The easyGWAS platform is a powerful online resource for computing, storing, sharing, and comparing the results of GWAS in both inbred and outbred plant and animal species.
Abstract
The ever-growing availability of high-quality genotypes for a multitude of species has enabled researchers to explore the underlying genetic architecture of complex phenotypes at an unprecedented level of detail using genome-wide association studies (GWAS). The systematic comparison of results obtained from GWAS of different traits opens up new possibilities, including the analysis of pleiotropic effects. Other advantages that result from the integration of multiple GWAS are the ability to replicate GWAS signals and to increase statistical power to detect such signals through meta-analyses. In order to facilitate the simple comparison of GWAS results, we present easyGWAS, a powerful, species-independent online resource for computing, storing, sharing, annotating, and comparing GWAS. The easyGWAS tool supports multiple species, the uploading of private genotype data and summary statistics of existing GWAS, as well as advanced methods for comparing GWAS results across different experiments and data sets in an interactive and user-friendly interface. easyGWAS is also a public data repository for GWAS data and summary statistics and already includes published data and results from several major GWAS. We demonstrate the potential of easyGWAS with a case study of the model organism Arabidopsis thaliana, using flowering and growth-related traits.
INTRODUCTION
The growing number of high-quality genotypes and phenotypes for many plant and animal species has created a unique opportunity to improve our understanding of the genetic basis of complex traits and diseases. Over the last decade, genome-wide association studies (GWAS) have become a key technique for exploiting this wealth of data, by detecting associations between a phenotype of interest and genetic variants present in a group of individuals (Bush and Moore, 2012). Unlike classic linkage mapping, GWAS can survey hundreds of thousands or even millions of single nucleotide polymorphisms (SNPs), which in turn gives GWAS greater power to detect small effects (Cordell and Clayton, 2005). Moreover, GWAS offer a higher resolution than linkage mapping because of the larger number of recombination events that will have occurred in natural panels used for association mapping (Nordborg and Weigel, 2008).
The utility of GWAS has been demonstrated in a variety of plants and crops, including Arabidopsis thaliana (Atwell et al., 2010; Filiault and Maloof, 2012; Meijón et al., 2014), rice (Oryza sativa; Zhao et al., 2011), wheat (Triticum aestivum; Liu et al., 2015), and tomato (Solanum lycopersicum; Lin et al., 2014), and in various animal species, such as fruit flies (Mackay et al., 2012), mice (Kirby et al., 2010), and humans (Scott et al., 2007; Chasman et al., 2011; Freilinger et al., 2012). These studies are accompanied by a steady improvement in the level of detail of the genotypic information. While early studies used SNPs to represent entire genomic regions (Atwell et al., 2010), recent studies have provided large panels of whole-genome information in species such as Arabidopsis (1001 Genomes Consortium, 2016).
The wealth of information provided by independent GWAS, many of which were conducted on related traits, offers unique opportunities for comparing and integrating findings across experiments. This integration makes it possible to detect genes with pleiotropic effects on multiple traits and traits with a shared genetic basis and to replicate findings or increase statistical power in association mapping through meta-analyses of several studies (Franke et al., 2010; Chasman et al., 2011; Andreassen et al., 2013; Evangelou and Ioannidis, 2013; Pickrell et al., 2016).
To exploit the full potential of comparative GWAS analyses, we present easyGWAS, a powerful platform for computing, storing, sharing, and comparing the results of GWAS in both inbred and outbred plant and animal species. easyGWAS offers tools to conduct GWAS and, more importantly, makes available additional data and functionality to facilitate the in-depth annotation, analysis, publishing, and comparison of GWAS results. There are three main aspects of easyGWAS, which together make it a unique online resource. The first aspect is its use as a repository of results obtained from private and public GWAS, which are either computed in easyGWAS or imported from external tools such as PLINK (Purcell et al., 2007). The second key component is the functionality to conduct GWAS in a standardized way to ensure a maximum degree of comparability between studies. This leads to easyGWAS’s third key feature, which is its ability to compare the results of different GWAS and conduct meta-analyses. While existing GWAS online resources, such as EMMA (Kang et al., 2008), DGRP2 (Mackay et al., 2012), Matapax (Childs et al., 2012), and GWAPP (Seren et al., 2012) allow for the computation, analysis, and annotation of GWAS, they focus exclusively on a single species and, most importantly, do not provide functionality to compare and integrate the results of already conducted GWAS.
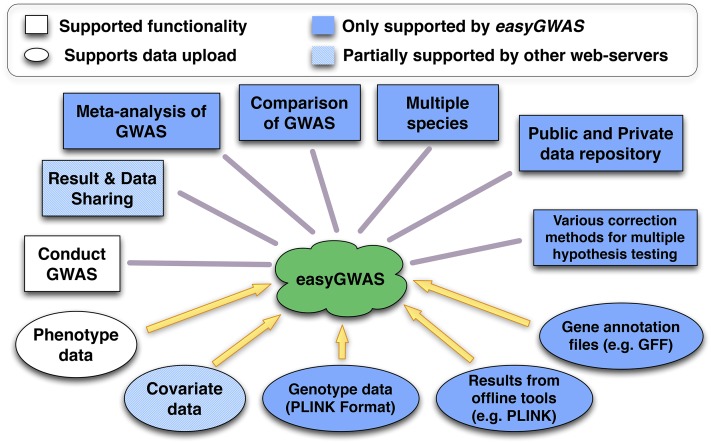
A summary of the main contributions of easyGWAS is highlighted in Figure 1. The platform is unique in five ways: (1) It is not limited to a certain species; (2) it allows users to upload and manage their own genotype and gene annotation data for a species of choice; (3) it supports the uploading of summary statistics of GWAS obtained from third-party tools; (4) it provides a variety of different methods to correct for multiple hypothesis testing; and, most importantly, (5) it integrates advanced methods for comparing results of GWAS across different data sets.
Figure 1.
Illustration of the Functionalities of easyGWAS in Comparison with Other Current Online GWAS Tools.
Squares illustrate supported functionalities, and ovals illustrate supported data types that can be uploaded to easyGWAS. White objects are supported by all available web servers, but hatched objects are only partially supported. Blue objects are only supported by easyGWAS.
Furthermore, easyGWAS supports state-of-the-art sharing and publishing functionalities: Users can choose between sharing the data and results associated to a genome-wide association study, either with a restricted set of collaborators or with the entire scientific community.
As a case study, we use easyGWAS on a large number of Arabidopsis accessions from the latest efforts of the 1001 Genomes Project (2016), in which we measured several flowering time and growth related traits. We integrate the results for different traits using the platform’s new comparison functionality. easyGWAS is available online at https://easygwas.ethz.ch.
RESULTS
Overview
easyGWAS consists of two main web components: a Data Repository and the GWAS Center. Each of them provides a user-friendly graphical front end that is divided into a publicly accessible area and a private area for registered users. The GWAS Center provides state-of-the-art methods to perform GWAS and meta-analyses, as well as an interactive results viewer for in-depth analysis of specific genomic regions. There is also an easy-to-use interface for interactive comparisons of the results of already computed GWAS or uploaded summary statistics.
Data Repository
The data repository comprises various functions related to data integration, storage, management, and representation. As mentioned above, the data repository is divided into a publicly accessible area and an area that is restricted to registered users. Published and publicly available data can be accessed via the Public Data view, while users’ private data are stored in a restricted and secure environment that can only be accessed through the Private Data view (Supplemental Figure 1 or at https://easygwas.ethz.ch/data/public/species/). Refer to Methods for details about data sets already integrated into the Public Data view. Data shared between collaborators or with subsets of other users will be displayed in the Private Data view. The data repository contains general information, meta-information, and graphics about the species, data sets, phenotypes, covariates, and samples. The Download Manager gives access to all publicly available data sets in the widely used PLINK format (Purcell et al., 2007). Users can easily make their private data available to the scientific community at a later time point. The Upload Manager supports users in the secure integration of new genotype, phenotype, covariate, or gene annotation data into easyGWAS. Furthermore, easyGWAS supports the automatic import of public phenotypes from AraPheno (https://arapheno.1001genomes.org), a central repository for population-scale phenotype data from Arabidopsis (Seren et al., 2016). Users can also upload their custom summary statistics of GWAS performed in different environments, for example, from offline analyses with PLINK (Purcell et al., 2007) or other third-party tools (Kang et al., 2010; Lippert et al., 2011; Rakitsch et al., 2013; Azencott et al., 2013; Sugiyama et al., 2014; Llinares-López et al., 2015), for visualization, subsequent meta-analysis, or comparison with GWAS results that have already been deposited in easyGWAS. Detailed descriptions of the different views can be found in the supplemental data (Supplemental Figures 2 to 6 and Supplemental Text 1).
GWAS Center
The GWAS Center offers a variety of different methods related to computing, analyzing, and managing GWAS; meta-analyses; and comparisons of results. Nonregistered users can investigate and download the results of published and publicly available projects, while registered users can perform their private analysis on data for a given species. As mentioned above, a user can conduct (1) GWAS, (2) meta-analyses, and (3) comparisons of results. For each of these tasks, there are step-by-step workflows, also referred to as “wizards,” which facilitate and standardize their execution. Each wizard guides the user through all necessary steps, such as selecting a species, phenotypes, transformations, and appropriate algorithms. The wizard analyzes the user’s input at different steps and only suggests suitable algorithms and transformations (for example, it excludes transformations or algorithms that cannot be used for specific data types). To avoid an overload of the easyGWAS server, a user is limited to five concurrent experiments. Submitted experiments are distributed to different computation queues in the back-end and the user receives automatic email notifications after the computations have finished (see the Runtime Analysis section for performance comparisons). Details about implemented genome-wide association (GWA) mapping methods, meta-analysis methods, transformations, and genotype encodings can be found in Methods. Please see the supplemental data for additional details about the wizards (Supplemental Text 2 and Supplemental Figure 7) as well as the easyGWAS online FAQ (https://easygwas.ethz.ch/faq/).
By default, results are kept for 48 h in the user’s profile and trashed afterwards. To store experiments permanently, users can save and group individual experiments into GWAS projects (Supplemental Text 3 and Supplemental Figures 8 to 11). Private projects can be easily shared with other collaborators or users using the project sharing functionality (Supplemental Figure 10), which reduces the need to send large results and data files via e-mail to collaborators. In addition to restricted data sharing with others, users can make their private GWAS projects available to the scientific community. We have added a hand-curated inquiry step to control the quality of publicly available projects and data (Supplemental Figure 11).
Result Views
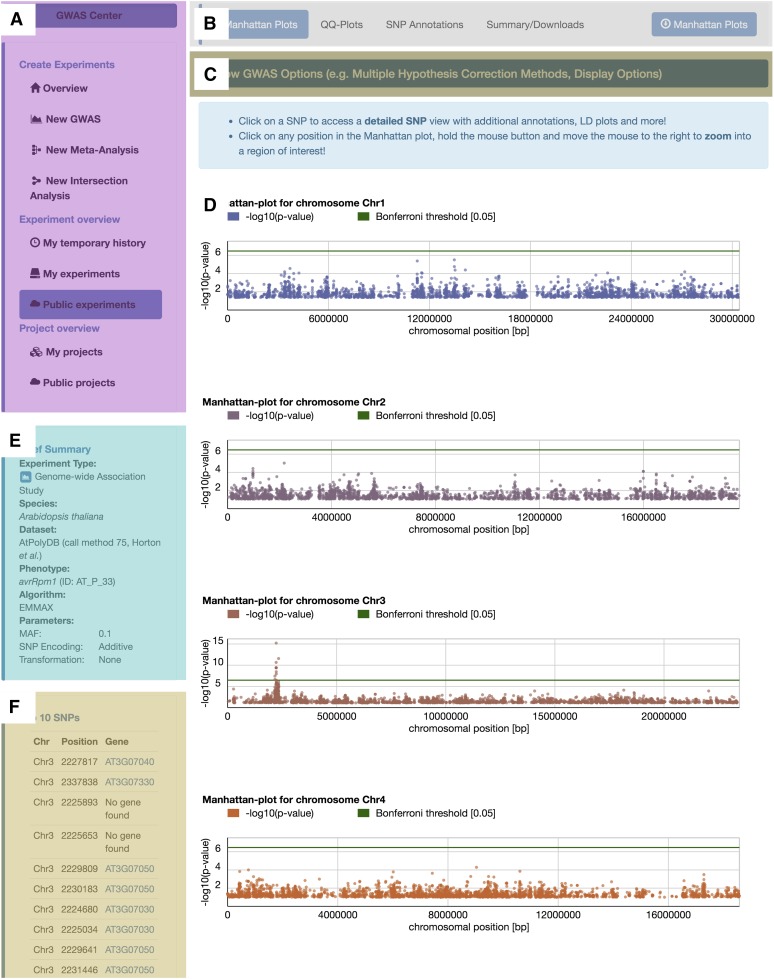
Results of computed GWAS and meta-analyses can be viewed by clicking on the experiment name in either the temporary or permanent history. The results window is divided into several panels, as illustrated in the screenshot shown in Figure 2 (Supplemental Text 4). Figure 2A shows the general “GWAS Center” menu panel. To navigate through different sections of the results, users can click on the different tabs shown in Figure 2B. The panels in Figures 2C and 2D are the default panels when displaying the results of each experiment. The panel in Figure 2D shows interactive Manhattan plots. The green line in each Manhattan plot illustrates the global multiple hypothesis correction threshold. Four widely used correction methods can be applied: Bonferroni (Abdi, 2007), Benjamini and Hochberg (1995), Benjamini and Yekutieli (2001), and Storey and Tibshirani (2003) (see Methods; Supplemental Text 10). By default, the conservative Bonferroni correction controls the family-wise error rate with a significance level of α = 5%, but the other methods also provide less stringent false discovery rate controls. The multiple hypothesis correction method and the significance level α (1, 5, or 10%) can be adjusted dynamically in the plotting options, which can be found in the panel in Figure 2C. All Manhattan plots contain interactive elements, such as the ability to zoom into regions of interest to display further information about the local genes. Manhattan plots can be downloaded as PDF files for use in manuscripts or presentations. The vertical panels on the left (Figure 2E and 2F) show a brief summary, including information about the selected species, data set, and selected settings. Additionally, the top 10 associated SNPs and their genes are ranked (if a gene annotation set was selected for the experiment).
Figure 2.
Screenshot of the easyGWAS Result Layout.
The screenshot shows the general layout of the easyGWAS result view.
(A) The “GWAS Center” menu with links to different wizards and experiment tables, e.g., to create new GWAS, meta-analysis, or comparative intersection experiments.
(B) A sub-menu for the GWAS results to navigate between Manhattan plots, QQ-plots, SNP annotations, or an experiment summary.
(C) General options to dynamically adjust the multiple hypothesis testing method or various different plotting options.
(D) The main results of a GWAS, meta-analysis, or intersection analysis. In this screenshot, Manhattan plots are shown.
(E) and (F) A brief summary of the most important experimental parameters is shown (E). This can be either a summary of a regular GWAS experiment, a meta-analysis, or a comparison of several GWAS. If available, the top 10 associated hits of a GWAS are shown in (F).
Moreover, each SNP in a Manhattan plot is clickable, such that users can obtain additional information about the alleles at that position, box plots about the phenotypic values for each allele, as well as a detailed map of the linkage disequilibrium (LD) pattern around the focal SNP (Figure 3; Supplemental Text 4). In addition, we also provide more detailed information about the selected variant, such as whether the variant is a missense mutation, frameshift, or stop codon. For this purpose, we automatically fetch the corresponding data using the Ensemble REST interface for the Variant Effect Predictor tool (Yates et al., 2014; McLaren et al., 2010, 2016).
Figure 3.
Screenshot of the easyGWAS Detailed SNP View.
The “Detailed SNP” view of a SNP gives more detailed information and annotation about the selected SNP and its close neighborhood, illustrated in this screenshot.
(A) A donut diagram with the allele distribution of the selected SNP.
(B) A box plot with the trait values for each allele is shown.
(C) The distribution of genes and the LD pattern around the focal SNP. The panel at the bottom shows more detailed annotations for the focal SNP, e.g., if a SNP is a missense variant, frameshift, or stop codon.
Figure 2B shows different clickable tabs that allow the user to explore all the results of the experiment. The second tab, “QQ-Plots,” renders quantile-quantile plots and provides information about the genomic control factor (Devlin and Roeder, 1999) (Supplemental Text 5).
The third tab, “SNP Annotations,” lists gene annotations for the top associated SNPs per chromosome. Users can dynamically change the number of top SNPs displayed or the multiple hypothesis correction method used and search for genes upstream or downstream of each SNP (Supplemental Text 5). For each SNP, we again redirect to the same detailed SNP view as described above.
The last tab in Figure 2B, “Summary/Downloads,” provides a detailed summary of the experiment, including all selected settings, information about the experiment’s owner, selected trait/covariate distributions, as well as associated publications (Supplemental Text 5). Adding too many covariates to regression-based models can easily lead to overfitting (Hastie et al., 2009). Therefore, it is important to find a good balance between goodness of fit R2 and model complexity. To this effect, three model selection parameters are implemented to measure the relative quality of each model: Akaike information criterion (AIC), Akaike information criterion with correction for finite sample sizes (AICc), and Bayesian information criterion (BIC) (Schwarz, 1978). A general distinction between AIC and AICc is that the latter gives preference to models with fewer parameters. The thought behind it is that a model with fewer parameters will generalize better. BIC is based on the assumption that a true model exists and is present among the models to be compared (Burnham and Anderson, 2002; Aho et al., 2014). It is important to note that neither of these criteria is equivalent to hypothesis testing. Instead, they simply provide guidance on which model to choose.
easyGWAS also provides variance-explained estimates for the null model when using linear mixed models, such as EMMAX (Kang et al., 2010) or FaST-LMM (Lippert et al., 2011) (Supplemental Text 6).
Comparative GWAS Intersection Analysis View
easyGWAS offers two types of comparative analyses of GWAS. The first type is the search for associations at the SNP or gene level that were found to be significant in more than one data set; in the following, we refer to this form of comparative search for intersecting association hits as “intersection analysis.” The second type includes meta-analyses that aggregate evidence from several data sets to find associations that are jointly supported by these data sets. Here, we describe easyGWAS’s Comparative Intersection Analysis View for intersection analyses. More information on the available meta-analysis approaches can be found in the “Meta-Analysis Methods” section in Methods.
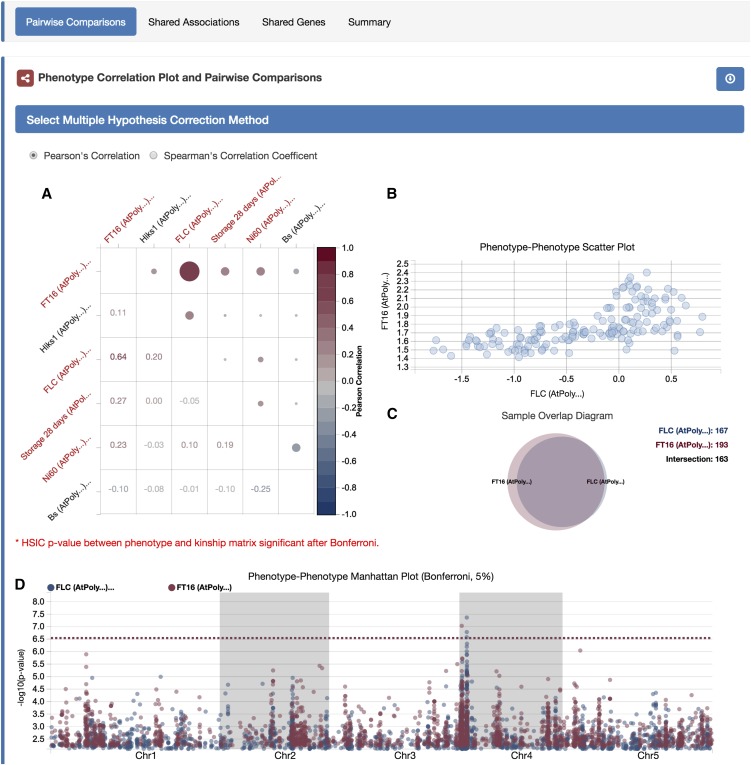
easyGWAS enables intersection analyses on the results of a set of GWAS and provides three comparison views: the “Pairwise Comparison View” for comparing results on pairs of GWAS including overlaid Manhattan plots; the “Shared Associations View,” which represents shared SNPs among the top-x associated SNPs of all GWAS; and the “Shared Genes View,” which lists all genes containing a significantly associated SNP in at least one GWAS. The pairwise comparison provides several dynamic visualizations to support the investigation of results for all possible pairs from a set of GWAS results (Figure 4; Supplemental Text 4).
Figure 4.
Screenshot of the easyGWAS Pairwise Comparison View.
The screenshot illustrates the general layout of the pairwise comparison view of different GWAS.
(A) A phenotype-phenotype correlation plot is shown, while phenotype names highlighted in red are significantly associated with population structure.
(B) Hovering over any correlation point in the plot will dynamically update the phenotype-phenotype scatterplot. The phenotype-phenotype scatter diagram plots the measurements of both phenotypes against each other.
(C) A Venn diagram is shown to illustrate the sample overlap between the two phenotypes.
(D) The Manhattan plots for both GWAS on top of each other.
Initial insights into whether traits might have a common genetic basis come from assessing how different traits in a population correlate with each other, as shown in easyGWAS’s phenotype-phenotype correlation plot. The cause for this correlation may be population structure; that is, systematic ancestry differences between different phenotypic classes. To make the user conscious of this source of confounding, easyGWAS highlights phenotypes that are significantly associated to kinship in red (for computational details, see Supplemental Text 7).
The phenotype-phenotype scatterplot (Figure 4B), sample overlap diagram (Figure 4C), and the phenotype-phenotype Manhattan plot (Figure 4D) are dynamically updated according to the position of the cursor on the correlation plot (Figure 4A). Users can dynamically investigate results from different GWAS experiments.
The second tab at the top of Figure 4, “Shared Associations,” shows all shared associations between selected experiments for the top x-associated SNPs per GWAS experiment as a chord diagram, where x is a parameter that can be chosen by the user dynamically (Supplemental Text 5). The third window gives an overview of all genes across all experiments that contained an associated hit (Supplemental Text 5). Again, users can dynamically change the multiple hypothesis correction method in all windows.
Runtime Analysis
We compared the runtimes of four popular genome-wide association tools and methods, including a linear regression (PLINK v1.0.7; Purcell et al., 2007), logistic regression (PLINK v1.0.7; Purcell et al., 2007), EMMAX (Kang et al., 2010), and FaST-LMM (Lippert et al., 2011), to those implemented in the easyGWASCore framework. We used real genotype data from the 1001 Genomes Project in Arabidopsis (1001 Genomes Consortium, 2016) and random continuous and binary phenotypes for the analysis. For all experiments, the number of SNPs varied from 10,000 to five million and the number of samples from 100 to 500. Data in PLINK format were used to conduct a fair comparison between all tools and methods. Real CPU runtime in seconds was reported over a single AMD Opteron CPU (2048 kb, 2600 MHz) with 512 GB of memory, running Ubuntu 12.04.5 LTS (Supplemental Figure 13). We observe that all algorithms implemented in easyGWAS, except for logistic regression, are at least as efficient as the tools to which we compared them. The results show that easyGWAS can compute GWAS with standard models such as linear regression within a few minutes for up to five million SNPs and up to 500 samples. More complex models, such as FaST-LMM or EMMAX, only take minutes for ∼100 samples and a few hours for up to 500 samples.
Case Study in Arabidopsis
To demonstrate the usability of easyGWAS, we analyzed nine flowering time- and growth-related traits of Arabidopsis. Phenotypes were scored for up to 936 of 1135 accessions (1001 Genomes Consortium, 2016; see Methods). We first used a standard linear mixed model (FaST-LMM by Lippert et al., 2011) to perform GWAS on these nine phenotypes while accounting for potential confounding effects due to population stratification (see Methods). All results, including Manhattan plots and QQ-plots, are publicly available online at https://easygwas.ethz.ch/gwas/myhistory/public/14/.
Next, we used the comparison functions of easyGWAS to combine the results for all traits. We identified a total of six significantly associated hits, when using Bonferroni correction to control the family-wise error rate at α = 5% (Supplemental Table 1). Multiple testing correction methods based on the false discovery rate (Storey and Tibshirani, 2003) are less conservative than Bonferroni correction and are often used in GWAS. Using Storey and Tibshirani’s approach (α = 5%), we identified a total of 254 significantly associated hits across all experiments, that is, associated with at least one phenotype. A total of 87 significantly associated hits are shared by at least two of the following five different phenotypes: flowering time as days until emergence of visible flowering buds in the center of the rosette from time of sowing (DTF1), flowering time as days until the inflorescence stem elongated to 1 cm (DTF2), flowering time as days until first open flower (DTF3), rosette leaf number (RL), and cauline leaf number (CL) (Supplemental Data Set 1).
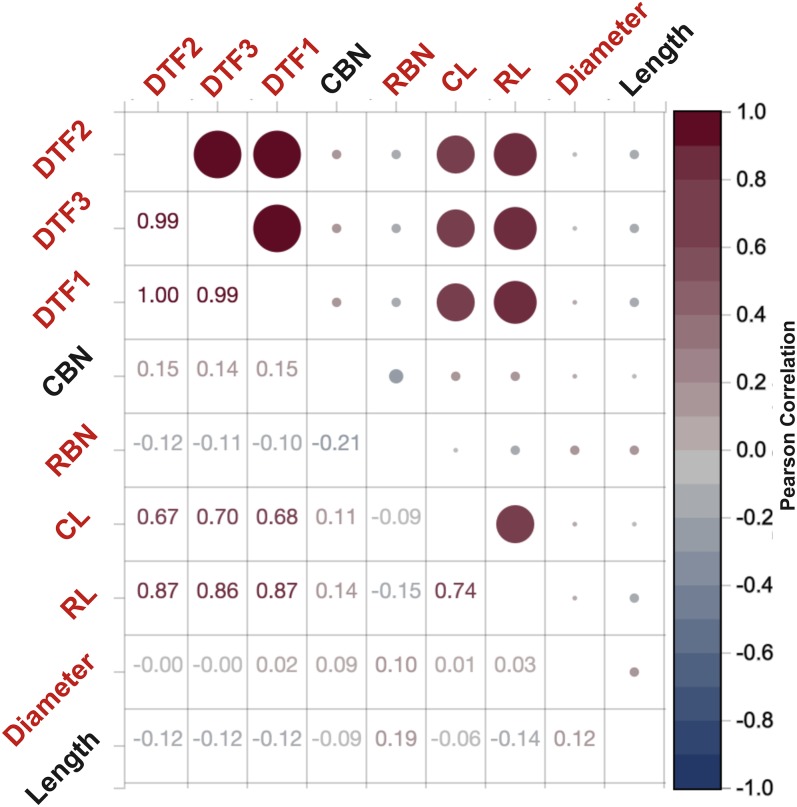
This is not surprising given that these five phenotypes are highly correlated with each other (Pearson’s r2 between 0.67 and 0.99), as shown in Figure 5 or online at: https://easygwas.ethz.ch/comparison/results/manhattan/view/2c8da231-96ff-4f28-a17e-fd0e3510d8e1/.
Figure 5.
Phenotype-Phenotype Correlation Plot for Case Study.
Phenotype-phenotype correlation plot showing the pairwise Pearson’s correlation coefficients between all phenotypes for the case study in Arabidopsis. Five of the phenotypes are highly correlated to each other: flowering time as days until emergence of visible flowering buds in the center of the rosette from time of sowing (DTF1); flowering time as days until the inflorescence stem elongated to 1 cm (DTF2); flowering time as days until first open flower (DTF3); rosette leaf number (RL); and cauline leaf number (CL). Phenotypes highlighted in red are significantly associated with population structure.
Of the 254 significant hits across all experiments using Storey and Tibshirani’s multiple hypothesis correction, 250 are located on chromosomes 4 and 5. Significantly associated hits are distributed across 30 different genes on four different chromosomes (1, 2, 4, and 5), as shown in the Shared Genes view in easyGWAS and in Supplemental Data Set 1. The 87 shared peaks include the dormancy regulator DOG1 (At5g45830), which has recently been shown to affect flowering time (Huo et al., 2016); FLOWERING LOCUS C (FLC; At5G10140), and FRIGIDA (At4G00650), which are linked to flowering time variation (Michaels and Amasino, 1999, 2001; Méndez-Vigo et al., 2013, 2016; Sanchez-Bermejo and Balasubramanian, 2016); and ANTHOCYANINLESS2 (ANL2; At4G00730), which is involved in root development (Kubo and Hayashi, 2011).
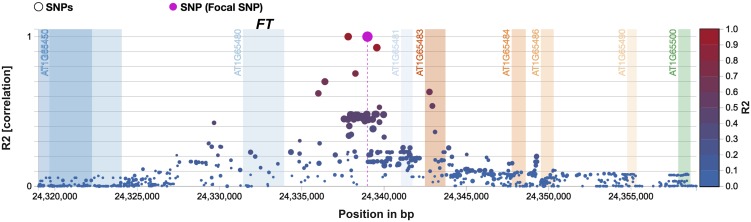
Furthermore, easyGWAS makes it possible to investigate the LD and gene information in close proximity to a focal SNP. For example, three significantly associated hits for the RL phenotype map to chromosome 1 and are in close proximity to the flowering regulator FLOWERING LOCUS T (FT; AT1G65480), as shown in Figure 6 and online using the easyGWAS Detailed SNP view: https://easygwas.ethz.ch/gwas/results/snp/detailed/57a7cf18-cb0f-408a-8954-49f94d1bfc47/Chr1/24338990/.
Figure 6.
Linkage Disequilibrium Plot for SNP Chr1:24338990 for Phenotype RL.
Three SNPs for the phenotype RL are significantly associated using Storey and Tibshirani’s correction for multiple hypothesis testing. These hits are in close proximity to the FT gene.
FT shows remarkably little variation within its coding sequence. However, recent quantitative trait loci fine-mapping efforts have highlighted the contribution of cis-regulatory polymorphisms in natural variation, including the flowering genes FT and FLC. For instance, a causal polymorphism was mapped not to the FT coding region, but to the FT promoter of the Est-1 FT allele, conferring delayed flowering relative to the Col-0 reference (Schwartz et al., 2009). A larger study found that FT promoter length varies and correlates with flowering time, while the FT coding sequence remains unchanged (Liu et al., 2014).
Similarly, FLC-dependent variation in flowering time often maps to promoter elements, resulting in expression differences between alleles, both in Arabidopsis and Brassica oleracea (Irwin et al., 2016; P. Li et al., 2014). Such variation caused by cis-regulatory polymorphisms is not limited to flowering time, as additional cases have been reported for (1) zinc homeostasis conferred by expression changes in the FRD3 MATE transporter (Pineau et al., 2012) and (2) sulfur homeostasis associated with the ATP sulfurylase ATPS1 (Koprivova et al., 2013). These studies underscore the potential contribution that cis-regulatory changes can make to natural variation, which is already reflected in the number of expression quantitative trait loci detected in selected recombinant populations (Cubillos et al., 2012).
Polymorphisms in noncoding regions can also result in phenotypic variation, as demonstrated by a naturally occurring SNP affecting splicing of the FLC antisense transcript COOLAIR in some Arabidopsis accessions (Li et al., 2015). It is conceivable that additional examples will be identified in the future, making full use of whole-genome sequences from thousands of accessions. Interestingly, the three SNPs reported by easyGWAS for the RL phenotype are found to overlap within several noncoding RNAs downstream of FT (At1NC09610, At1NC09620, and At1NC09630; Supplemental Figure 14) (Liu et al., 2012). This raises the possibility that these noncoding RNAs may act as enhancer elements and play roles in modulating FT expression level, perhaps in a mechanism similar to the PINOID/APOLO regulatory pair of loci, whereby the lncRNA APOLO regulates the expression of PINOID via chromatin looping (Ariel et al., 2014). As in any mapping project of EMS-induced mutations or natural variation, further analysis will help shed light on the mechanisms underlying the phenotype of interest.
Importantly, easyGWAS eliminates the need for complicated visualization scripts by providing them automatically and interactively. All plots, including Manhattan plots, LD plots, and the phenotype-phenotype correlation plot, can be explored by following the link to this public easyGWAS project: https://easygwas.ethz.ch/gwas/myhistory/public/14/. We also provide a list of links in Supplemental Table 2 to access the individual experiments directly.
DISCUSSION
We have introduced easyGWAS, a cloud platform that not only allows the computation, annotation, and subsequent analysis of GWAS, but, most importantly, also offers the unique feature of comparing results from GWAS across different experiments and data sets (Figure 1).
The constant increase of publicly available genotype and phenotype data (J.-Y. Li et al., 2014; 1001 Genomes Consortium, 2016) creates a demand for tools that enable biologists to compare the results of multiple GWAS in order to facilitate the identification of associations shared between related and/or correlated phenotypes. Such tools can lead to new biological insights, such as a common genetic architecture of related phenotypes or the seemingly unrelated functions of a gene due to pleiotropic effects. easyGWAS is currently the only tool that offers a variety of methods to compare GWAS. We have also described the publishing capabilities of easyGWAS that make it possible to share results between collaborators. Another salient feature is the novel interactive visualizations that aim to explore and compare the results of GWAS, not only for a single analysis but also across different data sets and samples.
While current web applications for GWAS are typically limited to a single species, such as Arabidopsis (Childs et al., 2012; Seren et al., 2012) or Drosophila melanogaster (Mackay et al., 2012), easyGWAS supports multiple species in a single platform. easyGWAS also provides an added functionality that is absent in the above-mentioned web applications: Users can upload private GWAS data sets for any species of choice (genotype, phenotype, and covariate data), custom gene annotation sets, as well as summary statistics from offline analyses; for example, from PLINK (Purcell et al., 2007) or from a custom user tool. While it is technically possible to also analyze human data, human genotype data must only be uploaded if this is explicitly allowed by the legal body governing data access.
We demonstrated some of the potential of easyGWAS in a case study in the model organism Arabidopsis on nine newly measured phenotypes and a population of 1135 recently sequenced lines (1001 Genomes Consortium, 2016). Conducting such an analysis without easyGWAS would be a time-consuming and cumbersome process. While performing standard GWAS is nowadays facilitated by many tools, such as PLINK (Purcell et al., 2007), EMMAX (Kang et al., 2010), and FaST-LMM (Lippert et al., 2011), these tools often ignore the annotation of the results. Web applications such as GWAPP (Seren et al., 2012) can be used to get on-the-fly annotations of GWAS results, but they are limited to a specific species only.
easyGWAS simplifies the execution of GWAS and the comparative analysis of their results, even for users who have never worked on GWAS before. It is important to note, however, that there are several errors that can be made and biases that can be introduced inadvertently when integrating results of different GWAS. For example, a common assumption in a meta-analysis is the independence of samples in the studies to be combined. If this assumption is violated, for example, because of overlap of a subset of individuals across studies, this may result in spurious associations (Lin and Sullivan, 2009; Zaykin and Kozbur, 2010). Similarly, in a comparative study, differences in sample size between data sets can have an adverse effect in an intersection analysis. In this case, true associations may be too weak to be detected in the smaller data sets, which will result in false negative findings when the results of the studies are intersected. Therefore, it is imperative that users of easyGWAS make themselves familiar with these pitfalls, which we summarize in Supplemental Table 3. To increase awareness of these issues, easyGWAS notifies each user of these potential problems before the submission of a comparative analysis.
In future releases, we will extend the application programming interface of easyGWAS that currently uses the Representational State Transfer (REST) web service to facilitate the exchange of phenotypic and genotypic data between different web platforms (Supplemental Text 11). This is especially important in order to keep not only public phenotypes but also genotypes synchronized between easyGWAS and other species-specific web applications, such as GWAPP (Seren et al., 2012) or AraPheno (Seren, Grimm et al., 2016). In addition, this application programming interface will allow users to acquire additional information and meta-information using their custom scripts or analysis pipelines. A similar feature that is already available is easyGWAS’s ability to automatically import public phenotypes from AraPheno, a central repository for population scale phenotype data from Arabidopsis (Seren et al., 2016). easyGWAS currently supports comparative analyses on one-dimensional phenotypes through intersection analyses and meta-analyses. In future work, we plan to integrate methods for mapping several phenotypes simultaneously in multitrait analyses (Korte et al., 2012; Lippert et al., 2014). Further plans include adding methods for multilocus mapping (Azencott et al., 2013; Llinares-López et al., 2015), and providing support for replicated phenotypic measurements as well as complex and high-dimensional data, such as methylome and transcriptome data.
METHODS
Genome-Wide Association Mapping Methods
The goal of a genome-wide association study is to detect genomic regions that are associated with a trait in a cohort of individuals. This is achieved by computing statistical associations between the trait and genetic variants in the form of SNPs. Of the many methods used to compute this statistical association (see Bush and Moore [2012] for a complete review), five have been implemented in easyGWAS: a Wilcoxon rank-sum test for homozygous genotype data; a linear regression and logistic regression for homozygous and heterozygous genotype data; and two linear mixed models, EMMAX (Kang et al., 2010) and FaST-LMM (Lippert et al., 2011), to account for confounding due to population stratification, family structure, and cryptic relatedness. The remainder of this subsection discusses these methods in detail.
As mentioned above, the Wilcoxon rank-sum test can only be used with biallelic SNPs. The easyGWAS wizard will not offer this method when the genotype data do not meet this requirement. This test measures the difference in the distributions of phenotypic values between the alleles, under the null hypothesis of equal distributions for both alleles.
In a linear regression there is an implicit assumption that the phenotype is normally (Gaussian) distributed and that it can be modeled as a linear (additive) combination of a set of terms, where one term is the list of genotypes of a given SNP and other (optional) terms may include covariates to correct for confounders.
Logistic regression makes a similar assumption about the additive effects of genotype and covariates, but unlike linear regression (which can be used for continuous phenotypes), logistic regression is applied in analyses where the phenotype is binary, for example, in case/control studies.
The default method for association testing in easyGWAS is the linear mixed model, which results in an improvement over linear regression in that the phenotype is measured as a sum of fixed and random effects. The linear mixed model (LMM) has historically been used to identify genetic associations between a phenotype of interest and a group of individuals when the relationship between the individuals is known. This model can effectively quantify the association of a genetic variant to the phenotype (fixed effect), while correcting for the familial structure in the data (random effect). However, the LMM has gained popularity in recent years in GWAS when there is no pedigree of the individuals. Although it may be tempting to assume that the individuals in a study are unrelated, that is, it may be a requirement in the collection or recruiting process, it is still possible that cryptic relatedness may be present in the data due to the fact that some individuals may share a common ancestry. If this relatedness is not taken into account, spurious associations may arise (Price et al., 2006). To correct for such confounding structure in the data, a LMM will estimate the genetic similarity between all pairs of individuals in the study. In easyGWAS, this similarity is modeled with the kinship matrix, which is estimated by computing the realized relationship matrix (Hayes et al., 2009) for both EMMAX (Kang et al., 2010) and FaST-LMM (Lippert et al., 2011). easyGWAS also enables users to include principal components as part of a linear regression, logistic regression, or LMM. When only population stratification is present, this strategy of adding principal components into the model has been shown to yield more power than a standard LMM (Zhao et al., 2007; Widmer et al., 2014).
In addition to the standard implementation of the above-mentioned algorithms, easyGWAS offers permutation-based versions of linear regression, logistic regression, and EMMAX. The added value of this additional functionality is that it allows for an empirical estimation of the true null distribution. Covariates can be easily added to any model (except for the Wilcoxon rank-sum test) to account for confounding effects, such as environmental factors or sex.
Finally, easyGWAS provides several means of encoding genotype data. Four genotype encodings have been implemented for heterozygous phenotypes to allow for the testing of different allelic effects. The standard encoding is based on what is known as the “additive model” where the major allele is encoded as 0, the heterozygous allele as 1, and the minor allele as 2 (Supplemental Table 4). However, the recessive genotype encoding encodes the major and heterozygous allele as 0 and the minor allele as 1. The dominant genotype encoding encodes the major allele as 0 and the remaining two alleles as 1, whereas the overdominant (or codominant) encoding encodes the major and minor allele as 0 and the heterozygous allele as 1 (Supplemental Table 4). All algorithms have been implemented in a custom C/C++ framework called easyGWASCore, which comes with Python interfaces to allow for easy integration into our web framework.
Transformation Methods
Several transformation methods have been implemented in easyGWAS to improve the normality of phenotypic measurements and covariates. Supplemental Text 8 provides additional details about the motivation behind the use of transformation methods, as well as detailed descriptions of each method. A summary of all possible transformations in easyGWAS and the type of measurements to which they can be applied is shown in Supplemental Table 5. In addition, a Shapiro-Wilk test is provided to test the null hypothesis if the data were drawn from a normal distribution (Shapiro and Wilk, 1965).
Meta-Analysis Methods
Historically, meta-analysis arose due to the need to pool results from different studies. Although its origins are rooted in combining the results from independent clinical trials, meta-analysis has been successfully used as a tool to combine or integrate the results from different GWAS. The rationale behind this is that, due to their limited sample size, GWAS are underpowered and the meta-analysis is a means to increase the overall power and to reduce false positives (Evangelou and Ioannidis, 2013).
easyGWAS implements a variety of different meta-analysis methods that can be used to combine results from several conducted GWAS on distinct sets of samples/cohorts. Five state-of-the-art algorithms are part of the easyGWASCore framework: (1) Fisher’s method to combine P values from several studies (Fisher, 1925); (2) Stouffer’s Z to combine Z-scores derived from the studies’ P values (Stouffer et al., 1949); (3) Stouffer’s weighted Z to weight each study based on the square root of the number of samples (Stouffer et al., 1949); (4) a fixed-effect model to combine effect estimates while assuming that they are fixed for each study (Borenstein et al., 2010, 2011); and (5) a random effect model that assumes that they arise randomly (Borenstein et al., 2010, 2011). See Supplemental Text 9 for more details about these methods, their assumptions, and guidelines on how to apply them.
Web Application Details
The easyGWAS web application is written in Python and builds upon the Python Django (https://www.djangoproject.com) and easyGWASCore frameworks running on an open-source Apache HTTP server (http://httpd.apache.org). Bootstrap (http://getbootstrap.com), a popular HTML5, CSS, and JavaScript (JS) framework, provides a modern and state-of-the-art front end. To create dynamic and interactive visualizations, we used the JS library D3.js (http://d3js.org). Asynchronous JavaScript and XML (AJAX) allows dynamic interactions and updates to the front end without reloading the whole webpage. To schedule and distribute long-running and file-system-intensive tasks across different computation nodes, such as performing GWAS or generating data for dynamic visualizations, the message-passing system RabbitMQ (http://rabbitmq.com) is used together with the asynchronous task queue Celery (http://celeryproject.org). Thus, easyGWAS is highly scalable and can be quickly extended to a larger number of users by adding additional computation nodes.
Reliable and fast data storage is essential for handling user-generated data as well as genotype data sets with hundreds of samples and potentially hundreds of thousands of genetic markers. Therefore, we developed a hybrid database model that is a combination of a PostgreSQL database, user-specific SQLite databases, and HDF5 files. General information about users, data sets, or GWA projects is stored in the PostgreSQL database. Genotype, phenotypes, covariate data, and results are stored in HDF5 files and linked to the user profiles in the PostgreSQL database. User-specific gene annotation sets are stored in user-specific SQLite database files such that efficient queries for different sets are ensured (Supplemental Figure 12). Currently, easyGWAS is running on an Ubuntu-based machine with 64 CPUs and 512 GB of memory.
Data Download, Upload, and Sharing
To simplify data handling, all publicly available data sets can be easily downloaded in the commonly used PLINK format (Purcell et al., 2007) using the easyGWAS download manager. To integrate newly sequenced genotype data, phenotypic measurements (in PLINK format), or custom gene annotation sets (in GFF format), the easyGWAS upload manager can be used by registered users (Supplemental Text 1 or online FAQ). Users can also create new species that are not already available in easyGWAS. Furthermore, easyGWAS supports the automatic import of public phenotypes from AraPheno, a central repository for population scale phenotype data from Arabidopsis thaliana (Seren et al., 2016). In addition, GWA projects can be easily shared with other registered users (Supplemental Text 3 or online FAQ).
Furthermore, we provide a REST web service that allows users to query and get data/meta-information from easyGWAS in a programmatic way, or simply via URLs in a web browser (Supplemental Text 11).
Publicly Available Data
As of December 2016, easyGWAS provides publicly available genotype, phenotype, and gene annotation data for the species Arabidopsis, Drosophila melanogaster, and Pristionchus pacificus. Additional data for other species will be added and included in the future.
For Arabidopsis, data sets from various studies have been integrated (Atwell et al., 2010; Cao et al., 2011; Horton et al., 2012; Long et al., 2013; Schmitz et al., 2013; 1001 Genomes Consortium, 2016; Seymour et al., 2016). The first data set (AtPolyDB) includes 1307 worldwide Arabidopsis accessions with a total of 214,051 SNPs genotyped with a 250k SNP chip (Horton et al., 2012). In addition, 107 binary, continuous, and categorical phenotypes have been integrated for a subset of these 1307 accessions (Atwell et al., 2010). The phenotypic data comprises (1) flowering-time-related traits, (2) defense-related traits, (3) ionomic traits, and (4) developmental-related traits. The second data set (80 genomes data) includes 80 accessions from the first phase of the 1001 Genomes Project in Arabidopsis (Cao et al., 2011). SNP data were retrieved from the original genome matrix from the 1001 Genomes website. All singletons and SNPs with incomplete information were removed, which resulted in a final subset of 1,438,752 SNPs. Third, we included 1135 samples and 6,973,565 non-singleton SNPs (1001 Genomes Data) from the final phase of the 1001 Genomes Project (1001 Genomes Consortium, 2016). Lastly, we included 372 in silico F1 hybrid genotypes generated from parental genome sequences (Cao et al., 2011) with a total of 204,753 SNPs (Seymour et al., 2016). We also integrated the TAIR9 and TAIR10 gene annotation sets.
For Drosophila, we integrated the Drosophila Genetic Reference Panel (DGRP) with a total number of 172 samples, 2,476,799 SNPs, and three phenotypes split into male and female, making a total of six (Harbison et al., 2004; Morgan and Mackay, 2006; Jordan et al., 2007; Mackay et al., 2012). Missing SNPs in the Drosophila genome are imputed using a majority allele imputation. Gene annotations were downloaded from the FlyBase website and integrated into easyGWAS (Attrill et al., 2016).
For the species P. pacificus, a total of 149 samples with 2,135,350 SNPs were integrated (McGaughran et al., 2016). The data set comes with two binary and one categorical phenotypes, as well as four categorical covariates.
Availability and Requirements of easyGWAS
The easyGWAS web application requires a modern internet browser that supports HTML5 and JavaScript (e.g., Google Chrome 47.0, Firefox 43.0). The web application can be accessed online at https://easygwas.ethz.ch.
Code for the algorithmically part of easyGWAS can be freely downloaded at https://github.com/dominikgrimm/easyGWASCore.
Data and Experimental Settings for the Arabidopsis Case Study
Phenotype Scoring
Seeds for 1135 Arabidopsis accessions (1001 Genomes Consortium, 2016) were surface-sterilized in 95% ethanol for 5 min and allowed to air-dry. After 6 d of stratification in the dark at 4°C in 0.1% agarose, seeds were distributed across 4800 pots as four replicates in a randomized block design, with each replicate corresponding to one block. Plants were grown in controlled growth chambers with the following settings: 16 h light/8 h darkness, 16°C constant temperature, 65% humidity. All trays within a block were moved to a new shelf and rotated 180°C every other day to minimize position effects. Flowering time was scored as days until the emergence of visible flowering buds in the center of the rosette from time of sowing (DTF1), days until the inflorescence stem elongated to 1 cm (DTF2), and days until first open flower (DTF3). For some accessions, the inflorescence stem did not reach 1 cm before scoring of DTF3. In addition, rosette leaf number (RL), cauline leaf number (CL), diameter of rosette (end point, after flowering) (diameter), rosette branch number (RBN), cauline leaf number (CBN), and length of main flowering stem (Length) were recorded (Supplemental Table 2).
Genome-Wide Association Mapping
All phenotypes were uploaded for the 1001 Genomes Arabidopsis data set (Supplemental Text 1 or online FAQ). GWAS were conducted for all phenotypes and the 1001 Genomes data set using the easyGWAS wizard (Supplemental Text 2 or online FAQ). All phenotypes were Box-Cox transformed (Box and Cox, 1964) to improve the measurements’ normality, except for RBN (no transformation) and CBN (square root transformation) (Supplemental Table 6). A minor allele frequency filter of 5% was applied for all experiments and a standard additive genotype encoding was chosen. To account for confounding due to population stratification and cryptic relatedness, we used FaST-LMM (Lippert et al., 2011), estimating the genetic similarity between all genotypes by computing the realized relationship kinship matrix (Hayes et al., 2009). All results are stored in a publicly accessible GWAS project and can be found at https://easygwas.ethz.ch/gwas/myhistory/public/14/.
Intersection Analysis of Genome-Wide Association Experiments
Results of all nine conducted GWAS experiments were compared using the easyGWAS comparison wizard to identify shared associations between phenotypes. Results are stored in a publicly available project and can be accessed via https://easygwas.ethz.ch/comparison/results/manhattan/view/2c8da231-96ff-4f28-a17e-fd0e3510d8e1/.
Accession Numbers
The following Arabidopsis Genome Initiative locus identifiers have been reported: DOG1 (At5g45830), FLC (At5G10140), FRIGIDA (At4G00650), ANL2 (At4G00730), FT (AT1G65480), ATPS1 (AT1G34355), and PINOID (AT2G34650). Sample information can be found online: https://easygwas.ethz.ch/data/public/samples/1/7/. All performed GWAS for the case study can be found at https://easygwas.ethz.ch/gwas/myhistory/public/14/.
Supplemental Data
Supplemental Figure 1. Public and Private Data Repository.
Supplemental Figure 2. Data Repository and Detailed Species View.
Supplemental Figure 3. Detailed Sample View.
Supplemental Figure 4. Detailed Phenotype View.
Supplemental Figure 5. Download Manager.
Supplemental Figure 6. Upload Manager.
Supplemental Figure 7. GWAS Wizard, Steps to Normalize Phenotypes.
Supplemental Figure 8. Temporary History.
Supplemental Figure 9. Save Experiments Permanently into GWAS Projects.
Supplemental Figure 10. easyGWAS Data Sharing Dialog.
Supplemental Figure 11. GWAS Project Publishing Inquiry Form.
Supplemental Figure 12. Schematics of the easyGWAS Architecture.
Supplemental Figure 13. Runtime Comparison between State-of-the-Art Tools and easyGWAS.
Supplemental Figure 14. Noncoding RNAs Downstream of FT (AT1G65480).
Supplemental Table 1. Case Study Results in Arabidopsis thaliana Using Bonferroni.
Supplemental Table 2. Phenotype Information for the Case Study.
Supplemental Table 3. Pitfalls When Conducting Intersection Analyses or Meta-Analyses of GWAS Results.
Supplemental Table 4. Available Genotype Encodings.
Supplemental Table 5. Transformation Methods.
Supplemental Table 6. Phenotype Information for the Case Study.
Supplemental Text 1. easyGWAS Data Repository.
Supplemental Text 2. easyGWAS Wizard.
Supplemental Text 3. easyGWAS GWAS History.
Supplemental Text 4. Step-by-Step Procedures to Reproduce the Content of Figures.
Supplemental Text 5. Tips on How to Access Certain Panels.
Supplemental Text 6. Variance Explained for Linear Mixed Models.
Supplemental Text 7. Procedure to Measure Dependence Between Phenotype and Population Structure.
Supplemental Text 8. Transformation Methods.
Supplemental Text 9. Meta-Analysis.
Supplemental Text 10. Correction Methods for Multiple Hypothesis Testing.
Supplemental Text 11. REST Interface.
Supplemental Data Set 1. Significantly Associated Phenotypes.
Supplementary Material
Acknowledgments
This study was funded by the Deutsche Forschungsgemeinschaft (SPP1529 ADAPTOMICS; D.W.), the ERC (AdG IMMUNEMESIS; D.W.), the Max Planck Society (D.W. and K.M.B.), and the Alfried Krupp Prize for Young University Teachers of the Alfried Krupp von Bohlen und Halbach-Stiftung (K.M.B.). We thank the IT departments at the Max Planck Campus Tübingen and the ETH Zürich for hosting easyGWAS and for their continued support.
AUTHOR CONTRIBUTIONS
D.G.G. and K.M.B. conceived and designed the study. D.G.G. implemented the back and front end of the web application. D.R., S.K., and B.G. contributed to the development of various modules. P.A.S. measured the phenotypes and contributed to the data analysis. W.Z. and C. Liu contributed the analysis of the biological results. O.S. and C. Lippert contributed to the development of the statistical analysis pipeline. D.G.G. performed all computational experiments for the case study. D.W. and B.S. gave statistical and biological advice for the development of the web application and the conducted experiments. D.G.G., D.R., and K.M.B. wrote the article with input from all authors.
Glossary
- GWAS
genome-wide association studies
- SNP
single nucleotide polymorphism
- GWA
genome-wide association
- LD
linkage disequilibrium
- LMM
linear mixed model
Footnotes
Articles can be viewed without a subscription.
References
- 1001 Genomes Consortium (2016). 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdi H. (2007). The Bonferonni and Šidák corrections for multiple comparisons. Encycl. Meas. Stat. 3: 103–107. [Google Scholar]
- Aho K., Derryberry D., Peterson T. (2014). Model selection for ecologists: the worldviews of AIC and BIC. Ecology 95: 631–636. [DOI] [PubMed] [Google Scholar]
- Andreassen O.A., et al. ;. International Consortium for Blood Pressure GWAS; Diabetes Genetics Replication and Meta-analysis Consortium; Psychiatric Genomics Consortium Schizophrenia Working Group (2013). Improved detection of common variants associated with schizophrenia by leveraging pleiotropy with cardiovascular-disease risk factors. Am. J. Hum. Genet. 92: 197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariel F., Jegu T., Latrasse D., Romero-Barrios N., Christ A., Benhamed M., Crespi M. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55: 383–396. [DOI] [PubMed] [Google Scholar]
- Attrill H., Falls K., Goodman J.L., Millburn G.H., Antonazzo G., Rey A.J., Marygold S.J.; FlyBase Consortium (2016). FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 44 (D1): D786–D792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S., et al. (2010). Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azencott C.-A., Grimm D., Sugiyama M., Kawahara Y., Borgwardt K.M. (2013). Efficient network-guided multi-locus association mapping with graph cuts. Bioinformatics 29: i171–i179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57: 289–300. [Google Scholar]
- Benjamini Y., Yekutieli D. (2001). The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 29: 1165–1188. [Google Scholar]
- Borenstein M., Hedges L.V., Higgins J.P.T., Rothstein H.R. (2010). A basic introduction to fixed-effect and random-effects models for meta-analysis. Res. Synth. Methods 1: 97–111. [DOI] [PubMed] [Google Scholar]
- Borenstein M., Hedges L.V., Higgins J.P.T., Rothstein H.R. (2011). Introduction to Meta-Analysis. (Chichester, UK: John Wiley & Sons; ). [Google Scholar]
- Box G.E.P., Cox D.R. (1964). An analysis of transformations. J. R. Stat. Soc. B 26: 211–252. [Google Scholar]
- Burnham K.P., Anderson D.R. (2002). Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach. (New York: Springer Science & Business Media; ). [Google Scholar]
- Bush W.S., Moore J.H. (2012). Chapter 11: Genome-wide association studies. PLOS Comput. Biol. 8: e1002822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao J., et al. (2011). Whole-genome sequencing of multiple Arabidopsis thaliana populations. Nat. Genet. 43: 956–963. [DOI] [PubMed] [Google Scholar]
- Chasman D.I., et al. (2011). Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat. Genet. 43: 695–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs L.H., Lisec J., Walther D. (2012). Matapax: an online high-throughput genome-wide association study pipeline. Plant Physiol. 158: 1534–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordell H.J., Clayton D.G. (2005). Genetic association studies. Lancet 366: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Cubillos F.A., et al. (2012). Expression variation in connected recombinant populations of Arabidopsis thaliana highlights distinct transcriptome architectures. BMC Genomics 13: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B., Roeder K. (1999). Genomic control for association studies. Biometrics 55: 997–1004. [DOI] [PubMed] [Google Scholar]
- Evangelou E., Ioannidis J.P.A. (2013). Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet. 14: 379–389. [DOI] [PubMed] [Google Scholar]
- Filiault D.L., Maloof J.N. (2012). A genome-wide association study identifies variants underlying the Arabidopsis thaliana shade avoidance response. PLoS Genet. 8: e1002589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R.A. (1925). Statistical Methods for Research Workers. (Genesis Publishing; ). [Google Scholar]
- Franke A., et al. (2010). Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat. Genet. 42: 1118–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freilinger T., et al. ; International Headache Genetics Consortium (2012). Genome-wide association analysis identifies susceptibility loci for migraine without aura. Nat. Genet. 44: 777–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harbison S.T., Yamamoto A.H., Fanara J.J., Norga K.K., Mackay T.F.C. (2004). Quantitative trait loci affecting starvation resistance in Drosophila melanogaster. Genetics 166: 1807–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hastie, T., Tibshirani, R., Friedman, J. (2009). The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed. (New York: Springer). [Google Scholar]
- Hayes B.J., Visscher P.M., Goddard M.E. (2009). Increased accuracy of artificial selection by using the realized relationship matrix. Genet. Res. 91: 47–60. [DOI] [PubMed] [Google Scholar]
- Horton M.W., et al. (2012). Genome-wide patterns of genetic variation in worldwide Arabidopsis thaliana accessions from the RegMap panel. Nat. Genet. 44: 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo H., Wei S., Bradford K.J. (2016). DELAY OF GERMINATION1 (DOG1) regulates both seed dormancy and flowering time through microRNA pathways. Proc. Natl. Acad. Sci. USA 113: E2199–E2206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J.A., Soumpourou E., Lister C., Ligthart J.-D., Kennedy S., Dean C. (2016). Nucleotide polymorphism affecting FLC expression underpins heading date variation in horticultural brassicas. Plant J. 87: 597–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan K.W., Carbone M.A., Yamamoto A., Morgan T.J., Mackay T.F. (2007). Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biol. 8: R172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., Sul J.H., Service S.K., Zaitlen N.A., Kong S.Y., Freimer N.B., Sabatti C., Eskin E. (2010). Variance component model to account for sample structure in genome-wide association studies. Nat. Genet. 42: 348–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H.M., Zaitlen N.A., Wade C.M., Kirby A., Heckerman D., Daly M.J., Eskin E. (2008). Efficient control of population structure in model organism association mapping. Genetics 178: 1709–1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A., et al. (2010). Fine mapping in 94 inbred mouse strains using a high-density haplotype resource. Genetics 185: 1081–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivova A., Giovannetti M., Baraniecka P., Lee B.-R., Grondin C., Loudet O., Kopriva S. (2013). Natural variation in the ATPS1 isoform of ATP sulfurylase contributes to the control of sulfate levels in Arabidopsis. Plant Physiol. 163: 1133–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A., Vilhjálmsson B.J., Segura V., Platt A., Long Q., Nordborg M. (2012). A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat. Genet. 44: 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo H., Hayashi K. (2011). Characterization of root cells of anl2 mutant in Arabidopsis thaliana. Plant Sci. 180: 679–685. [DOI] [PubMed] [Google Scholar]
- Li J.-Y., Wang J., Zeigler R.S. (2014). The 3,000 rice genomes project: new opportunities and challenges for future rice research. Gigascience 3: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Filiault D., Box M.S., Kerdaffrec E., van Oosterhout C., Wilczek A.M., Schmitt J., McMullan M., Bergelson J., Nordborg M., Dean C. (2014). Multiple FLC haplotypes defined by independent cis-regulatory variation underpin life history diversity in Arabidopsis thaliana. Genes Dev. 28: 1635–1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Tao Z., Dean C. (2015). Phenotypic evolution through variation in splicing of the noncoding RNA COOLAIR. Genes Dev. 29: 696–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D.Y., Sullivan P.F. (2009). Meta-analysis of genome-wide association studies with overlapping subjects. Am. J. Hum. Genet. 85: 862–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin T., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet. 46: 1220–1226. [DOI] [PubMed] [Google Scholar]
- Lippert C., Casale F.P., Rakitsch B., Stegle O. (2014). LIMIX: genetic analysis of multiple traits. bioRxiv, 10.1101/003905. [Google Scholar]
- Lippert C., Listgarten J., Liu Y., Kadie C.M., Davidson R.I., Heckerman D. (2011). FaST linear mixed models for genome-wide association studies. Nat. Methods 8: 833–835. [DOI] [PubMed] [Google Scholar]
- Liu J., Jung C., Xu J., Wang H., Deng S., Bernad L., Arenas-Huertero C., Chua N.H. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24: 4333–4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Adrian J., Pankin A., Hu J., Dong X., von Korff M., Turck F. (2014). Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat. Commun. 5: 4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Wang L., Mao S., Liu K., Lu Y., Wang J., Wei Y., Zheng Y. (2015). Genome-wide association study of 29 morphological traits in Aegilops tauschii. Sci. Rep. 5: 15562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llinares-López F., Grimm D.G., Bodenham D.A., Gieraths U., Sugiyama M., Rowan B., Borgwardt K. (2015). Genome-wide detection of intervals of genetic heterogeneity associated with complex traits. Bioinformatics 31: i240–i249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q., et al. (2013). Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat. Genet. 45: 884–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay T.F., et al. (2012). The Drosophila melanogaster genetic reference panel. Nature 482: 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaughran A., Rödelsperger C., Grimm D.G., Meyer J.M., Moreno E., Morgan K., Leaver M., Serobyan V., Rakitsch B., Borgwardt K.M., Sommer R.J. (2016). Genomic profiles of diversification and genotype-phenotype association in island nematode lineages. Mol. Biol. Evol. 33: 2257–2272. [DOI] [PubMed] [Google Scholar]
- McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R., Thormann A., Flicek P., Cunningham F. (2016). The Ensembl variant effect predictor. Genome Biol. 17: 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., Cunningham F. (2010). Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 26: 2069–2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijón M., Satbhai S.B., Tsuchimatsu T., Busch W. (2014). Genome-wide association study using cellular traits identifies a new regulator of root development in Arabidopsis. Nat. Genet. 46: 77–81. [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B., Gomaa N.H., Alonso-Blanco C., Picó F.X. (2013). Among- and within-population variation in flowering time of Iberian Arabidopsis thaliana estimated in field and glasshouse conditions. New Phytol. 197: 1332–1343. [DOI] [PubMed] [Google Scholar]
- Méndez-Vigo B., Savic M., Ausín I., Ramiro M., Martín B., Picó F.X., Alonso-Blanco C. (2016). Environmental and genetic interactions reveal FLOWERING LOCUS C as a modulator of the natural variation for the plasticity of flowering in Arabidopsis. Plant Cell Environ. 39: 282–294. [DOI] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13: 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan T.J., Mackay T.F.C. (2006). Quantitative trait loci for thermotolerance phenotypes in Drosophila melanogaster. Heredity (Edinb.) 96: 232–242. [DOI] [PubMed] [Google Scholar]
- Nordborg M., Weigel D. (2008). Next-generation genetics in plants. Nature 456: 720–723. [DOI] [PubMed] [Google Scholar]
- Pickrell J.K., Berisa T., Liu J.Z., Ségurel L., Tung J.Y., Hinds D.A. (2016). Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 48: 709–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau C., Loubet S., Lefoulon C., Chalies C., Fizames C., Lacombe B., Ferrand M., Loudet O., Berthomieu P., Richard O. (2012). Natural variation at the FRD3 MATE transporter locus reveals cross-talk between Fe homeostasis and Zn tolerance in Arabidopsis thaliana. PLoS Genet. 8: e1003120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 38: 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A.R., Bender D., Maller J., Sklar P., de Bakker P.I.W., Daly M.J., Sham P.C. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakitsch B., Lippert C., Stegle O., Borgwardt K. (2013). A Lasso multi-marker mixed model for association mapping with population structure correction. Bioinformatics 29: 206–214. [DOI] [PubMed] [Google Scholar]
- Sanchez-Bermejo E., Balasubramanian S. (2016). Natural variation involving deletion alleles of FRIGIDA modulate temperature-sensitive flowering responses in Arabidopsis thaliana. Plant Cell Environ. 39: 1353–1365. [DOI] [PubMed] [Google Scholar]
- Schmitz R.J., Schultz M.D., Urich M.A., Nery J.R., Pelizzola M., Libiger O., Alix A., McCosh R.B., Chen H., Schork N.J., Ecker J.R. (2013). Patterns of population epigenomic diversity. Nature 495: 193–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz C., Balasubramanian S., Warthmann N., Michael T.P., Lempe J., Sureshkumar S., Kobayashi Y., Maloof J.N., Borevitz J.O., Chory J., Weigel D. (2009). Cis-regulatory changes at FLOWERING LOCUS T mediate natural variation in flowering responses of Arabidopsis thaliana. Genetics 183: 723–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz G.E. (1978). Estimating the dimension of a model. Ann. Stat. 6: 461–464. [Google Scholar]
- Scott L.J., et al. (2007). A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 316: 1341–1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü., Grimm D., Fitz J., Weigel D., Nordborg M., Borgwardt K., Korte A. (2016). AraPheno: a public database for Arabidopsis thaliana phenotypes. Nucleic Acids Res. pii: gkw986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seren Ü., Vilhjálmsson B.J., Horton M.W., Meng D., Forai P., Huang Y.S., Long Q., Segura V., Nordborg M. (2012). GWAPP: a web application for genome-wide association mapping in Arabidopsis. Plant Cell 24: 4793–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour D.K., Chae E., Grimm D.G., Martín Pizarro C., Habring-Müller A., Vasseur F., Rakitsch B., Borgwardt K.M., Koenig D., Weigel D. (2016). Genetic architecture of nonadditive inheritance in Arabidopsis thaliana hybrids. Proc. Natl. Acad. Sci. USA 113: E7317–E7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro S.S., Wilk M.B. (1965). An analysis of variance test for normality (complete samples). Biometrika 52: 591–611. [Google Scholar]
- Storey J.D., Tibshirani R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 100: 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stouffer, S.A., Suchman, E.A., Devinney, L.C., Star, S.A., Williams, R.M., Jr. (1949). The American Soldier: Adjustment during Army Life. Studies in Social Psychology in World War II, Vol. 1. (Oxford, UK: Princeton University Press). [Google Scholar]
- Sugiyama M., Azencott C., Grimm D., Kawahara Y., Borgwardt K. (2014). Multi-task feature selection on multiple networks via maximum flows. In Proceedings of the 2014 SIAM International Conference on Data Mining. (Society for Industrial and Applied Mathematics; ), pp. 199–207. [Google Scholar]
- Widmer C., Lippert C., Weissbrod O., Fusi N., Kadie C., Davidson R., Listgarten J., Heckerman D. (2014). Further improvements to linear mixed models for genome-wide association studies. Sci. Rep. 4: 6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates A., Beal K., Keenan S., McLaren W., Pignatelli M., Ritchie G.R., Ruffier M., Taylor K., Vullo A., Flicek P. (2014). The Ensembl REST API: ensembl data for any language. Bioinformatics 31: 143–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin D.V., Kozbur D.O. (2010). P-value based analysis for shared controls design in genome-wide association studies. Genet. Epidemiol. 34: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., Aranzana M.J., Kim S., Lister C., Shindo C., Tang C., Toomajian C., Zheng H., Dean C., Marjoram P., Nordborg M. (2007). An Arabidopsis example of association mapping in structured samples. PLoS Genet. 3: e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K., et al. (2011). Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa. Nat. Commun. 2: 467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.