Abstract
Background
The fungal genus Aspergillus is of critical importance to humankind. Species include those with industrial applications, important pathogens of humans, animals and crops, a source of potent carcinogenic contaminants of food, and an important genetic model. The genome sequences of eight aspergilli have already been explored to investigate aspects of fungal biology, raising questions about evolution and specialization within this genus.
Results
We have generated genome sequences for ten novel, highly diverse Aspergillus species and compared these in detail to sister and more distant genera. Comparative studies of key aspects of fungal biology, including primary and secondary metabolism, stress response, biomass degradation, and signal transduction, revealed both conservation and diversity among the species. Observed genomic differences were validated with experimental studies. This revealed several highlights, such as the potential for sex in asexual species, organic acid production genes being a key feature of black aspergilli, alternative approaches for degrading plant biomass, and indications for the genetic basis of stress response. A genome-wide phylogenetic analysis demonstrated in detail the relationship of the newly genome sequenced species with other aspergilli.
Conclusions
Many aspects of biological differences between fungal species cannot be explained by current knowledge obtained from genome sequences. The comparative genomics and experimental study, presented here, allows for the first time a genus-wide view of the biological diversity of the aspergilli and in many, but not all, cases linked genome differences to phenotype. Insights gained could be exploited for biotechnological and medical applications of fungi.
Electronic supplementary material
The online version of this article (doi:10.1186/s13059-017-1151-0) contains supplementary material, which is available to authorized users.
Keywords: Aspergillus, Genome sequencing, Comparative genomics, Fungal biology
Background
The genus Aspergillus is one of the best studied genera of filamentous fungi, largely because of the medical (A. fumigatus, A. terreus), food spoilage (A. flavus, A. parasiticus), and industrial (A. niger, A. aculeatus, A. oryzae) relevance of some of its species, in addition to the fundamental studies in the model fungus A. nidulans that have contributed broadly to our understanding of eukaryotic cell biology and molecular processes. Aspergilli can grow in a wide range of niches, mainly in soils and on dead matter, and some are also capable of colonizing living animal or plant hosts and, in total, approximately 350 species have been identified in this genus [1]. The broad relevance and economic importance of the genus has pushed it to the forefront of fungal research, with one of the largest academic and industrial research communities [2].
Aspergillus species are characterized by the unifying feature of the “aspergillum,” an asexual reproductive structure. The aspergilli form a broad monophyletic group, but show large taxonomic divergence with respect to morphology [3] and phylogenetic distance [4]. Genome sequences for three aspergilli [4–6] were among the first to be reported from filamentous fungi and were soon followed by an additional five genomes [7–10]. This has resulted in many genomic, comparative genomic, and post-genomic studies covering a wide variety of topics [11, 12], largely due to the size of the Aspergillus research community. These studies were facilitated by genome resources for this genus, such as CADRE [13] and AspGD [14], in which gene curation and functional annotation of reference species were combined with synteny and orthology analysis. The inclusion of these genomes in MycoCosm [15, 16] enabled comparison to sister and more distant genera. These studies also revealed substantial genomic variations between these species and raised questions about the evolution of various aspects of fungal biology within the genus.
In this study, ten novel genome sequences of the genus Aspergillus were generated, namely A. luchuensis, A. aculeatus, A. brasiliensis, A. carbonarius, A. glaucus, A. sydowii, A. tubingensis, A. versicolor, A. wentii, and A. zonatus. These species were chosen primarily to provide better coverage of the whole genus, to complement the already available genome sequences of A. clavatus, A. fischeri, A. flavus, A. fumigatus, A. nidulans, A. niger, A. oryzae, and A. terreus, and to allow more detailed data mining of the industrially relevant section Nigri (A. luchuensis, A. aculeatus, A. brasiliensis, A. carbonarius, A. niger, A. tubingensis). Additional species from the section Nidulantes were included because of the high divergence of the genome sequence of A. nidulans from the other Aspergillus genomes, and A. sydowii because of its marine life-style and being a pathogen of Gorgonian corals [17]. We demonstrate that this combined set of genomes provides a highly valuable dataset for comparative and functional genomics. This study was performed as a global consortium effort with different researchers addressing different topics as subgroups of the consortium. Where possible, experimental data were generated to examine inferences from the genomic differences and to provide an unprecedented comparative analysis of variation and functional specialization within a fungal genus. The paper is organized in topic based sections, covering:
General comparison of the genomes and phylogenomics
Asexual and sexual reproduction
Primary carbon catabolism
Plant biomass degradation
Secondary metabolism
Stress response
Transporters
Flavohemoglobins and nitric oxide sensitivity
Signal transduction pathway genes
DNA protein organisation and methylation
Results and discussion
General comparison of the genomes and phylogenomics
The genome sequences were generated at the Joint Genome Institute (JGI) and compared to other fungal genomes in JGI’s MycoCosm database [15] with respect to genome size, structure, and gene content (Table 1, Fig. 1, Additional file 1). All Aspergillus genomes are similar in size (29–36 Mb) and GC content (48–53%), while smaller genomes are present in the Onygenales (Table 1, Additional file 1A). The number of predicted genes in the aspergilli is in the range of 9113–13,553, which is generally higher than that observed for the Onygenales (<10,000; Additional file 1B). Larger differences were found for the repetitive portion of the genome, which is in the range of 100 kb to 1.3 Mb for the aspergilli. Genomes of the Aspergillaceae have an average repeat content of 2–3% (Additional file 1C).
Table 1.
Comparison of genome features of 34 ascomycete genomes
| Species | Strain | Class | Order | Section | Genome size (Mb) | GC % | Contigs (n) | Reference |
|---|---|---|---|---|---|---|---|---|
| Aspergillus niger | ATCC1015 | Eurotiomycetes | Eurotiales | Nigri | 35 | 50.3 | 24 | [7] |
| Aspergillus niger | CBS513.88 | Eurotiomycetes | Eurotiales | Nigri | 34 | 50.5 | 470 | [10] |
| Aspergillus luchuensis | CBS106.47 | Eurotiomycetes | Eurotiales | Nigri | 37 | 49.1 | 318 | This study |
| Aspergillus tubingensis | CBS134.48 | Eurotiomycetes | Eurotiales | Nigri | 35 | 49.2 | 87 | This study |
| Aspergillus brasiliensis | CBS101740 | Eurotiomycetes | Eurotiales | Nigri | 36 | 50.5 | 290 | This study |
| Aspergillus carbonarius | CBS141172 | Eurotiomycetes | Eurotiales | Nigri | 36 | 51.7 | 1346 | This study |
| Aspergillus aculeatus | CBS172.66 | Eurotiomycetes | Eurotiales | Nigri | 35 | 50.9 | 851 | This study |
| Aspergillus versicolor | CBS795.97 | Eurotiomycetes | Eurotiales | Nidulantes | 33 | 50.1 | 58 | This study |
| Aspergillus sydowii | CBS593.65 | Eurotiomycetes | Eurotiales | Nidulantes | 34 | 50.0 | 133 | This study |
| Aspergillus nidulans | FGSC A4 | Eurotiomycetes | Eurotiales | Nidulantes | 30 | 50.4 | 82 | [4] |
| Aspergillus flavus | NRRL3557 | Eurotiomycetes | Eurotiales | Flavi | 37 | 48.4 | 765 | [9] |
| Aspergillus oryzae | RIB40 | Eurotiomycetes | Eurotiales | Flavi | 38 | 48.3 | 28 | [5] |
| Aspergillus terreus | NIH2624 | Eurotiomycetes | Eurotiales | Terrei | 29 | 52.9 | 267 | [14] |
| Aspergillus fumigatus | Af293 | Eurotiomycetes | Eurotiales | Fumigati | 29 | 49.8 | 19 | [6] |
| Aspergillus fischeri | NRRL181 | Eurotiomycetes | Eurotiales | Fumigati | 33 | 49.4 | 976 | [8] |
| Aspergillus clavatus | NRRL1 | Eurotiomycetes | Eurotiales | Clavati | 28 | 49.2 | 143 | [8] |
| Aspergillus glaucus | CBS516.65 | Eurotiomycetes | Eurotiales | Aspergillus | 30 | 49.4 | 433 | This study |
| Aspergillus wentii | CBS141173 | Eurotiomycetes | Eurotiales | Cremei | 31 | 48.0 | 118 | This study |
| Penicillium chrysogenum | unknown | Eurotiomycetes | Eurotiales | Chrysogena | 31 | 48.8 | 299 | This study |
| Penicillium rubens | Wisconsin 54-1255 | Eurotiomycetes | Eurotiales | Chrysogena | 32 | 49.0 | 242 | [18] |
| Penicillium digitatum | PHI26 | Eurotiomycetes | Eurotiales | Penicillium | 26 | 48.9 | 304 | [19] |
| Aspergillus zonatus | CBS506.65 | Eurotiomycetes | Eurotiales | Unclassified | 29 | 48.3 | 460 | This study |
| Talaromyces marneffei | ATCC18224 | Eurotiomycetes | Eurotiales | Talaromyces | 29 | 46.7 | 524 | Unpublished |
| Talaromyces stipitatus | ATCC10500 | Eurotiomycetes | Eurotiales | Talaromyces | 36 | 46.1 | 896 | Unpublished |
| Coccidioides immitis | RS | Eurotiomycetes | Onygenales | 29 | 46.0 | 10 | [20] | |
| Coccidioides posadasii | C735 delta SOWgp | Eurotiomycetes | Onygenales | 27 | 46.6 | 55 | [20] | |
| Uncinocarpus reesii | 1704 | Eurotiomycetes | Onygenales | 22 | 48.7 | 582 | [20] | |
| Microsporum canis | CBS113480 | Eurotiomycetes | Onygenales | 23 | 47.5 | 309 | [21] | |
| Trichophyton rubrum | CBS118892 | Eurotiomycetes | Onygenales | 23 | 48.3 | 622 | [21] | |
| Histoplasma capsulatum | NAm1 | Eurotiomycetes | Onygenales | 33 | 46.2 | 2867 | [20] | |
| Paracoccidioides brasiliensis | Pb03 | Eurotiomycetes | Onygenales | 29 | 44.5 | 552 | [22] | |
| Trichoderma reesei | QM6a | Sordariomycetes | Hypocreales | 33 | 52.8 | 131 | [23] | |
| Neurospora crassa | OR74A | Sordariomycetes | Sordariales | 41 | 48.3 | 411 | [24] | |
| Saccharomyces cerevisiae | S288C | Saccharomycetes | Saccharomycetales | 12 | 38.3 | 16 | [25] |
The novel genomes generated in this study are in bold. The order of species follows the taxonomic organization (Fig. 1a)
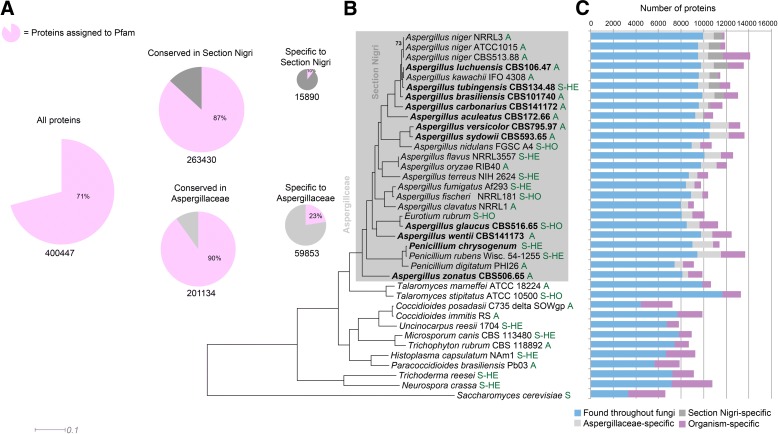
Fig. 1.
Genome overview of Aspergillus and comparative species. a Core genes, based on MCL clustering of protein sequences, and cluster membership in section Nigri and the Aspergillaceae. Pink area of pie charts indicates proteins assignable to one or more Pfam domain; white or gray areas indicate proteins with no Pfam domain. The majority of proteins conserved in Section Nigri, Aspergillaceae, or the full set of comparative fungi could be assigned to a Pfam. Contrastingly, most clade-specific proteins (occurring only in Section Nigri or Aspergillaceae) were not assignable to any Pfam. b Maximum likelihood phylogeny inferred from 149 conserved protein sequences. Organisms newly sequenced for this study are indicated in bold. All bootstrap values are 100 except where indicated. Section Nidulantes is inferred to be a sister group to section Nigri, in contrast to previous studies. Letters in green behind the strain numbers indicate the reproductive state: A asexual, S-HO sexual homothallic, S-HE sexual heterothallic. c Protein conservation, inferred from MCL clustering of proteins, indicates that the majority of proteins in Aspergillaceae have homologs in other fungi, bars showing number of proteins being aligned to individual species to the left-hand side in (b). Some 21% of proteins are specific to the Aspergillaceae, while 14% of proteins in section Nigri are specific to that clade. Organism-specific proteins make up 9% for the Aspergillaceae as a whole and 8% for section Nigri
A substantial proportion (80% on average) of the identified genes in Aspergillaceae genomes have homologs in other lineages of fungi, indicating that around 20% are specific to Aspergillaceae. Many such genes are specific to a clade, e.g. the genomes of section Nigri contain an average of ~1800 Nigri-specific genes (Fig. 1a). The number of unique genes per species differs significantly, even between the three A. niger genomes included in the analysis (Fig. 1b and c). However, different approaches to genome annotation and gene calling may have contributed to these differences, given that not all genomes were sequenced at the same time or by the same technology, nor analyzed with the same software. Therefore, the true number of unique genes is likely to be lower. There is also the possibility that some apparent differences are caused by genes missing due to gaps in the different genome sequences. Protein sequence identity within the Aspergillaceae ranges from 64% (e.g. between A. zonatus and A. versicolor) to 99% in A. niger strains NRRL3 and ATCC 1015, which compares to the average 68% amino acid identity reported in previous studies, and is similar to the divergence between mammals and fish [3].
A genome-wide phylogenetic analysis was conducted to determine the relationship between the newly sequenced genomes and other related species present in MycoCosm. This resulted in a highly supported phylogram providing insights into taxonomic relationships within Aspergillus and between Aspergillus and related genera. With the introduction of the single name nomenclature for fungal anamorph and teleomorph forms, there is discussion over whether to split Aspergillus or to maintain the genus as one. One argument for splitting is the indication that the genus is paraphyletic. In previous taxonomic studies on Aspergillaceae, up to five genes were studied and deeper nodes in these phylograms were either poorly or not supported [26, 27]. An important finding is that our analysis using 149 conserved protein sequences (Fig. 1b) places almost all Aspergillus species in a monophyletic clade that is sister to a clade containing Penicillium species. The only exception was A. zonatus, which is consistent with studies suggesting that A. zonatus is incorrectly classified in Aspergillus and is instead most closely related to Penicilliopsis clavariiformis [26]. The genus Aspergillus has traditionally been classified into a number of subgenera and sections based on morphological and phylogenetic analysis, although precise relationships for many groups have remained uncertain [19]. Our analysis indicates that the subgenus Circumdati appears to be polyphyletic, with A. flavus, A. oryzae, and A. terreus forming one clade and the black aspergilli (sect. Nigri) forming a separate clade. Aspergillus versicolor, A. sydowii, and A. nidulans, all members of the section Nidulantes, were positioned between these two clades. Aspergillus wentii occupied a basal position in the subgenus Aspergillus near to A. glaucus and A. ruber, consistent with the xerophilic nature of other species in this subgenus. Within the section Nigri, A. luchuensis (previously referred to as A. foetidus or A. acidus [28]) and A. tubingensis were sister species related to A. niger, while A. brasiliensis occupied a more basal position in agreement with other phylogenetic analyses [1, 29].
Asexual and sexual reproduction
One reason for the widespread occurrence of Aspergillus species is their ability to produce prolific numbers of mitotically derived conidia, which are dispersed by wind and colonize new substrates. These asexual spores are produced from conidiophores, which have a characteristic structure in most aspergilli consisting of a stalk with a vesicle bearing phialides from which chains of conidia radiate outwards [30, 31]. About 64% of Aspergillus species are known to reproduce only by asexual means, but the remainder can also reproduce sexually, forming structures known as cleistothecia within which meiotically derived ascospores are produced [32]. Where sexual reproduction occurs, it involves either homothallic (self-fertility) or heterothallic (obligate out-crossing) breeding systems, with different types of cleistothecia produced according to taxonomic grouping [33, 34]. The diversity of reproductive modes and their associated morphologies in the aspergilli provide excellent opportunities to study the evolutionary genetics of asexual and sexual reproduction in fungi.
Asexual reproduction
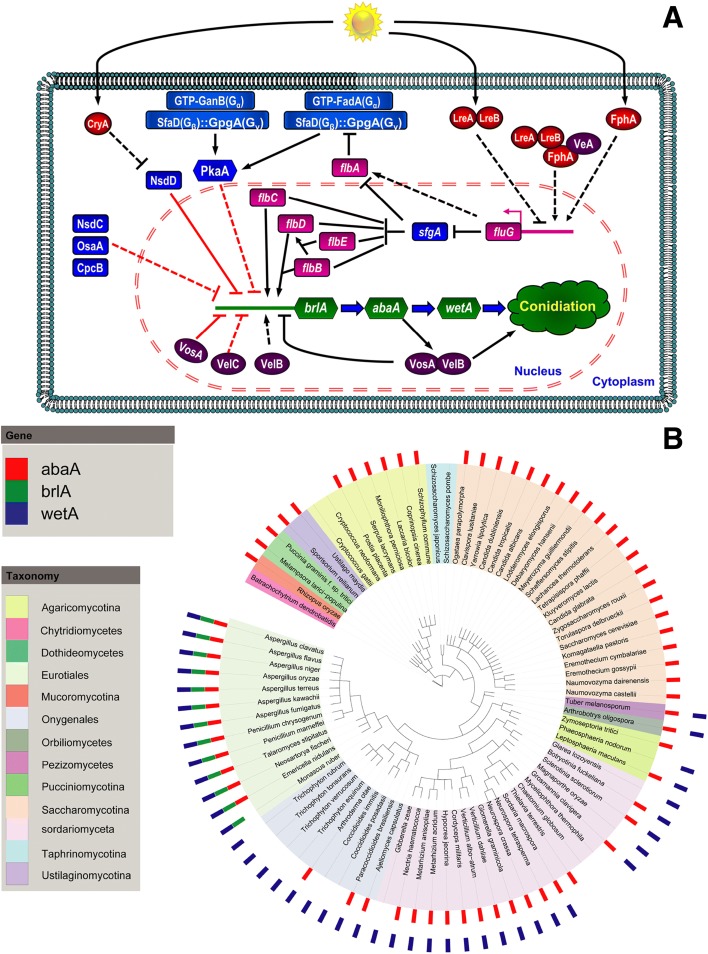
The regulation of asexual sporulation (conidiation) has been extensively studied in the model fungus A. nidulans. Many regulatory genes have been identified and can be divided into central regulators, upstream activators, negative regulators, light-responsive, and velvet regulators (Fig. 2a) [35]. A central regulatory pathway (brlA, abaA, and wetA) controls conidiation-specific gene expression and asexual developmental processes [36]. Proper activation of brlA requires upstream developmental activators (encoded by fluG, flbA-E) [30] and removal of repression by several negative regulators (including SfgA, VosA, NsdD, and two G-protein signaling pathways; see “Signal transduction pathway” section below) [35, 37]. Light also regulates expression of asexual developmental genes, including brlA and upstream activators, with several photoreceptors identified such as one red-light (FphA) and three blue-light receptors [38]. Moreover, velvet regulators can interact with each other and with non-velvet proteins to control development and sporogenesis [39]. Comparison of the conidiation system of A. nidulans to other aspergilli revealed that the regulatory genes are highly conserved, suggesting similar triggers and signaling of the conidiation regulatory pathways in this genus (Additional file 2). The central regulatory pathway (brlA → abaA → wetA) is present in all Aspergillus and Penicillium genomes, whereas only one or two elements of this pathway were found in the Ascomycotina in general, so making this a central conserved feature of the aspergilli and penicillia (Fig. 2b). Normally, these and other regulatory genes were only present in single copy. However, some absences and occurrences of gene duplication were observed (Additional file 2). Notably, Monascus ruber lacks an abaA homolog, correlating with a change in conidiation form [40], whereas A. oryzae and A. carbonarius have two copies of abaA and wetA. Regarding upstream developmental activators, almost half of the species, including A. flavus, A. fumigatus, and A. terreus, possess two copies of the fluG gene, suggesting more differentiated regulation of development in these species. Two copies of the blue light responsive lreA and lreB transcription factor genes were found in A. carbonarius and related experimental work showed this species had the highest levels of asexual sporulation in blue light (relative to white light) of all tested aspergilli (Additional files 2 and 3). Finally, multiple copies of the fphA red light receptor were found in some species, although in this case there was no clear correlation with the extent of asexual sporulation under different light treatments (Additional file 4).
Fig. 2.
Regulatory pathway of asexual sporulation in A. nidulans. a Central regulators, upstream activators, negative regulators, velvet regulators, and light-responsive regulators are illustrated by green, light purple, blue, dark purple, and red icons, respectively. b Distribution of central regulators for asexual sporulation in 85 fungi. These fungi are representatives from the phyla Ascomycota, Basidiomycota, Chytridiomycota, and Zygomycota. Their genome protein sequences were searched for homologs of AbaA, BrlA, and WetA by BlastP using sequences of A. nidulans AbaA, BrlA, and WetA as queries. Details of these fungi are presented in Central Regulator Strain Information. As shown, BrlA seems to be limited to the Eurotiales group, suggesting a specific role for conidiation in Eurotiales fungi. By contrast, WetA is widely distributed in Pezizomycotina fungi, which suggests a general function for the synthesis of cell wall layers to make conidia mature and impermeable. Surprisingly, AbaA is widespread being found in the phyla Ascomycota, Basidiomycota, and Zygomycota, suggesting that AbaA is involved not only in conidial development, but also has other general functions in fungal development
Sexual reproduction
The apparently asexual nature of the majority of Aspergillus species has precluded the use of the sexual cycle as a tool for strain improvement and genetic analysis in them. However, there is accumulating evidence that many such supposedly asexual species have the potential for sexual reproduction if the correct mating partners and environmental conditions to induce sexuality can be identified [32, 33]. The Aspergillus species included in this study were typical in that the majority of them have no known sexual cycle (Additional file 5). To investigate whether the asexual species may have a cryptic sexual cycle, the presence and expression of both mating-type (MAT) genes conferring sexual identity [41] and a pheromone signaling pathway involved with mating [42] were explored. All of the presumed asexual species were found to contain either a MAT1-1 alpha gene or MAT1-2 gene encoding a high-mobility group domain protein, with adjacent gene synteny conserved across all species. This is consistent with heterothallism, involving the presence of either MAT1-1 or MAT1-2 idiomorphs [41]. All of the presumed asexual species also contain ppgA homologs encoding a pheromone precursor, and preA and preB homologs encoding pheromone receptors (Additional file 5). These species were grown under conditions conducive to sexual reproduction in the aspergilli and gene expression was monitored by reverse transcriptase polymerase chain reaction (RT)-PCR. It was found that the MAT1-1 or MAT1-2, ppgA, preA, and preB genes were expressed in all of the asexual species in the same way as known sexual species, as shown by the detection of spliced transcripts (although exact expression levels were not determined) (Additional file 6). This suggests that key genes for sexual reproduction may be active in the asexual species. These results are of importance as they suggest the possibility of inducing the sexual cycle in species of biotechnological importance such as A. niger, as has recently been achieved for the penicillin producer P. chrysogenum [43]. This could accelerate cell-engineering efforts, because sexual crosses would become possible. These results also bring into question the supposed predominance of asexuality in the aspergilli. However, it has to be considered that at least 75 other genes are required for completion of the sexual cycle in the aspergilli [33]. Mutation or lack of expression in any of these genes may be responsible for asexuality if fixed within populations, although previous genome studies found no evidence for gene loss or mutation in over 200 genes implicated in fungal mating in A. oryzae and A. niger [4, 10].
One other outstanding question concerning the evolution of sex in Aspergillus species is whether the ancestral state was homothallism or heterothallism. It has been argued that homothallic fungi are derived from heterothallic ancestors [44]. However, given the predominance of homothallic species among the aspergilli and initial analysis of MAT gene organization, it has been suggested that homothallism was ancestral in the aspergilli with subsequent conversion to heterothallism [4, 45]. One key piece of supporting evidence was that heterothallism was presumed to be rare in the aspergilli. However, our findings indicate that many supposedly asexual Aspergillus species are in fact heterothallic in nature, and this is true across the wide taxonomic diversity of aspergilli under study (Fig. 1b). This indicates that heterothallism is widespread in the aspergilli, with a conserved organization and synteny of the mating-type (MAT) locus observed in the current study. This also contrasts with the variation in organization of MAT loci observed in homothallic Aspergillus species [4, 46–48]. Overall, our results strongly support the hypothesis that the aspergilli are evolved from a heterothallic ancestor, with homothallic species arising due to divergent forms of MAT gene arrangement as previously reported for Cochliobolus species [44]. This conclusion is of significance for future attempts to induce sexual reproduction in asexual species and possible genetic manipulation of MAT genes to induce sexuality [41].
Primary carbon metabolism
Fungi are well-known for their ability to exploit a wide range of simple and complex carbon substrates for growth [49, 50]. Differences in their primary metabolism may partly help to explain the diversity of fungal phenotypes. We experimentally and genetically analyzed three traits of fungal primary metabolism in the aspergilli to examine possible links to genome diversity, namely carbon source utilization, presence of alternative oxidases, and organic acid production.
Carbon source utilization
The genomes of 19 aspergilli were analyzed for copy numbers of carbon catabolic genes and compared for growth on 11 different monomeric and dimeric carbon sources (Fig. 3a, Additional files 6 and 7). A nearly complete correlation was found between the presence of genes predicted to encode catabolic enzymes and the ability to grow on a specific carbon source. Exceptions to this were the inability of A. niger to grow on D-galactose and lactose, but it is known that the relevant transport and germination triggers for these carbon sources are not active in A. niger spores [51–53] (Fig. 3a). Furthermore, A. clavatus and A. zonatus can grow on sucrose despite having no apparent invertases in their genomes. This could be due to genes missing in the assembly, the activity of α-glucosidases [54], non-specific enzymes, or a so far unknown type of invertase possibly found outside the standard GH32 Carbohydrate Active enZyme (CAZy) invertase family.
Fig. 3.
Experimental and in silico analysis of central metabolism. Clustering of the species was performed based on gpdA and orthologs from all genomes. a Catabolism of 11 sugars correlated to the number of isoenzymes investigated for each of the 11 catabolic pathways. A cross line marks all conditions where not growth was observed. b Copy numbers of (putative) alternative oxidases (aox) compared to growth assays on plates with agents inhibiting either the standard oxidative phosphorylation (KCN) or Aox (SHAM). c Correlation of putative isoenzymes for activities involved in organic acid formation and produced organic acids
In general, extra copies of a catabolic pathway, or steps thereof, did not confer more efficient carbon utilization under the conditions tested, possibly due to differential expression or altered substrate specificity of the paralogs. It was further observed that gene copy number of specific enzymes was generally correlated with phylogenetic relationships, in contrast to the increased levels of genetic diversity and horizontal gene transfer observed for secondary metabolism among closely related species (see below).
Alternative oxidase
A characteristic feature of fungal mitochondria is the presence of an additional terminal oxidase, the alternative oxidase (Aox), alongside the cytochrome-dependent one. Aox catalyzes the reduction of oxygen to water, bypassing respiratory complexes III and IV of the cytochrome-mediated electron transport chain. All examined species showed the presence of at least one alternative oxidase (aox) gene (Fig. 3b). Growth experiments confirmed that all Aspergillus species grew well in the presence of potassium cyanide (KCN), which inhibits the cytochrome pathway (Additional file 8). Addition of salicylhydroxamic acid (SHAM), which inhibits Aox, as well as KCN stunted or eliminated growth of nearly all species except A. niger and A. aculeatus. In general, the section Nigri exhibited higher tolerance to SHAM and KCN and grew faster than other aspergilli. As with the primary metabolism above, this increased growth could not be explained by changes in gene copy numbers as the Nigri had similar or lower aox copy numbers than other species (Fig. 3b).
Organic acid production
Organic acid production by A. niger is a process of major industrial importance. We examined the number of putative isoenzymes associated with citric acid and gluconic acid production in each of the 19 genomes and measured the concentration of relevant organic acids produced on three different growth media (Fig. 3c, Additional file 9). This revealed a close link between the number of isoenzymes and phylogeny, most markedly for the section Nigri, in which a series of extra isoenzymes relevant to citrate and gluconic acid production was found, correlating with increased acid production (Additional files 9A and B). This is in accord with this group being prolific producers of these two organic acids and suggests the evolution of specialized enzymes for this purpose. Indeed, transcriptome analysis of A. niger, A. oryzae, A. fumigatus, and A. nidulans showed that orthologs of citrate synthase, pyruvate dehydrogenase, isocitrate dehydrogenase, and malate dehydrogenase were highly expressed in A. niger relative to the other species (Additional file 10C), consistent with metabolic specialization.
Plant biomass degradation
Genomic comparison
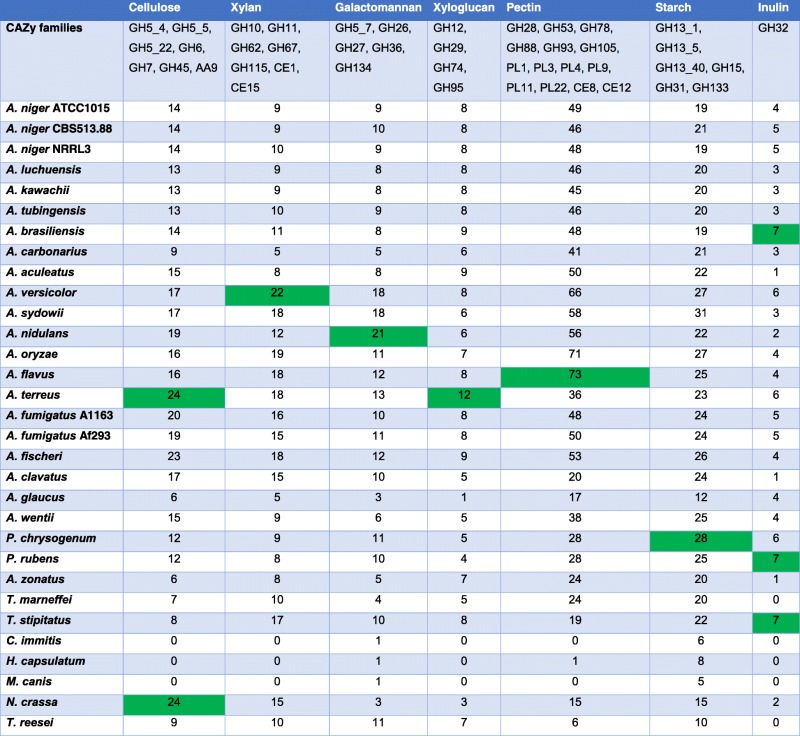
Several aspergilli have a long history of use as producers of plant polysaccharide modifying and degrading enzymes [55, 56]. Their broad ability to degrade these substrates helps to explain their presence in many different habitats, employing what has been considered a generalist lifestyle with respect to carbon utilization. This means that they do not specialize in any particular plant component, but have the ability to degrade all plant biomass polysaccharides [57, 58]. However, this has so far only been investigated for a relatively small number of species. Therefore, in this study we evaluated plant biomass degrading ability across a larger set of Aspergillus species and compared this ability to other ascomycete fungi. Comparison of the number of genes per CAZy family involved in plant biomass degradation revealed strong differences among the species investigated (Table 2, Additional file 10), although species from the same section generally contained similar numbers of genes. This comparison was based on the family (and subfamily where possible) designations set by the CAZy database (www.cazy.org) [59]. Exceptions to this were, for example, the lower number of (hemi-) cellulases for A. carbonarius compared to other species of the section Nigri and that A. versicolor is particularly enriched in inulinases compared to other species of the section Nidulantes (Table 2). Interestingly, the two most established enzyme producers of this genus, A. niger and A. oryzae, do not contain the highest number of genes required for degradation of any of the polysaccharides, with the exception of pectin for which the highest number was found in the A. oryzae genome (Table 2). The itaconic acid producer A. terreus has the highest number of putative cellulases and xyloglucanases, while A. versicolor and A. nidulans contain the highest numbers of putative xylanases and mannanases, respectively. Gene numbers related to the degradation of the storage polysaccharides starch and inulin are, in general, higher in the sister genera of Aspergillus (e.g. Penicillium).
Table 2.
Gene numbers related to degradation of different plant-based polysaccharides detected in genomes of selected ascomycete fungi
Only families that can be assigned to a specific polysaccharide, based on currently available data, are included. Highest numbers for any of the polysaccharides are highlighted in green
When comparing specific enzymes, highly variable gene numbers were observed for GH5_7 and GH26 endomannanases, GH32 endoinulinases, GH28 endorhamnogalacturonases, GH43 α-L-arabinofuranosidases, and GH115 α-glucuronidases (Additional file 11). Only some species contain genes encoding CE15 glucuronoyl esterases, GH45 endoglucanases, and PL11 rhamnogalacturonan lyases. While most of the feruloyl esterase encoding genes are included in the CAZy database, comparison of genes encoding these enzymes also revealed high variability, confirming the considerable diversity in feruloyl esterase activities recently reported for a smaller set of aspergilli [60]. This diversity is in part due to the absence of specific orthologs in some species, resulting in differences in the overall gene set per species (Additional file 11).
High variability was also observed in the number of genes encoding oxidative enzymes in the genomes analyzed (Additional file 10), which can also be involved in plant biomass degradation. Numbers in families AA1 (containing laccase), AA3_1 (cellobiose dehydrogenase), AA7 (containing vanillyl alcohol oxidase), and AA9 (lytic polysaccharide monooxygenases) are in the ranges of 5–18, 1–4, 0–7, and 3–12, respectively, but in general the variation within each section is relatively low.
Correlation of gene content to growth and enzyme production
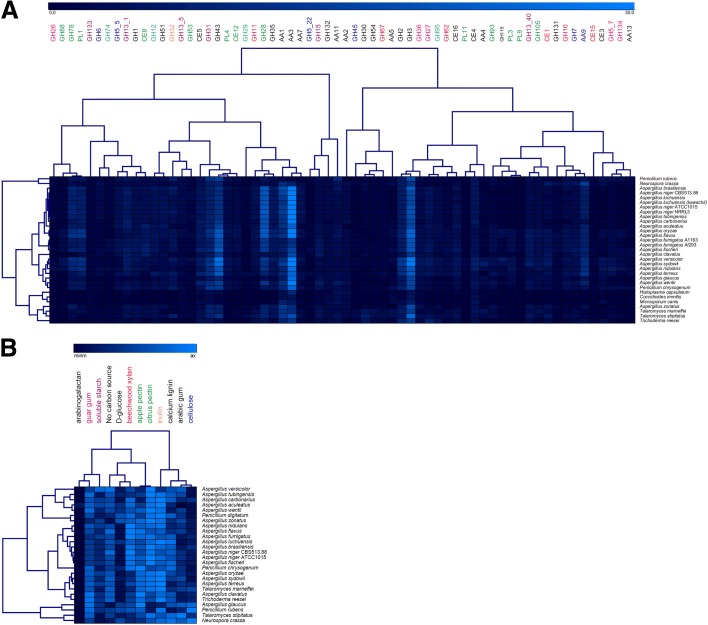
To assess any correlation between CAZy gene content and growth, strain profiling was performed by determining growth on 33 carbon sources which contained plant-related monosaccharides, oligosaccharaides, and polysaccharides as well as crude plant-based substrates, and no-carbon source and casein as controls (Additional file 8). Full profiles can be found at [61]. A hierarchical clustering (HCL) correlation analysis was performed on the number of genes per CAZy family for the tested species. Figure 4a shows that overall the evolution of CAZy families is similar to the taxonomic relationship of the species (compare to Fig. 1a). In this analysis, families that become more or less abundant simultaneously group together. No clear overall clustering of CAZy families related to the same substrate could be observed. This is likely due to the presence of multiple polymers in any plant biomass, requiring the production of enzymes acting on different substrates. In addition, commercial substrates used are unlikely to be identical to those encountered in nature and may therefore limit the observed specificity. In some cases, two to three enzyme families related to the same substrate did cluster, such as GH88/GH78/PL1 and PL3/PL9 (pectin), GH7/AA9 (cellulose), and GH27/GH36 and GH5_7/GH134 (galactomannan). In contrast, when the ability of these fungi to grow on polysaccharides (Additional file 8) was quantified and used for HCL, the taxonomic organization was largely lost (Fig. 4b). This suggests that despite conserved CAZome evolution, the ability of an individual species to degrade a specific substrate is dependent on other factors such as the regulation of gene expression or the transport of monomers. Therefore, as observed recently for eight aspergilli [57], no clear correlation between CAZome, taxonomy, and enzyme sets produced on plant biomass substrates could be detected. A possible explanation for this is that under laboratory conditions only complete disappearance of a CAZy family would result in loss of function. However, the persistence of the CAZy genes clearly indicates an evolutionary pressure to maintain them in the genomes, which could be due to the more complex natural environment. A better understanding of fungal biotopes, including the actual components of the available substrates they consume in nature, will likely reveal a better correlation between genome content and carbohydrate preference of each species.
Fig. 4.
HCL (a) the number of genes per CAZy family related to plant biomass degradation in the tested genomes and (b) the relative growth of the tested species on polymeric substrates. Color coding in (a) refers to the polysaccharide these families act on. Black families either contain multiple activities or are active on multiple polysaccharides. For the other colors: pink = guar gum (galactomannan), purple = starch, red = xylan, green = pectin, orange = inulin, dark blue = cellulose, pale blue = xyloglucan
To assess the impact on growth arising from differences in the number of genes encoding oxidative enzymes, 20 aspergilli were grown in minimal medium (MM) with 1% cotton seed hulls over 14 days and were assayed for laccase and cellobiose dehydrogenase activities (Additional file 12). The highest laccase activity was observed for A. fischeri and low activities were detected for A. oryzae, A. wentii, and A. versicolor. No laccase activity could be detected for 11 of the aspergilli. The highest cellobiose dehydrogenase levels were measured for A. fumigatus, A. wentii, A. tubingensis, and A. terreus. No cellobiose dehydrogenase activity was detected for five aspergilli. Thus, there was no clear correlation between these observed enzyme activities and the number of genes in the respective genomes (Additional file 10).
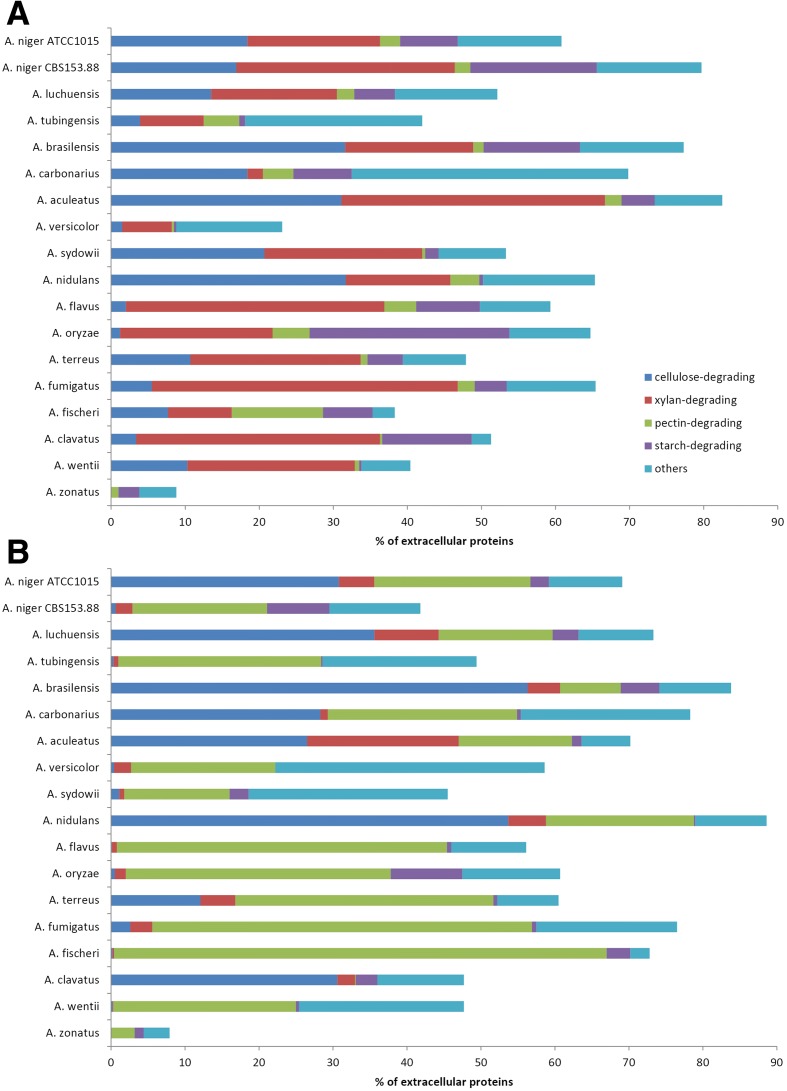
Recently, Benoit et al. compared the production of plant biomass degrading enzymes by eight aspergilli during growth on wheat bran or sugar beet pulp [57]. This work was extended in the present study to include the newly sequenced species, using the same culture conditions and data analysis as in the previous study. These substrates provide useful comparative datasets because both contain cellulose, but differ in that wheat bran is rich in xylan, while sugar beet pulp contains large amounts of pectin and xyloglucan. The results demonstrated a large variation in enzyme production between the aspergilli, even for species within the same section (Fig. 5). Most species produced predominantly cellulose-active and xylan-active enzymes during growth on wheat bran, but the ratio between these two enzyme groups varied strongly in some species (Fig. 5a). Some unusual profiles were also observed, such as the high relative amount of different pectinases produced by A. fischeri, the low amount of xylanases for A. carbonarius, and the high amount of amylases for A. oryzae. On sugar beet pulp, nearly all species produced high amounts of pectinases, appropriate to the composition of the substrate (Fig. 5b). An exception to this was A. clavatus, where genome analysis had already revealed a strong reduction in genes encoding pectin-active enzymes [57]. A. clavatus compensates by high production of cellulases, whereas cellulases are almost absent in the profiles of several other species (e.g. A. wentii, A. fischeri, A. flavus, A. tubingensis). The high production of xylanases by A. aculeatus and to a lesser extent by A. luchuensis was also unexpected.
Fig. 5.
Comparative proteomics of aspergilli during growth on wheat bran (a) and sugar beet pulp (b). Results are summarized by the polysaccharide that the enzymes act upon. Only CAZy families specific for one polysaccharide are included in the comparison. Cellulose-active enzymes: GH6, GH7, GH12, GH45, GH74, AA9; xylan-active enzymes: CE1, CE15, GH10, GH11, GH62, GH67, GH115; pectin-active enzymes: CE8, CE12, GH28, GH35, GH51, GH53, GH54, GH78, GH88, GH93, GH105, PL1, PL3, PL4, PL9, PL11; starch-active enzymes: GH13, GH15; other (CAZy families with multiple activities or minor activities on the substrates): CE16, GH1, GH2, GH3, GH5, GH26, GH27, GH31, GH32, GH36, GH43, GH95. Extracellular proteins following trypsin digestion were analyzed by LC-MS/MS using a LTQ-Orbitrap Velos mass analyzer (Thermo-Fisher). Quantification was based on MS precursor ion signal. Extracted ion chromatograms were used to determine the peptide area value associated to each identified precursor ion. A protein area value was calculated as the average of the three most intense, distinct peptides assigned to a protein. The amounts of proteins associated with each enzyme activity were expressed as percentage of the amount of total extracellular proteins present in each culture condition
The variation in detected enzymes in the proteomic data of the species investigated here far exceeded that which could be explained by genome variation, consistent with previous results from the first eight Aspergillus species to be genome sequenced [57]. This suggests that the observed variation is mainly caused by regulatory changes, rather than simply by gene copy number. Since all species contain the same set of conserved regulators for cellulase, xylan, and pectin degradation (data not shown), they presumably have a different fine-tuning of the regulatory system. This could be due to a different range of target genes for their regulators or different functioning of them, as was recently demonstrated for the major (hemi-) cellulolytic regulator XlnR [62]. The exoproteome analysis reported here confirms our recent observation [57] that the enzyme cocktails produced by these fungi in response to the same plant biomass are highly different and will therefore also have different efficiencies in saccharification of plant-derived biomass.
Secondary metabolism
The ability to produce secondary metabolites (SM) is an important feature of many fungi and certainly of the aspergilli and related fungi. These metabolites are involved in many processes in nature, such as communication between fungi and competition for nutrients and can be pathogenicity factors. Some of these metabolites have become widely used as antibiotics. In this section, we highlight the diversity of SM production in the aspergilli and the role of cytochrome P450 (CYP) genes in secondary metabolism.
Secondary metabolite biosynthesis gene clusters in the aspergilli
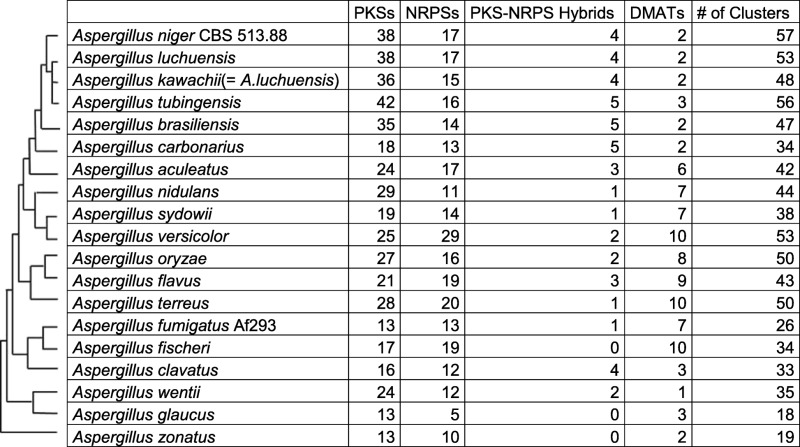
Polyketide synthases (PKSs), non-ribosomal peptide synthetases (NRPSs), or dimethylallyl tryptophan transferases (DMATs) are involved in the biosynthesis of the vast majority of SM products [63, 64]. The PKS and NRPS genes are frequently clustered together with other pathway genes, such as DMAT genes, involved with processing of the initial product to form the final SM, thereby forming a linked gene cluster. The high degree of conservation of these proteins and their specific domain structure allow reliable in silico prediction of their genes. Thus, the Aspergillus genomes were searched to gain insights into their evolution and potential to produce SMs. The number of individual PKS, NRPS, and DMAT genes was found to vary considerably from 21 in A. glaucus to 66 in A. tubingensis and A. versicolor (Table 3). These large variations do not correlate with genome size, suggestsing that the number of SM genes and their associated clusters depends on the lifestyle and the corresponding requirements of each species.
Table 3.
Number of predicted SM biosynthesis genes and number of associated clusters in the genomes
Aspergillus species were ordered according to their phylogenetic relationships (see Fig. 1)
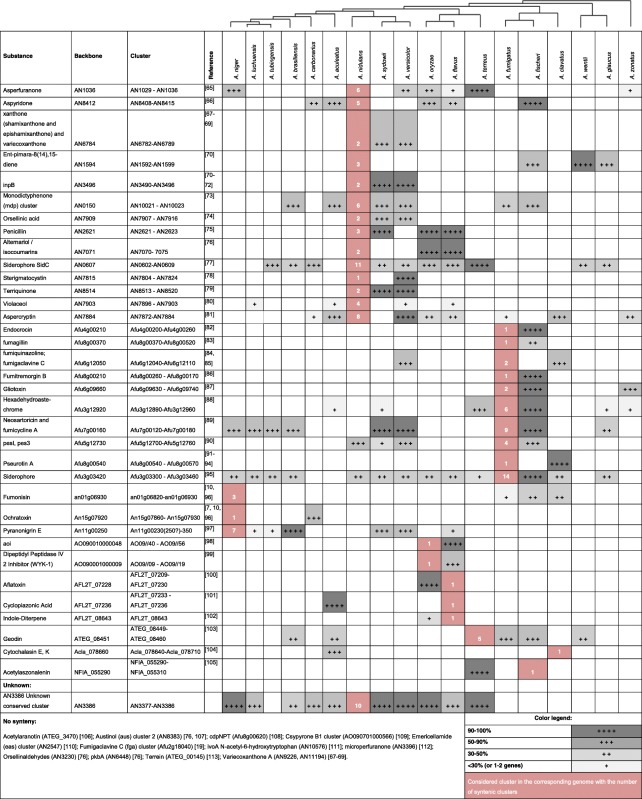
To date about 50 non-redundant SM gene clusters have been experimentally characterized in the aspergilli. We scanned the 19 genomes for regions syntenic with these known clusters to examine their distribution and thereby determine the potential of the various species for production of SMs. Thirty-six clusters appeared to be conserved with respect to gene content and order, with 23 clusters conserved in more than one genome (Table 4). By contrast, 12 clusters showed no conservation of gene content and order (Table 4). Interestingly, in total, we defined 44 unique PKS, NRPS, or DMAT genes that have no orthologs across all the analyzed 19 species.
Table 4.
Conservation of experimentally characterized clusters in the 18 Aspergillus species
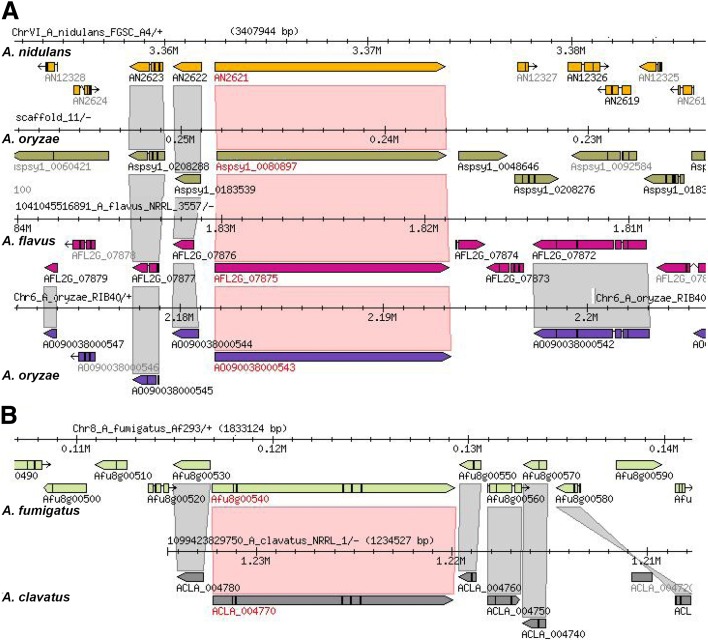
The degree of similarity between clusters from different species can provide an indication of potential similarities in the products that they synthesize, with clusters having very high similarity (90–100%) normally coding for the same SM. Genome data from the present study therefore allowed us to predict the production of certain SMs. This can be exemplified for the production of pseurotin and penicillin. Pseurotin was previously described for A. fumigatus [64, 91–93], A. clavatus [114, 115], A. oryzae [116], A. leporis, and A. nomius [117] but so far not for other Aspergillus species. The gene cluster in A. fumigatus was already known, but here we identified the conserved pseurotin gene cluster in the A. clavatus genome. There was no indication of a pseurotin gene cluster in A. oryzae, although its production was reported for this species. However, the particular strain reported to produce pseurotin is not available and may have been misidentified. In another example, analysis of the penicillin gene cluster known to be present in A. nidulans [75], A. oryzae [118], and A. flavus revealed a single additional species, A. sydowii, where this cluster is well conserved indicating the potential for penicillin production (Fig. 6).
Fig. 6.
Conservation of penicillin (a) and pseurotin (b) biosynthesis gene clusters in Aspergillus species
In addition, we found examples of clusters with partially conserved synteny (50–90%; Table 4). The SM products of such clusters can be expected to be similar but not identical. For instance, fumiquinazolins were observed in A. fumigatus [119], whereas in A. versicolor and A. clavatus there are cottoquinazolins [120] and tryptoquivalins [121], respectively, which is in agreement with the partial conservation and synteny of the corresponding clusters (Table 4). Similarly, geodin produced by A. terreus is related to sulochrins produced by A. fumigatus (3-O-methylsulochrin) and A. wentii (sulochrin and isosulochrin), consistent with moderate conservation of the respective gene clusters (Table 4).
In A. nidulans, three gene clusters (AN6784, AN9226, and AN11194) are responsible for the biosynthesis of xanthone (shamixanthone and epishamixanthone) and variecoxanthone. It was observed that the shamixanthone cluster (AN6784) is reasonably conserved, with five of nine A. nidulans cluster genes being syntenic in A. sydowii and A. versicolor (Table 4). However, the genes for the other clusters are unique to A. nidulans, which is known to produce a much more varied profile of these compounds than A. sydowii and A. versicolor. Meanwhile, the lack of production of particular SMs by some strains and/or isolates was often confirmed by the genome data. Aspergillus versicolor and A. nidulans produce sterigmatocystin [122], whereas the closely related A. sydowii CBS 593.65 does not. This can now be explained by the complete lack of the corresponding gene cluster in A. sydowii, whereas the sterigmatocystin cluster is perfectly conserved between A. versicolor and A. nidulans (Table 4). Thus, our bioinformatic analysis overall supports and provides a genetic basis for previous experimental data.
Analysis of the genome data also allowed us to predict gene clusters that might be responsible for the biosynthesis of experimentally detected SMs for which the genetic basis had previously been unknown. For example, the three black Aspergillus strains, A. brasiliensis CBS101740, A. luchuensis CBS106.47, and A. tubingensis CBS134.48, all produce naptho-gamma-pyrones (Table 4) [123]. The gene cluster for the related neosartoricin/fumicycline of A. fumigatus and A. fischeri has previously been characterized and may be analogous to the naptho-gamma-pyrone cluster in species of the section Nigri, given that both gene clusters occur in the same conserved syntenic region across all these species.
Finally, the analysis led to the discovery of previously unknown, but well conserved, clusters such as that with an AN3386 backbone (Table 4), providing exciting leads for future cluster analysis.
CYPomes and supporting cytochrome P450 reductases
CYP genes are also of interest for secondary metabolism. More than 2300 CYP sequences (PF00067) were identified in the 19 aspergilli examined (data not shown). These numbers are comparable with analysis of the CYPome for A. nidulans in the previous CADRE genome annotation [124]. Some of the CYP families were further analyzed in the present study relating to their possible use as antifungals and roles in fungal secondary metabolism, namely: CYP51 (sterol 14α-demethylase) [125], CYP61 (22-sterol desaturase) [126], CYP56 (dityrosine synthesis) [127], CYP53 (benzoate 4-hydroxylase) [128], CYP630 (oleic acid ω-hydroxylase) [129], Ppo (fatty acid oxygenases or oxylipin generating dioxygenases) [130], and a supporting enzyme CYP reductase (CPR) as well as CYP505, CYP-CPR fusion protein (fatty acid hydroxylase) [131].
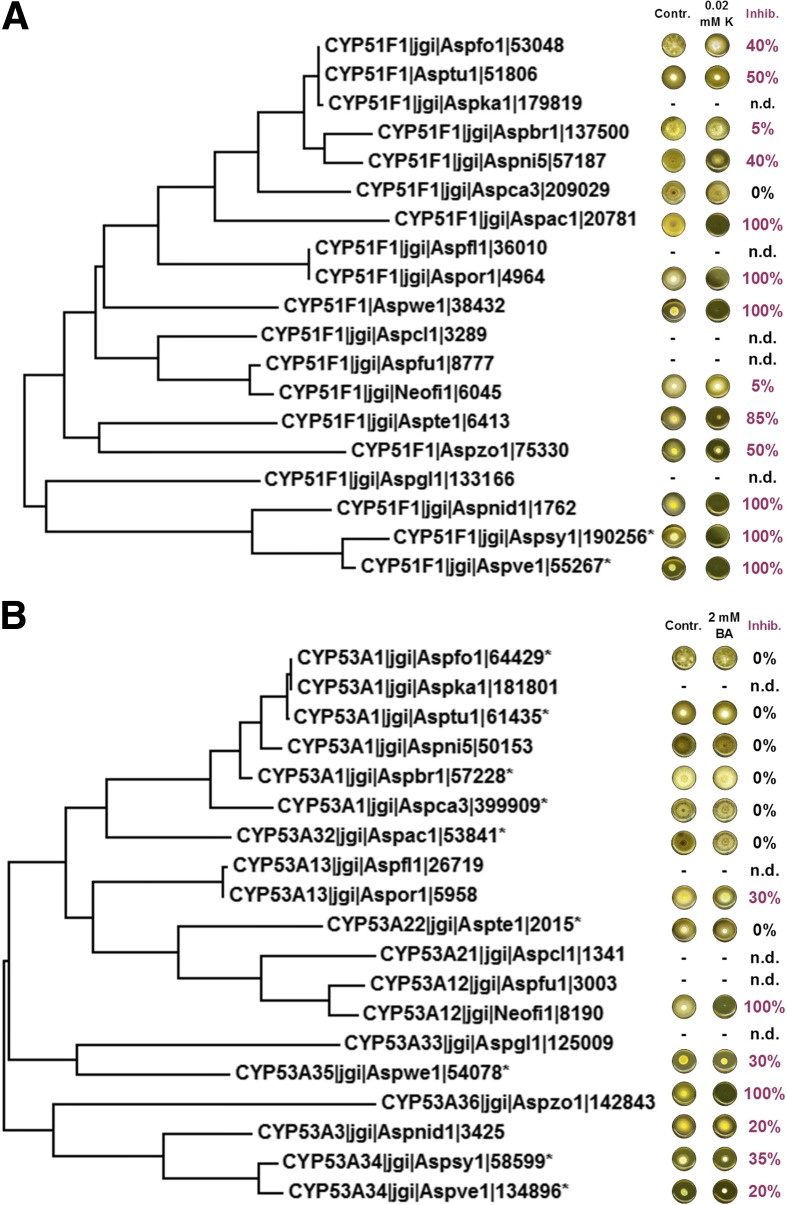
Most aspergilli were found to contain two copies of CYP51, whereas only a single copy of CYP53 was present in all species. The fact that CYP53 is unique to filamentous fungi makes it an attractive candidate drug target against fungal pathogens [131]. A series of inhibition experiments with ketoconazole (for CYP51) and benzoate (for CYP53) revealed complementary inhibition patterns for these CYPs, suggesting that simultaneous treatment with both drugs may be effective (Fig. 7). CYP61 was always present as a single ortholog, while surprisingly, CYP56 was not found in 11 species (Additional file 13A). As a part of an evolutionarily conserved gene cluster including CYP630, in most species a paralog, CPR2, was identified for the conserved main electron donor CPR1 (Additional file 13B). This cluster may be of importance when the main ß-oxidation pathway is overwhelmed by other substrates such as host terpenoid compounds [129].
Fig. 7.
Molecular phylogenetic tree of (a) CYP51F1 (CYP51B) family and inhibition of growth on MBFA agar supplemented by 0.02 mM ketoconazole (K) and (b) CYP53 family and inhibition of growth on 2 mM benzoic acid (BA). Inhibition of growth (Inhib.) as per cent of the control without supplementation (Contr.) is shown in relation to phylogenetic trees, with growth culture plates shown to the right-hand side of the accompanying tree branch. Key to Aspergillus species: Aspfo = A. luchuensis (formerly A. foetidus); Asptu = A. tubingensis; Aspka = A. luchuensis (A. kawachii); Aspbr = A. brasiliensis; Aspni = A. nidulans; Aspca = A. carbonarius; Aspac = A. aculeatus; Aspfl = A. flavus; Aspor = A. oryzae; Aspwe = A. wentii; Aspcl; = A. clavatus; Aspfu = A. fumigatus; Neofi = A. fischeri; Aster = A. terreus; Aszo = A. zonatus; Asgl = A. glaucus; Aspsy = A. sydowii; Aspve = A. versicolor
One to four copies of CYP-CPR fusion proteins were found in Aspergillus species. The presence of multiple copies may allow each particular fusion protein to function in a specific SM pathway. For the second type of fusion protein, haem peroxidase-CYP, orthologs of PpoA (CYP6001A subfamily) and PpoC (CYP6001C) were found in regions of colinearity in all genomes (except for A. zonatus and A. kawachii, respectively), implying conservation of biological function. Additional copies of haem peroxidase-CYP fusion proteins were present for some genomes in three different subgroups with homology either to PpoB from A. nidulans, PpoD from A. niger and A. flavus, or PpoB from A. fumigatus and A. flavus (Additional file 14). Specifically, the CYP6002A subfamily consists of PpoB from A. nidulans clustered together with orthologs from A. sydowii, A. versicolor, A. wentii, A. oryzae, A. terreus, A. clavatus, and A. fischeri. The CYP6003B subfamily consists of PpoD from A. niger and A. flavus and orthologs from A. glaucus, A. oryzae, A. terreus, A. tubingensis, A. luchuensis, A. kawachii, and A. brasilensis. Finally, the CYP6001C subfamily comprises PpoB from A. fumigatus and A. flavus together with ortholgs from A. glaucus, A. terreus, A. oryzae, and A. fischeri.
Stress response
Filamentous fungi are exposed to a complex set of abiotic or biotic stresses in natural environments as well as in industrial processes where they are used to produce various biotechnological products. Effective stress perception and signal transduction mechanisms are necessary for adaptation and survival regardless of whether rapid or slow adaptation responses are required.
The complex and robust stress defense systems of the aspergilli [132] were investigated using a wide array of stress physiology experimental and comparative genomics tools.
The stress tolerances of the aspergilli were screened under stress conditions including illumination, oxidative (H2O2, menadione), hyperosmotic (sorbitol, NaCl), heavy metal (Cd(II)), and cell wall integrity (calcofluor white, Congo Red) stress. The aspergilli were also exposed to selected antimycotics that interfere with cell wall (caspofungin) and cell membrane (voriconazole, amphotericin B) composition and structures. Stress sensitivities were tested in both liquid and surface cultures and the complete stress dataset with photographs is also available in the Fungal Stress Database [133]. A more extensive description of the data can be found in Additional files 3, 15, 16, and 17.
Illumination stress
Light-induced conidiation is common for most of the analyzed aspergilli. While light receptors and regulators were found to be conserved in most species (Additional file 17), rewiring of the response to the receptors and signal transduction pathways appears to have occurred in many species with respect to conidiation at different wavelengths. For example, blue or red light induced conidiation occurs in certain species but not others, while some species (e.g. A. versicolor) required specifically white light to induce sporulation. Interestingly, the industrially important black aspergilli and the opportunistic pathogens A. fumigatus and A. flavus showed little response to illumination (Fig. 8, Additional file 17).
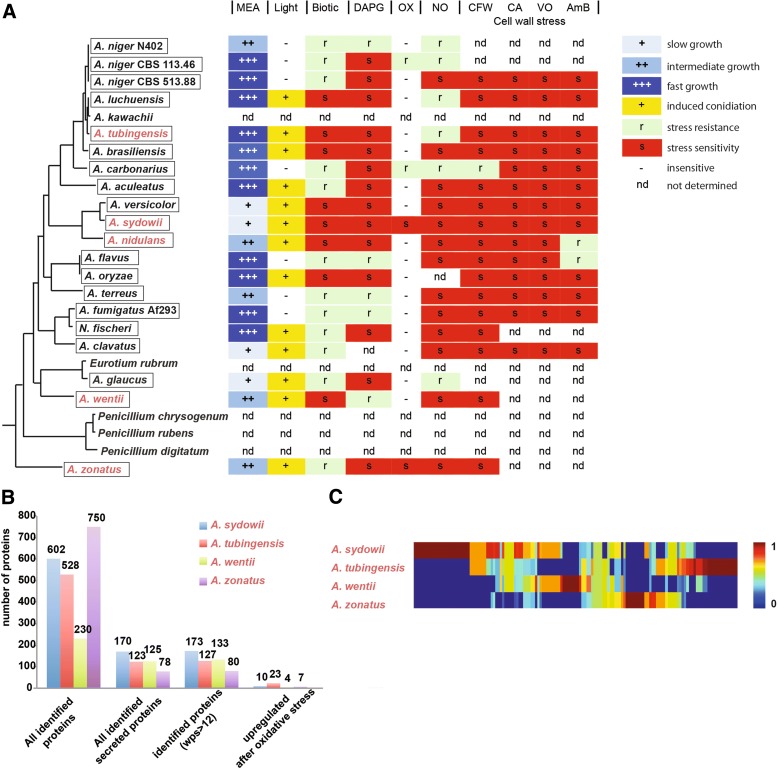
Fig. 8.
a Phylogenetic tree of Aspergillus species and summarized phenotypes from this study (modified from [308]). All Aspergillus species were grown on malt extract agar (MEA) (blue) at 30 °C. Yellow squares stand for induced conidiation in white light. Abiotic and biotic stress determinants were colored in green for resistance (r), red for sensitive (s), no color for insensitive (-) and not determined (nd) different abiotic and biotic stressors: 2,4-diacetylphloroglucinol (DAPG), Hydrogen peroxide (OX), nitric oxide (NO), calcofluor white (CFW), caspofungin (CA), voriconazole (VO), amphotericin B (AmB), and Aspergillus-Pseudomonas fluorescens co-cultivation (Biotic). Species used in this study are framed in black. b Qualitative descriptive statistics of the exoproteome of four selected Aspergillus strains. Exoproteomes were enriched from filtered culture supernatants (Aspergillus MM, 30 °C) and analyzed by shotgun proteomics (LC-MS). Proteins were identified using draft genomic databases of the respective strains. For prediction of subcellular localization of the proteins the programs “SignaIP” and “WoLF PSORT” were used. For further analysis, sequences with an extracellular score >12 (WoLF PSORT) were defined as putatively secreted proteins (Additional file 18). Proteins with higher spectral count values after 0.2 mM hydrogen peroxide treatment (“upregulated after oxidative stress”) were selected statistically using MARVIS Filter (s/l > 0.5). c Exoproteomic heterogeneity in the genus Aspergillus. Comparative MARVIS cluster analysis of PFAM domains predicted in identified exoproteins (WoLF PSORT columns in bar chart B) of four selected Aspergillus species (WoLF PSORT: extracellular score >12) (Additional file 19). Colors represent the normalized frequency of occurrence of PFAM-domains in the respective exoproteome. Indicated by the color scale on the right, the color scale ranges from blue (no domain) via green (few domains) to red (more domains). Each column of the cluster image resembles one PFAM-domain. The proteome data is available at http://wwwuser.gwdg.de/~hkusch/GBIO_DeVries/
Oxidative stress
In solid media experiments, A. brasiliensis showed outstanding menadione tolerance, which correlated with the emergence of two putative new-type SodA superoxide dismutases (Fig. 9). In liquid cultures, A. carbonarius and A. niger CBS113.46 were especially resistant to high oxidative stress provided by hydrogen peroxide while A. zonatus and A. sydowii were the most sensitive species (Fig. 8). In the case of A. carbonarius, the remarkable H2O2 tolerance was coupled to the co-occurrance of two C-type—one group cIII and one group cIV—catalases (Additional file 15). The catalase-encoding catC gene (coding for group cIV catalase; Additional file 15) has been lost in some Aspergillus genomes including A. fischeri, A. clavatus, and A. fumigatus. In the latter species, the easC gene for ergot synthesis (a group cIII catalase gene, Additional file 15) [134] appears to have taken over protective functions against oxidative stress (Additional files 15 and 17).
Fig. 9.
Linking species-level differences in selected Aspergillus stress-defense proteins to major stress tolerance phenotypes. The remarkable Cd(II) tolerance of A. fumigatus and A. sydowii (a), the outstanding menadione sodium bisulfite (MSB) tolerance of A. brasiliensis (b) and the osmophility of A. glaucus and A. wentii (c) were attributed to variations in the cadmium transporting P-type ATPases (a), Cu/Zn-superoxide dismutases (b), and NAD-dependent glycerol-3-phosphate dehydrogenases (c), respectively. In this Figure, the stress sensitivities of A. nidulans are also presented for comparison (a–c), although this species lacks any Pca1 ortholog (Additional file 15). In the dendograms, putative orthologs of baker’s yeast Pca1 cadmium transporting P-type ATPase (a), Aspergillus nidulans SodA Cu/Zn-superoxide dismutase (b), and A. nidulans GfdA/B NAD-dependent glycerol-3-phosphate dehydrogenases (c) are shown. In (c), S. cerevisiae Gpd1/2 paralogs are also presented. Putative A. sydowii ( ) and A. fumigatus (
) and A. fumigatus ( ) Pca1 orthologs (a) and putative A. glaucus (
) Pca1 orthologs (a) and putative A. glaucus ( ) and A. wentii (
) and A. wentii ( ) GfdA orthologs (c) are indicated by symbols presented in the parentheses. Hypothetical new-type SodA enzymes found in A. brasiliensis are indicated by red lines in (b). Pca1, SodA, and GfdA/B orthologs are indicated by four-letter species name identifiers (listed in Additional file 15) and locus IDs as found in AspGD (http://www.aspergillusgenome.org/) for the aspergilli. The relevant JGI locus IDs are listed in Additional file 16. The Gpd1/2 paralogs of budding yeast are from the Saccharomyces Genome Database (http://www.yeastgenome.org/). Photographs on the tress tolerance/sensitivity phenotypes were taken from the Fungal Stress Database (http://www.fung-stress.org/). Further details of the phylogenetic and evolutionary calculations and more information on the stress response proteins can be found in Additional file 15
) GfdA orthologs (c) are indicated by symbols presented in the parentheses. Hypothetical new-type SodA enzymes found in A. brasiliensis are indicated by red lines in (b). Pca1, SodA, and GfdA/B orthologs are indicated by four-letter species name identifiers (listed in Additional file 15) and locus IDs as found in AspGD (http://www.aspergillusgenome.org/) for the aspergilli. The relevant JGI locus IDs are listed in Additional file 16. The Gpd1/2 paralogs of budding yeast are from the Saccharomyces Genome Database (http://www.yeastgenome.org/). Photographs on the tress tolerance/sensitivity phenotypes were taken from the Fungal Stress Database (http://www.fung-stress.org/). Further details of the phylogenetic and evolutionary calculations and more information on the stress response proteins can be found in Additional file 15
Hyperosmotic stress
One of the most interesting outcomes of the stress studies was that osmophily was more widespread in the genus Aspergillus than previously thought. Experimental evidence was obtained for osmophily in A. glaucus, A. wentii, A. versicolor, A. sydowii, and A. oryzae (Additional file 3). In the case of A. glaucus and A. wentii, osmophily correlated with the lack of GfdB, a putative NAD+-dependent glycerol-3-phosphate dehydrogenase, with a predicted function in glycerol metabolism (Fig. 9; Additional file 15). This is consistent with previous work in A. nidulans showing that elimination of gfdA, which also encodes a putative glycerol metabolic enzyme, led to an osmoremediable phenotype in the presence of 1 M NaCl (Additional files 3 and 15) [135].
Heavy metal stress
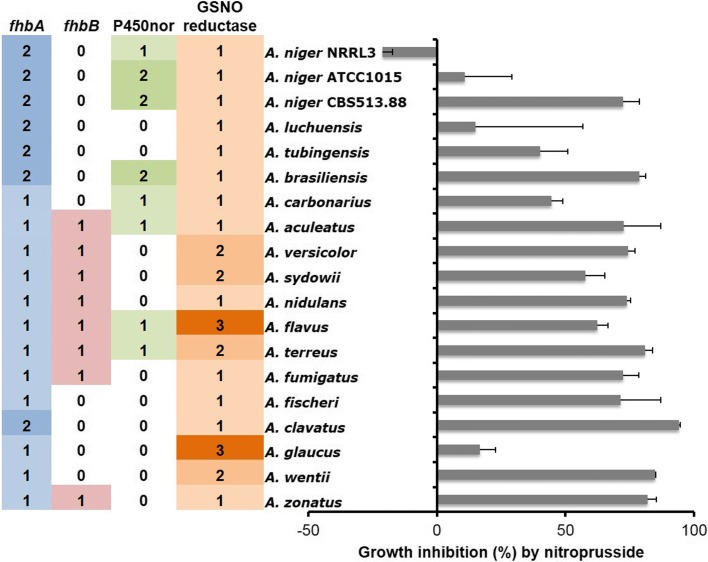
Cd(II) tolerance was clearly correlated with the presence of a Pca1-type Cd(II) transporter in the studied genomes (Fig. 9, Additional files 3 and 15). Not surprisingly, A. sydowii, A. fumigatus, A. wentii, A. versicolor, and A. terreus, all harboring a Pca1 ortholog, were the most tolerant to Cd(II). Interestingly, the important opportunistic human pathogen A. fumigatus also possessed high Cd(II) tolerance, but it was remarkably sensitive to oxidative, osmotic, and cell wall integrity stress when compared to other aspergilli (Fig. 9, Additional files 3 and 15).
Cell wall integrity stress and sensitivity to antimycotics
The cell walls of the aspergilli consist of chitin, β-1,3-glucan, β-1,6-glucan, α-1,3-glucan, galactomannan, and galactosaminogalactan. Genes required for the synthesis and remodeling of these polymers were found in all sequenced Aspergillus genomes, although a high degree of redundancy was observed (Additional file 17). The number of chitin synthases was in the range of 9–15 (noting that S. cerevisiae only harbors three chitin synthases). The number of chitin remodeling, chitin-glucan crosslinking, and β-glucan synthesis enzymes also differed considerably (21–31, 10–18, and 15–31, respectively). No such extreme differences were observed with respect to galactomannan and galactosaminogalactan synthesis. However, there was no correlation between the number of cell wall genes and overall susceptibility to the cell wall acting drugs calcofluor white, Congo Red, and caspofungin (Fig. 8, Additional files 15 and 17). In addition, voriconazole and amphotericin B, which affect the composition and structure of cell membranes, also showed species-specific effects (Additional file 17). Therefore, it is likely that numerous strategies have evolved in different Aspergillus species to counteract the presence of these antifungals, as has previously been suggested [136].
Transporters
The complex mixtures of nutrients found in natural growth substrates require transport across cell membranes in order to be assimilated into the fungal cell. To analyze genes that encode transporters, 19 Aspergillus genomes were compared to 16 genomes of other fungi, resulting in the detection of up to 120 different transporter-related Pfam motifs in at least one species. The total number of transporters identified in the genomes varied dramatically from 301 transporter-encoding open reading frames (ORFs) in Schizosaccharomyces pombe to 1601 in A. versicolor. The number of ORFs contributing to the transportome of members of the Eurotiomycetes was variable, the lowest number being 810 in A. clavatus, which is still more than twice the number found in the yeasts S. pombe or Saccharomyces cerevisiae. The majority of transporter types (as denoted by the presence of the Pfam motifs) are present in similar numbers in the genomes of all the fungi included in this study. The Major Facilitator Superfamily, which contributes the largest number of ORFs to the transportome, is also the transporter class showing the highest level of expansion in the Eurotiomyces, with the ABC transporter superfamily also showing a noticeable increase. This expansion of the transporter capacity of the fungi correlates with the ability to use a greater diversity of substrates. The redundancy of most transport systems renders the study of fungal transporters a complex topic where much remains to be discovered. Some subclasses of transporters are discussed in more detail below.
The Aspergillus sugar transportome
Sugars are not only important nutrients and structural precursors but are also signaling molecules that trigger changes in gene regulation. Sugar transport and sensing are therefore crucial functions in fungal cells and considerable progress in understanding the molecular bases of these processes has been made in studies on yeast [137]. Most sugar transporters belong to the Major Facilitator Superfamily (MFS) [138, 139] of secondary transporters (secondary active transporters and facilitators), which currently comprises 82 named classes [140, 141]. In order to investigate the sugar transporter complements of diverse filamentous fungi, we focused on those proteins from Aspergillus species that were classified as members of the PFAM family of “Sugar and other transporters” (PF00083). Most, but not all, members of this family catalyze sugar transport, with their substrates including hexoses, pentoses, disaccharides, alpha-glucosides, inositol, and quinic acid as well as organic and inorganic ions.
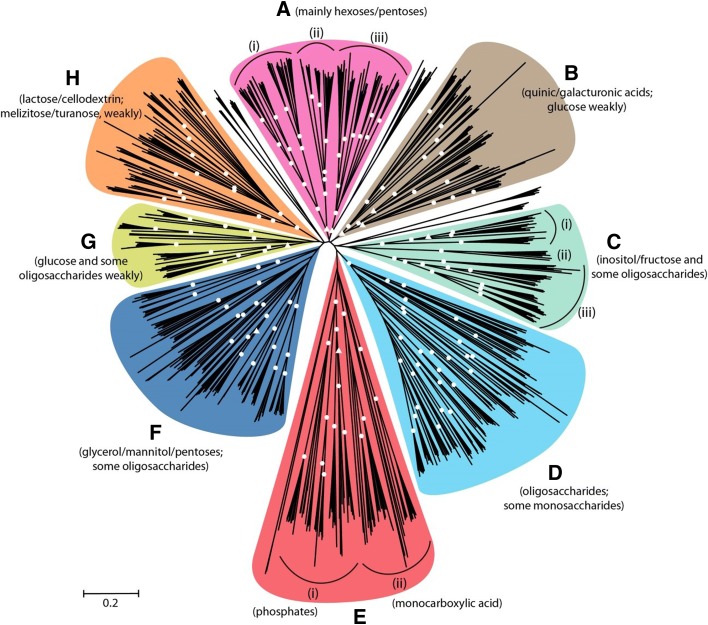
Using the annotated genome data from eight previously available Aspergillus species, a total of 940 putative PF00083 transporters were identified after manual filtering and reannoation to yield greater consensus to the conserved 12 transmembrane domain topology. From phylogenetic analysis, the vast majority of sequences were found to fall into eight major clades (A to H; Fig. 10). Although support for the very basal branches is generally low, the subclades within them have high bootstrap support.
Fig. 10.
Overview of sugar transporters. The evolutionary history of 940 PF00083 sequences identified in the genomes of A. clavatus, A. flavus, A. fumigatus, A. nidulans, A. niger, A. oryzae, A. terreus, and A. fischeri was inferred using the neighbor-joining method. The optimal tree is shown. Nodes with 50–69% (∆) or 70–100% (○) bootstrap support (1000 replicates) are indicated. Evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All ambiguous positions were removed for each sequence pair. Evolutionary analyses were conducted in MEGA6. A small number of sequences could not be assigned to the main clades identified; referring to the smallest genome analyzed (A. clavatus), these correspond to loci ACLA023780, ACLA026890, ACLA031920, and ACLA041390. Potential substrates for members of the clades are suggested based on experimentally determined transporter functions
This initial analysis was extended to include an additional 1422 sequences corresponding to the PF00083 complements of the novel Aspergillus genomes, along with those of two fungi for which experimental evidence of function has been obtained for a number of PF00083 members: N. crassa (Sordariomycete) and S. cerevisiae. For comparison, the complements of P. chrysogenum, T. marneffei, T. reesei, and two animal pathogens (Histoplasma capsulatum and Microsporum canis) were also included. Additional files 20 and 21 present a summary and details, respectively, of the numbers and distribution of PF00083 members found in each of the clades and the function(s) identified for some members of those clades. Considerable diversity in the numbers of PF00083 members was found, with a threefold difference existing between those species having the largest and smallest complements (A. sydowii vs. A. clavatus). Even species from the same sections do not have very similar numbers of loci, for example: members of the Nigri range from 83 to 113. Of the Nidulanti, A. sydowii (180) and A. versicolor (171) have the highest numbers of loci whereas the number present in A. nidulans (113) is similar to that observed in A. niger and P. chrysogenum. The three genome-sequenced A. niger strains (consisting of an enzyme producer, a citric acid producer, and a laboratory strain) all have similar PF00083 complements. Perhaps surprisingly, T. reesei (Sordariomycete), a well-known producer of plant cell wall degrading enzymes, only has 64 members. Interestingly, A. clavatus has a very similar complement size (63) but there is a notable enrichment of T. reesei members in clade D and a reduction in clade G compared to A. clavatus. A. terreus (123) and A. wentii (120) also have similar numbers of PF00083 members but clade D is threefold enriched in the former, while A. wentii, along with A. sydowii and A. versicolor, has the highest number of members of clade F. To evaluate transporter function in the different clades, we combined data from the literature with results from studies involving the functional complementation of S. cerevisiae sugar transporter mutants with 43 putative transporters from A. niger (Additional file 22).
Clade A comprises three distinct subclades (bootstrap values >90%) and experimental data demonstrated the transport of certain monosaccharides for a number of its members. Both high-affinity and low-affinity glucose transporters of A. nidulans (HxtC, HxtE, MstA, MstC, and MstE), A. niger (MstA, MstC), and N. crassa (Hgt-1, Glt-1) are found in subclade A(i), as well as two broad-specificity transporters of glucose/pentoses (An29-2, Xyt-1) [138, 139, 142–147]. Paralogous high-affinity glucose transporters (MstA and MstC) differing in their patterns of expression are present in A. nidulans [144]. This duplication is also found in the other two Nidulantes (in fact A. sydowii has three loci, as does N. crassa) but is absent in all the other aspergilli analyzed here. A. glaucus lacks MstA/MstC homologs. Analysis of the A. niger transporters indicated the ability to transport D-glucose, D-galactose, and D-mannose for all tested proteins, while all but one were also able to transport D-fructose (Additional file 22).
An N. crassa transporter (Lat-1) of broad sugar substrate specificity (arabinose and other sugars) and a closely related arabinose-specific transporter from Myceliophthora thermophila (MtLat-1) map to subclade A(ii) [148], as does the HxtA protein of A. nidulans [149]. The tested A. niger transporters of this group showed broad specificity, covering the monosaccharaides and oligosaccharides D-glucose, D-mannose, D-fructose, D-galactose, maltose, melizitose, and turanose (Additional file 22).
A. nidulans and N. crassa xylose transporters [150, 151] occur in subclade A(iii). While one of the tested A. niger transporters from this subclade appeared to be specific for D-glucose, the others are more similar in their specificities to the members of subclade A(i) (Additional file 22). One member is also able to transport the disaccharides maltose and trehalose. Clade A might therefore be considered to comprise three diverse groupings of predominantly pentose/hexose transporters.
Clade B contains quinic acid [152, 153] and D-galacturonic acid (GAT-1, GatA) [154, 155] transporters. GatA homologs are present in most aspergilli except section Fumigati, A. clavatus, and A. zonatus. By contrast, no GAT-1 homologs are present in section Nigri, A. glaucus, or A. zonatus. Analysis of A. niger members of this clade only revealed the ability to transport D-glucose for some members (Additional file 22).
Clade C comprises three major subclades supported by very high bootstrap values. Subclade C(i) contains homologs of the S. cerevisiae inositol transporters Itr1 and Itr2 [156] and the putative Cryptococcus neoformans inositol transporters [157]. Subclade C(ii) members show homology to the fructose transporters Fsy1 and Frt1 identified in S. pastorianus (carlsbergensis) and Botrytis cinerea, respectively [158, 159]. Clade C(iii) comprises homologs corresponding to “group 2” putative inositol transporters [157] that are related to the putative inositol transporter Hgt19 of Candida albicans [160]. The A. niger members of this clade revealed highly diverse abilities to transport monosaccharides and/or oligosaccharides (Additional file 22).
Clade D incorporates oligosaccharide transporters including the A. oryzae maltose permease MalP and its homolog [161], homologs of the melibiose transporter MBT1 of Colletotrichum graminicola [162], the maltose transporters of S. cerevisiae (Mal11, Mal31, Mal61, and Mph3) [137], and MRT of Metarhizium robertsii [163]. Similar to Clade C, highly diverse abilities of the tested A. niger members of this clade were observed (Additional file 22).
Clade E can be divided into two main subclades: E(i) contains homologs of phosphate transporters [164–166] and the S. cerevisiae transporter (GIT1) of glycerophosphoinositol and glycerophosphocholine [167], while members of E(ii) share homology with the S. cerevisiae monocarboxylic acid/selenite transporter Jen1 [168]. No A. niger members of this clade have been characterized.
Clade F contains the yeast glycerol transporter Stl1 [169], the N. crassa pentose permease XAT-1 [145], and the xylose specific permease An25 [151]. Several of the A. niger members of this clade were able to transport oligosaccharides, while others appeared to be specific for glycerol or mannitol (Additional file 22).
No homology to a functionally characterized transporter has been observed for any members of clade G. However, one of the tested A. niger members was able to transport D-glucose while another transported maltose, trehalose, and melizitose.
Clade H contains the A. nidulans lactose permeases LacpA and LacpB [170, 171] and N. crassa cellodextrin transporters [172, 173]. One of the tested A. niger members was able to transport melizitose and turanose (Additional file 22).
It is unclear if the widespread occurrence of broad-specificity transporters in A. niger, as indicated by the functional complementation studies in S. cerevisiae, is specific to this species or common among a wider range of fungi. A transportome containing a diversity of broad-specificity sugar transporters is congruent with a saprobic lifestyle where degradation of natural substrates provides complex mixtures of monosaccharides, disaccharides, and trisaccharides.
Amino acid transporters
Phylogenetic analysis (Additional file 23) makes it possible to propose which paralogs are responsible for the uptake of aromatic amino acids (two clades, shown in purple) and those responsible for the uptake of basic amino acids (shown in red). This analysis suggests that, in some cases, specificity arose prior to the divergence of the Saccharomycotina and the Pezizimycotina (e.g. for basic and di-carboxylic amino acids, and for proline) while in other cases the amino acid specificity evolved later. The S. cerevisiae-specific subclade shown in light orange is suggested to be a group of broad specificity permeases (excluding proline) based on their similarity to N. crassa pmg and its possible ortholog from Trichoderma harzianum [174, 175]. The clade clustering with Tat3 of Saccharomyces pastorianus (deep purple) [176] may indicate a specificity for aromatic amino acids, while the ability to transport cystine is indicated by the Candida glabrata gene YAT [177]. A clade, shown in blue contains the well-characterized proline permease of A. nidulans ([178] and references therein), clustered with its ortholog, Put4 from S. cerevisiae; similarly the S. cerevisiae aspartate/glutamate transporter Dip5 is an outgroup of a clade shown in green which contains the isofuctional ortholog of A. nidulans [179].
A recent study [180] has allowed us to predict that the smaller clade (shown in red) represents lysine-specific permeases while the larger clade (also shown in red; including AN8279) probably transports arginine and other basic amino acids. Interestingly, AN8279, possessing a Ser residue in TMS10, resembles the T456S mutant of Can1, suggesting that AN8279 is a high affinity and high capacity transporter for both arginine and lysine. A detailed study of the substrate specificity determinants of the A. nidulans PrnB transporter suggested that substrate specificity of this family is largely determined by residues in certain key transmembrane domains [178, 181].
The aspergilli screened were found to possess variable numbers of AAAP (Amino Acid/Auxin Permease) family members (TCDB 2.A.18) [140] and some S. cerevisiae AAAP proteins have been described as bi-directional vacuolar amino acid transporters, with some shown to be broad specificity permeases accepting aromatic and aliphatic amino acids [182]. All aspergilli were found to possess at least one methionine transporter.
Urea transporters
Urea is taken up by fungi and plants through a group of active urea/H+ symporters, belonging to the sodium:solute symporter family (SSS), as typified by A. nidulans UreA [183, 184] With the exception of A. zonatus, which has only one homolog, the Aspergillus genomes were found to contain a range of 2–7 UreA homologs, most of which are predicted to have 15 transmembrane segments (TMSs) (Table 5). The phylogenetic relationships of the UreA homologs suggest that two duplication events occurred before the different Aspergillus species diverged from a common ancestor, giving rise to two clades (Additional file 24). The first sub-clade, which contains UreA, also contains an ortholog from each of the fungi analyzed, suggesting conservation of urea transport function. The second clade contains members that have not yet been functionally characterized.
Table 5.
Number of UreA homologs found in the analyzed aspergilli. In A. niger, A. luchuensis, and A. fumigatus, the same number of homologs were found in each of the two sequenced strains
| Species | UreA homologs (n) | Species | UreA homologs (n) |
|---|---|---|---|
| A. niger | 4 | A. oryzae | 3 |
| A. luchuensis | 4 | A. flavus | 3 |
| A. tubingensis | 3 | A. terreus | 4 |
| A. brasiliensis | 4 | A. fumigatus | 3 |
| A. carbonarius | 4 | A. fischeri | 3 |
| A. aculeatus | 2 | A. clavatus | 3 |
| A. versicolor | 7 | A. glaucus | 4 |
| A. sydowii | 5 | A. wentii | 5 |
| A. nidulans | 4 | A. zonatus | 1 |
Amino acid residues important for the binding, recognition, and/or translocation of urea and for the sorting of UreA to the membrane were recently identified [185]. All of these residues are conserved in all UreA orthologs of the genus, with the exception of A. aculeatus where an alanine (Ala163 in UreA) is substituted by a serine. This is remarkable since there is a Ser present at the corresponding position in characterized UreA orthologs of S. cerevisiae, C. albicans, and P. involutus [185].
Nitrate, nitrite, and ammonium transporters
All sequenced aspergilli were found to possess at least one copy of the nrtA gene encoding a Nitrate Nitrite Porter (NNP, TCDB 2.A.1.8) [140] protein for nitrate/nitrite transport [186–188]. This conserved nitrate transporter-encoding gene is located in a cluster with genes for nitrite reductase and nitrate reductase. Aspergillus nidulans, A. fumigatus, and A. fischeri also have a second nitrate transporter located outside the cluster. In A. nidulans, it has been shown that the two transporters, NrtA (in the cluster with nitrate and nitrite reductases) and NrtB, are expressed under identical conditions but have different affinities for nitrate and nitrite, suggesting a kinetic flexibility that may confer an advantage to the species growing in its natural habitat in soil where substrate concentrations can vary rapidly by orders of magnitude [189, 190]. NrtA displays low affinity, high capacity while NrtB is a high affinity, low capacity protein. In species with a single NNP, the protein more closely resembles NrtA rather than NrtB using a Smith-Waterman algorithm for pairwise comparison (Additional file 25).
Nitrite transport can also be performed by proteins of the formate-nitrite transporter (FNT) family (PF01226; TCDB 2.A.44) [140]. Bioinformatic analysis in the present study revealed that the majority of the aspergilli contain one gene encoding an FNT family protein, but a few species (A. flavus, A. oryzae, A. sidowii, A. fischeri, and A. versicolor) have two such genes. In A. nidulans, the FNT protein NitA has been shown to be a high affinity but low capacity nitrite transporter compared to NNP proteins and is expressed under the same conditions as the NNP proteins [191].
The aspergilli also contain one to six members of the proton-dependent oligopeptide transporter (PTR) family (PF00854: TCDB 2.A.17) [140] that includes the Arabidopsis thaliana nitrate transporter, Nrt1.1 (CHL1). However, absence of the nitrate binding residues identified in Nrt1.1 (H356 and Y388) [192, 193] implies that nitrate transport by PTR family proteins in the aspergilli is unlikely.
The aspergilli usually possess three or four members of the ammonium transporter/methylammonium permease/Rhesus factor family (AMT/MEP/Rh family) (PF00909; TCDB 1.A.11.2) [140], although the A. zonatus genome contains five genes encoding such proteins. In A. nidulans, these genes are differentially regulated with MeaA being the principle ammonium transporter in conditions where ammonium is non-limiting. By contrast, MepA is expressed under nitrogen limiting and nitrogen starvation conditions and MepB is only expressed during complete nitrogen starvation [194]. MepA and MepB display higher substrate affinity than MeaA [194].
The inorganic nitrogen transport capability emphasized by the present study most likely reflects the natural habitat of Aspergillus species in the soil where nitrate, nitrite, and ammonium form the principle inorganic salts of nitrogen. The diversity of transporters detected is of applied significance as it indicates flexibility in terms of the inorganic nitrogen sources that can be provided for growth of biotechnologically important aspergilli. However, for nitrate utilization (and by extension nitrite utilization), uptake of nitrate has generally been considered a “bottleneck” and so the manipulation of gene copy number in strains could be a goal for strain improvement. On undefined industrial media used for growing fungi, there may be scope for higher biomass yields and faster growth by manipulating inorganic nitrogen transporters.
Nucleobase and nucleoside transporters
There are two main fungal families specific for nucleobase and one for nucleoside uptake as described below. In addition, a fourth family, the Equilibrative Nucleoside Transporter (ENT)-like family (2.A.57) [140], with a similarity to nucleoside transporters of protozoa and metazoa, is also present in fungi but has not been characterized [195]. Some of these transporters (e.g. NCS1 and AzgA; see below) are ubiquitous in fungi but absent in mammals, which makes them promising candidates for designing novel targeted pharmacologic agents against pathogenic fungi [196].
Nucleobase Ascorbate Transporter family
Homologs of the Nucleobase Ascorbate Transporter (NAT) family (or Nucleobase Cation Symporter NCS2 family, 2.A.40) [140] are present in all domains of life and most fungi [197–199]. Members are thought to consist of 14 transmembrane domains (TMD) and have extended cytoplasmic N-tails and C-tails [200]. Fungal NATs can be further classified intro three distinct subfamilies based on their specificity and primary sequences. First are the “canonical” UapA/C group [201, 202], which are uric acid/xanthine/H+ symporters [203–205]. UapC-like homologs were found to be syntenic and present in all species of aspergilli in the present study (and duplicated in some) [206], whereas UapA paralogues were present only in A. nidulans and in A. zonatus. UapA-like transporters are present outside the aspergilli (e.g. in N. crassa and in members of the Pezizales, a basal group of the Pezizomycotina), so the present data perhaps contradict the suggestion that UapC was the ancestral gene and UapA had arisen by a recent duplication [207]. Second, UapD members were detected in section Nigri, but no data supporting their function as nucleobase transporters have yet been reported [205]. Finally, the AzgA group comprises H+ symporters specific for adenine, hypoxanthine, and guanine, and this group was present in all of the sequenced aspergilli [204, 208, 209]. This family, despite no evident similarity in primary amino acid sequence, seems ancestrally related to the NAT family [209].
Nucleobase Cation Symport 1 family
Nucleobase Cation Symport 1 (NCS1) family (2.A.39) [140] proteins are H+ (or Na+ in some bacteria) symporters specific for the uptake of purines, pyrimidines, thiamine, hydantoin, pyridoxine, and related metabolites in prokaryotes, fungi, and some plants. All members contain 12 TMDs and extended cytoplasmic tails. NCS1 transporters of the aspergilli and other dikarya can be divided into two distinct subfamilies, Fur and Fcy, both of which are probably the result of independent horizontal transfers from prokaryotes [207]. Fur homologs in the aspergilli and penicillia have presviously been shown to exhibit patterns of gene loss and duplication [195]. Homologs of FurD (uracil) and FurA (allantoin) were the only ones found to be present in all analyzed species in the present study. Convergent evolution has occurred within the Fur family several times, so that prediction of substrate specificities without functional assays is impossible [195]. The phylogenetic relations of these transporters are shown in Additional file 26.
The Fcy family of A. nidulans includes an H+/cytosine-purine symporter (FcyB) [210, 211] and recently identified cryptic transporters for either adenine or guanine [212]. Orthologs and paralogs of Fcy were found to be present in most of the examined aspergilli and penicillia (Additional file 27).
Concentrative Nucleoside Transporters family
The Concentrative Nucleoside Transporters (CNT) family (2.A.41) [140] is exemplified by CntA of A. nidulans, which functions as a general H+/nucleoside symporter [195]. One Cnt-like homolog is present in most of the genomes investigated, with the homologs being extremely well conserved and fully syntenic, with variations only in the N-tail. However, they are not present in A. zonatus and A. aculeatus, but some evidence for remnants (pseudogenes) was observed. No homologs were found in S. pombe, S. cerevisiae, plants, protozoa, and archaea. The presence of an ortholog of CntA in C. albicans [213] indicates a possible conservation of function throughout the Ascomycotina.
Flavohemoglobins and nitric oxide (NO) sensitivity
Nitric oxide (NO) is a gaseous radical with a short half-life and a high reactivity. For these reasons, fungi have evolved a diverse set of strategies to cope with NO [214]. Flavohemoglobins are a family of conserved proteins in bacteria and fungi which have the ability to detoxify NO via an NADPH, FAD, and O2-dependent conversion of NO to NO3 – [215]. Flavohemoglobins have been previously characterized in A. nidulans and A. oryzae, where two proteins have been identified [216–218]. In A. nidulans, it was found that deletion of both flavohemoglobins fhbA and fhbB resulted in increased sensitivity to NO [216, 219]. In A. oryzae, FhbA was localized in the cytosol, while FhbB showed mitochondrial localization [218], although it was also found that FhbA contains a predicted motif for secretion in A. niger and some other aspergilli [217]. The cytosolic flavohemoglobin (fhbA) is regulated by nitrogen source and the mitochondrial one during development [216, 219]. A cytochrome P450 nor (nitric oxide reductase) that reduces NO to N2O was found in Fusarium oxysporum [220]. This P450nor is not widespread in aspergilli, being present only in six species (Fig. 11). A S-nitrosoglutathione (GSNO) reductase converts GSNO into ammonia and oxidized glutathione (GSSG) in Cryptococcus neoformans and Magnaporthe oryzae [221, 222]. GSNO red is present in all Aspergillus species sequenced in this work. Some of the strains have up to three copies of this gene (Fig. 11).
Fig. 11.
Inhibition of growth by NO. The table depicts the number of homologs of the flavohaemoglobin genes (fhbA and fhbB), the P450 nitric oxide reductase, and the S-nitrosoglutathione (GSNO) reductase in each species. Strains are listed in the same order as in the phylogenetic tree shown in Fig. 1. Strains were grown in the presence or in the absence of the NO-releasing compound nitroprusside. Growth was monitored at different time intervals. The graph shows the percentage of growth in the presence of 128 mM nitroprusside compared to the control samples grown in the absence of nitroprusside. The time and temperature of growth was optimized for each strain and it was as follows (species name, strain, temperature, and incubation time for growth inhibition calculation): A. glaucus: 22 °C for 72 h; A. tubingensis: 30 °C for 96 h; A. zonatus: 30 °C for 96 h; A. brasiliensis: 30 °C for 72 h; A. versicolor: 30 °C for 96 h; A. sydowii: 30 °C for 96 h; A. niger ATCC1015: 30 °C for 48 h; A. luchuensis: 30 °C for 72 h; A. niger NRRL3: 30 °C for 72 h; A. wentii: 30 °C for 96 h; A. niger CBS 513.88: 30 °C for 96 h; A. fischeri: 30 °C for 96 h; A. terreus: 30 °C for 96 h; A. flavus: 30 °C for 96 h; A. fumigatus: 30 °C for 96 h; A. clavatus: 30 °C for 96 h; A. nidulans: 30 °C for 96 h; A. carbonarius: 30 °C for 72 h; A. aculeatus: 30 °C for 96 h
In silico analysis of flavohemoglobins
A search for homologs of flavohemoglobins in the genomes of the test aspergilli revealed that all these species contain at least one gene encoding flavohemoglobin A (Additional file 28). Five species contain two copies of flavohemoglobin A and eight species contain one fhbA and one fhbB in their genomes. Phylogenetic analysis confirmed that flavohemoglobins A and B form two well-defined clusters (data not shown). The duplication of flavohemoglobin A could have happened after the separation of the A. nidulans–A. versicolor–A. sydowii cluster, which groups separately from the rest of the aspergilli fhbA genes. Interestingly, the fhbB genes of these three species also cluster together.
NO sensitivity
Most of the Aspergillus species were sensitive to high concentrations of the NO-releasing compound sodium nitroprusside (Fig. 11, Additional files 3 and 28). However, some were resistant to NO, in particular A. glaucus, A. niger (NRRL3 and ATCC1015), and A. luchuensis. Other strains, such as A. tubingensis and A. carbonarius, showed an intermediate sensitivity to NO. We could not find any correlation between the sensitivity to NO displayed in our growth tests and the number of copies or the type of genes involved in the detoxification of NO. The presence of one or two types of flavohemoglobins (cytosolic or mitochondrial) and the fact that all species having two copies of fhbA do not have the other flavohemoglobin (fhbB) is intriguing. Although no association between number and/or type of flavohemoglobin and sensitivity to NO is evident, it may reflect an adaptation to different lifestyles. However, all species contain three to six copies of NO detoxification genes with at least two types of mechanisms (Fig. 11). For example, in A. fumigatus detoxification of NO does not seem to be required for full virulence [223, 224] and it contains only three genes for the NO detoxification (two flavohemoglobins and one GSNO reductase) (Fig. 11).
Signal transduction pathway genes
Gene family members for four different protein classes involved in signal transduction in Eurotiomycetes were identified using a protein domain-based approach. In total 11,120 proteins were identified belonging to 29 Eurotiomycetes species, clustered into 765 orthologous groups. The analysis of orthologous groups produced a set of genes that could not be clustered with any other and are regarded here as putative species-specific genes. Of the 29 Eurotiomycetes genomes, 25 contain species-specific genes (Fig. 12a) that may be involved in particular traits of these species. G-protein-coupled receptor (GPCRs) and kinases contribute most to these genomic novelties and thus constitute candidates for further functional studies aiming at understanding the differences among them (Additional file 29).
Fig. 12.
a Proportion of species-specific genes of the different signal transduction protein-classes in Eurotiomycetes. b Number of protein phosphatases in fungi. Eurotiomycetes appear in red
G-protein receptors and subunits
Heterotrimeric G-proteins have universal roles as signaling proteins in eukaryotes. Each G-protein heterotrimer is composed of α, β, and γ subunits that interact with the plasma-membrane, in association with a GPCR. Most filamentous fungi have three conserved Gα subunits (I, II, and III), one Gβ protein, and one Gγ protein. The number of predicted GPCRs varies widely, with a larger number identified in ascomycete than in basidiomycete fungi [225–227]. Results of the domain-based identification of G-proteins and GPCR orthologs in the eurotiomycetes are illustrated in Additional file 28. Gα proteins were found to be highly conserved. The Gα group I (fadA) regulates multiple pathways; most filamentous fungi in this study have a single copy of this gene (Additional files 29 and 30). In contrast, Group II Gα proteins (ganA) are less conserved than the other Gα proteins and up to three copies were found in one species (T. stipitatus) (Additional file 30). The Group II Gα proteins play a positive role in germination of conidia, possibly through cAMP signaling in carbon sensing [228, 229]. The last Gα belongs to group III (ganB) and was found as a single copy in all eurotiomycetes where it is thought to act as a negative regulator (Additional files 2, 29, and 30). Additionally, we found three different Gα groups of orthologs (Clusters FungiJGICTBE21135 only present in A. niger ATCC 1015, FungiJGICTBE20122 present in A. flavus NRRL 3557 and in A. oryzae RIB40, and FungiJGICTBE14195 present in A. flavus NRRL 3557 (Additional files 29 and 30) that appear to be encode genes that contain the Gα domain. There was also one copy of the Gβ negative regulator of asexual reproduction (sfaD) in each genome (Additional files 2, 30, and 31) [230]. Finally, all Eurotiomycetes genomes contained a single highly conserved Gγ gene [231].
Serine-arginine protein kinases
Members of the serine-arginine protein kinases (SRPKs) family are known to phosphorylate serine within RS (arginine/serine rich) regions [232]. As previously reported in A. nidulans, N. crassa, and dermatophytes, there is high genome expansion of SRPKs in filamentous fungi [21]. Interestingly, the A. nidulans SRPK types were spread in seven clusters of orthologous genes, whereas only two SRPK types have been found in N. crassa (Additional file 32). However, A. nidulans is the only eurotiomycete with all seven SRPK types. Most of the SRPK members identified in Eurotiomycetes belong to the types A and F, while the SrpkB and SrpkD types were less conserved in the eurotiomycete genomes. It has been suggested that SRPKs of filamentous fungi are involved in the molecular response to a broad range of environmental stimuli, although these signaling pathways are currently poorly understood [233]. Since the main biological role for SRPKs is the regulation of messenger RNA (mRNA) processing, it is conceivable to speculate that the expansion of SRPK in the eurotiomycetes could reflect the greater importance of splicing in filamentous fungi than in S. cerevisiae, whose genes generally do not contain introns. A core set of 48 kinases is present in all eurotiomycetes and in other species of Ascomycota analyzed (N. crassa, F. oxysporum, T. reesei, S. cerevisiae, P. pastoris, and S. pombe), which suggests that these kinases have a critical role in the biology of those organisms. Components of mitogen-activated protein (MAP) kinases are classical examples of conserved kinases [234] and are involved in cell polarity and cell wall maintenance (Kin1, BckA, MkkA, MpkA), mating (SteC, Ste7, Ste 20), and osmotic stress sense (HogA, SskB, PbsA) [233].
In contrast to the above conserved pathways, there is growing evidence that filamentous fungi might have evolved unique kinases. Indeed, a block of kinases that appear to be specific to filamentous fungi have been identfied, named filamentous fungal kinases (Ffk from A to J). These show low sequence identity with kinases of S. cerevisiae, Caenorhabditis elegans, Drosophila melanogaster, and human [233].
Since single deletions of FfkA, FfkB, FfkD, FfkE, FfkF, FfkG, FfkH, and FfkI from A. nidulans were not lethal and did not give a different phenotype [233], little is known about the functionality of these kinases [99]. The present study found a similar group of kinases among the Eurotiomycetes that do not appear to have orthologs in the yeast species analyzed (Table 6) [233]. Overall, P. pastoris, S. cerevisiae, and S. pombe encode fewer kinase orthologs than the other fungal species analyzed, with 98, 126, and 101 copies, respectively, which is almost half the number of kinases predicted in the P. rubens Wisconsin (205). Conversely, there is a core set of 31 kinases (Additional file 33) that do not appear in any of the eurotiomycetes, but were retained in S. cerevisiae, P. pastoris, and/or S pombe. Twenty-five kinases are essential in A. nidulans [233] and all the analyzed eurotiomycetes have at least one copy of these genes, suggesting they are essential across the tested species.
Table 6.
Filamentous fungal kinases without orthologs in model yeast species
| Cluster | A. nidulans kinase locus tag | Domain | A. nidulans name | Picpa1 | Sacce1 | SacceM3707_1 | Schpo1 |
|---|---|---|---|---|---|---|---|
| FungiJGICTBE4492 | AN10485 | Pkinase | Stk21 | 0 | 0 | 0 | 0 |
| FungiJGICTBE1792 | AN6339 | Pkinase | Pod6 | 0 | 0 | 0 | 0 |
| FungiJGICTBE4614 | AN1560 | Pkinase | PlkA | 0 | 0 | 0 | 0 |
| FungiJGICTBE4599 | AN4717 | Pkinase | PkaB | 0 | 0 | 0 | 0 |
| FungiJGICTBE3865 | AN0822 | Pkinase | KfsA | 0 | 0 | 0 | 0 |
| FungiJGICTBE1403 | AN4936 | Pkinase | Prp4 | 0 | 0 | 0 | 1 |
| FungiJGICTBE4249 | AN7537 | Pkinase | Ppk33 | 0 | 0 | 0 | 1 |
| FungiJGICTBE2868 | AN3101 | HiskA | PhkB | 0 | 0 | 0 | 1 |
| FungiJGICTBE1795 | AN3102 | HisKA | PhkA | 0 | 0 | 0 | 2 |
| FungiJGICTBE4245 | AN6044 | Pkinase | NpkA | 0 | 0 | 0 | 1 |
| FungiJGICTBE4202 | AN3065 | Pkinase | CmkB | 0 | 0 | 0 | 1 |
| FungiJGICTBE3486 | AN0699 | Pkinase | Cak1 | 0 | 0 | 0 | 1 |
| FungiJGICTBE668 | AN4479 | HisKA | NikA | 2 | 0 | 0 | 0 |
| FungiJGICTBE164 | AN11032 | Pkinase | SepL | 1 | 0 | 0 | 1 |
| FungiJGICTBE3739 | AN4385 | Pkinase | SepH | 1 | 0 | 0 | 1 |
| FungiJGICTBE3573 | AN10800 | BCDHK_Adom3 | pkpA | 1 | 0 | 0 | 1 |
| FungiJGICTBE2991 | AN11101 | Pkinase | Gin4 | 1 | 0 | 0 | 1 |
| FungiJGICTBE3299 | AN5728 | Pkinase | Stk22 | 0 | 1 | 1 | 1 |
| FungiJGICTBE2936 | AN10515 | Pkinase | PrK1 | 0 | 1 | 1 | 1 |
| FungiJGICTBE598 | AN1665 | Pkinase | stk51 | 1 | 1 | 1 | 0 |
| FungiJGICTBE878 | AN2412 | Pkinase | CmkA | 1 | 2 | 2 | 0 |
| FungiJGICTBE3318 | AN7563 | Pkinase | ChkC | 1 | 1 | 1 | 0 |
Protein phosphatases
Protein phosphatases are classified into the following main groups: (1) classical Ser/Thr phosphatases: phosphoprotein phosphatases (PPP) family and protein phosphatase Mg2+ or Mn2+ dependent (PPM) family; (2) protein Tyr phosphatase (PTP superfamily); and (3) Asp-based protein phosphatases with DXDXT/V catalytic signature (CPDs) [235]. Ssu72, a different type of protein phosphatases, is regarded as separate, although it shares some similarity with the PTP Superfamily [236]. We found, on average, 8.8 PPPs, 8.37 PPMs, 5.26 CPDs, and 15.3 PTPs in the fungi included in this study (Fig. 12b). The actual numbers varied considerably between species, especially for the PTPs. Ssu72, is an exception, showing a single member in all species. PTP is the largest phosphatase superfamily presented here, including classical PTPs [236], dual-specificity phosphatases (DSPases), that have an important role in intracellular transduction pathways and regulate the mitogen-activated protein kinase (MAPK) pathway and cell cycle progression, Cdc25 type phosphatases, either regulating cell cycle progression [237], and low molecular weight phosphatases [236]. The increased and variable repertoire of protein kinases and phosphatases in Aspergillus could have helped in the establishment of several complex morphological traits, as well, as sophisticated responses to stress and the evolution of pathways that can help in the pathogenic process.
DNA protein organisation and methylation
Histones of the aspergilli
Five classes of histones have been identified in eukaryotes, comprising linker histone H1 and core histones H2A, H2B, H3, and H4. Bioinformatic analysis of the newly available Aspergillus genome sequences revealed the presence of all of these families of histones, with overall evidence of conservation throughout the aspergilli. However, there were some differences between species, the functional implications of which are unclear, as follows.
All analyzed Aspergillus genomes were found to contain an H1 histone gene, following manual curation. The sequences are highly similar at the N-terminus and within the globular domain (residues 18–93 in A. nidulans, Additional file 34). They diverge in the C-terminal domain, the maximal divergence being at the boundary of the globular domain with the C-terminal domain, due mainly to sequence insertions.
All Aspergillus genomes also contain a highly conserved H2B histone.
As previously reported [238], in the aspergilli, as in other ascomycetes, the extant H2A histone corresponds to the H2AX variant, rather than the canonical H2A form. This was confirmed for all aspergilli in the databases, where the diagnostic motif SQEL is conserved at the carboxy terminus. In all aspergilli, as reported previously for A. nidulans and N. crassa [239, 240], the H2AX gene is transcribed divergently from the H2B gene. In addition, all analyzed Aspergillus genomes have an H2A.Z ortholog. While nothing is known about the role of this histone in any of the Pezizomycotina, the cognate gene is not essential in N. crassa, S. cerevisiae, or S. pombe [241, 242].
All analyzed genomes contain an H3 gene, with complete amino acid sequence conservation for all species. While we have previously reported the presence of genes with significant amino acid similarity with histone H3 in A. terreus and A. oryzae [238], no other H3 paralogs were found in the other aspergilli nor in any of the other eurotiomycete genomes. The paralog in A. terreus appears real with two typical filamentous fungal introns but we could not find evidence for its expression in publicly available data. In the six sequenced strains of A. oryzae, as well as in the genome of the strongly related species A. sojae and A. parasiticus [243–245], a duplication has occurred of the central part of the functional H3 gene but no legitimate start-codon could be identified. There is no similarity among these sequences downstream of the H3-like sequence [238], which suggests that this “gene” results from a non-functional duplication of part of the canonical H3-encoding gene. This duplication is, however, absent in the ten currently sequenced strains of A. flavus [246]. The occurrence of a genetic feature common to A. sojae, A. parasiticus, and A. oryzae but absent from A. flavus (and A. nomius) is intriguing given the fact that A. oryzae are generally considered domesticated variants of A. flavus [9].
All species also contain the specific centromeric H3 histone (naming varies in different organisms, here named CenH3), with the motif LPFAR in helix α1 conserved in all species. Alignment of the CenH3 sequences (Additional file 34) shows the typical CenH3 divergence in the N-terminal region, which is a highly variable region in different species of D. melanogaster [247], confirming previous observations [238]. Additional file 34 also shows comparative phylogenies of the CenH3 histone and of the other sequence-variable histone, H1. CenH3 histone divergence has been proposed to be instrumental in driving speciation [247].
All genomes have two H4 encoding genes, one transcribed divergently from the H3 encoding gene, denoted H4.I, and a second one, denoted H4.II, unlinked to it. The H3-linked and unlinked H4 paralogs are almost identical at the amino acid sequence but the intron organization of the paralogs differs. Both H4 genes share an intron that splits the Arg4 codon. In H4.II, this is the only intron (in Aspergillus and Penicillium) and it is always at least twice as big as that in the H4.I paralog, often >200–350 nt. The H4.I gene generally has a second intron interrupting the Arg96 codon. This 3′ intron must have been lost in A. zonatus, A. glaucus, A. rubrum, and also in Penicillium paxilli, since it is present in the eight sequenced species that belong to other families of Eurotiales (Talaromyces, Thermoascus, Monascus, Byssochlamys, and Thermomyces) and in Onygenales. In addition, the amino acid at position 2 is a Ser or Thr in both I and II paralogs. Note that in a given species, paralogs I and II are always identical at this position (Ser for A. flavus, A. glaucus, A. nidulans, A. oryzae, and A. sydowii and A. versicolor, A. rubrum, and Thr for the other sequenced aspergilli). These species belong to different taxonomic sections within the aspergilli, suggesting that concerted evolution (possibly by gene conversion) could be responsible for the residue at this position. Similarly, at position 65, there is almost always a Thr in paralog I, which is a Ser in paralog II. At position 69, there is always a Gly in paralog I and a Ser in paralog II.
We have reported previously the presence of a small reading frame with significant similarity to a central part of histone H4 in the genome of A. nidulans [238]. We have now found similar reading frames in most Aspergillus genomes. This include species where it was previously overlooked or where it was erroneously annotated. The newly recognized “third” histone 4 gene invariably contains seven introns. As a consequence, this gene has considerably more intronic sequences than coding sequences; in A. nidulans for instance, the 357 nt long coding sequence (36%) is interrupted by 642 nt of intronic sequences (64%). The additional putative H4 gene is absent from A. glaucus and E. rubrum as well as from Penicillium species, but present in other sequenced Eurotiales species as well as in the Onygenales (Additional file 34).
Putative DNA methylases
Two putative DNA methylases have been described in the ascomycetes. They were identified through work relating to the phenomena of MIP (Masc1, Ascobolus immersus) and RIP (RID1, N. crassa) as inactivation of the masc1 and rid‐1 prevent MIP and RIP, respectively [248, 249].
There are, however, serious doubts as to whether masc1 and rid-1 encode actual DNA methylases. First, no DNA methylase activity could be detected in vitro with the purified proteins [248, 249]. Second, all DNA methylation in N. crassa, even methylation subsequent to RIP, depends on another protein, DIM2 [250]. Third, a homolog of Masc1 and RID1 was reported for A. nidulans (DmtA), a species for which DNA methylation has never been detected [251, 252] (Reyes Domínguez and Scazzocchio, unpublished data). Similar results were obtained for A. flavus [253]. Interestingly, DmtA is essential for meiosis in A. nidulans [251].
All the sequenced aspergilli were found to have Masc1/RID1 homologs at syntenic positions. A phylogentic tree shows DIM2 as outgroup to all other sequences (Additional file 35), while RID1 of N. crassa clusters with the two Talaromyces species, which, in the absence of other sequences in the tree, may simply mean that the latter have diverged less from an ancestral homolog. The pervasive presence of the DmtA orthologs may indicate that many Aspergillus and Penicillium “asexual” species are in fact sexual (see earlier “Asexual and sexual reproduction” section).
A DNA methylase with in vitro enzyme activity, Masc2, has been described in A. immersus [254] and BLAST analysis revealed a new protein family present in all aspergilli in syntenic positions. The A. nidulans protein includes from residue 322 to 586 a putative (incomplete) C‐5 cytosine DNA methylase domain while two (incomplete) SNF_2N domains occur from residues 1305 to 1470 and 1627 to 1818. No function is known for this protein or its homologs, but orthologs are present in all available sequences of Penicillium and Talaromyces, and in A. immersus and Tuber melanosporum, members of Pezizales, a basal order of the Pezizomycotina. Interestingly, in N. crassa, the fungus where DNA methylation has been more thoroughly studied, no protein with the same architecture is present nor was one found in most other Sordariomycetes.
Conclusions
Data presented in the present study emphasize the high genomic and functional diversity present within the genus Aspergillus. While strong conservation was observed for central biological functions (e.g. sporulation, aspects of cell wall structure, and histones), high diversity was apparent for many other physiological traits, such as carbon utilization, secondary metabolism, and stress response. While most aspergilli can occupy a broad range of habitats, our data suggest that evolution has generated considerable diversity within the genus to allow their exploitation of and persistence in different habitats.
The detailed analyses described in this paper will serve as a reference for variation in other fungal species. The unique position of this genus in the genomics of filamentous fungi, with genome sequences for a large number of species, will become even more evident with the future completion of the currently ongoing Aspergillus whole genus genome project in which more than 300 genomes from species of this genus will be sequenced. The data of our study will provide a backbone to rapidly perform detailed genomic comparisons and functional studies in these new genomes.
Methods
Genome sequencing, assembly, and annotation
Genome sequencing and assembly
The majority of genomes and transcriptomes in this study (A. luchuensis, A. brasiliensis, A. glaucus, A. sydowii, A. tubingensis, A. wentii, A. versicolor, and A. zonatus,) were sequenced with Illumina and assembled with AllPathsLG [255], except for A. aculeatus and A. carbonarius, which were sequenced using a combination of 454 and Sanger platforms and assembled with Newbler (Roche). For all standard fragment Illumina libraries, 500 ng–1 μg of genomic DNA was sheared using the Covaris E210 (Covaris) and sized selected for 270 bp using Agencourt Ampure Beads (Beckman Coulter). The DNA fragments were treated with end repair, A-tailing, and adapter ligation using the TruSeq DNA Sample Prep Kit (Illumina) and purified using Agencourt Ampure Beads (Beckma Coulter). These were sequenced on HiSeq in 2 × 150 bp read format and assembled using AllPathsLG with long-mate paired-end libraries, which were built using several approaches.
CLIP (Cre-Lox Inverse PCR) libraries were used for all Illumina sequenced genomes in this study except for A. zonatus and A. glaucus. For CLIP libraries, 10–15 μg of genomic DNA were sheared using the Hydroshear® and gel size selected for 4–5 kb. The fragments were end-repaired (NEB) and ligated with biotinylated Illumina compatible adapters containing loxP (IDT). The adapter ligated DNA fragments were circularized via recombination by a Cre excision reaction (NEB). The circularized DNA was digested using four base cutter restriction enzymes followed by self-ligation (NEB). The self-ligated products were immobilized on beads and inverse PCR was used to enrich for the final library (NEB). For A. zonatus and A. glaucus, ligation-free paired-end (LFPE) fragments were used. LFPE fragments were generated using the 5500 SOLiD Mate-Paired Library Construction Kit (SOLiD®). A total of 15 μg of genomic DNA was sheared using the Covaris g-TUBETM (Covaris) and gel size selected for 5 kb. The sheared DNA was end-repaired and ligated with biotinylated internal linkers. The DNA was circularized using intra-molecular hybridization of the internal linkers. The circularized DNA was treated with plasmid safe to remove non-circularized products. The circularized DNA was nick translated and treated with T7 exonuclease and S1 nuclease to generate fragments containing internal linkers with genomic tags on each end. The mate pair fragments were A-tailed and purified using Strepavidin bead selection (Invitrogen). The purified fragments were ligated with Illumina adaptors and amplified using ten cycles of PCR with Illumina primers (Illumina) to generate the final library. Quantitative PCR (qPCR) was used to determine the concentration of the libraries and were sequenced on the Illumina Hiseq in format 2 × 100 bp reads.
All Illumina reads from both types of libraries for each genome were assembled using AllPathsLG.
The genomes of A. aculeatus and A. carbonarius were sequenced using a combination of 454 standard, 454 paired-end, and Sanger fosmid libraries and assembled with Newbler. For 454 standard libraries, genomic DNA was fragmented by nebulization and purified using Ampure beads (Beckman Coulter). The ends of the fragments were treated with end repair and ligated with biotinylated adapters using either the Titanium (for A. aculeatus) or GS FLX (for A. carbonarius) Library Prep Kits (454). Adapter ligated products were immobilized on beads and treated with nick repair. Single strands were eluted to generate the final library. For 454 paired-end libraries, genomic DNA was sheared to 10 kb (for A. aculeatus) and 3 kb (for A. carbonarius) using the Hydroshear®. The ends of the fragments were ligated with biotinylated adapters containing loxP. The adapter ligated DNA fragments were gel-size selected and circularized via recombination by a Cre excision reaction (NEB). The circularized DNA was randomly sheared using the Covaris E210 (Covaris). The sheared fragments were end-repaired (NEB), ligated with 454 adapters (Roche), and immobilized on strepavidin beads followed by PCR using a biotinylated primer (Roche). The PCR product was immobilized and single-strand DNA libraries were isolated using an alkaline treatment. Sanger fosmid libraries for A. aculeatus and A. carbonarius were prepared from 20 ug of genomic DNA, sheared using the Hydroshear® and size selected for 40 kb using a pulse field gel. The long inserts were cloned into pCC1FOS vector. The hybrid 454-Sanger assemblies were produced using Newbler (Roche).
Transcriptome sequencing and assembly
For all transcriptomes in this study, except for A. aculeatus and A. carbonarius, stranded complementary DNA (cDNA) libraries were generated using the Illumina mRNA sample preparation kit (Illumina). mRNA was purified from 10 μg of total RNA using magnetic beads containing poly-T oligos. mRNA was fragmented using divalent cations and high temperature. The fragmented RNA was reverse transcribed using random hexamers and SSII (Invitrogen) followed by second strand synthesis. The fragmented cDNA was treated with end-pair, A-tailing, adapter ligation, and ten cycles of PCR. qPCR was used to determine the concentration of the libraries. Libraries were sequenced on the Illumina Hiseq in 2 × 100 bp read format. Illumina reads were de novo assembled into consensus sequences using Rnnotator (v. 2.5.2 or later) [256]. For A. aculeatus, cDNA libraries were generated using the cDNA Rapid Library Preparation Kit (Roche). mRNA was purified from total RNA using Absolutely mRNA™ purification kit (Stragene) and chemically fragmented using high heat. The fragmented RNA was reversed transcribed using random hexamers and AMV RT followed by second strand synthesis. The cDNA fragments were treated with end repair and ligated with 454 adapters. For A. carbonarius, mRNA was purified from total RNA using Absolutely mRNA™ purification kit (Stratagene). mRNA was reverse transcribed with SuperScriptIII using dT15VN2 primer. cDNA was synthesized with E. coli DNA Ligase, E. coli DNA polymerase I, and E. coli RnaseH (Invitrogen). cDNA was nebulized and gel purified to generate fragment sizes between 500–800 bp. The fragments were end-repaired, adaptor ligated, and made into single-strand DNA libraries using the GS FLX Titanium library kit. In both cases, 454 reads were filtered and screened for quality and contamination to include removal of ribosomal RNA, low-quality, and low-complexity reads. The remaining reads were assembled into contigs using Newbler (v2.3-PreRelease-6/30/2009) with default parameters.
Genome annotation
Genome assembly scaffolds were masked with RepeatMasker [257], RepBase library repeats [258], and repeats frequently detected (>150 times) by RepeatScout [259]. A combination of gene predictors was run on the masked assemblies: ab initio Fgenesh [260] and GeneMark [261]; homology-based Fgenesh + [260] and Genewise [262] seeded by BLASTx [263] alignments against the NCBI NR database; and transcriptome-based assemblies. In addition to protein-coding genes, tRNAs were predicted using tRNAscan-SE [264]. Predicted proteins were functionally annotated using SignalP [265] to predict signal peptides, TMHMM [266] for transmembrane domains, InterProScan [267] for protein domains, and protein alignments to NCBI NR, SwissProt, KEGG [268] for metabolic pathways, and KOG [269] for eukaryotic clusters of orthologs. Interpro and SwissProt hits were used to map Gene Ontology terms [270]. For each genomic locus, the best representative gene model was selected based on a combination of protein homology and EST support, which resulted in the final set of models used in this work.
Protein conservation and phylogeny
To assess protein conservation, we performed MCL clustering [271] with an inflation parameter of 2.0 on the 400,447 protein sequences from 37 predicted full fungal proteomes (25 from the Aspergillaceae, 12 ascomycete outgroups). Some 65,450 protein clusters (or families) resulted, with average size 6.12 including singletons and 19.40 excluding singletons, largest cluster containing 2179 proteins, and 47,245 singletons. Membership in a cluster was used to determine whether a protein was conserved across the comparative organism set. We considered a protein conserved if its homologs were found at least once in all members of an organism subset (e.g. section Nigri or Aspergillaceae). A protein was considered specific to an organism subset if it was found in at least one organism of the subset, but not in any organisms outside the subset.
Pfam domains [272] were assigned using v. 22 of the database and a TimeLogic Decypher machine and an e-value of 10–4.
The phylogenetic tree was generated from a separate cluster run which differed from the previous one by the addition of non-ascomycete outgroups Rhizopus oryzae and Batrachochytrium dendrobatidis (JGI portals Rhior3 and Batde5, respectively; omitted from Fig. 1). Protein sequences with single copies in each organism were aligned using MAFFT [273], trimmed to well-aligned regions using Gblocks [274] with default parameters. RAxML [275] was then run using the PROTMIXWAG model and B. dendrobatidis set as an outgroup to generate the phylogenetic tree shown in Fig. 1. All branches received maximum support, except for that between A. niger strains NRRL2 and ATCC 1015 (73%; Fig. 1b).
Identification of clusters of orthologous genes
Clusters of orthologous genes were identified using OrthMCL with an inflation value of 1.5 [276]. All other parameters, e.g., BLAST search, were used in the default settings.
Identification/annotation of specific gene sets
Genes involved in asexual and sexual reproduction
The annotation pipeline of putative genes involved in asexual and sexual sporulation involved a two-step procedure of identification and annotation. Genes previously identified from A. nidulans were used as references (Additional files 2 and 5). The identification step of asexual genes was performed by searching the reference protein sequences against Aspergillus overall protein sequences with phmmer of HMMER 3.0 (http://hmmer.org/). Hits that passed MSV, Bias, Vit, and Fwd filters (see HMMER User’s Guide, http://eddylab.org/) were then subject to annotation involving BlastP comparisons against the database of non-redundant protein sequences [277] (Additional file 2). The identification of sexual genes was performed by searching protein databases available at the JGI (http://genome.jgi.doe.gov/) or AspGD (http://www.aspergillusgenome.org/) websites by BLASTP interrogation. Hits that had a minimum E value of 1e–70 were considered significant except for analysis of the relatively short PpgA protein where less stringent criteria were applied (Additional file 5). Synteny analysis (SYBIL) was then performed using the Jaccard Orthologous cluster option at AspGD or synteny option at the JGI website. Based on high levels of similarity and/or a large functional homogeneity of the hits, predicted proteins were annotated as corresponding functional proteins.
Genes involved in organic acid production
For determination of isoenzymes in production of organic acids, a list of known and predicted isoenzymes from A. niger CBS 513.88 was used as the seeding set. Using a BLAST-comparison, all genes above a cutoff value of 80% identity and 80% coverage were recorded as isoenzymes for the individual species.
Prediction and synteny analysis of aspergilli SM clusters
Protein and genomic sequences of aspergilli were downloaded from AspGD (http://www.aspgd.org/). Detection of PKS, NRPS, and DMATs and further prediction of respective SM gene clusters were made with SMIPS and CASSIS tools, correspondingly (https://sbi.hki-jena.de/cassis/) Synteny analysis was performed with the help of an Aspergillus 21-way comparative database asp2_v9 powered by Sybil. Conservation of clusters was calculated as the percentage of genes in the query cluster conserved in the syntenic counterpart. Confirmation of the identity of different secondary metabolites from the aspergili were checked by ultra high performance liquid chromatography–diode array detection–high resolution mass spectrometric detection (UHPLC-DAD-HRMS) [278].
Analysis of CYP proteins
Evolutionary analyses were conducted in MEGA5 [279]. The analysis involved 19 amino acid sequences. If protein size is marked by a star, the sequence was manually curated or sequences are incomplete. Following MUSCLE alignment, the evolutionary history was inferred by using the maximum likelihood method based on the JTT matrix-based model [280]. There was a total of 496 (CYP51F1) or 510 (CYP53) positions in the final dataset. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. All positions containing gaps and missing data were eliminated. Additional copies of CYP51F2 and CYP51F4 are excluded from the tree.
Aspergillus sugar transportome
The assignment of a protein to the “Sugar and other transporters” PFAM family PF00083 is based on its level of similarity to a Hidden Markov Model (HMM). The HMM for PF00083 is derived from a seed alignment of 33 proteins chosen to be the core representatives of that family [281, 282]. The latter includes proteins for which either transport of a specific sugar or related substrate has been experimentally demonstrated, or there is very strong indirect evidence that such a transport function exists, e.g. genetic evidence. Despite their considerable diversity in primary structure, PF00083 members exhibit a common topology of 12 membrane-spanning domains (TMSs) divided into two groups of six. Transmembrane (TM) domain predictions were obtained for all the PF00083-annotated proteins studied using TMHMM v2 [283]. While the majority of protein sequences exhibited 2 × 6 TM topology, we manually reannotated some loci to yield greater consensus with the 12 TM topology and a very small number were excluded from further analysis due to the detection of fewer than three TM domains. The resulting collection of amino acid sequences was aligned by MUSCLE and a neighbor-joining (NJ) tree was generated using MEGA6 software [284]. Cluster analysis was also performed using the orthology detection tool “Proteinortho”) [285] and the results obtained found to be concordant with the identifications made in the NJ tree.
Amino acid and urea transporters
The high-affinity methionine transporter of S. cerevisiae (MUP1) was used to identify several close homologs in all Aspergillus species. The methionine transporters of S. cerevisiae belong to the L-type amino acid transporter family (LAT; TCDB 2.A.3.8) [140] and exhibit low similarity to YATs [286]. A. nidulans UreA [[183, 184] was used to search the fungal genomes analyzed in this study for the presence of urea-like transporters (Table 5).
The S. cerevisiae proteins Tat1, Tat2, Tat3, Gap1, Hip1, Gnp, Agp1, Agp2, Agp3, Bap2, Bap3, Sam3, Mmp1, Lyp1, Alp1, Can1, Dip5, Put4, and Ssy1 obtained from the Saccharomyces Genome Database [287] were used as in silico probes for BlastP and in several cases, TBlastN searches in the available Aspergillus genomes at either the AspGD [14] or the JGI [15] databases. In addition, the homologues obtained for A. nidulans were used as in silico probes in a second BlastP search round. We have also identified a number of pseudo genes, either because their coding sequence contains frameshift and/or early chain termination mutations (ANID_4604, ANID_8556, ANID_8659, AFL2G_12189, AO090103000195, AO090005000021, AFL2G_06574, AO090026000738). In three other paralogues (ANID_5024, Aspbr1_139443, and Aspwe1_115075), the coding sequences lack a significant part of a functional YAT transporter [288]. These could be also considered pseudo genes or may have been recruited to a different function. Interestingly, no transcripts at all are detected (Jbrowse, [14]) for ANID_5024 and the adjacent gene (ANID_5023) in complete medium, while the genes on either side are transcribed under these conditions.
Alignment was done with MAFFT, Auto mode, Blosum62. Curation was carried out with BMGE. The tree shown was obtained using Blosum30 as a similarity matrix, but a topologically identical tree was obtained using the more stringent Blosum62 similarity matrix default setting. The maximum likelihood rooted tree was obtained with PhyML. The final tree with collapsed branches was drawn with FigTree.
Nucleobase transporters
The alignment of these putative transporters was performed using MAFFT [289]; curation was with BMGE [290] with a Blosum30 substitution matrix. Phylogeny was performed with PhyML [291] and re-drawing of the tree with FigTree [292].
Identification of protein class members involved in signal transduction
We employed a domain-based approach for the identification of proteins for each class, i.e. G-proteins, GPCRs, kinases, and phosphatases, following the general approach described by Perez-Rodriguez et al. [293] and using the pipeline available at [294]. The most important step is the identification of a protein domain or block that is characteristic of the protein class/family of interest and that is represented as a HMM, either available in the public database PFAM or created in house. Then specific rules are built accounting for the domain architecture of the different families/classes, in an exclusive way, i.e. a protein can belong to one and only one family/class, which requires the introduction of forbidden domains to resolve ties (Additional file 36). The HMMs used for the identification of protein classes of interest are available as (Additional file 37). Classification results are available in Additional file 31.
G-proteins
We studied the three sub-units of G-protein complexes: alpha, beta, and gamma. G-alpha and G-gamma had already well-described, characteristic, and specific domains in the PFAM database [282] with accession numbers PF00503 and PF00631, respectively. The G-beta subunit has several copies of the WD40 repeat (PF00400); however, this domain is not specific for G-beta proteins. We assesed the accuracy of our classification comparing our results against those presented in Li et al. [227]. The G-gamma famly is always found as a single member in each species. However, following our domain-based apporach we could not find the corresponding protein in A. niger CBS 513.88, A. oryzae, and S. pombe. Further inspection revealed that the protein is not annotated in the Aspergillus genomes, but we found a genomic region with the information for this gene, using the A. nidulans protein sequence as bait in a BLAST search (Additional file 37). In the case of S. pombe, the G-gamma subunit appears to be missing from the current version of the genome sequence.
GPCRs
For the identification of GPCRs, we took as gold standard the list provided by Li et al. [227] for A. nidulans. For some of the families (Stm1-related, GprK-like, PTH11, MG00532-like, material available under request) we built specific HMMs able to retrieve all of the known members of the respective family in A. nidulans and then applied them to the other species. Only in the case of PTH11 did we identify more members than the ones listed in the gold standard.
Kinases
We used as gold standard the identification and classification of kinases in A. nidulans from de Souza et al. [233]. All kinase classes have characteristic and specific domains available in PFAM, except the TOR kinases. We built an HMM specific for TOR kinases as described in Additional files 37 and 38.
Classical Ser/Thr phosphatases (PPP and PTP)
All PPPs in A. nidulans have the PFAM domain Metallophos (PF00149), but this domain also appears in other proteins. We retrieved all A. nidulans proteins that had a hit to the Metallophos domain and inferred the phylogeny, reveling that PPPs form a distinct clade in this evolutionary tree (Additional file 39), thus allowing us to build a PPP-specific HMM, named PP2Ac. In the case of the PPM family there were already specific PFAM domains that could be used to develop classification rules (Additional file 36).
PTPs
For most PTP superfamily members in [236] there were already specific domains in PFAM, i.e. Y_phosphatase, Y_phosphatase2, Y_phosphatase3, DSPc, and LMWPc. AN3941, belonging to the CDC25 subclass, was the unique PTP with no specific Pfam domain. Thus, we built a specific HMM (CTBECDC25) for this protein phosphatase. Although SSU72 proteins share sequence similarity with the PTP superfamily, they are classified as a distinct type of protein phosphatases. In that case, we used this PFAM domain Ssu72 Pfam for their identification.
Asp-based protein phosphatases
Aspartate-based catalysis phosphatases, which share the DXDXT/V catalytic signature (CPDs), had a specific domain found in the PFAM database, i.e. NIF (PF03031).
Histones of the aspergilli
Initial searches for histone H1 by BlastP failed to locate matches in some Aspergillus databases, while in other species the gene model was incorrect. Therefore, manual curation was required to establish its presence in A. oryzae and correction of the amino acid sequences was required for A. brasiliensis, A. kawachii, A. sydowii, A. terreus, A. versicolor, and A. niger CBS513.88. The NCBI database for Aspergillus sojae was interrogated to search for histone proteins in this species. Manual corrections of the H3 histone gene models were required for A. niger CBS 513.88, A. wentii, and A. aculeatus.
Data access
Genome assembly and annotations are available at at the JGI fungal genome portal MycoCosm [15] and have been deposited at DDBJ/EMBL/GenBank under the following accessions: A. aculeatus ATCC16872 - MRCK00000000, A. brasiliensis CBS101740 - LJXV00000000, A. carbonarius ITEM5010 - AHIG00000000, A. foetidus CBS106.47 - MRBP00000000, A. glaucus CBS516.65 - LSTL00000000, A. sydowii CBS593.65 - MRCH00000000, A. tubingensis CBS134.48 - LJXU00000000, A. versicolor CBS583.65 - MRBN00000000, A. wentii CBS141173 (DTO134E9) - LJSE00000000, and A. zonatus CBS506.65 - MRBM00000000. Links to the raw genome and transcriptome data of these species are available in Additional file 1D. This also lists the genbank (if available) and JGI accession numbers of the genomes used for comparison as well as the papers in which they have been published.
Proteomics data of the wheat bran and sugar beet cultures are available via ProteomeXchange with identifier PXD005563. Proteomics data for the stress experiments are available at http://wwwuser.gwdg.de/~hkusch/GBIO_DeVries/.
Expression of mating-type and pheromone-signaling pathway genes
Representative asexual species (A. luchuensis (A. acidus, A. foetidus), A. aculeatus, A. brasilliensis, A. carbonarius, A. clavatus, A. niger, A. sydowii, A. versicolor, A. wentii, A. zonatus), together with some control sexual species (A. flavus, A. fumigatus, A. tubingensis), were selected for experimental work to assess possible expression of mating-type (MAT) and pheromone-signaling pathway (ppgA, preA, and preB) genes. Identified MAT1-1, MAT1-2, ppgA, preA, and preB sequences were used to design primers for (RT)-PCR investigations (primer sequences available on request). Primers were designed ideally either side of an intron to allow differentiation of genomic or cDNA products by resulting product sizes. Genomic DNA was extracted from all species using a Nucleospin® Plant II kit (Macherey-Nagel). Cultures were grown in Aspergillus complete media (ACM) broth [46] at 28 °C with shaking for two days. Cultures were harvested by filtration through a sterile miracloth filter (Millipore) and washed with a pH 7.5 phosphate buffer solution, then freeze-dried and ground under liquid nitrogen into powder by motor and pestle. After quantification, 50 ng was used as a DNA template for subsequent PCR. To evaluate possible transcription of genes, RNA was extracted from cultures of all test species grown on solid media under conditions known to be favourable for sex in the aspergilli. Nylon filter membranes (pore size 0.2 μm) were placed over the surface of 5 cm Petri dishes containing 10 mL of oat meal agar (N.B. This media was used for all species except for A. versicolor, which did not grow well on this substrate. ACM agar was instead used for A. versicolor). A total of 50 μL of a spore suspension (containing 1 × 105 conidia) was spread over the surface and cultures incubated in the dark at 32 °C. Plates were then sealed with two layer of Parafilm after 15–18 h of incubation [46]. Mycelia were then scraped off the surface of the filters after either four or eight days, dried using sterile paper tissue, and then flash frozen under liquid nitrogen. For most species, RNA extracted was then extracted using a TRizol method according to manufacturer’s instructions (Thermo Fisher). However, a CTAB method (Heather Darbyshir pers comm) was used for RNA extraction from members of the black aspergilli taxonomic grouping. For both methods, a Nucleospin® RNA extraction kit (Macherey-Nagel) was used for clean-up of the extracted RNA. After quantification, the extracted RNA was checked on agarose gel for purity and integrity of RNA. CDNA synthesis from all extracted RNA for all of species was achieved using SuperscriptTM III; 500 ng of total RNA was used for cDNA synthesis, and then 1 μL of cDNA was used for subsequent amplifications. Final PCR was performed using a Phusion High Fidelity PCR kit according to manufacturer’s instructions (New England Biolabs). One microliter of each genomic DNA and cDNA from four-day-old and eight-day-old cultures were used as a template for amplification of MAT1-1, MAT1-2, ppgA, preA, and preB genes. In addition, expression of the actA (actin) gene was monitored as a control “housekeeping” gene. PCR products were resolved on 2.0–2.5% agarose gels. Successful amplification of all mating-type and pheromone-signaling pathway genes was achieved for for all species except for the MAT1-2 gene of A. versicolor, which could not be amplified despite use of multiple primer sets and this gene was therefore excluded from the analysis.
Growth profiling on different nutrient sources
For growth profiling, all strains were grown on MM [295] containing monosaccharides/oligosaccharides, polysaccharides, and crude substrates at 25 mM, 1%, and 3% final concentration, respectively. Cultures for proteomics were performed in MM with 1% wheat bran or 1% sugar beet pulp, as previously described [57], and proteomics was performed as described in that paper. For laccase and cellobiose dehydrogenase activity, the 20 aspergilli were grown in duplicate in liquid shaken cultures at 30 °C, 120 rpm. A total of 250 mL baffled erlenmeyers filled with 50 mL of MM [295] and supplemented with 10 g of cotton seed hulls were inoculated with 2 × 106 spores/mL final concentration.
Stress tolerance studies
In the stress tolerance studies, 18 Aspergillus strains with fully sequenced fungal genomes representing 17 species were included (Additional files 3 and 4). For the production of conidia, normally the malt extract—mycological pepton sporulation agar medium (1.5% agar)—recommended by CBS-KNAW (http://www.fung-stress.org/) was used. The sporulation medium for A. glaucus was also supplemented with 1.0 M NaCl. Unless otherwise indicated, asexual sporulation was carried out at 25 °C in the dark for six days. A. fumigatus and A. nidulans were also sporulated on the CBS-KNAW medium at 37 °C and on nitrate minimal medium agar plates (NMM, solidified with 1.8% agar) [296] at 37 °C. Microbial growth and stress sensitivity assays were performed on NMM agar plates (1.8% VWR International Agar powder for bacteriology, ID number 20 767.298), which were incubated at 25 (5 or 10 d) or 37 °C (5 d) in the dark and were supplemented with the stress-initiating agents H2O2 (oxidative stress), menadione sodium bisulfite (MSB, oxidative stress), NaCl (ionic osmotic stress), sorbitol (non-ionic osmotic stress), Congo Red (cell wall integrity stress), or CdCl2 (heavy metal stress) as required (Additional files 3 and 4; [133]). We typically performed two to three sets of experiments (often more) with spores harvested from independent sporulations for each stress condition. Each set contained 4-4 replicates. We took photos of the most representative colonies.
Stress agar plates were inoculated and, after completing the physiological experiments, the fungal colonies were photographed as described before [297]. Fungal growth was characterized either by colony diameters or by scoring them by eye on a 0–10 scale (Additional files 3 and 4). Stress sensitivities were quantified by percentage decreases in the colony diameters in comparison to controls or by decreases in the growth scores. Based on the growth inhibition values, all species tested were ranked according to their stress tolerances (Additional file 4). Physiological data have been stored in the Fungal Stress Database (FSD; http://www.fung-stress.org/) at CBS-KNAW. Stress response proteins were identified and annotated in the newly sequenced Aspergillus genomes following the procedure described previously [298] and newly identified orthologs were inserted into the Fungal Stress Response Database (FSRD) - version 2 [298] (FSRDv_2; http://internal.med.unideb.hu/fsrd2/?p=consortium). The phylogeny of selected well-characterized stress response proteins and their orthologs was studied following the procedure described in Additional file 3.
Enzyme assays
For analysis of oxidative enzymes, the 20 aspergilli were grown in duplicate in liquid shaken cultures at 30 °C, 120 rpm. A total of 250 mL baffled erlenmeyers filled with 50 mL of medium were inoculated with to a final concentration of 2 × 106 spores/mL. MM (for 1000 L: 6 g NaNO3, 1.5 g KH2PO4, 0.5 g KCl, 0.5 g MgSO4.7H2O, trace elements, CuSO4 0.02 g, pH 6.0) was supplemented with 10 g of cotton seed hulls as the carbon source. Laccase activity in the culture supernatant was assayed by monitoring the oxidation of 500 μM ABTS at 420 nm to the respective radical (ε420 = 36 mM-1 cm-1) [299], in the presence of 50 mM sodium tartrate pH 4.0 at 30 °C.
The CDH activity was determined at 30 °C, using 0.2 mM 2,6-dichlorophenol indophenol (DCPIP) in 50 mM sodium acetate buffer, pH 5, in the presence of cellobiose 10 mM as the substrate [300]. The decrease in absorption of DCPIP (ε = 68,000 M-1 cm-1) was monitored at 520 nm for 30 s.
Functionality of P450 proteins and NO sensitivity
Functionality of P450 proteins was tested by growing mycelia of aspergilli on MBFA agar supplemented with 0.02 mM ketoconazole or 2 mM benzoic acid. Inhibition of growth (Inhib.) as a percentage of the control without supplementation (Contr.) was determined. For nitric oxide sensitivity, all 19 species were cultivated in liquid MM containing 1% glucose and 10 mM ammonium tartrate in microtiter plates. Only medium used for A. niger CBS 513.88 was supplemented with a vitamin solution (80 mg/L p-aminobenzoic acid, 50 mg/L thiamin HCl, 1 mg/L biotin, 400 mg/L inositol, 100 mg/L nicotinic acid, 200 mg/L calcium pantothenate, 100 mg/L riboflavin, 50 mg/L pyridoxine). The liquid medium was supplemented with different concentrations of sodium nitroprusside (SNP): 0 (control samples), 16, 64, or 128 mM. A total of 1 × 104 spores of each organism in a final volume of 150 μL per well was used as a replicate. Four replicates were done per organism per treatment. All fungi were incubated at 30 °C except for A. glaucus, which was grown at 22 °C. Absorbance was recorded at 492 nm every 24 h in an Axis expert plus plate reader. Percentage of growth inhibition was calculated comparing absorbance values at 0 versus 128 mM SNP. Growing time for calculations was in the range of 48–96 h depending on each organism (see Fig. 11 legend). Final values shown are the average of two independent experiments with four replicates per experiment.
Functional analysis of sugar transporters
cDNA clones of the transporter genes were amplified by PCR and cloned into pYEX-BX using DNA ligase according to the manufacturer’s instructions. Sequence analysis was used to verify the correct clones. S. cerevisiae tester strains used were: KY73 [301], EB.VW4000 [302], MaDH4 [303], SEY6210 [304], CMY1050 [305]. Plasmids were derived from pYEX-BX (Clontech) [306]. Plasmid transformations of yeast cells were carried out according to the quick and easy TRAFO protocol [307]. Ten milliliters of YNB + 2% maltose supplemented as described above, was inoculated with a single yeast colony and grown overnight (16 h) at 30 °C and 250 rpm in an orbital shaker. OD500 measurements were taken for all cultures. The cells were centrifuged at 10,000 rpm for 10 s and, after the supernatant was removed, resuspended in MilliQ water. Cells were washed twice in this manner. A final centrifugation step (10,000 rpm for 10 s) was followed by resuspension in sufficient MilliQ water to result in a cell suspension with an OD of 0.1 (approx. 104 cells/mL). NUNC 96-well optical bottom plates polymer base microtitre plates, containing media as described above, were inoculated with approximately 102 cells to a final volume of 100 uL in a sterile manner. Plates were sealed using breathable seals and incubated at 28 °C. Growth was monitored by OD590 measurements taken at 0, 24, 48, 72, 96, and 120 h time points.
Acknowledgements
Members of the University of Debrecen were supported financially by the Hungarian Scientific Research Fund (grant K100464 to IP) as well as by the TÁMOP 4.2.1./B-09/1/KONV-2010-0007 and SROP-4.2.2.B-15/1/KONV-2015-0001 projects, which were supported by the European Union, co-financed by the European Social Fund. EO was a recipient of a Campus Hungary Programme scholarship to the CBS-KNAW Fungal Biodiversity Centre. The authors aknowledge support from the Federal Ministery of Education and Research (BMBF) BioFung and the Deutsche Forschungsgemeinschaft (DFG) to GB, ES, and AAB. The work conducted by the U.S. Department of Energy Joint Genome Institute is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. FC was supported by the Major Program of National Natural Science Foundation of China (No. 31330059). WC was supported by the National Natural Science Foundation of China (No. 31601446) and the Fundamental Research Funds for the Central Universities (Nos. 2662014BQ051 and 2662015QC003). TB and IB were supported by grants of the Dutch Technology Foundation STW, Applied Science division of NWO, and the Technology Program of the Ministry of Economic Affairs UGC 14270 to RPdV. PAvK, MaA, and DvRU were supported by the Netherlands Technology Foundation STW (LGC 7393). We acknowledge financial support from the FWF (M01693-B22) to DCa.
This work was in part supported by the Intelligent Synthetic Biology Center of Global Frontier Project (2011-0031955) funded by the Ministry of Education, Science and Technology grants to JHY and Science Foundation Ireland 13/CDA/2142 to OB. NS was supported by a scholarship from the Kurdistan Regional Government of Iraq. HSP was supported by the National Research Foundation of Korea (NRF) grant founded by the Korean Government (no. 2016010945).
The work at UW-Madison (JHY) was supported by the Intelligent Synthetic Biology Center of Global Frontier Project (2011-0031955) funded by the Ministry of Science, ICT and Future Planning.
PSD, RH, and RH were supported by the British Mycological Society and the Biotechnology and Biological Sciences Research Council (UK). ES and AAB were supported by CRC 1127 ChemBioSys and CRC-Transregio FungiNet by Deutsche Forschungsgemeinschaft (DFG). JCF was supported by Agilent Technologies (Agilent Thought Leader Award #2871) and the Novo Nordisk Foundation (Grant NNF 13OC0005201). LK was supported by the Hungarian Scientific Research Fund (NN116519) and a Bólyai János Research Fellowship. The sugar transporter analysis was supported by grants AGL2011-29925 and AGL2015-66131- AGL2015-66131-C2-2-R (MINECO/FEDER). BRO was supported by the Irving S. Johnson fund of the Kansas University endowment. We are grateful to FAPESP (The State of São Paulo Research Foundation) for the financial support (2014/06923-6 to FMS; 2012/20549-4 to ARLD; 2011/08945-9 and 2014/11766-7 to JVCO; 2012/19040-0 and 2014/10351-8 to CAU). We are grateful to the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) and Conselho Nacional de Desenvolvimento Cientifico e Tecnologico (CNPq) for the financial support (310186/2014-5 and 442333/2014-5 to FMS; 441912/2014-1 to ARLD). We acknowledge the financial support of the Dirección General de Apoyo al Personal Académico, UNAM, projects IN 225710 and IN219813, and The Slovenian Research Agency (P1-0207, P1-0391, and J4-7162). KS was supported by the Slovenian Research Agency (P1-0207) and KS and NK were supported by the Slovenian Research Agency (J4-7162). TCV and MRA acknowledge support from the Villium Foundation, Project VKR023437. GB was supported by a grant from the Deutsche Forschungsgemeinschaft (BR1502/11-2). GB and RF were supported by a grant from the Deutsche Forschungsgemeinschaft (FOR1334/2). AL was supported by St. Petersburg State University, St. Petersburg, Russia (grant no. 15.61.951.2015).
The Authors thank A Gazdag, R Mohácsi, and LG Tóth (University of Debrecen) for contributing to the experimental work; and V Szabó and T Koszó (University of Debrecen) for the literature search during the construction of FSD and FSRD, respectively. The authors thank C Toft for her help in conducting the phylogenetic analysis of the sugar transporters. The authors thank DG Panaccione for providing the A. fumigatus easC mutant strain prior to publication and DR Nelson for naming CYP51 and CYP53 family representatives according to CYP nomenclature. The authors thank Arle Kruckeberg (yeast strain KY73; plasmid pYEX-BX), Barbara Bakker (yeast strain EB.VW4000), Remi Lemoine (yeast strain MaDH4), Jeremy Thorner (yeast strain SEY6210), and Ian Dawes (CMY1050) for kindly providing material used in this study.
Availability of data and materials
The datasets supporting the conclusions of this article are included within the article and its Additional File(s) or available through DDBJ/EMBL/GenBank under the following accessions: A. aculeatus ATCC16872 - MRCK00000000, A. brasiliensis CBS101740 - LJXV00000000, A. carbonarius ITEM5010 - AHIG00000000, A. foetidus CBS106.47 - MRBP00000000, A. glaucus CBS516.65 - LSTL00000000, A. sydowii CBS593.65 - MRCH00000000, A. tubingensis CBS134.48 - LJXU00000000, A. versicolor CBS583.65 - MRBN00000000, A. wentii CBS141173 (DTO134E9) - LJSE00000000, and A. zonatus CBS506.65 - MRBM00000000. Links to the raw genome and transcriptome data of these species are available in Additional file 1D. This also lists the genbank (if available) and JGI accession numbers of the genomes used for comparison as well as the papers in which they have been published. Proteomics data of the wheat bran and sugar beet cultures are available via ProteomeXchange with identifier PXD005563. Proteomics data for the stress experiments are available at http://wwwuser.gwdg.de/~hkusch/GBIO_DeVries/.
Authors’ contributions
Genome sequencing and general analysis: CiC and CYN performed sequencing. Ala, EL, and Ali perfomed transcriptome analysis. HS, KLB, ALa, and AC performed genome assembly. AK, RR, ASa, and IVG performed genome annotation. KB and IVG coordinated the genome sequencing. GCC, GS, and JW facilitated hosting of the genomes in AspGD. JH, RR, and RAS performed phylogenomic analysis and wrote this part of the manuscript. RBT and DA took part in merging and critically reviewing the manuscript. AFL, AG, ASu, GM, and GP collaborated the genome sequencing and preliminary annotation data. Asexual and sexual reproduction: PSD coordinated the section. WC, FC, HSP, and JHY performed bioinformatic analysis. NSS, RHa, RHo, and PSD performed bioinformatic analysis and experimental work. PSD and JHY wrote this section of the manuscript. Primary carbon metabolism: MRA and BS coordinated the section, analyzed data, and wrote this section of the manuscript. AH, KMO, and PP performed experiments and analyzed the acid production data. LK, EF, MoA, ÁPM, and ES planned and carried out experiments and evaluated data related to alternative oxidases. PMe analyzed genes related to trehalose synthesis. Plant biomass degradation: IB and RPdV coordinated this section. BH and MH performed the CAZy analysis of the genomes. ALe, ER, FP, and AT performed bioinformatic analysis and experimental work. AW, GAO, KSH, and MRM performed experimental work. MZ performed the orthology analysis. RPdV wrote this section of the manuscript. Transporters: PAvK, MaA, TS, DU, AR, and JV planned and carried out experiments and evaluated data relating to the functional characterization of the 43 A. niger MFS encoding genes. SU and EK performed bioinformatics analysis and wrote the section on nitrate, nitrite, and ammonium transporters. SA, CG, CS, VS, and MF did the bio-informatics, (manual) gene modeling and phylogenetic analyses for the amino acid transporters section, which was written by CS, CG, and SA. GD and CS performed the bioinformatics and phylogenetic analyses for the nucleobase transporters and wrote this section. AR and MS performed the analysis on the urea transporters and wrote this section. MF checked intron-exon organisation of the amino acid, nucleobase and urea transporters. Secondary metabolism: AAB, BO, JCF, and ES coordinated the section. ESh designed and performed bioinformatics analyses. ESh and JCF compared and analyzed bioinformatic and experimental evidence. NK, KS, and GA performed bioinformatic and experimental work for cytochromes P450 and aegerolysin-like proteins. AG, ASu, and GP contribute to mycotoxins biosynthetic cluster annotation in black aspergilli. All authors contributed to discussions and wrote this section of the manuscript. Stress response: EO, VR, NvdW, MZ, ZK, and MM constructed the FSDatabase and FSRDatabase. HK, SABS, SF, ÖB, GHB, AFJR, ELL, JPO, VM, NM, MM, DCa, SR, JR, RF, GB, EO, MM, TE, RPdV, and IP performed genome comparison, phylogeny, evolutionary analysis, design of experiments, and interpretation of data. HK, SF, EL, JPO, NM, SR, JR, and EO carried out experiments. HK, SABS, SF, and GHB performed proteome analysis. HK, SABS, DC, VM, GHB, IP, and MM wrote this section of the manuscript. Signal transduction: CAU, CaC, RACdS, ARLD, IM, DMRP, Rru, FMS, JVCO, and GHG analyzed the signal transduction pathways. This section was written by GHG. DNA binding proteins: CS, MF, and LH did the bio-informatics, (manual) gene modeling and phylogenetic analyses of the DNA binding proteins, and wrote this section. Project leaders: RPdV, PSD, PAvK, JW, SB, and IVG. RPdV coordinated the project. RPdV and PSD merged the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare that they have no competing interests.
Ethics approval and consent to participate
No ethical approval was needed for this study.
Abbreviations
- AA
Auxiliary activity
- AAAP
Amino acid/auxid permease
- AspGD
Aspergillus Genome Database
- CAZy
Carbohydrate Active enzyme database
- CE
Carbohydrate esterase
- CYP
Cytochrome P450
- GH
Glycoside hydrolase
- GPCR
G-protein-coupled receptor
- H1
Histone 1
- H2
Histone 2
- H3
Histone 3
- H4
Histone 4
- JGI
Joint Genome Institute
- MFS
Major Facilitator Superfamily
- NO
Nitric oxide
- PL
Polysaccharide lyase
- SM
Secondary metabolite
- SSS
Solute symporter family
- TMD
Transmembrane domain
Additional files
Genome statistics and information. (XLSX 97 kb)
Summary of putative genes related to asexual sporulation in tested strains. (XLSX 20 kb)
Stress response in Aspergilli. (XLS 619 kb)
Summary of putative genes related to mating and pheromone signalling in tested strains. (XLSX 12 kb)
Expression of mating and pheromone-signalling pathway genes in representative asexual aspergilli. (PDF 488 kb)
Overview of number of putative orthologs/paralogs in the compared fungal species related to primary carbon metabolism. (PDF 237 kb)
Comparative growth profiling of the aspergilli and selected other fungi. (PDF 8.65 kb)
Effect of SHAM and KCN on fungal growth and sporulation. (PDF 252 kb)
Organic acid production. (XLSX 40 kb)
CAZy comparison of the species. (XLSX 30 kb)
Orthology analysis of CAZy genes related to plant biomass degradation. (XLSX 148 kb)
Laccase and cellobiose dehydrogenase (CDH) activities of the selected aspergilli. (XLSX 11 kb)
Molecular phylogenetic tree of CYP56 family (A), and CPR2 and CYP630 family (B). (PDF 402 kb)
Molecular phylogenetic tree of CYP6001 - CYP6003 families (PpoA - PpoD) and colinearity of the genetic surroundings. (PDF 279 kb)
Extended analysis of stress tolerance of the aspergilli. (PDF 706 kb)
Orthologs of selected stress response proteins. (XLS 76 kb)
Extended analysis of stress tolerance of the aspergilli (2). (PDF 1842 kb)
Identified secreted proteins (A. sydowii, A. tubingensis, A. wentii, A. zonatus). (XLSX 34 kb)
Strain specific PFAM domains predicted for identified secreted proteins (A. sydowii, A. tubingensis, A. wentii, A. zonatus). (XLSX 12 kb)
Distribution of putative sugar transporters. (XLSX 17 kb)
Detailed overview of putative sugar transporters in the aspergilli. (XLSX 10790 kb)
Functional analysis of A. niger sugar transporters by complementation of S. cerevisiae transport mutant strains. (XLSX 12 kb)
Maximum likelihood tree of YAT transporters in the aspergilli. (PDF 89 kb)
Phylogenetic relationships of UreA homologs. (PDF 108 kb)
CLUSTAL O(1.2.3) multiple sequence alignment of NNP nitrate transporters. (PDF 149 kb)
Maximum likelihood rooted tree of Fur-like transporters of the aspergilli and some other members of the Eurotiales (Penicillium, Talaromyces). (PDF 537 kb)
Maximum likelihood rooted phylogeny of the FCY-like transporters of the aspergilli and some penicillia. (PDF 324 kb)
Number of flavohemoglobins and protein IDs found in the genome sequences of the Aspergillus species. (PDF 79 kb)
Species-specific signal transduction pathway genes in Eurotiomycetes. (XLSX 14 kb)
Predicted G-proteins and GPCRs proteins in the Eurotiomycetes. (XLSX 16 kb)
Classification and Clusters of signal transduction related genes. (XLSX 1577 kb)
Distribution of SRPKs in Eurotiomycetes. (XLSX 11 kb)
Kinases presented in at least one yeast species and with no orthologous genes in other eurotiomycetes. (PDF 117 kb)
Histones of the Aspergilli. (PDF 526 kb)
Maximal likelihood phylogeny of MASC1/RID1 orthologs. (PDF 115 kb)
Introduction of forbidden domains for the HMM analysis of signal transduction genes. (TXT 806 bytes)
HMM file used for the identification of protein classes of interest related to signal transduction. (HMMS 4334 kb)
Unannotated genes related to signal transduction. (XLSX 8 kb)
Phylogeny of the Methallophos domain in A. nidulans. (PDF 96 kb)
Contributor Information
Ronald P. de Vries, Email: r.devries@cbs.knaw.nl
Robert Riley, Email: rwriley@lbl.gov.
Ad Wiebenga, Email: a.wiebenga@cbs.knaw.nl.
Guillermo Aguilar-Osorio, Email: gao@unam.mx.
Sotiris Amillis, Email: samillis@biol.uoa.gr.
Cristiane Akemi Uchima, Email: cris.uchima@gmail.com.
Gregor Anderluh, Email: gregor.anderluh@ki.si.
Mojtaba Asadollahi, Email: assadollahi.m@gmail.com.
Marion Askin, Email: marion.askin@csiro.au.
Kerrie Barry, Email: kwbarry@lbl.gov.
Evy Battaglia, Email: evy.battaglia@cbs.knaw.nl.
Özgür Bayram, Email: ozgur.bayram@nuim.ie.
Tiziano Benocci, Email: t.benocci@cbs.knaw.nl.
Susanna A. Braus-Stromeyer, Email: sbraus@gwdg.de
Camila Caldana, Email: camila.caldana@bioetanol.org.br.
David Cánovas, Email: davidc@us.es.
Gustavo C. Cerqueira, Email: gustavo@broadinstitute.org
Fusheng Chen, Email: chenfs@mail.hzau.edu.cn.
Wanping Chen, Email: chenwanping@mail.hzau.edu.cn.
Cindy Choi, Email: Cindy.choi@roche.com.
Alicia Clum, Email: aclum@lbl.gov.
Renato Augusto Corrêa dos Santos, Email: renatoacsantos@gmail.com.
André Ricardo de Lima Damásio, Email: adamasio@unicamp.br.
George Diallinas, Email: diallina@biol.uoa.gr.
Tamás Emri, Email: emri.tamas@science.unideb.hu.
Erzsébet Fekete, Email: kicsizsoka@yahoo.com.
Michel Flipphi, Email: drir.michelflipphi@gmail.com.
Susanne Freyberg, Email: susi.freyberg@gmail.com.
Antonia Gallo, Email: antonia.gallo@ispa.cnr.it.
Christos Gournas, Email: Christos.Gournas@ulb.ac.be.
Rob Habgood, Email: stxrhh@exmail.nottingham.ac.uk.
Matthieu Hainaut, Email: matthieu.hainaut@afmb.univ-mrs.fr.
María Laura Harispe, Email: mlharispe@gmail.com.
Bernard Henrissat, Email: bernard.henrissat@afmb.univ-mrs.fr.
Kristiina S. Hildén, Email: kristiina.s.hilden@helsinki.fi
Ryan Hope, Email: stxrjh@exmail.nottingham.ac.uk.
Abeer Hossain, Email: abeer.hossain@ddna-biotech.com.
Eugenia Karabika, Email: me01519@cc.uoi.gr.
Levente Karaffa, Email: karaffa.levente@science.unideb.hu.
Zsolt Karányi, Email: karanyi.zsolt@med.unideb.hu.
Nada Kraševec, Email: nada.krasevec@ki.si.
Alan Kuo, Email: akuo@lbl.gov.
Harald Kusch, Email: harald.kusch@med.uni-goettingen.de.
Kurt LaButti, Email: klabutti@lbl.gov.
Ellen L. Lagendijk, Email: Elagendijk@koppert.nl
Alla Lapidus, Email: piterlabs@gmail.com.
Anthony Levasseur, Email: anthony.levasseur@univ-amu.fr.
Erika Lindquist, Email: EALindquist@lbl.gov.
Anna Lipzen, Email: alipzen@lbl.gov.
Antonio F. Logrieco, Email: antonio.logrieco@ispa.cnr.it
Andrew MacCabe, Email: andrew@iata.csic.es.
Miia R. Mäkelä, Email: miia.r.makela@helsinki.fi
Iran Malavazi, Email: imalavazi@ufscar.br.
Petter Melin, Email: petter.melin@icloud.com.
Vera Meyer, Email: vera.meyer@tu-berlin.de.
Natalia Mielnichuk, Email: natmiel@yahoo.com.
Márton Miskei, Email: mm@internal.med.unideb.hu.
Ákos P. Molnár, Email: molnar.akos@science.unideb.hu
Giuseppina Mulé, Email: giuseppina.mule@ispa.cnr.it.
Chew Yee Ngan, Email: Chewyee.Ngan@jax.org.
Margarita Orejas, Email: morejas@iata.csic.es.
Erzsébet Orosz, Email: orosz.zsoka@gmail.com.
Jean Paul Ouedraogo, Email: jpaul_oued@yahoo.fr.
Karin M. Overkamp, Email: Karin.overkamp@ddna-biotech.com
Hee-Soo Park, Email: phsoo97@gmail.com.
Giancarlo Perrone, Email: giancarlo.perrone@ispa.cnr.it.
Francois Piumi, Email: francois.piumi@jouy.inra.fr.
Peter J. Punt, Email: peter.punt@ddna-biotech.com
Arthur F. J. Ram, Email: A.F.J.Ram@biology.leidenuniv.nl
Ana Ramón, Email: anaramon@fcien.edu.uy.
Stefan Rauscher, Email: Stefan.Rauscher@gmx.de.
Eric Record, Email: eric.record@univ-amu.fr.
Diego Mauricio Riaño-Pachón, Email: diego.riano@bioetanol.org.br.
Vincent Robert, Email: v.robert@cbs.knaw.nl.
Julian Röhrig, Email: julian.roehrig@novartis.com.
Roberto Ruller, Email: roberto.ruller@bioetanol.org.br.
Asaf Salamov, Email: aasalamov@lbl.gov.
Nadhira S. Salih, Email: sbxns1@nottingham.ac.uk
Rob A. Samson, Email: r.samson@cbs.knaw.nl
Erzsébet Sándor, Email: karaffa@agr.unideb.hu.
Manuel Sanguinetti, Email: msanguinetti@fcien.edu.uy.
Tabea Schütze, Email: tabea.schuetze@tu-berlin.de.
Kristina Sepčić, Email: Kristina.Sepcic@bf.uni-lj.si.
Ekaterina Shelest, Email: ekaterina.shelest@leibniz-hki.de.
Gavin Sherlock, Email: gsherloc@stanford.edu.
Vicky Sophianopoulou, Email: vicky@bio.demokritos.gr.
Fabio M. Squina, Email: fabio.squina@bioetanol.org.br
Antonia Susca, Email: antonella.susca@ispa.cnr.it.
Richard B. Todd, Email: rbtodd@ksu.edu
Adrian Tsang, Email: adrian.tsang@concordia.ca.
Shiela E. Unkles, Email: su@st-andrews.ac.uk
Nathalie van de Wiele, Email: n.vdwiele@bio-aware.com.
Diana van Rossen-Uffink, Email: Diana.vanRossen@baseclear.nl.
Juliana Velasco de Castro Oliveira, Email: juliana.velasco@bioetanol.org.br.
Tammi C. Vesth, Email: tcve@bio.dtu.dk
Jaap Visser, Email: j.visser@cbs.knaw.nl.
Jae-Hyuk Yu, Email: jyu1@wisc.edu.
Miaomiao Zhou, Email: miaomiaozhou88@hotmail.com.
Mikael R. Andersen, Email: mr@bio.dtu.dk
David B. Archer, Email: david.archer@nottingham.ac.uk
Scott E. Baker, Email: scott.baker@pnnl.gov
Isabelle Benoit, Email: i.benoit@cbs.knaw.nl.
Axel A. Brakhage, Email: axel.brakhage@leibniz-hki.de
Gerhard H. Braus, Email: gbraus@gwdg.de
Reinhard Fischer, Email: reinhard.fischer@kit.edu.
Jens C. Frisvad, Email: jcf@bio.dtu.dk
Gustavo H. Goldman, Email: ggoldman@usp.br
Jos Houbraken, Email: j.houbraken@cbs.knaw.nl.
Berl Oakley, Email: boakley@ku.edu.
István Pócsi, Email: pocsi.istvan@science.unideb.hu.
Claudio Scazzocchio, Email: c.scazzocchio@imperial.ac.uk.
Bernhard Seiboth, Email: bernhard.seiboth@tuwien.ac.at.
Patricia A. vanKuyk, Email: pavankuyk@yahoo.com.au
Jennifer Wortman, Email: jwortman@serestherapeutics.com.
Paul S. Dyer, Email: paul.dyer@nottingham.ac.uk
Igor V. Grigoriev, Email: ivgrigoriev@lbl.gov
References
- 1.Samson RA, Visagie CM, Houbraken J, Hong SB, Hubka V, Klaassen CH, et al. Phylogeny, identification and nomenclature of the genus Aspergillus. Stud Mycol. 2014;78:141–73. doi: 10.1016/j.simyco.2014.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scazzocchio C. Aspergillus, a multifacted genus In: Schaechter M, editor. Encyclopaedia of Microbiology. Amsterdam: Elsevier; 2009. p. 401-420.
- 3.Geiser DM, Samson RA, Varga J, Rokas A, Witiak SM. A review of molecular phylogenetics in Aspergillus, and prospects for a robust genus-wide phylogeny. In: Varga J, Samson RA, editors. Aspergillus in the genomic era. Wageningen: Wageningen Academic Publishers; 2008. pp. 17–32. [Google Scholar]
- 4.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–15. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 5.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–61. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 6.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–6. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 7.Andersen MR, Salazar MP, Schaap PJ, van de Vondervoort PJ, Culley D, Thykaer J, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;21:885–97. doi: 10.1101/gr.112169.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Payne GA, Nierman WC, Wortman JR, Pritchard BL, Brown D, Dean RA, et al. Whole genome comparison of Aspergillus flavus and Aspergillus oryzae. Med Mycol. 2006;44(S1):9–11. doi: 10.1080/13693780600835716. [DOI] [PubMed] [Google Scholar]
- 10.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat Biotechnol. 2007;25:221–31. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 11.Benoit I, Malavazi I, Goldman G, Baker SE, de Vries RP. Aspergillus - Genomics of a cosmopolitan fungus. In: Horwitz BA, Mukherjee PK, Mukherjee M, Kubicek CP, editors. Genomics of soil- and plant-associated fungi. Heidelberg: Springer; 2013. pp. 89–126. [Google Scholar]
- 12.Rokas A, Payne G, Fedorova ND, Baker SE, Machida M, Yu J, et al. What can comparative genomics tell us about species concepts in the genus Aspergillus? Stud Mycol. 2007;59:11–7. doi: 10.3114/sim.2007.59.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mabey Gilsenan J, Cooley J, Bowyer P. CADRE: the Central Aspergillus Data REpository 2012. Nucleic Acids Res. 2012;40:D660–6. doi: 10.1093/nar/gkr971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arnaud MB, Cerqueira GC, Inglis DO, Skrzypek MS, Binkley J, Chibucos MC, et al. The Aspergillus Genome Database (AspGD): recent developments in comprehensive multispecies curation, comparative genomics and community resources. Nucleic Acids Res. 2012;40:D653–9. doi: 10.1093/nar/gkr875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42:D699–704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, et al. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res. 2012;40:D26–32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alker AP, Smith GW, Kim K. Characterization of Aspergillus sydowii (Thom et Church), a fungal pathogen of Caribbean sea fan corals. Hydrobiol. 2001;460:105–11. doi: 10.1023/A:1013145524136. [DOI] [Google Scholar]
- 18.van den Berg MA, Albang R, Albermann K, Badger JH, Daran J-M, Driessen AJM, et al. Genome sequencing and analysis of the filamentous fungus Penicillium chrysogenum. Nat Biotech. 2008;26:1161–8. doi: 10.1038/nbt.1498. [DOI] [PubMed] [Google Scholar]
- 19.Marcet-Houben M, Ballester AR, de la Fuente B, Harries E, Marcos JF, Gonzalez-Candelas L, et al. Genome sequence of the necrotrophic fungus Penicillium digitatum, the main postharvest pathogen of citrus. BMC Genomics. 2012;13:646. doi: 10.1186/1471-2164-13-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharpton TJ, Stajich JE, Rounsley SD, Gardner MJ, Wortman JR, Jordar VS, et al. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 2009;19:1722–31. doi: 10.1101/gr.087551.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martinez DA, Oliver BG, Graser Y, Goldberg JM, Li W, Martinez-Rossi NM, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. mBio. 2012;3:e00259–12. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desjardins CA, Champion MD, Holder JW, Muszewska A, Goldberg J, Bailao AM, et al. Comparative genomic analysis of human fungal pathogens causing paracoccidioidomycosis. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26:553–60. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 24.Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422:859–68. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 25.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, et al. Life with 6000 genes. Science. 1996;274:546–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 26.Houbraken J, Samson RA. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol. 2011;70:1–51. doi: 10.3114/sim.2011.70.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pitt JI, Taylor JW. Aspergillus, its sexual states and the new International Code of Nomenclature. Mycologia. 2014;106:1051–62. doi: 10.3852/14-060. [DOI] [PubMed] [Google Scholar]
- 28.Houbraken J, de Vries RP, Samson RA. Modern taxonomy of biotechnologically important Aspergillus and Penicillium species. Adv Appl Microbiol. 2014;86:199–249. doi: 10.1016/B978-0-12-800262-9.00004-4. [DOI] [PubMed] [Google Scholar]
- 29.Perrone G, Susca A, Cozzi G, Ehrlich K, Varga J, Frisvad JC, et al. Biodiversity of Aspergillus species in some important agricultural products. Stud Mycol. 2007;59:53–66. doi: 10.3114/sim.2007.59.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Etxebeste O, Garzia A, Espeso EA, Ugalde U. Aspergillus nidulans asexual development: making the most of cellular modules. Trends Microbiol. 2010;18:569–76. doi: 10.1016/j.tim.2010.09.007. [DOI] [PubMed] [Google Scholar]
- 31.Krijgsheld P, Bleichrodt R, van Veluw GJ, Wang F, Muller WH, Dijksterhuis J, et al. Development in Aspergillus. Stud Mycol. 2013;74:1–29. doi: 10.3114/sim0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dyer PS, O’Gorman CM. A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr Opin Microbiol. 2011;14:649–54. doi: 10.1016/j.mib.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 33.Dyer PS, O’Gorman CM. Sexual development and cryptic sexuality in fungi: insights from Aspergillus species. FEMS Microbiol Rev. 2012;36:165–92. doi: 10.1111/j.1574-6976.2011.00308.x. [DOI] [PubMed] [Google Scholar]
- 34.Scazzocchio C. Aspergillus genomes: secret sex and the secrets of sex. Trends Genet. 2006;22:521–5. doi: 10.1016/j.tig.2006.08.004. [DOI] [PubMed] [Google Scholar]
- 35.Park HS, Yu JH. Genetic control of asexual sporulation in filamentous fungi. Curr Opin Microbiol. 2012;15:669–77. doi: 10.1016/j.mib.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 36.Adams TH, Wieser JK, Yu JH. Asexual sporulation in Aspergillus nidulans. Microbiol Mol Biol Rev. 1998;62:35–54. doi: 10.1128/mmbr.62.1.35-54.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee MK, Kwon NJ, Choi JM, Lee IS, Jung S, Yu JH. NsdD is a key repressor of asexual development in Aspergillus nidulans. Genetics. 2014;197:159–73. doi: 10.1534/genetics.114.161430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayram O, Braus GH, Fischer R, Rodriguez-Romero J. Spotlight on Aspergillus nidulans photosensory systems. Fungal Genet Biol. 2010;47:900–8. doi: 10.1016/j.fgb.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 39.Bayram O, Braus GH. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev. 2012;36:1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- 40.Wong HC, Chien CY. Ultrastructural studies of the conidial anamorphs of Monascus. Mycologia. 1986;78:593–9. doi: 10.2307/3807771. [DOI] [Google Scholar]
- 41.Dyer PS, Inderbitzin P, Debuchy R. Mating-type structure, function, regulation and evolution in the Pezizomycotina. In: Wendland J, editor. Growth, differentiation and sexuality. Volume I. The Mycota. 3. Switzerland: Springer International Publishing; 2016. pp. 351–85. [Google Scholar]
- 42.Debuchy R, Berteaux-Lecellier V, Silar P. Mating systems and sexual morphogenesis in ascomycetes. In: Borkovich KA, Ebbole DJ, editors. Cellular and molecular biology of filamentous fungi. Washington, DC: ASM Press; 2010. pp. 501–35. [Google Scholar]
- 43.Böhm J, Hoff B, O’Gorman CM, Wolfers S, Klix V, Binger D, et al. Sexual reproduction and mating-type-mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci U S A. 2013;110:1476–81. doi: 10.1073/pnas.1217943110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yun SH, Berbee ML, Yoder OC, Turgeon BG. Evolution of the fungal self-fertile reproductive life style from self-sterile ancestors. Proc Natl Acad Sci U S A. 1999;96:5592–7. doi: 10.1073/pnas.96.10.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geiser DM, Pitt JI, Taylor JW. Cryptic speciation and recombination in the aflatoxin-producing fungus Aspergillus flavus. Proc Natl Acad Sci U S A. 1998;95:388–93. doi: 10.1073/pnas.95.1.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paoletti M, Seymour FA, Alcocer MJ, Kaur N, Calvo AM, Archer DB, et al. Mating type and the genetic basis of self-fertility in the model fungus Aspergillus nidulans. Curr Biol. 2007;17:1384–9. doi: 10.1016/j.cub.2007.07.012. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez-Prado JH, Moore GG, Horn BW, Carbone I. Characterization and population analysis of the mating-type genes in Aspergillus flavus and Aspergillus parasiticus. Fungal Genet Biol. 2008;45:1292–9. doi: 10.1016/j.fgb.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 48.Rydholm C, Dyer PS, Lutzoni F. DNA sequence characterization and molecular evolution of MAT1 and MAT2 mating-type loci of the self-compatible ascomycete mold Neosartorya fischeri. Eukaryot Cell. 2007;6:868–74. doi: 10.1128/EC.00319-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Flipphi M, Sun J, Robellet X, Karaffa L, Fekete E, Zeng AP, et al. Biodiversity and evolution of primary carbon metabolism in Aspergillus nidulans and other Aspergillus spp. Fungal Genet Biol. 2009;46(Suppl 1):S19–44. doi: 10.1016/j.fgb.2008.07.018. [DOI] [PubMed] [Google Scholar]
- 50.Khosravi C, Benocci T, Battaglia E, Benoit I, de Vries RP. Sugar catabolism in Aspergillus and other fungi related to the utilization of plant biomass. Adv Appl Microbiol. 2014;90:1–28. doi: 10.1016/bs.aambs.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Fekete E, de Vries RP, Seiboth B, vanKuyk PA, Sandor E, Metz B, et al. D-Galactose uptake is nonfunctional in the conidiospores of Aspergillus niger. FEMS Microbiol Lett. 2012;329:198–203. doi: 10.1111/j.1574-6968.2012.02524.x. [DOI] [PubMed] [Google Scholar]
- 52.Hayer K, Stratford M, Archer DB. Structural features of sugars that trigger or support conidial germination in the filamentous fungus Aspergillus niger. Appl Environ Microbiol. 2013;79:6924–31. doi: 10.1128/AEM.02061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hayer K, Stratford M, Archer DB. Germination of Aspergillus niger conidia is triggered by nitrogen compounds related to L-amino acids. Appl Environ Microbiol. 2014;80:6046–53. doi: 10.1128/AEM.01078-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Geber A, Williamson PR, Rex JH, Sweeney EC, Bennett JE. Cloning and characterization of a Candida albicans maltase gene involved in sucrose utilization. J Bacteriol. 1992;174:6992–6. doi: 10.1128/jb.174.21.6992-6996.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Culleton H, McKie V, de Vries RP. Physiological and molecular aspects of degradation of plant polysaccharides by fungi: What have we learned from Aspergillus? Biotechnol J. 2013;8:884–94. doi: 10.1002/biot.201200382. [DOI] [PubMed] [Google Scholar]
- 56.de Vries RP, Visser J. Aspergillus enzymes involved in degradation of plant cell wall polysaccharides. Microb Mol Biol Rev. 2001;65:497–522. doi: 10.1128/MMBR.65.4.497-522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Benoit I, Culleton H, Zhou M, DiFalco M, Aguilar-Osorio G, Battaglia E, et al. Closely related fungi employ diverse enzymatic strategies to degrade plant biomass. Biotechnol Biofuels. 2015;8:107. doi: 10.1186/s13068-015-0285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Coutinho PM, Andersen MR, Kolenova K, vanKuyk PA, Benoit I, Gruben BS, et al. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet Biol. 2009;46(Suppl 1):S161–9. doi: 10.1016/j.fgb.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 59.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mäkelä MR, Jiménez Barboza LA, de Vries RP, Hildén KS. Production of feruloyl esterases by Aspergillus species. In: de Vries RP, Benoit I, Andersen MR, editors. Aspergillus and Penicillium in the post-genomic era. Norfolk: Caister Academic Press; 2016. pp. 129–44. [Google Scholar]
- 61.The fung-growth database. Linking growth to genome. http://www.fung-growth.org/.
- 62.Klaubauf S, Narang HM, Post H, Zhou M, Brunner K, Mach-Aigner AR, et al. Similar is not the same: Differences in the function of the (hemi-)cellulolytic regulator XlnR (Xlr1/Xyr1) in filamentous fungi. Fungal Genet Biol. 2014;72:73–81. doi: 10.1016/j.fgb.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 63.Brakhage AA. Regulation of fungal secondary metabolism. Nat Rev Microbiol. 2013;11:21–32. doi: 10.1038/nrmicro2916. [DOI] [PubMed] [Google Scholar]
- 64.Wiemann P, Keller NP. Strategies for mining fungal natural products. J Ind Microbiol Biotechnol. 2014;41:301–13. doi: 10.1007/s10295-013-1366-3. [DOI] [PubMed] [Google Scholar]
- 65.Chiang YM, Szewczyk E, Davidson AD, Keller N, Oakley BR, Wang CC. A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J Am Chem Soc. 2009;131:2965–70. doi: 10.1021/ja8088185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bergmann S, Schumann J, Scherlach K, Lange C, Brakhage AA, Hertweck C. Genomics-driven discovery of PKS-NRPS hybrid metabolites from Aspergillus nidulans. Nat Chem Biol. 2007;3:213–7. doi: 10.1038/nchembio869. [DOI] [PubMed] [Google Scholar]
- 67.Sanchez JF, Entwistle R, Hung JH, Yaegashi J, Jain S, Chiang YM, et al. Genome-based deletion analysis reveals the prenyl xanthone biosynthesis pathway in Aspergillus nidulans. J Am Chem Soc. 2011;133:4010–7. doi: 10.1021/ja1096682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schätzle MA, Husain SM, Ferlaino S, Muller M. Tautomers of anthrahydroquinones: enzymatic reduction and implications for chrysophanol, monodictyphenone, and related xanthone biosyntheses. J Am Chem Soc. 2012;134:14742–5. doi: 10.1021/ja307151x. [DOI] [PubMed] [Google Scholar]
- 69.Simpson TJ. Genetic and biosynthetic studies of the fungal prenylated xanthone shamixanthone and related metabolites in Aspergillus spp. revisited. Chem Bio Chem. 2012;13:1680–8. doi: 10.1002/cbic.201200014. [DOI] [PubMed] [Google Scholar]
- 70.Bromann K, Toivari M, Viljanen K, Vuoristo A, Ruohonen L, Nakari-Setälä T. Identification and characterization of a novel diterpene gene cluster in Aspergillus nidulans. PLoS One. 2012;7 doi: 10.1371/journal.pone.0035450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bergmann S, Funk AN, Scherlach K, Schroeckh V, Shelest E, Horn U, et al. Activation of a silent fungal polyketide biosynthesis pathway through regulatory cross talk with a cryptic nonribosomal peptide synthetase gene cluster. Appl Environ Microbiol. 2010;76:8143–9. doi: 10.1128/AEM.00683-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeh HH, Ahuja M, Chiang YM, Oakley CE, Moore S, Yoon O, et al. Resistance gene-guided genome mining: Serial promoter exchanges in Aspergillus nidulans reveal the biosynthetic pathway for fellutamide B, a proteasome inhibitor. ACS Chem Biol. 2016;11:2275–84. doi: 10.1021/acschembio.6b00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chiang YM, Szewczyk E, Davidson AD, Entwistle R, Keller NP, Wang CC, et al. Characterization of the Aspergillus nidulans monodictyphenone gene cluster. Appl Environ Microbiol. 2010;76:2067–74. doi: 10.1128/AEM.02187-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schroeckh V, Scherlach K, Nutzmann HW, Shelest E, Schmidt-Heck W, Schuemann J, et al. Intimate bacterial-fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans. Proc Natl Acad Sci U S A. 2009;106:14558–63. doi: 10.1073/pnas.0901870106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacCabe AP, Riach MB, Unkles SE, Kinghorn JR. The Aspergillus nidulans npeA locus consists of three contiguous genes required for penicillin biosynthesis. EMBO J. 1990;9:279–87. doi: 10.1002/j.1460-2075.1990.tb08106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ahuja M, Chiang YM, Chang SL, Praseuth MB, Entwistle R, Sanchez JF, et al. Illuminating the diversity of aromatic polyketide synthases in Aspergillus nidulans. J Am Chem Soc. 2012;134:8212–21. doi: 10.1021/ja3016395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Eisendle M, Oberegger H, Zadra I, Haas H. The siderophore system is essential for viability of Aspergillus nidulans: functional analysis of two genes encoding l-ornithine N 5-monooxygenase (sidA) and a non-ribosomal peptide synthetase (sidC) Mol Microbiol. 2003;49:359–75. doi: 10.1046/j.1365-2958.2003.03586.x. [DOI] [PubMed] [Google Scholar]
- 78.Brown DW, Yu JH, Kelkar HS, Fernandes M, Nesbitt TC, Keller NP, et al. Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc Natl Acad Sci U S A. 1996;93:1418–22. doi: 10.1073/pnas.93.4.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bouhired S, Weber M, Kempf-Sontag A, Keller NP, Hoffmeister D. Accurate prediction of the Aspergillus nidulans terrequinone gene cluster boundaries using the transcriptional regulator LaeA. Fungal Genet Biol. 2007;44:1134–45. doi: 10.1016/j.fgb.2006.12.010. [DOI] [PubMed] [Google Scholar]
- 80.Gerke J, Bayram O, Feussner K, Landesfeind M, Shelest E, Feussner I, et al. Breaking the silence: protein stabilization uncovers silenced biosynthetic gene clusters in the fungus Aspergillus nidulans. Appl Environ Microbiol. 2012;78:8234–44. doi: 10.1128/AEM.01808-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chiang YM, Ahuja M, Oakley CE, Entwistle R, Asokan A, Zutz C, et al. Development of genetic dereplication strains in Aspergillus nidulans results in the discovery of aspercryptin. Angew Chem Int Ed Engl. 2016;55:1662–5. doi: 10.1002/anie.201507097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lim FY, Hou Y, Chen Y, Oh JH, Lee I, Bugni TS, et al. Genome-based cluster deletion reveals an endocrocin biosynthetic pathway in Aspergillus fumigatus. Appl Environ Microbiol. 2012;78:4117–25. doi: 10.1128/AEM.07710-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin HC, Chooi YH, Dhingra S, Xu W, Calvo AM, Tang Y. The fumagillin biosynthetic gene cluster in Aspergillus fumigatus encodes a cryptic terpene cyclase involved in the formation of beta-trans-bergamotene. J Am Chem Soc. 2013;135:4616–9. doi: 10.1021/ja312503y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ames BD, Liu X, Walsh CT. Enzymatic processing of fumiquinazoline F: a tandem oxidative-acylation strategy for the generation of multicyclic scaffolds in fungal indole alkaloid biosynthesis. Biochemistry. 2010;49:8564–76. doi: 10.1021/bi1012029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.O’Hanlon KA, Gallagher L, Schrettl M, Jochl C, Kavanagh K, Larsen TO, et al. Nonribosomal peptide synthetase genes pesL and pes1 are essential for fumigaclavine C production in Aspergillus fumigatus. Appl Environ Microbiol. 2012;78:3166–76. doi: 10.1128/AEM.07249-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Maiya S, Grundmann A, Li SM, Turner G. The fumitremorgin gene cluster of Aspergillus fumigatus: identification of a gene encoding brevianamide F synthetase. ChemBioChem. 2006;7:1062–9. doi: 10.1002/cbic.200600003. [DOI] [PubMed] [Google Scholar]
- 87.Gardiner DM, Howlett BJ. Bioinformatic and expression analysis of the putative gliotoxin biosynthetic gene cluster of Aspergillus fumigatus. FEMS Microbiol Lett. 2005;248:241–8. doi: 10.1016/j.femsle.2005.05.046. [DOI] [PubMed] [Google Scholar]
- 88.Yin WB, Baccile JA, Bok JW, Chen Y, Keller NP, Schroeder FC. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J Am Chem Soc. 2013;135:2064–7. doi: 10.1021/ja311145n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chooi YH, Fang J, Liu H, Filler SG, Wang P, Tang Y. Genome mining of a prenylated and immunosuppressive polyketide from pathogenic fungi. Org Lett. 2013;15:780–3. doi: 10.1021/ol303435y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Hanlon KA, Cairns T, Stack D, Schrettl M, Bignell EM, Kavanagh K, et al. Targeted disruption of nonribosomal peptide synthetase pes3 augments the virulence of Aspergillus fumigatus. Infect Immun. 2011;79:3978–92. doi: 10.1128/IAI.00192-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Igarashi Y, Yabuta Y, Sekine A, Fujii K, Harada K, Oikawa T, et al. Directed biosynthesis of fluorinated pseurotin A, synerazol and gliotoxin. J Antibiot (Tokyo) 2004;57:748–54. doi: 10.7164/antibiotics.57.748. [DOI] [PubMed] [Google Scholar]
- 92.Maiya S, Grundmann A, Li X, Li SM, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. ChemBioChem. 2007;8:1736–43. doi: 10.1002/cbic.200700202. [DOI] [PubMed] [Google Scholar]
- 93.Vödisch M, Scherlach K, Winkler R, Hertweck C, Braun HP, Roth M, et al. Analysis of the Aspergillus fumigatus proteome reveals metabolic changes and the activation of the pseurotin A biosynthesis gene cluster in response to hypoxia. J Proteome Res. 2011;10:2508–24. doi: 10.1021/pr1012812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wiemann P, Guo CJ, Palmer JM, Sekonyela R, Wang CC, Keller NP. Prototype of an intertwined secondary-metabolite supercluster. Proc Natl Acad Sci U S A. 2013;110:17065–70. doi: 10.1073/pnas.1313258110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Robinson SL, Panaccione DG. Chemotypic and genotypic diversity in the ergot alkaloid pathway of Aspergillus fumigatus. Mycologia. 2012;104:804–12. doi: 10.3852/11-310. [DOI] [PubMed] [Google Scholar]
- 96.Frisvad JC, Larsen TO, Thrane U, Meijer M, Varga J, Samson RA, et al. Fumonisin and ochratoxin production in industrial Aspergillus niger strains. PLoS One. 2011;6 doi: 10.1371/journal.pone.0023496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Awakawa T, Yang XL, Wakimoto T, Abe I. Pyranonigrin E: a PKS-NRPS hybrid metabolite from Aspergillus niger identified by genome mining. ChemBioChem. 2013;14:2095–9. doi: 10.1002/cbic.201300430. [DOI] [PubMed] [Google Scholar]
- 98.Nakazawa T, Ishiuchi K, Praseuth A, Noguchi H, Hotta K, Watanabe K. Overexpressing transcriptional regulator in Aspergillus oryzae activates a silent biosynthetic pathway to produce a novel polyketide. ChemBioChem. 2012;13:855–61. doi: 10.1002/cbic.201200107. [DOI] [PubMed] [Google Scholar]
- 99.Imamura K, Tsuyama Y, Hirata T, Shiraishi S, Sakamoto K, Yamada O, et al. Identification of a gene involved in the synthesis of a dipeptidyl peptidase IV inhibitor in Aspergillus oryzae. Appl Environ Microbiol. 2012;78:6996–7002. doi: 10.1128/AEM.01770-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yu J, Chang PK, Ehrlich KC, Cary JW, Bhatnagar D, Cleveland TE, et al. Clustered pathway genes in aflatoxin biosynthesis. Appl Environ Microbiol. 2004;70:1253–62. doi: 10.1128/AEM.70.3.1253-1262.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chang PK, Horn BW, Dorner JW. Clustered genes involved in cyclopiazonic acid production are next to the aflatoxin biosynthesis gene cluster in Aspergillus flavus. Fungal Genet Biol. 2009;46:176–82. doi: 10.1016/j.fgb.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 102.Zhang S, Monahan BJ, Tkacz JS, Scott B. Indole-diterpene gene cluster from Aspergillus flavus. Appl Environ Microbiol. 2004;70:6875–83. doi: 10.1128/AEM.70.11.6875-6883.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nielsen MT, Nielsen JB, Anyaogu DC, Holm DK, Nielsen KF, Larsen TO, et al. Heterologous reconstitution of the intact geodin gene cluster in Aspergillus nidulans through a simple and versatile PCR based approach. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qiao K, Chooi YH, Tang Y. Identification and engineering of the cytochalasin gene cluster from Aspergillus clavatus NRRL 1. Metab Eng. 2011;13:723–32. doi: 10.1016/j.ymben.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yin WB, Grundmann A, Cheng J, Li SM. Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J Biol Chem. 2009;284:100–9. doi: 10.1074/jbc.M807606200. [DOI] [PubMed] [Google Scholar]
- 106.Guo CJ, Yeh HH, Chiang YM, Sanchez JF, Chang SL, Bruno KS, et al. Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis. J Am Chem Soc. 2013;135:7205–13. doi: 10.1021/ja3123653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lo HC, Entwistle R, Guo CJ, Ahuja M, Szewczyk E, Hung JH, et al. Two separate gene clusters encode the biosynthetic pathway for the meroterpenoids austinol and dehydroaustinol in Aspergillus nidulans. J Am Chem Soc. 2012;134:4709–20. doi: 10.1021/ja209809t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Inglis DO, Binkley J, Skrzypek MS, Arnaud MB, Cerqueira GC, Shah P, et al. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 2013;13:91. doi: 10.1186/1471-2180-13-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Seshime Y, Juvvadi PR, Kitamoto K, Ebizuka Y, Fujii I. Identification of csypyrone B1 as the novel product of Aspergillus oryzae type III polyketide synthase CsyB. Bioorg Med Chem. 2010;18:4542–6. doi: 10.1016/j.bmc.2010.04.058. [DOI] [PubMed] [Google Scholar]
- 110.Chiang YM, Szewczyk E, Nayak T, Davidson AD, Sanchez JF, Lo HC, et al. Molecular genetic mining of the Aspergillus secondary metabolome: discovery of the emericellamide biosynthetic pathway. Chem Biol. 2008;15:527–32. doi: 10.1016/j.chembiol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Birse CE, Clutterbuck AJ. Isolation and developmentally regulated expression of an Aspergillus nidulans phenol oxidase-encoding gene, ivoB. Gene. 1991;98:69–76. doi: 10.1016/0378-1119(91)90105-K. [DOI] [PubMed] [Google Scholar]
- 112.Andersen MR, Nielsen JB, Klitgaard A, Petersen LM, Zachariasen M, Hansen TJ, et al. Accurate prediction of secondary metabolite gene clusters in filamentous fungi. Proc Natl Acad Sci U S A. 2013;110:E99–107. doi: 10.1073/pnas.1205532110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zaehle C, Gressler M, Shelest E, Geib E, Hertweck C, Brock M. Terrein biosynthesis in Aspergillus terreus and its impact on phytotoxicity. Chem Biol. 2014;21:719–31. doi: 10.1016/j.chembiol.2014.03.010. [DOI] [PubMed] [Google Scholar]
- 114.Botha CJ, Legg MJ, Truter M, Sulyok M. Multitoxin analysis of Aspergillus clavatus-infected feed samples implicated in two outbreaks of neuromycotoxicosis in cattle in South Africa. Onderstepoort J Vet Res. 2014;81. Art. #848, 6 pages. [DOI] [PubMed]
- 115.Zutz C, Gacek A, Sulyok M, Wagner M, Strauss J, Rychli K. Small chemical chromatin effectors alter secondary metabolite production in Aspergillus clavatus. Toxins (Basel) 2013;5:1723–41. doi: 10.3390/toxins5101723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shaaban M, El-Metwally MM, Nasr H. A new diketopiperazine alkaloid from Aspergillus oryzae. Nat Prod Res. 2014;28:86–94. doi: 10.1080/14786419.2013.841687. [DOI] [PubMed] [Google Scholar]
- 117.Frisvad JC, Samson RA. Neopetromyces gen. nov. and an overview of teleomorphs of Aspergillus subg. Circumdati. Stud Mycol. 2000;45:201–7. [Google Scholar]
- 118.Marui J, Ohashi-Kunihiro S, Ando T, Nishimura M, Koike H, Machida M. Penicillin biosynthesis in Aspergillus oryzae and its overproduction by genetic engineering. J Biosci Bioeng. 2010;110:8–11. doi: 10.1016/j.jbiosc.2010.01.001. [DOI] [PubMed] [Google Scholar]
- 119.Tamiya H, Ochiai E, Kikuchi K, Yahiro M, Toyotome T, Watanabe A, et al. Secondary metabolite profiles and antifungal drug susceptibility of Aspergillus fumigatus and closely related species, Aspergillus lentulus, Aspergillus udagawae, and Aspergillus viridinutans. J Infect Chemother. 2015;21:385–91. doi: 10.1016/j.jiac.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 120.Fremlin LJ, Piggott AM, Lacey E, Capon RJ. Cottoquinazoline A and cotteslosins A and B, metabolites from an Australian marine-derived strain of Aspergillus versicolor. J Nat Prod. 2009;72:666–70. doi: 10.1021/np800777f. [DOI] [PubMed] [Google Scholar]
- 121.Clardy J, Springer JP, Buchi G, Matsuo K, Wightman R. Letter: Tryptoquivaline and tryptoquivalone, two tremorgenic metabolites of Aspergillus clavatus. J Am Chem Soc. 1975;97:663–5. doi: 10.1021/ja00836a045. [DOI] [PubMed] [Google Scholar]
- 122.Jurjevic Z, Peterson SW, Solfrizzo M, Peraica M. Sterigmatocystin production by nine newly described Aspergillus species in section Versicolores grown on two different media. Mycotoxin Res. 2013;29:141–5. doi: 10.1007/s12550-013-0160-4. [DOI] [PubMed] [Google Scholar]
- 123.Nielsen KF, Mogensen JM, Johansen M, Larsen TO, Frisvad JC. Review of secondary metabolites and mycotoxins from the Aspergillus niger group. Anal Bioanal Chem. 2009;395:1225–42. doi: 10.1007/s00216-009-3081-5. [DOI] [PubMed] [Google Scholar]
- 124.Kelly DE, Krasevec N, Mullins J, Nelson DR. The CYPome (Cytochrome P450 complement) of Aspergillus nidulans. Fungal Genet Biol. 2009;46(Suppl 1):S53–61. doi: 10.1016/j.fgb.2008.08.010. [DOI] [PubMed] [Google Scholar]
- 125.Yoshida Y, Aoyama Y, Noshiro M, Gotoh O. Sterol 14-demethylase P450 (CYP51) provides a breakthrough for the discussion on the evolution of cytochrome P450 gene superfamily. Biochem Biophys Res Commun. 2000;273:799–804. doi: 10.1006/bbrc.2000.3030. [DOI] [PubMed] [Google Scholar]
- 126.Kelly SL, Lamb DC, Corran AJ, Baldwin BC, Parks LW, Kelly DE. Purification and reconstitution of activity of Saccharomyces cerevisiae P450 61, a sterol delta 22-desaturase. FEBS Lett. 1995;377:217–20. doi: 10.1016/0014-5793(95)01342-3. [DOI] [PubMed] [Google Scholar]
- 127.Briza P, Kalchhauser H, Pittenauer E, Allmaier G, Breitenbach M. N, N’-Bisformyl dityrosine is an in vivo precursor of the yeast ascospore wall. Eur J Biochem. 1996;239:124–31. doi: 10.1111/j.1432-1033.1996.0124u.x. [DOI] [PubMed] [Google Scholar]
- 128.Jawallapersand P, Mashele SS, Kovacic L, Stojan J, Komel R, Pakala SB, et al. Cytochrome P450 monooxygenase CYP53 family in fungi: comparative structural and evolutionary analysis and its role as a common alternative anti-fungal drug target. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Novak M, Lah L, Sala M, Stojan J, Bohlmann J, Komel R. Oleic acid metabolism via a conserved cytochrome P450 system-mediated omega-hydroxylation in the bark beetle-associated fungus Grosmannia clavigera. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Brodhun F, Feussner I. Oxylipins in fungi. FEBS J. 2011;278:1047–63. doi: 10.1111/j.1742-4658.2011.08027.x. [DOI] [PubMed] [Google Scholar]
- 131.Podobnik B, Stojan J, Lah L, Krasevec N, Seliskar M, Rizner TL, et al. CYP53A15 of Cochliobolus lunatus, a target for natural antifungal compounds. J Med Chem. 2008;51:3480–6. doi: 10.1021/jm800030e. [DOI] [PubMed] [Google Scholar]
- 132.Miskei M, Karányi Z, Pócsi I. Annotation of stress-response proteins in the aspergilli. Fungal Genet Biol. 2009;46 Suppl 1:S105–20. [DOI] [PubMed]
- 133.Fungal Stress Database. http://www.fung-stress.org/.
- 134.Goetz KE, Coyle CM, Cheng JZ, O’Connor SE, Panaccione DG. Ergot cluster-encoded catalase is required for synthesis of chanoclavine-I in Aspergillus fumigatus. Curr Genet. 2011;57:201–11. doi: 10.1007/s00294-011-0336-4. [DOI] [PubMed] [Google Scholar]
- 135.Fillinger S, Ruijter G, Tamas MJ, Visser J, Thevelein JM, d’Enfert C. Molecular and physiological characterization of the NAD- dependent glycerol 3-phosphate dehydrogenase in the filamentous fungus Aspergillus nidulans. Mol Microbiol. 2001;39:145–57. doi: 10.1046/j.1365-2958.2001.02223.x. [DOI] [PubMed] [Google Scholar]
- 136.Ouedraogo JP, Hagen S, Spielvogel A, Engelhardt S, Meyer V. Survival strategies of yeast and filamentous fungi against the antifungal protein AFP. J Biol Chem. 2011;286:13859–68. doi: 10.1074/jbc.M110.203588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Horák J. Regulations of sugar transporters: insights from yeast. Curr Genet. 2013;59:1–31. doi: 10.1007/s00294-013-0388-8. [DOI] [PubMed] [Google Scholar]
- 138.Yan N. Structural biology of the major facilitator superfamily transporters. Annu Rev Biophys. 2015;44:257–83. doi: 10.1146/annurev-biophys-060414-033901. [DOI] [PubMed] [Google Scholar]
- 139.Chen LQ, Cheung LS, Feng L, Tanner W, Frommer WB. Transport of sugars. Annu Rev Biochem. 2015;84:865–94. doi: 10.1146/annurev-biochem-060614-033904. [DOI] [PubMed] [Google Scholar]
- 140.Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. The transporter classification database. Nucleic Acids Res. 2014;42:D251–8. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Saier MHJ, Beatty JT, Goffeau A, Harley KT, Heijne WH, Huang SC, et al. The major facilitator superfamily. J Mol Microbiol Biotechnol. 1999;1:257–79. [PubMed] [Google Scholar]
- 142.Dos Reis TF, Menino JF, Bom VL, Brown NA, Colabardini AC, Savoldi M, et al. Identification of glucose transporters in Aspergillus nidulans. PLoS One. 2013;8 doi: 10.1371/journal.pone.0081412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Forment JV, Flipphi M, Ramon D, Ventura L, Maccabe AP. Identification of the mstE gene encoding a glucose-inducible, low affinity glucose transporter in Aspergillus nidulans. J Biol Chem. 2006;281:8339–46. doi: 10.1074/jbc.M508198200. [DOI] [PubMed] [Google Scholar]
- 144.Forment JV, Flipphi M, Ventura L, Gonzalez R, Ramon D, et al. High-affinity glucose transport in Aspergillus nidulans is mediated by the products of two related but differentially expressed genes. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Li J, Lin L, Li H, Tian C, Ma Y. Transcriptional comparison of the filamentous fungus Neurospora crassa growing on three major monosaccharides D-glucose, D-xylose and L-arabinose. Biotechnol Biofuels. 2014;7:31. doi: 10.1186/1754-6834-7-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.vanKuyk PA, Diderich JA, MacCabe AP, Hererro O, Ruijter GJG, Visser J. Aspergillus niger mstA encodes a high-affinity sugar/H+ symporter which is regulated in response to extracellular pH. Biochem J. 2004;379:375–83. doi: 10.1042/bj20030624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xie X, Wilkinson HH, Correa A, Lewis ZA, Bell-Pedersen D, Ebbole DJ. Transcriptional response to glucose starvation and functional analysis of a glucose transporter of Neurospora crassa. Fungal Genet Biol. 2004;41:1104–19. doi: 10.1016/j.fgb.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 148.Li J, Xu J, Cai P, Wang B, Ma Y, Benz JP, et al. Functional analysis of two L-arabinose transporters from filamentous fungi reveals promising characteristics for improved pentose utilization in Saccharomyces cerevisiae. Appl Environ Microbiol. 2015;81:4062–70. doi: 10.1128/AEM.00165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Wei H, Vienken K, Weber R, Bunting S, Requena N, Fischer R. A putative high affinity hexose transporter, hxtA, of Aspergillus nidulans is induced in vegetative hyphae upon starvation and in ascogenous hyphae during cleistothecium formation. Fungal Genet Biol. 2004;41:148–56. doi: 10.1016/j.fgb.2003.10.006. [DOI] [PubMed] [Google Scholar]
- 150.Colabardini AC, Ries LN, Brown NA, Dos Reis TF, Savoldi M, Goldman MH, et al. Functional characterization of a xylose transporter in Aspergillus nidulans. Biotechnol Biofuels. 2014;7:46. doi: 10.1186/1754-6834-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Du J, Li S, Zhao H. Discovery and characterization of novel d-xylose-specific transporters from Neurospora crassa and Pichia stipitis. Mol Biosyst. 2010;6:2150–6. doi: 10.1039/c0mb00007h. [DOI] [PubMed] [Google Scholar]
- 152.Giles NH, Case ME, Baum J, Geever R, Huiet L, Patel V, et al. Gene organization and regulation in the qa (quinic acid) gene cluster of Neurospora crassa. Microbiol Rev. 1985;49:338–58. doi: 10.1128/mr.49.3.338-358.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Whittington HA, Grant S, Roberts CF, Lamb H, Hawkins AR. Identification and isolation of a putative permease gene in the quinic acid utilization (QUT) gene cluster of Aspergillus nidulans. Curr Genet. 1987;12:135–9. doi: 10.1007/BF00434668. [DOI] [PubMed] [Google Scholar]
- 154.Benz JP, Protzko RJ, Andrich JM, Bauer S, Dueber JE, Somerville CR. Identification and characterization of a galacturonic acid transporter from Neurospora crassa and its application for Saccharomyces cerevisiae fermentation processes. Biotechnol Biofuels. 2014;7:20. doi: 10.1186/1754-6834-7-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sloothaak J, Schilders M, Schaap PJ, de Graaff LH. Overexpression of the Aspergillus niger GatA transporter leads to preferential use of D-galacturonic acid over D-xylose. AMB Express. 2014;4:66. doi: 10.1186/s13568-014-0066-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Nikawa J, Tsukagoshi Y, Yamashita S. Isolation and characterization of two distinct myo-inositol transporter genes of Saccharomyces cerevisiae. J Biol Chem. 1991;266:11184–91. [PubMed] [Google Scholar]
- 157.Xue C, Liu T, Chen L, Li W, Liu I, Kronstad JW, et al. Role of an expanded inositol transporter repertoire in Cryptococcus neoformans sexual reproduction and virulence. mBio. 2010;1:e00084–10. doi: 10.1128/mBio.00084-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Doehlemann G, Molitor F, Hahn M. Molecular and functional characterization of a fructose specific transporter from the gray mold fungus Botrytis cinerea. Fungal Genet Biol. 2005;42:601–10. doi: 10.1016/j.fgb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 159.Goncalves AM, Silva CS, Madeira TI, Coelho R, de Sanctis D, San Romao MV, et al. Endo-beta-D-1,4-mannanase from Chrysonilia sitophila displays a novel loop arrangement for substrate selectivity. Acta Crystallogr D Biol Crystallogr. 2012;68:1468–78. doi: 10.1107/S0907444912034646. [DOI] [PubMed] [Google Scholar]
- 160.Fan J, Chaturvedi V, Shen SH. Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J Mol Evol. 2002;55:336–46. doi: 10.1007/s00239-002-2330-4. [DOI] [PubMed] [Google Scholar]
- 161.Hasegawa S, Takizawa M, Suyama H, Shintani T, Gomi K. Characterization and expression analysis of a maltose-utilizing (MAL) cluster in Aspergillus oryzae. Fungal Genet Biol. 2010;47:1–9. doi: 10.1016/j.fgb.2009.10.005. [DOI] [PubMed] [Google Scholar]
- 162.Lingner U, Munch S, Sode B, Deising HB, Sauer N. Functional characterization of a eukaryotic melibiose transporter. Plant Physiol. 2011;156:1565–76. doi: 10.1104/pp.111.178624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fang W, St Leger RJ. Mrt, a gene unique to fungi, encodes an oligosaccharide transporter and facilitates rhizosphere competency in Metarhizium robertsii. Plant Physiol. 2010;154:1549–57. doi: 10.1104/pp.110.163014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Bun-Ya M, Nishimura M, Harashima S, Oshima Y. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol Cell Biol. 1991;11:3229–38. doi: 10.1128/MCB.11.6.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dick CF, Dos-Santos AL, Meyer-Fernandes JR. Inorganic phosphate uptake in unicellular eukaryotes. Biochim Biophys Acta. 2014;1840:2123–7. doi: 10.1016/j.bbagen.2014.03.014. [DOI] [PubMed] [Google Scholar]
- 166.Yadav V, Kumar M, Deep DK, Kumar H, Sharma R, Tripathi T, et al. A phosphate transporter from the root endophytic fungus Piriformospora indica plays a role in phosphate transport to the host plant. J Biol Chem. 2010;285:26532–44. doi: 10.1074/jbc.M110.111021. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 167.Patton-Vogt JL, Henry SA. GIT1, a gene encoding a novel transporter for glycerophosphoinositol in Saccharomyces cerevisiae. Genetics. 1998;149:1707–15. doi: 10.1093/genetics/149.4.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.McDermott JR, Rosen BP, Liu Z. Jen1p: a high affinity selenite transporter in yeast. Mol Biol Cell. 2010;21:3934–41. doi: 10.1091/mbc.E10-06-0513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ferreira C, van Voorst F, Martins A, Neves L, Oliveira R, Kielland-Brandt MC, et al. A member of the sugar transporter family, Stl1p is the glycerol/H+ symporter in Saccharomyces cerevisiae. Mol Biol Cell. 2005;16:2068–76. doi: 10.1091/mbc.E04-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fekete E, Karaffa L, Seiboth B, Fekete E, Kubicek CP, Flipphi M. Identification of a permease gene involved in lactose utilisation in Aspergillus nidulans. Fungal Genet Biol. 2012;49:415–25. doi: 10.1016/j.fgb.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 171.Fekete E, Orosz A, Kulcsar L, Kavalecz N, Flipphi M, Karaffa L. Characterization of a second physiologically relevant lactose permease gene (lacpB) in Aspergillus nidulans. Microbiology. 2016;162:837–47. doi: 10.1099/mic.0.000267. [DOI] [PubMed] [Google Scholar]
- 172.Cai P, Wang B, Ji J, Jiang Y, Wan L, Tian C, et al. The putative cellodextrin transporter-like protein CLP1 is involved in cellulase induction in Neurospora crassa. J Biol Chem. 2015;290:788–96. doi: 10.1074/jbc.M114.609875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Galazka JM, Tian C, Beeson WT, Martinez B, Glass NL, Cate JH. Cellodextrin transport in yeast for improved biofuel production. Science. 2010;330:84–6. doi: 10.1126/science.1192838. [DOI] [PubMed] [Google Scholar]
- 174.Margolis-Clark E, Hunt I, Espinosa S, Bowman BJ. Identification of the gene at the pmg locus, encoding system II, the general amino acid transporter in Neurospora crassa. Fungal Genet Biol. 2001;33:127–35. doi: 10.1006/fgbi.2001.1273. [DOI] [PubMed] [Google Scholar]
- 175.Vasseur V, Van Montagu M, Goldman GH. Trichoderma harzianum genes induced during growth on Rhizoctonia solani cell walls. Microbiology. 1995;141(Pt 4):767–74. doi: 10.1099/13500872-141-4-767. [DOI] [PubMed] [Google Scholar]
- 176.Omura F, Hatanaka H, Nakao Y. Characterization of a novel tyrosine permease of lager brewing yeast shared by Saccharomyces cerevisiae strain RM11-1a. FEMS Yeast Res. 2007;7:1350–61. doi: 10.1111/j.1567-1364.2007.00310.x. [DOI] [PubMed] [Google Scholar]
- 177.Yadav AK, Bachhawat AK. CgCYN1, a plasma membrane cystine-specific transporter of Candida glabrata with orthologues prevalent among pathogenic yeast and fungi. J Biol Chem. 2011;286:19714–23. doi: 10.1074/jbc.M111.240648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Gournas C, Evangelidis T, Athanasopoulos A, Mikros E, Sophianopoulou V. The Aspergillus nidulans proline permease as a model for understanding the factors determining substrate binding and specificity of fungal amino acid transporters. J Biol Chem. 2015;290:6141–55. doi: 10.1074/jbc.M114.612069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Apostolaki A, Erpapazoglou Z, Harispe L, Billini M, Kafasla P, Kizis D, et al. AgtA, the dicarboxylic amino acid transporter of Aspergillus nidulans, is concertedly down-regulated by exquisite sensitivity to nitrogen metabolite repression and ammonium-elicited endocytosis. Eukaryot Cell. 2009;8:339–52. doi: 10.1128/EC.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ghaddar K, Krammer EM, Mihajlovic N, Brohee S, Andre B, Prevost M. Converting the yeast arginine can1 permease to a lysine permease. J Biol Chem. 2014;289:7232–46. doi: 10.1074/jbc.M113.525915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Gournas C, Prevost M, Krammer EM, Andre B. Function and regulation of fungal amino acid transporters: Insights from predicted structure. Adv Exp Med Biol. 2016;892:69–106. doi: 10.1007/978-3-319-25304-6_4. [DOI] [PubMed] [Google Scholar]
- 182.Russnak R, Konczal D, McIntire SL. A family of yeast proteins mediating bidirectional vacuolar amino acid transport. J Biol Chem. 2001;276:23849–57. doi: 10.1074/jbc.M008028200. [DOI] [PubMed] [Google Scholar]
- 183.Abreu C, Sanguinetti M, Amillis S, Ramon A. UreA, the major urea/H+ symporter in Aspergillus nidulans. Fungal Genet Biol. 2010;47:1023–33. doi: 10.1016/j.fgb.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 184.Pateman JA, Dunn E, Mackay EM. Urea and thiourea transport in Aspergillus nidulans. Biochem Genet. 1982;20:777–90. doi: 10.1007/BF00483973. [DOI] [PubMed] [Google Scholar]
- 185.Sanguinetti M, Amillis S, Pantano S, Scazzocchio C, Ramon A. Modelling and mutational analysis of Aspergillus nidulans UreA, a member of the subfamily of urea/H(+) transporters in fungi and plants. Open Biol. 2014;4:140070. doi: 10.1098/rsob.140070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Unkles SE, Karabika E, Symington VF, Cecile JL, Rouch DA, Akhtar N, et al. Alanine scanning mutagenesis of a high-affinity nitrate transporter highlights the requirement for glycine and asparagine residues in the two nitrate signature motifs. Biochem J. 2012;447:35–42. doi: 10.1042/BJ20120631. [DOI] [PubMed] [Google Scholar]
- 187.Unkles SE, Rouch DA, Wang Y, Siddiqi MY, Glass AD, Kinghorn JR. Two perfectly conserved arginine residues are required for substrate binding in a high-affinity nitrate transporter. Proc Natl Acad Sci U S A. 2004;101:17549–54. doi: 10.1073/pnas.0405054101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zheng H, Wisedchaisri G, Gonen T. Crystal structure of a nitrate/nitrite exchanger. Nature. 2013;497:647–51. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Unkles SE, Zhou D, Siddiqi MY, Kinghorn JR, Glass AD. Apparent genetic redundancy facilitates ecological plasticity for nitrate transport. EMBO J. 2001;20:6246–55. doi: 10.1093/emboj/20.22.6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Wang Y, Li W, Siddiqi Y, Symington VF, Kinghorn JR, Unkles SE, et al. Nitrite transport is mediated by the nitrite-specific high-affinity NitA transporter and by nitrate transporters NrtA, NrtB in Aspergillus nidulans. Fungal Genet Biol. 2008;45:94–102. doi: 10.1016/j.fgb.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 191.Unkles SE, Symington VF, Kotur Z, Wang Y, Siddiqi MY, Kinghorn JR, et al. Physiological and biochemical characterization of AnNitA, the Aspergillus nidulans high-affinity nitrite transporter. Eukaryot Cell. 2011;10:1724–32. doi: 10.1128/EC.05199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Parker JL, Newstead S. Molecular basis of nitrate uptake by the plant nitrate transporter NRT1.1. Nature. 2014;507:68–72. doi: 10.1038/nature13116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Sun J, Bankston JR, Payandeh J, Hinds TR, Zagotta WN, Zheng N. Crystal structure of the plant dual-affinity nitrate transporter NRT1.1. Nature. 2014;507:73–7. doi: 10.1038/nature13074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Monahan BJ, Askin MC, Hynes MJ, Davis MA. Differential expression of Aspergillus nidulans ammonium permease genes is regulated by GATA transcription factor AreA. Eukaryot Cell. 2006;5:226–37. doi: 10.1128/EC.5.2.226-237.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Hamari Z, Amillis S, Drevet C, Apostolaki A, Vagvolgyi C, Diallinas G, et al. Convergent evolution and orphan genes in the Fur4p-like family and characterization of a general nucleoside transporter in Aspergillus nidulans. Mol Microbiol. 2009;73:43–57. doi: 10.1111/j.1365-2958.2009.06738.x. [DOI] [PubMed] [Google Scholar]
- 196.Lougiakis N, Gavriil ES, Kairis M, Sioupouli G, Lambrinidis G, Benaki D, et al. Design and synthesis of purine analogues as highly specific ligands for FcyB, a ubiquitous fungal nucleobase transporter. Bioorg Med Chem. 2016;24:5941–52. doi: 10.1016/j.bmc.2016.09.055. [DOI] [PubMed] [Google Scholar]
- 197.Diallinas G. Understanding transporter specificity and the discrete appearance of channel-like gating domains in transporters. Front Pharmacol. 2014;5:207. doi: 10.3389/fphar.2014.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Diallinas G, Gournas C. Structure-function relationships in the nucleobase-ascorbate transporter (NAT) family: lessons from model microbial genetic systems. Channels (Austin) 2008;2:363–72. doi: 10.4161/chan.2.5.6902. [DOI] [PubMed] [Google Scholar]
- 199.Gournas C, Papageorgiou I, Diallinas G. The nucleobase-ascorbate transporter (NAT) family: genomics, evolution, structure-function relationships and physiological role. Mol Biosyst. 2008;4:404–16. doi: 10.1039/b719777b. [DOI] [PubMed] [Google Scholar]
- 200.Alguel Y, Amillis S, Leung J, Lambrinidis G, Capaldi S, Scull NJ, et al. Structure of eukaryotic purine/H(+) symporter UapA suggests a role for homodimerization in transport activity. Nat Commun. 2016;7:11336. doi: 10.1038/ncomms11336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Koukaki M, Vlanti A, Goudela S, Pantazopoulou A, Gioule H, Tournaviti S, et al. The nucleobase-ascorbate transporter (NAT) signature motif in UapA defines the function of the purine translocation pathway. J Mol Biol. 2005;350:499–513. doi: 10.1016/j.jmb.2005.04.076. [DOI] [PubMed] [Google Scholar]
- 202.Pantazopoulou A, Diallinas G. The first transmembrane segment (TMS1) of UapA contains determinants necessary for expression in the plasma membrane and purine transport. Mol Membr Biol. 2006;23:337–48. doi: 10.1080/09687860600738239. [DOI] [PubMed] [Google Scholar]
- 203.Goudela S, Karatza P, Koukaki M, Frillingos S, Diallinas G. Comparative substrate recognition by bacterial and fungal purine transporters of the NAT/NCS2 family. Mol Membr Biol. 2005;22:263–75. doi: 10.1080/09687860500093016. [DOI] [PubMed] [Google Scholar]
- 204.Goudela S, Reichard U, Amillis S, Diallinas G. Characterization and kinetics of the major purine transporters in Aspergillus fumigatus. Fungal Genet Biol. 2008;45:459–72. doi: 10.1016/j.fgb.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 205.Krypotou E, Scazzocchio C, Diallinas G. Functional characterization of NAT/NCS2 proteins of Aspergillus brasiliensis reveals a genuine xanthine-uric acid transporter and an intrinsically misfolded polypeptide. Fungal Genet Biol. 2015;75:56–63. doi: 10.1016/j.fgb.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 206.Krypotou E, Diallinas G. Transport assays in filamentous fungi: kinetic characterization of the UapC purine transporter of Aspergillus nidulans. Fungal Genet Biol. 2014;63:1–8. doi: 10.1016/j.fgb.2013.12.004. [DOI] [PubMed] [Google Scholar]
- 207.Krypotou E, Evangelidis T, Bobonis J, Pittis AA, Gabaldon T, Scazzocchio C, et al. Origin, diversification and substrate specificity in the family of NCS1/FUR transporters. Mol Microbiol. 2015;96:927–50. doi: 10.1111/mmi.12982. [DOI] [PubMed] [Google Scholar]
- 208.Cecchetto G, Amillis S, Diallinas G, Scazzocchio C, Drevet C. The AzgA purine transporter of Aspergillus nidulans. Characterization of a protein belonging to a new phylogenetic cluster. J Biol Chem. 2004;279:3132–41. doi: 10.1074/jbc.M308826200. [DOI] [PubMed] [Google Scholar]
- 209.Krypotou E, Lambrinidis G, Evangelidis T, Mikros E, Diallinas G. Modelling, substrate docking and mutational analysis identify residues essential for function and specificity of the major fungal purine transporter AzgA. Mol Microbiol. 2014;93:129–45. doi: 10.1111/mmi.12646. [DOI] [PubMed] [Google Scholar]
- 210.Krypotou E, Kosti V, Amillis S, Myrianthopoulos V, Mikros E, Diallinas G. Modeling, substrate docking, and mutational analysis identify residues essential for the function and specificity of a eukaryotic purine-cytosine NCS1 transporter. J Biol Chem. 2012;287:36792–803. doi: 10.1074/jbc.M112.400382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Vlanti A, Diallinas G. The Aspergillus nidulans FcyB cytosine-purine scavenger is highly expressed during germination and in reproductive compartments and is downregulated by endocytosis. Mol Microbiol. 2008;68:959–77. doi: 10.1111/j.1365-2958.2008.06198.x. [DOI] [PubMed] [Google Scholar]
- 212.Sioupouli G, Lambrinidis G, Mikros E, Amillis S, Diallinas G. Cryptic purine transporters in Aspergillus nidulans reveal the role of specific residues in the evolution of specificity in the NCS1 family. Mol Microbiol. 2016; in press. [DOI] [PubMed]
- 213.Loewen SK, Ng AM, Mohabir NN, Baldwin SA, Cass CE, Young JD. Functional characterization of a H+/nucleoside co-transporter (CaCNT) from Candida albicans, a fungal member of the concentrative nucleoside transporter (CNT) family of membrane proteins. Yeast. 2003;20:661–75. doi: 10.1002/yea.1000. [DOI] [PubMed] [Google Scholar]
- 214.Canovas D, Marcos JF, Marcos AT, Strauss J. Nitric oxide in fungi: is there NO light at the end of the tunnel? Curr Genet. 2016;62:513–8. doi: 10.1007/s00294-016-0574-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Gardner PR, Gardner AM, Martin LA, Salzman AL. Nitric oxide dioxygenase: an enzymic function for flavohemoglobin. Proc Natl Acad Sci U S A. 1998;95:10378–83. doi: 10.1073/pnas.95.18.10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Schinko T, Berger H, Lee W, Gallmetzer A, Pirker K, Pachlinger R, et al. Transcriptome analysis of nitrate assimilation in Aspergillus nidulans reveals connections to nitric oxide metabolism. Mol Microbiol. 2010;78:720–38. doi: 10.1111/j.1365-2958.2010.07363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.te Biesebeke R, Levasseur A, Boussier A, Record E, van den Hondel CA, Punt PJ. Phylogeny of fungal hemoglobins and expression analysis of the Aspergillus oryzae flavohemoglobin gene fhbA during hyphal growth. Fungal Biol. 2010;114:135–43. doi: 10.1016/j.mycres.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 218.Zhou S, Fushinobu S, Kim SW, Nakanishi Y, Maruyama J, Kitamoto K, et al. Functional analysis and subcellular location of two flavohemoglobins from Aspergillus oryzae. Fungal Genet Biol. 2011;48:200–7. doi: 10.1016/j.fgb.2010.08.011. [DOI] [PubMed] [Google Scholar]
- 219.Marcos AT, Ramos MS, Marcos JF, Carmona L, Strauss J, Canovas D. Nitric oxide synthesis by nitrate reductase is regulated during development in Aspergillus. Mol Microbiol. 2016;99:15–33. doi: 10.1111/mmi.13211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Nakahara K, Tanimoto T, Hatano K, Usuda K, Shoun H. Cytochrome P-450 55A1 (P-450dNIR) acts as nitric oxide reductase employing NADH as the direct electron donor. J Biol Chem. 1993;268:8350–5. [PubMed] [Google Scholar]
- 221.de Jesus-Berrios M, Liu L, Nussbaum JC, Cox GM, Stamler JS, Heitman J. Enzymes that counteract nitrosative stress promote fungal virulence. Curr Biol. 2003;13:1963–8. doi: 10.1016/j.cub.2003.10.029. [DOI] [PubMed] [Google Scholar]
- 222.Zhang Z, Wang J, Chai R, Qiu H, Jiang H, Mao X, et al. An S-(hydroxymethyl)glutathione dehydrogenase is involved in conidiation and full virulence in the rice blast fungus Magnaporthe oryzae. PLoS One. 2015;10 doi: 10.1371/journal.pone.0120627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Lapp K, Vodisch M, Kroll K, Strassburger M, Kniemeyer O, Heinekamp T, et al. Characterization of the Aspergillus fumigatus detoxification systems for reactive nitrogen intermediates and their impact on virulence. Front Microbiol. 2014;5:469. doi: 10.3389/fmicb.2014.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Philippe B, Ibrahim-Granet O, Prevost MC, Gougerot-Pocidalo MA, Sanchez Perez M, Van der Meeren A, et al. Killing of Aspergillus fumigatus by alveolar macrophages is mediated by reactive oxidant intermediates. Infect Immun. 2003;71:3034–42. doi: 10.1128/IAI.71.6.3034-3042.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Han KH, Prade RA. Osmotic stress-coupled maintenance of polar growth in Aspergillus nidulans. Mol Microbiol. 2002;43:1065–78. doi: 10.1046/j.1365-2958.2002.02774.x. [DOI] [PubMed] [Google Scholar]
- 226.Lafon A, Han KH, Seo JA, Yu JH, d’Enfert C. G-protein and cAMP-mediated signaling in aspergilli: a genomic perspective. Fungal Genet Biol. 2006;43:490–502. doi: 10.1016/j.fgb.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 227.Li L, Wright SJ, Krystofova S, Park G, Borkovich KA. Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol. 2007;61:423–52. doi: 10.1146/annurev.micro.61.080706.093432. [DOI] [PubMed] [Google Scholar]
- 228.Chang MH, Chae KS, Han DM, Jahng KY. The GanB Galpha-protein negatively regulates asexual sporulation and plays a positive role in conidial germination in Aspergillus nidulans. Genetics. 2004;167:1305–15. doi: 10.1534/genetics.103.025379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Lafon A, Seo JA, Han KH, Yu JH, d’Enfert C. The heterotrimeric G-protein GanB(a)-SfaD(b)-GpgA(g) is a carbon source sensor involved in early cAMP-dependent germination in Aspergillus nidulans. Genetics. 2005;171:71–80. doi: 10.1534/genetics.105.040584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Rosen S, Yu JH, Adams TH. The Aspergillus nidulans sfaD gene encodes a G protein beta subunit that is required for normal growth and repression of sporulation. EMBO J. 1999;18:5592–600. doi: 10.1093/emboj/18.20.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Downes GB, Gautam N. The G protein subunit gene families. Genomics. 1999;62:544–52. doi: 10.1006/geno.1999.5992. [DOI] [PubMed] [Google Scholar]
- 232.Giannakouros T, Nikolakaki E, Mylonis I, Georgatsou E. Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J. 2011;278:570–86. doi: 10.1111/j.1742-4658.2010.07987.x. [DOI] [PubMed] [Google Scholar]
- 233.De Souza CP, Hashmi SB, Osmani AH, Andrews P, Ringelberg CS, Dunlap JC, et al. Functional analysis of the Aspergillus nidulans kinome. PLoS One. 2013;8 doi: 10.1371/journal.pone.0058008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Widmann C, Gibson S, Jarpe MB, Johnson GL. Mitogen-activated protein kinase: conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79:143–80. doi: 10.1152/physrev.1999.79.1.143. [DOI] [PubMed] [Google Scholar]
- 235.Moorhead GB, Trinkle-Mulcahy L, Ulke-Lemee A. Emerging roles of nuclear protein phosphatases. Nat Rev Mol Cell Biol. 2007;8:234–44. doi: 10.1038/nrm2126. [DOI] [PubMed] [Google Scholar]
- 236.Son S, Osmani SA. Analysis of all protein phosphatase genes in Aspergillus nidulans identifies a new mitotic regulator, fcp1. Eukaryot Cell. 2009;8:573–85. doi: 10.1128/EC.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Lyon MA, Ducruet AP, Wipf P, Lazo JS. Dual-specificity phosphatases as targets for antineoplastic agents. Nat Rev Drug Discov. 2002;1:961–76. doi: 10.1038/nrd963. [DOI] [PubMed] [Google Scholar]
- 238.Scazzocchio C, Ramón A. Chromatin in the genus Aspergillus. In: Osmani S, Goldman G, editors. The Aspergilli: Genomics, medical applications, biotechnology, and research methods. Boca Raton, FL: CRC Press; 2008. pp. 321–42. [Google Scholar]
- 239.Ehinger A, Denison SH, May GS. Sequence, organization and expression of the core histone genes of Aspergillus nidulans. Mol Gen Genet. 1990;222:416–24. doi: 10.1007/BF00633848. [DOI] [PubMed] [Google Scholar]
- 240.Hays SM, Swanson J, Selker EU. Identification and characterization of the genes encoding the core histones and histone variants of Neurospora crassa. Genetics. 2002;160:961–73. doi: 10.1093/genetics/160.3.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Santisteban MS, Kalashnikova T, Smith MM. Histone H2A.Z regulats transcription and is partially redundant with nucleosome remodeling complexes. Cell. 2000;103:411–22. doi: 10.1016/S0092-8674(00)00133-1. [DOI] [PubMed] [Google Scholar]
- 242.Ahmed S, Dul B, Qiu X, Walworth NC. Msc1 acts through histone H2A.Z to promote chromosome stability in Schizosaccharomyces pombe. Genetics. 2007;177:1487–97. doi: 10.1534/genetics.107.078691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Linz JE, Wee J, Roze LV. Aspergillus parasiticus SU-1 genome sequence, predicted chromosome structure, and comparative gene expression under aflatoxin-inducing conditions: evidence that differential expression contributes to species phenotype. Eukaryot Cell. 2014;13:1113–23. doi: 10.1128/EC.00108-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Nikkuni S, Nakajima H, Hoshina SI, Ohno M, Suzuki C, Kashiwagi Y, et al. Evolutionary relationships among Aspergillus oryzae and related species based on the sequences of 18S rRNA genes and internal transcribed spacers. J Gen Appl Microbiol. 1998;44:225–30. doi: 10.2323/jgam.44.225. [DOI] [PubMed] [Google Scholar]
- 245.Sato A, Oshima K, Noguchi H, Ogawa M, Takahashi T, Oguma T, et al. Draft genome sequencing and comparative analysis of Aspergillus sojae NBRC4239. DNA Res. 2011;18:165–76. doi: 10.1093/dnares/dsr009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Faustinelli PC, Wang XM, Palencia ER, Arias RS. Genome sequences of eight Aspergillus flavus spp. and one A. parasiticus sp., isolated from peanut seeds in Georgia. Genome Announc. 2016;4:e00278–16. doi: 10.1128/genomeA.00278-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Malik HS, Vermaak D, Henikoff S. Recurrent evolution of DNA-binding motifs in the Drosophila centromeric histone. Proc Natl Acad Sci U S A. 2002;99:1449–54. doi: 10.1073/pnas.032664299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Freitag M, Williams RL, Kothe GO, Selker EU. A cytosine methyltransferase homologue is essential for repeat-induced point mutation in Neurospora crassa. Proc Natl Acad Sci U S A. 2002;99:8802–7. doi: 10.1073/pnas.132212899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Malagnac F, Wendel B, Goyon C, Faugeron G, Zickler D, Rossignol JL, et al. A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure. Cell. 1997;91:281–90. doi: 10.1016/S0092-8674(00)80410-9. [DOI] [PubMed] [Google Scholar]
- 250.Kouzminova E, Selker EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO J. 2001;20:4309–23. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lee DW, Freitag M, Selker EU, Aramayo R. A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS One. 2008;3 doi: 10.1371/journal.pone.0002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Li Destri Nicosia MG, Brocard-Masson C, Demais S, Hua Van A, Daboussi MJ, Scazzocchio C. Heterologous transposition in Aspergillus nidulans. Mol Microbiol. 2001;39:1330–44. doi: 10.1111/j.1365-2958.2001.02323.x. [DOI] [PubMed] [Google Scholar]
- 253.Liu SY, Lin JQ, Wu HL, Wang CC, Huang SJ, Luo YF, et al. Bisulfite sequencing reveals that Aspergillus flavus holds a hollow in DNA methylation. PLoS One. 2012;7 doi: 10.1371/journal.pone.0030349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Malagnac F, Gregoire A, Goyon C, Rossignol JL, Faugeron G. Masc2, a gene from Ascobolus encoding a protein with a DNA-methyltransferase activity in vitro, is dispensable for in vivo methylation. Mol Microbiol. 1999;31:331–8. doi: 10.1046/j.1365-2958.1999.01177.x. [DOI] [PubMed] [Google Scholar]
- 255.Gnerre S, Maccallum I, Przybylski D, Ribeiro FJ, Burton JN, Walker BJ, et al. High-quality draft assemblies of mammalian genomes from massively parallel sequence data. Proc Natl Acad Sci U S A. 2011;108:1513–8. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Martin J, Bruno VM, Fang Z, Meng X, Blow M, Zhang T, et al. Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-Seq reads. BMC Genomics. 2010;11:663. doi: 10.1186/1471-2164-11-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Smit AFA, Hubley R, Geen P. RepeatMasker Open-3.0. 1996-2010. http://www.repeatmasker.org.
- 258.Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–7. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 259.Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–8. doi: 10.1093/bioinformatics/bti1018. [DOI] [PubMed] [Google Scholar]
- 260.Salamov AA, Solovyev VV. Ab initio gene finding in Drosophila genomic DNA. Genome Res. 2000;10:516–22. doi: 10.1101/gr.10.4.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 2008;18:1979–90. doi: 10.1101/gr.081612.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Birney E, Durbin R. Using GeneWise in the Drosophila annotation experiment. Genome Res. 2000;10:547–8. doi: 10.1101/gr.10.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 264.Lowe TM, Eddy SR. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;25:955–64. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 266.Melen K, Fagerlund R, Franke J, Kohler M, Kinnunen L, Julkunen I. Importin alpha nuclear localization signal binding sites for STAT1, STAT2, and influenza A virus nucleoprotein. J Biol Chem. 2003;78:28193–200. doi: 10.1074/jbc.M303571200. [DOI] [PubMed] [Google Scholar]
- 267.Zdobnov EM, Apweiler R. InterProScan--an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–8. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 268.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–4. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Koonin EV, Fedorova ND, Jackson JD, Jacobs AR, Krylov DM, Makarova KS, et al. A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol. 2004;5:R7. doi: 10.1186/gb-2004-5-2-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–84. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–30. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–52. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 275.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.NCBI BLAST tool 2.4.0. http://blast.ncbi.nlm.nih.gov/Blast.cgi.
- 278.Klitgaard A, Iversen A, Andersen MR, Larsen TO, Frisvad JC, Nielsen KF. Aggressive dereplication using UHPLC-DAD-QTOF: screening extracts for up to 3000 fungal secondary metabolites. Anal Bioanal Chem. 2014;406:1933–43. doi: 10.1007/s00216-013-7582-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–82. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 281.PFAM Domain PF00083. http://pfam.xfam.org/family/PF00083/alignment/seed/jalview.
- 282.Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, Boursnell C, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.DTU. TMHMM Server v. 2.0. 2015. http://www.cbs.dtu.dk/services/TMHMM/.
- 284.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Lechner M, Findeiss S, Steiner L, Marz M, Stadler PF, Prohaska SJ. Proteinortho: detection of (co-)orthologs in large-scale analysis. BMC Bioinformatics. 2011;12:124. doi: 10.1186/1471-2105-12-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Isnard AD, Thomas D, Surdin-Kerjan Y. The study of methionine uptake in Saccharomyces cerevisiae reveals a new family of amino acid permeases. J Mol Biol. 1996;262:473–84. doi: 10.1006/jmbi.1996.0529. [DOI] [PubMed] [Google Scholar]
- 287.Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–4. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Vangelatos I, Vlachakis D, Sophianopoulou V, Diallinas G. Modelling and mutational evidence identify the substrate binding site and functional elements in APC amino acid transporters. Mol Membr Biol. 2009;26:356–70. doi: 10.1080/09687680903170546. [DOI] [PubMed] [Google Scholar]
- 289.Katoh K, Kuma K-i, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–8. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–21. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 292.FigTree v1.4.2. http://tree.bio.ed.ac.uk/software/figtree/.
- 293.Pérez-Rodriguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res. 2010;38:D822–7. doi: 10.1093/nar/gkp805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Bitbucket PlnTFDB. https://bitbucket.org/diriano/plntfdb.
- 295.de Vries RP, Burgers K, van de Vondervoort PJI, Frisvad JC, Samson RA, Visser J. A new black Aspergillus species, A. vadensis, is a promising host for homologous and heterologous protein production. Appl Environ Microbiol. 2004;70:3954–9. doi: 10.1128/AEM.70.7.3954-3959.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Barratt RW, Johnson GB, Ogata WN. Wild-type and mutant stocks of Aspergillus nidulans. Genetics. 1965;52:233–46. doi: 10.1093/genetics/52.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Yin WB, Reinke AW, Szilágyi M, Emri T, Chiang YM, Keating AE, et al. bZIP transcription factors affecting secondary metabolism, sexual development and stress responses in Aspergillus nidulans. Microbiology. 2013;159:77–88. [DOI] [PMC free article] [PubMed]
- 298.Karányi Z, Holb I, Hornok L, Pócsi I, Miskei M. FSRD: fungal stress response database. Database (Oxford) 2013;2013:bat037. doi: 10.1093/database/bat037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Sigoillot JC, Herpoël I, Frasse P, Moukha S, Lesage-Meessen L, Asther M. Laccase production by a monokaryotic strain of Pycnoporus cinnabarinus derived from a dikaryotic strain. World J Microbiol Biotechnol. 1999;15:481–4. doi: 10.1023/A:1008986809395. [DOI] [Google Scholar]
- 300.Henriksson G, Sild V, Szabo IJ, Pettersson G, Johansson G. Substrate specificity of cellobiose dehydrogenase from Phanerochaete chrysosporium. Biochim Biophys Acta. 1998;1383:48–54. doi: 10.1016/S0167-4838(97)00180-5. [DOI] [PubMed] [Google Scholar]
- 301.Kruckeberg AL, Ye L, Berden JA, Dam K. Functional expression, quantification and cellular localization of the hxt2 hexose transporter of Saccharomyces cerevisiae tagged with the green fluorescent protein. Biochem J. 1999;339:299–307. doi: 10.1042/bj3390299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464:123–8. doi: 10.1016/S0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- 303.Noiraud N, Maurousset L, Lemoine R. Identification of a mannitol transporter, AgmaT1, in celery phloem. Plant Cell. 2001;13:695–705. doi: 10.1105/tpc.13.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Emr SD, Scheckman R, Flessel MC, Thorner J. An Mfα1-SUC2 (σ-factor-invertase) gene fusion for study of protein localisation and gene expression in yeast. Proc Natl Acad Sci U S A. 1983;80:7080–4. doi: 10.1073/pnas.80.23.7080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Salema-Oom M, Pinto VV, Gonçalves P, Spencer-Martins I. Maltotriose utilization by industrial Saccharomyces strains: characterization of a new member of the a-glucoside transporter family. Appl Environ Microbiol. 2005;71:5044–9. doi: 10.1128/AEM.71.9.5044-5049.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Macreadie IG, Horaitis O, Verkuylen AJ, Savin KW. Improved shuttle vectors for cloning and high-level Cu(2+)-mediated expression of foreign genes in yeast. Gene. 1991;104:107–11. doi: 10.1016/0378-1119(91)90474-P. [DOI] [PubMed] [Google Scholar]
- 307.Gietz RD, Schiestl RH. Quick and easy yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:35–7. doi: 10.1038/nprot.2007.14. [DOI] [PubMed] [Google Scholar]
- 308.Gibbons JG, Rokas A. The function and evolution of the Aspergillus genome. Trends Microbiol. 2013;21:14–22. doi: 10.1016/j.tim.2012.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets supporting the conclusions of this article are included within the article and its Additional File(s) or available through DDBJ/EMBL/GenBank under the following accessions: A. aculeatus ATCC16872 - MRCK00000000, A. brasiliensis CBS101740 - LJXV00000000, A. carbonarius ITEM5010 - AHIG00000000, A. foetidus CBS106.47 - MRBP00000000, A. glaucus CBS516.65 - LSTL00000000, A. sydowii CBS593.65 - MRCH00000000, A. tubingensis CBS134.48 - LJXU00000000, A. versicolor CBS583.65 - MRBN00000000, A. wentii CBS141173 (DTO134E9) - LJSE00000000, and A. zonatus CBS506.65 - MRBM00000000. Links to the raw genome and transcriptome data of these species are available in Additional file 1D. This also lists the genbank (if available) and JGI accession numbers of the genomes used for comparison as well as the papers in which they have been published. Proteomics data of the wheat bran and sugar beet cultures are available via ProteomeXchange with identifier PXD005563. Proteomics data for the stress experiments are available at http://wwwuser.gwdg.de/~hkusch/GBIO_DeVries/.