The inhibitor endosin 16 defines a distinct pathway for endomembrane sorting via regulation of the small GTPase RabA that is essential for the establishment of cell polarity.
Abstract
The endomembrane system is an interconnected network required to establish signal transduction, cell polarity, and cell shape in response to developmental or environmental stimuli. In the model plant Arabidopsis thaliana, there are numerous markers to visualize polarly localized plasma membrane proteins utilizing endomembrane trafficking. Previous studies have shown that the large ARF-GEF GNOM plays a key role in the establishment of basal (rootward) polarity, whereas the apically (shootward) polarized membrane proteins undergo sorting via different routes. However, the mechanism that maintains apical polarity is largely unknown. Here, we used a chemical genomic approach and identified the compound endosidin 16 (ES16), which perturbed apically localized plasma membrane proteins without affecting basal polarity. We demonstrated that ES16 is an inhibitor for recycling of apical, lateral, and nonpolar plasma membrane proteins as well as biosynthetic secretion, leaving the basal proteins as the only exceptions not subject to ES16 inhibition. Further evidence from pharmaceutical and genetic data revealed that ES16 effects are mediated through the regulation of small GTPase RabA proteins and that RabA GTPases work in concert with the BIG clade ARF-GEF to modulate the nonbasal trafficking. Our results reveal that ES16 defines a distinct pathway for endomembrane sorting routes and is essential for the establishment of cell polarity.
INTRODUCTION
Endomembrane trafficking plays an essential role in maintaining cellular homeostasis and signal transduction and in response to environmental stimuli. The membrane trafficking pathways start from the endoplasmic reticulum (ER) then go through the Golgi apparatus to different destinations including vacuoles/lysosomes, endosomes, and the plasma membrane (PM) (Morita and Shimada, 2014). The plant endomembrane system is mostly conserved among eukaryotes but displays complex characteristics. The trans-Golgi network (TGN) is a unique subcellular structure, which is a sorting center that integrates upstream cargos from secretory vesicles, the plasma membrane, and other organelles. Recent evidence indicates that the TGN functions as an early endosome compartment, adding to the complexity of sorting mechanisms in plant cells.
Auxin transporters have been used to study sorting mechanisms due to their fundamental importance in development and cell viability. Auxin transporters, including the PIN-FORMED (PIN) efflux carriers (Friml et al., 2002; Blilou et al., 2005; Xu and Scheres, 2005; Zádníková et al., 2010), the influx transporter AUXIN RESISTANT1 (AUX1) (Swarup et al., 2004), and a subset of ATP BINDING CASSETTE SUBFAMILY B proteins (Geisler and Murphy, 2006), are localized at the plasma membrane in a polar manner. The polarity is at the apical (shootward), basal (rootward) (Baskin et al., 2010), or lateral PM, and it is largely unknown how protein polarity is established and maintained. Current research points to a model in which polar localization of PINs at the PM is orchestrated by clathrin-mediated endocytosis (Kitakura et al., 2011), polar recycling (Geldner et al., 2003), lipid raft dependent restriction of lateral diffusion (Willemsen et al., 2003; Jaillais and Gaude, 2008), and protein phosphorylation (Huang et al., 2010; Zhang et al., 2010). Recent publications have revealed that the trafficking of apically localized PM auxin transporters is distinct from their basal sorting routes. The ADP ribosylation factor guanine nucleotide exchange factor (ARF-GEF) GNOM is the only protein thus far known to guide basal recycling (Geldner et al., 2003). In the GNOM weak allele mutant gnomR5 or after prolonged treatment with Brefeldin A (BFA), a fungal toxin targeting ARF-GEFs, there is a complete basal-to-apical shift of PIN1, but the polarity of apically localized PIN2 or AUX1 is mostly unchanged (Kleine-Vehn et al., 2006, 2008b). When tracking vesicle movement with photoconvertible PIN2-EosFP, BFA-induced endomembrane bodies move to the apical PM but not to the basal PM (Kleine-Vehn et al., 2008b). This pattern suggests that in the presence of functionally compromised GNOM, vesicle movement is redirected toward apical trafficking. Based on these observations, we hypothesized that there were specific components regulating apical sorting.
We performed a screen of a 360-compound bioactive chemical library containing inhibitors of endomembrane trafficking to identify small molecules that interfered with the apical trafficking. The library was generated previously from a screen of 46,418 diverse molecules that inhibited tobacco (Nicotiana tabacum) pollen germination in vitro, which is strongly dependent on exocytosis and endocytosis (Drakakaki et al., 2011). We identified the compound endosidin 16 (ES16) that perturbed recycling of apical, lateral, and nonpolar plasma membrane proteins without affecting the basal polarity. Using pharmacological approaches, we confirmed that ES16 works in concert with the BIG clade ARF-GEFs to modulate the nonbasal trafficking. Using genetic and biochemical evidence, we further demonstrated that ES16 functions through the regulation of small GTPase RAB GTPASE A2A (RabA2A). Our results revealed an essential and previously uncharacterized regulatory mechanism for endomembrane sorting and the establishment of polarity.
RESULTS
ES16 Inhibits Apical Polarity without Disturbing Basal Polarity
To begin to understand the mechanisms of apical trafficking, we addressed two key questions. First, does an endomembrane trafficking pathway exist that is specific for apical targeting? Second, how does the cell distinguish apical and basal recycling and then regulate their convergence at the PM?
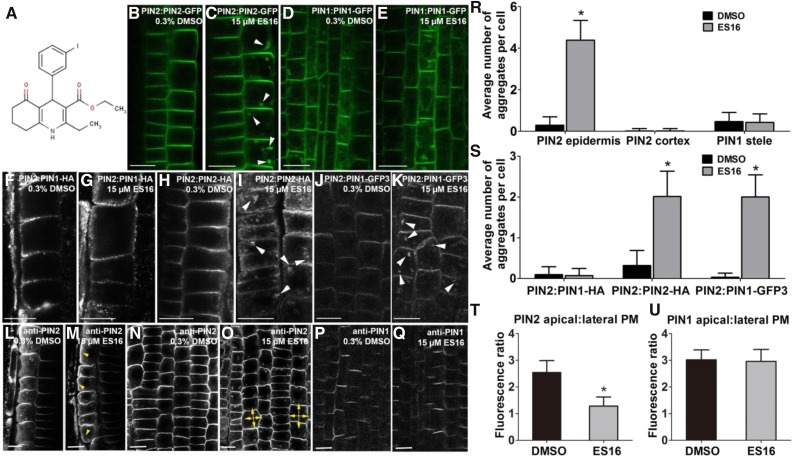
To address the questions, we designed a chemical screen to identify compounds that induce endomembrane bodies only of apically localized PIN2 without interfering with basal PIN1 localization (Benková et al., 2003). Endomembrane bodies are chemically induced alterations in endomembrane compartments that partially or fully inhibit trafficking at specific compartments. Such bodies can be the result of perturbations in processes such a vesicle budding and fusion leading to the formation of aberrant endomembrane compartments (Drakakaki et al., 2011). In this screen, we found that the compound ES16 (Figure 1A) selectively modified PIN2 apical PM targeting in epidermal cells of Arabidopsis (Arabidopsis thaliana) roots without affecting the localization of PIN1 and PIN2, which are basally localized in cortical cells (Figures 1B to 1E and 1R). To further explore ES16 phenotypes, we examined ES16 treatment of other basally localized auxin transporters, including PIN3 (Benková et al., 2003; Marhavý et al., 2013) and PIN7 (Teale et al., 2006) in the stele region as well as PIN5 localized in the ER (Mravec et al., 2009). Our results indicated that ES16 did not interfere with the membrane targeting or polarity of these auxin transporters (Supplemental Figures 1A to 1C and 1E to 1G).
Figure 1.
ES16 Selectively Interferes with Apically Localized Plasma Membrane Protein.
(A) The chemical structure of ES16.
(B) and (C) Images of root cells from PIN2:PIN2-GFP line after treatment with either DMSO (B) or ES16 (C). White arrowheads indicate the ES16 induced aggregates are observed in epidermal cells but are absent from the adjacent cortex cells.
(D) and (E) Root stele cells from the PIN1:PIN1-GFP line are insensitive to ES16 treatment.
(F) and (G) PIN1 basal localization is largely unchanged in the PIN1:PIN1-HA line. Cells were detected by immunostaining with anti-HA antibody.
(H) and (I) ES16-induced agglomerations are observed in immunolocalized PIN2:PIN2-HA seedlings with anti-HA antibody.
(J) and (K) ES16 could also induce aggregates in PIN2:PIN1-GFP3 cells.
(L) to (O) ES16 treatment reduced apical polarity of PIN2 and increased lateral localization (yellow arrowheads in [M], and double yellow arrows in [O] represent lateral diffusion of PIN2 signal).
(P) and (Q) PIN1 basal polarity remained unchanged after ES16 treatment. Images were detected by immunostaining with either anti-PIN2 ([L] to [O]) or anti-PIN1 ([P] and [Q]) antibodies in Col seedlings.
(R) and (S) Quantification of aggregates induced by ES16 compared with DMSO control. Quantification was calculated by measuring the number of aggregates in each cell; 200 cells from 10 seedlings were chosen for quantification per treatment per genotype.
(T) and (U) Quantification of polar to lateral plasma membrane signal as shown from (L) to (R); 300 cells from 10 seedlings were selected for quantification. The significance of difference was calculated by a two-tailed Student’s t test, and the asterisks represent P < 0.01. Images are representative from three repeats.
Images from (B) to (E) are from live imaging, while the ones from (F) to (R) are from immunostaining. Error bars represent sd. Bars = 10 µm.
The lack of effect of ES16 on basal PIN1 could be explained by inability of the compound to penetrate to deeper cell layers of the root compared with PIN2 in the outer epidermal layer. We compared PIN1 and PIN2 localization by immunofluorescence in a line expressing HA-tagged PIN1 (PIN1-HA) under the PIN2 promoter resulting in PIN1 expression in epidermal cells. In an untreated control, PIN1 displayed basal localization, as published previously (Wisniewska et al., 2006) (Figure 1F). Significantly, ES16 had no effect on basal PIN1-HA in epidermal cells (Figure 1G) in the same cell layer where ES16 resulted in the formation of bodies containing PIN2-HA (Figures 1H, 1I, and 1S). Thus, we concluded that the lack of effect of ES16 on basal polarity was a property of the basal polarity pathway rather than differential transport of the compound in epidermal and cortical cells.
This conclusion was supported by the use of a line expressing PIN1-GFP3 under the PIN2 promoter. In PIN2:PIN1-GFP3, GFP is inserted in the hydrophilic loop of PIN1 resulting in apical (rather than basal) localization of PIN1-GFP in the epidermis (Wisniewska et al., 2006). Thus, PIN1-GFP3 was in the epidermal layer and apically localized. Under these conditions, apically localized PIN1 was sensitive to ES16, resulting in endomembrane bodies (Figures 1J, 1K, and 1S). This indicated that ES16 inhibition of apical polarity (and the lack effect on basal polarity) was a property of the apical and basal pathways and not of PIN1 protein itself. Therefore, the bioactivity of ES16 appeared to be specific for the apical pathway.
To understand its direct effect on polar localization, we quantified the effect of ES16 on apical polarity by measuring the ratio of polar-to-lateral PM signal. ES16 treatment resulted in strong lateral diffusion of PIN2 as revealed by immunostaining (Figures 1L to 1O) and significantly decreased the apical-to-lateral localization ratio (Figure 1T). By contrast, PIN1 polarity was not altered significantly when detected under similar treatment conditions (Figures 1P, 1Q, and 1U). These results indicated that ES16 not only induced endomembrane bodies containing PIN2 but also inhibited PIN2 apical polarity resulting in increased isodiametric PM localization.
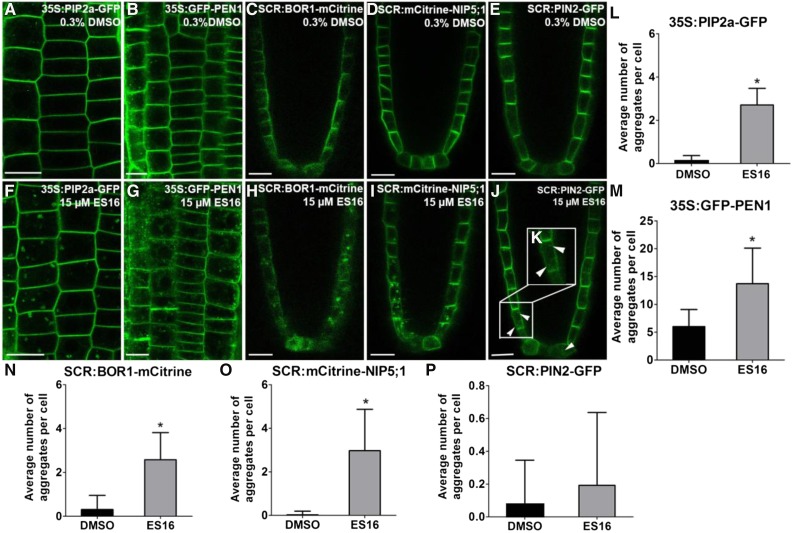
As further evidence that ES16 action was dependent upon protein polarity and not tissue specificity, we examined the auxin transporter AUX1, which is nonpolar in cells of the epidermis and lateral root cap but is enriched at the apical PM in protophloem cells (Swarup et al., 2001; Kleine-Vehn et al., 2006). Consistent with previous results, ES16 treatment induced bodies of AUX1 in protophloem cells (Supplemental Figures 1D and 1H), again indicating an effect on apical targeting. This was confirmed using a line where PIN2 was expressed under the endodermis-specific SCARECROW (SCR) promoter, resulting in apolar signal in the quiescent center but basal localization in the initials and proendodermal cells (Alassimone et al., 2010). ES16 induced small bodies in cells near the quiescent center where the polarity of PIN2 was not obvious; however, it did not interfere with basal localization of PIN2 in the upper, more developed cells (Figures 2E, 2J, and 2P, white arrowheads indicate bodies near the quiescent center, which are highlighted in the enlarged Figure 2K), confirming that ES16 did not target basal polarity irrespective of the tissue layer.
Figure 2.
ES16 Also Disturbs Nonpolar and Lateral Localization of Plasma Membrane Proteins.
(A) to (D) Images of root cells from 35S:PIP2a-GFP (A), 35S:GFP-PEN1(B), SCR:BOR1-mCitrine (C), SCR:mCitrine-NIP5;1 (D), or SCR:PIN2-GFP (E) after 0.3% DMSO treatment for 1 h.
(E) to (H) Images of root cells from 35S:PIP2a-GFP (F), 35S:GFP-PEN1(G), SCR:BOR1-mCitrine (H), SCR:mCitrine-NIP5;1 (I), or SCR:PIN2-GFP ([J] and [K]) after 15 µM ES16 treatment for 1 h.
(K) Zoom-in image of the cells close to the quiescent center where the polarity of PIN2 is not obvious.
(L) to (P) Quantification of aggregates induced by ES16 compared with DMSO control. Quantification is calculated by measuring the number of aggregates in each cell; 200 cells from 10 seedlings were chosen for quantification per treatment per genotype. Note that the y axis scales for (M) and (P) differ from those of (L), (N), and (O). The significance of difference was calculated by a two-tailed Student’s t test, and the asterisks represent P < 0.01. Images are representative from three repeats. Error bars represent sd. Bars = 10 µm.
Overall, we concluded from these experiments that ES16 reduced the targeting or maintenance of proteins enriched in the apical PM without affecting basal PM protein polarization. Our results also suggest that apical and basal trafficking occur via distinct mechanisms that are not limited by tissue differentiation.
Basal Trafficking Is a Distinct Sorting Route
To understand in more detail the effects of ES16 on other PM trafficking pathways and to gain more insight into the uniqueness of the basal and apical polarity pathways, we examined ES16 effects on nonpolar and especially laterally localized PM proteins, for which the mechanisms of localization are not well understood. Interestingly, ES16 induced bodies containing the nonpolar localized aquaporin PLASMA MEMBRANE INTRINSIC PROTEIN 2a (PIP2a) (Li et al., 2011) (Figures 2A, 2F, and 2L) and the syntaxin PENETRATION1 (PEN1) (Collins et al., 2003) (Figures 2B, 2G, and 2M). Normally, PEN1 is localized primarily to the PM in a nonpolar manner and also displays endosome localization. After ES16 treatment, bodies containing PEN1 were significantly increased compared with the control (Figures 2B, 2G, and 2M), indicating that ES16 inhibited nonpolar PM localization.
We then examined the effect of ES16 on laterally polarized PM proteins using two marker lines, SCR:BOR1-mCitrine and SCR:mCitrine-NIP5;1 (Alassimone et al., 2010). REQUIRES HIGH BORON1 (BOR1) and NOD26-LIKE INTRINSIC PROTEIN5;1 (NIP5;1) are both boron transporters. When driven by the SCR promoter, BOR1 shows lateral polarity toward the stele layer, whereas NIP5;1 displays lateral polarity in the opposite direction toward the cortex (Alassimone et al., 2010). ES16 treatment induced bodies containing these two transporters (Figures 2C, 2H, 2D, 2I, 2N, and 2O), whereas it did not disrupt basal localized PIN2 in the same endodermis cells (Figures 2E, 2J, and 2P). We concluded that ES16 not only interfered with apical localization of PIN2 but also with nonpolar and lateral localization of PM proteins. Biologically and mechanistically, the results challenged our hypothesis that apical trafficking is regulated through unique components that are distinct from other PM routes. Instead, our results indicated that basal trafficking is distinct from the apical, nonpolar, and lateral polarity pathways, which may share common or closely related components that are targeted by ES16.
Basal Trafficking May Utilize TGN Subdomains or Bypass the TGN
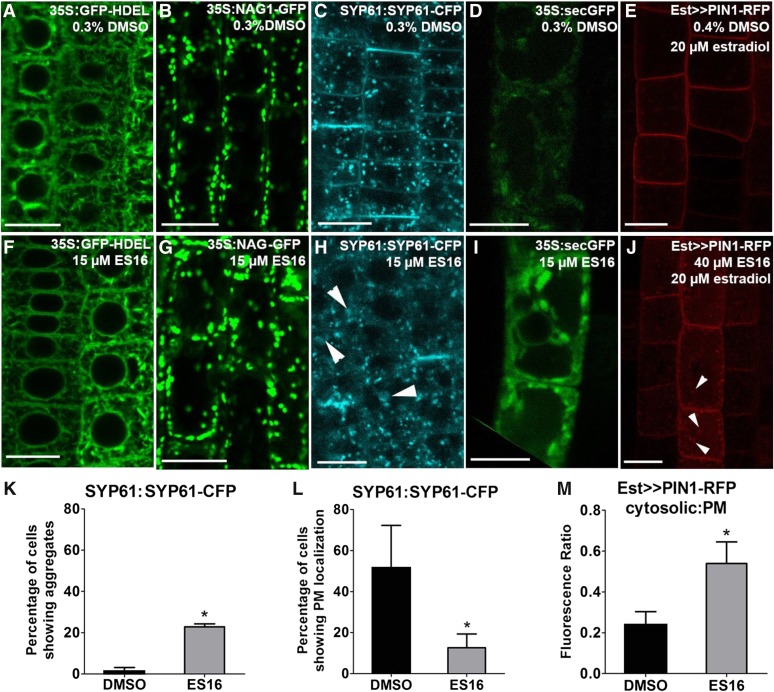
Beyond PM targeting, we next focused on identifying additional endomembrane trafficking pathways perturbed by ES16. The compound did not significantly alter ER-to-Golgi trafficking; neither the ER marker GFP-HDEL (Ridge et al., 1999) (Figures 3A and 3F) nor the Golgi protein N-ACETYLGLUCOSAMINYL TRANSFERASE I (NAG; Grebe et al., 2003) (Figures 3B and 3G) was altered in localization or indicated aberrant organelle morphology. No detectable alterations in cytoskeleton morphology were observed (Supplemental Figures 2A, 2B, 2D, and 2E). However, ES16 strongly blocked the post-Golgi trafficking of TGN localized SYNTAXIN OF PLANTS61 (SYP61; Robert et al., 2008), which was found in enlarged bodies and had greatly reduced PM localization (Figures 3C, 3H, 3K, and 3L, white arrowheads indicate enlarged bodies). However, ES16 did not significantly interfere with the morphology of another TGN marker, VACUOLAR PROTON ATPASE A1 (VHA-a1; Dettmer et al., 2006) (Supplemental Figures 2C and 2F). Interestingly, this suggested that ES16 perturbed only a subfraction of TGN compartments or proteins. In summary, ES16 perturbed a subset of the TGN but not the ER or Golgi compartments.
Figure 3.
ES16 Blocks Post-Golgi Trafficking and Secretion.
(A) to (E) Images of 35S:GFP-HDEL (A), 35S:NAG-GFP (B), SYP61:SYP61-CFP (C), 35S:secGFP (D), and Est>>PIN1-RFP (E) lines after 0.3% DMSO treatment.
(F) to (J) Images of 35S:GFP-HDEL (F), 35S:NAG-GFP (G), SYP61:SYP61-CFP (H), 35S:secGFP (I), and Est>>PIN1-RFP (J) after ES16 treatment. The white arrowheads in (H) indicate the big aggregates induced by ES16. The white arrowheads in (J) represent the retention secretory vesicles within the cells after ES16 treatment.
(K) Quantification of the percentage of cells showing PM localization as in (C) and (H).
(L) Quantification of the percentage of cells showing intracellular aggregates as in (C) and (H). For (K) and (L), 20 seedlings with 30 cells per seedling per treatment were chosen for quantification.
(M) Quantification of cytosol to plasma membrane signal as shown in (E) and (J); 300 cells from 10 seedlings were selected for quantification. The significance of difference was calculated by a two-tailed Student’s t test, and the asterisks represent P < 0.01. Images are representative from three repeats. Error bars represent sd. Bars = 10 µm.
We next examined the secretory pathway using a secreted GFP (secGFP) (Zheng et al., 2004). This marker emits a greatly diminished GFP signal in the apoplast but a strong GFP signal in the cytoplasm due to differences in pH at these cellular locations. Under normal conditions, secGFP was transported through a default secretory pathway to the apoplast with only faint signal observed within the cell (Figure 3D). After ES16 treatment, the intracellular signal intensity was significantly increased (cf. Figures 3I and 3D), indicating that ES16 inhibited general secretion. We also analyzed another established marker line Est>>PIN1-RFP (Richter et al., 2014). In this estradiol-induced line, PIN1-RFP was found to be nonpolar at the PM rather than displaying basal polarity (Richter et al., 2014) (Figure 3E). ES16 did not inhibit the PM localization of PIN1 but strongly enhanced intracellular signal intensity and bodies within the cells (Figures 3E, 3J, and 3M, white arrowheads indicate bodies), suggesting that ES16 interfered with PIN1 biosynthetic secretion. This confirmed the results with secGFP that ES16 interfered with secretion. Overall, the data indicate that ES16 inhibited post-Golgi trafficking, a subset of the TGN and general secretion without interfering with cytoskeleton organization. Since basal trafficking is the only sorting route not affected by ES16, we hypothesized that basal sorting may use either non-ES16 sensitive TGN subcompartments or bypass the TGN.
ES16 Redirects Post-Golgi Trafficking to Vacuole Transport
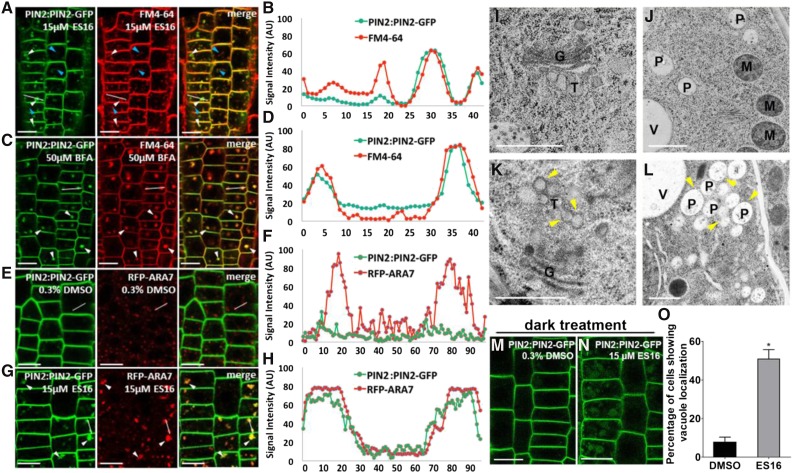
To explore the nature of aggregates induced by ES16 (ES16 bodies), we first tested the response of lyophilic dye FM4-64, which is widely used to track the dynamics of endomembrane trafficking (Bolte et al., 2004). Cotreatment of E16 together with FM 4-64 dye for 1 h resulted in only partial colocalization of PIN2 aggregates with the FM4-64-labeled vesicles (Figures 4A and 4B). This is in contrast with the fungal toxin BFA, which blocks the activities of ARF-GEFs by targeting the conserved Sec7 domain (Peyroche et al., 1999). BFA-induced PIN2 agglomerations almost completely colocalized with FM4-64 dye (Figures 4C and 4D), indicating that ES16 and BFA may be involved in different endomembrane sorting pathways. However, the PIN2 ES16 bodies colocalized with the enlarged prevacuolar compartment (PVC) marker ARA7 (Ueda et al., 2004) to a great extent (Figures 4E to 4H). Considering that ES16 also induced the disorganization of TGN protein SYP61 (Figures 3C, 3H, 3K, and 3L), we speculate that ES16 may interfere with TGN-to-PVC transport. To test the hypothesis, a transmission electron microscopy (TEM) experiment was performed to further examine the subcellular changes. In control samples, TGN could be clearly detected in close adjacency to the Golgi apparatus, with tubular vesicles budding from the trans-Golgi cisterna and the PVC separate from each other (Figures 4I and 4J; Supplemental Figures 3I and 3J). However, ES16 treatment altered both compartments. Clusters of vacuolated vesicles were observed either detached from or close to Golgi, which appeared to be TGN derived (Figure 4K; Supplemental Figure 3K, yellow arrowheads). Golgi morphology was not significantly altered (Figure 4K; Supplemental Figure 3K), consistent with our previous result from CLSM. ES16 also induced PVC clustering into large agglomerations (Figure 4L; Supplemental Figure 3L, yellow arrowheads), which could explain the enlarged ARA7 bodies. However, ES16 did not cause PVC fusion or prevent its fusion with the vacuole (Supplemental Figure 3L, blue arrowhead). To further explore the nature of PVC agglomerations, the dynamic response of ARA7 to ES16 was analyzed at different time points. Long-term treatment with ES16 resulted in less colocalization between ARA7 and PIN2 aggregates, with more PIN2 transferred to the vacuole and reduced signal intensity and size of ARA7 bodies after 3 h (Supplemental Figures 3A to 3H and 3M), indicating that the colocalization between PIN2 and ARA7 may be the transition stage to the vacuole. This was confirmed by the increased vacuole localization of PIN2 after ES16 treatment under dark conditions (Figures 4M to 4O). Taken together, our data suggest that ES16 treatment redirects post-Golgi trafficking to vacuole transport and the basal trafficking may bypass this route.
Figure 4.
ES16 Treatment Promotes Vacuole Transport.
(A) to (D) ES16 treatment induced partial colocalization between PIN2 aggregates and the FM4-64-labeled vesicles ([A] and [B]), while BFA leads to almost complete colocalization ([C] and [D]). White arrowheads indicate colocalized vesicles, whereas the blue arrowheads represent the FM4-64-specific vesicles not found in the green channel.
(E) to (H) ES16 treatment resulted in the colocalization between PIN2 and ARA7 to a high extent ([G] and [H]) compared with DMSO control ([E] and [F]). To quantify the colocalization, a line was drawn in the same position of both red and green channels and the signal intensity of each pixel was plotted against the relative distance in both channels ([B], [D], [F], and [H]).
(I) to (J) Transmission electron microscopy results from the cryofixed, freeze-substituted root tip sections after treatment with DMSO ([I] and [J]) or ES16 ([K] and [L]). After ES16 treatment, a cluster of vacuolated vesicles (yellow arrowheads) appeared to be derived from TGN. Golgi morphology was not significantly altered ([K] compared with [I]). ES16 also induced agglomerations of PVCs ([L], yellow arrowheads) without fusion with each other ([L], blue arrowhead). G, Golgi; T, TGN; P, PVC; M, mitochondrion; V, vacuole.
(M) to (O) ES16 promoted vacuole transport after 3 h dark treatment ([N] compared with [M]). Quantification was based on the ratio of the cells showing vacuole localization to total cells in the same seedling; 15 seedlings were quantified per treatment per repeat. The significance of difference was calculated by a two-tailed Student’s t test, and the asterisks represent P < 0.01. Images are representative of three repeats. Bars represent sd.
Bars = 10 µm in (A), (C), (E), (G), (M), and (N) and 500 nm in (I) to (L).
Basal Recycling Is ES16 Independent
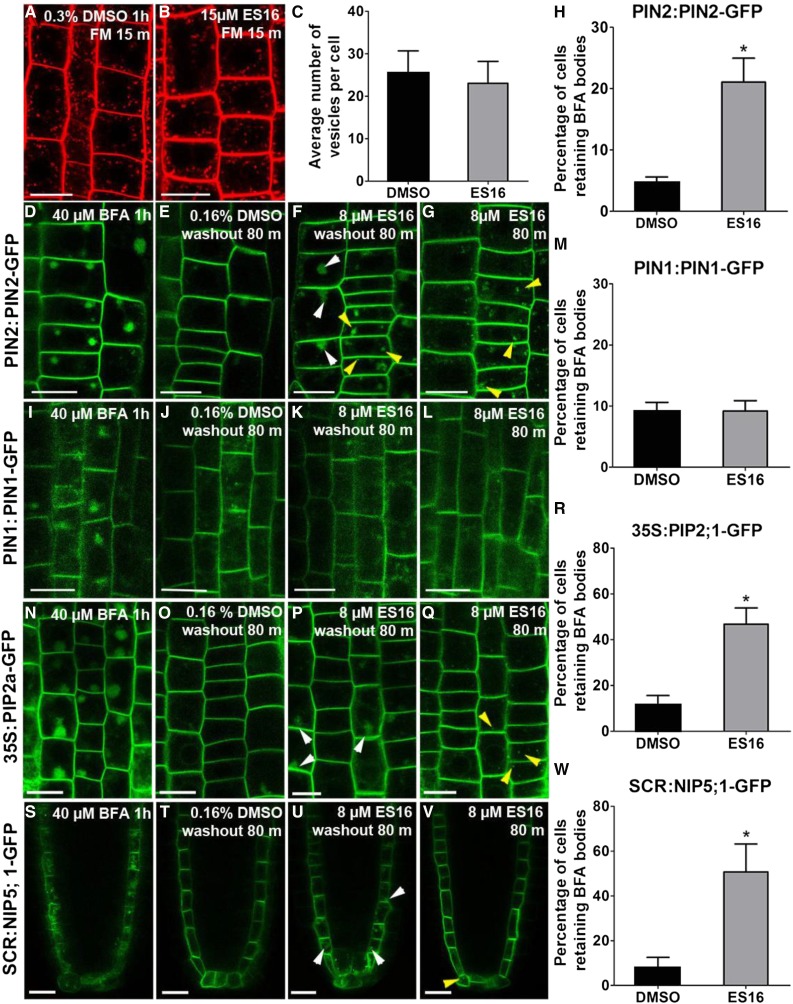
Given the effects of ES16 on the TGN, PVC, and PM, the chemical could have altered the dynamics of endocytosis and recycling. Although cotreatment of ES16 with FM4-64 lead to partial colocalization between PIN2 aggregates and FM dye (Figures 4A and 4B), pretreatment with ES16 did not inhibit the uptake of FM4-64 (Figures 5A to 5C), indicating that ES16 was not an endocytosis inhibitor. We next tested the effect of ES16 on recycling by performing BFA washout experiments (Hendricks et al., 1992). All PM proteins tested, including PIN1, PIN2, PIP2a, and NIP5;1, formed large bodies (BFA bodies) in response to BFA, which interferes with vesicle fusion resulting in aggregates of Golgi and TGN (Figures 5D, 5I, 5N, and 5S). When BFA was washed out, PM proteins were released from the BFA bodies and recycled back to the PM (Figures 5E, 5J, 5O, and 5T). However, washout in the presence of ES16 inhibited the recycling of apically polarized PIN2, nonpolarized PIP2a, and laterally polarized NIP5;1 (Figures 5F, 5P, 5U, 5H, 5R, and 5W) without affecting basally localized PIN1 recovery (Figures 5K and 5M). A low concentration of ES16 was used for BFA washout to minimize ES16 induced bodies (Figures 5F, 5G, 5Q, and 5V, yellow arrowheads represent ES16 aggregates), which were easily distinguishable from the much larger BFA bodies (white arrowheads in Figures 5F, 5P, and 5U indicate the leftover BFA bodies). We concluded that ES16-induced aggregates from PIN2, PIP2a, and NIP5;1 resulted from the inhibition of recycling rather than interference with endocytosis. This is also consistent with our previous result that inhibition of recycling by ES16 redirects post-Golgi trafficking to vacuole transport (Figure 4; Supplemental Figure 3). Overall, we concluded from these experiments that ES16 is a localization inhibitor of apically polarized, nonpolar, and laterally polarized PM proteins, but not basally polarized PM proteins. This result indicated that basally localized PM proteins such as PIN1 use an ES16 independent pathway for basal recycling, which could bypass the TGN.
Figure 5.
ES16 Inhibits Recycling but Not Endocytosis.
(A) and (B) Images of FM4-64 uptake after pretreatment with either 0.3% DMSO (A) or 15 µM ES16 (B).
(C) Quantification of internalization of FM4-64 dye after 15 min uptake as shown in (A) and (B). Quantification was based on counting the number of FM vesicles in each cell; 200 cells from 10 seedlings were chosen for quantification per treatment.
(D), (I), (N), and (S) Large aggregates induced by BFA in root cells of PIN2:PIN2-GFP (D), PIN1:PIN1-GFP (E), 35S:PIP2a-GFP (N), and SCR:NIP5;1-mCitrine (S) lines.
(E), (J), (O), and (T) Root cells after 80 min washout in the presence of 0.16% DMSO, with almost all the BFA bodies in the root cells recycled back to the PM.
(F), (K), (P), and (U) Washout in the presence of 8 µM ES16 showed reduced recycle efficiency of PIN2 (F), PIP2a (P), and NIP5;1 (U) but not PIN1 (K).
(G), (L), (Q), and (V) Small agglomerations of PIN2 (G), PIP2a (Q), and NIP5;1 (V) but not PIN1 (Q) in the presence of 8 µM ES16.
(H), (M), (R), and (W) Quantification of BFA washout efficiency. Quantification was calculated using the ratio of the cells retaining BFA bodies to total cells in the same seedling. Note that the y axis maximum values for (H) and (M) differ from those of (R) and (W). Images are representative from three repeats; 15 seedlings were quantified per treatment per repeat. A two-tailed Student’s t test was used to calculate significance and asterisks represent P < 0.01. Error bars represent sd. Bars = 10 µm.
Inhibition of Root Growth and Gravity Response Corresponds with ES16’s Influence on Endomembrane Recycling Pathways
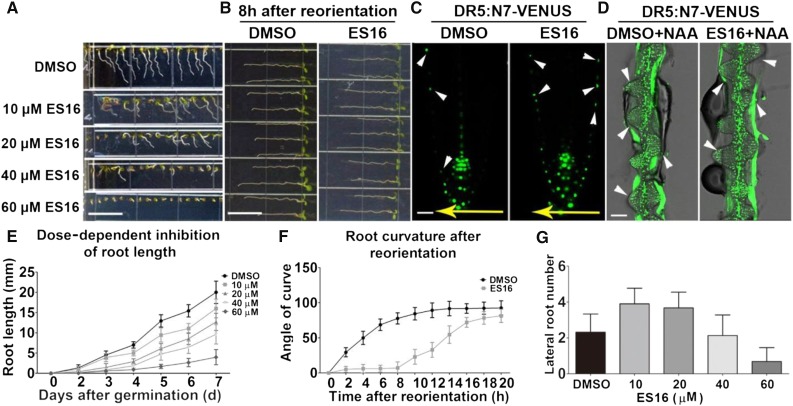
To understand the link between trafficking pathways affected by ES16 and early plant development, we analyzed ES16 activity in intact Arabidopsis seedlings. We observed dose-dependent inhibitory effects on root elongation and gravitropic response (Figures 6A and 6E). The alteration of gravitropism was further demonstrated by delayed root curvature after reorientation (Figures 6B and 6F) and impaired asymmetric auxin redistribution before significant curvature (Figure 6C). However, long-term incubation on ES16-supplemented medium resulted in loss of agravitropism (Figure 7), possibly due to instability of the compound after long-term exposure to light. Asymmetric auxin flow is a key signal in gravity response and results from differential degradation of PIN2 at the upper and lower sides of the root (Abas et al., 2006; Kleine-Vehn et al., 2008a). Considering that ES16 blocks PIN2 recycling and facilitates its vacuole trafficking, we proposed that the ES16-induced defect in seedlings could result from interference with PIN2 translocation after gravity induction. Interestingly, although ES16 treatment greatly altered root elongation and gravitropism, it did not block lateral root organogenesis. At 20 µM or less of ES16, lateral root number was slightly increased (Figure 6G), possibly being the secondary effect of main root inhibition. Reduced lateral root growth could be observed only at very high ES16 concentration (60 µM) (Figure 6G). The lack of ES16 inhibition of lateral roots was confirmed by combining ES16 and auxin treatments. Auxin stimulates lateral root initiation by activating division of pericycle cells (Fukaki and Tasaka, 2009). We reasoned that if ES16 actively blocked lateral root initiation then it would prevent auxin-stimulated lateral root development. In cotreatment experiments, ES16 failed to block 1-naphthaleneacetic acid (NAA)-induced lateral root development (Figure 6D, white arrowheads indicate lateral root initiation sites), confirming that ES16 did not target lateral root organogenesis. The lack of effect of ES16 on lateral root growth corresponded with its influence on endomembrane trafficking. PIN1 relocation from the transverse side of roots toward the primordia is essential for lateral root initiation (Benková et al., 2003). Both BFA treatment and mutation of GNOM, which inhibits PIN1 basal recycling, completely abolishes lateral root organogenesis (Geldner et al., 2003). The fact that ES16 strongly interfered with gravitropism but not lateral root growth is coincident with ES16 inducing intracellular bodies of PIN2 but not of PIN1. From these data we concluded that ES16-induced growth phenotypes were correlated with its effects on apical PIN2 recycling and lack of effect on basal PIN1 recycling.
Figure 6.
ES16 Inhibits Root Elongation and Gravitropic Response without Affecting Lateral Root Organogenesis.
(A) Growth phenotype of 5-d-old Col seedlings on different dosages of ES16 or DMSO.
(B) Root growth phenotype after 90° reorientation for 12 h on 0.4% DMSO- or 40 µM ES16-supplemented medium.
(C) Auxin redistribution as labeled by DR5:N7-VENUS in roots in response to gravity stimulus on 0.4% DMSO or 40 µM ES16 medium after reorientation for 90 min.
(D) Lateral root initiation visualized by DR5:N7-VENUS on medium supplemented with 10 µM NAA together with either 0.4% DMSO or 40 µM ES16.
(E) Quantification of root length on medium with different dosages of ES16 as shown in (A). Day 0 was scored when seeds started to germinate, and growth phenotypes were recorded every day up to 7 d; 80 seedlings were scored for each concentration.
(F) Time course of root curvature after gravity stimulation in the presence of either 0.4% DMSO or 40 µM ES16. Angles of curvature were measured every 2 h up to 20 h after reorientation.
(G) Quantification of lateral root number after 9 d on either DMSO medium or different dosages of ES16 medium. Lateral roots longer than 0.5 mm were chosen for monitoring; 80 seedlings were used for quantification for each concentration. Bars represent mean values with sd.
Bars =1 cm in (A), 20 µm in (C), and 50 µm in (D).
Figure 7.
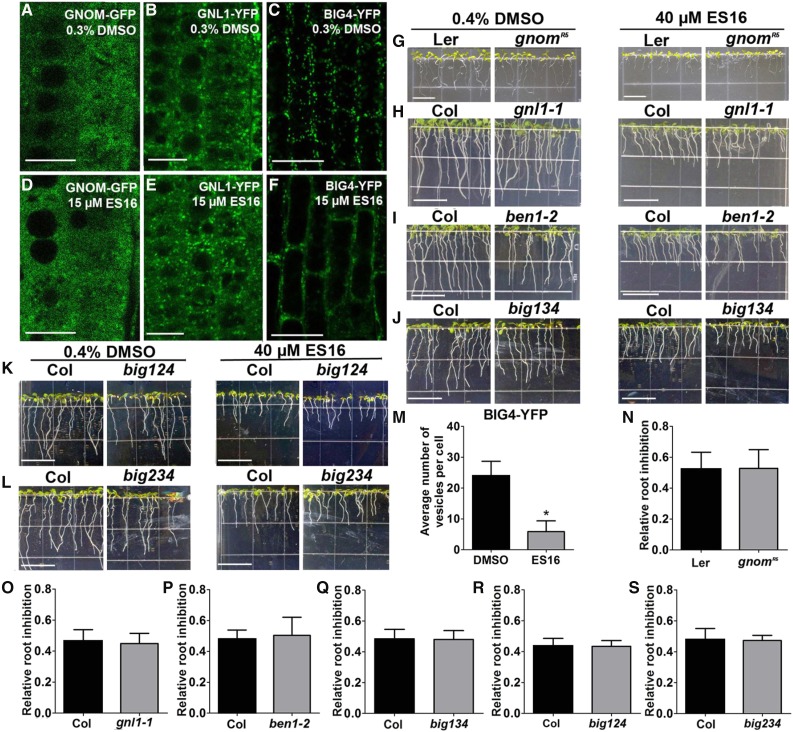
ES16 Does Not Directly Target the Large ARF-GEFs.
(A) to (F) ES16 treatment did not change the subcellular localization of GNOM ([A] and [D]) and GNL1 ([B] and [E]) but increased the cytosolic signal of BIG4 ([C] and [F]).
(G) to (L) Growth phenotypes of gnomR5, gnl1-1, ben1-2, and big134, big124, and big234 triple mutants and the BIG4-YFP overexpression seedlings together with the control on either DMSO- or ES16-supplemented medium.
(M) Quantification of vesicle number in YFP-BIG4 after ES16 treatment. Quantification was based on the number of vesicles in each cell; 200 cells from 10 seedlings were chosen for quantification per treatment.
(N) to (S) Quantification of root inhibition rate as shown from (G) to (L). Root inhibition was calculated as the ratio of mean root length on ES16 medium to that on DMSO medium. No significant difference was observed with root sensitivity between mutants and control seedlings; 100 roots were chosen for quantification. The significance of difference was calculated by a two-tailed Student’s t test, and the asterisks represent P < 0.01. Bars represent sd. Bars = 10 µm in (A) to (F) and 1 cm in (G) to (L).
ES16 Modulates Nonbasal Trafficking in Parallel with the BIG Clade ARF-GEFs
We examined potential mechanisms by which ES16 was selective for nonbasal recycling pathways. The large ARF-GEF GNOM is the main regulator of PIN1 basal recycling. GNOM belongs to the GBF clade of large ARF-GEFs in Arabidopsis, which includes two other ARF-GEFs, GNOM-LIKE1 (GNL1) and GNL2 (Anders and Jürgens, 2008). GNL1 shows ubiquitous expression and is vital for ER-to-Golgi trafficking (Richter et al., 2007; Teh and Moore, 2007), whereas GNL2 performs a pollen-specific function (Jia et al., 2009; Richter et al., 2011). Since ES16 did not block ER-to-Golgi transport or PIN1 basal recycling, we hypothesized that ES16 would not interfere with the GBF clade ARF-GEFs. As anticipated, when we examined the subcellular localization of GNOM and GNL1, they were insensitive to ES16 treatment (Figures 7A, 7B, 7D, and 7E). The GNOM weak allele mutants gnomR5 and gnl1 knockout mutants displayed root sensitivity similar to that of the control (Figures 7G, 7H, 7N, and 7O). This indicated that ES16 activity on apical and lateral polarity acted via a mechanism that was independent from the GBF clade ARF-GEFs.
The other clade of ARF-GEFs, the BIG clade proteins, localize on TGN, regulate late secretory trafficking and are essential for cell plate formation and lateral root initiation (Richter et al., 2014). Since ES16 treatment did not block lateral root formation (Figures 6D and 6G), we speculated that the ES16 sensitive pathway could also bypass BIG clade ARF-GEF-mediated trafficking. Surprisingly, short-term (2 h) treatment with ES16 increased the cytosolic distribution of BIG4-YFP and reduced the amount of BIG4-YFP in membrane organelles (Figures 7C, 7F, and 7M). To unravel the controversy, we also checked the growth sensitivity of seedlings defective in proteins in BIG clade ARF-GEFs. Knockout mutation of BEN1 (also known as BIG5 and MIN7) increased susceptibility to Pseudomonas syringae (Nomura et al., 2006; Tanaka et al., 2009) but did not change the sensitivity to ES16 (Figures 7I and 7P). Similarly, different combination of big triple mutants showed similar response to ES16 as the control seedlings (Figures 7J to 7L and 7Q to 7S), which is in contrast with the hypersensitivity of big3 mutants on BFA medium (Richter et al., 2014). We could not examine the big 1 big2 big3 big4 quadruple mutants as they are defective in male transmission (Richter et al., 2014). Based on this, we reasoned that the altered BIG4-YFP subcellular localization could have been due to ES16 significantly altering the TGN as indicated by the marker SYP61 (Figures 3C, 3H, 3K, and 3L) and the clustered vacuolated TGN shown in the TEM result (Figure 4K; Supplemental Figure 3K). This conclusion is also supported by the fact that seedlings after ES16 treatment still showed relatively normal lateral root growth (Figures 6D and 6G), whereas functionally compromised big mutants lacked any lateral root initiation (Richter et al., 2014). Considering the facts that all the ARF-GEF mutants we tested here displayed similar sensitivity on ES16 medium (Figures 7G to 7L and 7N to 7S) and that the ES16 bodies are different from BFA aggregates (Figures 4A to 4D), we concluded that ES16 does not directly target the large ARF-GEFs.
Since a previous study showed that the big mutants are defective in secretion and cell plate formation without interference with the basal polarity of PIN1 (Richter et al., 2014), we further examined whether the BIG proteins could also be involved in nonbasal trafficking, like ES16. For this purpose, we checked the sensitivity of basal PIN1, apical PIN2, and nonpolarized PEN1 after BFA treatment in different mutants. BIG3 mutation did not alter the PIN1 basal localization in engineered BFA-resistant GNOM (GN-ML-MYC) (Supplemental Figures 4A to 4D), as reported before (Richter et al., 2014). By contrast, apically localized PIN2 and nonpolarized PEN1 formed large aggregates after BFA treatment in the GN-ML-MYC background with or without BIG3 mutation (Supplemental Figures 4E to 4L), suggesting that basal trafficking is the sole pathway that is not affected in big mutants, similar to the effects of ES16. Since ES16 does not directly target the BIG proteins, ES16 may function in concert with the BIG clade ARF-GEFs for nonbasal trafficking. Considering that Golgi-localized GNOM and GNL1 (Teh and Moore, 2007; Naramoto et al., 2014) are insensitive to ES16 and that the TGN defects caused by BIG mutation or after ES16 treatment does not perturb PIN1 basal polarity, it is plausible to speculate that the basal recycling may either bypass the TGN but transport from Golgi to the PM or use different TGN domains insensitive to ES16 or BIG protein mutation.
A RabA GTPase-Dependent Pathway Is Responsible for the Nonbasal PM Recycling
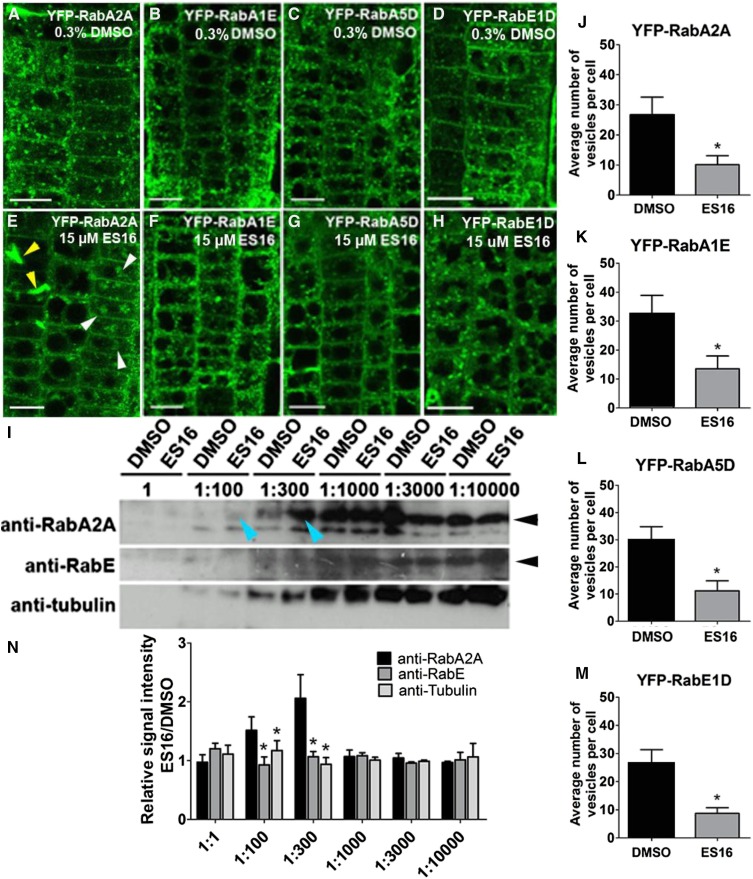
Since ES16 does not directly target the large ARF-GEFs, we hypothesized that it could affect a parallel secretory/recycling route used for apically and laterally localized proteins and general secretion. In mammalian cells, the Rab11 small G proteins are master regulators of post-Golgi transport and are essential for the establishment of apical polarity in epithelial cells (Welz et al., 2014). In the model plant Arabidopsis, a group of RabA GTPases are considered homologs of Rab11, and they can be further divided into six clades from RabA1 to RabA6 (Rutherford and Moore, 2002). Some members of the RabA2 and RabA3 clades are known to be involved in secretion and cell plate expansion (Chow et al., 2008). In mammalian cells, Rab11 and Rab8 work together in the GTPase activation cascade during the secretory process (Drakakaki et al., 2011; Doyle et al., 2015), and RabE GTPases are Arabidopsis homologs of Rab8. Therefore, we examined whether ES16 inhibition of nonbasal recycling may act through RabA and RabE GTPases by examining their subcellular localization. In control seedlings, the RabA and RabE GTPases, including RabA2A, RabA1E, RabA5D, and RabE1D, all displayed vesicular and PM localization. ES16 treatment significantly increased the cytosolic distribution and reduced the number of membrane-bound vesicles (Figures 8A to 8H and 8J to 8M). RabA2A showed the strongest alteration among the four GTPases tested. RabA2A increased in cytosolic signal, was found in large bodies, and displayed aberrant cell wall stubs. Aberrant cell walls were not observed for the other two RabA GTPases (cf. Figure 8E with Figures 8F and 8G, white arrowheads indicate bodies and yellow arrowheads indicate aberrant cell wall stunts). We also observed a significant reduction of vesicular membrane localization in RabE1D (Figures 8D, 8H, and 8M). However, other Rab GTPases tested, including RabC1, RabD2A, and RabD2B, did not display significant alteration of subcellular localization after ES16 treatment (Supplemental Figure 5), indicating that ES16 may alter trafficking directly or indirectly through RabA and RabE proteins.
Figure 8.
ES16 Functions through the Regulation of RabA GTPases.
(A) to (D) Subcellular localization of RabA2A (A), RabA1E (B), RabA5D (C), and RabE1D (D) after treatment with 0.3% DMSO.
(E) to (H) Subcellular localization of RabA2A (E), RabA1E (F), RabA5D (G), and RabE1D (H) after treatment with 15 µM ES16. White arrowheads in (E) represent large aggregates and the yellow arrowheads represent cell wall stubs.
(I) Pretreatment with ES16 specifically protected RabA2A but not RabE or TUBULIN from degradation in a DARTS assay. Blue arrowheads indicate 1:100 and 1:300 dilution of pronase. Black arrowheads indicate the correct mass of the corresponding proteins.
(J) to (M) Quantification of vesicle number after ES16 treatment as shown from (A) to (H). Quantification was based on the number of vesicles in each cell; 200 cells from 10 seedlings were chosen for quantification per treatment per genotype.
(N) Quantification of bands intensity as shown in (I). The image was first reversed by Image J, and the net signal intensity of bands from ES16-treated sample was normalized against the ones from DMSO-treated sample with the same dilution ratio of pronase. Three representative scanned immunoblots were used for quantification. A two-tailed Student’s t test was used for significance calculation. For (J) to (M), asterisks represent P < 0.01; for (N), asterisks represent P < 0.05. Images are representative of three repeats. Bars stand for sd. Bars = 10 µm.
To test the hypothesis, we performed the drug affinity responsive target stability (DARTS) assay (Lomenick et al., 2011). DARTS is a straightforward approach to evaluate compound-protein interaction based on the fact that upon compound binding, the target protein is presumably stabilized and protected from proteinase digestion (Lomenick et al., 2009, 2011). We examined RabA2A and RabE proteins for the DARTS assay because these two proteins showed strong alteration in vivo after ES16 treatment and there were available antibodies. Pretreatment with ES16 protected RabA2A from degradation at 100 and 300 times dilution of the protease mixture Pronase (Figure 8I, yellow arrowheads). The protection was specific to RabA2A; antibodies against TUBULIN or RabE in the same protein extraction resulted in no significant difference compared with the DMSO control (Figures 8I and 8N). This result indicated that RabA2A is stabilized specifically in the presence of ES16, suggesting that ES16 functions through RabA2A-dependent protein complex.
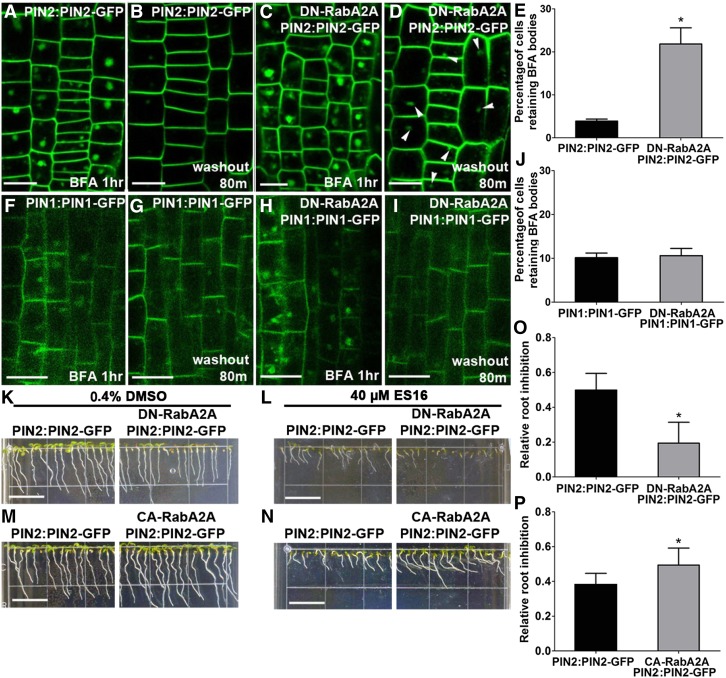
We reasoned that if ES16 defined a RabA GTPase-dependent pathway for recycling, then a dominant-negative mutation of RabA2A should phenocopy effect of ES16 on endomembrane trafficking. Indeed, a dexamethasone (DEX)-inducible dominant-negative mutation of RabA2A (S26N) induced ES16-like bodies of PIN2 but not PIN1 (Supplemental Figures 6A to 6D). Stable transgenic lines overexpression of DN-RabA2A also increased PIN2 but not PIN1 internalization, which colocalized with the DN-RabA2A vesicles (Supplemental Figures 6E to 6J), similar to ES16. In addition, BFA washout experiments using the same dominant-negative line (DN-RabA2A) inhibited recycling of only PIN2 but not PIN1 (Figures 9A to 9J), indicating that PIN1 recycling did not require RabA2A regulation. We also analyzed the growth phenotype of lines overexpressing constitutively active RabA2A (CA-RabA2A) or DN-RabA2A on ES16 medium. The genetic results were consistent with the endomembrane phenotype in that the DN-RabA2A lines were hypersensitive to ES16, whereas the CA-RabA2A lines showed resistance (Figures 9K to 9P). To exclude the possibility that the high amount accumulation of dominant-negative RabA2A may result in unexpected secondary effects, we further confirmed the data using the DEX-inducible DN-RabA2A lines. Our results showed that low dosage of induction (5 µM) did not produce discernable phenotypic defects but resulted in hypersensitivity on ES16 medium (Supplemental Figures 7C, 7D, 7I, and 7J), while the seedlings without induction did not display a significant difference (Supplemental Figures 7B and 7J). Increased concentrations of DEX slightly inhibited main root growth of both control and DN-RabA2A seedlings on DMSO medium (Supplemental Figures 7A, 7C, 7E, 7G, and 7I) but led to significantly enhanced sensitivity of DN-RabA4A on ES16 (Supplemental Figures 7D, 7F, 7H, and 7J), suggesting synergistic effects of ES16 with RabA2A mutation and confirming that the hypersensitivity of DN-RabA2A was not an artifact. Based on all the evidence, we concluded that RabA GTPases regulate recycling of apically polarized PIN2 in a manner distinct from basal PIN1. The cell biology and genetic evidence together indicated that ES16 inhibits this pathway through an effect on RabA GTPases and that the RabA and BIG GTPases may work in parallel to modulate the nonbasal trafficking.
Figure 9.
Overexpression of DN-RabA2A Phenocopies the Effects of ES16.
(A) and (C) BFA treatment (40 µM) induced large aggregates in PIN2:PIN2-GFP (A) and DN-RabA2A PIN2:PIN2-GFP (C) lines.
(B) and (D) BFA washout was delayed in DN-RabA2A PIN2:PIN2-GFP (D) compared with the control (B) seedlings.
(E) Quantification of BFA washout efficiency as shown from (A) to (D).
(F) and (H) Large aggregates induced by 40 µM BFA treatment in PIN1:PIN1-GFP (F) and DN-RabA2A PIN1:PIN1-GFP (H).
(G) and (I) BFA washout in DN-RabA2A PIN1:PIN1-GFP (G) and the control (I) line.
(F) Quantification of BFA washout efficiency as shown from (F) to (I). Quantification was calculated based on the ratio of cells retaining BFA bodies to total cells in the same seedling; 15 seedlings were quantified per treatment per repeat.
(K) and (L) DN-RabA2 PIN2:PIN2-GFP seedlings were hypersensitive on ES16 medium compared with the control plants.
(M) and (N) CA-RabA2A PIN2:PIN2-GFP seedlings were more resistant to ES16 treatment compared with the control plants.
(O) and (P) Quantification of root inhibition rate as shown from (K) to (N). Root inhibition was calculated as the ratio of mean root length on ES16 medium to that on DMSO medium. For each genotype per treatment, 100 roots are chosen for quantification. Images are representative of three repeats. Bars represent sd. A two-tailed Student’s t test was used for significance calculation. Asterisks represent P < 0.01. Bars = 10 µm in (A) to (D) and (F) to (I) and 1 cm in (K) to (N).
DISCUSSION
We searched for apical trafficking specific inhibitors based on evidence from previous publications reporting that apical recycling and the maintenance of apical polarity depend on distinct sorting routes from the basal recycling pathway (Kleine-Vehn et al., 2006, 2008b). We identified one compound, ES16, which selectively interfered with apically localized PM proteins without disturbing the targeting of basal PM proteins. ES16 also induced aggregates of nonpolarly localized PM proteins PIP2a and PEN1 as well as laterally polarized BOR1 and NIP5;1. Furthermore, ES16 was a strong inhibitor of biosynthetic secretion, as indicated by the block of secretion of secGFP and Est>>PIN1-RFP. Our results indicate that among the proteins examined, only the basal PIN1 recycling was the exception. Thus, ES16 defined apical, lateral, and general secretory pathways, but not basal polarity. Why is basal polar recycling unique from other sorting routes? The ARF-GEF GNOM is so far the only known essential upstream regulator for basal PIN1 recycling, as either prolonged BFA treatment or gnomR5 mutation leads to a complete basal-to-apical shift of PIN1 polarity (Kleine-Vehn et al., 2008b; Richter et al., 2010). GNOM belongs to the GBF clade in the large ARF-GEF family, which is conserved throughout eukaryotes (Anders and Jürgens, 2008). However, the unique feature of basal recycling by GNOM is plant specific; even its closest homolog GNL1 could not complement the PIN1 polarity defect in the gnom loss-of-function mutant (Richter et al., 2007), suggesting that the function of GNOM in polar recycling and developmental regulation has evolved de novo during plant evolution. To validate the hypothesis, it would be worthwhile to analyze how the GBF clade ARF-GEFs evolved during plant evolution. It would also be interesting to study the evolutionary correlation between GNOM and the PIN proteins as these auxin transporters are also plant specific (Bennett, 2015). Such parallel evolutionary analysis could provide insight into how and when the GNOM protein evolved to specialize in basal recycling, and such knowledge could contribute to an understanding of mechanisms.
ES16 does not inhibit ER-to-Golgi transport but significantly interferes with post-Golgi trafficking. Considering the fact that ES16 inhibits apical, nonpolar, and lateral polar recycling as well as biosynthetic secretion without perturbation of basal recycling and the fact that big mutants display similar TGN defects without affecting basal polarity, it is plausible that basal recycling either utilizes distinct TGN domains or occurs via a TGN-independent pathway. Since the Golgi-localized ARF-GEF GNOM is in a central position for this scenario, it would be interesting to investigate the downstream targets of GNOM and whether their translocation from Golgi to the PM upon activation requires the TGN. Since ES16 does not directly regulate BIG proteins, the potential targets RabA GTPases and BIG clade ARF-GEFs may work in concert with each other to modulate the post-Golgi nonbasal trafficking.
Our results suggest that ES16 perturbed endomembrane trafficking in a different way than BFA. This conclusion is supported by the different behavior of FM dye in response to the two compounds and confirmed by the different endomembrane organelle reorganization pattern observed in the TEM results. While BFA induces large fragmentation of TGN surrounded by Golgi apparatus (Richter et al., 2007), ES16 promoted TGN budding and detaching from Golgi and clustered both TGN and PVCs into agglomerations. As BFA induces aggregates of plasma membrane proteins regardless of their polarities, ES16 could be used as a valuable tool to dissect different polar trafficking pathways. Recently, another compound, endosidin 8 (ES8), was reported to define an early secretory pathway for PIN1 basal polarity establishment (Doyle et al., 2015). Thus, there are specific compounds now available to study basal and nonbasal pathways separately, which would be difficult through traditional mutagenesis analysis. Our data also suggest that the BIG clade ARF-GEFs are involved in nonbasal trafficking, similar to ES16. Thus, the integration of mutant analysis and a chemical genomic approach has great potential in dissecting different endomembrane trafficking pathways for the establishment of polarities.
ES16 mainly interferes with post-Golgi trafficking as evidenced by the mislocalization of SYP61, RabA2A, and BIG4 proteins and the disorganization of TGN and PVC in the TEM results. Although the subcellular localization of the other TGN marker VHA-a1 largely overlapped with SYP61, it showed only minor alteration. Even for BIG4 protein, although ES16 treatment increased the cytosolic signal, the compound only partially disrupted the protein function. BIG4 still retained its role in initiation of lateral roots in the presence of ES16, and the big triple mutants showed no change in terms of sensitivity to ES16. The different responses of TGN marker proteins to ES16 treatment could be explained by functional diversification of TGN subdomains. This hypothesis is supported by previous reports (Gu and Innes, 2012). KEEP ON GOING, a protein located at the TGN, is essential for the secretion of soluble defense proteins to the apoplast but not for the delivery of AUX1, PIN1, and BRI1 to the PM (Gu and Innes, 2012). Loss of function of ECHIDIA (ECH), a TGN-localized protein of unknown function, affects the secretion of polysaccharides and some specific proteins such as AUX1 but not that of other PM proteins (Boutté et al., 2013). Interestingly, although ECH and VHA-a1 colocalize, the interruption of VHA-a1 function with the V-ATPase inhibitor concanamycin A does not affect AUX1 secretion (Boutté et al., 2013; McFarlane et al., 2013). Moreover, the induced suppression of VHA-a1 during seed development has no effect on mucilage secretion, which is impaired in the ech mutant (McFarlane et al., 2013). This finding also suggests that VHA-a1 is not involved in secretion, which could explain why ES16, as a secretion and recycling inhibitor, has little effect on VHA-a1. For BIG4 protein, although localization largely overlaps with VHAa1, functionally compromised big mutants have no effect on SYP61 in the same TGN domain (Richter et al., 2014). By contrast, ES16 showed an effect on SYP61 rather than VHA-a1. Thus, the different sensitivities of TGN proteins to ES16 could reflect subdomain specification within the TGN and different trafficking pathways through the TGN could use different domains or proteins to modulate distinct sorting directions. For this scenario, ES16 could be used as a useful tool to dissect different TGN sorting pathways.
Our data indicate that ES16 functions through the regulation of RabA GTPases, mainly through the direct or indirect perturbation of RabA2A, although we could not exclude the possibility that other RabA GTPases may also be involved in the same process. RabA2 and RabA3 clade GTPases are known to localize at the TGN and to be essential for late secretory processes (Chow et al., 2008). RabA1d is involved in cell plate formation and oscillatory root hair formation (Berson et al., 2014). RabA4b interacts with PLANT U-BOX13 and PI4Kβ1/β2 and plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis (Antignani et al., 2015). Our data indicate that RabA2A is a factor that can distinguish between basal and apical recycling in endomembrane trafficking. Since ES16 redirects the post-Golgi secretory pathway (including secretion and recycling) to vacuole transport, it also would be interesting to explore whether the potential target RabA2A is in the center position of TGN sorting to either PM or vacuole transport and whether the basal recycling bypasses the process. Although beyond the scope of this report, to further characterize the molecular mechanism of RabA2A regulated membrane-sorting pathways, it will be necessary to identify the downstream effectors and test whether they can also differentiate between the basal and nonbasal PM trafficking pathways.
In summary, we identified ES16 as an inhibitor that separates basal recycling from other trafficking pathways. Our data represent an important starting point that directs questions about how basal recycling and the de novo function of GNOM were adapted during plant evolution. Our findings also raise questions about how the TGN integrates different upstream signals and uses functionally diversified TGN domains to direct sorting processes. Furthermore, our work highlights the fact that chemical biology offers the potential to dissect complicated plant endomembrane trafficking routes and is meeting with success in this regard (Robert et al., 2008; Hicks and Raikhel, 2009; Doyle et al., 2015; Zhang et al., 2016).
METHODS
Plant Material and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) marker lines PIN1:PIN1-GFP (Friml et al., 2002), PIN2:PIN2-GFP (Xu and Scheres, 2005), PIN3:PIN3-GFP (Zádníková et al., 2010), PIN7:PIN7-GFP (Blilou et al., 2005), PIN5:PIN5-GFP (Mravec et al., 2009), AUX1:AUX1-YFP (Swarup et al., 2004), PIN2:PIN1-HA (Wisniewska et al., 2006), PIN2:PIN2-HA (Wisniewska et al., 2006), PIN2:PIN1-GFP3 (Wisniewska et al., 2006), 35S:PIP2a-GFP (Cutler et al., 2000), 35S:GFP-PEN1 (Kwon et al., 2008), SCR:BOR1-mCitrine (Alassimone et al., 2010), SCR:mCitrine-NIP5;1 (Alassimone et al., 2010), SCR:PIN2-GFP (Alassimone et al., 2010), 35S:GFP-HDEL (Ridge et al., 1999), 35S: NAG1-GFP (Grebe et al., 2003), 35S:secGFP (Zheng et al., 2004), Est>>PIN1-RFP (Richter et al., 2014), SYP61:SYP61-CFP (Robert et al., 2008), VHA-a1-GFP (Dettmer et al., 2006), ABD2-GFP, GFP-MAP4 (Marc et al., 1998), DR5:N7-VENUS (Heisler et al., 2005), YFP-RabA2A (Chow et al., 2008), YFP-RabA1E (Geldner et al., 2009), YFP-RabA5D (Geldner et al., 2009), YFP-RabC1 (Geldner et al., 2009), YFP-RabD2A (Geldner et al., 2009), YFP-RabD2B (Geldner et al., 2009), YFP-RabE1d (Geldner et al., 2009), RFP-ARA7(Ueda et al., 2004), DEX-induced DN-RabA2A (Chow et al., 2008), BIG4-YFP (Richter et al., 2014), GNL1-YFP (Teh and Moore, 2007), and GNOM-GFP (Geldner et al., 2003) and mutant lines gnom (Geldner et al., 2004), gnl1-1 (Richter et al., 2007), ben1-2 (Tanaka et al., 2009), big3, big134, big 124, and big234 triple mutants (Richter et al., 2014), GN-ML-MYC, and big3 GN-ML-MYC mutants (Richter et al., 2014) were previously described. The colocalization between PIN2 and ARA7 was analyzed in the F2 line of a cross between PIN2:PIN2-GFP and RFP-ARA7. The DN-RabA2A was introduced into PIN1:PIN1-GFP and PIN2:PIN2-GFP backgrounds by crossing DEX-inducible DN-RabA2A with the two marker lines (Supplemental Figure 7). ES16 treatment was performed on F2 homozygous seedlings. Stable transgenic lines overexpressing DN- or CA-RabA2A were generated by Agrobacterium tumefaciens (strain GV3101) transformation with constitutive overexpression constructs (Figure 8). F3 homozygous seedlings were selected for treatment.
The seeds were stratified for 2 d in the dark at 4°C, and the seedlings were germinated and grown vertically in square plates containing 0.5× Murashige and Skoog (MS) medium and 0.8% phytoagar with 1% sucrose (pH 5.6) at 22°C in long-day photoperiod conditions (16 h light/8 h dark; Sylvania FO32/T35/ECO 3500K fluorescent tube light bulb). Columbia (Col-0) ecotype was used for immunostaining, FM4-64 uptake, and as wild-type control in seedling growth experiments, except for growth of gnom weak allele mutants, for which Landsberg erecta was used.
Molecular Cloning
The full-length cDNA sequence encoding RabA2A was amplified by PCR. Primers used for amplification were RabA2A-F, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGCGAGAAGACCGGACGAA-3′, and RabA2A-R, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTCAAGACGATGAGCAACAAGGCTTC-3′. The purified PCR product was first cloned into pDNOR201 by BP reaction and then integrated to destination vector pB7WGR2,0 (Karimi et al., 2002) after LR reaction. Point mutations to generate the DN- and CA-RabA2A were performed as reported before (Chow et al., 2008). The amino acid substitutions Ser-26 to Asn (S26N) for the DN form, and the Gln-71 to Leu (Q71L) for the CA form were created by overlapping PCR. Primers used to generate CA mutation were RabA2A-CA-F, 5′-ATGGGACACGGCTGGGTTAGAACGATACAGAGCCA-3′, and RabA2A-CA-R, 5′-TGGCTCTGTATCGTTCTAACCCAGCCGTGTCCCAT-3′. Primers used to generate DN mutation were RabA2A-DN-F, 5′-CTCCGGTGTCGGCAAGAATAATCTCCTCTCTAGAT-3′, and RabA2A-DN-R, 5′-ATCTAGAGAGGAGATTATTCTTGCCGACACCGGAG-3′.
Chemical Treatments
Stock solutions of 40 mM BFA (Sigma-Aldrich), 5 mM and 10 mM ES16 (Chembridge ID 5470964), 10 mM NAA (Sigma-Aldrich), 10 mM estradiol (Sigma-Aldrich), and 10 mM DEX (Sigma-Aldrich) were dissolved in DMSO. FM4-64 (5 mM; Invitrogen) was prepared in deionized water. ES16 was aliquoted for one-time use to eliminate freeze-thaw cycles. For short-term treatment, seedlings in different backgrounds were grown on 0.5× MS solid medium for 5 to 6 d and then five to eight seedlings per well were transferred to 24-well plates containing 0.5× MS liquid medium and chemicals (0.5× MS and 1% sucrose, pH 5.6). All the stock solutions were diluted in liquid 0.5× MS medium for treatment at the indicated concentrations with equal volumes of solvent added as the control treatment. All the short-term treatments with ES16 used dilutions from 5 mM stock solution and were performed within 3 h. Longer treatments were performed on solid medium since long-term treatment in liquid medium could result in ES16 precipitates. For all treatments with chemicals on solid medium, chemicals were added to 0.5× MS solid medium at 55°C before pouring the plates. ES16 and DEX were added from the 10 mM stock solutions in the 0.5× MS solid medium.
Confocal Microscopy and Image Quantification
To test the root length inhibition, different doses of ES16 or the same ratio of DMSO were added as indicated. To compare the root inhibition between different mutants and the control seedlings, 40 µM ES16 or the same volume of DMSO was added in the solid medium and plants were grown for 7 d. Seedling phenotypes were recorded using a flatbed scanner (Epson model 2450) and root length was measured by Image J software (imagej.nih.gov/ij/). For the quantification of the lateral root density, Col wild-type seedlings were germinated and grown on 0.5× MS solid medium containing different doses of ES16 as indicated or DMSO for 9 d and documented by the Epson scanner. Lateral roots with the length more than 0.5 mm were recorded. For testing the lateral root initiation rate on the DR5:N7-VENUS line, seedlings were first grown on 0.5× MS solid medium for 6 d and then transferred to solid medium containing 10 µM NAA and 40 µM ES16 or the equal volume of DMSO for 40 h before being imaged by confocal microscopy (Leica SP5). For dark treatment of PIN2:PIN2-GFP, seedlings were grown on 0.5× MS solid medium for 5 d and then transferred to liquid medium in the presence of 15 µM ES16 or DMSO for 3 h before imaging by confocal microscopy (Leica SP5). The vacuole translocation efficiency was quantified by scoring the ratio between the number of cells containing the vacuole signal and the total number of cells from the image in each seedling. For the time-lapse colocalization analysis between ARA7 and PIN2, 5-d-old seedlings were transferred to liquid MS containing 0.3% DMSO or 15 μM ES16 and incubated in dark conditions for 1, 2, 2.5, or 3 h. Quantitative colocalization and statistical analysis were performed using the Imaris colocalization software (version 8.1; Bitplane). PIN2 and ARA7 colocalization was quantified by Pearson correlation coefficient parameters performed in a selected region of interest of the images. For the BFA washout experiment, seedlings were pretreated with 40 µM BFA for 1 h, briefly washed in 0.5× MS liquid medium, and then transferred to 0.5× MS liquid medium containing 8 µM ES16 or DMSO for 80 min. An equal amount of ES16 was used as the control to distinguish between the leftover BFA bodies and the newly produced ES16 bodies. For quantification of BFA washout efficiency, at least 500 epidermal or stele cells or 300 endodermal cells were scored from at least 15 seedlings per treatment per replicate. The washout efficiency was quantified by scoring the ratio between the number of cells containing the leftover BFA bodies and the total number of cells from the image in each seedling. To analyze the gravitropic response at the starting point, DR5:N7-VENUS seedlings were vertically grown on 0.5× MS solid medium for 6 d then transferred to medium containing 40 µM ES16 or an equal volume of DMSO and rotated 90°. After 90-min reorientation, the symmetry of DR5 signal along the two sides of root was recorded by confocal microscopy (Leica SP5). The image was focused on the section where the two sides of VENUS signal could be observed. To measure the root curvature in response to gravity stimulus, Col-0 wild-type seedlings were first germinated and grown on 0.5× MS solid medium for 6 d, then transferred to medium containing 40 µM ES16 or equal volume of DMSO and rotated 90°. Root curvature was scored by imaging on a scanner (Epson) every 2 h for 20 h and then quantified by Image J. For the FM4-64 uptake experiment, 5-d-old Col seedlings were first pretreated with either 15 µM ES16 or equal amount of DMSO for 1 h, briefly washed in 0.5× MS liquid medium twice, and then transferred to liquid medium containing 5 µM FM4-64 in the presence of ES16 or DMSO for 15 m at room temperature before imaging by confocal microscopy. To visualize the FM4-64 stained cells, a laser line of 488 nm was used for excitation, and an emission wavelength of 600 to 700 nm was used for collecting the signal. For testing the biosynthetic secretion in the Est>>PIN1-RFP line, seedlings were first germinated and vertically grown on 0.5× MS solid medium for 6 d and then transferred to solid medium containing 20 µM estradiol together with either 40 µM ES16 or equal volume of DMSO for 7 h before recorded by confocal microscopy (Leica SP5). To quantify the average number of aggregates/bodies or vesicles per cell, 200 cells were selected per treatment per genotype. The aggregates or vesicles within the cell were manually selected and measured by the “Analyze Particles” function of Image J. Particles with a maximum diameter of fewer than two pixels were discarded during statistical analysis. After BFA washout, the number of remaining BFA bodies showed large variation between cells; accordingly the “ratio of cells retaining BFA bodies” was used to quantify BFA washout efficiency instead of the “number of aggregates,” which had been used to quantify ES16-induced agglomerations as described above.
Immunocytochemistry
Whole-mount immunostaining in Arabidopsis roots was performed as described previously (Fischer et al., 2006). To visualize the BFA bodies, mutants in different background as shown in Supplemental Figure 4 were first treated with 50 µM BFA for 1 h before fixation. Dilutions of primary antibodies applied were as follows: goat anti-PIN1, 1:50 (Santa Cruz Biotech, cat. no. sc-27163), rabbit anti-PIN2, 1:750 (Müller et al., 1998), rabbit anti-PEN1, 1:200 (Collins et al., 2003), mouse anti-HA, 1:250 (Sigma-Aldrich; H9658), and mouse anti-GFP, 1:250 (Clontech; J1-8, cat. no. 632381). Dilutions of secondary antibodies applied were as follows: goat anti-rabbit CY3-coupled, 1:300 (Kirkegaard and Perry Laboratories; cat. no. 078-15-061), goat anti-mouse TRITC-coupled, 1:300 (Jackson ImmunoResearch; cat. no. 115-025-075), and rabbit anti-goat CY3-coupled, 1:250 (Kirkegaard and Perry Laboratories; cat. no. 072-01-13-06).
TEM Analysis
Plant samples for TEM study are prepared using the approach of high-pressure freezing and frozen substitution. Briefly, root tips of Arabidopsis seedlings (days 5 to 7 after germination) were frozen with a high-pressure freezer (Leica EM PACT2), followed by dehydration and contrasting in acetone containing 0.2% uranylacetate for 24 h at −85°C, infiltration sequentially in 33, 66 and 100% HM20 resin diluted with ethanol, and embedding in 100% HM20 at −35°C. Finally, the HM20 polymerization was completed under UV illumination at −35°C. The sample blocks were cut into ultrathin sections, followed by poststaining with uranylacetate and lead citrate, and then images were collected with a transmission electron microscope (Hitachi HT7700) operating at 80 kV.
DARTS Assay
DARTS assay was performed as described (Lomenick et al., 2011) with minor changes. Briefly, protein was obtained from 8-d-old Col-0 seedlings using extraction buffer (1× PBS, 0.05% Triton X-100, 2 mM DTT, and 1× protease inhibitor mixture). The supernatant from 16,000g was used as the input. Protein extract (300 µL) was first pretreated with either 100 µM ES16 or equal volume of DMSO for 1 h at room temperature with gentle shaking, and then aliquoted into six samples each containing 50 µL. Different dilutions of pronase (Sigma-Aldrich) were added as indicated and incubated at room temperature for 30 min before addition of sample buffer and boiling to stop the reaction. Samples were loaded onto SDS-PAGE gels and immunoblotting was performed. Membranes were probed with either anti-RabA2A (1:2000) (Chow et al., 2008), anti-TUBULIN (1:10,000) (Sigma-Aldrich; cat. no. T6074) or anti-RabE (1:500) (Speth et al., 2009) antibodies. The secondary antibodies used were goat anti-rabbit HRP-coupled (Kirkegaard and Perry Laboratories; cat. no. 074-1506), goat anti-mouse HRP-coupled (Kirkegaard and Perry Laboratories; cat. no. 074-1806), and goat anti-chicken HRP-coupled (Kirkegaard and Perry Laboratories; cat. no. 14-24-06).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: RabA2A (At1g09630), RabA1E (At4g18430), RabA5D (At2g31680), RabE1D (At5g03520), RabC1 (At1g43890), RabD2A (At1g02130), RabD2B (At5g47200), PIN1 (At1g73590), PIN2 (At5g57090), PIN3 (At1g70940), PIN5 (At5g16530), PIN7 (At1g23080), AUX1 (At2g38120), GNOM (At1g13980), GNL1 (At5g39500), GNL2 (At5g19610), BIG1 (At4g38200), BIG2 (At3g60860), BIG3 (At1g01960), BIG4 (At4g35380), KN (At1g08560), VHA-a1 (At2g28520), SYP61 (At1g28490), NAG1 (At4g38240), BOR1 (At2g47160), NIP5;1 (At4g10380), PEN1 (At3g11820), PIP2a (At3g53420), and ARA7(At4g19640).
Supplemental Data
Supplemental Figure 1. ES16 does not interfere with PIN3, PIN7, and PIN5 but induces aggregates of AUX1.
Supplemental Figure 2. ES16 does not affect VHAa1 or cytoskeleton organization.
Supplemental Figure 3. ES16 promotes vacuole transport via the transition through PVC.
Supplemental Figure 4. BIG clade ARF-GEFs regulate nonbasal trafficking.
Supplemental Figure 5. ES16 is not a general Rab GTPase inhibitor.
Supplemental Figure 6. Overexpression of DN-RabA2A induces aggregates of PIN2 but not PIN1.
Supplemental Figure 7. ES16 shows synergistic effects with RabA2A mutation.
Supplementary Material
Acknowledgments
We thank Ian Moore, Shengyang He, and Paul Schutze-Lefert for kindly sharing the antibodies of RabA2A, RabE, and PEN1, respectively. We also thank the many researchers who kindly provided published Arabidopsis lines. Confocal laser scanning microscopy was performed at the Microscopy Core at the Center for Plant Cell Biology and Institute for Integrative Genome Biology at UC Riverside. We also thank Jocelyn Brimo for administrative support and Ivan Sotelo, Uyen Nguyen, and Maria Diaz for their technical support. We thank Jikui Song and Zhiming Zhang from the Biochemistry Department in UC Riverside for technique support. We thank Angela Wandinger-Ness and Jiri Friml for helpful discussions and critical reading of the manuscript. This work was supported by U.S. Department of Energy Grant DE-FG02-02ER15295 (to N.V.R. and G.R.H.). The TEM work was supported by Shenzhen Innovation of Science and Technology Commission JCYJ20140417105816348 and Natural Science Foundation of Guangdong Province 2015A030313856 (to J.W.).
AUTHOR CONTRIBUTIONS
R.L., G.R.H., and N.V.R. designed research. R.L., C.R.-F., J.W., W.v.d.V., and T.G. performed research. R.L., C.R.-F., J.W., and T.G. analyzed data. R.L. and G.R.H. wrote the article.
Glossary
- ER
endoplasmic reticulum
- PM
plasma membrane
- TGN
trans-Golgi network
- BFA
Brefeldin A
- PVC
prevacuolar compartment
- TEM
transmission electron microscopy
- NAA
1-naphthaleneacetic acid
- DARTS
drug affinity responsive target stability
- DEX
dexamethasone
- MS
Murashige and Skoog
References
- Abas L., Benjamins R., Malenica N., Paciorek T., Wiśniewska J., Moulinier-Anzola J.C., Sieberer T., Friml J., Luschnig C. (2006). Intracellular trafficking and proteolysis of the Arabidopsis auxin-efflux facilitator PIN2 are involved in root gravitropism. Nat. Cell Biol. 8: 249–256. Erratum. Nat. Cell Biol. 8: 424. [DOI] [PubMed] [Google Scholar]
- Alassimone J., Naseer S., Geldner N. (2010). A developmental framework for endodermal differentiation and polarity. Proc. Natl. Acad. Sci. USA 107: 5214–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders N., Jürgens G. (2008). Large ARF guanine nucleotide exchange factors in membrane trafficking. Cell. Mol. Life Sci. 65: 3433–3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antignani V., Klocko A.L., Bak G., Chandrasekaran S.D., Dunivin T., Nielsen E. (2015). Recruitment of PLANT U-BOX13 and the PI4Kβ1/β2 phosphatidylinositol-4 kinases by the small GTPase RabA4B plays important roles during salicylic acid-mediated plant defense signaling in Arabidopsis. Plant Cell 27: 243–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett T. (2015). PIN proteins and the evolution of plant development. Trends Plant Sci. 20: 498–507. [DOI] [PubMed] [Google Scholar]
- Baskin T.I., et al. (2010). Shootward and rootward: peak terminology for plant polarity. Trends Plant Sci. 15: 593–594. [DOI] [PubMed] [Google Scholar]
- Berson T., von Wangenheim D., Takáč T., Šamajová O., Rosero A., Ovečka M., Komis G., Stelzer E.H., Šamaj J. (2014). Trans-Golgi network localized small GTPase RabA1d is involved in cell plate formation and oscillatory root hair growth. BMC Plant Biol. 14: 252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bolte S., Talbot C., Boutte Y., Catrice O., Read N.D., Satiat-Jeunemaitre B. (2004). FM-dyes as experimental probes for dissecting vesicle trafficking in living plant cells. J. Microsc. 214: 159–173. [DOI] [PubMed] [Google Scholar]
- Boutté Y., Jonsson K., McFarlane H.E., Johnson E., Gendre D., Swarup R., Friml J., Samuels L., Robert S., Bhalerao R.P. (2013). ECHIDNA-mediated post-Golgi trafficking of auxin carriers for differential cell elongation. Proc. Natl. Acad. Sci. USA 110: 16259–16264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow C.M., Neto H., Foucart C., Moore I. (2008). Rab-A2 and Rab-A3 GTPases define a trans-golgi endosomal membrane domain in Arabidopsis that contributes substantially to the cell plate. Plant Cell 20: 101–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins N.C., Thordal-Christensen H., Lipka V., Bau S., Kombrink E., Qiu J.L., Hückelhoven R., Stein M., Freialdenhoven A., Somerville S.C., Schulze-Lefert P. (2003). SNARE-protein-mediated disease resistance at the plant cell wall. Nature 425: 973–977. [DOI] [PubMed] [Google Scholar]
- Cutler S.R., Ehrhardt D.W., Griffitts J.S., Somerville C.R. (2000). Random GFP:cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proc. Natl. Acad. Sci. USA 97: 3718–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dettmer J., Hong-Hermesdorf A., Stierhof Y.D., Schumacher K. (2006). Vacuolar H+-ATPase activity is required for endocytic and secretory trafficking in Arabidopsis. Plant Cell 18: 715–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle S.M., Haeger A., Vain T., Rigal A., Viotti C., Łangowska M., Ma Q., Friml J., Raikhel N.V., Hicks G.R., Robert S. (2015). An early secretory pathway mediated by GNOM-LIKE 1 and GNOM is essential for basal polarity establishment in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: E806–E815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drakakaki G., et al. (2011). Clusters of bioactive compounds target dynamic endomembrane networks in vivo. Proc. Natl. Acad. Sci. USA 108: 17850–17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer U., Ikeda Y., Ljung K., Serralbo O., Singh M., Heidstra R., Palme K., Scheres B., Grebe M. (2006). Vectorial information for Arabidopsis planar polarity is mediated by combined AUX1, EIN2, and GNOM activity. Curr. Biol. 16: 2143–2149. [DOI] [PubMed] [Google Scholar]
- Friml J., Benková E., Blilou I., Wisniewska J., Hamann T., Ljung K., Woody S., Sandberg G., Scheres B., Jürgens G., Palme K. (2002). AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108: 661–673. [DOI] [PubMed] [Google Scholar]
- Fukaki H., Tasaka M. (2009). Hormone interactions during lateral root formation. Plant Mol. Biol. 69: 437–449. [DOI] [PubMed] [Google Scholar]
- Geisler M., Murphy A.S. (2006). The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett. 580: 1094–1102. [DOI] [PubMed] [Google Scholar]
- Geldner N., Denervaud-Tendon V., Hyman D.L., Mayer U., Stierhof Y.D., Chory J. (2009). Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59: 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N., Richter S., Vieten A., Marquardt S., Torres-Ruiz R.A., Mayer U., Jürgens G. (2004). Partial loss-of-function alleles reveal a role for GNOM in auxin transport-related, post-embryonic development of Arabidopsis. Development 131: 389–400. [DOI] [PubMed] [Google Scholar]
- Geldner N., Anders N., Wolters H., Keicher J., Kornberger W., Muller P., Delbarre A., Ueda T., Nakano A., Jürgens G. (2003). The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 112: 219–230. [DOI] [PubMed] [Google Scholar]
- Grebe M., Xu J., Möbius W., Ueda T., Nakano A., Geuze H.J., Rook M.B., Scheres B. (2003). Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13: 1378–1387. [DOI] [PubMed] [Google Scholar]
- Gu Y., Innes R.W. (2012). The KEEP ON GOING protein of Arabidopsis regulates intracellular protein trafficking and is degraded during fungal infection. Plant Cell 24: 4717–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heisler M.G., Ohno C., Das P., Sieber P., Reddy G.V., Long J.A., Meyerowitz E.M. (2005). Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr. Biol. 15: 1899–1911. [DOI] [PubMed] [Google Scholar]
- Hendricks L.C., McClanahan S.L., Palade G.E., Farquhar M.G. (1992). Brefeldin A affects early events but does not affect late events along the exocytic pathway in pancreatic acinar cells. Proc. Natl. Acad. Sci. USA 89: 7242–7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks G.R., Raikhel N.V. (2009). Opportunities and challenges in plant chemical biology. Nat. Chem. Biol. 5: 268–272. [DOI] [PubMed] [Google Scholar]
- Huang F., Zago M.K., Abas L., van Marion A., Galván-Ampudia C.S., Offringa R. (2010). Phosphorylation of conserved PIN motifs directs Arabidopsis PIN1 polarity and auxin transport. Plant Cell 22: 1129–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y., Gaude T. (2008). Plant cell polarity: sterols enter into action after cytokinesis. Dev. Cell 14: 318–320. [DOI] [PubMed] [Google Scholar]
- Jia D.J., Cao X., Wang W., Tan X.Y., Zhang X.Q., Chen L.Q., Ye D. (2009). GNOM-LIKE 2, encoding an adenosine diphosphate-ribosylation factor-guanine nucleotide exchange factor protein homologous to GNOM and GNL1, is essential for pollen germination in Arabidopsis. J. Integr. Plant Biol. 51: 762–773. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kitakura S., Vanneste S., Robert S., Löfke C., Teichmann T., Tanaka H., Friml J. (2011). Clathrin mediates endocytosis and polar distribution of PIN auxin transporters in Arabidopsis. Plant Cell 23: 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Swarup R., Bennett M., Friml J. (2006). Subcellular trafficking of the Arabidopsis auxin influx carrier AUX1 uses a novel pathway distinct from PIN1. Plant Cell 18: 3171–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Leitner J., Zwiewka M., Sauer M., Abas L., Luschnig C., Friml J. (2008a). Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc. Natl. Acad. Sci. USA 105: 17812–17817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J., Dhonukshe P., Sauer M., Brewer P.B., Wiśniewska J., Paciorek T., Benková E., Friml J. (2008b). ARF GEF-dependent transcytosis and polar delivery of PIN auxin carriers in Arabidopsis. Curr. Biol. 18: 526–531. [DOI] [PubMed] [Google Scholar]
- Kwon C., et al. (2008). Co-option of a default secretory pathway for plant immune responses. Nature 451: 835–840. [DOI] [PubMed] [Google Scholar]
- Li X., Wang X., Yang Y., Li R., He Q., Fang X., Luu D.T., Maurel C., Lin J. (2011). Single-molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23: 3780–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., Jung G., Wohlschlegel J.A., Huang J. (2011). Target identification using drug affinity responsive target stability (DARTS). Curr. Protoc. Chem. Biol. 3: 163–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomenick B., et al. (2009). Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. USA 106: 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao Th., McCubbin A.G., Cyr R.J. (1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhavý P., Vanstraelen M., De Rybel B., Zhaojun D., Bennett M.J., Beeckman T., Benková E. (2013). Auxin reflux between the endodermis and pericycle promotes lateral root initiation. EMBO J. 32: 149–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFarlane H.E., Watanabe Y., Gendre D., Carruthers K., Levesque-Tremblay G., Haughn G.W., Bhalerao R.P., Samuels L. (2013). Cell wall polysaccharides are mislocalized to the Vacuole in echidna mutants. Plant Cell Physiol. 54: 1867–1880. [DOI] [PubMed] [Google Scholar]
- Morita M.T., Shimada T. (2014). The plant endomembrane system--a complex network supporting plant development and physiology. Plant Cell Physiol. 55: 667–671. [DOI] [PubMed] [Google Scholar]
- Mravec J., et al. (2009). Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature 459: 1136–1140. [DOI] [PubMed] [Google Scholar]
- Müller A., Guan C., Gälweiler L., Tänzler P., Huijser P., Marchant A., Parry G., Bennett M., Wisman E., Palme K. (1998). AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 17: 6903–6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naramoto S., Otegui M.S., Kutsuna N., de Rycke R., Dainobu T., Karampelias M., Fujimoto M., Feraru E., Miki D., Fukuda H., Nakano A., Friml J. (2014). Insights into the localization and function of the membrane trafficking regulator GNOM ARF-GEF at the Golgi apparatus in Arabidopsis. Plant Cell 26: 3062–3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura K., Debroy S., Lee Y.H., Pumplin N., Jones J., He S.Y. (2006). A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science 313: 220–223. [DOI] [PubMed] [Google Scholar]
- Peyroche A., Antonny B., Robineau S., Acker J., Cherfils J., Jackson C.L. (1999). Brefeldin A acts to stabilize an abortive ARF-GDP-Sec7 domain protein complex: involvement of specific residues of the Sec7 domain. Mol. Cell 3: 275–285. [DOI] [PubMed] [Google Scholar]
- Richter S., Geldner N., Schrader J., Wolters H., Stierhof Y.D., Rios G., Koncz C., Robinson D.G., Jürgens G. (2007). Functional diversification of closely related ARF-GEFs in protein secretion and recycling. Nature 448: 488–492. [DOI] [PubMed] [Google Scholar]
- Richter S., Müller L.M., Stierhof Y.D., Mayer U., Takada N., Kost B., Vieten A., Geldner N., Koncz C., Jürgens G. (2011). Polarized cell growth in Arabidopsis requires endosomal recycling mediated by GBF1-related ARF exchange factors. Nat. Cell Biol. 14: 80–86. [DOI] [PubMed] [Google Scholar]
- Richter S., Anders N., Wolters H., Beckmann H., Thomann A., Heinrich R., Schrader J., Singh M.K., Geldner N., Mayer U., Jürgens G. (2010). Role of the GNOM gene in Arabidopsis apical-basal patterning--From mutant phenotype to cellular mechanism of protein action. Eur. J. Cell Biol. 89: 138–144. [DOI] [PubMed] [Google Scholar]
- Richter S., Kientz M., Brumm S., Nielsen M.E., Park M., Gavidia R., Krause C., Voss U., Beckmann H., Mayer U., Stierhof Y.D., Jürgens G. (2014). Delivery of endocytosed proteins to the cell-division plane requires change of pathway from recycling to secretion. eLife 3: e02131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridge R.W., Uozumi Y., Plazinski J., Hurley U.A., Williamson R.E. (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 40: 1253–1261. [DOI] [PubMed] [Google Scholar]
- Robert S., Chary S.N., Drakakaki G., Li S., Yang Z., Raikhel N.V., Hicks G.R. (2008). Endosidin1 defines a compartment involved in endocytosis of the brassinosteroid receptor BRI1 and the auxin transporters PIN2 and AUX1. Proc. Natl. Acad. Sci. USA 105: 8464–8469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford S., Moore I. (2002). The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5: 518–528. [DOI] [PubMed] [Google Scholar]
- Speth E.B., Imboden L., Hauck P., He S.Y. (2009). Subcellular localization and functional analysis of the Arabidopsis GTPase RabE. Plant Physiol. 149: 1824–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., Friml J., Marchant A., Ljung K., Sandberg G., Palme K., Bennett M. (2001). Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev. 15: 2648–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R., et al. (2004). Structure-function analysis of the presumptive Arabidopsis auxin permease AUX1. Plant Cell 16: 3069–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka H., Kitakura S., De Rycke R., De Groodt R., Friml J. (2009). Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 19: 391–397. [DOI] [PubMed] [Google Scholar]
- Teale W.D., Paponov I.A., Palme K. (2006). Auxin in action: signalling, transport and the control of plant growth and development. Nat. Rev. Mol. Cell Biol. 7: 847–859. [DOI] [PubMed] [Google Scholar]
- Teh O.K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448: 493–496. [DOI] [PubMed] [Google Scholar]
- Ueda T., Uemura T., Sato M.H., Nakano A. (2004). Functional differentiation of endosomes in Arabidopsis cells. Plant J. 40: 783–789. [DOI] [PubMed] [Google Scholar]
- Welz T., Wellbourne-Wood J., Kerkhoff E. (2014). Orchestration of cell surface proteins by Rab11. Trends Cell Biol. 24: 407–415. [DOI] [PubMed] [Google Scholar]
- Willemsen V., Friml J., Grebe M., van den Toorn A., Palme K., Scheres B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15: 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisniewska J., Xu J., Seifertová D., Brewer P.B., Ruzicka K., Blilou I., Rouquié D., Benková E., Scheres B., Friml J. (2006). Polar PIN localization directs auxin flow in plants. Science 312: 883. [DOI] [PubMed] [Google Scholar]
- Xu J., Scheres B. (2005). Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. Plant Cell 17: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zádníková P., et al. (2010). Role of PIN-mediated auxin efflux in apical hook development of Arabidopsis thaliana. Development 137: 607–617. [DOI] [PubMed] [Google Scholar]
- Zhang C., et al. (2016). Endosidin2 targets conserved exocyst complex subunit EXO70 to inhibit exocytosis. Proc. Natl. Acad. Sci. USA 113: E41–E50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Nodzynski T., Pencík A., Rolcík J., Friml J. (2010). PIN phosphorylation is sufficient to mediate PIN polarity and direct auxin transport. Proc. Natl. Acad. Sci. USA 107: 918–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H., Kunst L., Hawes C., Moore I. (2004). A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 37: 398–414. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.