ-secretase-mediated processing of Notch and APPL. First, overexpression of the N™-SV reporter proteins in the wing induced vein truncation phenotypes that are totally dependent on γ-secretase activity. Second, not only γ-secretase activity, but also apoptosis contributes to the vein truncation phenotypes of the APPL-SV reporter transgene. This appears to be consistent with previous observations suggesting that human APP induces APP intracellular domain (AICD)-dependent cell death in Drosophila (Gunawardena and Goldstein, 2001; Wang et al., 2014). Since the γ-secretase-cleaved C-terminal fragment of APPL-SV contains the APPL intracellular domain (dAICD) in addition to the SV, dAICD might be responsible for the contribution of apoptosis. Third, both N™-SV and APPL-SV transduce Notch signaling activity in a dose-dependent manner. In support of this claim, reporter-induced vein phenotypes are enhanced by one copy of NAx-E2, which is a Notch GOF allele. Fourth, APPL-SV-induced phenotype is almost 2-fold greater than the GOF phenotype of N™-SV, probably due to differences in their RIP and the contribution of dAICD-dependent apoptosis.
Several γ-secretase associated proteins (GSAPs), in addition to the four main components of the γ-secretase complex, have been discovered (Chen et al., 2006; He et al., 2010; Teranishi et al., 2015; Wakabayashi et al., 2009; Zhou et al., 2005). These GSAP proteins, including TMP21, pigeon homologue protein, and proton myo-inositol cotransporter, regulate substrate selectivity and Aβ production. Therefore, these GSAPs can serve as therapeutic targets for the treatment of AD. These observations might indicate the existence of unknown GSAP proteins regulating Aβ production without affecting proteolytic cleavage of other γ-secretase substrates including Notch. Future work using the in vivo reporter system developed in this study will determine whether known and additional unknown GSAP proteins are required for modulation of γ-secretase activity, and how substrate specificity of γ-secretase is regulated. Our current reporter system can also be applied to screening of chemical compound libraries.
Supplementary Information
ACKNOWLEDGMENTS
We thank Jaeseob Kim, Eunkyung Bae, and Jungmo Kim for their comments on the manuscript; James Posakony, Kyuhyung Han, Spyros Artavanis-Tsakonas, and the Drosophila Genomics Resource Center for clones; and the Bloomington Drosophila Stock Center for the fly strains. This study was supported by NRF-2013R1A1A4A01011329 (S.J.).
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Bai X.C., Yan C., Yang G., Lu P., Ma D., Sun L., Zhou R., Scheres S.H., Shi Y. An atomic structure of human γ-secretase. Nature. 2015;525:212–217. doi: 10.1038/nature14892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey A.M., Posakony J.W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Bertet C., Li X., Erclik T., Cavey M., Wells B., Desplan C. Temporal patterning of neuroblasts controls Notch-mediated cell survival through regulation of Hid or Reaper. Cell. 2014;158:1173–1186. doi: 10.1016/j.cell.2014.07.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat Rev Genet. 2005;6:9–23. doi: 10.1038/nrg1503. [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Cagan R.L. Patterning the fly eye: the role of apoptosis. Trends Genet. 2003;19:91–96. doi: 10.1016/S0168-9525(02)00041-0. [DOI] [PubMed] [Google Scholar]
- Brand A.H., Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Brown M.S., Ye J., Rawson R.B., Goldstein J.L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–398. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- Cao X., Südhof T.C. A transcriptionally active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- Carmine-Simmen K., Proctor T., Tschäpe J., Poeck B., Triphan T., Strauss R., Kretzschmar D. Neurotoxic effects induced by the Drosophila amyloid-β peptide suggest a conserved toxic function. Neurobiol Dis. 2009;33:274–281. doi: 10.1016/j.nbd.2008.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F., Hasegawa H., Schmitt-Ulms G., Kawarai T., Bohm C., Katayama T., Gu Y., Sanjo N., Glista M., Rogaeva E., et al. TMP21 is a presenilin complex component that modulates γ-secretase but not ε-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- de Celis J.F., García-Bellido A. Roles of the Notch gene in Drosophila wing morphogenesis. Mech Dev. 1994a;46:109–122. doi: 10.1016/0925-4773(94)90080-9. [DOI] [PubMed] [Google Scholar]
- de Celis J.F., García-Bellido A. Modifications of the notch function by Abruptex mutations in Drosophila melanogaster. Genetics. 1994b;136:183–194. doi: 10.1093/genetics/136.1.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J.F. Positioning and differentiation of veins in the Drosophila wing. Int J Dev Biol. 1998;42:335–343. [PubMed] [Google Scholar]
- De Strooper B. Aph-1, Pen-2, and Nicastrin with Presenilin generate an active γ-Secretase complex. Neuron. 2003;38:9–12. doi: 10.1016/s0896-6273(03)00205-8. [DOI] [PubMed] [Google Scholar]
- De Strooper B., Vassar R., Golde T. The secretases: enzymes with therapeutic potential in Alzheimer disease. Nat Rev Neurol. 2010;6:99–107. doi: 10.1038/nrneurol.2009.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esler W.P., Wolfe M.S. A portrait of Alzheimer secretases--new features and familiar faces. Science. 2001;293:1449–1454. doi: 10.1126/science.1064638. [DOI] [PubMed] [Google Scholar]
- Fernandez-Funez P., de Mena L., Rincon-Limas D.E. Modeling the complex pathology of Alzheimer’s disease in Drosophila. Exp Neurol. 2015;274:58–71. doi: 10.1016/j.expneurol.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini M.E. Notch signaling: the core pathway and its posttranslational regulation. Dev Cell. 2009;16:633–647. doi: 10.1016/j.devcel.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Goedert M. NEURODEGENERATION. Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science. 2015;349:1255555. doi: 10.1126/science.1255555. [DOI] [PubMed] [Google Scholar]
- Greeve I., Kretzschmar D., Tschäpe J.A., Beyn A., Brellinger C., Schweizer M., Nitsch R.M., Reifegerste R. Age-dependent neurodegeneration and Alzheimer-amyloid plaque formation in transgenic Drosophila. J Neurosci. 2004;24:3899–3906. doi: 10.1523/JNEUROSCI.0283-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena S., Goldstein L.S. Disruption of axonal transport and neuronal viability by amyloid precursor protein mutations in Drosophila. Neuron. 2001;32:389–401. doi: 10.1016/s0896-6273(01)00496-2. [DOI] [PubMed] [Google Scholar]
- Han K., Manley J.L. Transcriptional repression by the Drosophila even-skipped protein: definition of a minimal repression domain. Genes Dev. 1993;7:491–503. doi: 10.1101/gad.7.3.491. [DOI] [PubMed] [Google Scholar]
- Hay B.A., Wolff T., Rubin G.M. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–2129. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- He G., Luo W., Li P., Remmers C., Netzer W.J., Hendrick J., Bettayeb K., Flajolet M., Gorelick F., Wennogle L.P., et al. Gamma-secretase activating protein is a therapeutic target for Alzheimer’s disease. Nature. 2010;467:95–98. doi: 10.1038/nature09325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herreman A., Serneels L., Annaert W., Collen D., Schoonjans L., De Strooper B. Total inactivation of γ-secretase activity in presenilin-deficient embryonic stem cells. Nat Cell Biol. 2000;2:461–462. doi: 10.1038/35017105. [DOI] [PubMed] [Google Scholar]
- Jeong S., Juhaszova K., Kolodkin A.L. The control of semaphorin-1a-mediated reverse signaling by opposing pebble and RhoGAPp190 functions in Drosophila. Neuron. 2012;76:721–734. doi: 10.1016/j.neuron.2012.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson R.L., Grenier J.K., Scott M.P. patched overexpression alters wing disc size and pattern: transcriptional and post-transcriptional effects on hedgehog targets. Development. 1995;121:4161–4170. doi: 10.1242/dev.121.12.4161. [DOI] [PubMed] [Google Scholar]
- Ju B.G., Jeong S., Bae E., Hyun S., Carroll S.B., Yim J., Kim J. Fringe forms a complex with Notch. Nature. 2000;405:191–195. doi: 10.1038/35012090. [DOI] [PubMed] [Google Scholar]
- Kim J., Irvine K.D., Carroll S.B. Cell recognition, signal induction, and symmetrical gene activation at the dorsal-ventral boundary of the developing Drosophila wing. Cell. 1995;82:795–802. doi: 10.1016/0092-8674(95)90476-x. [DOI] [PubMed] [Google Scholar]
- Louvi A., Artavanis-Tsakonas S. Notch signalling in vertebrate neural development. Nat Rev Neurosci. 2006;7:93–102. doi: 10.1038/nrn1847. [DOI] [PubMed] [Google Scholar]
- Lu B., Vogel H. Drosophila models of neurodegenerative diseases. Annu Rev Pathol. 2009;4:315–342. doi: 10.1146/annurev.pathol.3.121806.151529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L., Tully T., White K. Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for appl gene. Neuron. 1992;9:595–605. doi: 10.1016/0896-6273(92)90024-8. [DOI] [PubMed] [Google Scholar]
- Martin-Morris L.E., White K. The Drosophila transcript encoded by the β-amyloid protein precursor-like gene is restricted to the nervous system. Development. 1990;110:185–195. doi: 10.1242/dev.110.1.185. [DOI] [PubMed] [Google Scholar]
- McCarthy J.V., Twomey C., Wujek P. Presenilin-dependent regulated intramembrane proteolysis and γ-secretase activity. Cell Mol Life Sci. 2009;66:1534–1555. doi: 10.1007/s00018-009-8435-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas M., Hassan B.A. Amyloid precursor protein and neural development. Development. 2014;141:2543–2548. doi: 10.1242/dev.108712. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Selkoe D.J. The cell biology of β-amyloid precursor protein and presenilin in Alzheimer’s disease. Trends Cell Biol. 1998;8:447–453. doi: 10.1016/s0962-8924(98)01363-4. [DOI] [PubMed] [Google Scholar]
- Selkoe D.J., Wolfe M.S. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–221. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Stempfle D., Kanwar R., Loewer A., Fortini M.E., Merdes G. In vivo reconstitution of γ-secretase in Drosophila results in substrate specificity. Mol Cell Biol. 2010;30:3165–3175. doi: 10.1128/MCB.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi R.E. The genetics of Alzheimer disease. Cold Spring Harb Perspect Med. 2012;2:a006296. doi: 10.1101/cshperspect.a006296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teranishi Y., Inoue M., Yamamoto N.G., Kihara T., Wiehager B., Ishikawa T., Winblad B., Schedin-Weiss S., Frykman S., et al. Proton myo-inositol cotransporter is a novel γ-secretase associated protein that regulates Aβ production without affecting Notch cleavage. FEBS J. 2015;282:3438–3451. doi: 10.1111/febs.13353. [DOI] [PubMed] [Google Scholar]
- Wakabayashi T., Craessaerts K., Bammens L., Bentahir M., Borgions F., Herdewijn P., Staes A., Timmerman E., Vandekerckhove J., Rubinstein E., et al. Analysis of the γ-secretase interactome and validation of its association with tetraspanin-enriched microdomains. Nat Cell Biol. 2009;11:1340–1346. doi: 10.1038/ncb1978. [DOI] [PubMed] [Google Scholar]
- Wang X., Wang Z., Chen Y., Huang X., Hu Y., Zhang R., Ho M.S., Xue L. FoxO mediates APP-induced AICD-dependent cell death. Cell Death Dis. 2014;5:e1233. doi: 10.1038/cddis.2014.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J.B., Suyama K.L., Lee H.H., Scott M.P. Jelly belly: a Drosophila LDL receptor repeat-containing signal required for mesoderm migration and differentiation. Cell. 2001;107:387–398. doi: 10.1016/s0092-8674(01)00540-2. [DOI] [PubMed] [Google Scholar]
- Wharton K.A., Johansen K.M., Xu T., Artavanis-Tsakonas S. Nucleotide sequence from the neurogenic locus notch implies a gene product that shares homology with proteins containing EGF-like repeats. Cell. 1985;43:567–581. doi: 10.1016/0092-8674(85)90229-6. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Nadeau P., Song W., Donoviel D., Yuan M., Bernstein A., Yankner B.A. Presenilins are required for γ-secretase cleavage of β-APP and transmembrane cleavage of Notch-1. Nat Cell Biol. 2000;2:463–465. doi: 10.1038/35017108. [DOI] [PubMed] [Google Scholar]
- Zhou S., Zhou H., Walian P.J., Jap B.K. CD147 is a regulatory subunit of the γ-secretase complex in Alzheimer’s disease amyloid β-peptide production. Proc Natl Acad Sci USA. 2005;102:7499–7504. doi: 10.1073/pnas.0502768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


