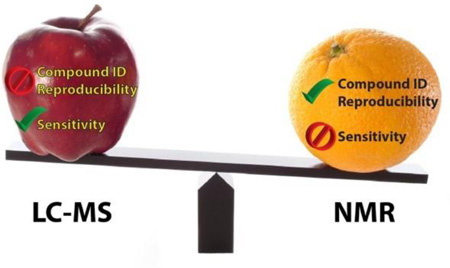
Abstract
The two leading analytical approaches to metabolomics are mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. Although currently overshadowed by MS in terms of numbers of compounds resolved, NMR spectroscopy offers advantages both on its own and coupled with MS. NMR data are highly reproducible and quantitative over a wide dynamic range and are unmatched for determining structures of unknowns. NMR is adept at tracing metabolic pathways and fluxes using isotope labels. Moreover, NMR is non-destructive and can be utilized in vivo. NMR results have a proven track record of translating in vitro findings to in vivo clinical applications.
Graphical Abstract
Introduction
The metabolic state of an organism depends on its genome, transcriptome, proteome, epigenome, microbiome, and exposome (environment). Thus, metabolomics, the study of small molecules (< 1,500 Da) in living systems, provides information with a high potential for accurately describing the physiological state of an organism. The two most successful approaches to determining the metabolic state of an organism have been mass spectrometry (MS) and nuclear magnetic resonance (NMR) spectroscopy. Several years ago, the number of publications utilizing the two approaches were comparable; more recently, however, MS-based metabolomics has clearly overtaken NMR-based metabolomics. This state of affairs prompted the organization of a workshop to review the current state of NMR-based metabolomics, to assess its strengths and weaknesses, and to envision its future potential. As reported here, this workshop (“NMR-Based Metabolomics,” held in the Discovery Building, Morgridge Institute for Research, Madison, Wisconsin, USA, on June 10, 2016) highlighted a number of benefits of NMR-based metabolomics that appear to be currently underappreciated. MS and NMR offer different strengths, which can be used synergistically. The workshop stressed the need for more extensive small molecule databases and improved standards at each step of a metabolomics study.
The metabolome
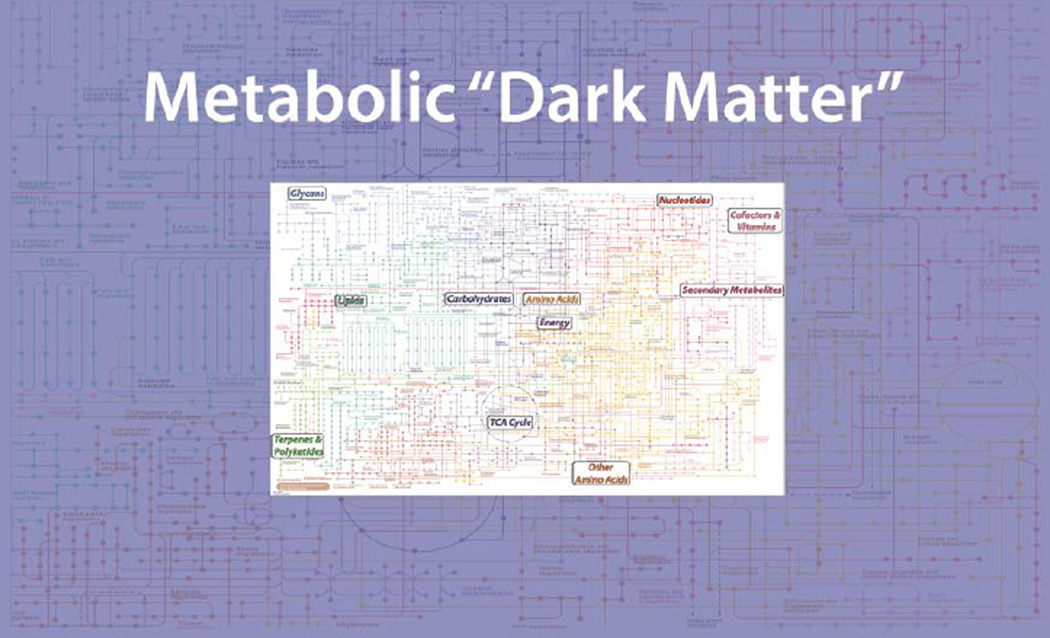
The two major fields of chemical research on biological small molecules, metabolomics and natural product discovery, have the similar goals of identifying and characterizing small molecules, either in their isolated active state (natural product chemistry) or as mixtures (metabolomics) [1]. The number of small molecules of importance to humans is far greater than those currently represented on metabolic charts, with the excess constituting current “metabolic dark matter” (Figure 1). The swapping of metabolites between pathways in humans and those of organisms in the human microbiome increases the network of relevant reactions by a staggering amount. The HMDB [2] lists 42,000 metabolites and the number of lipid variants is on the order of 100,000; thus, a lower limit of expected endogenous and exogenous human metabolites is around 150,000, but the actual number of metabolites could be much higher. Of this vast number of metabolites, only 1,500 may be identified from global profiling, 200–500 from targeted profiling, and far fewer are routinely subjected to quantitative analysis.
Figure 1.
Schematic representation of metabolic “dark matter”.
NMR and its advantages
Despite its lower sensitivity, NMR spectroscopy offers many unparalleled advantages over MS [3,4]. NMR offers a window into observing and rigorously quantifying all of the more abundant compounds present in biological fluids, cell extracts, and tissues without the need for elaborate sample preparation or fractionation. NMR offers advantages for compounds that are difficult to ionize or require derivatization for MS. NMR allows the identification of compounds with identical masses, including those with different isotopomer distributions. NMR is the mainstay for determining structures of unknown compounds. Through the use of stable isotope labels, NMR can be used to elucidate the dynamics and mechanisms of metabolite transformations and to explore the compartmentalization of metabolic pathways. NMR has advantages in drug screening [5]. Finally, site-specific NMR imaging and spectroscopy offer approaches for metabolic studies in living organisms.
Strategies for the identification of metabolites in complex mixtures from NMR data have been reviewed recently [6]. The most important nuclei in biomolecular NMR studies are 1H (proton), 13C, 15N, and 31P. Of these, 1H is the most sensitive followed by 31P; both are present at near 100% natural abundance. 31P NMR is useful for studies of cellular energy states in vivo and ex vivo, but a limitation is that the 31P signals from most phosphorylated compounds overlap. One-dimensional (1D) 1H NMR is the most widely used NMR approach in metabolomics. Signals are either binned and then analyzed or fitted to patterns of signals corresponding to the metabolites expected to be present in the mixture. The latter approach can be problematic in that many 1H signals overlap in ways that offer alternative fitting solutions, a problem that can be overcome by standardizing the analysis in terms of biofluid, solution conditions, data collection protocol, and by employing probabilistic fitting (Bayesil) [7]. 13C NMR signals cover a 200 ppm range compared with 10 ppm for 1H and as a consequence are better resolved; however, the low sensitivity of 13C (less by a factor of 8 or more) is compounded by its low natural abundance (1.1 %). Two dimensional (2D) NMR methods offer improved approaches for unambiguous identification of metabolites in mixtures. These 2D methods include 1H-1H COSY (correlated spectroscopy), 1H-1H TOCSY (total correlation spectroscopy), and 1H-13C HSQC (heteronuclear single-quantum correlation). A widely used software package (rNMR) matches regions of interest in spectra of standards to those in experimental mixtures for compound identifications [8]. Software is available for automating metabolite identification from combined TOCSY and HSQC data [9,10]. By setting tolerances for the matching of 1H and 13C signals, one can maximize compound identification while minimizing false positives [11]. This approach has been extended to a calculated confidence level for compound identifications from NMR data [12]. Another approach for connecting signals from individual compounds in mixtures is based on searching for statistical correlations among the intensities of NMR signals from various samples [13]. Nuclei present at low natural abundance 2H (deuteron), 13C, and 15N serve as ideal metabolic tracers [14].
Need for standards in NMR metabolomics
Standard NMR spectra and associated information on small biological molecules are available from freely-accessible databases, including HMDB [2], BMRB [15], TOCCATA [9], and COLMAR [10], but they still cover only a fraction of relevant compounds. A repository has been established for results of metabolomics studies from the NIH Common Fund Centers [16]. The Coordination of Standards in Metabolomics (COSMOS) Initiative is developing a robust data infrastructure and new data exchange standards (http://nmrml.org) for metabolomics data and metadata to support workflows metabolomics applications [17]. One of the COSMOS projects is a website (http://metabolomexchange.org) that federates data available from the leading metabolomics data repositories. Best practices and standards have been published for metabolic phenotyping of biological fluids [18,19]. An open-source platform for complete NMR metabolomics data handling (MVAPACK) has been developed as a step toward establishing best practices for the analysis of metabolic fingerprinting data [20].
Sample preparation
Certain biofluids, e.g. cerebrospinal fluid, require little or no preparation for NMR. Others, such as plasma contain proteins and lipids that interfere with NMR spectral quality. Treatment with methanol at solvent-to-serum ratio of 2:1 (v/v) has been shown to remove lipoproteins and minimize the loss of metabolites [21] enabling the detection of about 67 different compounds [22]. Another promising protocol utilizes the removal of protein by added silica nanoparticles [23].
Tagging
An approach for compounds with overlapping 1H signals or present at lower concentration is to tag them with an NMR-active label. Nitrogen-15 with attached hydrogen is an attractive tag because 2D 1H-15N signals can be acquired at high sensitivity without interference from signals from unlabeled compounds owing to the low natural abundance of 15N [24,25]. Such tags also provide a permanent positive charge for MS analysis.
Combining NMR and MS
As reviewed recently [26], advances in NMR- and MS-based metabolomics, including the combination of the two approaches, promise to greatly improve the identification and quantitation of compounds in mixtures. One example is the simultaneous analysis of DI-ESI-MS and 1D 1H NMR spectral data to yield accurate mass measurements and class separation scores [27]. Other approaches filter data from one approach against the other to increase the number of compounds confidently identified [28,29]. Another method identifies compounds by exploiting the principle that abundance/intensity ratios are relatively constant for the same metabolite in different samples [30]. Combined NMR and MS has advantages for isotope tracing experiments and metabolic flux analysis. MS generally quantifies isotopic labeling distributions but even with MS/MS often does not give the specific labeling position, which is available from NMR.
Quantification
If 1D 1H NMR peaks from a compound are well resolved with acceptable signal-to-noise, their intensities correlate linearly with its relative concentration. To determine absolute concentrations, one adds a standard of known concentration. The cross peak intensities of 2D 1H-13C HSQC spectra, however, do not correlate linearly with concentration. One can collect spectra of mixtures with known concentrations bracketing those of the unknowns and use these to determine factors that translate peak intensity to concentration [31]. Peak intensities in 2D 1H-13C HSQC spectra can also be converted to concentration by from the slopes generated by spectra utilizing different replicates of the pulse sequence module; linear extrapolation back to zero time of the peak intensities following the delays from one and two modules yields the “HSQC0 spectrum” whose peak intensities are proportional to concentration [32]. Spectral overlaps can be accounted for by methods such as FMLR (fast maximum-likelihood reconstruction) [33]. A new experiment (1H-13C QUIPU HSQC) aims to quantify in one map a complex mixture composed of low concentrated metabolites [34]. Another approach, one that requires full 13C labeling, achieves quantification through the collection of 13C-13C CT-TOCSY spectra and the application of analytical approximations based on the known carbon-backbone topologies [35].
Applications of metabolomics
Applications of metabolomics include disease diagnosis, monitoring the effects of medical interventions including drugs, detection of adulteration of food, and analysis of biochemical pathways and their perturbations resulting from mutations, aging, diet, exercise, or life style. A recent study showed how ex vivo 1D 1H NMR spectroscopy can be used for the simultaneous identification of quantification of coenzymes that report on cellular function [36]. Another study used this approach to investigate alterations in the energy/redox-metabolome in dopaminergic cells exposed to environmental/mitochondrial toxins [37]. Studies of the metabolomics of model organisms are both timely and important for understanding of their different biology [38]. Protocols have been described for studies of the metabolomics of bacteria [39] and plants [40]. Metabolomics, along with activity-guided fractionation followed by structural analysis, constitutes a powerful approach for identifying biologically active compounds for studies in chemical ecology [41]. Metabolomics is used regularly in drug discovery programs to uncover the efficacy, specificity, or toxicity of lead compounds [42]. Metabolomics can provide information on the in vivo mechanism of action and to eliminate compounds likely to cause problems with side effects [43]. Recent studies have utilized metabolomics to search for biomarkers for colon cancer [44] and multiple sclerosis [45].
Future technology
All technologies that increase NMR sensitivity are of extreme importance as are improvements in sample preparation [46]. Approaches to high sensitivity include NMR spectrometers with ultra-high-field magnets operating at 1H resonance frequencies of 1.2 GHz or higher. The first such systems are scheduled for delivery in 2017. Small high-temperature superconducting coils can maximize the signal per sample mass: a 13C-optimized 1.5-mm high temperature superconducting NMR probe has enabled novel 13C NMR studies of natural products [47], and this has been followed up with a 1H-13C dual-optimized NMR probe based on double-tuned high temperature superconducting resonators [48]. These probes take advantage of the excellent peak dispersion of 13C spectra [49], which can be augmented by further 2D 13C-13C experiments, such as INADEQUATE [50]
Hyperpolarization offers an approach for enhanced sensitivity with even higher potential. The underlying physics utilizes the magnetic moment of the unpaired electron, which is roughly 2800 times that of 13C and 6900 times that of 15N, to polarize nuclear spins. First demonstrated by Golman and coworkers [51], studies utilizing hyperpolarized 13C to increase sensitivity are becoming routine. An exciting advance is the discovery [52] of an efficient and inexpensive method for hyperpolarizing 15N spins at room temperature. Enhancements are on the order of >10,000 enabling the detection of NMR signals for over an hour [52]. It may become possible to use this approach to tag a range of compounds for metabolomics studies in vivo.
Conclusions
The workshop demonstrated that NMR-based metabolomics promises to continue to play an important role in the studies of complex mixtures of small biological molecules, their metabolic networks, and their interactions with biomacromolecules. Although the development of new and better methods continues to be an integral part, the field needs to focus on developing standardized, enlarged, and integrated databases of NMR data on small molecules as well as archives representing the NMR metabolic fingerprints of standard biological fluids and tissue extracts from humans and model organisms. Standardization of best practices for sample preparation, data collection and analysis should enhance the reproducibility of results within the metabolomics community, while at the same time avoiding the risk of adhering to methods and protocols that are suboptimal in a field that is very much in flux. In order to overcome the current skepticism in omics [53], it will be advisable for metabolomics to build the ideas of reproducibility and data sharing into every tool and database. Future work is expected to build upon core strengths of NMR spectroscopy, which includes its versatility and specificity in the form of 1D and higher dimensional spectra, its reproducibility, its quantitative ability, its capability for following chemical reactions and flux, its ability to identify compounds and deduce structures of unknowns, and its growing potential for collecting metabolomics data in vivo.
Highlights.
NMR offers advantages for metabolomics that may be currently underappreciated
In future, NMR-based metabolomics needs to focus on its inherent strengths
Reproducibility and data sharing are of key importance in technology development
Acknowledgments
The authors thank the Morgridge Institute for Research, Madison WI, for its financial support for the workshop on “NMR-Based Metabolomics”. Research of the authors received support from NIH R01GM109046 and P41GM103399 (to JLM); NIH R01 GM 066041 (to RB); NIH U24 (SECIM) DK097209-01A1 (to ASE and RB); NIH 1U24DK097209-01A1 and the Georgia Research Alliance (to ASE); NIH P41GM111135 (to HRE); NIH P20 RR-17675, P30GM103335, R01 CA163649-01A1, and R01 AI087668-01A1 (to RP); NIH R01GM085291 and P30CA015704 (to DR); and Genome Canada support (to DSW). ASE gratefully acknowledges Gary Patti for introducing the term “Metabolic Dark Matter”. JLM thanks Jing Fan for useful comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Robinette SL, Brüschweiler R, Schroeder FC, Edison AS. NMR in metabolomics and natural products research: two sides of the same coin. Acc Chem Res. 2012;45:288–297. doi: 10.1021/ar2001606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fan TW, Lane AN. Applications of NMR spectroscopy to systems biochemistry. Prog Nucl Magn Reson Spectrosc. 2016;92–93:18–53. doi: 10.1016/j.pnmrs.2016.01.005.. Comprehensive review of the role of NMR in metabolic research.
- 4. Nagana Gowda GA, Raftery D. Can NMR solve some significant challenges in metabolomics? J Magn Reson. 2015;260:144–160. doi: 10.1016/j.jmr.2015.07.014.. The authors present their perspectives on emerging trends in NMR-based metabolomics and NMR's continuing role in the field with an emphasis on recent and ongoing research from their laboratory.
- 5.Tiziani S, Kang Y, Choi JS, Roberts W, Paternostro G. Metabolomic high-content nuclear magnetic resonance-based drug screening of a kinase inhibitor library. Nat Commun. 2011;2:545. doi: 10.1038/ncomms1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bingol K, Bruschweiler-Li L, Li D, Zhang B, Xie M, Brüschweiler R. Emerging new strategies for successful metabolite identification in metabolomics. Bioanalysis. 2016;8:557–573. doi: 10.4155/bio-2015-0004.. Review of recent strategies for the identification of metabolites in complex biological mixtures.
- 7. Ravanbakhsh S, Liu P, Bjorndahl TC, Mandal R, Grant JR, Wilson M, Eisner R, Sinelnikov I, Hu X, Luchinat C, et al. Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS One. 2015;10:e0124219. doi: 10.1371/journal.pone.0124219.. The authors present a publicly-accessible system, BAYESIL, for the automated generation of a the most probable metabolic profile from 1H NMR spectra of serum or cerebrospinal fluid. BAYESIL provides quantitative NMR spectral profiling with an accuracy meeting or exceeding the performance of trained experts.
- 8.Lewis IA, Schommer SC, Markley JL. rNMR: open source software for identifying and quantifying metabolites in NMR spectra. Magn Reson Chem. 2009;47:s123–s126. doi: 10.1002/mrc.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bingol K, Bruschweiler-Li L, Li DW, Brüschweiler R. Customized metabolomics database for the analysis of NMR 1H-1H TOCSY and 13C-1H HSQC-TOCSY spectra of complex mixtures. Anal Chem. 2014;86:5494–5501. doi: 10.1021/ac500979g.. The authors introduce the TOCCATA datbase, which contains complete 1H and 13C chemical shift information on individual spin systems and isomeric states of common metabolites. The database supports identifications of compounds present in complex metabolic mixtures at 13C natural abundance from 2D 1H TOCSY and 2D 13C-1H HSQC-TOCSY spectra.
- 10. Bingol K, Li DW, Bruschweiler-Li L, Cabrera OA, Megraw T, Zhang F, Brüschweiler R. Unified and isomer-specific NMR metabolomics database for the accurate analysis of 13C-1H HSQC spectra. ACS Chem Biol. 2015;10:452–459. doi: 10.1021/cb5006382.. The Complex Mixture Analysis by NMR (COLMAR) 13C-1H HSQC database, which treats slowly exchanging isomers a metabolite as separate species, permits improved detection of compounds present at low concentration. The software is accssible from an interactive web interface.
- 11.Cui Q, Lewis IA, Hegeman AD, Anderson ME, Li J, Schulte CF, Westler WM, Eghbalnia HR, Sussman MR, Markley JL. Metabolite identification via the Madison Metabolomics Consortium Database. Nat Biotechnol. 2008;26:162–164. doi: 10.1038/nbt0208-162. [DOI] [PubMed] [Google Scholar]
- 12.Everett JR. A new paradigm for known metabolite identification in metabonomics/metabolomics: metabolite identification efficiency. Comput Struct Biotechnol J. 2015;13:131–144. doi: 10.1016/j.csbj.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wei S, Zhang J, Liu L, Ye T, Gowda GA, Tayyari F, Raftery D. Ratio analysis nuclear magnetic resonance spectroscopy for selective metabolite identification in complex samples. Anal Chem. 2011;83:7616–7623. doi: 10.1021/ac201625f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fan TW, Lane AN, Higashi RM. Stable Isotope Resolved Metabolomics Studies TIssue Slices. Bio Protoc. 2016;6 doi: 10.21769/bioprotoc.1730.. The authors present an elegant study of the effect of tumor microenvironment on metabolic flux.
- 15.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sud M, Fahy E, Cotter D, Azam K, Vadivelu I, Burant C, Edison A, Fiehn O, Higashi R, Nair KS, et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016;44:D463–D470. doi: 10.1093/nar/gkv1042.. The Metabolomics Workbench, part of the data coordinating effort of the National Institute of Health (NIH) Common Fund's Metabolomics Program and available at www.metabolomicsworkbench.org, is a public repository for metabolomics metadata and experimental data spanning various species and experimental platforms, metabolite standards, metabolite structures, protocols, tutorials, and training material and other educational resources. The Metabolomics Workbench provides data from the Common Fund's Metabolomics Resource Cores, metabolite standards, and analysis tools to the wider metabolomics community and seeks data depositions from metabolomics researchers across the world.
- 17. Salek RM, Neumann S, Schober D, Hummel J, Billiau K, Kopka J, Correa E, Reijmers T, Rosato A, Tenori L, et al. COordination of Standards in MetabOlomicS (COSMOS): facilitating integrated metabolomics data access. Metabolomics. 2015;11:1587–1597. doi: 10.1007/s11306-015-0810-y.. The authors announce an effort sponsored by the Framework Programme 7 EU Initiative 'coordination of standards in metabolomics' (COSMOS) to develop a robust data infrastructure and exchange standards for metabolomics data and metadata.
- 18. Emwas AH, Roy R, McKay RT, Ryan D, Brennan L, Tenori L, Luchinat C, Gao X, Zeri AC, Gowda GA, et al. Recommendations and Standardization of Biomarker Quantification Using NMR-Based Metabolomics with Particular Focus on Urinary Analysis. J Proteome Res. 2016;15:360–373. doi: 10.1021/acs.jproteome.5b00885.. The authors review published studies on NMR-based urine metabolic profiling with the aim of identifying key variables that may affect the results of metabolomics studies. From this survey, they identify a number of issues that require either standardization or careful accounting in experimental design and provide recommendations for urine collection, sample preparation and data acquisition.
- 19.Dona AC, Jimenez B, Schafer H, Humpfer E, Spraul M, Lewis MR, Pearce JT, Holmes E, Lindon JC, Nicholson JK. Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal Chem. 2014;86:9887–9894. doi: 10.1021/ac5025039. [DOI] [PubMed] [Google Scholar]
- 20. Worley B, Powers R. MVAPACK: a complete data handling package for NMR metabolomics. ACS Chem Biol. 2014;9:1138–1144. doi: 10.1021/cb4008937.. The authors report a newly developed open-source platform (MVAPACK) for complete NMR metabolomics data handling and describe its application to metabolic fingerprinting.
- 21.Nagana Gowda GA, Gowda YN, Raftery D. Massive glutamine cyclization to pyroglutamic acid in human serum discovered using NMR spectroscopy. Anal Chem. 2015;87:3800–3805. doi: 10.1021/ac504435b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nagana Gowda GA, Gowda YN, Raftery D. Expanding the limits of human blood metabolite quantitation using NMR spectroscopy. Anal Chem. 2015;87:706–715. doi: 10.1021/ac503651e.. Through investigations of pooled human serum, the authors developed a protocol for removing protein while minimizing the removal of metabolites, which enables the routine identification of 67 blood metabolites including amino acids, organic acids, carbohydrates, and heterocyclic compounds,. The proposed protocol, which utilizes 2:1 (v/v) methanol:serum to precipitate protein, was found to be far superior to the previously established acetonitrile treatment in retaining metabolites.
- 23. Zhang B, Xie M, Bruschweiler-Li L, Bruschweiler R. Nanoparticle-Assisted Removal of Protein in Human Serum for Metabolomics Studies. Anal Chem. 2016;88:1003–1007. doi: 10.1021/acs.analchem.5b03889.. The authors demonstrate how serum protein can be efficiently removed at physiological pH through attractive interactions with silica nanoparticles. The authors conclude that the combination of nanoparticle treatment and ultrafiltration has a minimal effect on the metabolite content and permits the collection of clean homo- and heteronuclear NMR spectra of the serum metabolome.
- 24.Tayyari F, Gowda GA, Gu H, Raftery D. 15N-cholamine--a smart isotope tag for combining NMR- and MS-based metabolite profiling. Anal Chem. 2013;85:8715–8721. doi: 10.1021/ac401712a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lane AN, Arumugam S, Lorkiewicz PK, Higashi RM, Laulhe S, Nantz MH, Moseley HN, Fan TW. Chemoselective detection and discrimination of carbonyl-containing compounds in metabolite mixtures by 1H-detected 15N nuclear magnetic resonance. Magn Reson Chem. 2015;53:337–343. doi: 10.1002/mrc.4199.. The authors describe a carbonyl-selective aminooxy probe incorporating 15N in the aminooxy functional group that specifically reacts with free keto and aldehyde functions, but not carboxylates. This probe enables the exclusive detection by 15N-edited NMR of metabolites that contain a free carbonyl function with differential identification of aminooxy adducts of ketones and aldehydes by their very different chemical shifts. By utilizing 2-bond or 3-bond 15N-1H couplings more than 30 carbonyl-containing compounds could be detected by the 15N-edited NMR analysis. As the aminooxy probe contains a permanently charged quaternary ammonium group, the adducts are also optimized for detection by mass spectrometry.
- 26. Bingol K, Brüschweiler R. Two elephants in the room: new hybrid nuclear magnetic resonance and mass spectrometry approaches for metabolomics. Curr Opin Clin Nutr Metab Care. 2015;18:471–477. doi: 10.1097/MCO.0000000000000206.. The authors review approaches for combining metabolomics data from mass spectrometry with those from NMR spectroscopy.
- 27.Marshall DD, Lei S, Worley B, Huang Y, Garcia-Garcia A, Franco R, Dodds ED, Powers R. Combining DI-ESI-MS and NMR datasets for metabolic profiling. Metabolomics. 2015;11:391–402. doi: 10.1007/s11306-014-0704-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bingol K, Brüschweiler R. NMR/MS Translator for the Enhanced Simultaneous Analysis of Metabolomics Mixtures by NMR Spectroscopy and Mass Spectrometry: Application to Human Urine. J Proteome Res. 2015;14:2642–2648. doi: 10.1021/acs.jproteome.5b00184.. The authors present an approach for increasing the number of metabolites detected in urine by combining information from NMR and MS data.
- 29. Bingol K, Bruschweiler-Li L, Yu C, Somogyi A, Zhang F, Brüschweiler R. Metabolomics beyond spectroscopic databases: a combined MS/NMR strategy for the rapid identification of new metabolites in complex mixtures. Anal Chem. 2015;87:3864–3870. doi: 10.1021/ac504633z.. In the SUMMIT MS/NMR appooach, the chemical formulas of the mixture components are first identified from accurate masses by MS and then all feasible structures (structural manifold) consistent with these chemical formulas are generated. Next, NMR spectra of each member of the structural manifold are predicted and compared with the experimental NMR spectra in order to identify the molecular structures that match the information obtained from both the MS and NMR techniques.
- 30.Gu H, Gowda GA, Neto FC, Opp MR, Raftery D. RAMSY: ratio analysis of mass spectrometry to improve compound identification. Anal Chem. 2013;85:10771–10779. doi: 10.1021/ac4019268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis IA, Schommer SC, Hodis B, Robb KA, Tonelli M, Westler WM, Sussman MR, Markley JL. Method for determining molar concentrations of metabolites in complex solutions from two-dimensional 1H-13C NMR spectra. Anal Chem. 2007;79:9385–9390. doi: 10.1021/ac071583z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu K, Ellinger JJ, Chylla RA, Markley JL. Measurement of Absolute Concentrations of Individual Compounds in Metabolite Mixtures by Gradient-Selective Time-Zero 1H-13C HSQC with Two Concentration References and Fast Maximum Likelihood Reconstruction Analysis. Anal Chem. 2011;83:9352–9360. doi: 10.1021/ac201948f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chylla RA, Hu K, Ellinger JJ, Markley JL. Deconvolution of two-dimensional NMR spectra by fast maximum likelihood reconstruction: application to quantitative metabolomics. Anal Chem. 2011;83:4871–4880. doi: 10.1021/ac200536b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mauve C, Khlifi S, Gilard F, Mouille G, Farjon J. Sensitive, highly resolved, and quantitative (1)H-(13)C NMR data in one go for tracking metabolites in vegetal extracts. Chem Commun (Camb) 2016;52:6142–6145. doi: 10.1039/c6cc01783e. [DOI] [PubMed] [Google Scholar]
- 35.Bingol K, Zhang F, Bruschweiler-Li L, Brüschweiler R. Quantitative analysis of metabolic mixtures by two-dimensional 13C constant-time TOCSY NMR spectroscopy. Anal Chem. 2013;85:6414–6420. doi: 10.1021/ac400913m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nagana Gowda GA, Abell L, Lee CF, Tian R, Raftery D. Simultaneous Analysis of Major Coenzymes of Cellular Redox Reactions and Energy Using ex Vivo 1H NMR Spectroscopy. Anal Chem. 2016;88:4817–4824. doi: 10.1021/acs.analchem.6b00442.. The authors show how 1H NMR can be used to simultaneously deterimine levels of key coenzymes: oxidized and reduced forms of nicotinamide adenine dinucleotide (NAD+ and NADH), oxidized and reduced forms of nicotinamide adenine dinucleotide phosphate (NADP+ and NADPH), and adenosine triphosphate (ATP) and its precursors, adenosine diphosphate (ADP) and adenosine monophosphate (AMP).
- 37.Lei S, Zavala-Flores L, Garcia-Garcia A, Nandakumar R, Huang Y, Madayiputhiya N, Stanton RC, Dodds ED, Powers R, Franco R. Alterations in energy/redox metabolism induced by mitochondrial and environmental toxins: a specific role for glucose-6-phosphate-dehydrogenase and the pentose phosphate pathway in paraquat toxicity. ACS Chem Biol. 2014;9:2032–2048. doi: 10.1021/cb400894a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Edison AS, Hall RD, Junot C, Karp PD, Kurland IJ, Mistrik R, Reed LK, Saito K, Salek RM, Steinbeck C, et al. The Time Is Right to Focus on Model Organism Metabolomes. Metabolites. 2016;6 doi: 10.3390/metabo6010008.. The authors propose a grand challenge for metabolomics studies of model organisms representing microbes, animals and plants in which all metabolites are identified and mapped onto metabolic pathways and quantitative metabolic models are developed. These should enable the analysis of metabolic pathways within the context of evolutionary metabolomics (phylometabolomics).
- 39.Halouska S, Zhang B, Gaupp R, Lei S, Snell E, Fenton RJ, Barletta RG, Somerville GA, Powers R. Revisiting Protocols for the NMR Analysis of Bacterial Metabolomes. J Integr OMICS. 2013;3:120–137. doi: 10.5584/jiomics.v3i2.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sumner LW, Lei Z, Nikolau BJ, Saito K. Modern plant metabolomics: advanced natural product gene discoveries, improved technologies, and future prospects. Nat Prod Rep. 2015;32:212–229. doi: 10.1039/c4np00072b. [DOI] [PubMed] [Google Scholar]
- 41. Edison AS, Clendinen CS, Ajredini R, Beecher C, Ponce FV, Stupp GS. Metabolomics and Natural-Products Strategies to Study Chemical Ecology in Nematodes. Integr Comp Biol. 2015;55:478–485. doi: 10.1093/icb/icv077.. As illustrated with results from studis of Caenorhabditis elegans and related free-living nematodes, this review describes the two complementary approaches used to identify biologically active compounds for studies in chemical ecology: activity-guided fractionation and metabolomics. A new liquid chromatography-mass spectrometry-based method called isotopic ratio outlier analysis is described that can be used as an adjunct to NMR spectroscopy.
- 42.Forgue P, Halouska S, Werth M, Xu K, Harris S, Powers R. NMR metabolic profiling of Aspergillus nidulans to monitor drug and protein activity. J Proteome Res. 2006;5:1916–1923. doi: 10.1021/pr060114v. [DOI] [PubMed] [Google Scholar]
- 43. Powers R. The current state of drug discovery and a potential role for NMR metabolomics. J Med Chem. 2014;57:5860–5870. doi: 10.1021/jm401803b.. The authors propose ways in which the inclusion of metabolomics as a routine component of the drug discovery process may improve its efficiency and effectiveness. Metabolomics can be used to eliminate compounds with potential efficacy and side effect problems while prioritizing well-behaved leads with druglike characteristics.
- 44.Chen C, Deng L, Wei S, Nagana Gowda GA, Gu H, Chiorean EG, Abu Zaid M, Harrison ML, Pekny JF, Loehrer PJ, et al. Exploring Metabolic Profile Differences between Colorectal Polyp Patients and Controls Using Seemingly Unrelated Regression. J Proteome Res. 2015;14:2492–2499. doi: 10.1021/acs.jproteome.5b00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gebregiworgis T, Nielsen HH, Massilamany C, Gangaplara A, Reddy J, Illes Z, Powers R. A Urinary Metabolic Signature for Multiple Sclerosis and Neuromyelitis Optica. J Proteome Res. 2016;15:659–666. doi: 10.1021/acs.jproteome.5b01111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ardenkjaer-Larsen JH, Boebinger GS, Comment A, Duckett S, Edison AS, Engelke F, Griesinger C, Griffin RG, Hilty C, Maeda H, et al. Facing and Overcoming Sensitivity Challenges in Biomolecular NMR Spectroscopy. Angew Chem Int Ed Engl. 2015;54:9162–9185. doi: 10.1002/anie.201410653.. The authors review strategies for overcomint the inherent low sensitivity of biomolecular NMR spectroscopy.
- 47.Ramaswamy V, Hooker JW, Withers RS, Nast RE, Brey WW, Edison AS. Development of a 13C-optimized 1.5-mm high temperature superconducting NMR probe. J Magn Reson. 2013;235:58–65. doi: 10.1016/j.jmr.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramaswamy V, Hooker JW, Withers RS, Nast RE, Edison AS, Brey WW. Development of a 1H-13C Dual-Optimized NMR Probe Based on Double-Tuned High Temperature Superconducting Resonators. Ieee Transactions on Applied Superconductivity. 2016;26. The authors report a novel NMR probe design simultaneously optimized for 1H and 13C detection. The probe employs novel double-tuned HTS resonators designed for optimal magnetic field homogeneity and minimal electric field that generate strong, uniform, and mutually orthogonal magnetic fields at the 1H and 13C NMR frequencies.
- 49.Clendinen CS, Lee-McMullen B, Williams CM, Stupp GS, Vandenborne K, Hahn DA, Walter GA, Edison AS. 13C NMR metabolomics: applications at natural abundance. Anal Chem. 2014;86:9242–9250. doi: 10.1021/ac502346h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clendinen CS, Pasquel C, Ajredini R, Edison AS. 13C NMR Metabolomics: INADEQUATE Network Analysis. Anal Chem. 2015;87:5698–5706. doi: 10.1021/acs.analchem.5b00867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ardenkjaer-Larsen JH, Fridlund B, Gram A, Hansson G, Hansson L, Lerche MH, Servin R, Thaning M, Golman K. Increase in signal-to-noise ratio of >10,000 times in liquid-state NMR. Proc Natl Acad Sci U S A. 2003;100:10158–10163. doi: 10.1073/pnas.1733835100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Theis T, Ortiz GX, Jr, Logan AW, Claytor KE, Feng Y, Huhn WP, Blum V, Malcolmson SJ, Chekmenev EY, Wang Q, et al. Direct and cost-efficient hyperpolarization of long-lived nuclear spin states on universal 15N2-diazirine molecular tags. Sci Adv. 2016;2:e1501438. doi: 10.1126/sciadv.1501438.. The authors demonstrate how a recently developed method (SABRE-SHEATH) can be used to directly hyperpolarize 15N2 magnetization and 15N2 singlet spin order, with signal decay time constants of 5.8 and 23 minutes, respectively. The approach generates >10,000-fold enhancements with detectable nuclear MR signals that last for over an hour from molecular tags that can be incorporated into a wide range of biomolecules.
- 53. Ioannidis JP, Greenland S, Hlatky MA, Khoury MJ, Macleod MR, Moher D, Schulz KF, Tibshirani R. Increasing value and reducing waste in research design, conduct, and analysis. Lancet. 2014;383:166–175. doi: 10.1016/S0140-6736(13)62227-8.. Potential solutions are proposed for the problems of irreproducible scientific studies including improvements in protocols and documentation, consideration of evidence from studies in progress, standardization of research efforts, and scientific workforce development.