Abstract
We report a set of brominated luciferins for bioluminescence imaging. These regioisomeric scaffolds were accessed using a common synthetic route. All analogs produced light with firefly luciferase, although varying levels of emission were observed. Differences in photon output were analyzed via computation and photophysical measurements. The “brightest” brominated luciferin was further evaluated in cell and animal models. At low doses, the analog outperformed the native substrate in cells. The remaining luciferins, while weak emitters with firefly luciferase, were inherently capable of light production and thus potential substrates for orthogonal mutant enzymes.
Keywords: luciferin, bioluminescence, imaging, Appel’s salt, luciferase
Graphical Abstract
Recent years have seen a surge of interest in accessing novel luciferins for bioluminescence imaging (BLI).[1–5] BLI relies on light generation via luciferase enzymes and luciferin small molecules.[6] These probes are routinely used in vitro and in vivo for monitoring diverse biological processes.[7] However, a limited supply of robust, light-emitting scaffolds has stymied efforts to visualize cellular communication and other multicomponent processes in vivo. We and others are attempting to fill this void with new luciferin architectures.[7,8] To date, these efforts have focused on analogs of D-luciferin, the native light-emitting substrate for firefly luciferase (Fluc, Figure 1A).[1,4]6] Fluc can tolerate a variety of modified substrates, although most are not processed as efficiently as D-luciferin, resulting in reduced photon production.[4,5,9] Improved light outputs have been achieved, in some cases, using mutant luciferases.[10,11]
Figure 1.
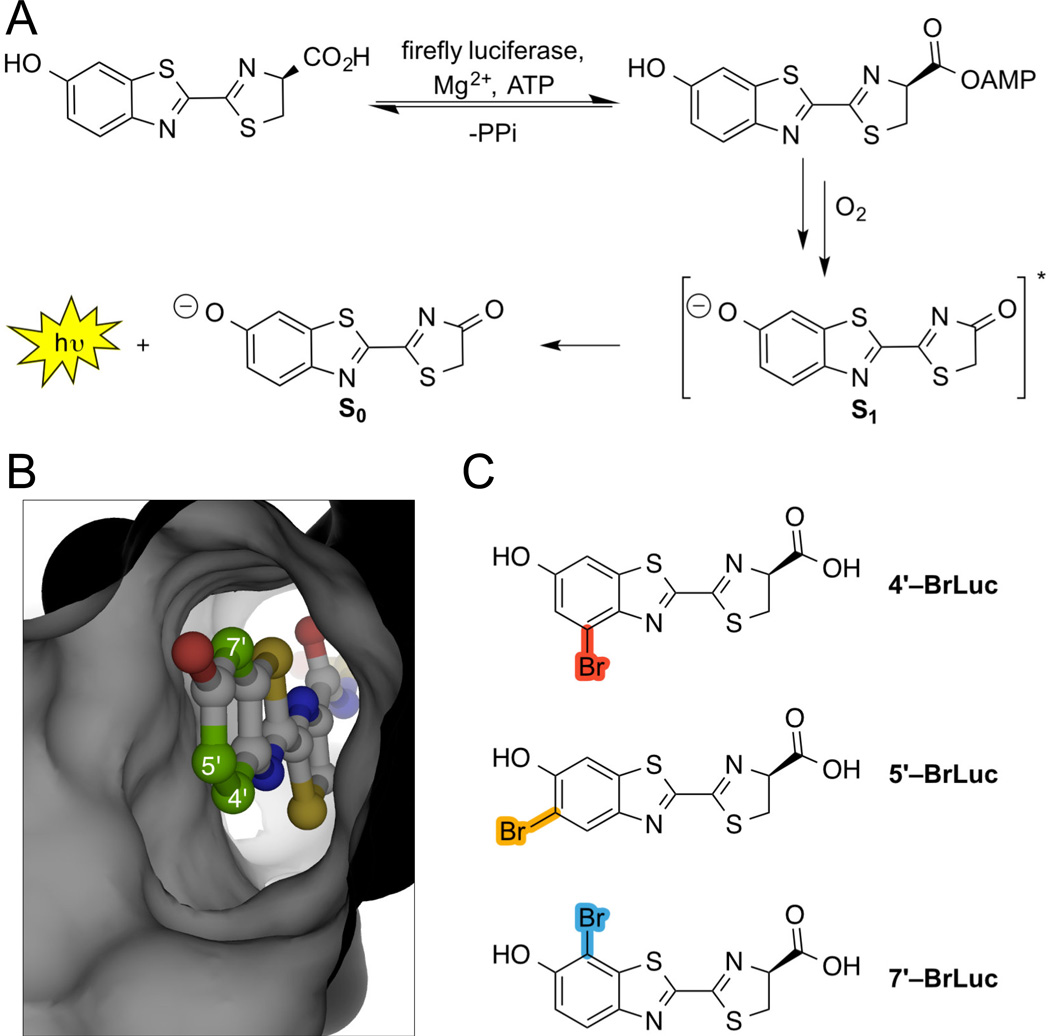
Bioluminescence imaging with luciferases and luciferins. (A) Fluc catalyzes the oxidation of D-luciferin. The reaction proceeds through an excited state (S1) oxyluciferin intermediate. Relaxation to the ground state (S0) results in photon release. (B) Crystal structure of a luciferin adenylate bound to Fluc (PDB: 4G36). Targeted positions on the luciferin core are highlighted. (C) Brominated analogs investigated in this study.
Efforts to develop new bioluminescent tools would benefit from a broad assortment of “bright” luciferins. Historically, such scaffolds have been difficult to access owing to their densely functionalized cores. Late-stage modifications to D-luciferin are also non-trivial, preventing the rapid production of new analogs. To address these issues, we aimed to prepare the bromo-substituted scaffolds (4′–BrLuc, 5′–BrLuc, and 7′–BrLuc) shown in Figure 1. Bromo groups would impart a modest steric perturbation to the luciferin core, but could serve as versatile chemical handles for donwstream cross-coupling reactions and novel probe development.[12–14] The luciferins would also likely be tolerated by Fluc or related mutants. We envisioned that the C5′ bromo appendage would “fit” into the active site, as a relatively large enzyme pocket juxtaposes this position (Figure 1B).[15] Fluc can also process many 5′-substituted luciferins.[16–19] Bromo groups at C4′ and C7′, by contrast, could present a steric barrier to Fluc utilization as these positions lie in close proximity to the enzyme backbone. Such substrates would be excellent candidates for engineered (orthogonal) luciferases, though, provided that they are capable of robust light emission.
Our selection of bromo substituents was predicated on the functional groups not interfering with photon production. To assess this parameter, we employed time-dependent density functional theory[20] (TDDFT) to analyze the adiabatic emissions of relevant oxyluciferins from the excited state (S1) to the ground state (S0). The performance of TDDFT for computing the intensities of such emissions is well established: the oscillator strength is proportional to the spontaneous emission rate from S1 and thus tracks with light output.[21,22] When we calculated oscillator strengths for a variety of known luciferins, we found that the values correlated with reported bioluminescent outputs (Table 1). For example, D-luciferin analogs lacking electron-donating groups at the 6′ position (and thus poor emitters) were predicted to have low oscillator strengths.[23–27] By contrast, analogs with 6′–amino substituents—and known light emitters—were predicted to have oscillator strengths on par with the native substrate. TDDFT analyses performed on 4′–BrLuc, 5′–BrLuc, and 7′–BrLuc suggested that the bromo substituents would minimally impact bioluminescent output (Table 1).
Table 1.
Comparison of calculated oscillator strengths and bioluminescence emission intensities.
| Compound | Name | Oscillator Strengtha |
Rel. BLIb |
|---|---|---|---|
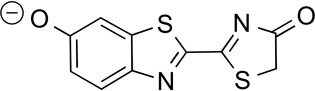 |
D–Luc | 100 | 100 |
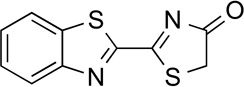 |
6'–deoxyLuc | 1.4 | <0.01c |
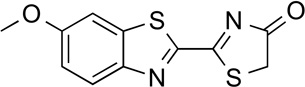 |
6'–methoxyLuc | 57.2 | <0.01 |
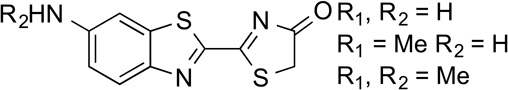 |
6'–aminoLuc 6'–MeNHLH2 6'–Me2NLH2 |
91.9 94.9 102.9 |
61 101 1 |
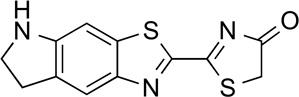 |
CycLuc1 | 127.6 | 38 |
 |
4'–BrLuc | 86.1 | 3.1 |
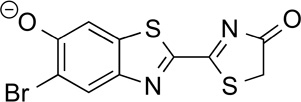 |
5'–BrLuc | 104 | 46.0 |
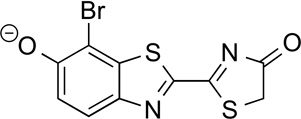 |
7'–BrLuc | 79.3 | 3.4 |
Calculated as a theoretical maximum.
Bioluminescence was measured using 100 µM luciferin and 1 µg Fluc.
No signal observed.
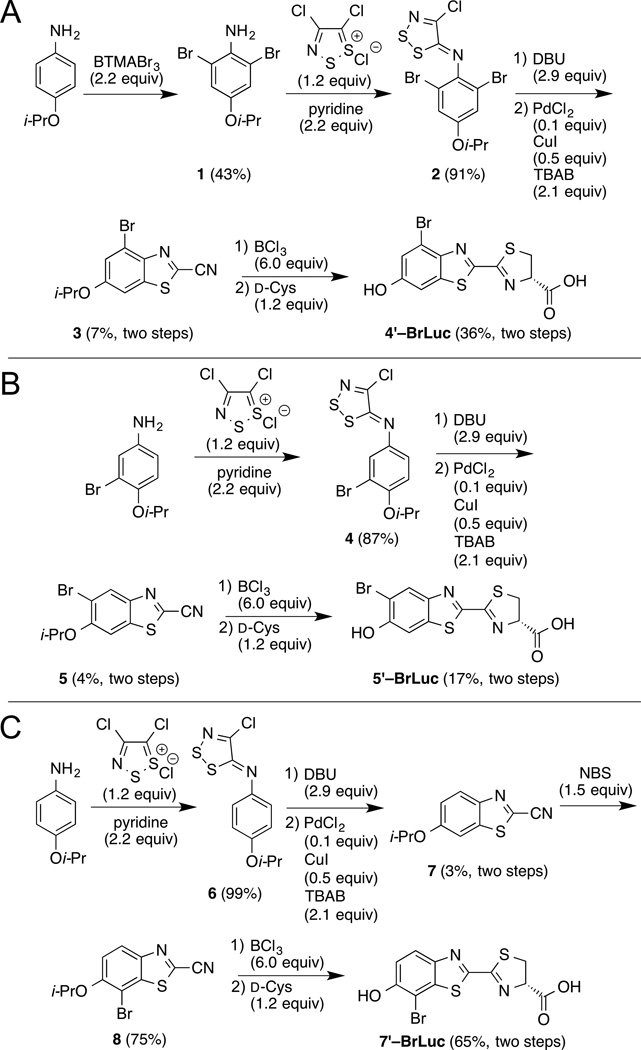
Encouraged by these results, we set out to synthesize the target brominated luciferins. The desired scaffolds all comprised benzothiazole cores; such structures are readily accessible from anilines and Appel’s salt chemistry (Scheme 1).[28] En route to 4′–BrLuc, the dibrominated aniline 1 was first condensed with Appel’s salt. The resulting dithiazole (2) was then fragmented and cyclized to provide cyanobenzothiazole 3 (Scheme 1A). The benzothiazole precursors to 5′–BrLuc and 7′–BrLuc were prepared analogously (Schemes 1B–C). In the latter case, the bromine substituent was installed post-cyclization. The desired luciferins 4′–BrLuc, 5′–BrLuc, and 7′–BrLuc were ultimately accessed via D-cysteine condensations with the appropriate cyanobenzothiazoles.
Scheme 1.
Synthesis of brominated luciferin analogs.
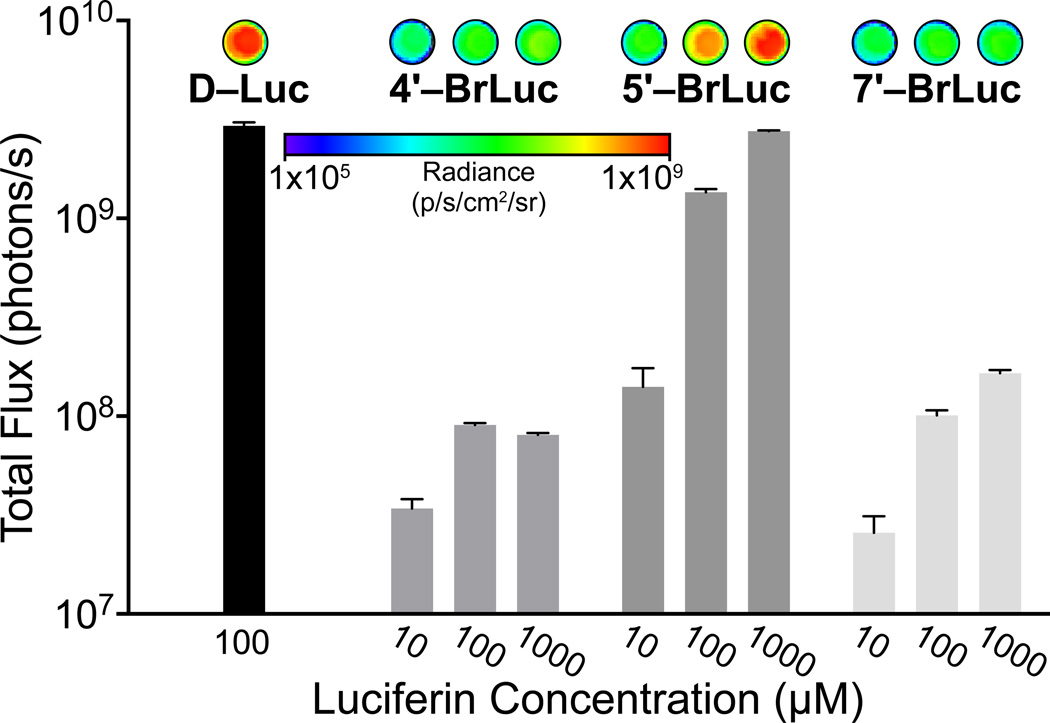
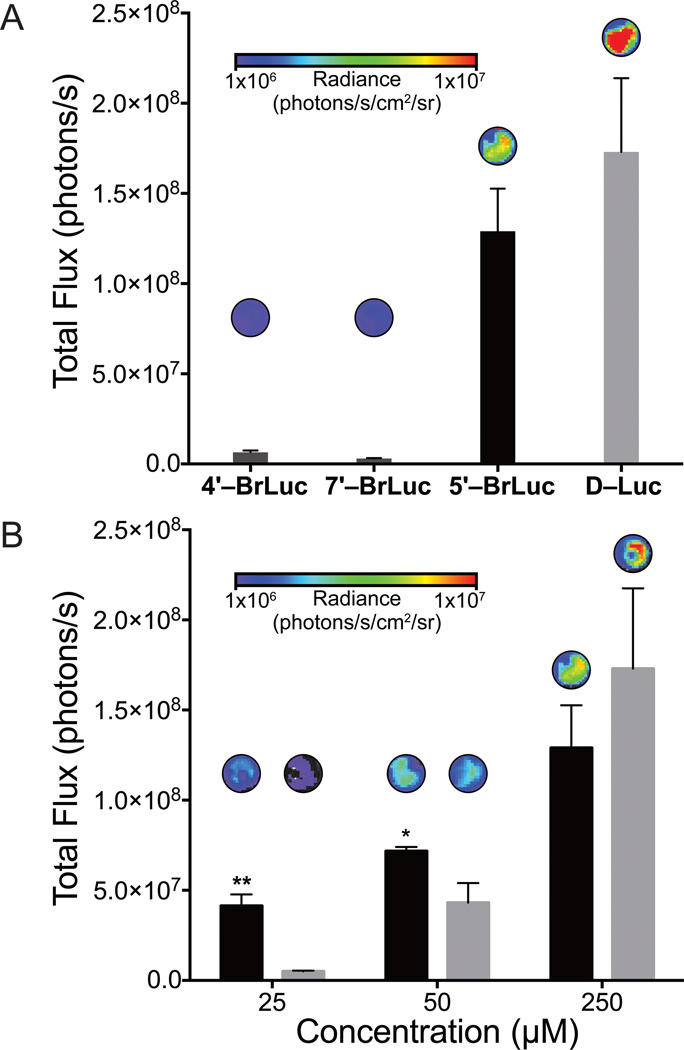
With the analogs in hand, we analyzed their light-emitting properties. The compounds were incubated with recombinant Fluc and bioluminescent photon production was measured. As shown in Figure 2, all analogs produced light in a dose-dependent fashion. Compared to the native substrate, though, only 5′–BrLuc exhibited robust levels of emission. As noted earlier, steric clashes may prevent efficient processing of 4′–BrLuc and 7′–BrLuc (Table 2). Interestingly, the apparent Km for 4′–BrLuc was on par with that for D-Luc, but the relative kcat was 50-fold reduced. This result indicates that the halogen atom at this position may have room to dock, but might interfere with catalysis. The enzymatic parameters for 5′–BrLuc, as expected, were similar to those of the native substrate. Bioluminescence spectra for the brominated luciferins were also red-shifted from D–Luc, while the fluorescence spectra were virtually identical (Figure S1). These results indicate that the analogs can access alternate excited state geometries and relaxation pathways in the Fluc active site.
Figure 2.
Differential bioluminescent photon production is observed with bromo luciferins and recombinant Fluc. Sample bioluminescence images are shown. Error bars represent the standard deviation of the mean for n = 3 experiments. Signal at 105 photons/s represents background luminescence.
Table 2.
Enzymatic parameters for Fluc-catalyzed light emission.
| Compound | Apparent Km (µM) | Relative kcata |
|---|---|---|
| D–Luc | 3.07 ± 0.80 | 100 ± 0.3 |
| 4'–BrLuc | 2.47 ± 0.92 | 1.8 ± 0.4 |
| 5'–BrLuc | 6.72 ± 0.31 | 104 ± 2.2 |
| 7'–BrLuc | 17.3 ± 8.8 | 1.9 ± 0.4 |
Expressed as percent relative to D–Luc
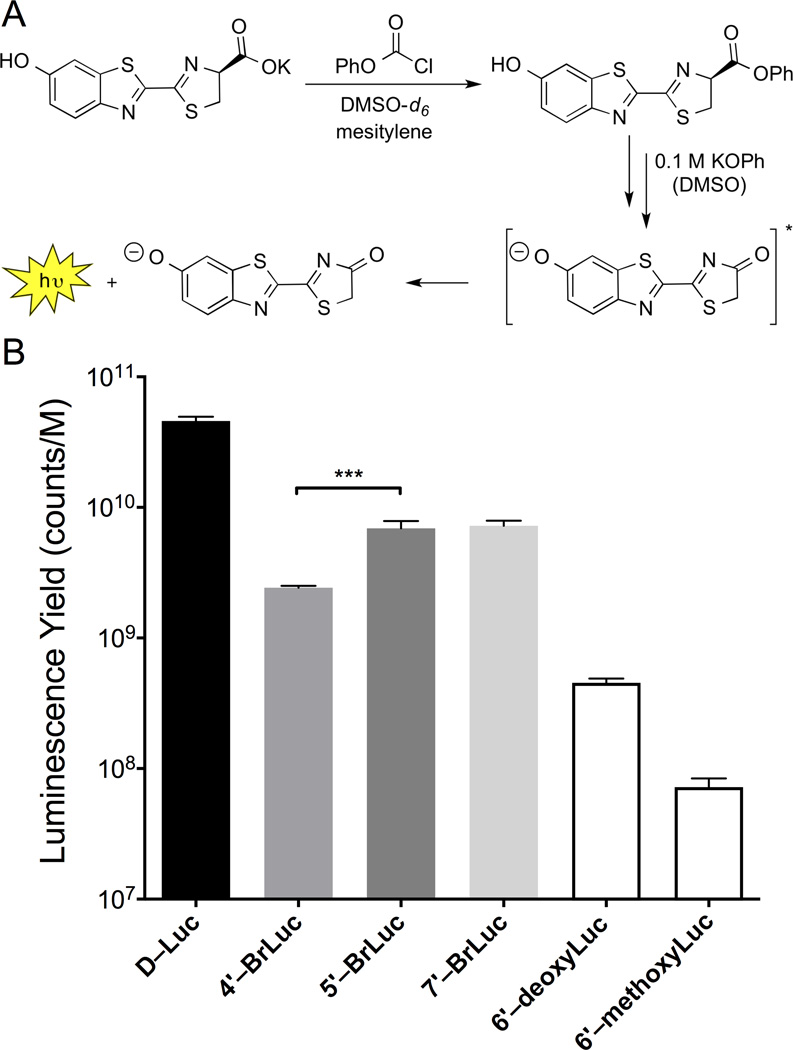
While photon measurements suggested that 5′–BrLuc was the best Fluc substrate, it was possible that the 4′ and 7′ isomers were just inherently weaker emitters (as suggested by TDDFT calculations, Table 1). To explore this possibility, we used a traditional chemiluminescence assay to measure each analog’s intrinisic ability to produce light (Figure 3).[29–31] This non-enzymatic process mimics the Fluc reaction itself, involving the formation of an activated ester intermediate, followed by oxidation (Figures S2–S3).[32] When the brominated analogs were subjected to the assay, light emission was observed at levels above 6′–deoxyLuc and 6′–methoxyLuc (known poor light emitters). More specifically, 5′–BrLuc and 7′–BrLuc exhibited nearly identical levels of chemiluminescence (Figure 3), while the photon emission value for 4′–BrLuc was slightly reduced. These results suggest that C7′- and C4′ modified luciferins are capable of bioluminescent light emission, provided that a mutant enzyme can be identified to accommodate them. It should also be noted that deviations between the computed oscillator strengths (Table 1) and the observed emission strengths (Figure 3) suggest the existence of fast, non-radiative deactivation pathways for the analogs. While an exhaustive study of such decay mechanisms is beyond our present scope, additional calculations revealed "dark" states that are energetically accessible from S1: (1) T1 triplet states that can mediate deactivation through intersystem crossing, and (2) twisted intramolecular charge-transfer (TICT) states with near-zero oscillator strengths due to broken pi-conjugation (see SI). In chemiluminescence experiments, these compounds would exhibit emission profiles from a range of torsional angles, only some of which are light emitting.
Figure 3.
Chemiluminescent light production with luciferin analogs. (A) Luciferin scaffolds were activated to form phenyl esters. Subsequent treatment with KOPh resulted in light emission. (B) Chemiluminescence observed with luciferin analogs. ***p < 0.001 (t-test).
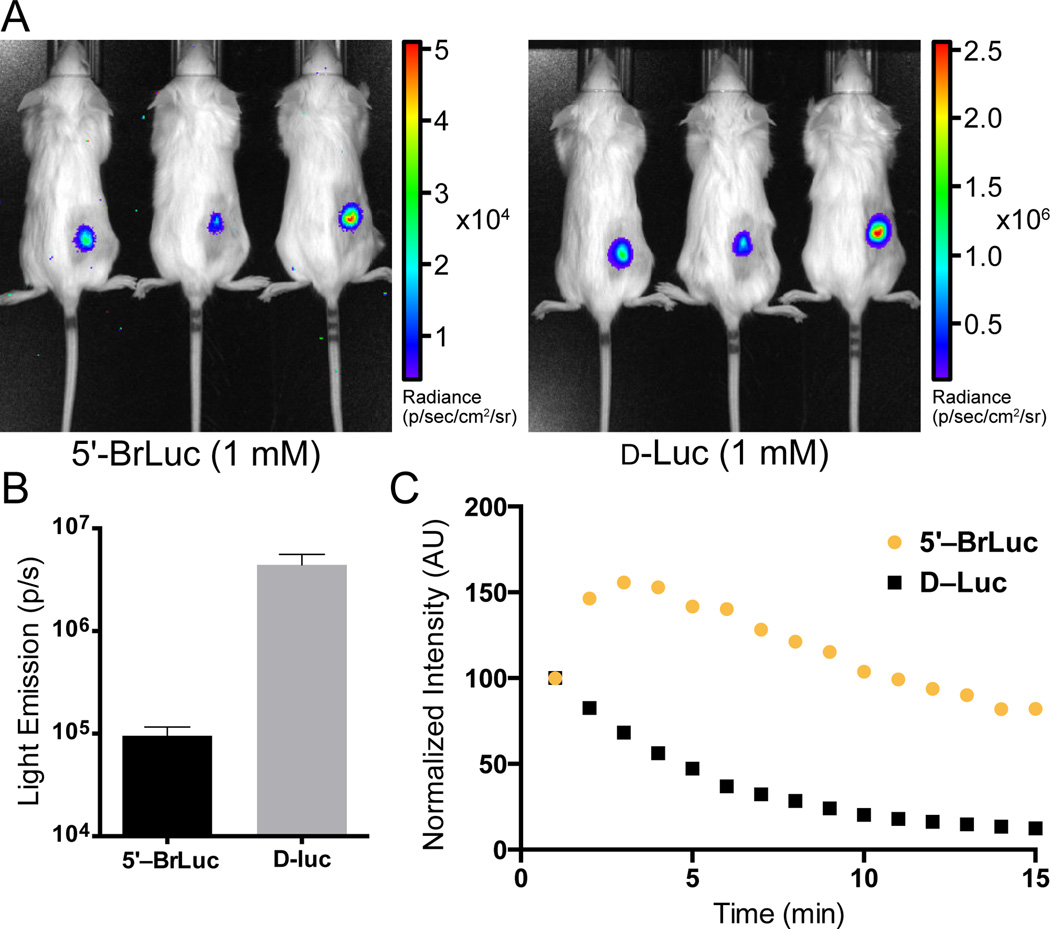
Our computational and experimental data suggested that 5′–BrLuc would be suitable for BLI in cells and tissues. Indeed, when the brominated compound was incubated with Fluc-expressing HEK293 cells, robust light emission was observed (Figure 4A). Minimal bioluminescence was produced when cultures were treated with 4′–BrLuc and 7′–BrLuc, consistent with in vitro data. Interestingly, light emission values from cells incubated with 5′–BrLuc were on par with those from D–Luc-treated cultures (Figure 4). At low doses, 5′–BrLuc even outperformed the native substrate, suggesting that the analog is more cell permeant.[[33] Similar trends were observed in vivo. When 5′–BrLuc was injected into mice bearing Fluc-expressing DB7 cells, light emission was observed (Figure 5A). The overall photon output was lower than that of a comparable dose of D-luciferin (Figure 5B). However, the light emission from mice treated with 5′–BrLuc was more sustained (Figure 5C).
Figure 4.
Differential photon production observed with Fluc-expressing cells treated with bromo luciferins. Compounds were administered to Fluc-expressing HEK293 cells (100,000 cells per well) in PBS (pH 7.4). Sample images are included above each bar. (A) Peak emission for all analogs at 100 µM. (B) Dose response comparison between 5′–BrLuc (black) and D–Luc (gray). For (A)–(B), error bars represent the standard deviation of the mean for n = 3 experiments. *p < 0.1, **p < 0.01 (t-test).
Figure 5.
In vivo imaging with a bromo luciferin analog. (A) Luciferins (100 µL of 1 mM solutions) were administered into mice bearing Fluc-expressing DB7 cells. Bioluminescent images were shown. (B) Quantification of the images shown in (A). Error bars represent the standard error in the mean for n = 3 measurements. (C) Bromo luciferin analog enables sustained imaging. Luciferins (100 µL of 1 mM solutions) were adminstered (i.v.) into a luciferase transgenic mouse and bioluminescence images were acquired over time. Data are representative of n = 3 independent experiments.
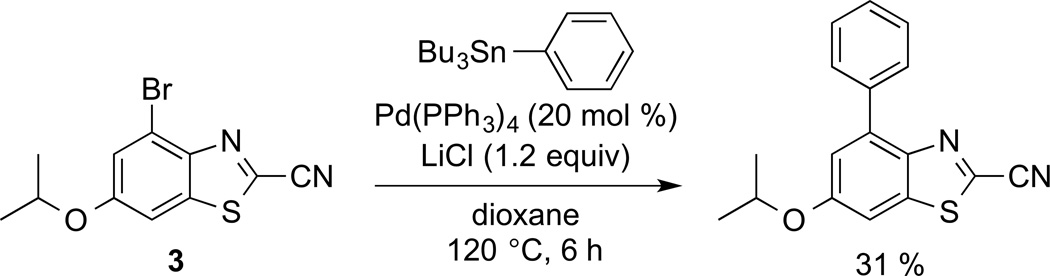
As noted previously, the brominated luciferins are useful entry points for new probe development, as they can be further modified using traditional cross-coupling reactions. As proof of principle, we used Stille coupling conditions to install a phenyl substituent on the luciferin core (Scheme 2). These conditions can be used at a late stage in luciferin synthesis, and should translate well across the analog series, enabling new families of probes to be readily accessed. We are also exploring other cross-coupling methodologies to derivatize luciferins with alkyl and other appendages, and these results will be published in due course.
Scheme 2.
Brominated luciferins can be dervativzed by Stille cross-coupling.
In conclusion, we developed a series of brominated luciferins that are versatile probes for bioluminescence. The analogs were syntheszied from a common synthetic route, and their light-emitting properties were investigated using a combination of photophysical assays. One of the probes enabled sensitive imaging in cultured cells and animals, while the others are candidates for orthogonal probe development. Importantly, these scaffolds can be further diversified to access next-generation bioluminescent tools.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (R01 GM107630 to J.A.P). and the Department of Energy (DE-SC0008694 to F.F.). C.M.R. was supported by a National Science Foundation Graduate Research Fellowship (DGE-1321846). D.C.M. was supported, in part, by a fellowship from Allergan. We thank members of the Overman, Jarvo, Esser-Kahn and Rychnovsky laboratories for providing reagents and equipment. We also thank Drs. John Greaves and Beniam Berhane for assistance with mass spec analyses. Last, we acknowledge the Laser Spectroscopy Faciltiy at University of California Irvine.
References
- 1.McCutcheon DC, Paley MA, Steinhardt RC, Prescher JA. J. Am. Chem. Soc. 2012;134:7604–7607. doi: 10.1021/ja301493d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Conley NR, Dragulescu Andrasi A, Rao J, Moerner WE. Angew. Chem. Int. Ed. 2012;51:3350–3353. doi: 10.1002/anie.201105653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kojima R, Takakura H, Ozawa T, Tada Y, Nagano T, Urano Y. Angew. Chem. Int. Ed. 2012;52:1175–1179. doi: 10.1002/anie.201205151. [DOI] [PubMed] [Google Scholar]
- 4.Mofford DM, Reddy GR, Miller SC. J. Am. Chem. Soc. 2014;136:13277–13282. doi: 10.1021/ja505795s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jathoul AP, Grounds H, Anderson JC, Pule MA. Angew. Chem. Int. Ed. 2014;53:13059–13063. doi: 10.1002/anie.201405955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraga H. Photochem. Photobiol. Sci. 2008;7:146–158. doi: 10.1039/b719181b. [DOI] [PubMed] [Google Scholar]
- 7.Paley MA, Prescher JA. MedChemComm. 2014;5:255–267. doi: 10.1039/C3MD00288H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adams ST, Miller SC. Curr. Opin. Chem. Biol. 2014;21:112–120. doi: 10.1016/j.cbpa.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuchimaru T, Iwano S, Kiyama M, Mitsumata S, Kadonosono T, Niwa H, Maki S, Kizaka-Kondoh S. Nat. Commun. 2016;7:11856. doi: 10.1038/ncomms11856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams ST, Mofford DM, Reddy GSKK, Miller SC. Angew. Chem. Int. Ed. 2016;55:4943–4946. doi: 10.1002/anie.201511350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harwood KR, Mofford DM, Reddy GR, Miller SC. Chem. Biol. 2011;18:1649–1657. doi: 10.1016/j.chembiol.2011.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Littke AF, Schwarz L, Fu GC. J. Am. Chem. Soc. 2002;124:6343–6348. doi: 10.1021/ja020012f. [DOI] [PubMed] [Google Scholar]
- 13.Farina V, Krishnamurthy V, Scott WJ. The Stille Reaction. Organic Reactions. 2004;50:1–652. [Google Scholar]
- 14.Nicolaou KC, Bulger PG, Sarlah D. Angew. Chem. Int. Ed. 2005;44:4442–4489. doi: 10.1002/anie.200500368. [DOI] [PubMed] [Google Scholar]
- 15.Sundlov JA, Fontaine DM, Southworth TL, Branchini BR, Gulick AM. Biochemistry. 2012;51:6493–6495. doi: 10.1021/bi300934s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takakura H, Kojima R, Ozawa T, Nagano T, Urano Y. ChemBioChem. 2012;13:1424–1427. doi: 10.1002/cbic.201200142. [DOI] [PubMed] [Google Scholar]
- 17.Reddy GR, Thompson WC, Miller SC. J. Am. Chem. Soc. 2010;132:13586–13587. doi: 10.1021/ja104525m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cali JJ, Daily W, Hawkins E, Klaubert D, Liu J. Luminogenic and Fluorogenic Compounds and Methods to Detect Molecules or Conditions. 2006/130551 A8. [Feb 1, 2007];U.S. Patent. 2011
- 19.Pirrung MC, Biswas G, De Howitt N, Liao J. Bioorg. Med. Chem. Lett. 2014;24:4881–4883. doi: 10.1016/j.bmcl.2014.08.048. [DOI] [PubMed] [Google Scholar]
- 20.Bauernschmitt R, Ahlrichs R. Chem. Phys. Lett. 1996;256:454–464. [Google Scholar]
- 21.Rappoport D, Furche F. J. Am. Chem. Soc. 2004;126:1277–1284. doi: 10.1021/ja037806u. [DOI] [PubMed] [Google Scholar]
- 22.Aquino AJA, Lischka H, Hättig C. J. Phys. Chem. A. 2005;109:3201–3208. doi: 10.1021/jp050288k. [DOI] [PubMed] [Google Scholar]
- 23.Van de Bittner GC, Dubikovskaya EA, Bertozzi CR, Chang CJ. Proc. Natl. Acad. Sci. USA. 2010;107:21316–21321. doi: 10.1073/pnas.1012864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porterfield WB, Jones KA, McCutcheon DC, Prescher JA. J. Am. Chem. Soc. 2015;137:8656–8659. doi: 10.1021/jacs.5b02774. [DOI] [PubMed] [Google Scholar]
- 25.Li J, Chen L, Du L, Li M. Chem. Soc. Rev. 2013;42:662–676. doi: 10.1039/c2cs35249d. [DOI] [PubMed] [Google Scholar]
- 26.White EH, Wörther H, Field GF, McElroy WD. J. Org. Chem. 1965;30:2344–2348. [Google Scholar]
- 27.White EH, Wörther H. J. Org. Chem. 1966;31:1484–1488. doi: 10.1021/jo01343a039. [DOI] [PubMed] [Google Scholar]
- 28.McCutcheon DC, Porterfield WB, Prescher JA. Org. Biomol. Chem. 2015;13:2117–2121. doi: 10.1039/c4ob02529f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White EH, Steinmetz MG, Miano JD, Wildes PD, Morland R. J. Am. Chem. Soc. 1980;102:3199–3208. [Google Scholar]
- 30.White EH, Rapaport E, Hopkins TA, Seliger HH. J. Am. Chem. Soc. 1969;91:2178–2180. doi: 10.1021/ja01036a093. [DOI] [PubMed] [Google Scholar]
- 31.Seliger HH, McElroy WD. Science. 1962;138:683–685. doi: 10.1126/science.138.3541.683. [DOI] [PubMed] [Google Scholar]
- 32.Shimomura O, Goto T, Johnson FH. Proc Natl Acad Sci USA. 1977;74:2799–2802. doi: 10.1073/pnas.74.7.2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans MS, Chaurette JP, Adams ST, Reddy GR, Paley MA, Aronin N, Prescher JA, Miller SC. Nat. Methods. 2014;11:393–395. doi: 10.1038/nmeth.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.