Abstract
Purpose
The etiology of maternal aging, a common cause of female factor infertility and a rate-limiting step in vitro fertilization (IVF) success, remains still unclear. Proteomic changes responsible for the impaired successful pregnancy outcome after IVF with aged blastocysts have not been yet evaluated. The objective of this prospective study was to employ proteomic techniques and bioinformatic tools to enlight differences at the protein level in blastocoel fluid of aged and younger woman.
Methods
Protein composition of human blastocoel fluid isolated by micromanipulation from 46 blastocysts of women aged <37 years (group A) and 29 of women aged ≥37 years (group B) have been identified by a shotgun proteomic approach based on high-resolution nano-liquid chromatography electrospray-ionization-tandem mass spectrometry (nLC-ESI-MS/MS) using label free for the relative quantification of their expression levels.
Results
The proteomic analysis leads to the identification and quantification of 148 proteins; 132 and 116 proteins were identified in groups A and B, respectively. Interestingly, the identified proteins are mainly involved in processes aimed at fine tuning embryo implantation and development. Among the 100 proteins commonly expressed in both groups, 17 proteins are upregulated and 44 downregulated in group B compared to group A. Overall, the analysis identified 33 proteins, which were increased or present only in B while 76 were decreased in B or present only in A.
Conclusions
Data revealed that maternal aging mainly affects blastocyst survival and implantation through unbalancing the equilibrium of the ubiquitin system known to play a crucial role in fine-tuning several aspects required to ensure successful pregnancy outcome.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-016-0842-x) contains supplementary material, which is available to authorized users.
Keywords: Blastocoel fluid, Aging, Embryo implantation, Proteomics, Shotgun
Introduction
Embryo implantation requires synchronous development of the blastocyst and the endometrium [1]. During the early stages of implantation, the blastocyst enters the uterine cavity apposes and adheres to the endometrial epithelium to initiate implantation. It is well established that abnormalities in the bidirectional embryo-endometrial dialog are one of the major cause of infertility resulting in implantation failure [2, 3]. Noteworthy, advancing maternal age represents one of the major causes of the altered embryo-endometrial communication [4]. As expected, several large studies have reported age-specific outcomes of IVF, and maternal age has clearly been associated with a decline in cumulative live birth rates in patients older than 37 [5, 6]. Unfortunately, the molecular changes that occur in the endometrium during the “window of receptivity” and the blastocyst contribution have been poorly investigated due to the difficulty and the ethical limitations in studying implantation in humans; thus, no clinically useful markers have been identified to assess embryo quality and positive pregnancy outcome [7, 8].
In the recent years, the development of a wealth of “omic” technologies has opened the way for identifying potential markers, although validation of these is still a major issue [9, 10]. In particular, proteomics is an emerging powerful technology in the identification of both biomarkers and disease targets, allowing for the detection and monitoring of differing conditions or treatments. Although the classic proteomics approach has had limited success in the field of embryology [11, 12], the new developments in mass spectrometry have been helpful and the sensitivity of this approach has enabled the development of new protocols that are capable of profiling the proteome of small groups of mouse embryos [13], and even of single human blastocysts [14–16]. Therefore, several studies have recently addressed the analysis of the proteome of the human preimplantation embryo, in particular the ability of the blastocyst to secrete and/or consume different proteins in surrounding medium (secretome), in order to communicate with the maternal endometrium and proceed with the implantation process [13–15, 17, 18]. Interestingly, degenerating embryos presented a different protein expression profile compared to that of developing blastocysts, with a significant alteration of numerous biomarkers. Besides surrounding medium, the cells of the blastocyst are exposed to the fluid of the blastocoel, an internal cavity that becomes the yolk sac of the developing embryo [19]. Thus, the blastocoel fluid is the natural environment of blastocyst growth and the assessment of its protein composition provides an opportunity to expand the knowledge of embryonic physiology, including the maternal-embryonic dialog at the time of implantation. From a clinical perspective, the analysis of blastocoel fluid is emerging as a crucial topic in the field of reproductive medicine aimed at identifying new molecular markers related to highest implantation potential. The ultimate goal is to improve blastocyst culture conditions and fertilization rate, as well as to develop an affordable and cost-effective non-invasive biochemical assay to diagnose blastocyst viability, thus facilitating the move to single embryo transfer. The major problem concerning the analysis of the blastocoel fluid relies on its limited bioavailability (1–8 nl) and, despite the fact that recent developments in mass spectrometry allow the identification and characterization of thousands of proteins from low microgram levels of protein extracted from tissues or cell cultures [16, 17, 20, 21], the low amounts of proteins extractable from blastocoel fluid are still a limiting step in this study.
Here, by a shotgun proteomic approach and label-free quantification, we report for the first time the blastocoel fluid proteins that are differentially expressed in 29 collected surplus aged and 46 non-aged blastocysts, taking 37-year-old women as the cut off parameter to categorized human blastocysts. The differences in the protein profile of blastocoel fluid of aged versus non-aged blastocysts rely on the identification of 109 differentially expressed proteins, which are mainly involved in processes aimed at fine tuning embryo implantation and development. Upon validation, the suggested key proteins may serve as potential biomarkers of embryo quality and may contribute towards the effectiveness of protocols for blastocyst culture management. .
Materials and methods
Patient population
Written informed consent was obtained from each patient and the study was approved by our local Ethics Commitee. All consenting patients undergoing fertility treatment at the Humanitas Fertility Center, Humanitas Research Hospital, Rozzano, Italy, with supernumerary blastocysts were included in the study irrespective of the woman’s age (range years 26–41), infertility diagnosis (female or male), ovarian stimulation protocol (use of GnRH agonist or antagonist protocols in combination with recombinant and/or highly purified urinary gonadotropins), or performance of standard IVF or intracytoplasmic sperm injection (ICSI). The study was part of a trial on blastocoel fluid collection in order to test the safety of this procedure on pregnancy outcome [22].
Blastocyst generation
Oocytes, sperm, and embryos were cultured in the SAGE sequential culture media system (SAGE, Pasadena, USA) at 37 °C in 5% CO2, 5%O2 in nitrogen. Oocytes retrieval was performed 36 h after the administration of recombinant human chorionic gonadotropin through transvaginal ultrasonography. Oocytes were inseminated using either standard IVF insemination or ICSI; after ICSI, individual oocytes were transferred to a single 25 μl droplets under oil in Cleavage Medium supplemented with 10% Serum Protein Substitute (SAGE, Pasadena, USA), while standard IVF-inseminated oocytes were fertilized in 35 ul Fertilization Medium droplets under oil and transferred on day 1 to the Cleavage Medium. Fertilization results were assessed at 16–20-h post-sperm insemination. On day 3, and in some cases on day 2, 2/3 embryos were selected for transfer to each patient in the study; the embryos in excess of the number appropriate for transfer were cultured in single 25 μl droplets under oil in the Blastocyst Medium up to day 5 or day 6, and were cryopreserved at the blastocyst stage. Human blastocysts were scored up 120 or 144 h after insemination according to the expansion of the blastocoel cavity and number and integrity of both the inner cell mass (ICM) and trophectoderm (TE).
Isolation of the blastocoel fluid and sample collection
Blastocoel fluids from 75 blastocysts obtained from 52 IVF patients were collected. The blastocoel fluid was isolated by micromanipulation using an Olympus X40 (200×) microscope with Narishige manipulators and pipettes (angle 35°, inner diameter 5.5 um) from Cook (Brisbane, Australia). A droplet of 5–10 μl Blastocyst Medium and a droplet of 3 μl water were prepared in an ICSI dish (Falcon-Corning brand, NY, USA) and covered with oil. The blastocysts were transferred to the droplet with culture medium and the dish was placed in the micromanipulator. The microinjection needle was loaded with water before sampling. The blastocyst was immobilized by the holding pipette with the ICM placed toward the holding pipette. The microinjection needle was then gently led in to blastocyst cavity, the fluid was aspirated until the blastocyst collapsed and the blastocoel fluid was transferred to the water droplet. After micromanipulation, the samples were transferred to a labeled PCR tube using a 10-μl pipette and immediately frozen at −80 °C. To obtain adequate protein material for analysis, the samples were collected and analyzed in two pools according on the patient’s age: group A (blastocoel fluid from 46 samples of women aged <37 years) and group B (29 samples of women aged ≥ to 37 years). Both groups were homogeneous for infertility factor, for dose of gonadotropin administrated for ovarian stimulation and for average of oocytes retrieved after induction.
Proteomic analysis
The proteins of the blastocoel fluid were lysed in a buffer containing 8 M urea, 0.1 M ammonium bicarbonate and the complete protease inhibitor cocktail (Roche, Basel, Switzerland). After reduction and derivatization, proteins were digested with sequence grade trypsin for 16 h at 37 °C using a protein:trypsin ratio of 1:20. The proteolytic digest was desalted using Zip-Tip C18 (Millipore) before mass spectrometric (MS) analysis [23, 24]. LC-ESI-MS/MS analysis was performed on a Dionex UltiMate 3000 HPLC System with a PicoFrit ProteoPrep C18 column (200 mm, internal diameter of 75 μm) (New Objective, USA). Gradient: 1% ACN in 0.1% formic acid for 10 min, 1–4% ACN in 0.1% formic acid for 6 min, 4–30% ACN in 0.1% formic acid for 147 min and 30–50% ACN in 0.1% formic for 3 min at a flow rate of 0.3 μl/min. The eluate was electrosprayed into an LTQ Orbitrap Velos (Thermo Fisher Scientific, Bremen, Germany) through a Proxeon nanoelectrospray ion source. The LTQ-Orbitrap was operated in positive mode in data-dependent acquisition mode to automatically alternate between a full scan (m/z 350–2000) in the Orbitrap (at resolution 60,000, AGC target 1,000,000) and subsequent CID MS/MS in the linear ion trap of the 20 most intense peaks from full scan (normalized collision energy of 35%, 10 ms activation). Isolation window: 3 Da, unassigned charge states: rejected, charge state 1: rejected, charge states 2+, 3+, 4+: not rejected; dynamic exclusion enabled (60 s, exclusion list size: 200) [25]. Data acquisition was controlled by Xcalibur 2.0 and Tune 2.4 software (Thermo Fisher Scientific, MA USA). A sample containing only culture medium, kept as a control (CTR), was also treated and analyzed.
Data analysis
Mass spectra were analyzed using MaxQuant software (version 1.3.0.5). The initial maximum allowed mass deviation was set to 20 ppm for monoisotopic precursor ions and 0.5 Da for MS/MS peaks. Enzyme specificity was set to trypsin, defined as C-terminal to arginine and lysine excluding proline, and a maximum of two missed cleavages were allowed. Carbamidomethylcysteine was set as a fixed modification, N-terminal acetylation, methionine oxidation, and asparagine/glutamine deamidation as variable modifications. The spectra were searched by the Andromeda search engine against the human Uniprot sequence database (release 22.01.2014). The reversed sequences of the target database were used as decoy database. Protein identification required at least one unique or razor peptide per protein group. Quantification in MaxQuant was performed using the built in XIC-based label-free quantification (LFQ) algorithm [26] using fast LFQ. False protein identifications (1 or 5%) [17] were estimated by searching MSMS spectra against the corresponding reversed-sequence (decoy) database. The minimum required peptide length was set to 9 amino acids. Statistical analyses were performed using the Perseus software (version 1.4.0.6). Five technical replicates were carried out for each group (A and B) and for the control. Only proteins present and quantified in at least 3 out of 5 technical repeats and not detected in the sample containing only the culture medium (CTR) were positively identified as blastocoel fluid components and used for statistical analyses. Many keratins known to be due to a contamination during sample handling were excluded from the analysis. The intensity of each protein (measured in at least 3 replicates in group A and group B) in each replicate is used for the t test. Proteins were considered differentially expressed if they were present only in women aged <37 years (group A) or women aged ≥37 years (group B) or showed significant t test difference (filtered for permutation-based False Discovery Rate of many independent hypotheses, cut-off at 5%) between the two groups. The proteins differently expressed in the two groups were clustered according to their functions using the DAVID platform [27] filtered for significant Gene Ontology terms: Biological Process (GOBP) and Molecular Function (GOMF), using a p value <0.05. Protein-protein interactions were analyzed by String [28]. Gene products previously experimentally detected at the protein level as secreted and/or in extracellular region and/or exosome and/or microvesicles were determined according to public available databases (www. neXtProt.org, www.exocarta.org, microvesicle.org, geneontology.org, www.proteinatlas.org), and dataset [29]. Gene products previously detected at the transcription level in blastocystis or embyonic tissue are listed Tables 1, 2, 3, and 4 and S2-according to microarray datasets and gene expression databases (http://genomewidepdb.proteomix.org or https://www.ncbi.nlm.nih.gov/geoprofiles) [16, 30]. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD003535
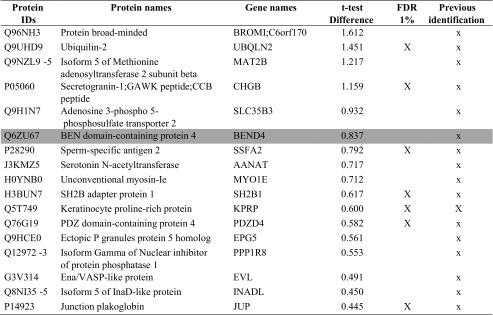
Table 1.
List of the proteins increased in group B
Proteins were considered increased or decreased if they showed significant t test difference (cut-off at 5% permutation-based False Discovery Rate) between group A and B. Previous identification: in the Table the proteins previously identified in human blastocoel fluid (16, 17) are indicated with a capital X, while x indicates gene products previously experimentally detected at the protein level as secreted and/or present in extracellular region and/or in exosome and/or in microvesicles according to public available databases and dataset (www.neXtProt.org, www.exocarta.org, microvesicle.org, geneontology.org, www.proteinatlas.org, 29) or at the gene expression level from microarray data (16, 30) or at the transcription level in blastocystis or embyonic tissue according to http://genomewidepdb.proteomix.org or https://www.ncbi.nlm.nih.gov/geoprofiles databases. The proteins ascribed to fertilization and embryo implantation or nucleic acid interaction categories of Fig. 3 are highlighted in light and dark gray, respectively. The table lists only the proteins present and quantified in at least 3 out of 5 technical repeats using a false positive rate of 0.05 and minimum peptide length 9 amino acids, reporting in a column the proteins identified with FDR 0.01
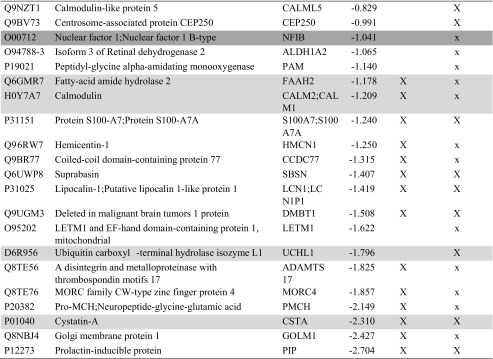
Table 2.
List of the proteins decreased in group B
Proteins were considered increased or decreased if they showed significant t test difference (cut-off at 5% permutation-based False Discovery Rate) between groups A and B. Previous identification: in the Table the proteins previously identified in human blastocoel fluid (16, 17) are indicated with a capital X, while x indicates gene products previously experimentally detected at the protein level as secreted and/or present in extracellular region and/or in exosome and/or in microvesicles according to public available databases and dataset (www.neXtProt.org, www.exocarta.org, microvesicle.org, geneontology.org, www.proteinatlas.org, 29) or at the gene expression level from microarray data (16, 30) or at the transcription level in blastocystis or embyonic tissue according to http://genomewidepdb.proteomix.org or https://www.ncbi.nlm.nih.gov/geoprofiles databases. The proteins ascribed to fertilization and embryo implantation or nucleic acid interaction categories of Fig. 3 are highlighted in light and dark gray, respectively. The table lists only the proteins present and quantified in at least 3 out of 5 technical repeats using a false positive rate of 0.05 and minimum peptide length 9 amino acids, reporting in a column the proteins identified with FDR 0.01
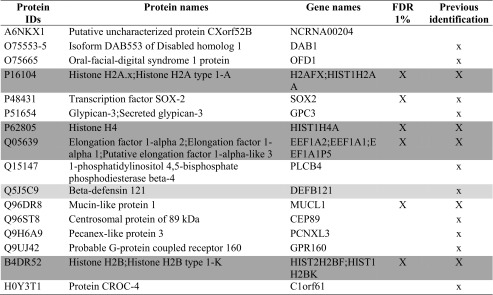
Table 3.
List of the proteins present only in group A
Previous identification: in the Table, the proteins previously identified in human blastocoel fluid (16, 17) are indicated with a capital X, while x indicates gene products previously experimentally detected at the protein level as secreted and/or present in extracellular region and/or in exosome and/or in microvesicles according to public available databases and dataset (www.neXtProt.org, www.exocarta.org, microvesicle.org, geneontology.org, www.proteinatlas.org, 29) or at the gene expression level from microarray data (16, 30) or at the transcription level in blastocystis or embyonic tissue according to http://genomewidepdb.proteomix.org or https://www.ncbi.nlm.nih.gov/geoprofiles databases. The proteins ascribed to fertilization and embryo implantation or nucleic acid interaction categories of Fig. 3 are highlighted in light and dark gray, respectively. The table lists only the proteins present and quantified in at least 3 out of 5 technical repeats using a false positive rate of 0.05 and minimum peptide length 9 amino acids, reporting in a column the proteins identified with FDR 0.01
Table 4.
List of the proteins present only in group B
Previous identification: in the Table the proteins previously identified in human blastocoel fluid (16, 17) are indicated with a capital X, while x indicates gene products previously experimentally detected at the protein level as secreted and/or present in extracellular region and/or in exosome and/or in microvesicles according to public available databases and dataset (www.neXtProt.org, www.exocarta.org, microvesicle.org, geneontology.org, www.proteinatlas.org, 29) or at the gene expression level from microarray data (16, 30) or at the transcription level in blastocystis or embyonic tissue according to http://genomewidepdb.proteomix.org or https://www.ncbi.nlm.nih.gov/geoprofiles databases. The proteins ascribed to fertilization and embryo implantation or nucleic acid interaction categories of Fig. 3 are highlighted in light and dark gray, respectively. The table lists only the proteins present and quantified in at least 3 out of 5 technical repeats using a false positive rate of 0.05 and minimum peptide length 9 amino acids, reporting in a column the proteins identified with FDR 0.01
Results
Proteomic profile of human blastocoel fluid isolated from women aged < and ≥37 years
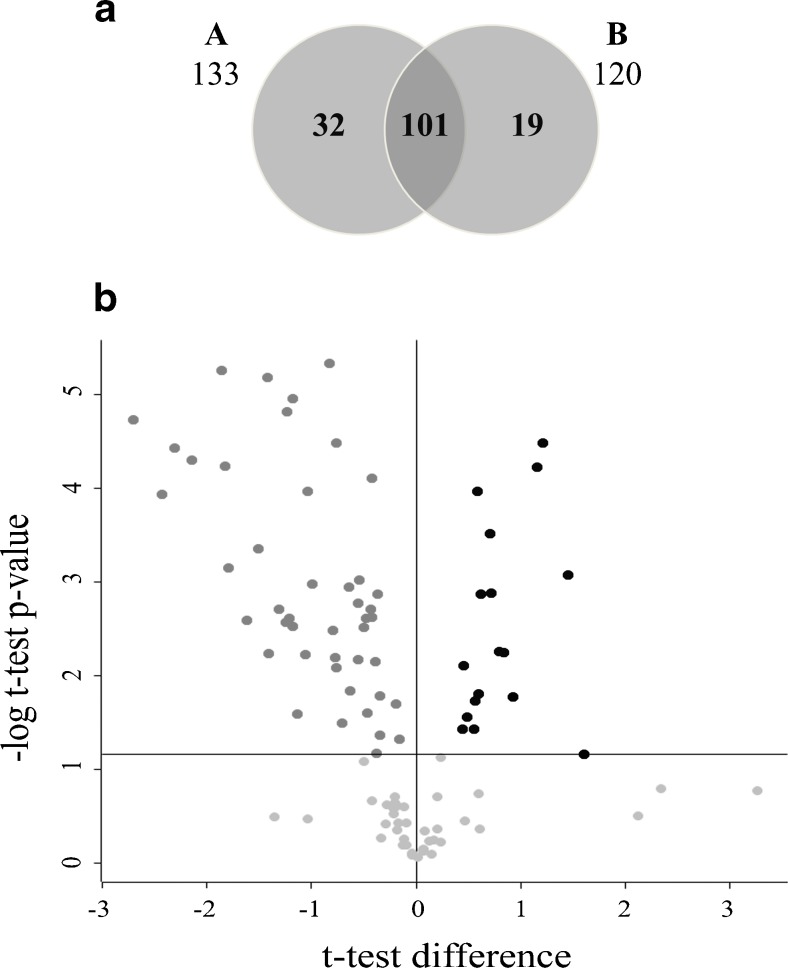
The aim of the study was to examine the protein composition of human blastocoel fluid isolated from blastocysts of women aged <37 years (group A) and women aged ≥37 years (group B) to enlighten differences, at the protein level, that can possibly explain blastocyst survival and implantation success known to be very different in the two groups. Blastocoel fluid was isolated by micromanipulation, and proteins were identified by a shotgun proteomic approach, using label free for the relative quantification of their expression levels. Proteins identified in the control (containing only culture medium) and keratins- likely due to a contamination during sample preparation were discarded from the analysis (Supplementary Table 1). The proteomic analysis leads to the identification and quantification of 148 proteins (Fig. 1); 132 and 116 proteins were identified in group A (Supplementary Table 2, average CV 1.2%) and B (Supplementary Table 3, average CV 1.3%), respectively (Fig. 1a). Among the 100 proteins commonly expressed in both groups, 17 proteins are upregulated and 44 downregulated in group B compared to group A (Fig. 1b). These proteins are listed in Tables 1 and 2, respectively, while Tables 3 and 4 report the proteins expressed only in A (32) or in B (16), respectively. Overall, the analysis identified 33 proteins, which were increased or present only in B while 76 were decreased in B or present only in A (Fig. 1 As reported in Tables 1, 2, 3, and 4, Supplementary Table 2 and 3), 98% of the differentially regulated proteins and 91% of the total proteins identified have been previously experimentally detected either at the protein level in blastocoel fluid, exosomes, microvesicles or extracellular region or at the gene expression level in microarray studies concerning blastocyst transcriptome and gene expression databases.
Fig. 1.
Proteomic profile of human blastocoel fluid isolated from women aged < (group A) and ≥ (group B) 37 years. a Venn diagram of the proteins identified in groups A and B. b Vulcano plot showing the proteins common to both groups differentially expressed in group B compared to group A. Common proteins were considered differentially expressed if they showed significant t test difference (cut-off at 5% permutation-based False Discovery Rate) between the two groups. Proteins up- and downregulated are indicated in black and dark gray, respectively. Proteins which are present at levels not statistically different between groups A and B are indicated in light gray below the horizontal threshold line
Up to now, only two papers addressed the issue of the protein content in the blastocoel fluid by a proteomic analysis: Jansen et al. [17] described the protein content of the blastocoel together with the medium and the cells, while Poli et al. [16], very recently, reported on the proteome of the blastocoel fluid combining the results from shot gun proteomics and gene expression analysis. The present study allows to identify 108 proteins never described before, adding unprecedented information on this important human body fluid.
Classification of the differentially expressed proteins based on bioinformatic analysis
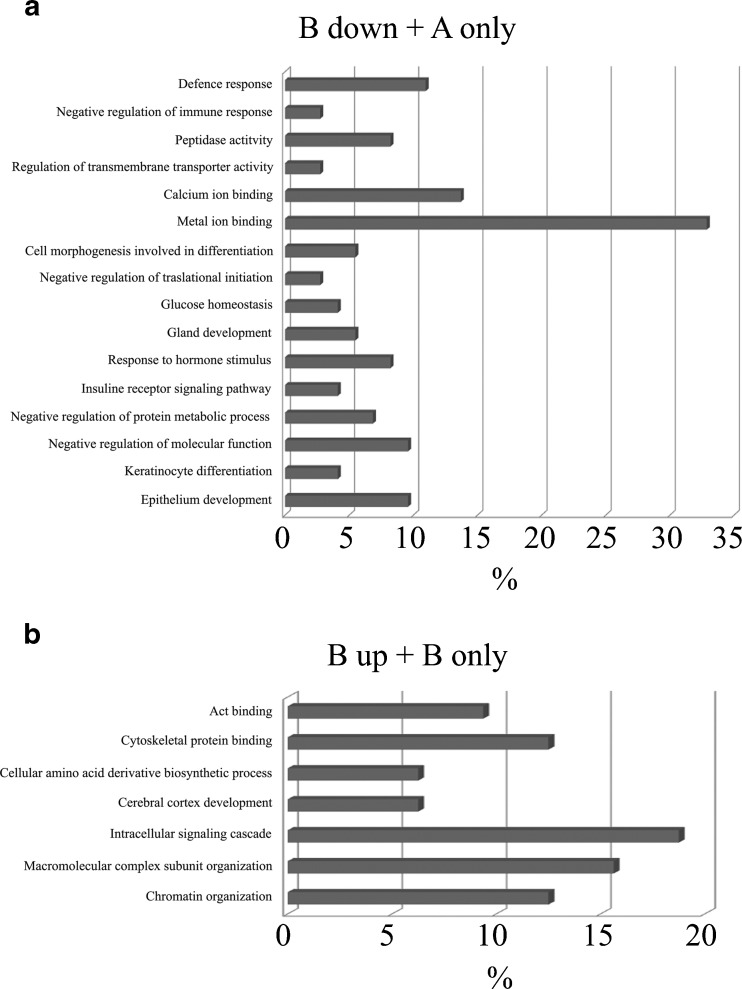
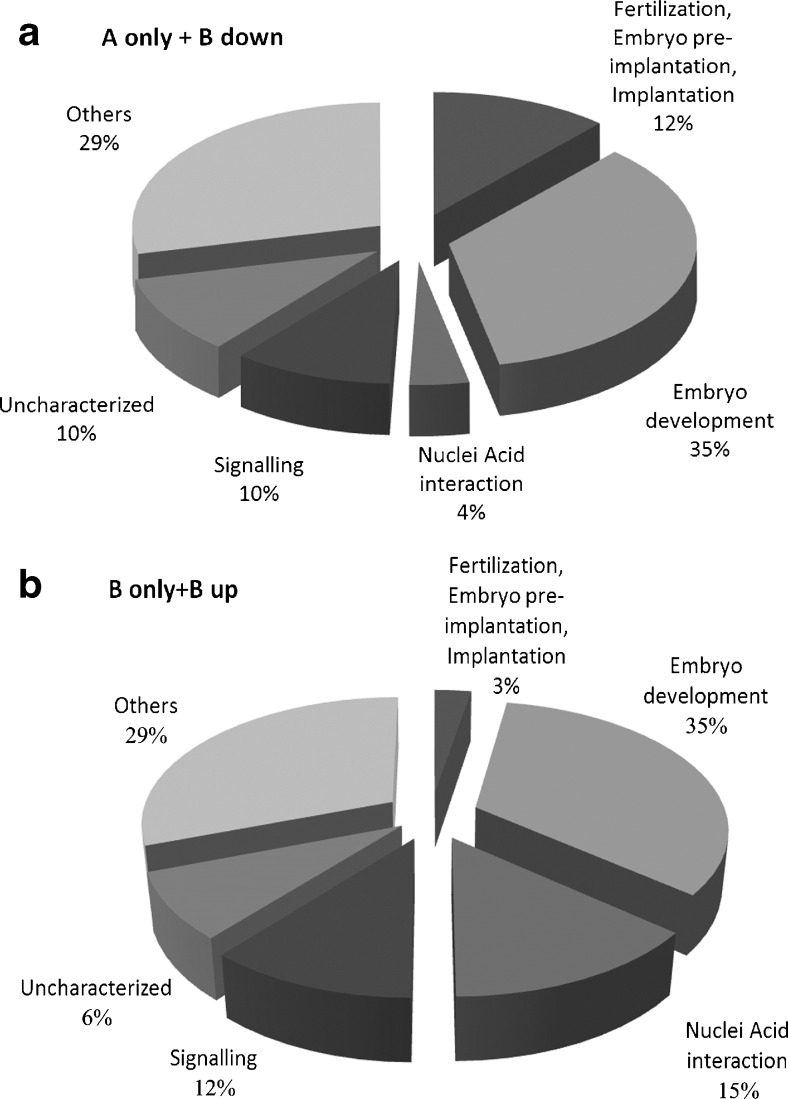
The proteins differentially expressed in the two groups were classified based on Gene Ontology Molecular Function and Biological Processes, as shown in Fig. 2. As a further step in the bioinformatic analysis, the proteins were manually grouped according to their function taking into account, besides GOMF and GOBP, also keywords and known annotations from the literature. It was thus possibile to ascribe each protein to 6 major groups: (1) fertilization and embryo implantation, (2) embryo development, (3) nucleic acid interaction, (4) signaling, (5) uncharacterized proteins, (6) proteins excluded from the groups listed above categorized as “other” (Fig. 3, Supplementary Table 4). Interestingly, comparing the analysis of proteins increased or only expressed in A (Fig. 3a) with the proteins increased or only expressed in B (Fig. 3b), the major differences can be observed in 2 categories: fertilization and embryo implantation (12% group A vs 2% group B) and nucleic acid interaction (4% group A vs 13% group B).
Fig. 2.
Classification of the proteins differentially expressed based on GOMF and GOBP. The proteins differentially expressed in A and B groups were classified based on molecular function (GOMF) and biological processes (GOBP). a Classification of proteins downregulated in B or present only in A. b Classification of proteins upregulated or present only in B
Fig. 3.
Functional analysis of the proteins differentially expressed in A and B. The proteins were manually grouped according to their function in biological processes taking into account, besides GOMF, and GOBP, also keywords and annotations known from literature (Supplementary Table 4). a Classification of proteins downregulated in B or present only in A. b Classification of proteins up regulated or present only in B
Discussion
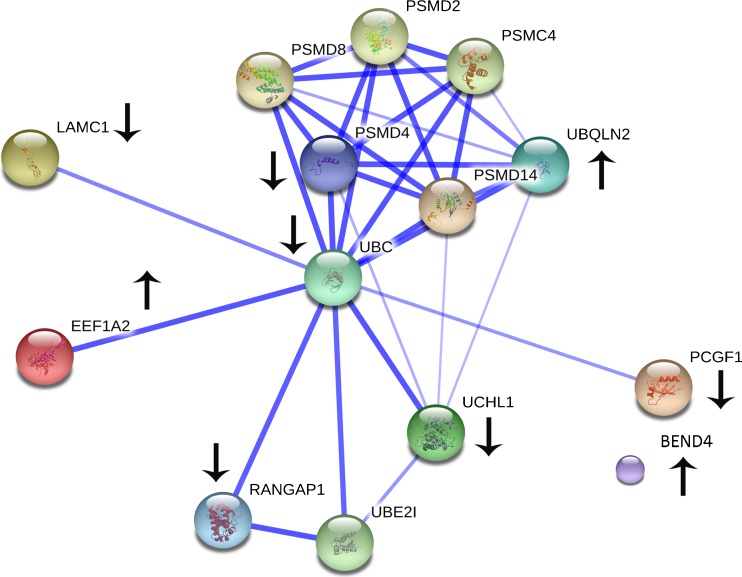
According to the promising results obtained by Jensen and collaborators and by Poli and coworkers [16, 17], to better delineate possible mechanisms for the effect of maternal aging on fertility and potential biomarkers for evaluating the blastocysts quality and improving pregnancy outcome rate in IVF patients, we examined the proteomic profile of blastocoel fluid in woman <37 year old (group A) or ≥37 year old (group B). Comparison between the two matched study groups demonstrated, as already known, that pregnancy rate was lower in group B. In this group, 4/12 (33.3%) pregnancies were obtained after frozen—thawed blastocyst transfer cycles versus group A (13/30 pregnancies, 43.3%). The shotgun proteomic approach allowed identifying 148 proteins, out of which 108 are described for the first time in the blastocoel fluid, and 109 are differentially expressed between the two groups. These proteins were functionally profiled using a suite of bioinformatics annotation tools and pathway databases, to obtain a comprehensive view of the impacted processes, pathways, functions, and networks related to maternal aging. The bioinformatic analysis showed that differential proteins participate in various biological processes, but the main perturbed networks refer to fertilization and embryo implantation, as well as to nucleic acid interactions (Fig. 3 and Table S4). Interestingly, many of the proteins differentially expressed are related to processes involving ubiquitin, that is largely known to play a crucial role in controlling several aspects required to ensure successful pregnancy outcome. In particular, the major molecular functions of these proteins consist of histone turnover and transcription factors, crucially required for cell cycle progression and proliferation. Therefore, it is presumed that alterations in their expression level could affect implantation potential by altering blastocyst homeostasis towards apoptosis and autophagy settings. The following paragraph details the known biological role in reproductive processes of ubiquitin-related proteins which have been identified as blastocoel fluid components in the present study and listed in Tables 1, 2, 3, and 4 and S2-S3. In addition, a network showing the known interactions among up- or downregulated ubiquitin-related proteins is shown in Fig. 4.
Fig. 4.
Protein-protein interaction analysis of the proteins differentially expressed in A and B involved in the ubiquitin system. The analysis was performed by STRING. Stronger associations are represented by thicker lines. Proteins increased or decreased in B are indicated by an arrow. PSMD8, PSMD4, PSMD2, and PSMD14 are isoforms of PSMD4 which was found decreased in B
In the recent years, increasing observations have demonstrated the role of ubiquitin system in reproductive processes, such as gametogenesis, modulation of steroid receptor concentrations, placental development and endo-metrial modification at the beginning of pregnancy [31]. These wide-range effects have led to extensive research and several studies recently demonstrated the increase of ubiquitylation in the uterus during embryo implantation; thus, the proper control and turnover of this key signaling protein activity suggest potential roles in controlling critical aspects of implantation [32–35]. This is also emphasized by the observation that ubiquitin is accumulated in trophoblast of expanding blastocysts [36], suggesting that dysregulation of ubiquitin system may be responsible for failure IVF outcome. In 2009, Katz-Jaffe and collaborators reviewed findings on potential protein biomarkers in the embryo secretome [18], identifying a crucial role for ubiquitin. In our study, a huge number of proteins related to ubiquitin system has been identified and differentially regulated among the two groups analyzed. The ubiquitinase UBC and Ubiquitin Carboxyl-terminal Hydrolase isozyme L1 (UCHL1) are downregulated in aged blastocysts, as well as proteins containing a RING finger domain that plays a key role in the ubiquitination pathway during embryonic development [37, 38] such as RING Finger protein 223 (RNF223) and Kelch-like protein 17 (KLHL17). Instead, the expression of the 26S Proteasome non-ATPase regulatory subunit 4 (PSMD4), a component of the 26S proteasome complex responsible for ubiquitylated proteins degradation, and the AN1-type zinc finger protein 6 (ZFAND6), a polyubiquitin binding protein involved in regulating NF-kappaB signaling [39], are completely switched off. Interestingly, several differential expressed proteins are substrate of Small Ubiquitin MOdifier (SUMO), which is also involved in regulating embryonic viability in mammals [40]. In particular, aged blastocysts lack Ran GTPaseactivating protein 1 (RANGAP1), that regulates pathways mainly involved in differentiation, apoptosis and cell cycle by altering protein function through changes in activity or cellular localization or by protecting substrates from ubiquitination [41]. Instead, they show increased expression of BEN domain-containing protein 4 that belongs to a family of proteins involved in histone deubiquitination. Accumulated evidence in the literature has currently demonstrated that the monoubiquitylation of histone H2A at lysine 119 is crucially associated with transcriptionally repressed chromatin, as a result of the repression activity of Polycomb group proteins [42–46]. The latter proteins are crucial developmental regulators that maintain transcriptional repression of hundreds of genes involved in development from early embryogenesis through birth to adulthood [37, 47–50], as well as in signaling or cancer development using chromatin-based epigenetic mechanisms [51, 52]. In particular, the Polycomb group RING finger protein 1 (PCGF1) acts as key cell growth regulator that promotes cell cycle progression and proliferation by transcriptional repression of the CDK cyclin inhibitor p21Waf1/Cip1 [53]. Interestingly, our study reveals that Polycomb group RING finger protein 1 (PCGF1) has not been detected in aged blastocysts, providing a further confirmation of the key role of ubiquitylation for initiation of pregnancy. Noteworthy, the importance of the crosstalk between Polycomb group proteins and histones has been recently demonstrated by the identification of the Jumonji protein JARID2, a component of the histone methyltransferase complex that regulates binding of the Polycomb repressive complex 2 to target genes in embryonic stem cells [49], as the most represented proteins in the secretome of the preimplantation human embryo able to predict positive pregnancy outcome [54]. Moreover, according to a recent observation describing the important role of histone H2A variants (H2A.X, H2A.Z and macroH2A) in genome remodeling after fertilization [55] and preimplantation embryo development [56], our study reveals that blastocoel fluid of aged woman undergoes a global change in histone composition, as it contains soluble histones (H2A.X, H2B and H4) that are completely absent in younger women.
Noteworthy, any excess of the positively charged histones can allow them to potentially associate non-specifically with negatively charged molecules such as DNA in the cell, resulting in deleterious effects on cell viability and genomic stability [57]. Indeed, recent studies have highlighted a link between increased soluble H2A.X and apoptosis induction [58, 59]. Excessive soluble H2A.X causes chromatin aggregation and inhibition of ongoing gene transcription, but it has also non-nucleosomal functions, specifically pro-apoptotic activities. Therefore, to avoid the problems associated with excess histone accumulation, most eukaryotic cells largely rely on the strict regulation of their histone protein levels and excess histones are degraded by phosphorylation and ubiquitylation-dependent proteolysis [60, 61]. Moreover, a very recent report suggests that histones may be targeted for degradation also via the chaperone-mediated autophagy pathway in mammalian cells [62]. In keeping with these observations, we identified in blastocoel fluid from aged blastocysts an increased amount of Ubiquilin-2 (UBQLN2), which is involved in regulation of different protein degradation mechanisms and pathways including ubiquitin-proteasome system and autophagy, a survival mechanism whose deregulation has been linked to non-apoptotic cell death through inducing cell cycle arrest [63, 64]. Noteworthy, increased autophagy of aged blastocysts is further confirmed by absence in the blastocoel fluid of Laminin subunit gamma-1 (LAMC1), which is crucially involved in protection from autophagy [65, 66].
Besides ubiquitin, our proteomic data reveals that also FAAH2, a key protein in the endocannabinoid system, is differentially expressed in young and aged women blastocoel fluid. Recently, comprehensive studies have indicated that endocannabinoids, a group of bioactive lipids including anandamide (AEA), are actively involved in the process of reproduction and pregnancy, from gametogenesis, uterine receptivity, early embryo-blastocyst development, oviductal transport and implantation through to birth [67–69]. As a consequence of its potential physiological impact, endocannabinoid signaling is finely regulated at multiple levels through several metabolic routes, including the main degradative enzyme fatty acid amide hydrolase (FAAH), that cleaves and inactivates AEA [70]. Changes in the activity and/or expression of FAAH determine significant fluctuations in AEA levels, which in turn can lead to success or failure of pregnancy [71]. In line with this, it has been reported that women in an IVF program who have successful implantation have low serum AEA levels associated with elevated levels of in their peripheral lymphocytes at 6 weeks gestation, whereas low FAAH [72, 73] and high AEA levels in blood are associated with spontaneous threatened miscarriage [74]. In keeping with these observations, our data revealed lower levels of FAAH2, an isoform highly expressed in higher placental mammals [75] in the blastocoel fluid of aged woman. Given the potential impact of this observation, additional studies will be devoted to validate this preliminary result.
In summary, by applying a proteomic approach to compare blastocoel fluid from <37-year-old and ≥37-year-old women blastocysts, we suggest that increased rate of implantation failure in aged IVF patients relies on the pro-apoptotic signature of their blastocysts, as a consequence of the dramatic dysregulation of mechanisms involved in the regulation of ubiquitin system as well as, potentially, in the control of a correct AEA tone. Noteworthy, several additional proteins not related to the ubiquitin system but equally involved in apoptotic processes have been found differentially regulated among the two groups, such as the Isoform 2 of Protein BEX3 (NGFRAP1) [76], Dermicin (DCD) [77], the Isoform 3 of Proline-rich AKT1 substrate 1 (AKT1S1) [78], Calmodulin (CALM) [79], and the eukaryotic translation elongation factor 1A (EEF1A) [80, 81], providing a further confirmation of the pro-apoptotic signature of aged blastocysts.
In conclusion, the results of our deep proteomic investigation provide valuable information on the molecular player involved in biological processes related to blastocyst features in aged women which, in turn, can be used as a starting point towards the identification of non-invasive tools to investigate and ameliorate blastocyst viability, and improving clinical outcome of IVF cycles.
Electronic supplementary material
Below is the link to the electronic supplementary material.
(PDF 94 kb)
(PDF 190 kb)
(PDF 102 kb)
(PDF 371 kb)
Acknowledgements
We thank Dr. Francesca Grassi Scalvini and Dr. Fabiana Santagata for their skillful technical assistance.
Authors’ roles
G.T., E.A., V.P., A.B., and P.E.L.S. designed the research. G.T., E.A., V.P., A.B., E.M., A.N., and S.N. performed the research. G.T., E.A., V.P., A.B., E.M.B., E.M., A.N., S.N., and M.M. analyzed the data. G.T., E.A., V.P., A.B., E.M.B., and M.M. wrote the paper.
Compliance with ethical standards
Funding
None declared.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Capsule
Maternal aging mainly affects blastocyst survival and implantation through unbalancing the equilibrium of theubiquitin system
Gabriella Tedeschi and Elena Albani contributed equally to this work.
References
- 1.Paiva P, Salamonsen LA, Manuelpillai U, Dimitriadis E. Interleukin 11 inhibits human trophoblast invasion indicating a likely role in the decidual restraint of trophoblast invasion during placentation. Biol Reprod. 2009;80(2):302–10. doi: 10.1095/biolreprod.108.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koot YE, Teklenburg G, Salker MS, Brosens JJ, Macklon NS. Molecular aspects of implantation failure. Biochim Biophys Acta. 2012;1822(12):1943–50. doi: 10.1016/j.bbadis.2012.05.017. [DOI] [PubMed] [Google Scholar]
- 3.Dimitriadis E, White CA, Jones RL, Salamonsen LA. Cytokines, chemokines and growth factors in endometrium related to implantation. Hum Reprod Update. 2005;11(6):613–30. doi: 10.1093/humupd/dmi023. [DOI] [PubMed] [Google Scholar]
- 4.Humm KC, Dodge LE, Wu LH, Penzias AS, Malizia BA, Sakkas D, et al. In vitro fertilization in women under 35: counseling should differ by age. J Assist Reprod Genet. 2015 doi: 10.1007/s10815-015-0570-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malizia BA, Hacker MR, Penzias AS. Cumulative live-birth rates after in vitro fertilization. N Engl J Med. 2009;360(3):236–43. doi: 10.1056/NEJMoa0803072. [DOI] [PubMed] [Google Scholar]
- 6.Luke B, Brown MB, Wantman E, Lederman A, Gibbons W, Schattman GL, et al. Cumulative birth rates with linked assisted reproductive technology cycles. N Engl J Med. 2012;366(26):2483–91. doi: 10.1056/NEJMoa1110238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aplin JD. Embryo implantation: the molecular mechanism remains elusive. Reprod Biomed Online. 2006;13(6):833–9. doi: 10.1016/S1472-6483(10)61032-2. [DOI] [PubMed] [Google Scholar]
- 8.Garrido-Gomez T, Dominguez F, Simon C. Proteomics of embryonic implantation. Handb Exp Pharmacol. 2010;198:67–78. doi: 10.1007/978-3-642-02062-9_5. [DOI] [PubMed] [Google Scholar]
- 9.Aydiner F, Yetkin CE, Seli E. Perspectives on emerging biomarkers for non-invasive assessment of embryo viability in assisted reproduction. Curr Mol Med. 2010;10(2):206–15. doi: 10.2174/156652410790963349. [DOI] [PubMed] [Google Scholar]
- 10.Bromer JG, Seli E. Assessment of embryo viability in assisted reproductive technology: shortcomings of current approaches and the emerging role of metabolomics. Curr Opin Obstetr Gynecol. 2008;20(3):234–41. doi: 10.1097/GCO.0b013e3282fe723d. [DOI] [PubMed] [Google Scholar]
- 11.Latham KE, Garrels JI, Chang C, Solter D. Analysis of embryonic mouse development: construction of a high-resolution, two-dimensional gel protein database. Appl Theoret Electrophoresis : Off J Int Electrophoresis Soc. 1992;2(6):163–70. [PubMed] [Google Scholar]
- 12.Shi CZ, Collins HW, Garside WT, Buettger CW, Matschinsky FM, Heyner S. Protein databases for compacted eight-cell and blastocyst-stage mouse embryos. Mol Reprod Dev. 1994;37(1):34–47. doi: 10.1002/mrd.1080370106. [DOI] [PubMed] [Google Scholar]
- 13.Katz-Jaffe MG, Linck DW, Schoolcraft WB, Gardner DK. A proteomic analysis of mammalian preimplantation embryonic development. Reproduction. 2005;130(6):899–905. doi: 10.1530/rep.1.00854. [DOI] [PubMed] [Google Scholar]
- 14.Katz-Jaffe MG, Gardner DK, Schoolcraft WB. Proteomic analysis of individual human embryos to identify novel biomarkers of development and viability. Fertil Steril. 2006;85(1):101–7. doi: 10.1016/j.fertnstert.2005.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Katz-Jaffe MG, Schoolcraft WB, Gardner DK. Analysis of protein expression (secretome) by human and mouse preimplantation embryos. Fertil Steril. 2006;86(3):678–85. doi: 10.1016/j.fertnstert.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Poli M, Ori A, Child T, Jaroudi S, Spath K, Beck M et al. Characterization and quantification of proteins secreted by single human embryos prior to implantation. EMBO Molec Med. 2015; 7(11): 1465–79. doi: 10.15252/emmm.201505344. [DOI] [PMC free article] [PubMed]
- 17.Jensen PL, Beck HC, Petersen J, Hreinsson J, Wanggren K, Laursen SB, et al. Proteomic analysis of human blastocoel fluid and blastocyst cells. Stem Cells Dev. 2013;22(7):1126–35. doi: 10.1089/scd.2012.0239. [DOI] [PubMed] [Google Scholar]
- 18.Katz-Jaffe MG, McReynolds S, Gardner DK, Schoolcraft WB. The role of proteomics in defining the human embryonic secretome. Mol Hum Reprod. 2009;15(5):271–7. doi: 10.1093/molehr/gap012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner RL, Beddington RS. Multi-lineage ‘stem’ cells in the mammalian embryo. J Cell Sci Suppl. 1988;10:11–27. doi: 10.1242/jcs.1988.Supplement_10.2. [DOI] [PubMed] [Google Scholar]
- 20.Chambery A, Colucci-D’Amato L, Vissers JP, Scarpella S, Langridge JI, Parente A. Proteomic profiling of proliferating and differentiated neural mes-c-myc A1 cell line from mouse embryonic mesencephalon by LC-MS. J Proteome Res. 2009;8(1):227–38. doi: 10.1021/pr800454n. [DOI] [PubMed] [Google Scholar]
- 21.Levin Y, Wang L, Ingudomnukul E, Schwarz E, Baron-Cohen S, Palotas A, et al. Real-time evaluation of experimental variation in large-scale LC-MS/MS-based quantitative proteomics of complex samples. J Chromatogr B Anal Technol Biomed Life Sci. 2009;877(13):1299–305. doi: 10.1016/j.jchromb.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 22.Levi-Setti PE, Menduni F, Smeraldi A, Patrizio P, Morenghi E, Albani E. Artificial shrinkage of blastocysts prior to vitrification improves pregnancy outcome: analysis of 1028 consecutive warming cycles. J Assist Reprod Genet. 2016;33(4):461–6. doi: 10.1007/s10815-016-0655-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vernocchi V, Morselli MG, Varesi S, Nonnis S, Maffioli E, Negri A, et al. Sperm ubiquitination in epididymal feline semen. Theriogenology. 2014;82(4):636–42. doi: 10.1016/j.theriogenology.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Maffioli EAF, Nonnis S, Santagata F, Negri A, DeLano FA, Santamaria MH, Kistler EB, Schmid-Schönbein GW, Tedeschi G. Analysis of rat plasma peptidome in hemorrhagic shock. Shock. 2015; 44 (Suppl 2:6).
- 25.Tamplenizza M, Lenardi C, Maffioli E, Nonnis S, Negri A, Forti S, et al. Nitric oxide synthase mediates PC12 differentiation induced by the surface topography of nanostructured TiO2. J Nanobiotechnol. 2013;11:35. doi: 10.1186/1477-3155-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–72. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 27.da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 28.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–52. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fraser KB, Moehle MS, Daher JP, Webber PJ, Williams JY, Stewart CA, et al. LRRK2 secretion in exosomes is regulated by 14-3-3. Hum Mol Genet. 2013;22(24):4988–5000. doi: 10.1093/hmg/ddt346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, et al. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development. 2011;138(17):3699–709. doi: 10.1242/dev.064741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bebington C, Doherty FJ, Fleming SD. The possible biological and reproductive functions of ubiquitin. Hum Reprod Update. 2001;7(1):102–11. doi: 10.1093/humupd/7.1.102. [DOI] [PubMed] [Google Scholar]
- 32.Bebington C, Bell SC, Doherty FJ, Fazleabas AT, Fleming SD. Localization of ubiquitin and ubiquitin cross-reactive protein in human and baboon endometrium and decidua during the menstrual cycle and early pregnancy. Biol Reprod. 1999;60(4):920–8. doi: 10.1095/biolreprod60.4.920. [DOI] [PubMed] [Google Scholar]
- 33.Bebington C, Doherty FJ, Fleming SD. Ubiquitin cross-reactive protein gene expression is increased in decidualized endometrial stromal cells at the initiation of pregnancy. Mol Hum Reprod. 1999;5(10):966–72. doi: 10.1093/molehr/5.10.966. [DOI] [PubMed] [Google Scholar]
- 34.Bebington C, Doherty FJ, Ndukwe G, Fleming SD. The progesterone receptor and ubiquitin are differentially regulated within the endometrial glands of the natural and stimulated cycle. Mol Hum Reprod. 2000;6(3):264–8. doi: 10.1093/molehr/6.3.264. [DOI] [PubMed] [Google Scholar]
- 35.Grzmil P, Altmann ME, Adham IM, Engel U, Jarry H, Schweyer S, et al. Embryo implantation failure and other reproductive defects in Ube2q1-deficient female mice. Reproduction. 2013;145(1):45–56. doi: 10.1530/REP-12-0054. [DOI] [PubMed] [Google Scholar]
- 36.Sutovsky P, Motlik J, Neuber E, Pavlok A, Schatten G, Palecek J, et al. Accumulation of the proteolytic marker peptide ubiquitin in the trophoblast of mammalian blastocysts. Cloning Stem Cells. 2001;3(3):157–61. doi: 10.1089/153623001753205115. [DOI] [PubMed] [Google Scholar]
- 37.Voncken JW, Roelen BA, Roefs M, de Vries S, Verhoeven E, Marino S, et al. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A. 2003;100(5):2468–73. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leeb M, Wutz A. Ring1B is crucial for the regulation of developmental control genes and PRC1 proteins but not X inactivation in embryonic cells. J Cell Biol. 2007;178(2):219–29. doi: 10.1083/jcb.200612127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fenner BJ, Scannell M, Prehn JH. Identification of polyubiquitin binding proteins involved in NF-kappaB signaling using protein arrays. Biochim Biophys Acta. 2009;1794(7):1010–6. doi: 10.1016/j.bbapap.2009.02.013. [DOI] [PubMed] [Google Scholar]
- 40.Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, et al. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9(6):769–79. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 41.Reverter D, Lima CD. Insights into E3 ligase activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex. Nature. 2005;435(7042):687–92. doi: 10.1038/nature03588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, et al. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell. 2004;7(5):663–76. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, et al. Role of histone H2A ubiquitination in Polycomb silencing. Nature. 2004;431(7010):873–8. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 44.Endoh M, Endo TA, Endoh T, Isono K, Sharif J, Ohara O, et al. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet. 2012;8(7):e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, et al. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol. 2014;21(6):569–71. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 46.Luo M, Zhou J, Leu NA, Abreu CM, Wang J, Anguera MC, et al. Polycomb protein SCML2 associates with USP7 and counteracts histone H2A ubiquitination in the XY chromatin during male meiosis. PLoS Genet. 2015;11(1):e1004954. doi: 10.1371/journal.pgen.1004954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qin J, Whyte WA, Anderssen E, Apostolou E, Chen HH, Akbarian S, et al. The polycomb group protein L3mbtl2 assembles an atypical PRC1-family complex that is essential in pluripotent stem cells and early development. Cell Stem Cell. 2012;11(3):319–32. doi: 10.1016/j.stem.2012.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441(7091):349–53. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 49.Pasini D, Cloos PA, Walfridsson J, Olsson L, Bukowski JP, Johansen JV, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–10. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 50.O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330–6. doi: 10.1128/MCB.21.13.4330-4336.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kerppola TK. Polycomb group complexes—many combinations, many functions. Trends Cell Biol. 2009;19(12):692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10(10):697–708. doi: 10.1038/nrn2731. [DOI] [PubMed] [Google Scholar]
- 53.Gong Y, Yue J, Wu X, Wang X, Wen J, Lu L, et al. NSPc1 is a cell growth regulator that acts as a transcriptional repressor of p21Waf1/Cip1 via the RARE element. Nucleic Acids Res. 2006;34(21):6158–69. doi: 10.1093/nar/gkl834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cortezzi SS, Garcia JS, Ferreira CR, Braga DP, Figueira RC, Iaconelli A, Jr, et al. Secretome of the preimplantation human embryo by bottom-up label-free proteomics. Anal Bioanal Chem. 2011;401(4):1331–9. doi: 10.1007/s00216-011-5202-1. [DOI] [PubMed] [Google Scholar]
- 55.Nashun B, Yukawa M, Liu H, Akiyama T, Aoki F. Changes in the nuclear deposition of histone H2A variants during pre-implantation development in mice. Development. 2010;137(22):3785–94. doi: 10.1242/dev.051805. [DOI] [PubMed] [Google Scholar]
- 56.Wu BJ, Dong FL, Ma XS, Wang XG, Lin F, Liu HL. Localization and expression of histone H2A variants during mouse oogenesis and preimplantation embryo development. Genet Molec Res : GMR. 2014;13(3):5929–39. doi: 10.4238/2014.August.7.8. [DOI] [PubMed] [Google Scholar]
- 57.Singh RK, Liang D, Gajjalaiahvari UR, Kabbaj MH, Paik J, Gunjan A. Excess histone levels mediate cytotoxicity via multiple mechanisms. Cell Cycle. 2010;9(20):4236–44. doi: 10.4161/cc.9.20.13636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Y, Tseng M, Perdreau SA, Rossi F, Antonescu C, Besmer P, et al. Histone H2AX is a mediator of gastrointestinal stromal tumor cell apoptosis following treatment with imatinib mesylate. Cancer Res. 2007;67(6):2685–92. doi: 10.1158/0008-5472.CAN-06-3497. [DOI] [PubMed] [Google Scholar]
- 59.Liu Y, Parry JA, Chin A, Duensing S, Duensing A. Soluble histone H2AX is induced by DNA replication stress and sensitizes cells to undergo apoptosis. Mol Cancer. 2008;7:61. doi: 10.1186/1476-4598-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh RK, Kabbaj MH, Paik J, Gunjan A. Histone levels are regulated by phosphorylation and ubiquitylation-dependent proteolysis. Nat Cell Biol. 2009;11(8):925–33. doi: 10.1038/ncb1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Singh RK, Paik J, Gunjan A. Generation and management of excess histones during the cell cycle. Front Biosci (Landmark Ed) 2009;14:3145–58. doi: 10.2741/3441. [DOI] [PubMed] [Google Scholar]
- 62.Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell. 2011;44(6):918–27. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- 63.Li Y, Lu J, Prochownik EV. Dual role for SUMO E2 conjugase Ubc9 in modulating the transforming and growth-promoting properties of the HMGA1b architectural transcription factor. J Biol Chem. 2007;282(18):13363–71. doi: 10.1074/jbc.M610919200. [DOI] [PubMed] [Google Scholar]
- 64.Benbrook DM, Long A. Integration of autophagy, proteasomal degradation, unfolded protein response and apoptosis. Exp Oncol. 2012;34(3):286–97. [PubMed] [Google Scholar]
- 65.Murray P, Edgar D. Regulation of programmed cell death by basement membranes in embryonic development. J Cell Biol. 2000;150(5):1215–21. doi: 10.1083/jcb.150.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carmignac V, Svensson M, Korner Z, Elowsson L, Matsumura C, Gawlik KI, et al. Autophagy is increased in laminin alpha2 chain-deficient muscle and its inhibition improves muscle morphology in a mouse model of MDC1A. Hum Mol Genet. 2011;20(24):4891–902. doi: 10.1093/hmg/ddr427. [DOI] [PubMed] [Google Scholar]
- 67.Taylor AH, Amoako AA, Bambang K, Karasu T, Gebeh A, Lam PM, et al. Endocannabinoids and pregnancy. Clin Chimica Acta; Int J Clin Chem. 2010;411(13-14):921–30. doi: 10.1016/j.cca.2010.03.012. [DOI] [PubMed] [Google Scholar]
- 68.Maccarrone M. Endocannabinoids: friends and foes of reproduction. Prog Lipid Res. 2009;48(6):344–54. doi: 10.1016/j.plipres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 69.Battista N, Bari M, Maccarrone M. Endocannabinoids and reproductive events in health and disease. Handb Exp Pharmacol. 2015;231:341–65. doi: 10.1007/978-3-319-20825-1_12. [DOI] [PubMed] [Google Scholar]
- 70.McKinney MK, Cravatt BF. Structure and function of fatty acid amide hydrolase. Annu Rev Biochem. 2005;74:411–32. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- 71.Rapino C, Battista N, Bari M, Maccarrone M. Endocannabinoids as biomarkers of human reproduction. Hum Reprod Update. 2014;20(4):501–16. doi: 10.1093/humupd/dmu004. [DOI] [PubMed] [Google Scholar]
- 72.Maccarrone M, Bisogno T, Valensise H, Lazzarin N, Fezza F, Manna C, et al. Low fatty acid amide hydrolase and high anandamide levels are associated with failure to achieve an ongoing pregnancy after IVF and embryo transfer. Mol Hum Reprod. 2002;8(2):188–95. doi: 10.1093/molehr/8.2.188. [DOI] [PubMed] [Google Scholar]
- 73.Maccarrone M, Valensise H, Bari M, Lazzarin N, Romanini C, Finazzi-Agro A. Relation between decreased anandamide hydrolase concentrations in human lymphocytes and miscarriage. Lancet. 2000;355(9212):1326–9. doi: 10.1016/S0140-6736(00)02115-2. [DOI] [PubMed] [Google Scholar]
- 74.Habayeb OM, Taylor AH, Finney M, Evans MD, Konje JC. Plasma anandamide concentration and pregnancy outcome in women with threatened miscarriage. Jama. 2008;299(10):1135–6. doi: 10.1001/jama.299.10.1135. [DOI] [PubMed] [Google Scholar]
- 75.Wei BQ, Mikkelsen TS, McKinney MK, Lander ES, Cravatt BF. A second fatty acid amide hydrolase with variable distribution among placental mammals. J Biol Chem. 2006;281(48):36569–78. doi: 10.1074/jbc.M606646200. [DOI] [PubMed] [Google Scholar]
- 76.Jiang JY, Xiong H, Cao M, Xia X, Sirard MA, Tsang BK. Mural granulosa cell gene expression associated with oocyte developmental competence. J Ovarian Res. 2010;3:6. doi: 10.1186/1757-2215-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Esposito G, Schiattarella GG, Perrino C, Cattaneo F, Pironti G, Franzone A, et al. Dermcidin: a skeletal muscle myokine modulating cardiomyocyte survival and infarct size after coronary artery ligation. Cardiovasc Res. 2015;107(4):431–41. doi: 10.1093/cvr/cvv173. [DOI] [PubMed] [Google Scholar]
- 78.Huang B, Porter G. Expression of proline-rich Akt-substrate PRAS40 in cell survival pathway and carcinogenesis. Acta Pharmacol Sin. 2005;26(10):1253–8. doi: 10.1111/j.1745-7254.2005.00184.x. [DOI] [PubMed] [Google Scholar]
- 79.O’Neill C, Li Y, Jin XL. Survival signaling in the preimplantation embryo. Theriogenology. 2012;77(4):773–84. doi: 10.1016/j.theriogenology.2011.12.016. [DOI] [PubMed] [Google Scholar]
- 80.Abbas W, Kumar A, Herbein G. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Front Oncol. 2015;5:75. doi: 10.3389/fonc.2015.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Migliaccio N, Ruggiero I, Martucci NM, Sanges C, Arbucci S, Tate R, et al. New insights on the interaction between the isoforms 1 and 2 of human translation elongation factor 1A. Biochimie. 2015 doi: 10.1016/j.biochi.2015.07.021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 94 kb)
(PDF 190 kb)
(PDF 102 kb)
(PDF 371 kb)