ABSTRACT
Influenza virus RNA-dependent RNA polymerase consists of three viral protein subunits: PA, PB1, and PB2. Protein-protein interactions (PPIs) of these subunits play pivotal roles in assembling the functional polymerase complex, which is essential for the replication and transcription of influenza virus RNA. Here we developed a highly specific and robust bimolecular luminescence complementation (BiLC) reporter system to facilitate the investigation of influenza virus polymerase complex formation. Furthermore, by combining computational modeling and the BiLC reporter assay, we identified several novel small-molecule compounds that selectively inhibited PB1-PB2 interaction. Function of one such lead compound was confirmed by its activity in suppressing influenza virus replication. In addition, our studies also revealed that PA plays a critical role in enhancing interactions between PB1 and PB2, which could be important in targeting sites for anti-influenza intervention. Collectively, these findings not only aid the development of novel inhibitors targeting the formation of influenza virus polymerase complex but also present a new tool to investigate the exquisite mechanism of PPIs.
IMPORTANCE Formation of the functional influenza virus polymerase involves complex protein-protein interactions (PPIs) of PA, PB1, and PB2 subunits. In this work, we developed a novel BiLC assay system which is sensitive and specific to quantify both strong and weak PPIs between influenza virus polymerase subunits. More importantly, by combining in silico modeling and our BiLC assay, we identified a small molecule that can suppress influenza virus replication by disrupting the polymerase assembly. Thus, we developed an innovative method to investigate PPIs of multisubunit complexes effectively and to identify new molecules inhibiting influenza virus polymerase assembly.
KEYWORDS: influenza virus polymerase, protein-protein interactions, BiLC, influenza inhibitor screening
INTRODUCTION
Influenza virus (IAV) is a negative single-stranded RNA (ssRNA) virus belonging to the Orthomyxoviridae family (1). It has caused annual epidemics and some pandemics, including the 1918 Spanish flu (caused by H1N1), 1957 Asian flu (H2N2), 1968 Hong Kong flu (H3N2), and 2009 swine flu (reassorted H1N1) pandemics (2, 3). Small-molecule therapeutics targeting the M2-ion channel (amantadine and rimantadine) or neuraminidase (oseltamivir and zanamivir) were effective in suppressing influenza virus replication (1). However, the emergence of drug-resistant variants calls for novel therapeutics against influenza virus (4, 5). Moreover, the outbreak of highly pathogenic avian virus (H5N1 or H7N9) also highlights the need to develop new ways to combat influenza virus infections (6). The RNA-dependent RNA polymerase (RdRp) complex of influenza virus, responsible for RNA synthesis, is a heterotrimeric complex composed of three subunits—PA, PB1, and PB2 (7). Since the structure and function relationships of influenza virus polymerase have been well illustrated, the protein-protein interactions (PPIs) between influenza virus polymerase subunits have been shown to be potential drug targets for structure-based drug design (8–12).
PPIs play essential roles in many biological activities, such as signaling transduction, host-pathogen recognition, cell-cell interaction, and so on. These activities have been shown to occur in cells via stable and dynamic interactions (13). Stable protein interactions occur constitutively in cells, whereas dynamic interactions occur transiently and are often too weak to be detected. The dynamic interactions often act as biological regulators which are correlated to clinical diseases, such as breast cancer and autoimmunity diseases (14–16). Altered interactions are often a useful indicator of breast cancer progression (15). Disrupting the PPIs can often provide new avenues for finding potential therapeutics (14, 17). To date, many methods have been developed to monitor the protein-protein interactions and to screen antagonists of PPIs (18). Coimmunoprecipitation (co-IP) or pulldown analyses can be used to detect stable interactions, but they have low sensitivity for detecting weak or transient interactions (19). Surface plasmon resonance (SPR) and isothermal titration calorimetry (ITC) methods needed to purify interaction proteins are labor-intensive and time-consuming. The yeast 2-hybrid (Y2H) method has traditionally been used to determine protein interactions. However, it is unable to quantitatively determine whether the interaction is in the nucleus or the cytoplasm. The need to develop a novel assay to quantify the strength of PPI effectively is urgent, especially for dynamic interactions, which are always critical for protein function.
The protein fragment complementation assay (PCA) based on the use of split green fluorescent protein (GFP) or firefly luciferase (Fluc) has often been used to investigate the PPIs in vitro and in vivo, which are representative of PPIs between the purified proteins in a tube or those that happen in the cytoplasm or nucleus of cells or in living animals/plants, respectively (20–25). The marine copepod Gaussia princeps has the smallest (∼19.9-kDa) known luciferase (Gluc) molecule, which does not require other cofactors for activation (26, 27). A codon-optimized Gluc molecule has been widely used as a reporter in cultured mammalian cells (28). The sensitivity of Gluc is up to 2,000-fold higher than that of Renilla reniformis luciferase (Rluc) or firefly luciferase (Fluc), which is encoded by an important reporter gene (29). Features of PCA, including the detected interactions, are fully reversible, and the readout is easily detected (30). Those allow effective high-throughput screening of PPIs of antagonists. Moreover, in order to screen PPI inhibitors more effectively, we have developed a Tet on-bimolecule fluorescence complementation (Tet on-BiLC) system, by combining the BiLC assay and Tet in an inducible expression system, which expresses targeted proteins controlled by an inducer. As result, it will also improve protein folding (31–33).
Here we developed the Tet on-BiLC assay to detect the influenza virus polymerase assembly and to screen novel therapeutics methods that inhibit influenza virus polymerase assembly. By combining in silico modeling data, we identified 8 molecules that bound to the hydrophobic patch of PB1c. We found that molecule 5 suppressed influenza virus replication by specifically disrupting the PB1-PB2 interaction. The Tet on-BiLC system was also used to construct a reporter cell line to illustrate the effects of the reported peptide inhibitors targeting the polymerase assembly and to investigate the mechanism of influenza virus polymerase assembly in vivo. Our new system provides us new tools to detect the PPIs and screen the antagonist of the PPIs.
RESULTS
Detecting the influenza virus polymerase interaction in vivo using the new BiLC assay.
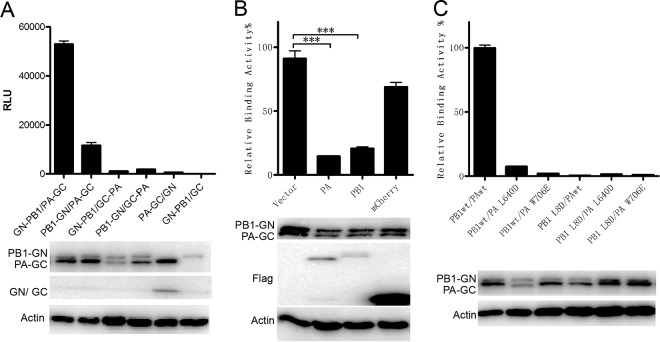
Previous studies have illustrated strong interactions between influenza virus polymerase subunits PA and PB1 or subunits PB1 and PB2 using co-IP, a pulldown assay, or the bimolecular fluorescence complementation (BiFC) assay. PCAs, including BiFC, were frequently used to reveal the dynamics of PPIs in vivo and in vitro (20, 22, 23). The full potential of PCA requires assays that are fully reversible and sensitive at subendogenous protein expression levels. In order to investigate the influenza virus polymerase assembly and screen inhibitors targeting the polymerase assembly more efficiently, we constructed a new PCA that meets these criteria, based on the Gaussia princeps luciferase enzyme, which is much more sensitive than Renilla luciferase (26). First, Gaussia luciferase was divided into two segments at position 109. The N terminus (GlucN, positions 17 to 109, named GN) and C terminus (GlucC, positions 110 to 185, named GC) were cloned onto lentivirus vector FG12 (34) with a cytomegalovirus (CMV) promoter (see Fig. S1 in the supplemental material), which is based on the Tet-on method (33), to ensure that expression of the fusion protein would be induced by doxycycline in a stable cell line. The newly developed BiLC assay was then named the Tet on-BiLC assay. To test the efficacy of the assay, interactions between influenza virus subunits were investigated. Since influenza virus PB1 interacted with PA and PB2 independently, PB1 was fused with the N or C terminus of GN by Gateway cloning (see Fig. S1A and C), and the product was named PB1-GN or GN-PB1, respectively, whereas PA was fused with the N or C terminus of GC and then the products, termed PA-GC or GC-PA, respectively, were coexpressed in 293T cells (see Fig. S1B and D). The results showed that the combinations of GN-PB1 and PA-GC fusion proteins had the highest luminescence signal, which confirmed previous findings (35, 36) showing that the N terminus of PB1 binds to the C terminus of PA (Fig. 1A; see also Fig. S2A and B). To test binding specificity, Flag-tagged wild-type (WT) PA or PB1 was coexpressed with GN-PB1 and PA-GC. Compared with the negative control (Flag-mCherry), Flag-tagged PA and PB1 outcompeted GN-PB1 or PA-GC and reduced the bioluminescence signal of the PA-PB1 interaction (Fig. 1B; see also Fig. S2C), suggesting that the PA-PB1 interaction detected using the BiLC assay was very specific. To further investigate the sensitivity and specificity of our assay, we examined the effects of PB1 L8D, PA L640D, and PA W706E mutations on the PA-PB1 interaction. As expected, mutations PB1 L8D, PA L640D, and PA W706E abolished the PA-PB1 interaction (Fig. 1C; see also Fig. S3A), which is in agreement with previous results determined by a pulldown assay (35, 36).
FIG 1.
The specificity and sensitivity of the BiLC assay is used to illustrate the PA-PB1 interaction. (A) The interactions between influenza virus PA and PB1 were detected using the BiLC assay. PA and PB1 were fused to the N or C terminus of GC and GN, respectively. The interactions between the indicated pairs of fusion proteins were detected using the BiLC assay. The relative luminescence units (RLU) were determined by the use of a microplate reader. (B) Plasmids expressing GN-PB1 and PA-GC fusion proteins were cotransfected with empty vector or with vector expressing Flag-PA, Flag-PB1, or Flag-mCherry. The RLU values of cell lysates were determined by the use of a microplate reader at 24 h posttransfection. The RLU value for samples containing vector was set to 100%. (C) The effects of site mutations on PA (L640D or W706E) or PB1 (L8D) on the interaction between PA and PB1 were detected using the BiLC assay. The expression of proteins was detected by Western blotting as indicated. Data are shown as means ± standard errors of the means (SEM). ***, P < 0.001 (Student's t test).
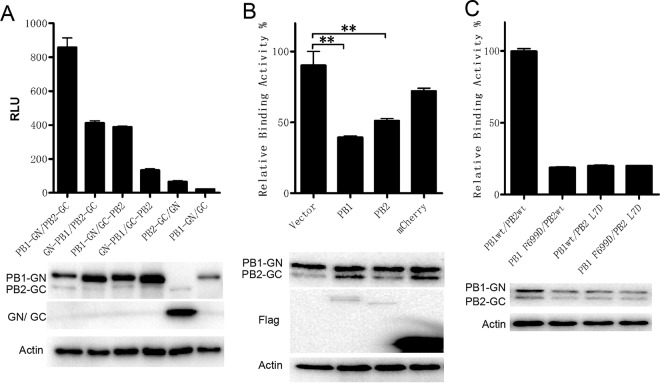
The interaction between PB1 and PB2 was also investigated using this BiLC assay. PB2 was fused with the N or C terminus of GC, termed PB2-GC or GC-PB2 (see Fig. S1B and D), and the products were then coexpressed with PB1-GN or GN-PB1 in 293T cells, respectively. The result showed significant interaction between PB1 and PB2 (Fig. 2A). The combination of PB1-GN and PB2-GC showed the highest level of signal, which is in agreement with previous reports showing that GFP on the N terminus of PB1 interfered with influenza virus polymerase activity (37, 38). Moreover, Flag-tagged PB1 or PB2 but not mCherry interfered with PB1-GN/PB2-GC complex bioluminescence specifically (Fig. 2B). More importantly, effects of mutants PB2 L7D and PB1 F699D on the PB1-PB2 interaction were tested. The mutant of PB2 L7D efficiently disrupted PB1-PB2 binding, and PB1 F699D interfered with PB1-PB2 binding, which was reduced by 80% (Fig. 2C; see also Fig. S3B), which is consistent with a previous report (39), indicating that there may be other regions responsible for the PB1-PB2 interaction (10, 11, 40). These results suggest that this BiLC assay is specific and sensitive in identifying the influenza virus polymerase interaction.
FIG 2.
The specificity and sensitivity of the BiLC assay were used to illustrate the PB1-PB2 interaction. (A) The interactions between influenza virus PB1 and PB2 were detected using the BiLC assay. PB2 and PB1 were fused to the N or C terminus of GC and GN, respectively. The interactions between the indicated pairs of fusion proteins were detected using the BiLC assay. The relative luminescence units (RLU) were determined by the use of a microplate reader. (B) Plasmids expressing PB1-GN and PB2-GC were cotransfected with empty vector or with vector expressing Flag-PB1, Flag-PB2, or Flag-mCherry. The RLU values of cell lysates were determined by the use of a microplate reader at 24 h posttransfection. The RLU value for samples containing vector was set to 100%. (C) The effects of site mutations on PB1 (F699D) or PB2 (L7D) on interactions between PB1 and PB2 were detected using the BiLC assay. The expression of proteins was detected by Western blotting as indicated. Data are shown as means ± SEM. **, P < 0.01 (Student's t test).
Finding novel small-molecule inhibitors of influenza virus polymerase assembly by combining in silico modeling and BiLC assay.
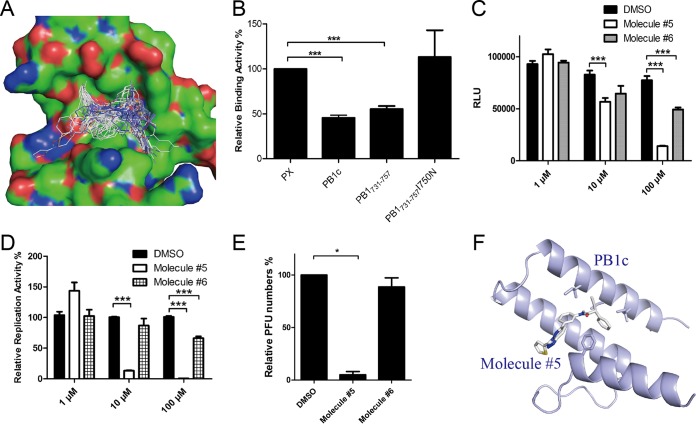
Previous studies showed that the assembly of the influenza virus polymerase complex could be a potential drug target for suppression of viral replication. Sugiyama et al. found that PB1c-PB2n interaction was mainly mediated by 2 residues on the hydrophobic patch of PB1c (39), although there may have been other regions on PB1 or PB2 that contributed to the PB1-PB2 interaction (10, 11, 40). And our previous studies showed that PB1731–757 bound to the hydrophobic patch of PB1c to inhibit influenza virus replication (12). These results indicated that the hydrophobic patch formed by PB1c could be a potential drug target for screening inhibitors. To investigate that conjecture, we combined in silico screening methods that we had developed previously (41, 42) and the BiLC method described above to find potential small molecules targeting the hydrophobic patch of PB1c (see Fig. S4A and B). Eight molecules that specifically target the PPIs from a target-focused library of ∼30,000 molecules were found to bind efficiently to the hydrophobic patch of PB1c (Fig. 3A).
FIG 3.
Finding novel anti-influenza virus molecules by combining in silico modeling and Teton-BiLC assay. (A) The PB1c binding model of the top 100 hits from in silico modeling. (B) Relative levels of binding activity between PB1c and PB2n in the presence of peptides PX, PB1c, PB1731–737, and PB1731–737I750N. PX and PB1c were used as negative and positive controls, respectively. (C) The inhibitory function of molecules 5 and 6 for PB1c-PB2n interactions detected using the BiLC assay. (D) IAV-Luc replication was inhibited by molecules 5 and 6. A549 cells were pretreated with molecules at 10 or 100 μM for 2 h. At 24 h postinfection, luminescence from cell supernatant was detected by the use of a microplate reader. (E) The inhibitory effects of molecules 5 and 6 on WSN33 virus detected by plaque assay. A549 cells were pretreated with molecules at 10 μM for 2 h. At 24 h postinfection, virus titers in supernatants were determined by plaque assay. Plaque numbers in samples with DMSO were set to 100%. (F) Structure model of molecule 5 binding to PB1c. Data are shown as means ± SEM. *, P < 0.05; ***, P < 0.001 (Student's t test).
In order to screen inhibitors targeting the PB1c-PB2n interaction more effectively, the Tet on-BiLC assay was used in 293T cells instead since luciferase is more sensitive than GFP. PB1c and PB2n were fused to both the N and C termini of GN and GC, respectively, to improve the sensitivity of the method (see Fig. S1). The level of the PB1c-PB2n interaction was much higher than that seen with the controls (see Fig. S4C). A PB1c-PB2n BiLC cell line was constructed and induced by doxycycline in a dose- and time-dependent manner (see Fig. S4D and E). In order to test whether this method can be used to detect the inhibitors of the PB1c-PB2n association, GFP-fused PB1c was coexpressed with the interaction pairs GN-PB1c and GC-PB2n. Figure 3B shows that PB1c disrupted the interaction between PB1c and PB2n. It was the same with PB1731–757 but not with its I750N mutants. The results were correlated with a previous report that PB1731–757 inhibited the PB1c-PB2n association by binding to PB1c whereas a PB1731–757I750N mutant did not (12). The results indicated that the Tet on-BiLC assay can be used to screen inhibitors of the PB1-PB2 association.
We then verified the inhibitory function of these 8 molecules with respect to the PB1c-PB2n interaction using the BiLC assay (see Materials and Methods). The result showed that 4 of the 8 molecules inhibited PB1c-PB2n at 100 μM and that molecule 5 impaired the PB1c-PB2n interaction even at 10 μM (Fig. 3C; see also Fig. S5), while molecules 3 and 8 had cell toxicity.
Furthermore, IAV-Luc with a luciferase reporter on the NA gene was used to verify the inhibitory function of molecules 5 and 6 with respect to influenza virus replication. We showed that molecule 5 suppressed virus replication by more than 80% at 10 μM and that molecule 6 inhibited virus replication by about 30% at 100 μM (Fig. 3D), while the other molecules were unable to suppress influenza virus replication at 10 μM (see Fig. S6). In agreement with the results, molecule 5 inhibited replication of WSN33 virus by more than 90% as detected using the plaque assay (Fig. 3E; see also Fig. S7B). It had no cytotoxicity for cells at 10 μM (see Fig. S7C). In order to understand the inhibitory mechanism of molecule 5 with respect to the PB1c-PB2n interaction, a model of binding of molecule 5 to the PB1c′ hydrophobic patch was constructed (Fig. 3F; see also Fig. S7A). Similarly to the binding of PB2n with PB1c, molecule 5 was predicted to interact with F695, F703, and I750 on PB1c's hydrophobic patch. These results indicated that our BiLC assay can be used to screen novel inhibitors targeting influenza virus polymerase activity.
Molecule 5 inhibits PB1-PB2 assembly specifically.
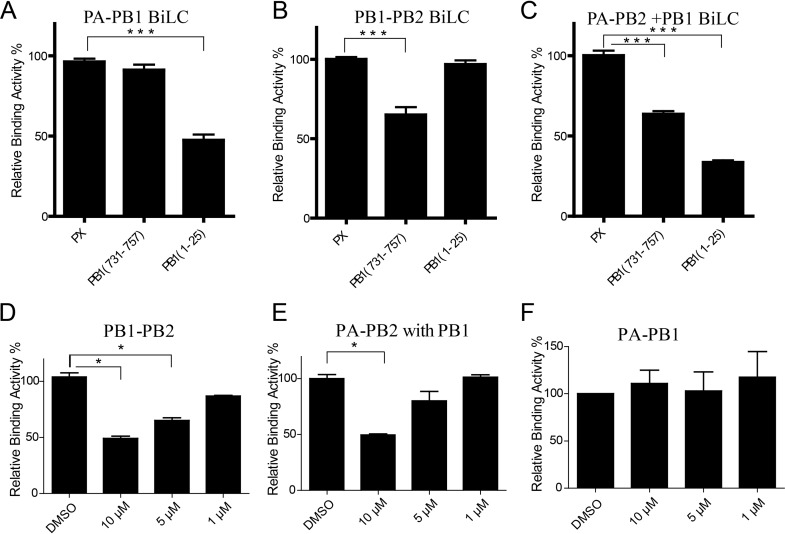
In order to investigate the specificity inhibitory function of molecule 5 with respect to the PB1c-PB2n interaction, its effects on PB1-PB2 and PA-PB1 interactions and even heterotrimer assembly were further detected by our BiLC assay. Since previous studies found that PB11–25 could suppress the PA-PB1 interaction by interacting with PA and that PB1731–757 could suppress the PB1-PB2 interaction by binding to PB1c (12, 43–45), PB11–25 and PB1731–757 were used as controls to test whether the BiLC method could be used to illustrate and identify potential inhibitors that target influenza virus polymerase assembly. First, PB11–25 and PB1731–757 were cloned onto the N terminus of GFP, which facilitates the expression of the peptide. PX, an unrelated peptide from a Borna disease virus phosphoprotein, was used as a negative control (44). Then, peptide-GFP-expressing plasmids were cotransfected with PA-GC- and GN-PB1-expressing plasmids into 293T cells. We found that PB11–25 did inhibit the PA-PB1 interaction efficiently but not PB1731–757 (Fig. 4A), which is in agreement with reports from previous studies that PB11–25 bound to PA whereas PB1731–757 did not (44). Similarly, we also tested the inhibitory effects of PB1731–757 on the PB1-PB2 interaction. As expected, PB1731–757 interfered with the PB1-PB2 interaction but PB11–25 did not (Fig. 4B).
FIG 4.
Testing the inhibitory functions of influenza A virus polymerase-derived peptides with respect to influenza virus polymerase interaction via BiLC. (A to C) Plasmids expressing GN-PB1 and PA-GC (A), PB1-GN and PB2-GC (B), or PA-GC, PB2-GN, and Flag-PB1 (C) were cotransfected with plasmids expressing GFP-fused PX, PB1731–757, or PB11–25, respectively. The RLU values of cell lysates were determined by the use of a microplate reader at 24 h posttransfection. The RLU value of samples containing PX was set to 100%. (D to F) The inhibitory function of molecule 5 with respect to PB1-PB2 interaction (D), PA-PB2 interaction in the presence of PB1 (E), and PA-PB1 interaction (F) detected using the BiLC assay. Data are shown as means ± SEM. *, P < 0.05; ***, P < 0.001 (Student's t test).
To test the inhibitory effects of PB11–25 and PB1731–757 on the PA-PB2 interaction, we also investigated the PA-PB2 interaction using the BiLC method. PB2 was fused onto the N or C terminus of GN (see Fig. S1) and coexpressed with PA-GC or GC-PA. The results showed that there was no interaction between PA and PB2 (see Fig. S8). To improve the coverage of the detection, PA was fused with GN and PB2 with GC. However, interactions between PA and PB2 were still undetectable. The results correlate with the fact that there is no direct interaction between PA and PB2 observed from the crystal structure of influenza virus polymerase (10, 11). PA and PB2 were packed in close proximity, as indicated from the cryo-electron microscopy (CryoEM) structure of the viral ribonucleoprotein (vRNP) complex (46). Furthermore, previous reports found that PB1 was the core subunit since it firmly binds to PA and PB2 via the N and C termini, respectively (10, 11, 47). It has been shown that PA and PB1 first form a heterodimer in cytoplasm and then form the active heterotrimer complex with PB2 in the nucleus in vitro (37). So it was interesting to investigate whether there was an interaction between PA and PB2 when PB1 was coexpressed in vivo using our BiLC assay. To address this issue, PB1-Flag was coexpressed with PA-GC and PB2-GN in 293T cells. Interestingly, we found there were interaction signals between PA and PB2 when PB1 was coexpressed, which indicated that the PA-PB2 interaction requires the PB1 protein in vivo (Fig. 5A), and the interaction signals reached their highest level when GC and GN were fused to the C termini of PA and PB2, respectively (see Fig. S9A). We then tested the inhibitory functions of PB11–25-GFP and PB1731–757-GFP with respect to assembly of PA-PB2 in the presence of PB1, representing the heterotrimer complex assembly. We discovered that both PB11–25-GFP and PB1731–757-GFP suppressed the heterotrimer complex assembly (Fig. 4C). As shown in Fig. 4D to F, in agreement with the results showing that molecule 5 binds to the hydrophobic patch of PB1c, molecule 5 suppressed PB1-PB2 interaction and heterotrimer assembly (Fig. 4D and E) whereas it was unable to disrupt the PA-PB1 interaction at 10 μM (Fig. 4F). The data reveal that our methods can be used for screening inhibitors targeting influenza virus polymerase assembly specifically and effectively.
FIG 5.
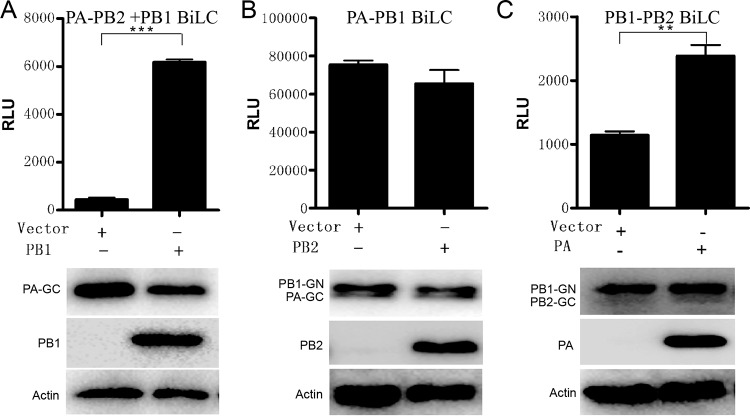
Illustration showing the assembly mechanism of influenza virus RNA-dependent RNA polymerase (RdRp) using BiLC. Plasmids expressing PA-GC and PB2-GN (A), GN-PB1 and PA-GC (B), or PB1-GN and PB2-GC (C) were cotransfected with empty vector or with vector expressing Flag-PB1 (A), Flag-PB2 (B), or Flag-PA (C), respectively. The RLU values of cell lysates were determined by the use of a microplate reader at 24 h posttransfection. The levels of protein expression were detected by Western blotting as indicated. Data are shown as means ± SEM. **, P < 0.01; ***, P < 0.001 (Student's t test).
Illustrating the assembly of influenza virus polymerase subunits in vivo using the BiLC assay.
We have found that the PA-PB2 interaction is based on the presence of PB1 in vivo (Fig. 5A), and it has been shown in vitro that PB1 dimerizes with PA in the cytosol and is then imported into the nucleus as a dimer and separately forms a heterotrimer complex with the PB2 subunit, which was imported into the nucleus with Hsp70 from the cytosol (37). However, it is unclear whether the interaction between PA and PB1 had been influenced by the presence of PB2 or whether that between PB1 and PB2 was influenced by PA in vivo. To address these issues, Flag-tagged PA with PB2-GC and GN-PB1 or Flag-tagged PB2 was coexpressed with PA-GC or PB1-GN in 293T cells, respectively. Gaussia luciferase activity was measured 24 h posttransfection. The results showed that the presence of PA improved the PB1-PB2 interaction, whereas PB2 did not interfere with the PA-PB1 interaction no matter what kind of fusion pattern was used (Fig. 5B and C; see also Fig. S9B and C). Our in vivo interaction results were in agreement with previous results detected in vitro: PA and PB1 first formed a dimer in the cytoplasm and then formed the active heterotrimer complex with PB2 in the nucleus (37). The results indicated that the new BiLC assay can also be used to investigate the mechanism of influenza virus polymerase assembly in cell lines.
Constructing the BiLC-reporter cell line used for detecting the inhibitors of influenza virus polymerase assembly.
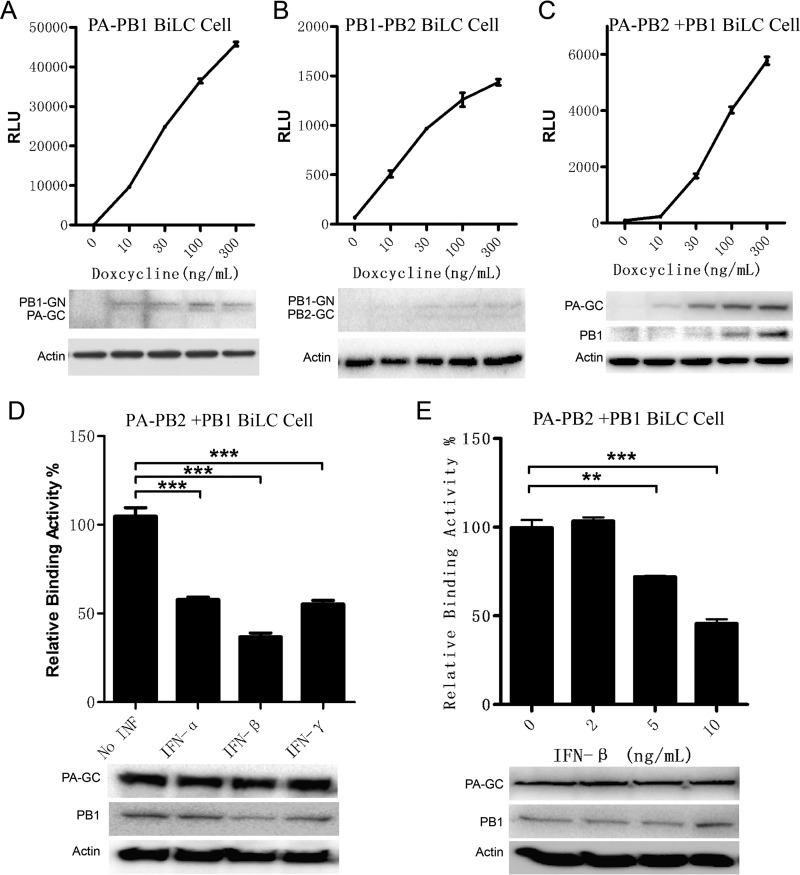
To monitor influenza virus polymerase assembly more efficiently and to facilitate the screening of small molecules targeting the polymerase assembly, stable cell lines carrying PA-GC and PB1-GN were constructed using a lentiviral system. Plasmids expressing PA-GC and PB1-GN were packaged with third-generation packaging plasmids into HIV-1 lentivirus (see Materials and Methods), offering optimal biohazard safety and improved efficiency compared to the packaging system using two plasmids (48). Then, 293T cells with Tet on expression were infected by the lentivirus as described above. The stable cell line, named the PA-PB1 reporter cell line, was obtained after puromycin and blasticidin S deaminase (BSD) coselection. Figure 6A shows that the binding activity between PA and PB1 was detected in a doxycycline-dose-dependent manner. The expression levels of PA-GC and PB1-GN were also shown to be dose dependent (Fig. 6A), which indicated that the PA-PB1 reporter cell line can be used to detect the assembly of PA and PB1.
FIG 6.
Construction of Tet on-BiLC stable cell lines for detecting the interactions between influenza virus polymerase subunits. 293T stable cell lines expressing GN-PB1 and PA-GC (A), PB1-GN and PB2-GC (B), and PA-GC, PB2-GN, and Flag-PB1 (C) were constructed using the Tet on system. The stable cell lines were induced by doxycycline, and RLU values of cell lysates were determined by the use of a microplate reader at 24 h postinduction. The levels of protein expression were detected by Western blotting as indicated. (D) The inhibitory functions of interferon alpha (IFN-α), IFN-β, and IFN-γ with respect to the influenza virus polymerase assembly were detected with the stable cell line expressing PA-GC, PB2-GN, and PB1-Flag. The stable cell lines were incubated with the indicated interferons (IFN-α and IFN-γ, 1,000 U/ml; IFN-β, 10 ng/ml) for 24 h and then induced by doxycycline (300 ng/ml) for another 24 h. The RLU values of cell lysates were determined by the use of a microplate reader at 24 h postinduction. The levels of protein expression were detected by Western blotting as indicated. (E) The dose-dependent effects of IFN-β on the influenza virus polymerase assembly were detected with the stable cell line expressing PA-GC, PB2-GN, and PB1-Flag. Data are shown as means ± SEM. **, P < 0.01; ***, P < 0.001 (Student's t test).
Using the same method, the PB1-PB2 stable cell line was set up to detect the PB1-PB2 interaction efficiently. As expected, the PB1-PB2 interaction was closely monitored via doxycycline induction (Fig. 6B). The PA-PB2 stable cell line in the presence of PB1 (named the PA-PB2+PB1 BiLC cell line) was also set up to detect the PA-PB2 interaction which is dependent on the presence of PB1 (Fig. 6C).
We investigated how to use the stable cell line for detecting influenza virus polymerase assembly modulators, since interferons can induce expression of at least 300 kinds of interferon-stimulated genes (ISGs) and some ISGs may act directly on influenza virus proteins (49–51). Here we tested the inhibitory function of interferons with respect to the PA-PB2+PB1 BiLC reporter cells. As expected, the results showed that interferons alpha, beta, and gamma suppressed the interaction signal efficiently (Fig. 6D). And the inhibitory effect of IFN-β on the PA-PB2+PB1 interaction signal occurred in a dose-dependent manner (Fig. 6E). It appears that IFNs can induce ISGs, inhibiting expression of influenza virus proteins or disrupting assembly of influenza virus polymerase subunits.
Our results not only revealed the mechanism of influenza virus polymerase assembly in vivo but also provided a novel strategy for screening for small-molecule inhibitors as drug leads targeting influenza virus polymerase assembly.
DISCUSSION
Here we developed a sensitive and specific assay to interrogate the mechanism of and to screen inhibitors for dynamic protein-protein interactions (PPI) controlling the assembly of multisubunit protein complexes. Influenza virus polymerase was taken as an example in this study since the domains involved in the interactions have been extensively characterized and the structure and function of influenza virus polymerase are highly conserved (9–11, 47). Influenza virus polymerase assembly is a potential drug target to combat influenza virus replication (8). Although small-molecule inhibitors or peptide inhibitors were found to suppress influenza virus replication by targeting polymerase assembly (44, 52, 53), the methods used to illustrate or screen these inhibitors, such as co-IP, pulldown assays, and enzyme-linked immunosorbent assays (ELISA), are labor-intensive and time-consuming. The Gaussia luciferase activity-based BiLC assay allowed us to screen PPI inhibitors using a high-throughput method.
Actually, BiLC assay can be used for screening inhibitors targeting other PPIs. Inhibitors such as PB11–25 and PB1481–515 have been shown to suppress different strains of influenza virus (12, 43), indicating that inhibitory compounds targeting the interaction surface of influenza virus polymerase likely have broad efficacy. Here PB1c and PB2n were used to mimic the PB1-PB2 interaction since the PB1c-PB2n interaction mainly mediates the PB1-PB2 interaction and, more importantly, the PB1c-PB2n interaction is much stronger than the PB1-PB2 interaction (Fig. 2A; see also Fig. S4C). By combining the data with in silico screening, we found inhibitors derived from large amounts of compounds. At last, we obtained a lead compound (molecule 5) that could substantially inhibit influenza virus replication. However, further computer-based drug design/modification is clearly needed to improve the inhibitory effects of this lead compound.
Prior to this current study, the assembly of influenza virus polymerase was revealed largely by using pulldown assay and colocalization analysis. However, the dynamic PPIs underlying the assembly of the influenza virus polymerase in vivo remain unclear (37). Hemerka et al. first reported weak binding between PA and PB2 (using the BiFC assay) (19). Our results revealed that the PA-PB2 interaction is dependent on the presence of a PB1 subunit (Fig. 5A). These results are in agreement with the fact that, based on the crystal structure of the influenza polymerase complex, PA and PB2 are in very close proximity but do not interact directly. In our BiLC disruption assay, due to instability of influenza virus polymerase subunits, it appears that the expression levels of the proteins were reduced by degradation due to the fact that the interactions were disrupted (Fig. 1B and C).
In conclusion, our Tet on-BiLC system provides a new and effective tool to screen antagonists targeting multisubunit protein complexes involving weak or transient PPIs inside living cells and can facility drug development.
MATERIALS AND METHODS
Cells and plasmids.
HEK293T, A549, and MDCK cells were cultured with Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS). The compounds were purchased from J&K Scientific Ltd. Plasmids expressing PX, PB11–25, PB1731–757, PB1731–757I750N, and PB1c-GFP fusion proteins were described elsewhere (12). WSN33 virus-derived PB1, PA, and PB2 were cloned onto pGateway-Flag (Thermo Fisher) expressing Flag fusion proteins. BiLC vectors were cloned from ENTRY vectors by Gateway cloning. IAV-Luc was kindly provided by Chen Ling (Guangzhou Biomedicine and Health Institute, Chinese Academy of Science) and propagated in chicken eggs.
BiLC assay.
HEK293T cells were transfected by polyethylenimine (PEI; Sigma). At 24 h posttransfection, cells were lysed and levels of luminescence were measured using a Renilla luciferase assay kit (E2820; Promega) and a microplate reader (Bio-Tek) following the instructions of the manufacturers. A 293T (PA/PB1/PB2) BiLC stable cell line was induced with doxycycline for 24 h or was treated with IFN-α, IFN-β, or IFN-γ (PeproTech) at the indicated concentrations for 12 h followed by induction with doxycycline for 24 h. Luminescence levels were measured using a Renilla luciferase assay kit and a microplate reader.
Site mutation.
Plasmids with certain site mutations were amplified from templates with PfuUltra (Stratagene) and then digested with DpnI (NEB). Clones with positive results were selected by sequencing.
Lentivirus packaging assay.
Tet on-3G and PA/PB1/PB2 BiLC expression vectors are based on FG12 lentiviral vector (34, 48). Pseudolentiviruses were generated by the use of the third lentiviral packaging system in 293T cells (48). Tet on-3G stable-expression cells were generated in 293T cells first, and then TRE-PA/PB1/PB2 BiLC vectors were stably delivered.
Western blotting.
Cells were lysed by the use of lysis buffer from a Renilla luciferase assay kit (E2820; Promega). The cell lysates were centrifuged at 15,000 × g for 10 min at 4°C, and the resulting supernatant was boiled with SDS loading buffer. Proteins were transferred to polyvinylidene difluoride (PVDF) membranes (Millipore) followed by further incubation with rabbit anti-Gluc antibody (E8023S; NEB), anti-Flag horseradish peroxidase (HRP)-conjugated antibody (Sigma), or anti-beta actin antibody (A00730-40; GenScript). A LumiGlo chemiluminescent substrate system (Millipore) was used for protein detection.
In silico modeling.
From 28,900 compounds listed in the J&K compound database as potentially targeting PPIs, we selected three subsets (using similarity, machine learning methods [MLM], and the Rule of Four) for virtual screening. Initially, we generated the three-dimensional (3D) structures of those compounds using OpenBabel software (54). And we optimized the target structure of PB1c (PDB code 2ZTT) by the use of Molecular Graphics Laboratory (MGL) tools (mgl.scripps.edu). The binding pocket was selected to be the interface of PB1c and PB2n with a search space size of 18Å by 18 Å by 20 Å. After preprocessing, a PB1 C-terminal fragment and the 28,900 potential molecules were submitted to AutoDock Vina (55) for the first-round virtual screening using default parameters. These compounds were ranked according to their affinity to PB1c. The top 1,000 compounds were selected for recomputation using more-exhaustive parameters. (The value corresponding to the exhaustiveness of the global conformational search was increased to 12 in contrast to the former value of 8.) The docking result was reranked by Cyscore (42), and the final compounds were selected manually among the top 200 compounds as the structures with the best complementarity to PB1c.
Plaque assay.
The compounds were diluted in dimethyl sulfoxide (DMSO) and added into medium containing A549 cells at different concentrations for 2 h before infection. Cells were washed once using phosphate-buffered saline (PBS) and were then infected with WSN33 or IAV-Luc (56) at a multiplicity of infection (MOI) of 0.01 and incubated with the compounds for 1 h. The cells were then washed once using PBS and incubated with the compounds for 24 h. Supernatants were taken for analysis of luciferase activity by the use of a microplate reader or plaque assay. Plaque assays were performed on MDCK cells. The plaques were counted after staining with crystal violet at 3 days postinfection. The luciferase activity or the plaque number from the group treated with DMSO only was set to 100%.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the CAMS Initiative for Innovative Medicine (no. 2016-I2M-1-005), a Ministry of Health grant (no. 201302018), the National Major Scientific and Technological Special Project for “Significant New Drugs Development” (no. 2015ZX09102023), a National Natural Science Foundation of Key Projects grant (no. 81590765) to F. Xiao-Feng Qin, National Natural Science Foundation of China (no. 31500145) and PUMC Youth Fund (no. 3332016125) to Chunfeng Li, the Institutional Research Fund for Thousand Talents Program at Chinese Academy of Medical Sciences and the research special fund for public welfare industry of health (no. 201302018), a National Natural Science Foundation of China grant (no. 31170832) to Genhong Cheng, National Basic Research Program of China (no. 2015CB910501) and National Natural Science Foundation of China (no. 31671371) grants to Taijiao Jiang, and a National Natural Science Foundation of China grant (no. 31401130) to Yang Cao.
We thank Ling Chen for providing IAV-Luc and members of our laboratories for helpful discussions and technical assistance.
F.X.-F.Q., G.C., and T.J. jointly designed and directed the research. C.L. and Z.W. performed the major part of the experiments; Y.C. performed computational analysis; L.W., J.J., Z.C., and T.D. performed supporting experiments and contributed to manuscript revision. F.X.-F.Q. and C.L. analyzed data and wrote the paper.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.02282-16.
REFERENCES
- 1.Salomon R, Webster RG. 2009. The influenza virus enigma. Cell 136:402–410. doi: 10.1016/j.cell.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tumpey TM, Belser JA. 2009. Resurrected pandemic influenza viruses. Annu Rev Microbiol 63:79–98. doi: 10.1146/annurev.micro.091208.073359. [DOI] [PubMed] [Google Scholar]
- 3.Hampson AW, Mackenzie JS. 2006. The influenza viruses. Med J Aust 185:S39–43. [DOI] [PubMed] [Google Scholar]
- 4.de Jong JC, Beyer WE, Palache AM, Rimmelzwaan GF, Osterhaus AD. 2000. Mismatch between the 1997/1998 influenza vaccine and the major epidemic A(H3N2) virus strain as the cause of an inadequate vaccine-induced antibody response to this strain in the elderly. J Med Virol 61:94–99. [PubMed] [Google Scholar]
- 5.Du X, Dong L, Lan Y, Peng Y, Wu A, Zhang Y, Huang W, Wang D, Wang M, Guo Y, Shu Y, Jiang T. 2012. Mapping of H3N2 influenza antigenic evolution in China reveals a strategy for vaccine strain recommendation. Nat Commun 3:709. doi: 10.1038/ncomms1710. [DOI] [PubMed] [Google Scholar]
- 6.Su S, Bi Y, Wong G, Gray GC, Gao GF, Li S. 2015. Epidemiology, evolution, and recent outbreaks of avian influenza virus in China. J Virol 89:8671–8676. doi: 10.1128/JVI.01034-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boivin S, Cusack S, Ruigrok RW, Hart DJ. 2010. Influenza A virus polymerase: structural insights into replication and host adaptation mechanisms. J Biol Chem 285:28411–28417. doi: 10.1074/jbc.R110.117531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boltz DA, Aldridge JR Jr, Webster RG, Govorkova EA. 2010. Drugs in development for influenza. Drugs 70:1349–1362. doi: 10.2165/11537960-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Resa-Infante P, Jorba N, Coloma R, Ortin J. 2011. The influenza virus RNA synthesis machine: advances in its structure and function. RNA Biol 8:207–215. doi: 10.4161/rna.8.2.14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reich S, Guilligay D, Pflug A, Malet H, Berger I, Crepin T, Hart D, Lunardi T, Nanao M, Ruigrok RW, Cusack S. 2014. Structural insight into cap-snatching and RNA synthesis by influenza polymerase. Nature 516:361–366. doi: 10.1038/nature14009. [DOI] [PubMed] [Google Scholar]
- 11.Pflug A, Guilligay D, Reich S, Cusack S. 2014. Structure of influenza A polymerase bound to the viral RNA promoter. Nature 516:355–360. doi: 10.1038/nature14008. [DOI] [PubMed] [Google Scholar]
- 12.Li C, Ba Q, Wu A, Zhang H, Deng T, Jiang T. 2013. A peptide derived from the C-terminus of PB1 inhibits influenza virus replication by interfering with viral polymerase assembly. FEBS J 280:1139–1149. doi: 10.1111/febs.12107. [DOI] [PubMed] [Google Scholar]
- 13.Nooren IM, Thornton JM. 2003. Diversity of protein-protein interactions. EMBO J 22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loregian A, Marsden HS, Palu G. 2002. Protein-protein interactions as targets for antiviral chemotherapy. Rev Med Virol 12:239–262. doi: 10.1002/rmv.356. [DOI] [PubMed] [Google Scholar]
- 15.Taylor IW, Linding R, Warde-Farley D, Liu Y, Pesquita C, Faria D, Bull S, Pawson T, Morris Q, Wrana JL. 2009. Dynamic modularity in protein interaction networks predicts breast cancer outcome. Nat Biotechnol 27:199–204. doi: 10.1038/nbt.1522. [DOI] [PubMed] [Google Scholar]
- 16.Li S, Wang L, Berman M, Kong YY, Dorf ME. 2011. Mapping a dynamic innate immunity protein interaction network regulating type I interferon production. Immunity 35:426–440. doi: 10.1016/j.immuni.2011.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivanov AA, Khuri FR, Fu H. 2013. Targeting protein-protein interactions as an anticancer strategy. Trends Pharmacol Sci 34:393–400. doi: 10.1016/j.tips.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Berggård T, Linse S, James P. 2007. Methods for the detection and analysis of protein-protein interactions. Proteomics 7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 19.Hemerka JN, Wang D, Weng Y, Lu W, Kaushik RS, Jin J, Harmon AF, Li F. 2009. Detection and characterization of influenza A virus PA-PB2 interaction through a bimolecular fluorescence complementation assay. J Virol 83:3944–3955. doi: 10.1128/JVI.02300-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu CD, Chinenov Y, Kerppola TK. 2002. Visualization of interactions among bZIP and Rel family proteins in living cells using bimolecular fluorescence complementation. Mol Cell 9:789–798. doi: 10.1016/S1097-2765(02)00496-3. [DOI] [PubMed] [Google Scholar]
- 21.Hu CD, Grinberg AV, Kerppola TK. 2006. Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Curr Protoc Cell Biol 29:21.3:21.3.1–21.3.21. doi: 10.1002/0471143030.cb2103s29. [DOI] [PubMed] [Google Scholar]
- 22.Eglen RM. 2002. Enzyme fragment complementation: a flexible high throughput screening assay technology. Assay Drug Dev Technol 1:97–104. doi: 10.1089/154065802761001356. [DOI] [PubMed] [Google Scholar]
- 23.Kerppola TK. 2009. Visualization of molecular interactions using bimolecular fluorescence complementation analysis: characteristics of protein fragment complementation. Chem Soc Rev 38:2876–2886. doi: 10.1039/b909638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leng W, Pang X, Xia H, Li M, Chen L, Tang Q, Yuan D, Li R, Li L, Gao F, Bi F. 2013. Novel split-luciferase-based genetically encoded biosensors for noninvasive visualization of Rho GTPases. PLoS One 8:e62230. doi: 10.1371/journal.pone.0062230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piehler J. 2005. New methodologies for measuring protein interactions in vivo and in vitro. Curr Opin Struct Biol 15:4–14. doi: 10.1016/j.sbi.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 26.Chopra A. 2004. Gaussia princeps luciferase. Molecular Imaging and Contrast Agent Database (MICAD), Bethesda, MD. [Google Scholar]
- 27.Remy I, Michnick SW. 2006. A highly sensitive protein-protein interaction assay based on Gaussia luciferase. Nat Methods 3:977–979. doi: 10.1038/nmeth979. [DOI] [PubMed] [Google Scholar]
- 28.Hossain MJ, Perez S, Guo Z, Chen LM, Donis RO. 2010. Establishment and characterization of a Madin-Darby canine kidney reporter cell line for influenza A virus assays. J Clin Microbiol 48:2515–2523. doi: 10.1128/JCM.02286-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu W, Zhou J, Qin K, Du N, Liu L, Yu Z, Zhu Y, Tian W, Wu X, Shu Y. 2011. A reporter system for assaying influenza virus RNP functionality based on secreted Gaussia luciferase activity. Virol J 8:29. doi: 10.1186/1743-422X-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Morell M, Czihal P, Hoffmann R, Otvos L, Aviles FX, Ventura S. 2008. Monitoring the interference of protein-protein interactions in vivo by bimolecular fluorescence complementation: the DnaK case. Proteomics 8:3433–3442. doi: 10.1002/pmic.200700739. [DOI] [PubMed] [Google Scholar]
- 31.Corbel SY, Rossi FM. 2002. Latest developments and in vivo use of the Tet system: ex vivo and in vivo delivery of tetracycline-regulated genes. Curr Opin Biotechnol 13:448–452. doi: 10.1016/S0958-1669(02)00361-0. [DOI] [PubMed] [Google Scholar]
- 32.Pluta K, Luce MJ, Bao L, Agha-Mohammadi S, Reiser J. 2005. Tight control of transgene expression by lentivirus vectors containing second-generation tetracycline-responsive promoters. J Gene Med 7:803–817. doi: 10.1002/jgm.712. [DOI] [PubMed] [Google Scholar]
- 33.Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A 89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin XF, An DS, Chen IS, Baltimore D. 2003. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A 100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.He X, Zhou J, Bartlam M, Zhang R, Ma J, Lou Z, Li X, Li J, Joachimiak A, Zeng Z, Ge R, Rao Z, Liu Y. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123–1126. doi: 10.1038/nature07120. [DOI] [PubMed] [Google Scholar]
- 36.Obayashi E, Yoshida H, Kawai F, Shibayama N, Kawaguchi A, Nagata K, Tame JR, Park SY. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127–1131. doi: 10.1038/nature07225. [DOI] [PubMed] [Google Scholar]
- 37.Deng T, Sharps J, Fodor E, Brownlee GG. 2005. In vitro assembly of PB2 with a PB1-PA dimer supports a new model of assembly of influenza A virus polymerase subunits into a functional trimeric complex. J Virol 79:8669–8674. doi: 10.1128/JVI.79.13.8669-8674.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fodor E, Smith M. 2004. The PA subunit is required for efficient nuclear accumulation of the PB1 subunit of the influenza A virus RNA polymerase complex. J Virol 78:9144–9153. doi: 10.1128/JVI.78.17.9144-9153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sugiyama K, Obayashi E, Kawaguchi A, Suzuki Y, Tame JR, Nagata K, Park SY. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J 28:1803–1811. doi: 10.1038/emboj.2009.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Biswas SK, Nayak DP. 1996. Influenza virus polymerase basic protein 1 interacts with influenza virus polymerase basic protein 2 at multiple sites. J Virol 70:6716–6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bu C, Peng B, Cao Y, Wang X, Chen Q, Li J, Shi G. 2015. Novel and selective acetylcholinesterase inhibitors for Tetranychus cinnabarinus (Acari: Tetranychidae). Insect Biochem Mol Biol 66:129–135. doi: 10.1016/j.ibmb.2015.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Cao Y, Li L. 2014. Improved protein-ligand binding affinity prediction by using a curvature-dependent surface-area model. Bioinformatics 30:1674–1680. doi: 10.1093/bioinformatics/btu104. [DOI] [PubMed] [Google Scholar]
- 43.Li C, Wu A, Peng Y, Wang J, Guo Y, Chen Z, Zhang H, Wang Y, Dong J, Wang L, Qin FX, Cheng G, Deng T, Jiang T. 2014. Integrating computational modeling and functional assays to decipher the structure-function relationship of influenza virus PB1 protein. Sci Rep 4:7192. doi: 10.1038/srep07192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ghanem A, Mayer D, Chase G, Tegge W, Frank R, Kochs G, Garcia-Sastre A, Schwemmle M. 2007. Peptide-mediated interference with influenza A virus polymerase. J Virol 81:7801–7804. doi: 10.1128/JVI.00724-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wunderlich K, Mayer D, Ranadheera C, Holler AS, Manz B, Martin A, Chase G, Tegge W, Frank R, Kessler U, Schwemmle M. 2009. Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication. PLoS One 4:e7517. doi: 10.1371/journal.pone.0007517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Torreira E, Schoehn G, Fernandez Y, Jorba N, Ruigrok RW, Cusack S, Ortin J, Llorca O. 2007. Three-dimensional model for the isolated recombinant influenza virus polymerase heterotrimer. Nucleic Acids Res 35:3774–3783. doi: 10.1093/nar/gkm336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohtsu Y, Honda Y, Sakata Y, Kato H, Toyoda T. 2002. Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol Immunol 46:167–175. doi: 10.1111/j.1348-0421.2002.tb02682.x. [DOI] [PubMed] [Google Scholar]
- 48.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L. 1998. A third-generation lentivirus vector with a conditional packaging system. J Virol 72:8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu SY, Sanchez DJ, Aliyari R, Lu S, Cheng G. 2012. Systematic identification of type I and type II interferon-induced antiviral factors. Proc Natl Acad Sci U S A 109:4239–4244. doi: 10.1073/pnas.1114981109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu SY, Aliyari R, Chikere K, Li G, Marsden MD, Smith JK, Pernet O, Guo H, Nusbaum R, Zack JA, Freiberg AN, Su L, Lee B, Cheng G. 2013. Interferon-inducible cholesterol-25-hydroxylase broadly inhibits viral entry by production of 25-hydroxycholesterol. Immunity 38:92–105. doi: 10.1016/j.immuni.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schneider WM, Chevillotte MD, Rice CM. 6 February 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muratore G, Goracci L, Mercorelli B, Foeglein A, Digard P, Cruciani G, Palu G, Loregian A. 2012. Small molecule inhibitors of influenza A and B viruses that act by disrupting subunit interactions of the viral polymerase. Proc Natl Acad Sci U S A 109:6247–6252. doi: 10.1073/pnas.1119817109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chase G, Wunderlich K, Reuther P, Schwemmle M. 2011. Identification of influenza virus inhibitors which disrupt of viral polymerase protein-protein interactions. Methods 55:188–191. doi: 10.1016/j.ymeth.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 54.O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. 2011. Open Babel: an open chemical toolbox. J Cheminform 3:33. doi: 10.1186/1758-2946-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trott O, Olson AJ. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan W, Dong Z, Li F, Meng W, Feng L, Niu X, Li C, Luo Q, Li Z, Sun C, Chen L. 2013. Visualizing influenza virus infection in living mice. Nat Commun 4:2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.