Abstract
Ocean acidification is a major threat for marine life but seagrasses are expected to benefit from high CO2. In situ (long-term) and transplanted (short-term) plant incubations of the seagrass Cymodocea nodosa were performed near and away the influence of volcanic CO2 vents at Vulcano Island to test the hypothesis of beneficial effects of CO2 on plant productivity. We relate, for the first time, the expression of photosynthetic, antioxidant and metal detoxification-related genes to net plant productivity (NPP). Results revealed a consistent pattern between gene expression and productivity indicating water origin as the main source of variability. However, the hypothesised beneficial effect of high CO2 around vents was not supported. We observed a consistent long- and short-term pattern of gene down-regulation and 2.5-fold NPP decrease in plants incubated in water from the vents and a generalized up-regulation and NPP increase in plants from the vent site incubated with water from the Reference site. Contrastingly, NPP of specimens experimentally exposed to a CO2 range significantly correlated with CO2 availability. The down-regulation of metal-related genes in C. nodosa leaves exposed to water from the venting site suggests that other factors than heavy metals, may be at play at Vulcano confounding the CO2 effects.
Rising atmospheric carbon dioxide (CO2) is increasing the concentration of dissolved inorganic carbon (DIC) in the ocean causing ocean acidification. Projections forecast that ocean acidification will continue and that by the end of the century the mean global ocean surface pH will decrease up to 0.4 units1, profoundly affecting marine systems1,2. Biological responses associated with ocean acidification range from changes in organismal physiology and behaviour up to changes in population structure, with major global implications for the entire ecosystem functioning and the goods and services provided1.
The increase in seawater DIC and CO2 associated with ocean acidification may favour carbon-limited species, such as seagrasses, by increasing both the passive diffusion of CO2 and the efficiency of RuBisCO carboxylation over photorespiration3, lowering the energetic photosynthetic requirements and consequently increasing photosynthetic production2,4. Physiological acclimation to ocean acidification has been widely documented in seagrasses and includes changes in photosynthetic rates, metabolism, growth and survival (e.g. refs 5, 6, 7). However, experimental evidence for increased seagrass productivity as a response to elevated CO2 levels and ocean acidification is inconclusive, and particularly scarce over long-time scale. Indeed, recent meta-analysis did not identify significant effects of ocean acidification on seagrass photosynthesis8.
The modulation of gene expression plays a central role in plant plasticity and adaptation to environmental changes9, since physiological machinery and metabolic pathways are coordinated at the genetic level by an array of regulatory genes, which are also affected by environmental stimuli10. In the last decade, molecular techniques for studying gene expression have been increasingly recognized as a powerful tool for physiological research to assess the acclimation responses and adaptive potential of marine organisms to ocean acidification (see ref. 11 for a review). Gene expression can be used to assess the role that plasticity and long-term adaptation to high CO2 play in altering specific metabolic pathways related with the physiological response, but also the fast acclimation and reversibility capacity to short-term acute disturbances (see ref. 12 for a review). The analysis of the expression levels of targeted genes may thus provide new understanding of molecular changes that accompany alterations in physiological states13,14.
Gene expression studies in seagrasses are yet scarce15 and have been mainly focused on the acclimation and adaptation potential to temperature (in Zostera spp.16,17,18) and light availability (in Zostera marina19 and in Posidonia oceanica20,21). Only one study looked at modulation in the gene expression of seagrasses to ocean acidification, by comparing the expression of antioxidant and stress-related genes in P. oceanica plants growing under the long-term influence of CO2 vents with plants from a control site22. This study targeted stress-related and antioxidant genes to infer the physiological state of plants near vents, since up-regulation of antioxidant genes is a response to a higher cellular antioxidant need resulting from the formation of reactive oxygen species (ROS) associated with increased metabolism23. However, so far no study integrated gene expression and physiological responses of seagrasses exposed to high CO2.
The combination of molecular and physiological tools may allow the identification of ecologically important gene-physiology interactions and advance our insights into how organisms respond to environmental pressure at different scales, from the fast and versatile modulation of gene expression up to the integrative physiological response. Unfortunately, research on seagrass ecophysiology and ecogenomics is not quite integrated yet24 and a better understanding of seagrass photosynthetic plasticity and gene-regulation is needed25.
Responses of organisms and communities to environmental changes span across different temporal scales, from short-term acclimation to adaptation over multiple generations. Prediction of marine organisms responses to ocean acidification has been primarily inferred by investigating short-term acclimation plastic responses in relatively short-term (from hours up to few months), single generation experiments8,11. Only few experiments tested the long-term potential for gradual acclimation or adaptation26 revealing that trans-generational and evolutionary adaptation can partly mitigate adverse effects of ocean acidification and highlighting the necessity of long-term experimentation11,27. A major experimental limitation for evaluating species’ long-term phenotypic plasticity and adaptive potential is to replicate the temporal scale at which ocean acidification occurs11.
Naturally CO2 enriched sites, such as submarine volcanic CO2 vents, have been used to study the species and communities potential to face ocean acidification. The use of CO2 vents provides the enormous advantage of testing organisms and communities that have been exposed for long-term over multiple generations to high CO228, which is not feasible in experimental studies. Even though the physico-chemical composition of seeping fluids in volcanic vents is not exclusively CO2 and varies among vents29, the presence of other factors that may confound the effects of CO2, such as heavy metals or sulphides, have been disregarded due to the low concentrations observed at the seagrass meadows30. Studies conducted in volcanic CO2 vents revealed that seagrasses have successfully adapted to live under permanently high CO2 levels31,32, but support for the stimulation of photosynthetic performance and productivity of seagrasses near the vents is scarce and inconclusive. So far, studies have focused on community responses by evaluating the biodiversity and richness (e.g. ref. 33), the community structure (e.g. ref. 31) or the community metabolism (e.g. ref. 34). For the species targeted in this work, the seagrass Cymodocea nodosa, there is evidence of an increase in chlorophyll a content and maximum electron transport rate but low biomass production in meadows growing in acidified areas near CO2 vents34.
The aim of this study was to evaluate the long- and short-term responses of the seagrass Cymodocea nodosa to high CO2 to test the hypothesis of beneficial effects of ocean acidification on seagrass productivity, taking advantage of the high CO2 environment existing near the submerged volcanic vents at Vulcano Island (Sicily, Italy).
A combination of molecular and physiological techniques was used to relate, for the first time, the expression of target genes involved in plant metabolism (i.e. photosynthetic, carbon assimilation, antioxidant and metal-related genes) to the net plant productivity (NPP) to investigate if molecular and physiological responses are coordinated. Antioxidant and metal-related genes were also targeted to evaluate the existence of a gene activation to deal with oxidative stress and heavy metal toxicity of plants near the vents in relation to plants away from the venting influence.
A set of plant incubations with C. nodosa specimens and water from sites under (CO2 site) and away (Reference site) the influence of CO2 vents at Vulcano Island was performed at the same depth. Differences in the gene expression and productivity between plants exposed for a long-time to high CO2 (CO2 plants) and present-day levels of CO2 (Reference plants) were investigated (in situ incubations) to assess if plasticity and long-term adaptation to high CO2 affected specific metabolic pathways and productivity response of C. nodosa. The short-term acclimation responses to CO2 and reversibility capacity of C. nodosa were assessed investigating the gene expression and productivity of plants from one site (CO2 or Reference) incubated with water from the other site (transplant incubations).
To validate the observed effects of high CO2 on productivity and evaluate the adequacy of volcanic CO2 vents as natural laboratories, a set of controlled experimental incubations of C. nodosa plants were conducted in Ria Formosa (Portugal) under a wide range of CO2 concentrations.
Results
Seawater parameters
A description of the seawater parameters and irradiance during the incubations conducted both in Vulcano (Italy) and Ria Formosa (Portugal) is given in Table 1. In Vulcano, mean pCO2 concentrations (and associated parameters of the DIC system) were significantly higher in the CO2 site than in Reference site (p < 0.05, df = 6). In accordance with descriptions from previous studies29,30,34 conducted in this site, seawater chemical variability near the venting site (CO2 site) was higher than in the Reference site, particularly regarding the inorganic carbon system. Indeed, occasional extreme high values of pCO2 and DIC were recorded in the CO2 site (Table 1).
Table 1. Irradiance and seawater physico-chemical parameters during the incubations.
| Vulcano (Italy) | Ria Formosa (Portugal) | ||
|---|---|---|---|
| Irradiance (μmol quanta m−2 s−1) | Midday: 1137 ± 65 | 517–1326 | |
| Afternoon: 539 ± 107 | |||
| Reference site | CO2 site | ||
| S | 37.5 ± 0.0 | 37.5 ± 0.0 | 36.3 ± 0.3 |
| T (°C) | 19.8 ± 0.5 | 20.3 ± 1.0 (20.2 ± 0.8) | 25.7 ± 1.6 |
| pH (NBS) | 8.179 ± 0.058 | 7.985 ± 0.088 (7.896 ± 0.193) | 7.706–8.387 |
| TA (μmol · kg SW−1) | 2545 ± 7 | 2577 ± 1 (2579 ± 12) | 2466 ± 77 |
| DIC (μmol · kg SW−1) | 2244 ± 40 | 2377 ± 37 (2417 ± 86) | 2018–2373 |
| pCO2 (μatm) | 427 ± 68 | 737 ± 158 (1004 ± 549) | 245–1465 |
| CO2 (μmol · kg SW−1) | 14 ± 2 | 23 ± 6 (32 ± 17) | 7–38 |
| HCO3− (μmol · kg SW−1) | 2012 ± 61 | 2197 ± 63 (2250 ± 117) | 1685–2227 |
| CO32− (μmol · kg SW−1) | 218 ± 23 | 156 ± 32 (136 ± 49) | 98–359 |
Data are mean ± sd or min–max values recorded in the water. Irradiance was measured 0.5 m above the incubation chambers. Data in parenthesis include one extreme high value recorded in the CO2 site in Vulcano.
Vulcano Incubations
Gene expression
Expression stability of reference genes (RGs): Four RGs (Eukaryotic initiation factor 4A (eIF4A), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH), 18S ribosomal RNA (18S) and Ubiquitin (UBI)), belonging to different functional classes, were tested for stability in the different experimental conditions (Table 2). The three algorithms applied (BestKeeper, geNorm and NormFinder) agreed in suggesting the eIF4A as the best RG (see Supplementary Tables S1, S2 and S3 in Appendix 1, Supplementary Information). However, we decided to include GAPDH as RG, indicated by geNorm, and also the 18S, the second best RG according to NormFinder and geNorm, for target gene expression normalization (Supplementary Table S4). The use of three RGs allowed for a more reliable normalization of gene expression data.
Table 2. List of reference genes (RGs) and genes of interest (GOIs) analysed in Cymodocea nodosa.
| Gene symbol | Protein name | Gene Ontology |
|---|---|---|
| Reference genes (RGs) | ||
| 18S | Ribosomal RNA 18S | Translation |
| eIF4A | Eukaryotic initiation factor 4A | Translation/Protein biosynthesis |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase | Glycolysis |
| UBI | Ubiquitin | Ubiquitin-dependent protein catabolic process |
| Genes of interest (GOIs) | ||
| psaJ | Photosystem I reaction center subunit IX | Photosynthesis |
| psaC | Photosystem I iron-sulfur center | Photosynthesis |
| psbA | Photosystem II protein D1 | Photosynthesis |
| psbD | Photosystem II protein D2 | Photosynthesis |
| LHCA1 | Photosystem I Light Harvesting Complex gene 1 | Photosynthesis |
| FD | Ferredoxin, chloroplastic | Electron transport chain |
| rbcL | RuBisCO large subunit | Carbon dioxide fixation |
| ATPA | ATP synthase subunit alpha | ATP biosynthetic process |
| PEPC | Phosphoenolpyruvate carboxylase | Carbon dioxide fixation |
| SUS | Sucrose synthase | Sucrose metabolic process |
| BCA | Beta carbonic anhydrase | Carbon utilization |
| SOD | Copper/zinc superoxide dismutase | Response to oxidative stress |
| CAT | Catalase | Response to oxidative stress |
| APX7 | L-ascorbate peroxidase 7, chloroplastic | Response to oxidative stress |
| APX6 | L-ascorbate peroxidase 6 | Response to oxidative stress |
| LBP | Luminal binding protein | Response to oxidative stress |
| GSH-S | Glutathione synthase | Glutathione biosynthetic process |
| GR | Glutathione reductase | Glutathione metabolic process |
| MTP | Metal tolerance protein | Ion transport |
| MT | Metallothionein | Cellular metal ion homeostasis |
Gene names, gene encoding protein names and gene ontology are given.
Genes of interest (GOIs) expression level: Relative expression levels of 20 GOIs involved in photosynthesis, carbon dioxide fixation and metabolic carbon assimilation pathways, antioxidant and metal responses were analysed in C. nodosa plants from both Reference and CO2 sites after in situ and transplanted incubations (Table 2).
Major changes in gene expression patterns were observed in plants long-term growing near the vents (in situ incubations in CO2 site) with respect to plants from the Reference site (in situ incubations in Reference site) (Fig. 1a), as confirmed by the multivariate analysis of similarity (ANOSIM), which revealed a certain degree of separation between the two sites (Global R: 0.370, p = 0.098, Supplementary Table S5). Overall, expression levels of genes involved in light-dependent reaction of photosynthesis, carbon dioxide fixation and carbon assimilation pathways, were reduced in plants long-term growing in the CO2 site compared to plants from Reference site (Fig. 1a). According to univariate REST analysis, a significant down-regulation was observed for the subunit psbA of the Photosystems II, the Photosystem I Light Harvesting Complex gene 1 (LHCA1), the Beta carbonic anhydrase (BCA), and the Sucrose synthase (SUS) (P(H1): 0.000), as well as the large subunit of RuBisCO (rbcL) (P(H1): 0.037). The transcripts for the Ferredoxin (FD) and Phosphoenolpyruvate carboxylase (PEPC) were close to the significance level (P(H1): 0.058 for both) (Supplementary Table S6). The same pattern was found for genes involved in the antioxidant response and metal detoxification (Fig. 1a). The expression of genes involved in reactive oxygen species (ROS) detoxification enzymes (antioxidant system), such as Superoxide dismutase (SOD) and Ascorbate peroxidases (APX7 and APX6), was significantly reduced, as were genes involved in glutathione metabolism (GSH-S and GR) and luminal binding protein (LBP) (P(H1): 0.000). Down-regulation of metal tolerance and metal detoxification proteins (MTP and MT) was also significant (P(H1): 0.000). The general down-regulation pattern observed points to an overall reduction of metabolic processes related with photosynthesis, oxidative (ROS) and metal detoxification response in plants growing near the CO2 vents with respect to plants from the Reference site.
Figure 1. Relative expression of photosynthetic, antioxidant and metal detoxification-related genes.
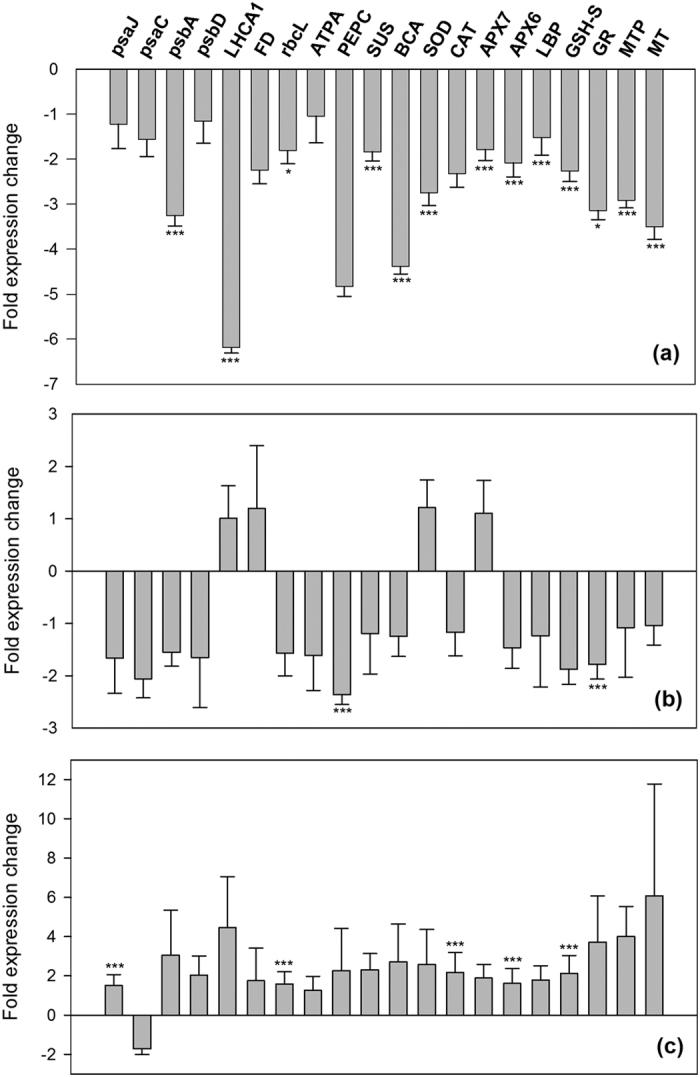
(a) Bars indicate fold expression changes of CO2 plants incubated in CO2 water with respect to Reference plants incubated in Reference water. (b) Bars indicate fold expression changes of Reference plants incubated in CO2 water with respect to Reference plants incubated in Reference water. (c) Bars indicate fold expression changes of CO2 plants incubated in Reference water with respect to CO2 plants incubated in CO2 water. Error bars represent standard error. (*) P(H1) < 0.05; (***) P(H1) < 0.001.
When specimens of C. nodosa from the Reference site were incubated in water from the CO2 site (transplant incubation), expression of almost all the GOIs analysed was reduced (Fig. 1b) also indicating an overall reduction of metabolic processes related with photosynthesis and antioxidant response when compared to in situ conditions (i.e. plants from the Reference site incubated in water from the Reference site). Significant down-regulation was also detected for the enzymes Phosphoenolpyruvate carboxylase (PEPC) and Glutathione reductase (GR) (P(H1): 0.000) (Supplementary Table S6).
On the contrary, when plants growing in the CO2 site were incubated in water from the Reference site, a clear gene up-regulation pattern was observed on the metabolic paths linked to photosynthesis as well as oxidative (ROS) and metal detoxification to acclimate to the new conditions (Fig. 1c). Although multivariate differences in gene expression were not significant (Global R: 0.370, p = 0.200, Supplementary Table S5), univariate REST analysis revealed a significant up-regulation of the photosynthesis-related genes psaJ, and rbcL, as well as the antioxidant-related genes CAT, APX6 and GSH-S (P(H1): 0.000) (Fig. 1c). Several GOIs analysed (psbA, L, LHCA1, SUS, BCA, APX7,GR, MTP) were also close to the significance level (P(H1) < 0.1) (Supplementary Table S6).
Net plant productivity
In Vulcano, the net plant productivity (NPP) of C. nodosa was significantly affected by the origin of the water where plants were incubated (p < 0.01). NPP was significantly lower when plants were incubated with water from the CO2 site, both in plants long-term growing near the CO2 vents (i.e. CO2 plants in situ incubations) or away from them but incubated with water from the CO2 site (i.e. Reference plants, REF, transplant incubations) (Fig. 2).
Figure 2. Net plan productivity (NPP) of in situ and transplanted incubations of Cymodocea nodosa plants.
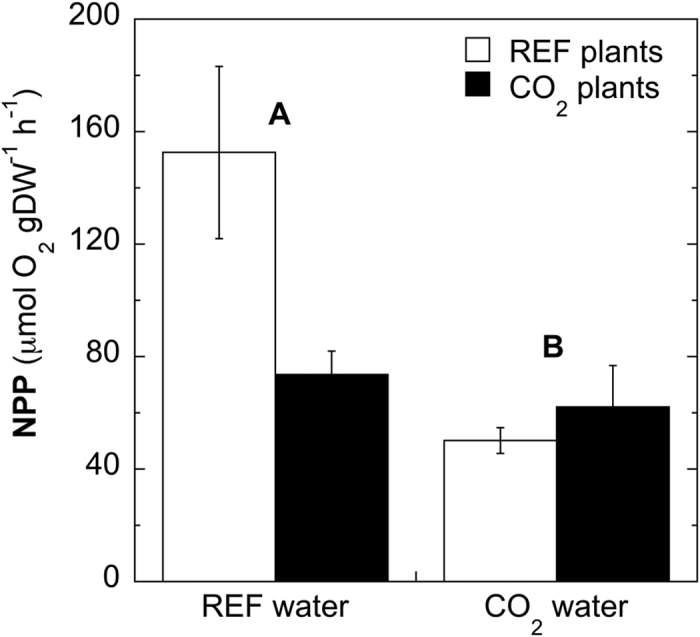
Letters (A,B) indicate significant differences between incubations conducted with water from the Reference and CO2 site. Bars represent means ± sem.
Plants collected from both sites did not differ significantly in their exposure to different CO2 concentrations in water (i.e. no significant interaction “plant origin” x “water” was detected, F1,25 = 2.698, p = 0.113, see ANOVA results in Supplementary Table S7).
Gene expression and productivity
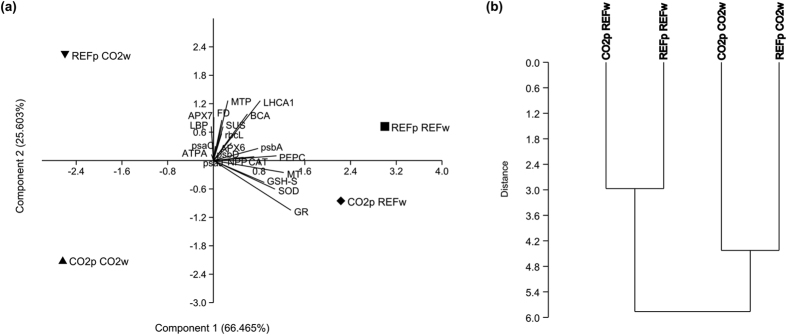
A PCA and cluster analysis and a correlation analysis were performed to link gene expression and productivity results. The PCA and Cluster analysis, conducted considering the combined contribution of all GOIs analysed and NPP, clearly highlighted two groups (PCA component 1 explaining 66.5% of the total variance) defined by the origin of the water where plants were incubated (i.e. water from Reference vs. CO2 site) (Fig. 3). These two groups matched the patterns described in Figs 1 and 2. A general up-regulation of photosynthetic, antioxidant and metal detoxification-related genes and higher NPP was found in plants long-term growing in the Reference site (in situ incubations) and plants from CO2 site incubated in water from the Reference site (transplant incubations). Reversely, plants long-term growing in the CO2 site (in situ incubations) and plants from the Reference site incubated in water from the CO2 site (transplant incubations) showed a generalized gene down-regulation and lower NPP. The biplot in the PCA identify GR, MT, PEPC, SOD, GSH-S, LHCA1 and psbA as the genes most contributing to this separation (for detailed information on the contribution of all GOIs and NPP see Supplementary Table S8).
Figure 3. PCA (a) and Cluster analysis (b) conducted considering the combined contribution of all GOIs analysed and NPP on in situ and transplanted incubations of Cymodocea nodosa plants.
REFp REFw: in situ incubations of Reference plants in Reference water; REFp CO2w: transplant incubations of Reference plants in CO2 water; CO2p CO2w: in situ incubations of CO2 plants in CO2 water; CO2p REFw: transplant incubations of CO2 plants in Reference water.
Direct correlations were found between gene expression and productivity (Spearman’s rank correlation). Significant correlations were detected between NPP and expression of photosynthetic (psaJ, psbA, LHCA1 ρ = 0.49, p < 0.01), and carbon metabolism-related genes (PEPC, ρ = 0.59, p < 0.001) as well as antioxidant-related genes (SOD, GR ρ = 0.59, p < 0.001 and GSH.S ρ = 0.39, p < 0.05) (Supplementary Table S9).
Ria Formosa Incubations
In Ria Formosa, where there are no volcanic vents, a set of in situ incubations of C. nodosa plants were performed under the same physico-chemical conditions with CO2 levels experimentally controlled (Table 1). NPP of C. nodosa plants in the Ria Formosa incubations showed a significant positive correlation with DIC (adjusted R2 = 0.42, p < 0.01) (Fig. 4).
Figure 4. Net plan productivity (NPP) vs. dissolved inorganic carbon (DIC) on incubations of Cymodocea nodosa conducted in Ria Formosa.
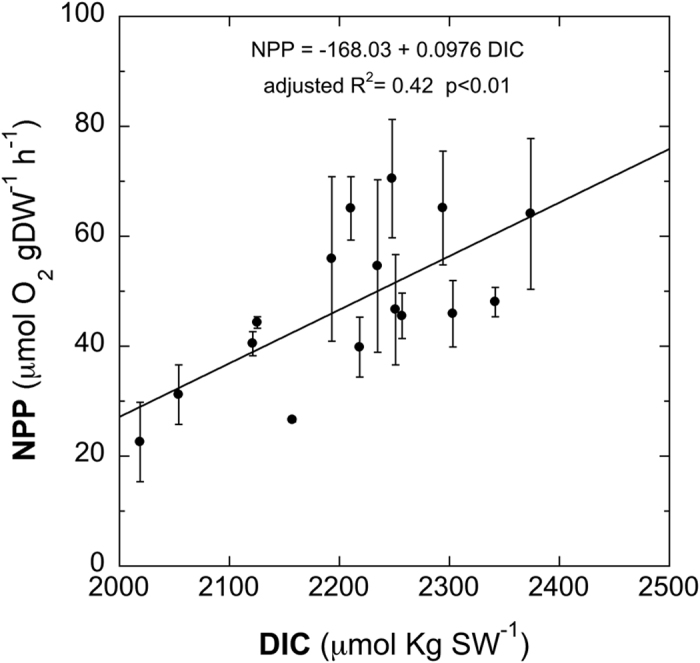
Dots represent mean ± sem. Linear regression equation, adjusted R2 and p-value are also indicated.
Discussion
Our results showed that both the net plant productivity (NPP) and the gene expression of C. nodosa were significantly lower in plants incubated in CO2–rich water from Vulcano CO2 vents when compared to Reference water. Plants living in the venting site showed a general reduction in the expression of genes involved in various metabolic processes, particularly those related with the light-dependent reactions of photosynthesis, carbon fixation and metabolic carbon assimilation when compared to plants from the Reference site. The same pattern was observed for genes involved in the oxidative stress antioxidant response and metal detoxification. This generalized gene down-regulation was paralleled by a reduction in plant metabolism, which resulted in 2.5 fold decrease of NPP in plants naturally growing under acidified water near the vents. Broad-scale decreases in the expression of genes related to key cellular processes, including metabolism and stress response, affecting physiological processes have been already described in different marine taxa in response to ocean acidification (ref. 35 and references therein). During long-term CO2 enrichment, down-regulation of RuBisCo has been described in C3 plants as a photosynthetic acclimation response (ref. 25 and references therein). A reduction in global metabolism has been also described in many marine organisms under high CO2 conditions8. A general down-regulation of photosynthesis-related genes has been described in seagrasses of the genus Zostera in response to temperature increases and global warming17, but current knowledge on seagrasses gene expression patterns under high CO2 conditions is still very limited. The only other available study described variable patterns on expression of stress-related genes in the seagrass Posidonia oceanica growing near the volcanic CO2 vents of Panarea and Ischia Islands (Italy)22. In agreement with our results, these authors reported down-regulation of several metal-detoxification and antioxidant genes and suggested there was no need for activation of these metabolic defence mechanisms due to the low heavy metal-stress and oxidative damage. Seagrass species show many convergent adaptive features to allow life in the marine environment (see refs 36 and 37), however different lineages can have different response to specific biotic and abiotic factors. We have analysed and identified reference genes (RGs) that are stable in C. nodosa exposed to volcanic CO2 vents and the expression level of the genes of interest (GOIs). The pioneer information reported here can represent a baseline for understanding adaptive features and stress responses in this species and in the Cymodoceaceae family in general.
The short-term acclimation responses of C. nodosa to high CO2, evaluated through the transplant incubations, agreed with the molecular and productivity long-term responses measured in situ. Reference plants incubated in water from the CO2 site experienced a generalized, drastic down-regulation in the gene expression of several photosynthetic, antioxidant and metal-related genes that globally reduced plant metabolism and resulted in a significant NPP decrease. On the contrary, plants from CO2 site incubated in Reference water significantly up-regulated the expression of those genes and moderately increased NPP.
Globally, we identified a coordinated gene expression pattern among photosynthetic antioxidant and metal-detoxification genes in all incubations conducted in Vulcano. We also detected a fast response of gene expression over physiology and global metabolism in C. nodosa leading to significant differences in plant productivity when incubated in water with different CO2 concentrations. Significant positive correlations were detected between gene expression and productivity for specific genes encoding proteins related with the efficiency of the photosynthetic process, such as the Photosystem I reaction center (psaJ) and the Photosystem II protein D1 (psbA), the Photosystem I light harvesting complex (LHCA1), and the carbon fixation (Phosphoenolpyruvate carboxylase, PEPC). Significant correlations were also detected between oxidative stress-related genes (Superoxide dismutase, SOD, Glutathione reductase, GR, and Glutathione synthase, GSH-S) and productivity. These results support the use of molecular tools as early indicators of potential stress or physiological readjustments in response to environmental change38 since metabolic pathways and physiological machinery are primarily gene-coordinated10. They may thus provide earliest evidence of organism responses before than morphological and physiological indicators39. Work is still needed to untangle the scaling-up control from genes, over protein expression and biochemistry up to physiology in seagrasses, their energetic implications and the metabolic consequences.
The contrasting response pattern recorded in Vulcano between plants long-term growing at each site (in situ incubations) could be attributed to local adaptation and differential selection of specific genotypes under acidified conditions40. However, preliminary results of a population genetic analysis carried out in Vulcano using microsatellite markers, at the same time and sampling sites than the present study, showed no genetic differentiation and high gene flow between plants from CO2 and Reference sites (Silva et al. in prep.). Consequently, results point to a low contribution of local adaptation to the observed differences between plants from the two sites. Organisms may also be able to cope with conditions found within the CO2 vents through plastic responses at different levels, from molecular (i.e. gene expression) to physiological, morphological and behavioural, without genotypic changes occurring through selection41. The large photosynthetic plasticity attributed to C. nodosa, in terms of photosynthetic response capacity42,43, could thus explain the differences found between specimens in Vulcano sites and the rapid acclimation mechanisms triggered during the transplant incubations.
The observed decrease of C. nodosa productivity under the influence of the high CO2 vents in Vulcano island appears to contradict the common assumption that seagrasses will increase their productivity in a foreseen high CO2 world. Contrastingly, the pattern obtained in Ria Formosa incubations, where CO2 availability was the only factor tested, significantly correlated with DIC availability. This latter pattern supports the hypothesis that seagrasses are C-limited at current CO2 concentrations4,32,44 and that they may benefit from higher DIC availability, enhancing photosynthetic rates and productivity if no other limiting factors, such as irradiance, nutrients or temperature, are present7. The photosynthetic plasticity attributed to C. nodosa42,43 could once more explain, via acclimation to local conditions, the differences found between specimens from distant locations long-term growing under different environmental pressure (i.e. high CO2 exposure in Vulcano vs. present day CO2 exposure (ca. 380 μmol pCO2) in Ria Formosa) when exposed to high CO2. However, the sharp NPP decrease observed in C. nodosa growing at present day CO2 levels in Vulcano (Reference plants) when incubated in CO2-rich seawater (i.e. transplants) is difficult to justify, particularly when considering the photosynthetic carbon-limited physiology of this species44,45 and the significant correlation of NPP to CO2 showed by Ria Formosa plants short-term exposed to a CO2 range.
The PCA analysis revealed a consistent pattern between gene expression and productivity indicating water origin as the main source of variability among all the incubations conducted in Vulcano. Results point to the possibility that other environmental factors related to volcanic vent emissions may correlate with the high CO2 in Vulcano46. Since plants from both CO2 and Reference sites in Vulcano were growing and incubated under similar environmental conditions (i.e. irradiance, temperature, salinity (Table 1) and nutrients47) only differing in their exposure to the venting seeping fluid, its specific physico-chemical composition may be the cause of the particular response of C. nodosa.
Submarine CO2 vents share the common trait of CO2 being the main gas emitted. However, the particular physico-chemical composition of seeping fluids is site specific and varies widely among vents29. The seeping fluid in Vulcano has been characterized as a CO2 source (97–99%) with sulphide (H2S) concentrations of about 270 μM48 and minor concentrations of trace elements and toxic metals (e.g. Cd, Co, Cr, Cu, Mn)46. Negative and toxic effects of H2S and heavy metals on the photosynthetic apparatus, metabolism and survival of marine organisms, including seagrasses, have been previously described49,50. The down-regulation of metal detoxification genes (MTP and MT) recorded in C. nodosa leaves when incubated in water from CO2 site suggests no significant damage associated with heavy metals. However, attenuation, and even inhibition, of cytochromes functioning has been reported at concentrations of H2S as low as 100 μM compromising the photosynthetic output of photosystems51,52. This is a likely explanation for the significant down-regulation detected in the photosynthesis-related genes, particularly in the photosystems I and II, and the lower productivity of C. nodosa plants incubated with water from the venting site in Vulcano. Unfortunately, no data on seawater sulphide or metal concentration during the experimental period are available. At present, a major knowledge gap exists regarding the combined effects of high CO2 and other drivers associated with Global Climate Change on marine benthic communities, including those dominated by seagrasses. The particular chemical composition of seeping fluids at each venting site may recreate different foreseen multi-drivers scenarios associated with Climate Change (i.e. hypoxia, high temperature, sulphide and metal presence, anthropogenic pressure)1.
In conclusion, we related, for the first time, gene expression with net plant productivity in the seagrass species, Cymodocea nodosa, exposed to high CO2 in volcanic CO2 vents. We found that the gene expression and productivity responses of C. nodosa to high CO2 at Vulcano vents were coupled indicating water origin as the main source of variability among all the incubations. However, the hypothesis that high CO2 environment near the vents is beneficial to plant productivity was not supported. The consistent and unexpected long- and short-term pattern of gene down-regulation and NPP decrease observed in C. nodosa when incubated with water from the vent site suggests that environmental factors, other than increased inorganic carbon availability, may be at a play affecting photosynthetic metabolism and decreasing productivity. The down-regulation of metal detoxification genes suggests no significant damage associated with heavy metals. Further research is needed to clarify if sulphide or other components of seeping fluids may explain our observations. Natural submarine CO2 vents provide unique experimental conditions to evaluate the acclimation and adaptive potential of species and communities to scenarios where ocean acidification occurs in combination with other environmental factors.
Material and Methods
Site Description
This study was conducted in May 2013 in Vulcano Island (Sicily, Italy) where submarine gas emissions date from late 18th century53. The experimental site was located in Levante Bay, a shallow bay under the influence of a submarine volcanic vent46. A pH gradient is observed along the coast, with the lowest values near the venting emission points (pH 5.65) increasing to normal values (pH 8.1) at about 400 m away from the vents30. Fluctuations in DIC and pH along this gradient have been described depending on wind direction and consequent water mass movement30.
We selected two sites, characterized by different pH regimes, where the seagrass Cymodocea nodosa grows30,34,46. The low pH/high CO2 site (CO2) is located approximately 200 m from the vent at 2 m depth and 50 m from the shore. The Reference site (REF), away from the vent influence, is located approximately 450 m from the vent at 2.5 m deep and 40 m from the shore.
In order to test the productivity results obtained in Vulcano Island, we conducted a set of incubations at an independent control site. Specimens of C. nodosa growing in Ria Formosa (Portugal), a lagoon system with no venting influence, were incubated following the same methodological approach used in Vulcano, i.e. in situ NPP estimates through O2 determination in the same incubation chambers used in Vulcano.
Seawater parameters
During the incubations, underwater irradiance (Licor Li-192SA) above the incubation chambers, salinity (VWR CO310), temperature (CheckTemp1, Roth), and pH (Thermo Scientific Orion) were monitored in both Vulcano (Italy) and Ria Formosa (Portugal). Alkalinity, for characterization of the DIC system, was measured by final point potentiometric titration and accuracy (~5 μmol·Kg SW−1) was checked using CRM batch #126 (Dickson laboratory, Scripps). DIC system parameters were calculated with the CO2SYS program54 using the dissociation constants of Mehrbach et al. (1973) refitted by Dickson and Millero (1987) (references included in the CO2SYS).
Plant incubations
Cymodocea nodosa plants, each unit consisting of one rhizome with 6–7 shoots and corresponding roots, and water were collected at each donor site (CO2 and Reference) and quickly transported (less than 5 minutes) to an intermediate incubation location, halfway of donor sites, where all incubations took place at 2 m depth. Plants were gently cleaned from sediment and epiphytes and placed into the incubation chambers with water from their own collection site (in situ incubations) or from the other site (transplant incubations).
For each type of incubation (in situ or transplants) two runs were conducted, one at midday (around 12:00 h) and one in the afternoon (around 16:00 h). At each run, four incubation chambers from each site were incubated together and thus they were exposed to the same irradiance. Light measurements (Licor sensor) were done at the incubation location along the incubation time. No significant differences were found between the midday (t-test, p = 0.7) or the afternoon (p = 0.2) irradiances during the two consecutive days of incubations.
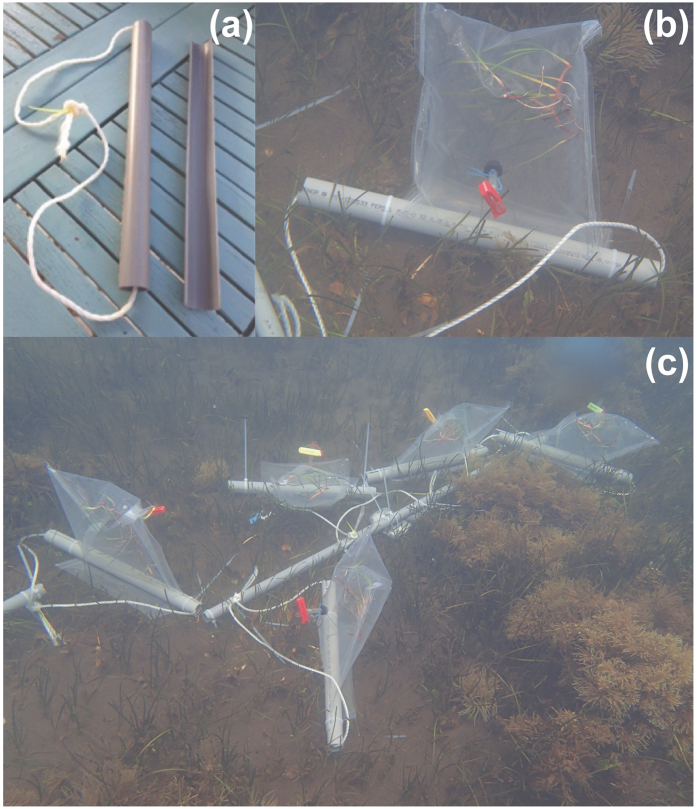
Incubation chambers consisted of a gas-tight polyethylene plastic bag (30 cm long, 20 cm wide, 2.5 L approx.) with one sampling port to withdraw water samples (Fig. 5b). Chambers were sealed with a clamp made of a polyvinyl chloride (PVC) half-cylinder adjustable to a PVC rigid bar (Fig. 5a). Chambers were mounted and fixed in a steel bar and incubated together at 2 m depth (Fig. 5c). The positive buoyancy and flexible nature of the plastic bags allowed propagation of external turbulence to the interior of the chamber to prevent too thick surface boundary layers55. Incubation time ranged between 1–2 h to avoid within chamber O2 saturation effects56. Water samples were taken at the beginning and end of each incubation and rapidly preserved for oxygen determination (see below). Plants were weighted and measured for NPP normalization and a shoot subsample was collected for gene expression analysis (see below).
Figure 5. Incubation chambers.
(a) Detail of closing system, (b) deployed incubation chamber and (c) running set of incubations.
In Ria Formosa, in situ incubations (n = 64) were conducted at 2 m depth to measure the productivity of C. nodosa to varying CO2 concentrations. Seawater used in the incubations was collected from the field and bubbled with air at different concentrations of CO2, ranging from ~250 up to ~1500 μatm (pCO2) (i.e. ~2000 to ~2400 μmol Kg SW−1 DIC and 8.39 to 7.70 pH (NBS scale)), thus covering the range found in Vulcano and the predicted CO2 scenarios for the end of the century1 without modifying any other seawater parameters. Water and biomass samples were processed as in Vulcano.
Gene expression
Material collection, RNA extraction and cDNA preparation
One shoot from three randomly chosen incubation chambers was collected at the end of both in situ and transplants incubations. Apical (youngest) shoots were avoided and only middle parts of the youngest fully mature leaves (2° or 3° rank leaves) of intermediate shoots were selected to standardize leaf developmental stages. Immediately after collection, leaves were blotted dry with tissue paper and preserved in RNAlater© (Ambion, Life technologies) to protect RNA from degradation. The whole procedure, from collection to fixing, lasted less than 5 minutes. After one night at 4 °C, samples were stored at −20 °C until RNA extraction. RNA was extracted from C. nodosa leaves with the Aurum™ Total RNA Mini Kit (BIO-RAD) following manufacturer’s instructions. The full protocol for RNA extraction and cDNA preparation is described in Appendix 2, Supplementary Information.
Gene selection and RT-qPCR
Reverse Transcription-quantitative Polymerase Chain Reaction (RT-qPCR) analysis was performed to investigate differences in expression levels of genes belonging to major pathways affected by changes in CO2 availability (i.e. photosynthesis, carbon assimilation pathways) that can be directly correlated with plant productivity (psaJ, psaC, psbA, psbD, LHCA1, FD, rbcL, ATPA, PEPC, SUS and BCA). Additionally, some studies have related high-CO2 environments with antioxidant and oxidative stress responses in land plants, and more recently in other seagrass species growing close to natural volcanic CO2 vents22, so a number of genes involved in free-radical detoxification and oxidative-stress response were also targeted (SOD, CAT, APX7, APX6, LBP, GSH-S and GR). Finally, since the presence of metals and toxic trace elements has been shown in fluid emissions near some vents, also two genes related to metal detoxification (MTP and MT) were selected. Specific primer pairs were established and tested for 8 putative reference genes (RGs) and 20 genes of interest (GOIs). (The full RT-qPCR protocol and gene description for selected genes are available in Appendix 2 and Supplementary Table S10.
Net plant productivity
Net plant productivity (NPP) was estimated following the methodology described in ref. 56. Initial and final water samples were collected from each chamber with plastic syringes (100 mL) at the beginning and end of the incubations, respectively. Once collected, syringes were quickly transported to shore for sample fixation (2 m away from the incubation site). For oxygen fixation, Exetainer vials (Labco Limited) (12 mL) (n = 3) were filled with water from each syringe. Dissolved oxygen concentration (O2) was determined spectrophotometrically by the modified Winkler method according to the protocol described in ref. 57. Net plant productivity (NPP, μmol O2 g DW−1 h−1) rates were calculated by the difference between final and initial oxygen concentration normalized by the incubation time, the volume of water and the biomass incubated.
Statistics
Normality (Shapiro) and homoscedasticity (Levene) tests were run to test parametric assumptions. Differences in seawater parameters between sites in Vulcano were tested with paired t-tests.
For gene expression, multivariate differences in gene expression among different treatments were tested by an analysis of similarity (ANOSIM), with Primer 6 (v.6.1.12). To inspect the significant gene regulation in the different conditions (in situ incubations and transplants) relative to controls, we used the method developed in REST 2009 Software version 2.0.1358. REST 2009 Software provide a means for determining the mean output and a p value for the likelihood of upregulation or downregulation using a hypothesis test. Statistical significance was set at p < 0.05.
To investigate the effect of CO2 on productivity (NPP) of C. nodosa plants long- (in situ incubations) and short-term (transplants incubations) exposed to high CO2 water from CO2 vents, an ANCOVA analysis considering two factors, the “origin” of the plants (plants from CO2 or Reference site) and the “water” used in the incubation (water from CO2 or Reference site), and “irradiance” (as a continuous covariate) was performed in order to account for any potential effect of irradiance on NPP. ANCOVA analysis indicated a highly significant main effect of the “water” (p = 0.002) whereas the effects of “irradiance” or “origin” were not significant. A significant three-way (irradiance, origin, water) interaction was also revealed (p = 0.03), but the post-hoc tests did not provide a clear interpretation of the occurrence of any other relevant processes, i.e. it was not clear what explanatory factors needed to be retained in the model. We then tested for a model simplification using an Akaike’s information criterion (AIC) test among 3-way and 2-way factorial and additive possible ANCOVA and ANOVA models. The 2-way ANOVA (two fixed crossed factors) with “origin” and “water” as explanatory factors produced the lowest AIC value, that is, the best fit and explanatory power indicating that a simplification of the ANCOVA model was justified59. We retained the two-way ANOVA (two fixed crossed factors) as the most adequate model to interpret the NPP results. Vulcano NPP data were log-transformed to meet parametric assumptions.
Likewise, at Ria Formosa the incubations were carried out along the day with changing irradiances. A multiple regression analysis including irradiance and DIC as explanatory variables was performed to investigate the effect of CO2 on productivity (NPP) of C. nodosa plants. The explanatory power of DIC was highly significant (p = 0.01) as opposed to irradiance (p = 0.2) and the best AIC fit was obtained with DIC as single explanatory variable. For this reason we performed a linear regression with DIC as single explanatory variable.
To analyse the combined response of gene expression patterns and NPP on plants from in situ and transplant incubations, a Principal Component Analysis (PCA) and a Cluster analysis were also performed with the software PAST (v.3.03). To link gene expression to productivity a Spearman’s Rank correlation analysis was conducted between the NPP and expression of each gene measured (n = 20 genes) considering the mean gene expression of each incubation type (n = 4 ranks, i.e. in situ and transplant incubations for both Reference and CO2 plants). Unless otherwise indicated, data were analysed using R60.
Additional Information
How to cite this article: Olivé, I. et al. Linking gene expression to productivity to unravel long- and short-term responses of seagrasses exposed to CO2 in volcanic vents. Sci. Rep. 7, 42278; doi: 10.1038/srep42278 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
We thank the ESF COST Action (ES0906) “Seagrass Productivity: From Genes to Ecosystem Management” for the funds provided and all the participants of the Training School “Effects of CO2/Ocean Acidification on Seagrass Meadows” held at Vulcano (Aeolian Islands, Italy), 6–11 May 2013. We particularly thank the local organizer, Salvatrice Vizzini. This paper is a contribution to the Portuguese Foundation for Science and Technology (FCT) project HighGrass (PTDC/MAR-EST/3687/2012), the main funding source for this study, and Italian MIUR Flagship project RITMARE (NRP 2011–2013). The study also received funds from FCT project UID/Multi/04326/2013. IO and MMC were supported by FCT post-doctoral (SFRH/BPD/71129/2010) and PhD (SFRH/BD/64590/2009) fellowships, respectively. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no competing financial interests.
Author Contributions I.O., J.S. and R.S. conceived, designed and coordinated the study. I.O., J.S., M.M.C., C.L. and M.R. carried out the field work. C.L., M.R. and G.P. conducted genetic lab and data analysis. I.O., M.M.C. and J.S. conducted productivity lab and data analysis. I.O., J.S., G.P. and R.S. drafted the manuscript. I.O. wrote the manuscript. All authors reviewed the manuscript and gave final approval.
References
- Pörtner H. O. et al. Ocean systems in Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part A: Global and Sectoral Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel of Climate Change (eds Field C. B. et al.) Ch. 6, 411–484 (Cambridge University Press, 2014). [Google Scholar]
- Brodie J. et al. The future of the northeast Atlantic benthic flora in a high CO2 world. Ecol. Evol. 4, 2787–2798, doi: 10.1002/ece3.1105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. P., Ainsworth E. A., Rogers A. & Ort D. R. Rising atmospheric carbon dioxide: Plants FACE the Future. Annu. Rev. Plant Biol. 55, 591–628, doi: 10.1146/annurev.arplant.55.031903.141610 (2004). [DOI] [PubMed] [Google Scholar]
- Beer S. & Koch E. W. Photosynthesis of seagrasses and marine macroalgae in globally changing CO2 environments. Mar. Ecol. Prog. Ser. 141, 199–204, doi: 10.3354/meps141199 (1996). [DOI] [Google Scholar]
- Zimmerman R. C., Kohrs D. G., Steller D. L. & Alberte R. S. Impacts of CO2 enrichment on productivity and light requirements of eelgrass. Plant Physiol. 115, 599–607, doi: 10.1104/pp.115.2.599 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. E. & Fourqurean J. W. Effects of in situ CO2 enrichment on the structural and chemical characteristics of the seagrass Thalassia testudinum. Mar. Biol. 160, 1465–1475, doi: 10.1007/s00227-013-2199-3 (2013). [DOI] [Google Scholar]
- Ow Y. X., Collier C. J. & Uthicke S. Responses of three tropical seagrass species to CO2 enrichment. Mar. Biol. 162, 1005–1017, doi: 10.1007/s00227-015-2644-6 (2015). [DOI] [Google Scholar]
- Kroeker K. J. et al. Impacts of ocean acidification on marine organisms: quantifying sensitivities and interaction with warming. Glob. Chang. Biol. 19, 1884–1896, doi: 10.1111/gcb.12179 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Witt T. J., Sih A. & Wilson D. S. Costs and limits of phenotypic plasticity. Trends Ecol. Evol. 13, 77–81, doi: 10.1016/S0169-5347(97)01274-3 (1998). [DOI] [PubMed] [Google Scholar]
- Pigliucci M. How organisms respond to environmental changes: from phenotypes to molecules (and vice versa). Trends Ecol. Evol. 11, 168–173, doi: 10.1016/0169-5347(96)10008-2 (1996). [DOI] [PubMed] [Google Scholar]
- Sunday J. M. et al. Evolution in an acidifying ocean. Trends Ecol. Evol. 29, 117–125, doi: 10.1016/j.tree.2013.11.001 (2014). [DOI] [PubMed] [Google Scholar]
- Lopez-Maury L., Marguerat S. & Bahler J. Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet. 9, 583–593, doi: 10.1038/nrg2398 (2008). [DOI] [PubMed] [Google Scholar]
- Gracey A. Y. Interpreting physiological responses to environmental change through gene expression profiling. J. Exp. Biol. 210, 1584, doi: 10.1242/jeb.004333 (2007). [DOI] [PubMed] [Google Scholar]
- Evans T. G. & Hofmann G. E. Defining the limits of physiological plasticity: how gene expression can assess and predict the consequences of ocean change. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 367, 1733, doi: 10.1098/rstb.2012.0019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey P. A. et al. The emergence of molecular profiling and omics techniques in seagrass biology; furthering our understanding of seagrasses. Funct. Integr. Genomics 16, 465–480, doi: 10.1007/s10142-016-0501-4 (2016). [DOI] [PubMed] [Google Scholar]
- Franssen S. U. et al. Transcriptomic resilience to global warming in the seagrass Zostera marina, a marine foundation species. Proc. Nat. Acad. Sci. 108, 19276–19281, doi: 10.1073/pnas.1107680108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa S. I. et al. Expressed sequence tags from heat-shocked seagrass Zostera noltii (Hornemann) from its southern distribution range. Mar. Genom. 4, 181–188, doi: 10.1016/j.margen.2011.04.003 (2011). [DOI] [PubMed] [Google Scholar]
- Winters G., Nelle P., Fricke B., Rauch G. & Reusch T. B. H. Effects of a simulated heat wave on photophysiology and gene expression of high- and low-latitude populations of Zostera marina. Mar. Ecol. Prog. Ser. 435, 83–95, doi: 10.3354/meps09213 (2011). [DOI] [Google Scholar]
- Salo T., Reusch T. B. H. & Boström C. Genotype-specific responses to light stress in eelgrass Zostera marina, a marine foundation plant. Mar. Ecol. Prog. Ser. 519, 129–140, doi: 10.3354/meps11083 (2015). [DOI] [Google Scholar]
- Dattolo E. et al. Acclimation to different depths by the marine angiosperm Posidonia oceanica: transcriptomic and proteomic profiles. Front. Plant Sci. 4, doi: 10.3389/fpls.2013.00195 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dattolo E. et al. Response of the seagrass Posidonia oceanica to different light environments: Insights from a combined molecular and photo-physiological study. Mar. Environ. Res. 101, 225–236, doi: 10.1016/j.marenvres.2014.07.010 (2014). [DOI] [PubMed] [Google Scholar]
- Lauritano C. et al. Response of key stress-related genes of the seagrass Posidonia oceanica in the vicinity of submarine volcanic vents. Biogeosciences 12, 4185–4194, doi: 10.5194/bg-12-4185-2015 (2015). [DOI] [Google Scholar]
- Timmins-Schiffman E. et al. Shotgun proteomics reveals physiological response to ocean acidification in Crassostrea gigas. BMC Genomics 15, 951, doi: 10.1186/1471-2164-15-951 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procaccini G. et al. Seagrass ecophysiology meets ecological genomics: are we ready? Mar. Ecol. 33, 522–527, doi: 10.1111/j.1439-0485.2012.00518.x (2012). [DOI] [Google Scholar]
- Koch M., Bowes G., Ross C. & Zhang X.-H. Climate change and ocean acidification effects on seagrasses and marine macroalgae. Glob. Chang. Biol. 19, 103–132, doi: 10.1111/j.1365-2486.2012.02791.x (2013). [DOI] [PubMed] [Google Scholar]
- Godbold J. A. & Calosi P. Ocean acidification and climate change: advances in ecology and evolution. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 368, doi: 10.1098/rstb.2012.0448 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gattuso J. P. et al. Contrasting futures for ocean and society from different anthropogenic CO2 emissions scenarios. Science 349, doi: 10.1126/science.aac4722 (2015). [DOI] [PubMed] [Google Scholar]
- Garilli V. et al. Physiological advantages of dwarfing in surviving extinctions in high-CO2 oceans. Nat. Clim. Change. 5, 678–682, doi: 10.1038/nclimate2616 (2015). [DOI] [Google Scholar]
- Dando P. R., Stüben D. & Varnavas S. P. Hydrothermalism in the Mediterranean Sea. Prog. Oceanogr. 4, 333–367, doi: 10.1016/S0079-6611(99)00032-4 (1999). [DOI] [Google Scholar]
- Boatta F. et al. Geochemical survey of Levante Bay, Vulcano Island (Italy), a natural laboratory for the study of ocean acidification. Mar. Pollut. Bull. 73, 485–494, doi: 10.1016/j.marpolbul.2013.01.029 (2013). [DOI] [PubMed] [Google Scholar]
- Hall-Spencer J. M. et al. Volcanic carbon dioxide vents show ecosystem effects of ocean acidification. Nature 454, 96–99, doi: 10.1038/nature07051 (2008). [DOI] [PubMed] [Google Scholar]
- Russell B. D. et al. Future seagrass beds: Can increased productivity lead to increased carbon storage? Mar. Pollut. Bull. 73, 463–469, doi: 10.1016/j.marpolbul.2013.01.031 (2013). [DOI] [PubMed] [Google Scholar]
- Kroeker K. J., Micheli F., Gambi M. C. & Martz T. R. Divergent ecosystem responses within a benthic marine community to ocean acidification. Proc. Nat. Acad. Sci. 108, 14515–14520, doi: 10.1073/pnas.1107789108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostolaki E. T., Vizzini S., Hendriks I. E. & Olsen Y. S. Seagrass ecosystem response to long-term high CO2 in a Mediterranean volcanic vent. Mar. Environ. Res. 99, 9–15, doi: 10.1016/j.marenvres.2014.05.008 (2014). [DOI] [PubMed] [Google Scholar]
- Stillman J. H. & Paganini A. W. Biochemical adaptation to ocean acidification. J. Exp. Biol. 218, 1946–1955, doi: 10.1242/jeb.115584 (2015). [DOI] [PubMed] [Google Scholar]
- Wissler L. et al. Back to the sea twice: identifying candidate plant genes for molecular evolution to marine life. BMC Evol. Biol. 11, 1–13, doi: 10.1186/1471-2148-11-8 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen J. L. et al. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530, 331–335, doi: 10.1038/nature16548 (2016). [DOI] [PubMed] [Google Scholar]
- Hoffmann A. A. & Daborn P. J. Towards genetic markers in animal populations as biomonitors for human-induced environmental change. Ecol. Lett. 10, 63–76, doi: 10.1111/j.1461-0248.2006.00985.x (2007). [DOI] [PubMed] [Google Scholar]
- Macreadie P. I., Schliep M. T., Rasheed M. A., Chartrand K. M. & Ralph P. J. Molecular indicators of chronic seagrass stress: A new era in the management of seagrass ecosystems? Ecol. Indic. 38, 279–281, doi: 10.1016/j.ecolind.2013.11.017 (2014). [DOI] [Google Scholar]
- Pespeni M. H., Chan F., Menge B. A. & Palumbi S. R. Signs of adaptation to local pH conditions across an environmental mosaic in the California Current Ecosystem. Integr. Comp. Biol. 53, 857–870, doi: 10.1093/icb/ict094 (2013). [DOI] [PubMed] [Google Scholar]
- Pigliucci M. Phenotypic Plasticity: Beyond Nature and Nurture (Johns Hopkins University Press, 2001). [Google Scholar]
- Olesen B., Enríquez S., Duarte C. M. & Sand-Jensen K. Depth-acclimation of photosynthesis, morphology and demography of Posidonia oceanica and Cymodocea nodosa in the Spanish Mediterranean Sea. Mar. Ecol. Prog. Ser. 236, 89–97, doi: 10.3354/meps236089 (2002). [DOI] [Google Scholar]
- Olivé I., Vergara J. J. & Pérez-Lloréns J. L. Photosynthetic and morphological photoacclimation of the seagrass Cymodocea nodosa to season, depth and leaf position. Mar. Biol. 160, 285–297, doi: 10.1007/s00227-012-2087-2 (2013). [DOI] [Google Scholar]
- Invers O., Zimmerman R. C., Alberte R. S., Pérez M. & Romero J. Inorganic carbon sources for seagrass photosynthesis: an experimental evaluation of bicarbonate use in species inhabiting temperate waters. J. Exp. Mar. Biol. Ecol. 265, 203–217, doi: 10.1016/S0022-0981(01)00332-X (2001). [DOI] [Google Scholar]
- Beer S. & Waisel Y. Some photosynthetic carbon fixation properties in seagrasses. Aquat. Bot. 7, 129–138, doi: 10.1016/0304-3770(79)90017-2 (1979). [DOI] [Google Scholar]
- Vizzini S. et al. Trace element bias in the use of CO2-vents as analogues for low-pH environments: Implications for contamination levels in acidified oceans. Estuar. Coast. Shelf Sci. 134, 19–30, doi: 10.1016/j.ecss.2013.09.015 (2013). [DOI] [Google Scholar]
- Johnson R. V., Brownlee C., Milazzo M. & Hall-Spencer M. J. Marine microphytobenthic assemblage shift along a natural shallow-water CO2 gradient subjected to multiple environmental stressors. J. Mar. Sci. Eng. 3, doi: 10.3390/jmse3041425 (2015). [DOI] [Google Scholar]
- Sedwick P. & Stuben D. Chemistry of shallow submarine warm springs in an arc-volcanic setting: Vulcano Island, Aeolian Archipelago, Italy. Mar. Chem. 53, 147–161, doi: 10.1016/0304-4203(96)00020-5 (1996). [DOI] [Google Scholar]
- Prange J. A. & Dennison W. C. Physiological responses of five seagrass species to trace metals. Mar. Pollut. Bull. 41, 327–336, doi: 10.1016/S0025-326X(00)00126-0 (2000). [DOI] [Google Scholar]
- Holmer M. & Bondgaard E. J. Photosynthetic and growth response of eelgrass to low oxygen and high sulfide concentrations during hypoxic events. Aquat. Bot. 70, 29–38, doi: 10.1016/S0304-3770(00)00142-X (2001). [DOI] [Google Scholar]
- Brack W. & Frank H. Chlorophyll a fluorescence: A tool for the investigation of toxic effects in the photosynthetic apparatus. Ecotoxicol. Environ. Saf. 40, 34–41, doi: 10.1006/eesa.1997.1639 (1998). [DOI] [PubMed] [Google Scholar]
- Oren A., Padan E. & Malkin S. Sulfide inhibition of Photosystem II in cyanobacteria (blue-green algae) and tobacco chloroplasts. Biochim. Biophys. Act Bioenergetics 546, 270–279, doi: 10.1016/0005-2728(79)90045-8 (1979). [DOI] [PubMed] [Google Scholar]
- Inguaggiato S. et al. Total CO2 output from Vulcano island (Aeolian Islands, Italy). Geochemistry, Geophysics, Geosystems 13, doi: 10.1029/2011gc003920 (2012). [DOI] [Google Scholar]
- MS Excel Program Developed for CO2 System Calculations. ORNL/CDIAC-105a. Carbon Dioxide Information Analysis Center, Oak Ridge National Laboratory, U.S. Department of Energy, Oak Ridge, Tennessee (2006).
- Barrón C. & Duarte C. M. Dissolved organic matter release in a Posidonia oceanica meadow. Mar. Ecol. Prog. Ser. 374, 75–84, doi: 10.3354/meps07715 (2009). [DOI] [Google Scholar]
- Olivé I., Silva J., Costa M. M. & Santos R. Estimating seagrass community metabolism using benthic chambers: The effect of incubation time. Estuar. Coast. 39, 138–144, doi: 10.1007/s12237-015-9973-z (2016). [DOI] [Google Scholar]
- Labasque T., Chaumery C., Aminot A. & Kergoat G. Spectrophotometric Winkler determination of dissolved oxygen: re-examination of critical factors and reliability. Mar. Chem. 88, 53–60, doi: 10.1016/j.marchem.2004.03.004 (2004). [DOI] [Google Scholar]
- Pfaffl M. W., Horgan G. W. & Dempfle L. Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 75, 30 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawley M. J. Analysis of Covariance In The R Book. Ch. 12, 489–509 (John Wiley & Sons, Ltd, 2007). [Google Scholar]
- R. Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/ (2013).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.