Abstract
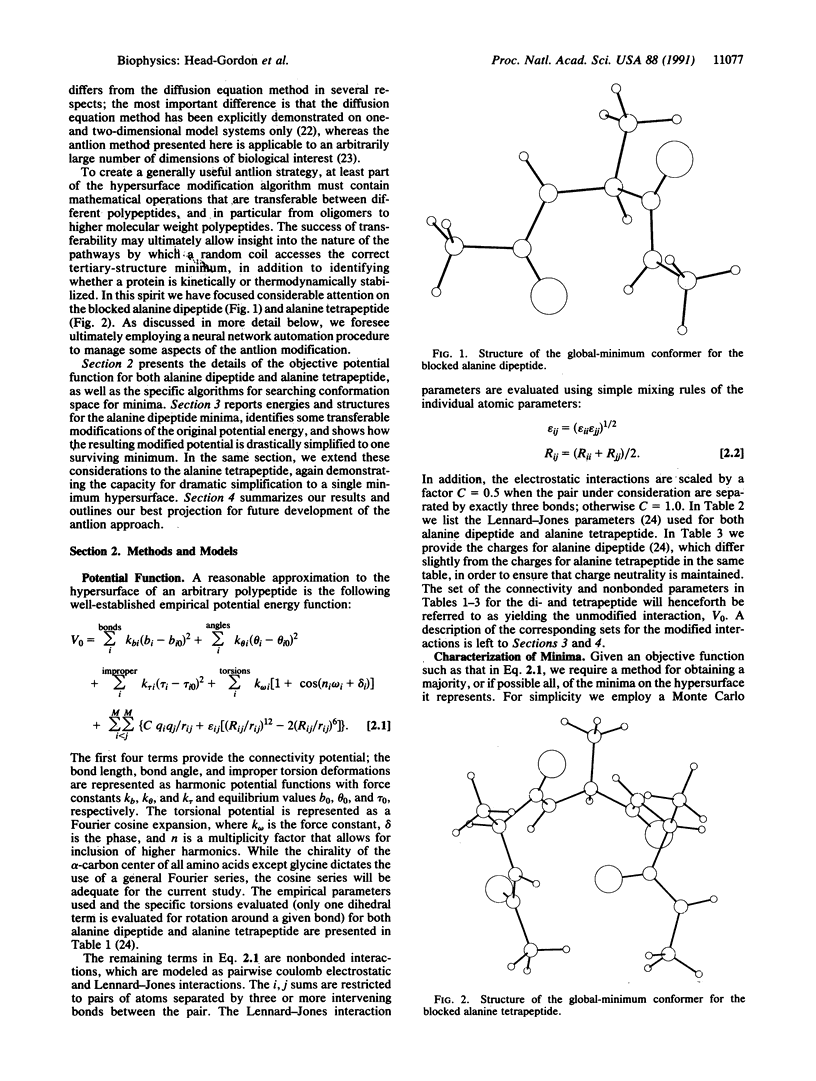
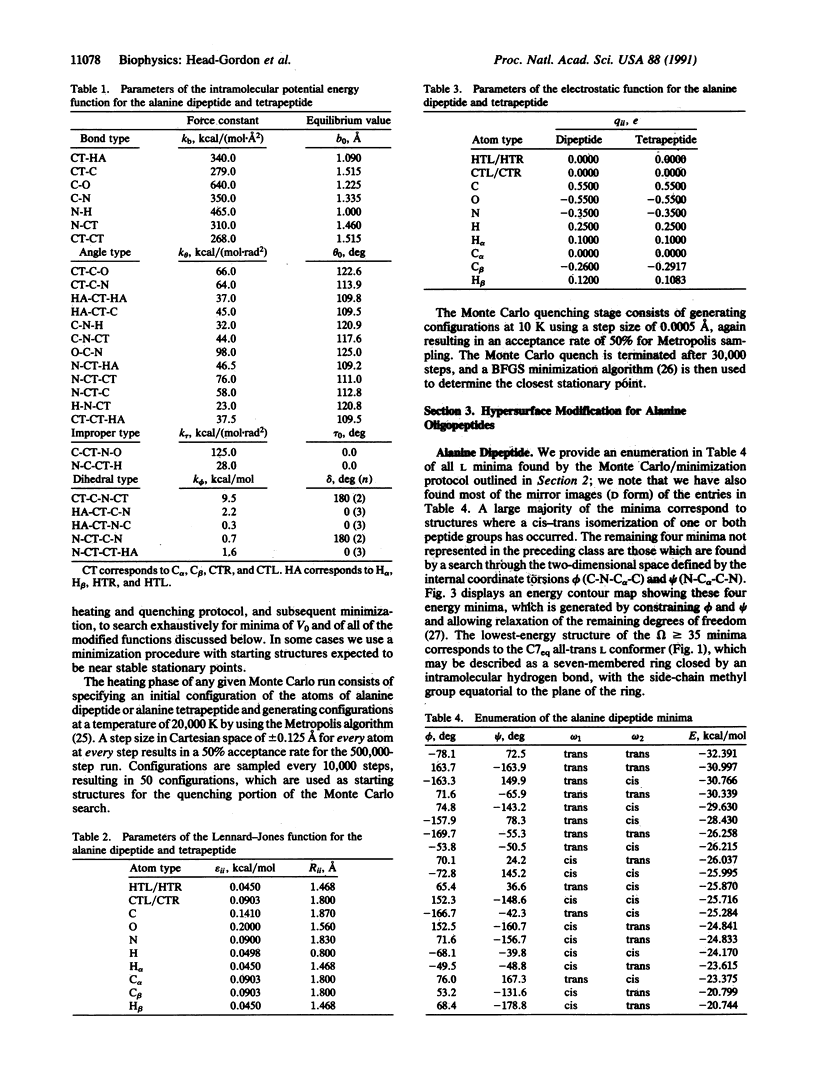
Locating the native structure of a given protein is a task made difficult by the complexity of the potential energy hypersurface and by the huge number of local minima it contains. We have explored a strategy (the "antlion" method) for hypersurface modification that suppresses all minima but that of the native structure. Transferrable penalty functions with general applicability for modifying a hypersurface to retain the desired minimum are identified, and two blocked oligopeptides (alanine dipeptide and tetrapeptide) are used for specific numerical illustration of the dramatic simplification that ensues. In addition, an intermediary role for neural networks to manage some aspects of the antlion strategy applied to large polypeptides and proteins is introduced.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Anfinsen C. B., Scheraga H. A. Experimental and theoretical aspects of protein folding. Adv Protein Chem. 1975;29:205–300. doi: 10.1016/s0065-3233(08)60413-1. [DOI] [PubMed] [Google Scholar]
- Argos P. Analysis of sequence-similar pentapeptides in unrelated protein tertiary structures. Strategies for protein folding and a guide for site-directed mutagenesis. J Mol Biol. 1987 Sep 20;197(2):331–348. doi: 10.1016/0022-2836(87)90127-6. [DOI] [PubMed] [Google Scholar]
- Bohr H., Bohr J., Brunak S., Cotterill R. M., Lautrup B., Nørskov L., Olsen O. H., Petersen S. B. Protein secondary structure and homology by neural networks. The alpha-helices in rhodopsin. FEBS Lett. 1988 Dec 5;241(1-2):223–228. doi: 10.1016/0014-5793(88)81066-4. [DOI] [PubMed] [Google Scholar]
- Creighton T. E. Toward a better understanding of protein folding pathways. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5082–5086. doi: 10.1073/pnas.85.14.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrichs M. S., Wolynes P. G. Toward protein tertiary structure recognition by means of associative memory hamiltonians. Science. 1989 Oct 20;246(4928):371–373. doi: 10.1126/science.246.4928.371. [DOI] [PubMed] [Google Scholar]
- Holley L. H., Karplus M. Protein secondary structure prediction with a neural network. Proc Natl Acad Sci U S A. 1989 Jan;86(1):152–156. doi: 10.1073/pnas.86.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honeycutt J. D., Thirumalai D. Metastability of the folded states of globular proteins. Proc Natl Acad Sci U S A. 1990 May;87(9):3526–3529. doi: 10.1073/pnas.87.9.3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. On the use of sequence homologies to predict protein structure: identical pentapeptides can have completely different conformations. Proc Natl Acad Sci U S A. 1984 Feb;81(4):1075–1078. doi: 10.1073/pnas.81.4.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneller D. G., Cohen F. E., Langridge R. Improvements in protein secondary structure prediction by an enhanced neural network. J Mol Biol. 1990 Jul 5;214(1):171–182. doi: 10.1016/0022-2836(90)90154-E. [DOI] [PubMed] [Google Scholar]
- Kolinski A., Skolnick J., Yaris R. Monte Carlo simulations on an equilibrium globular protein folding model. Proc Natl Acad Sci U S A. 1986 Oct;83(19):7267–7271. doi: 10.1073/pnas.83.19.7267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian N., Sejnowski T. J. Predicting the secondary structure of globular proteins using neural network models. J Mol Biol. 1988 Aug 20;202(4):865–884. doi: 10.1016/0022-2836(88)90564-5. [DOI] [PubMed] [Google Scholar]
- RAMACHANDRAN G. N., RAMAKRISHNAN C., SASISEKHARAN V. Stereochemistry of polypeptide chain configurations. J Mol Biol. 1963 Jul;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Scheraga H. A., Paine G. H. Conformational energy calculations on polypeptides and proteins: use of a statistical mechanical procedure for evaluating structure and properties. Ann N Y Acad Sci. 1986;482:60–68. doi: 10.1111/j.1749-6632.1986.tb20937.x. [DOI] [PubMed] [Google Scholar]
- Stillinger FH. Role of potential-energy scaling in the low-temperature relaxation behavior of amorphous materials. Phys Rev B Condens Matter. 1985 Sep 1;32(5):3134–3141. doi: 10.1103/physrevb.32.3134. [DOI] [PubMed] [Google Scholar]


