Significance
The transition path time in folding reactions—the time required to cross through the high-energy transition states—offers a powerful tool for studying folding mechanisms because it is uniquely sensitive to the microscopic motions along the transition paths that dominate the folding dynamics. We used transition path times to probe helix formation in nucleic acids and elucidate differences between the formation of canonical G:C and A:T base pairs. Stem-loop hairpins of the same size but increased G:C content had faster transition path times, revealing that the conformational diffusion coefficient determining the timescale for molecular dynamics was higher for formation of G:C base pairs than A:T base pairs. Such sequence dependence, not previously observed, reflects differences in the internal friction for reconfiguring the bases.
Keywords: folding, DNA hairpins, energy landscapes, optical tweezers
Abstract
The conformational diffusion coefficient, D, sets the timescale for microscopic structural changes during folding transitions in biomolecules like nucleic acids and proteins. D encodes significant information about the folding dynamics such as the roughness of the energy landscape governing the folding and the level of internal friction in the molecule, but it is challenging to measure. The most sensitive measure of D is the time required to cross the energy barrier that dominates folding kinetics, known as the transition path time. To investigate the sequence dependence of D in DNA duplex formation, we measured individual transition paths from equilibrium folding trajectories of single DNA hairpins held under tension in high-resolution optical tweezers. Studying hairpins with the same helix length but with G:C base-pair content varying from 0 to 100%, we determined both the average time to cross the transition paths, τtp, and the distribution of individual transit times, PTP(t). We then estimated D from both τtp and PTP(t) from theories assuming one-dimensional diffusive motion over a harmonic barrier. τtp decreased roughly linearly with the G:C content of the hairpin helix, being 50% longer for hairpins with only A:T base pairs than for those with only G:C base pairs. Conversely, D increased linearly with helix G:C content, roughly doubling as the G:C content increased from 0 to 100%. These results reveal that G:C base pairs form faster than A:T base pairs because of faster conformational diffusion, possibly reflecting lower torsional barriers, and demonstrate the power of transition path measurements for elucidating the microscopic determinants of folding.
Structure formation in biological macromolecules like proteins and nucleic acids is a complex, dynamic process. Physically, it is described in the context of energy landscape theory as a diffusive search in conformational space for the minimum-energy structure (1–3). The speed at which this search takes place at the microscopic level is set by the conformational diffusion coefficient, D. Because D plays a key role in determining the kinetics of the folding, as encapsulated in the well-known expression for rates derived by Kramers (4), it is one of the fundamental physical determinants of folding phenomena: it connects the thermodynamics encoded in the energy landscape to the dynamics of conformational changes. The properties of D relate to such key issues as internal friction in the polymer chain (5–8), roughness in the energy landscape (9–12), projection of the full phase-space for conformational dynamics onto experimental reaction coordinates (13), and “speed limits” for folding transitions (14).
Despite its importance, D remains challenging to determine experimentally. Several studies have found D from the reconfiguration times for unfolded proteins or polypeptides (15–19), using fluorescence probes to measure interactions between specific locations on the polymer chain. However, few studies have measured D across the energy barrier separating the folded and unfolded states (12, 20–22), which is not necessarily the same as the value within the unfolded state because D can be position dependent (13, 23). It is the value of D within the barrier region that is most relevant for describing the kinetics of folding. Unfortunately, standard approaches for determining D from the folding rates, such as via Kramers’ kinetic theory (4), tend to suffer from high experimental uncertainty: rates are very sensitive to barrier heights, but errors in barrier height measurements can be quite large (24).
Recently, an alternate approach for determining D based on measurements of the duration of transition paths has been developed (24, 25). Transition paths are the brief portions of a folding trajectory during which the molecule passes through the transition states and undergoes the most significant parts of the structural changes (Fig. 1). In contrast to the situation with folding rates—which are exponentially more sensitive to the barrier height, ΔG‡, than to D (4)—the average transition path time, τtp, is far more sensitive to D than to ΔG‡. For a harmonic barrier in a 1D energy landscape, τtp is given in the high-barrier limit (ΔG‡ > 2kBT) by the following:
| [1] |
where κb is the barrier stiffness, β is the inverse thermal energy, and γ is Euler’s constant (26, 27). As a result, τtp provides a much more sensitive measure of D than does the folding rate (8, 24, 28). Moreover, if the individual transit times (ttp) can themselves be measured, in addition to the average transition path time, then D can also be found independently from the distribution of ttp, PTP(t): in the same limit as above, PTP(t) decays exponentially at long times as follows:
| [2] |
where ωK = βDκb (27).
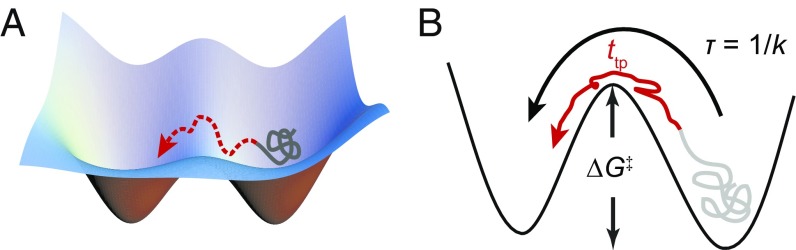
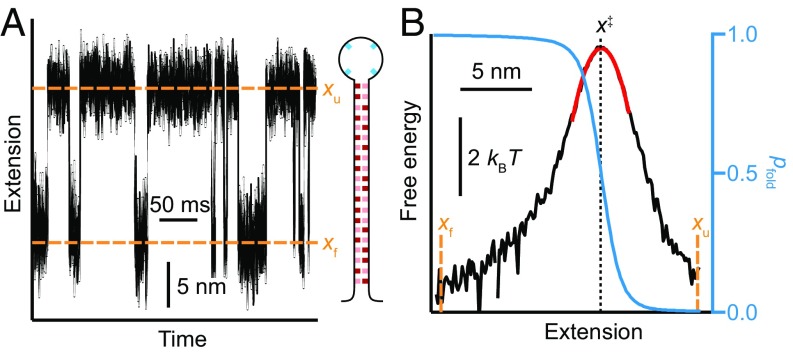
Fig. 1.
Folding transition paths. (A) Transition paths represent the brief portion of folding trajectories (red) where the energy barrier is crossed. (B) Whereas the mean first-passage time (τ) across the barrier is mostly spent waiting for a sufficiently large thermal fluctuation to overcome the barrier height, ΔG‡, the time to cross the transition path (ttp) is much shorter than τ and depends primarily on the rate of diffusion over the landscape, D, rather than ΔG‡.
Advances in single-molecule methods now allow transition times to be measured directly, opening up more detailed and direct studies of conformational diffusion. Fluorescence spectroscopy studies have measured τtp for both proteins (8, 29) and nucleic acids (30), permitting questions such as the origin of internal friction to be investigated in greater detail (28). More recently, force spectroscopy experiments using optical tweezers (31) have measured both τtp (22) and the information-rich distribution PTP(t) (25), as well as other transition path properties such as occupancy statistics (32, 33). Here, we investigate the sequence dependence of conformational diffusion in nucleic acid duplex formation by measuring transition paths in DNA hairpins held under tension in optical tweezers. Nucleic acid hairpins provide a powerful model system for studying folding phenomena because they have been studied extensively at the single-molecule level by experiment (34–39), theoretical and computational models have described experimental results quantitatively (38, 40–42), and hairpin folding can be manipulated predictably through sequence changes (43). We used measurements of τtp and PTP(t) from the folding of hairpins having different stem sequences to determine whether the two canonical Watson–Crick base pairs, A:T (containing two hydrogen bonds) and G:C (containing three), give rise to sequence dependence in D. By applying Eq. 1 to measurements of τtp and fitting PTP(t) to Eq. 2, we found that the conformational diffusion during A:T formation was in fact distinctly slower than for G:C formation.
Results
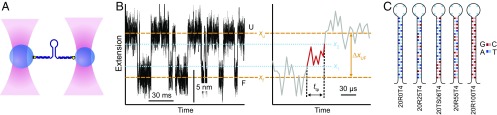
Single DNA hairpins that fold as two-state systems (37, 43) were attached to polystyrene beads via kilobase-long DNA handles as described previously (37), and their extension was measured while held under tension in high-resolution dual-beam optical traps at constant trap separation (Fig. 2A). We kept the stiffness of each trap high (0.56–0.63 for one trap and 0.75–1.1 pN/nm for the other) to obtain maximal time resolution as measured from the instrument response time (25), 6–9 μs (Fig. S1 and Methods). The applied load was such that the hairpins fluctuated rapidly in equilibrium between folded and unfolded states around the force where each state was occupied equally, F1/2, allowing transition paths across the barrier to be observed directly (Fig. 2B) similar to previous work (25). We measured five different hairpins having G:C content ranging from 0 to 100% but identical stem length: hairpins 20R0/T4, 20R25/T4, 20R55/T4, 20R100/T4, and 20TS06/T4 from refs. 37 and 43. These hairpins have been well characterized previously (37, 43) and have important differences, such as an energy barrier in hairpin 20TS06/T4 that is both higher and stiffer than in the other hairpins. The hairpin sequences are listed in Table S1 and illustrated in Fig. 2C.
Fig. 2.
Force spectroscopy measurements of transition paths in DNA hairpin folding. (A) Schematic of assay: a single DNA hairpin is attached at each end via double-stranded handles to beads held in optical traps. (B, Left) Segment of extension trajectory for hairpin 20R100/T4, showing repeated transitions between the folded state (F) at extension xf and the unfolded state (U) at xu (dashed lines). (Right) Single transition from the trajectory, revealing the transition path across the barrier region (red). The transition time, ttp, is measured between boundaries x1 and x2 bracketing the barrier region (dotted lines). (C) Sequences of hairpins studied with varying G:C content.
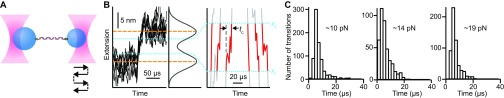
Fig. S1.
Instrumental response time. (A) The response time of the optical tweezers to changes in extension, tc, was measured using a reference construct consisting of DNA handles only. The construct was held at about 10, 14, and 19 pN, and one trap was jumped abruptly back and forth to cause the extension to change by an amount equivalent to the extension change in the folding of the hairpins. (B) Extension trajectories of the handle-only reference construct (black) were analyzed in the same way as the folding/unfolding transitions, measuring tc from individual transitions (red) as the time required to move between the boundaries x1 and x2 (dotted lines). Back-and-forth motion in the trajectories reflects the diffusive motion of the bead. (C) The distribution of response times was peaked near 4–6 μs for the different forces, with an average of 6 ± 1 (20 pN) to 9 ± 1 (10 pN) μs.
Table S1.
DNA hairpins
| Hairpin name | Sequence | G:C, % | No. transitions | |
| U | R | |||
| 20R100T4 | CGCCGCGGGCCGGCGCGCGG–(T)4–CCGCGCGCCGGCCCGCGGCG | 100 | 20,700 | 20,705 |
| 20R55T4 | GAGTCAACGTCTGGATCCTG–(T)4–CAGGATCCAGACGTTGACTC | 55 | 18,262 | 18,292 |
| 30R50T4 | GAGTCAACGTACTGATCACGCTGGATCCTA–(T)4–TAGGATCCAGCGTGATCAGTACGTTGACTC | 50 | 24,591 | 24,600 |
| 20TS06T4 | GCCGGCTATTATTTATATTC–(T)4–GAATATAAATAATAGCCGGC | 35 | 6,816 | 6,824 |
| 20R25T4 | AAGTTAACATCTAGATTCTA–(T)4–TAGAATCTAGATGTTAACTT | 25 | 26,702 | 26,719 |
| 20R0T4 | TATTATATATTAATATATAA–(T)4–TTATATATTAATATATAATA | 0 | 24,811 | 24,949 |
The sequence of the DNA hairpins and their GC content. Number of transitions shown for unfolding (U) and refolding (R).
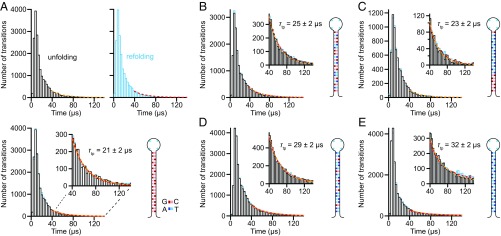
We identified individual transition paths from the extension trajectories as those parts of the trajectory crossing between the boundaries x1 and x2 bracketing the barrier region. For consistency when comparing the different hairpins, x1 and x2 were defined based on the extension change between the folded and unfolded states, ΔxFU, being located at 1/4ΔxFU above the folded state and 1/4ΔxFU below the unfolded state, respectively (Fig. 2B, dotted lines). We then measured individual transit times (ttp) directly from the trajectory as the time required to pass between x1 and x2, or vice versa (25). The hairpin stem lengths were all equal, to ensure that ΔxFU was similar for all hairpins and hence that artifactual differences in ttp owing to travel over different distances were minimized. The transit time distributions PTP(t) for unfolding and refolding (Fig. 3, unfolding, black; refolding, cyan) were the same within error for each hairpin, as expected from time-reversal symmetry (27).
Fig. 3.
Distribution of individual transit times. (A) Distributions for the time to cross the barrier region during transition paths for unfolding (black) and refolding (cyan) of hairpin 20R100/T4. The exponential tails of the distributions are fit to Eq. 2 (unfolding, yellow; refolding, red). (Lower) The two distributions are virtually identical; (Inset) exponential tails. (B–E) Unfolding (black) and refolding (cyan) distributions, with fits to exponential tails (Insets; unfolding, yellow; refolding, red), for hairpins (B) 20R55/T4, (C) 20TS06/T4, (D) 20R25/T4, and (E) 20R0/T4.
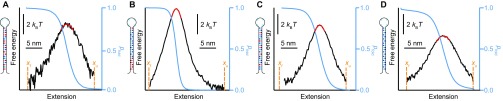
We next reconstructed the shape of the energy barrier to determine the barrier height and stiffness, which are needed to find D from Eqs. 1 and 2. The barriers were reconstructed from equilibrium trajectories of the molecular extension using a method based on the committor statistics (44); as we showed previously using DNA hairpins, this method allows barrier shapes to be determined without the need to deconvolve instrumental compliance effects (24, 43, 45–47). We measured the extension at F1/2 (Fig. 4A), but this time at constant force, using a passive force clamp (48) to avoid feedback loop artifacts (49). We calculated the splitting probability, pfold (Fig. 4B, blue), as described in Methods and then used Eq. 3 to reconstruct the barrier profile (Fig. 4B, black). The barrier height was found directly from this reconstructed landscape, and the curvature was measured from a harmonic fit to the barrier top (Fig. 4B, red). Repeating these calculations for each hairpin sequence (Fig. S2), we found that ΔG‡ varied a few kBT with changing sequence but κb was roughly constant, with the exception of hairpin 20TS06/T4, which had a barrier that was noticeably sharper (Table S2).
Fig. 4.
Landscape reconstruction. (A) Extension trajectory at constant force for hairpin 20R100/T4. (B) Energy landscape profile of the barrier between the folded and unfolded states (black) reconstructed from the empirical splitting probability, pfold (blue), showing a harmonic fit to the barrier top (red).
Fig. S2.
Energy landscape reconstructions from pfold. Energy landscapes (black) reconstructed from pfold (blue) using Eq. 3, with harmonic fit to the barrier peak (red) to determine curvature, for hairpins (A) 20R55/T4, (B) 20TS06/T4, (C) 20R25/T4, and (D) 20R0/T4.
Table S2.
Energy landscape parameters from pfold analysis
| Hairpin | ΔG‡, kBT | κb, kBT/nm2 | ΔG′‡tp, kBT |
| 20R100T4 | 5.0 ± 0.2 | 0.27 ± 0.03 | 2.7 ± 0.1 |
| 20R55T4 | 4.4 ± 0.2 | 0.25 ± 0.04 | 2.5 ± 0.1 |
| 30R50T4 | 9.1 ± 0.1 | 0.29 ± 0.02 | 4.3 ± 0.2 |
| 20TS06T4 | 6.4 ± 0.2 | 0.4 ± 0.1 | 4.1 ± 0.2 |
| 20R25T4 | 4.8 ± 0.2 | 0.29 ± 0.03 | 2.3 ± 0.1 |
| 20R0T4 | 3.8 ± 0.2 | 0.24 ± 0.02 | 2.0 ± 0.1 |
Errors represent SEM.
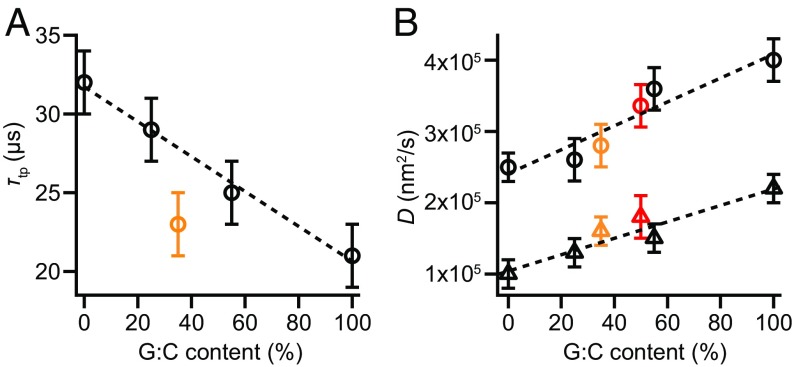
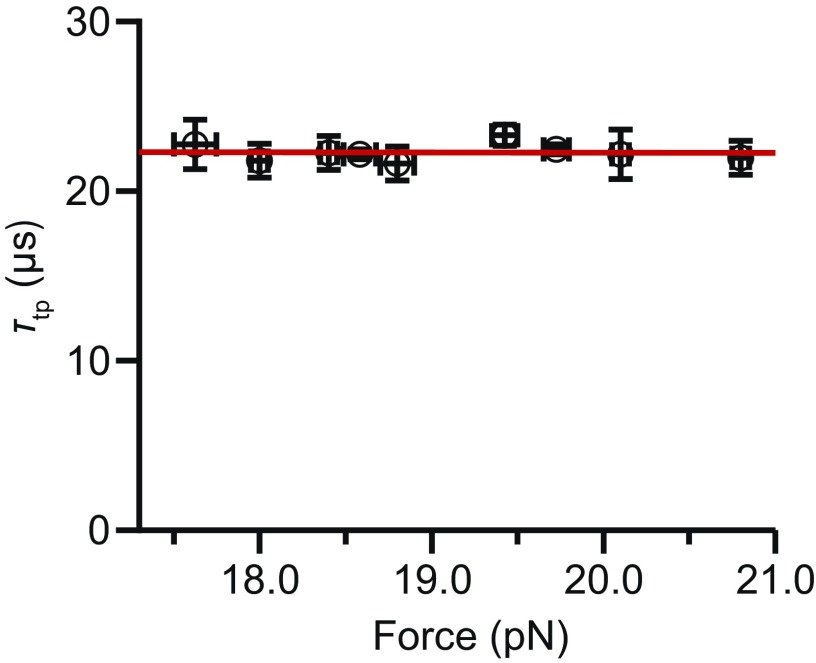
Finally, we found D for each of the hairpins studied by combining the individual transit time measurements with the landscape analysis. We calculated D in two independent ways for each hairpin: via Eq. 1 using τtp, averaging over all transitions (Fig. 5A), and by fitting the exponential tail of PTP(t) to Eq. 2 (Fig. 3, Insets). For these calculations, we used the height of the barrier measured with respect to the barrier region boundaries x1 and x2, rather than the full activation barrier, as argued recently (50). The results, based on 13,000–53,000 transitions for each of the five hairpins studied, show a linear rise of D with G:C content (Fig. 5B). Numerical values for all results are listed in Table S3. Note that hairpin unfolding force also correlates linearly with G:C content (37), hence the diffusion coefficient might be varying with force instead of G:C content. We verified that D did not have any appreciable force dependence by measuring transition paths for hairpin 20R100/T4 at difference forces, over the range of forces (∼3 pN) within which sufficient transitions could be detected to calculate τtp reliably (Fig. S3). Determining D from Eq. 1, we found that force had little, if any, effect on D—less than 10% of the effect observed from changing the sequence—confirming that the differences in D did indeed arise primarily from sequence differences.
Fig. 5.
G:C dependence of τtp and D. (A) The average transition path time decreases linearly with increasing G:C content, with the exception of hairpin 20TS06/T4 (orange). (B) D increases linearly with G:C content for all hairpins, including hairpin 20TS06/T4 (orange), as determined from τtp (circles) and from fitting the exponential tail of PTP(t) (triangles). Error bars represent SEM, averaging over both folding and unfolding data for all molecules measured. Data points in red represent values for hairpin 30R50/T4 derived from measurements in ref. 25.
Table S3.
Numerical results for each hairpin
| Hairpin name | τtp, μs | D from τtp, ×105 nm2/s | D from PTP(t), ×105 nm2/s | ||||||
| U | R | Avg | U | R | Avg | U | R | Avg | |
| 20R100T4 | 21 ± 2 | 21 ± 2 | 21 ± 2 | 4.1 ± 0.3 | 4.1 ± 0.3 | 4.1 ± 0.3 | 2.2 ± 0.2 | 2.1 ± 0.2 | 2.2 ± 0.2 |
| 20R55T4 | 24 ± 1 | 25 ± 1 | 25 ± 2 | 3.5 ± 0.3 | 3.6 ± 0.3 | 3.6 ± 0.3 | 1.5 ± 0.2 | 1.4 ± 0.2 | 1.5 ± 0.2 |
| 30R50T4 | 27 ± 2 | 28 ± 2 | 28 ± 2 | 3.5 ± 0.3 | 3.4 ± 0.3 | 3.5 ± 0.3 | 1.8 ± 0.3 | 1.8 ± 0.3 | 1.8 ± 0.2 |
| 20TS06T4 | 23 ± 2 | 22 ± 2 | 23 ± 2 | 3.1 ± 0.3 | 3.1 ± 0.3 | 3.1 ± 0.3 | 1.5 ± 0.2 | 1.6 ± 0.2 | 1.6 ± 0.2 |
| 20R25T4 | 29 ± 1 | 29 ± 1 | 29 ± 2 | 2.5 ± 0.3 | 2.6 ± 0.3 | 2.6 ± 0.3 | 1.3 ± 0.2 | 1.3 ± 0.2 | 1.3 ± 0.2 |
| 20R0T4 | 31 ± 2 | 32 ± 2 | 32 ± 2 | 2.5 ± 0.2 | 2.5 ± 0.2 | 2.5 ± 0.2 | 1.0 ± 0.1 | 1.0 ± 0.2 | 1.0 ± 0.2 |
Results for unfolding transitions (U), refolding transition (R), and the average (Avg). Errors represent SEM.
Fig. S3.
Force dependence of τtp. τtp for the hairpin 20TS06T4 over a force range of 3 pN, in which the hairpin changed from mostly folded to mostly unfolded. Transition path time varied very little with the force for the hairpin (slope, 0.02 ± 0.02 μs/pN), indicating little to no change in D with force.
Discussion
For all hairpins but one, τtp decreased linearly with increasing G:C content, being 50% longer for hairpins with only A:T base pairs compared with hairpins with only G:C base pairs. The exception, hairpin TS06/T4, had a shorter transition time than expected based on the G:C content and was almost as fast as hairpin 20R100/T4. However, this hairpin was designed to have a different barrier than the others: the barrier is both higher and stiffer. The stiffer barrier increases the speed for crossing the transition paths much more than the higher barrier height reduces it (Eq. 1). Indeed, when we look at D for this hairpin instead of τtp (Fig. 5B), we find that it lies on the same linear curve as for the other hairpins—the effect of the stiffer barrier is no longer apparent. This result shows that analyzing the average transition time alone, without also characterizing D, may sometimes be misleading: it is D that is the more fundamental descriptor of the folding, because τtp is affected by factors like the shape of the barrier.
From Fig. 5, we see that there are distinct differences between the diffusion coefficients for A:T and G:C base-pair formation during folding. G:C base pairs are thus not only more stable, because of their extra hydrogen bond, but they also undergo conformational fluctuations faster. To confirm that the sequence-dependent values for D obtained from these five hairpins represent a basic property of the base pairs, we performed the same analysis on previous measurements of a hairpin with a stem that is 50% longer, hairpin 30R50/T4 (25). Notably, the values for D calculated from Eqs. 1 and 2 (Fig. 5B, red) matched very well the results expected based on the G:C content of the hairpin, given the G:C dependence of D found from the shorter hairpins.
The qualitative trend in D (i.e., a linear increase in D with G:C content) is the same whether D is found from τtp via Eq. 1 (Fig. 5B, circles) or the fit to the exponential tail via Eq. 2 (Fig. 5B, triangles), as shown by replotting the results in terms of relative changes (Fig. S4). However, the quantitative results differ: the values found from τtp are consistently about twofold higher. One possible source of this discrepancy is that the assumptions used in the analyses—that the folding involves simple 1D diffusion over a harmonic barrier—are not completely correct. The barriers do differ significantly from a purely harmonic profile: whereas the very tops of the barriers are well approximated as harmonic, regions further away are not (Fig. 4B, black). Such anharmonicity can alter the properties of the transition paths (51), with flatter energy profiles (as in the regions away from the barrier top) yielding longer transit times (27), consistent with a longer tail in PTP(t) suggesting slower diffusion. Brownian dynamics simulations of motions transitions across an anharmonic barrier like that in Fig. 4B did indeed find that all transitions were slower than for a harmonic barrier with the same height and curvature (Fig. S5), but D implied by the exponential tail of the distribution decreased about 35% more than did D implied by τtp. These simulations thus suggest that barrier anharmonicity accounts for up to one-half of the observed discrepancy between the two estimates of D. The rest of the discrepancy presumably arises from contributions from other factors, such as deviations from 1D behavior that distort PTP(t) from expectations, or the mechanical coupling of the hairpins to the linkers and beads, which can alter kinetic properties (24, 52–55) including transition times (24, 55). Such effects are expected to be relatively small, however: previous analysis of some of the same DNA hairpins found that the conditional transition path probability matched expectations for 1D diffusion over the measured landscapes quite well (32), and our measurements are done in a limit where the mechanical coupling artifacts are small (56).
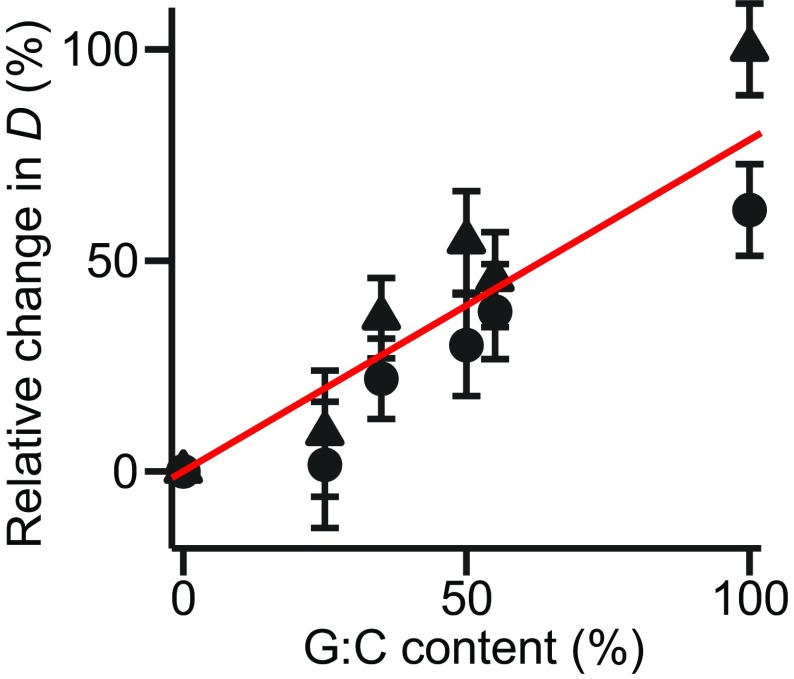
Fig. S4.
Relative change in D with G:C content. The change in D relative to the result at 0 G:C content, based on Eq. 1 (circles) or Eq. 2 (triangles).
Fig. S5.
Transit time distributions from Brownian dynamics simulations. The transit times observed in simulations of diffusive motion over a 1D anharmonic energy profile similar to that in Fig. 4B (red) are longer than those (black) found for a harmonic energy profile having the same barrier height and curvature. The timescale for the exponential decay of the distribution for the anharmonic barrier increased 35% more than did the average transition path time, compared with the results for the harmonic barrier, leading to a discrepancy between D calculated from Eq. 1 versus Eq. 2 for the anharmonic barrier.
What might account for the difference in diffusion coefficient between A:T and G:C base pairs? Because D is inversely proportional to the friction coefficient, one possible explanation is that the internal friction during base-pair formation is different for the different base pairs. We speculate that the sequence dependence of D may result from sequence-dependent differences in the torsion angle dynamics during folding, motivated by recent computational simulations indicating that torsion angle isomerization effects are a key source of internal friction in proteins (57, 58) and earlier work on RNA hairpins showing that torsion angle isomerization can cause dramatic changes in rates (41). It is known, for example, that G can adopt the syn conformation of the glycosidic bond between the base and the backbone more readily than other nucleotides, such as in Z-DNA (59). The energy barrier for syn/anti isomerization is predicted to be lower for purines than pyrimidines (60), and the isomerization rate was observed to be faster for G than A (61). Calculations of dinucleoside energies in torsion angle hyperspace also found generally broader energy minima for G:C-paired dinucleosides than A:T-paired ones (62). Such results suggest that G may be more dynamic in torsion angle space, allowing it to find the right torsion angle faster and hence raising the diffusion coefficient for G:C formation compared with A:T formation. If this picture is correct, we can estimate the difference in the torsion angle energy barriers using the theory of Zwanzig (9), treating the problem as diffusion over a set of periodic “microbarriers” between each base pair, each of which is too small and too close together to resolve on its own. The factor of ∼2 difference for D between A:T and G:C would then reflect a torsional barrier that is just marginally higher for A:T, by about 1 kBT.
This work provides a first look at the sequence dependence of conformational diffusion, showing how measurements of individual transit times can be used to probe the fundamental properties that govern folding dynamics. Our results suggest that the kinetic models widely used to describe nucleic acid duplex formation (35, 38, 40) can now be refined, replacing the constant microscopic rates typically assumed with rates that are sequence dependent, so as to reflect the twofold difference in D between G:C and A:T base-pair formation. An interesting implication of the sequence dependence of D is that D should be position dependent along the duplex, leading to changes in the shape of the transition paths (51). Such position dependence has been previously sought but not reliably observed (44, 63); it may soon be detectable through analysis of transition path shapes.
Methods
Samples and Measurements.
DNA hairpin constructs containing hairpins of the desired sequence connected at each terminus to double-stranded DNA handles were prepared as described previously (25). Hairpin constructs were attached to avidin- and anti-digoxigenin–labeled polystyrene beads, respectively, via biotin and digoxigenin labels at the ends of the handles to create dumbbells (Fig. 2A). For measurement, dumbbells were put in 50 mM Mops, pH 7.5, 200 mM KCl, with an oxygen-scavenging system [8 mU/μL glucose oxidase, 20 mU/μL catalase, 0.01% (wt/vol) d-glucose] to prevent oxidative damage. Constant-force measurements were made using a passive force clamp (48), measuring in the linear-stiffness region of one trap (stiffness, 0.3 pN/nm) and the zero-stiffness region of the other trap, sampling at 20–50 kHz and filtering on-line at the Nyquist frequency. Constant-position measurements were made with stiffness 0.56–0.63 pN/nm in one trap and 0.75–1.1 pN/nm in the other, sampling data at 124–400 kHz and again filtering on-line at the Nyquist frequency. Additional details about the properties of the hairpins (unfolding forces, rates, barrier location, etc.) may be found in previous work (37, 43). Owing to the effect of compliance (48), the extension change observed during unfolding/refolding transitions in measurements at constant trap position was shorter than that in experiments at constant force.
Energy Barrier Reconstructions.
The shape of the energy barrier for each hairpin was reconstructed from the extension trajectories using the committor, pfold, as described previously (44). Briefly, pfold(x) was calculated empirically from the trajectory at each value of the extension, and then the landscape was reconstructed using the following:
| [3] |
This method recovers the shape of the landscape in the region between the folded and unfolded states, allowing the barrier height and stiffness to be measured directly from the reconstruction. Note that Eq. 3 assumes that the diffusion coefficient is constant along the reaction coordinate, and the method depends on extension being a good reaction coordinate (else the barrier height and curvature will appear to be broader than they actually are). Previous work on some of the same hairpins discussed here, however, showed that extension was a good reaction coordinate (32). Furthermore, landscapes reconstructed by Eq. 3 were found to agree well with those reconstructed from the inverse Boltzmann transform (which makes no assumptions about D), suggesting not only that the correct barrier height and curvature are being reliably recovered (within experimental error) but also that any effects from the small sequence dependence of D do not noticeably alter the landscape reconstruction (44). To reduce noise in the numerical differentiation, pfold(x) was first smoothed in a 1-nm window with a boxcar filter. The barrier height within just the barrier region (used for calculating D from Eq. 1), ΔG‡tp (values listed in Table S2), was found by averaging the energy difference between the top of the barrier and the energy at the two boundaries defining the barrier region, x1 and x2.
Instrument Response Time.
The instrument response time was measured over the range of forces studied here (10–20 pN) similarly to previous work (25), using constructs that were the same as those for the hairpin measurements but without any hairpin between the handles. The traps were moved apart suddenly to cause a change in the extension (Fig. S1) equivalent to the extension change seen in the hairpin measurements, and the resulting motion of the beads was measured. Individual response trajectories were analyzed in a similar way as the folding/unfolding transitions (Fig. S1B), obtaining the distribution of times required to cross between boundaries x1 and x2 separated by the same distance as in the hairpin trajectories. The average relaxation time varied from 6 ± 1 μs at 20 pN to 9 ± 1 μs at 10 pN.
Transition Time and Diffusion Coefficient Analysis.
Transitions were identified as those parts of the trajectories traversing between the boundaries x1 and x2 separating the folded and unfolded states, respectively, from the transition region, as described previously (25). Briefly, the boundaries were chosen to bracket the middle half of the distance between the folded and unfolded states (ΔxUF), so that x1 = xf + 1/4ΔxUF and x2 = xu − 1/4ΔxUF, where xf and xu are the positions of the folded and unfolded states, respectively (Fig. 2B, Inset). An adaptive smoothing algorithm was used to mitigate the effects of noise in the data. First, an initial estimate of the transit time for a given transition, ttp, was found by fitting the transition path trajectory to a logistic function. The trajectory was then median-filtered in a window equal to one-half this initial estimate, and ttp was measured directly from the smoothed trajectory as the time required to move between x1 and x2, interpolating linearly between the sampled points where necessary to obtain the best estimate of the crossing time at each boundary (25).
The instrument response time was roughly one-third to one-quarter the average transition time for each hairpin. The finite response time would be expected to alter the observed transition times only for the most rapid transitions, those having transit times close to the response time limit. To avoid any distortion of the shape of the transit time distribution PTP(t) at small times arising from effects of the finite response time, when fitting PTP(t) to Eq. 2 to determine D, we used only the long-duration tail of the distribution, involving transit times 40 μs or longer (severalfold larger than the instrument response time). We verified that the fitting results did not depend on the precise choice of the fitting bound, being the same (within error) for choices between 30 and 50 μs, suggesting that the finite time response does not distort the exponential tail of PTP(t).
We note that care must be taken when calculating D from constant-position measurements to account for compliance corrections to the distance changes that are measured, corrections that arise from the changes in the force during the trajectory (48). Although the time taken to cross the barrier region is not affected by compliance corrections, the apparent distance traversed (i.e., the width of the barrier) is reduced, because the beads move less than the ends of the hairpin. The curvature of the barrier thus appears larger than it truly is, and D is artificially lowered. We account for the compliance correction when calculating D by using the barrier curvature obtained from constant-force measurements, where there is no compliance correction and the motions of the beads match the motions of the hairpin.
Brownian Dynamics Simulations.
Simulations of transition paths arising from diffusive motion over a 1D potential surface were performed following a procedure described previously (64). Briefly, simulations were based on a rescaled, discretized Langevin equation with a random Gaussian force of mean 0. Simulations were run for an anharmonic potential similar to the energy profile in Fig. 4B, rescaled proportionately in energy and distance to reduce the simulation time, and for a harmonic potential with the same barrier height and curvature as the anharmonic potential.
Acknowledgments
We thank Arthur Firmino for performing the Brownian dynamics simulations. This work was supported by the Natural Sciences and Engineering Research Council of Canada, the National Research Council, and Alberta Innovates Technology Futures.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1611602114/-/DCSupplemental.
References
- 1.Bryngelson JD, Wolynes PG. Spin glasses and the statistical mechanics of protein folding. Proc Natl Acad Sci USA. 1987;84(21):7524–7528. doi: 10.1073/pnas.84.21.7524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14(1):70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Dill KA, MacCallum JL. The protein-folding problem, 50 years on. Science. 2012;338(6110):1042–1046. doi: 10.1126/science.1219021. [DOI] [PubMed] [Google Scholar]
- 4.Hanggi P, Talkner P, Borkovec M. Reaction-rate theory—50 years after Kramers. Rev Mod Phys. 1990;62(2):251–341. [Google Scholar]
- 5.Hagen SJ. Solvent viscosity and friction in protein folding dynamics. Curr Protein Pept Sci. 2010;11(5):385–395. doi: 10.2174/138920310791330596. [DOI] [PubMed] [Google Scholar]
- 6.Cellmer T, Henry ER, Hofrichter J, Eaton WA. Measuring internal friction of an ultrafast-folding protein. Proc Natl Acad Sci USA. 2008;105(47):18320–18325. doi: 10.1073/pnas.0806154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Borgia A, et al. Localizing internal friction along the reaction coordinate of protein folding by combining ensemble and single-molecule fluorescence spectroscopy. Nat Commun. 2012;3:1195. doi: 10.1038/ncomms2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung HS, Eaton WA. Single-molecule fluorescence probes dynamics of barrier crossing. Nature. 2013;502(7473):685–688. doi: 10.1038/nature12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zwanzig R. Diffusion in a rough potential. Proc Natl Acad Sci USA. 1988;85(7):2029–2030. doi: 10.1073/pnas.85.7.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyeon C, Thirumalai D. Can energy landscape roughness of proteins and RNA be measured by using mechanical unfolding experiments? Proc Natl Acad Sci USA. 2003;100(18):10249–10253. doi: 10.1073/pnas.1833310100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Volk M, Milanesi L, Waltho JP, Hunter CA, Beddard GS. The roughness of the protein energy landscape results in anomalous diffusion of the polypeptide backbone. Phys Chem Chem Phys. 2015;17(2):762–782. doi: 10.1039/c4cp03058c. [DOI] [PubMed] [Google Scholar]
- 12.Solanki A, Neupane K, Woodside MT. Single-molecule force spectroscopy of rapidly fluctuating, marginally stable structures in the intrinsically disordered protein α-synuclein. Phys Rev Lett. 2014;112(15):158103. doi: 10.1103/PhysRevLett.112.158103. [DOI] [PubMed] [Google Scholar]
- 13.Best RB, Hummer G. Coordinate-dependent diffusion in protein folding. Proc Natl Acad Sci USA. 2010;107(3):1088–1093. doi: 10.1073/pnas.0910390107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubelka J, Hofrichter J, Eaton WA. The protein folding “speed limit.”. Curr Opin Struct Biol. 2004;14(1):76–88. doi: 10.1016/j.sbi.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 15.Hagen SJ, Hofrichter J, Szabo A, Eaton WA. Diffusion-limited contact formation in unfolded cytochrome c: Estimating the maximum rate of protein folding. Proc Natl Acad Sci USA. 1996;93(21):11615–11617. doi: 10.1073/pnas.93.21.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapidus LJ, Eaton WA, Hofrichter J. Measuring the rate of intramolecular contact formation in polypeptides. Proc Natl Acad Sci USA. 2000;97(13):7220–7225. doi: 10.1073/pnas.97.13.7220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nettels D, Gopich IV, Hoffmann A, Schuler B. Ultrafast dynamics of protein collapse from single-molecule photon statistics. Proc Natl Acad Sci USA. 2007;104(8):2655–2660. doi: 10.1073/pnas.0611093104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmad B, Chen Y, Lapidus LJ. Aggregation of α-synuclein is kinetically controlled by intramolecular diffusion. Proc Natl Acad Sci USA. 2012;109(7):2336–2341. doi: 10.1073/pnas.1109526109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranno A, et al. Quantifying internal friction in unfolded and intrinsically disordered proteins with single-molecule spectroscopy. Proc Natl Acad Sci USA. 2012;109(44):17800–17806. doi: 10.1073/pnas.1117368109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neupane K, et al. Transition path times for nucleic acid folding determined from energy-landscape analysis of single-molecule trajectories. Phys Rev Lett. 2012;109(6):068102. doi: 10.1103/PhysRevLett.109.068102. [DOI] [PubMed] [Google Scholar]
- 21.Yu H, et al. Energy landscape analysis of native folding of the prion protein yields the diffusion constant, transition path time, and rates. Proc Natl Acad Sci USA. 2012;109(36):14452–14457. doi: 10.1073/pnas.1206190109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu H, et al. Protein misfolding occurs by slow diffusion across multiple barriers in a rough energy landscape. Proc Natl Acad Sci USA. 2015;112(27):8308–8313. doi: 10.1073/pnas.1419197112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chahine J, Oliveira RJ, Leite VBP, Wang J. Configuration-dependent diffusion can shift the kinetic transition state and barrier height of protein folding. Proc Natl Acad Sci USA. 2007;104(37):14646–14651. doi: 10.1073/pnas.0606506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woodside MT, Lambert J, Beach KSD. Determining intrachain diffusion coefficients for biopolymer dynamics from single-molecule force spectroscopy measurements. Biophys J. 2014;107(7):1647–1653. doi: 10.1016/j.bpj.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neupane K, et al. Direct observation of transition paths during the folding of proteins and nucleic acids. Science. 2016;352(6282):239–242. doi: 10.1126/science.aad0637. [DOI] [PubMed] [Google Scholar]
- 26.Chung HS, Louis JM, Eaton WA. Experimental determination of upper bound for transition path times in protein folding from single-molecule photon-by-photon trajectories. Proc Natl Acad Sci USA. 2009;106(29):11837–11844. doi: 10.1073/pnas.0901178106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chaudhury S, Makarov DE. A harmonic transition state approximation for the duration of reactive events in complex molecular rearrangements. J Chem Phys. 2010;133(3):034118. doi: 10.1063/1.3459058. [DOI] [PubMed] [Google Scholar]
- 28.Chung HS, Piana-Agostinetti S, Shaw DE, Eaton WA. Structural origin of slow diffusion in protein folding. Science. 2015;349(6255):1504–1510. doi: 10.1126/science.aab1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung HS, McHale K, Louis JM, Eaton WA. Single-molecule fluorescence experiments determine protein folding transition path times. Science. 2012;335(6071):981–984. doi: 10.1126/science.1215768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Truex K, Chung HS, Louis JM, Eaton WA. Testing landscape theory for biomolecular processes with single molecule fluorescence spectroscopy. Phys Rev Lett. 2015;115(1):018101. doi: 10.1103/PhysRevLett.115.018101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie DB, Woodside MT. Probing the structural dynamics of proteins and nucleic acids with optical tweezers. Curr Opin Struct Biol. 2015;34:43–51. doi: 10.1016/j.sbi.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Neupane K, Manuel AP, Lambert J, Woodside MT. Transition-path probability as a test of reaction-coordinate quality reveals DNA hairpin folding is a one-dimensional diffusive process. J Phys Chem Lett. 2015;6(6):1005–1010. doi: 10.1021/acs.jpclett.5b00176. [DOI] [PubMed] [Google Scholar]
- 33.Neupane K, Manuel AP, Woodside MT. Protein folding trajectories can be described quantitatively by one-dimensional diffusion over measured energy landscapes. Nat Phys. 2016;12(7):700–703. [Google Scholar]
- 34.Bonnet G, Krichevsky O, Libchaber A. Kinetics of conformational fluctuations in DNA hairpin-loops. Proc Natl Acad Sci USA. 1998;95(15):8602–8606. doi: 10.1073/pnas.95.15.8602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ansari A, Kuznetsov SV, Shen Y. Configurational diffusion down a folding funnel describes the dynamics of DNA hairpins. Proc Natl Acad Sci USA. 2001;98(14):7771–7776. doi: 10.1073/pnas.131477798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liphardt J, Onoa B, Smith SB, Tinoco I, Jr, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292(5517):733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 37.Woodside MT, et al. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc Natl Acad Sci USA. 2006;103(16):6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manosas M, Collin D, Ritort F. Force-dependent fragility in RNA hairpins. Phys Rev Lett. 2006;96(21):218301. doi: 10.1103/PhysRevLett.96.218301. [DOI] [PubMed] [Google Scholar]
- 39.Anthony PC, et al. Electrostatics of nucleic acid folding under conformational constraint. J Am Chem Soc. 2012;134(10):4607–4614. doi: 10.1021/ja208466h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cocco S, Marko JF, Monasson R. Slow nucleic acid unzipping kinetics from sequence-defined barriers. Eur Phys J E Soft Matter. 2003;10(2):153–161. doi: 10.1140/epje/e2003-00019-8. [DOI] [PubMed] [Google Scholar]
- 41.Hyeon C, Thirumalai D. Forced-unfolding and force-quench refolding of RNA hairpins. Biophys J. 2006;90(10):3410–3427. doi: 10.1529/biophysj.105.078030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kuznetsov SV, Ansari A. A kinetic zipper model with intrachain interactions applied to nucleic acid hairpin folding kinetics. Biophys J. 2012;102(1):101–111. doi: 10.1016/j.bpj.2011.11.4017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Woodside MT, et al. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314(5801):1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Manuel AP, Lambert J, Woodside MT. Reconstructing folding energy landscapes from splitting probability analysis of single-molecule trajectories. Proc Natl Acad Sci USA. 2015;112(23):7183–7188. doi: 10.1073/pnas.1419490112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta AN, et al. Experimental validation of free-energy-landscape reconstruction from non-equilibrium single-molecule force spectroscopy measurements. Nat Phys. 2011;7(8):631–634. [Google Scholar]
- 46.Engel MC, Ritchie DB, Foster DAN, Beach KSD, Woodside MT. Reconstructing folding energy landscape profiles from nonequilibrium pulling curves with an inverse Weierstrass integral transform. Phys Rev Lett. 2014;113(23):238104. doi: 10.1103/PhysRevLett.113.238104. [DOI] [PubMed] [Google Scholar]
- 47.Woodside MT, Block SM. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu Rev Biophys. 2014;43:19–39. doi: 10.1146/annurev-biophys-051013-022754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Greenleaf WJ, Woodside MT, Abbondanzieri EA, Block SM. Passive all-optical force clamp for high-resolution laser trapping. Phys Rev Lett. 2005;95(20):208102. doi: 10.1103/PhysRevLett.95.208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Elms PJ, Chodera JD, Bustamante CJ, Marqusee S. Limitations of constant-force-feedback experiments. Biophys J. 2012;103(7):1490–1499. doi: 10.1016/j.bpj.2012.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pollak E. Transition path time distribution and the transition path free energy barrier. Phys Chem Chem Phys. 2016;18(41):28872–28882. doi: 10.1039/c6cp05052b. [DOI] [PubMed] [Google Scholar]
- 51.Makarov DE. Shapes of dominant transition paths from single-molecule force spectroscopy. J Chem Phys. 2015;143(19):194103. doi: 10.1063/1.4935706. [DOI] [PubMed] [Google Scholar]
- 52.Wen J-D, et al. Force unfolding kinetics of RNA using optical tweezers. I. Effects of experimental variables on measured results. Biophys J. 2007;92(9):2996–3009. doi: 10.1529/biophysj.106.094052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chang J-C, de Messieres M, La Porta A. Effect of handle length and microsphere size on transition kinetics in single-molecule experiments. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87(1):012721. doi: 10.1103/PhysRevE.87.012721. [DOI] [PubMed] [Google Scholar]
- 54.Nam G-M, Makarov DE. Extracting intrinsic dynamic parameters of biomolecular folding from single-molecule force spectroscopy experiments. Protein Sci. 2016;25(1):123–134. doi: 10.1002/pro.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cossio P, Hummer G, Szabo A. On artifacts in single-molecule force spectroscopy. Proc Natl Acad Sci USA. 2015;112(46):14248–14253. doi: 10.1073/pnas.1519633112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neupane K, Woodside MT. Quantifying instrumental artifacts in folding kinetics measured by single-molecule force spectroscopy. Biophys J. 2016;111(2):283–286. doi: 10.1016/j.bpj.2016.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.de Sancho D, Sirur A, Best RB. Molecular origins of internal friction effects on protein-folding rates. Nat Commun. 2014;5:4307. doi: 10.1038/ncomms5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Echeverria I, Makarov DE, Papoian GA. Concerted dihedral rotations give rise to internal friction in unfolded proteins. J Am Chem Soc. 2014;136(24):8708–8713. doi: 10.1021/ja503069k. [DOI] [PubMed] [Google Scholar]
- 59.Wang AH, Hakoshima T, van der Marel G, van Boom JH, Rich A. AT base pairs are less stable than GC base pairs in Z-DNA: The crystal structure of d(m5CGTAm5CG) Cell. 1984;37(1):321–331. doi: 10.1016/0092-8674(84)90328-3. [DOI] [PubMed] [Google Scholar]
- 60.Foloppe N, Hartmann B, Nilsson L, MacKerell AD., Jr Intrinsic conformational energetics associated with the glycosyl torsion in DNA: A quantum mechanical study. Biophys J. 2002;82(3):1554–1569. doi: 10.1016/S0006-3495(02)75507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nishikawa S, Huang H, Jordan F. Structural effect of nucleotides on syn−anti glycosyl isomerization kinetics by ultrasonic relaxation methods. J Phys Chem B. 2000;104(6):1391–1394. [Google Scholar]
- 62.Borkar A, Ghosh I, Bhattacharyya D. Structure and dynamics of double helical DNA in torsion angle hyperspace: A molecular mechanics approach. J Biomol Struct Dyn. 2010;27(5):695–712. doi: 10.1080/07391102.2010.10508582. [DOI] [PubMed] [Google Scholar]
- 63.Chodera JD, Pande VS. Splitting probabilities as a test of reaction coordinate choice in single-molecule experiments. Phys Rev Lett. 2011;107(9):098102. doi: 10.1103/PhysRevLett.107.098102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim WK, Netz RR. The mean shape of transition and first-passage paths. J Chem Phys. 2015;143(22):224108. doi: 10.1063/1.4936408. [DOI] [PubMed] [Google Scholar]