Abstract
The unsatisfactory outcomes of typical multiple cytotoxic chemotherapeutic combination therapies used to treat patients have fostered a need for new unconventional combinations of therapeutic agents. Among the candidates, siRNA has been widely discussed and tested. However, the right time right place co-delivery of siRNA with other types of active ingredients is challenging because of the possible differences among their physiochemical and pharmacodynamics properties. To accomplish a synergistic cytotoxic effect, a nano-assembly was thus designed to co-deliver siRNA with other therapeutic agents. An siRNA, targeting pro-survival gene for the p75 neurotrophin receptor, and an organelle-fusing peptide, targeting mitochondria, were layered onto a nano-template by charge-charge interaction, followed by a layer of CD44 targeting ligand. The formulated triple-functional nanomedicine was efficiently internalized by the CD44 expressing triple-negative breast cancer cells. The encapsulated siRNA and the pro-apoptotic peptide were released inside cells, silencing the intended prosurvival gene and inducing apoptosis by fusing the mitochondrial membrane, respectively. A synergistic effect was achieved by this three-agent combination. The design of the developed multi-functional nanomedicine could be generalized to deliver other siRNA and drugs for a maximum therapeutic combination with minimal off-targeting effects.
Keywords: nanoplatform, siRNA therapy, triple negative breast cancer, active targeting, KLA peptide
Graphical abstract
The right time right place co-delivery of unconventional therapeutic agents was fulfilled by layering siRNA, mitochondria-fusing peptide and targeting ligand onto a nano-template through simple charge-charge interaction. The formulated triple-functional nanomedicine (TFN) was efficiently internalized by the CD44 expressing triple-negative breast cancer cells and accomplished a superior synergistic therapeutic effect with minimum off-targeting effect.
1. Introduction
Triple-negative breast cancer (TNBC) is an aggressive subtype of breast cancer with limited treatment options.[1] Patients with TNBCs have a very poor prognosis and a significantly shorter relapse-free period due to the ineffectiveness of chemotherapy. Without the three most common types of receptors known to fuel most breast cancer growth – estrogen, progesterone, and HER-2, there is currently no available target therapies for TNBC.[2, 3] TNBC is capable of evading treatment via the overexpression of specific genes involving pro-survival and anti-apoptotic pathways.[4, 5] Therefore, a combination therapy that simultaneously inhibits the pro-survival pathway and triggers apoptosis could be an effective candidate to treat TNBCs.
An overexpression of neurotrophin receptor (p75NTR), which is a member of the tumor necrosis factor receptor superfamily, in breast cancer has been documented as responsible for the promotion of tumor growth. The inhibition of this pro-survival gene by small interference RNA (siRNA) results in diminished tumor growth in preclinical animal models.[6–9]
On the other hand, mitochondria, the cell’s powerhouse, are not only indispensable energy generators, but are also master regulators of danger signals like the intrinsic apoptotic pathway.[10–12] Because the dysfunction of mitochondria triggers cell death signaling cascades and mitochondria-dependent apoptosis, mitochondria are regarded as an important therapeutic target in cancer therapy.[13, 14] An amphipathic antimicrobial peptide, KLAKLAKKLAKLAK, termed KLA14, has shown to disrupt bacterial cell membranes, but has a limited toxic effect against the plasma membranes of eukaryotic cells. Nonetheless, once KLA is inside eukaryotic cells, it disrupts the mitochondrial membrane and initiates apoptotic cell death.[15, 16] Since KLA14 is not permeable to the plasma membrane of eukaryotic cells, a proper facilitated intracellular delivery method is required to acquire an optimal mitochondria lysing activity.[17–19] In a prior study, we reported that an arginine-rich cell penetrating peptide increased the cellular uptake and therapeutic potency of KLA14 in vivo.[20]
However, a successful co-delivery of an siRNA with a peptide drug can be challenging. Both therapeutic ingredients have to be assembled into the same vehicle and simultaneously delivered to sensitize specific cancer cells, sparing the healthy cells. siRNA is a fully negatively charged molecule and the KLA peptide is a lysine rich positively charged peptide. The difference in their charges allows the nanomedicine to be fabricated together using a previously developed layer-by-layer (LbL) strategy.[21–25] The LbL fabrication method is a gentle and simple layering procedure based on the charge–charge interactions between the positively and the negatively charged polymers.[26–28] The formulated nano-assembly can then be sealed with a layer of hyaluronic acid (HA) for the active targeting of TNBC cells. HA is a natural ligand for cell-surface receptor CD44, which is a well-defined biomarker for various cancer cells and TNBCs with tumor-initiating and stem-like phenotypes.[29–31] Therefore, HA has been widely utilized for active cancer targeting through CD44 receptor-mediated endocytosis.[32–34]
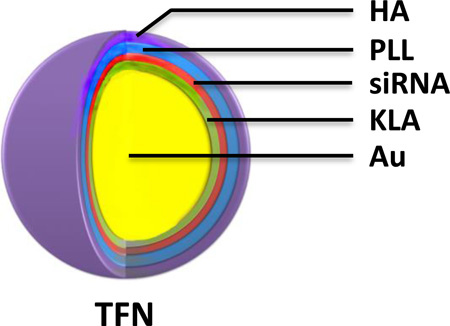
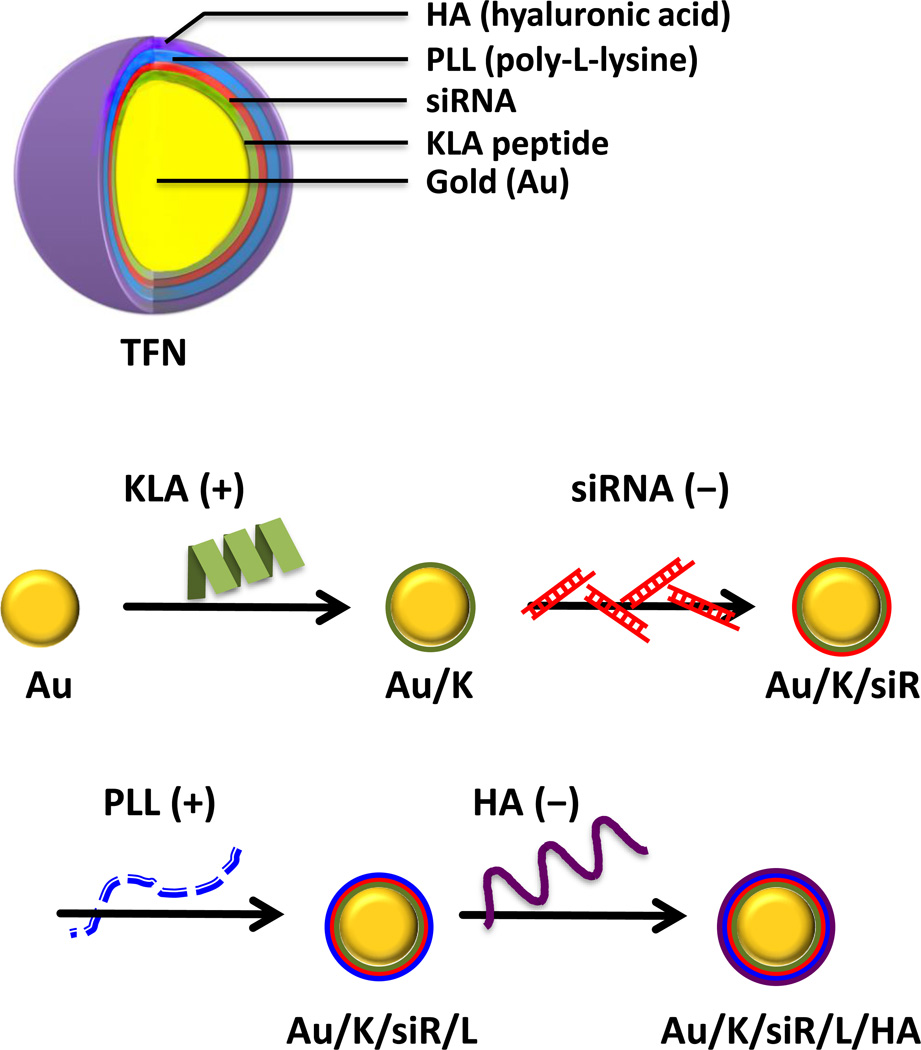
In this study, a triple-functional nanomedicine (TFN) was fabricated by layering three biocompatible pharmaceutical components on gold nanoparticles (AuNPs): (1) a positively charged fusing peptide for mitochondrial disruption, (2) a negatively charged siRNA for silencing the p75NTR tumorigenic gene, and (3) a negatively charged HA for CD44 targeting after a positively charged poly-L-lysine (PLL) filler layer (Figure 1). The prepared TFNs were specifically picked up by CD44 expressing TNBC cells, and released the active siRNA and KLA therapeutics to achieve a superior synergistic effect with minimum off-target effect.
Figure 1.
Schematic illustration of triple-functional nanomedicine (TFN) preparation. Polyelectrolytes, including two layers of active therapeutic ingredients (KLA peptide and siRNA), one layer of spacing polymer (PLL), and one layer of targeting ligand (HA), were sequentially assembled through the charged-charge interaction.
2. Results and Discussion
AuNPs were selected as the cores for their uniform size, electronic features, and feasibility of surface modification.[35] TFN was fabricated following a previously optimized siRNA layering platform.[21–23] The negatively charged AuNP which contain citrate layer[36] was layered by alternating the charged molecules, i.e. KLA (+), siRNA (−), PLL (+) and HA (−) (Figure 1). However, during the first layer of coating, it was found that the 14-mer KLA peptide (KLA14) did not have enough positive charges to form a stable monolayer on AuNPs. To secure enough positive charges, two extended KLA peptides, 21-mer (KLA21: KLAKLAKKLAKLAKKLAKLAK) and 28-mer (KLA28: KLAKLAKKLAKLAKKLAKLAKKLAKLAK), were prepared. The cytotoxicity of the KLA peptide analogues was validated in MDA-MB231 cells. The original KLA14 was non-cytotoxic up to 200 µM because of poor permeability; whereas, the IC50 of KLA21 and KLA28, which have higher numbers of positive charges, were around 20 and 10 µM, respectively (Supporting Information, Figure S1). It was found that KLA28 formed a better coating without causing aggregation; therefore it was selected for the subsequent LbL depositions. The fabrication of the other layers was done smoothly and required no further chemical modifications. Although the increased positive charges could further benefit fabrication, the increased cytotoxicity could cause a concern for undesired side-toxicities to other organs and tissues. Therefore, a final coating of HA, which is a natural ligand for CD44, is essential to maximize the specific cancer therapeutic effect while sparing normal cells.
In brief, to assemble the TFN, negatively charged AuNPs in water were dropped into the positively charged KLA28 peptide solution to create the first coating layer. The reaction solution was incubated for 30 min and then the coated particles were spun down by centrifugation. After several washes with ultra-pure water, the KLA-coated AuNPs were added to the negatively charged anti-p75 neurotrophin receptor (p75NTR) siRNA solution. After incubation, free unbound siRNAs were removed by centrifugation and the particles were re-suspended in ultra-pure water. As shown in Figure 1, by repeating these procedures with the proceeding layers of positively charged PLL and negatively charged HA, TFN with four layers of polymers – including two layers of active therapeutic ingredients (KLA peptide and siRNA), one layer of targeting ligand (HA), and one layer of spacing polymer (PLL) – were successfully fabricated onto the surface of Au through electrostatic interactions.
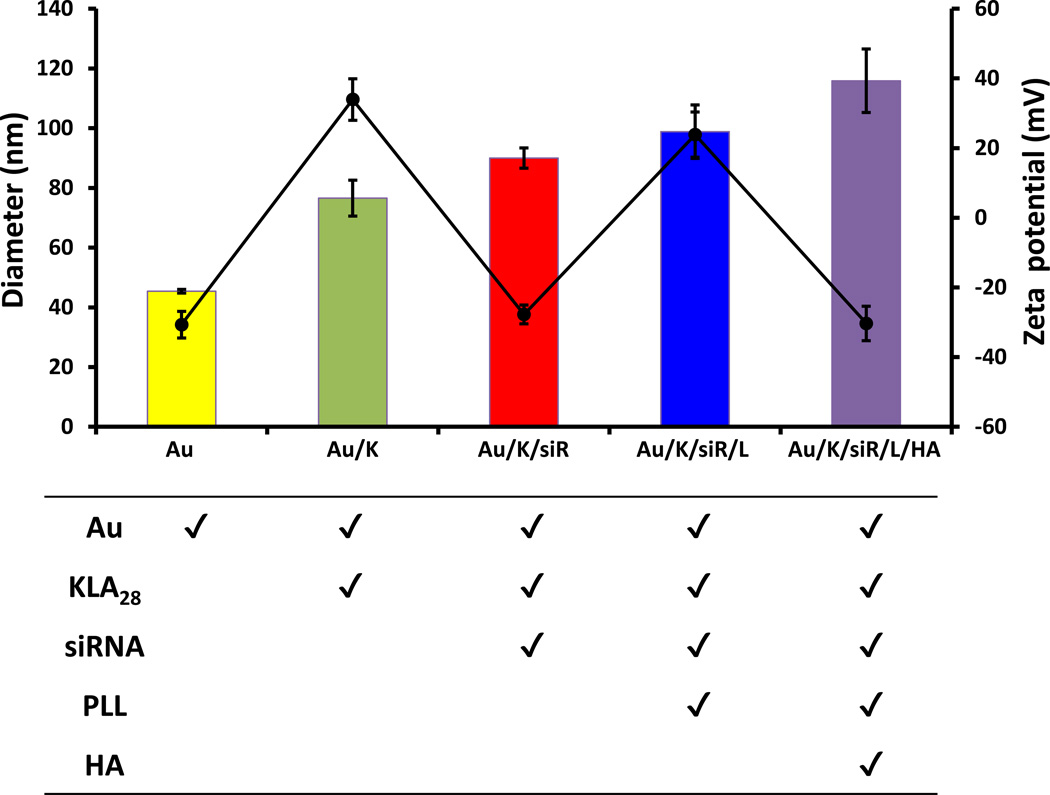
Hydrodynamic diameters of the formulated nanocomplexes were measured by dynamic light scattering (DLS) after each layer of coating (Figure 2). The size of the initial bare AuNPs was 40 nm. The particle size increased steadily with the number of layers added (Au/K: 77 nm; Au/K/siR: 90 nm; Au/K/siR/L: 99 nm; Au/K/siR/L/HA: 116 nm). The initial zeta potential of bare AuNPs was −31 mV. The KLA28 coating brought the surface charge up to about +34 mV, while the subsequent siRNA layer dragged it down to about −28 mV again. The final charged of the assembled TFN was about −30 mV. The zigzag pattern of the surface zeta potential of each layer supported the successful layering of the alternately charged components (Figure 2). In addition, the size distribution of prepared particle showed a single sharp peak with low polydispersity index (PDI < 0.28) (Supporting Information, Figure S2).
Figure 2.
Characterization of TFN. The average size (solid color bar) and zeta potential (black circle) of each multilayered nanocomplex were measured by dynamic light scattering (DLS). All measurements were done in triplicate and the data were shown as mean ± s.d.
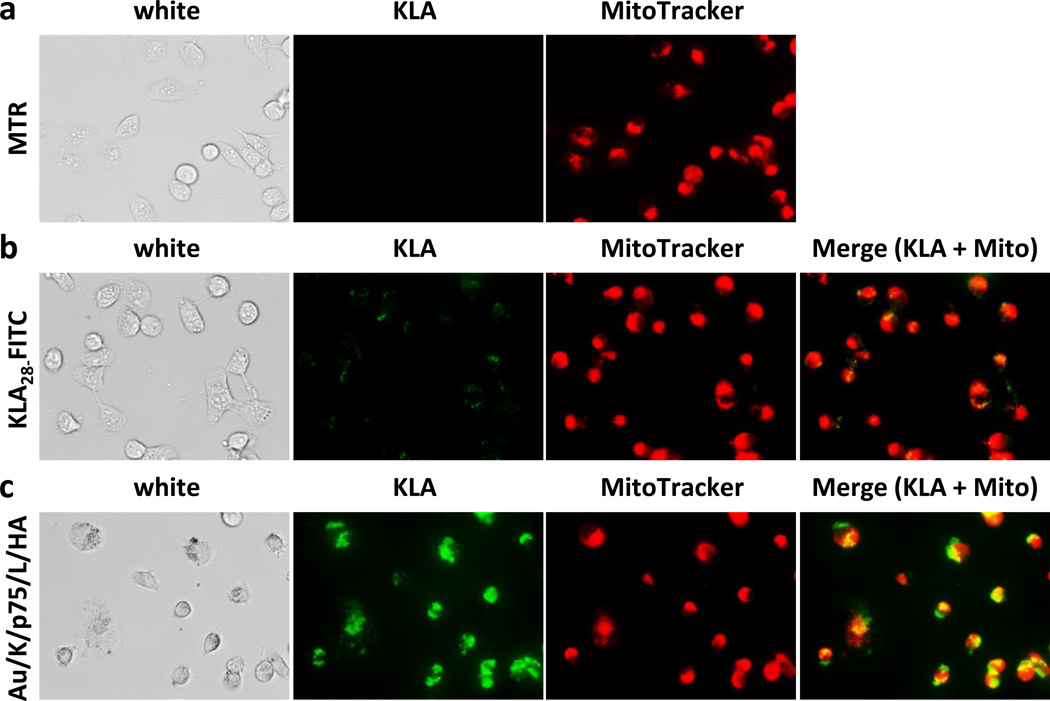
The uptake efficiency and subcellular distribution of free KLA28 peptide versus the formulated TFN was tracked in MDA-MB231 cells. KLA28 (5 µM) tagged with fluorescein (FITC) showed poor membrane translocation, while TFN (KLA28-FITC: 1.6 µM) was picked up by cells quickly (Figure 3). A much higher FITC signal was seen in TFN treated cells, although the absolute amount of KLA28 in TFN was 3-fold lower than that of the free KLA28. Once internalized, KLA28 released from TFN was distributed to mitochondria, confirmed by a merged mitochondrial fluorescence signal (yellow, Figure 3c). It was noticed that the initial fluorescent signal of the intact TFN was low due to the quenching effect of AuNP; however, the quenched fluorescence signal was restored when the fluorochromes were released from the AuNP core.[21]
Figure 3.
Intracellular distribution of KLA peptide. MDA-MB231 cells were stained with MitoTracker Red alone (1 µM) for 30 min (a), or treated MitoTracker together with free KLA28-FITC (5 µM) (b) or with the formulated Au/K/siR-p75/L/HA (KLA28-FITC: 1.6 µM, siR-p75: 0.12 µM) for 4 h (c). Merged fluorescence signal (yellow color) indicates the mitochondrial sub-localization of KLA peptide.
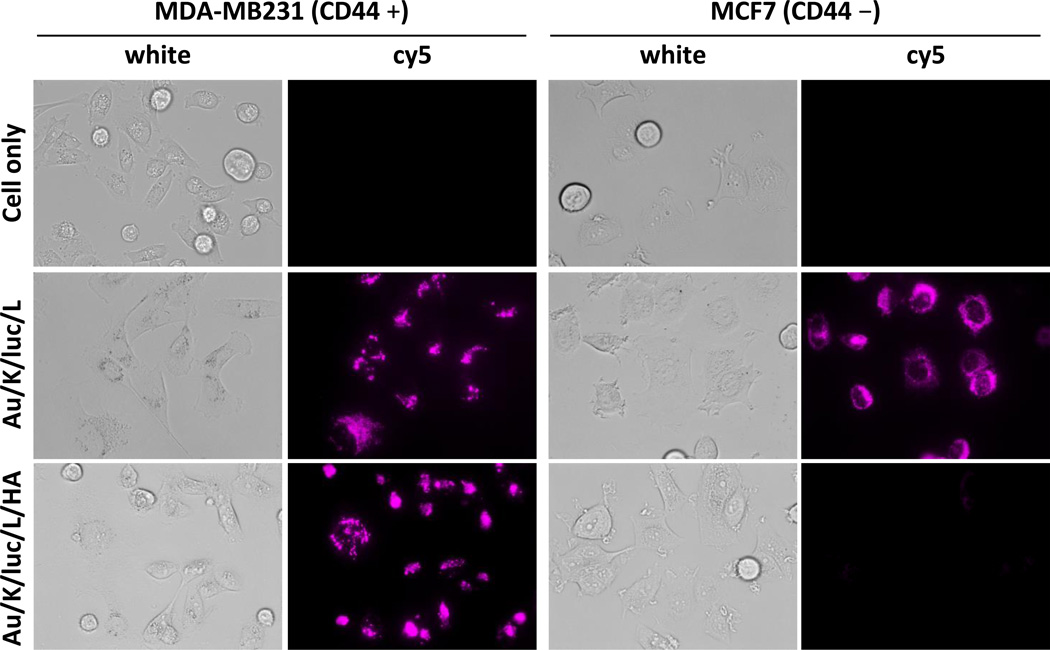
Next, to investigate the CD44 specific intracellular delivery of therapeutic ingredients, nanocomplexes with or without HA, targeting ligand, were tested in two different breast cancer cell lines, triple negative CD44+ MDA-MB231 and ER+ CD44− MCF7. Differential CD44 expression level in MDA-MB231 and MCF7 was confirmed using immunofluorescence staining (Supporting Information, Figure S3). Cellular uptake of targeted and untargeted nanocomplexes were tracked by fabricating a cy5 labeled model siRNA (Figure 4). As expected, the targeted TFN, which contains HA, can only be seen in high CD44 expressing MDA-MB231 cells, and not low CD44 expressing MCF7 cells. A diffused and spotty fluorescence signal was observed in the cytoplasm of MDA-MB231 cells. In contrast, unlike the results seen in HA-containing particle treated cells, the siRNA fluorescent signal was found in all tested cells when the final HA targeting layer was not included in the particle (Figure 4). Data obtained from observing the particle’s cellular uptake indicate that the positively charged particles which have PLL as the final layer were picked up by all cells without selectivity, while the HA coated particles were only internalized by CD44 expressing cells.
Figure 4.
Target-specific delivery of siRNA to CD44 expressing cells. Images of released siRNA, which were labeled with a cy5 reporter, from non-HA or HA-layered nanocomplex (KLA28: 1.6 µM, siR-luc-cy5: 0.12 µM) were visualized using fluorescence microscopy with cy5 filter after 12 h incubation in MDA-MB231 and MCF7 cells.
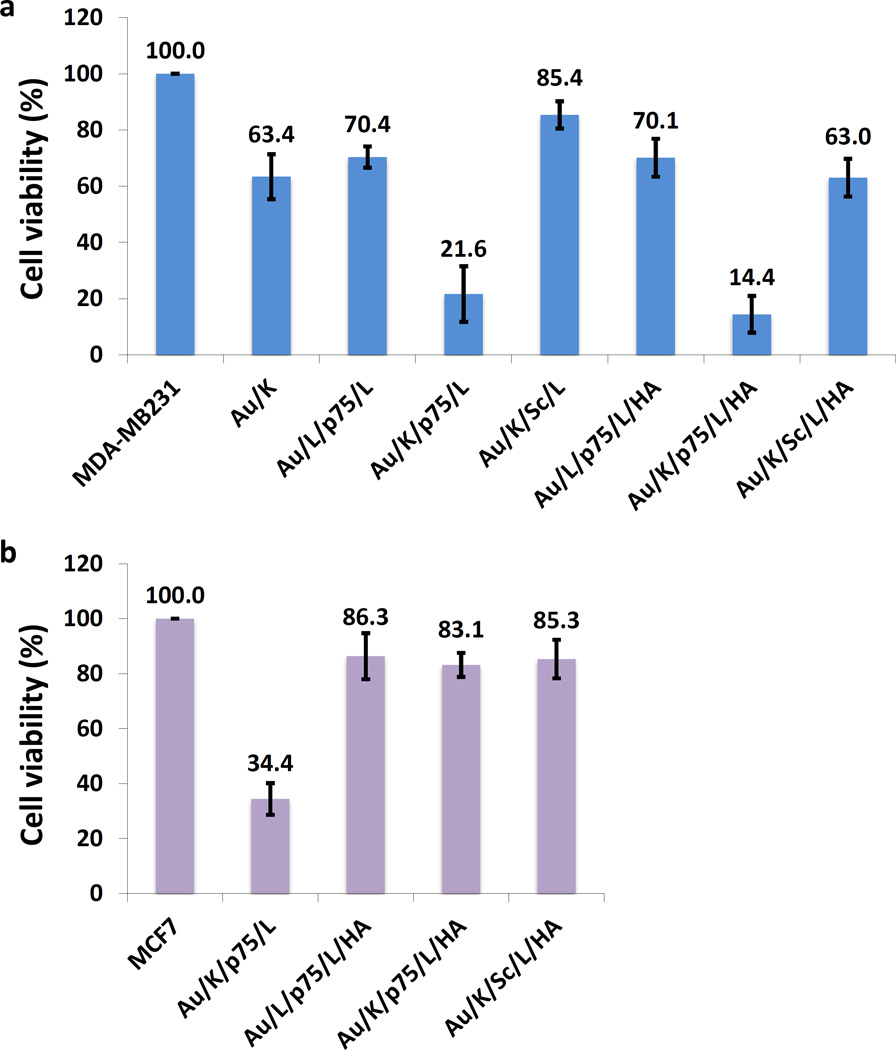
The cell-specific therapeutic effect of TFN was then investigated by including several controls. As shown in Figure 5, particles incorporated with either the KLA28 or p75NTR siRNA alone showed only moderate cell toxicity (63% or 70% viable, respectively); while the KLA28 and p75NTR siRNA combined particle demonstrated a significant effect in killing cells (22% viable). A scramble siRNA, with the same length but randomized sequence, was similarly formulated and included in the study as a negative control to confirm the sequence dependent gene silencing effect. The cytotoxic effect of the particle with a scrambled siRNA has dramatically reduced (85% viable). This data confirmed the synergistic therapeutic effect from KLA28 and p75NTR siRNA, and the advantage of combining these two non-typical anticancer agents. The data also confirm that CD44 targeted delivery (14% viable) is more beneficial than the non-targeted delivery (Figure 5a). Conversely, the combined effect of TFN was not achieved (83% viable) in MCF7 (CD44 −) cells due to the absence of HA and CD44 binding (Figure 5b). Other targeted negative controls with single active ingredient, such as Au/L/p75/L/HA and Au/K/Sc/L/HA, couldn’t obtain preferred cell cytotoxicity, either. This data further confirms the importance of simultaneous delivery of the siRNA and the peptide drug. TFN’s cell-specific selectivity was also confirmed by fluorescence microscope in both cells (Supporting Information, Figure S4). The cells with fluorescence signal of KLA28-FITC were very sick as evidenced by their morphology.
Figure 5.
Specific synergistic cytotoxic effect induced by TFN in CD44 positive TNBC cells. Cell viability was evaluated two days after treatment with various nanocomplexes (KLA28: 1.6 µM, siR-p75 or siR-Sc: 0. 12 µM) using CellTiter solution in (a) MDA-MB231 and (b) MCF7 cells. The cell viability of the untreated MDA-MB231 and MCF7 cells were set as 100%. Each experiment was performed three times and the results were expressed as the median change in cell viability ± s.d.
The TFN-mediated silencing of the corresponding p75NTR gene in the protein level was further confirmed by western blot analysis (Figure 6). Consistent with cell viability results (Figure 5), the expression level of p75NTR decreased dramatically when p75NTR targeting siRNA coated nanocomplexes (both Au/K/p75/L and Au/K/p75/L/HA) were used in MDA-MB231 cells (Figure 6a). This decrease was seen only after treatment with non-specific Au/K/p75/L and not with Au/K/p75/L/HA because of the lack of CD44 assisted uptake in MCF7 cells (Figure 6b). Once again, the scramble siRNA in nanocomplex did not affect the expression of p75NTR, indicating high specificity from the p75NTR targeted TFN to CD44 positive TNBCs.
Figure 6.
Gene silencing effect of TFN in CD44 positive TNBC cells. Protein expression of p75NTR after treatment with various nanocomplexes (KLA28: 1.6 µM, siR-p75 or siR-Sc: 0.12 µM) was evaluated by western blot in (a) MDA-MB231 and (b) MCF7 cells.
TFN could also be fabricated with different orders of layers, for example, the positively charged PLL instead of KLA, could be used to initiate the layering, and then followed with siRNA, KLA and HA. Interestingly, a slightly higher therapeutic efficiency was observed when KLA was used to prime the particles; therefore most particles were prepared by using KLA as the starting layer.
3. Conclusion
In summary, the triple-functional nanomedicine (TFN) has demonstrated a superior synergistic cytotoxic effect to treat TNBCs. TFN integrates two unconventional therapeutic ingredients, an organelle-specific therapy and a nucleic acid therapy, into a controllable vehicle by simple LbL fabrication. By adding a targeting layer, TFN harness active drugs to a specific cell type. In contrast to the current combination therapies that administered drugs may not simultaneously arrive at the tumor, the co-assembled TFN transported all active ingredients to specific cancer cells and achieved the desired synergistic effect. In this study, two non-typical drug candidates, acting on the pro-survival inhibition and apoptotic induction, have shown significant effects. The KLA28 destroyed the cell’s energy source, the mitochondria, while the siRNA inhibited the specific p75NTR expression, a key player in tumorigenesis. As TFN are conveniently fabricated with various functional building blocks, which could be exchanged easily with minimum modification, the possible combinations of therapeutic agents and targeting ligands are enormous. This concept of TFN may have a major impact in formulating combination therapy.
4. Experimental Section
Peptide Synthesis
Peptides (0.1 mmole) were synthesized on a PS3 peptide synthesizer (Protein Technologies, Tucson, AZ) employing the Fmoc strategy on rink amide resin. For the synthesis of FITC-conjugated peptides, Lys(ivDde) was introduced at the peptide C-termini. After elongation, the ivDde protection group was selectively removed by 2% (v/v) hydrazine in DMF. FITC (0.4 mmole) was then conjugated to the lysine’s side chain in the presence of N’N-diisoproylethyleamine 20% (v/v) in DMSO (5 mL). All peptides were purified by reverse-phase HPLC to >98 % purity and the expected molecular weights were confirmed by MALD-TOF mass spectrometer.
Preparation and characterization of multilayered nanocomplexes
Using previous modified LbL fabrication method,[21–25] therapeutic ingredients, siRNA and KLA peptide, were successfully deposited onto the surface of AuNPs (size: 40 nm, BB International, Cardiff, UK). All siRNAs were obtained from Sigma-Aldrich (St. Louis, MO). The sequences of siR-p75NTR against p75 neurotrophin receptor[9] are sense strand: 5’-AUGCCUCCUUGGCACCUCCdTdT-3’, antisense strand: 5’-GGAGGUGCCAAGGAGGCAUdTdT-3’, and of control nonsnese siRNA (siR-Sc) are sense strand: 5’-GCUGACCCUGAAGUUCAUCdTdT-3’, antisense strand: 5’-GAUGAACUUCAGGGUCAGCdTdT-3’. The sequences of siR-luc against luciferase[37] are sense strand: 5’-UUAAUCAGAGACUUCAGGCGGUdTdT-3’, antisense strand: 5’-ACCGCCUGAAGUCUCUGAUUAAdTdT-3’. AuNPs (3.15 × 109 particles in 0.7 mL) were added dropwisely onto a KLA28 solution (62.5 nmole, 0.5 mL) in ultra-pure water (Life Technologies, Carlsbad, CA). After incubating for 30 min in the dark with gentle shaking, the solution was centrifuged for 30 min at 16,100 g using a micro centrifuge (Eppendorf, Hauppauge, NY). The supernatant was removed, and the gel-like deep red pellet was re-suspended with pure water and centrifuged for 30 min at 16,100 g. KLA coated AuNPs were stored in pure water after additional wash. Next the polyelectrolyte layer was deposited by adding KLA coated AuNPs (in 0.5 mL of pure water) to the siRNA solution (4.0 nmole, 0.5 mL). The reaction solution was incubated in the dark for 30 min with gentle shaking, followed by three washes. The deposition procedures were repeated sequentially with poly-L-lysine (PLL, Sigma-Aldrich) (Mw = 15,000~30,000 g mol−1, PLL solution = 0.5 mL of 5 mg mL−1 in pure water) and sodium hyaluronate (HA, Lifecore Biomedical, Chaska, MN) (Mw = 100~150 KDa, HA solution = 0.5 mL of 8 mg mL−1 in pure water) to have a total of 4 layers of polyelectrolytes (KLA, siRNA, PLL and HA). The sizes and zeta potentials of each AuNPs in water were measured using a ZetaPALS (Brookhaven, Holtsville, NY) according to the manufacturer’s instructions. The amount of siRNA and KLA in TFN was calculated by measuring the concentration of siRNA or FITC-KLA in the supernatant before and after the coating using a spectrophotometer (Cary 60 UV-Vis, Agilent, Santa Clara, CA). The prepared TNF was stored in water at 4 °C.[23]
Cell lines
MDA-MB231 (triple negative) and MCF7 (ER+) human breast cancer cells were purchased from ATCC (Manassas, VA) and cultured in DMEM medium (Mediatech Inc., Manassas, VA) supplemented with 2 mM L-glutamine, 100 U mL−1 penicillin, 100 mg mL−1 streptomycin, and 10 % heat-inactivated fetal bovine serum (Sigma-Aldrich) in a humidified atmosphere of 5 % CO2 at 37 °C.
Mitochondrial localization of KLA peptide and CD44 expression in breast cancer cells
To investigate the subcellular localization of the KLA peptide with or without layering onto nanocomplex, MDA-MB231 cells were collected by trypsinization, counted, and plated in a 96-well black clear-bottom culture plate (Corning Life Sciences, Pittston, PA) at a density of 5 × 103 cells per well. After one day, KLA28-FITC (5 µM) or Au/K/p75/L/HA (KLA28-FITC: 1.6 µM) were added and incubated for 4 h. Cells were then washed with phosphate buffered saline (PBS) and stained with MitoTracker Red (1 µM, Life Technologies) for 30 min at 37 °C according to the manufacturer’s protocol. After two washes with PBS, cells were imaged under EVOS FL Auto Cell Imaging System (Life Technologies).
To examine the CD44 expression level in live cells, MDA-MB231 cells were collected by trypsinization, counted, and plated in a 96-well black clear-bottom culture plate (Corning Life Sciences) at a density of 5 × 103 (or 1.0 × 104 of MCF7) cells per well. One day later, cells were incubated with Alexa 488-conjugated CD44 antibody (Life Technologies) with a 1:50 volume of the dye-conjugated antibody for 30 min at 37 °C according to the manufacturer’s protocol. After two washes with FluroBrite DMEM (Life Technologies), cells were imaged using an EVOS FL Auto Cell Imaging System (Life Technologies).
Live cell imaging for cellular uptake of nanocomplex
For fluorescence imaging, a cy5 fluorochrome was tagged on the 5' end of the sense siR-luc and FITC was conjugated with KLA28 peptide. Briefly, MDA-MB231 cells were seeded on a 96-well black clear-bottom culture plate (Corning Life Sciences) at a density of 5.0 × 103 (or 1.0 × 104 of MCF7) cells per well. After one day, the culture medium was replaced with Au/K/luc/L or Au/K/luc/L/HA particles (KLA28: 1.6 µM, siR-luc: 0.12 µM) containing medium, and further cultured for 12 h. Cells were then washed twice with PBS and cultured in phenol red-free medium and imaged with an EVOS FL Auto Cell Imaging System (Life Technologies).
Cancer therapeutic effect of nanocomplexes
A cell proliferation assay was performed to assess cell viability after treatment with various nanocomplexes. Briefly, MDA-MB231 cells were collected by trypsinization, counted, and plated in a 96-well culture plate at a density of 5 × 103 (or 1.0 × 104 of MCF7) cells per well. One day later, the culture medium was replaced with various nanocomplexes (KLA28: 1.6 µM, siR-p75 or siR-Sc: 0.12 µM) containing medium. After 12 h incubation, cells were washed twice with PBS and then further cultured for 48 h. At day 3, 20 µL CellTiter solution (Promega, Madison, WI) was added to each well after replacing with fresh medium and incubated for an additional 3 h, and then the absorbance of the solution was measured at 490 nm using a plate reader (Tecan, Mannedorf, Switzerland).
To assess the cytotoxicity of KLA peptide analogues, different concentrations (ranging from 0 to 200 µM) of various KLA peptides (KLA14, KLA21 and KLA28) were treated for 12 h instead of nanocomplexes, and then followed the same methods.
Western blot analysis
MDA-MB231 cells were collected by trypsinization, counted, and plated in a 6-well culture plate (BD Falcon, San Jose, CA) at a density of 1.5 × 105 (or 3.0 × 105 of MCF7) cells per well. One day later, the culture medium was replaced with various nanocomplexes (KLA28: 1.6 µM, siR-p75 or siR-Sc: 0.12 µM) containing medium. After 12 h incubation, cells were washed twice with PBS and then further cultured for 48 h. At day 3, cells were lysed (2 wells for each group) and proteins extracted in a buffer containing 50 mM Tris–HCl (pH, 7.4), 1% NP-40, 0.25% sodium deoxycholate, 150 mM NaCl, 1 mM EDTA and protease inhibitor cocktail. The protein concentration was measured using the BCA Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA). Equal amounts of protein (30 ~ 70 µg per sample) were resolved over 10~12% polyacrylamide-SDS gels and transferred to a nitrocellulose membrane. The membranes were blocked for 2 h at room temperature (RT) with 5% bovine serum albumin (Sigma-Aldrich) in 0.1% Tween-20-PBS. Blocked membranes were then incubated with anti-p75NTR (clone D8A8) antibody (1:1,000; Cell Signaling Technology, Boston, MA) or anti-β-actin antibody (1:5,000; Sigma-Aldrich) overnight at 4 °C and then washed three times (5 min each) with PBS containing 0.1% Tween-20. The membranes were incubated with HRP-conjugated anti-rabbit IgG secondary antibody (1:5,000; cell Signaling Technology) for p75NTR, and with HRP-conjugated anti-mouse IgG secondary antibody (1:5,000; Cell Signaling Technology) for β-actin for 1 h at RT. The immunoreactive proteins on the membranes were visualized with In-Vivo F PRO (Bruker, Billerica, MA) after treatment with LumiGLO Reagent and Peroxide (Cell Signaling Technology).
Supplementary Material
Acknowledgments
We thank Dr. Barbara L. Hempstead for the helpful discussion and Myung Shin Han for preparing the drawing. This study was supported in part by NIH CA135312 and DOD W81XWH-11-1-0442.
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Foulkes WD, Smith IE, Reis-Filho JS. N Engl J Med. 2010;363:1938. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 2.Dent R, Trudeau M, Pritchard KI, Hanna WM, Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P, Narod SA. Clin Cancer Res. 2007;13:4429. doi: 10.1158/1078-0432.CCR-06-3045. [DOI] [PubMed] [Google Scholar]
- 3.Andre F, Zielinski CC. Ann Oncol. 2012;23(Suppl 6):vi46. doi: 10.1093/annonc/mds195. [DOI] [PubMed] [Google Scholar]
- 4.Lee MJ, Ye AS, Gardino AK, Heijink AM, Sorger PK, MacBeath G, Yaffe MB. Cell. 2012;149:780. doi: 10.1016/j.cell.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT. ACS Nano. 2013;7:9571. doi: 10.1021/nn4047925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reis-Filho JS, Steele D, Di Palma S, Jones RL, Savage K, James M, Milanezi F, Schmitt FC, Ashworth A. Mod Pathol. 2006;19:307. doi: 10.1038/modpathol.3800542. [DOI] [PubMed] [Google Scholar]
- 7.Verbeke S, Meignan S, Lagadec C, Germain E, Hondermarck H, Adriaenssens E, Le Bourhis X. Cell Signal. 2010;22:1864. doi: 10.1016/j.cellsig.2010.07.014. [DOI] [PubMed] [Google Scholar]
- 8.Adriaenssens E, Vanhecke E, Saule P, Mougel A, Page A, Romon R, Nurcombe V, Le Bourhis X, Hondermarck H. Cancer Res. 2008;68:346. doi: 10.1158/0008-5472.CAN-07-1183. [DOI] [PubMed] [Google Scholar]
- 9.Vanhecke E, Adriaenssens E, Verbeke S, Meignan S, Germain E, Berteaux N, Nurcombe V, Le Bourhis X, Hondermarck H. Clin Cancer Res. 2011;17:1741. doi: 10.1158/1078-0432.CCR-10-1890. [DOI] [PubMed] [Google Scholar]
- 10.Fulda S, Galluzzi L, Kroemer G. Nat Rev Drug Discov. 2010;9:447. doi: 10.1038/nrd3137. [DOI] [PubMed] [Google Scholar]
- 11.Galluzzi L, Kepp O, Kroemer G. Nat Rev Mol Cell Biol. 2012;13:780. doi: 10.1038/nrm3479. [DOI] [PubMed] [Google Scholar]
- 12.Weinberg SE, Chandel NS. Nat Chem Biol. 2015;11:9. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Green DR, Kroemer G. Science. 2004;305:626. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 14.Fu B, Long W, Zhang Y, Zhang A, Miao F, Shen Y, Pan N, Gan G, Nie F, He Y, Zhang J, Teng G. Sci Rep. 2015;5:8029. doi: 10.1038/srep08029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Javadpour MM, Juban MM, Lo WC, Bishop SM, Alberty JB, Cowell SM, Becker CL, McLaughlin ML. J Med Chem. 1996;39:3107. doi: 10.1021/jm9509410. [DOI] [PubMed] [Google Scholar]
- 16.Ellerby HM, Arap W, Ellerby LM, Kain R, Andrusiak R, Rio GD, Krajewski S, Lombardo CR, Rao R, Ruoslahti E, Bredesen DE, Pasqualini R. Nat Med. 1999;5:1032. doi: 10.1038/12469. [DOI] [PubMed] [Google Scholar]
- 17.Agemy L, Friedmann-Morvinski D, Kotamraju VR, Roth L, Sugahara KN, Girard OM, Mattrey RF, Verma IM, Ruoslahti E. Proc Natl Acad Sci U S A. 2011;108:17450. doi: 10.1073/pnas.1114518108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cieslewicz M, Tang J, Yu JL, Cao H, Zavaljevski M, Motoyama K, Lieber A, Raines EW, Pun SH. Proc Natl Acad Sci U S A. 2013;110:15919. doi: 10.1073/pnas.1312197110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen WH, Xu XD, Luo GF, Jia HZ, Lei Q, Cheng SX, Zhuo RX, Zhang XZ. Sci Rep. 2013;3:3468. doi: 10.1038/srep03468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law B, Quinti L, Choi Y, Weissleder R, Tung CH. Mol Cancer Ther. 2006;5:1944. doi: 10.1158/1535-7163.MCT-05-0509. [DOI] [PubMed] [Google Scholar]
- 21.Lee SK, Han MS, Asokan S, Tung CH. Small. 2011;7:364. doi: 10.1002/smll.201001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SK, Tung CH. Adv Funct Mater. 2013;23:3488. doi: 10.1002/adfm.201202777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee SK, Tung CH. Methods Mol Biol. 2016;1372:113. doi: 10.1007/978-1-4939-3148-4_9. [DOI] [PubMed] [Google Scholar]
- 24.Lee SK, Han MS, Tung CH. Small. 2012;8:3315. doi: 10.1002/smll.201200751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee SK, Mortensen LJ, Lin CP, Tung CH. Nat Commun. 2014;5:5216. doi: 10.1038/ncomms6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jewell CM, Lynn DM. Adv Drug Deliv Rev. 2008;60:979. doi: 10.1016/j.addr.2008.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reum N, Fink-Straube C, Klein T, Hartmann RW, Lehr CM, Schneider M. Langmuir. 2010;26:16901. doi: 10.1021/la103109b. [DOI] [PubMed] [Google Scholar]
- 28.Suma T, Miyata K, Anraku Y, Watanabe S, Christie RJ, Takemoto H, Shioyama M, Gouda N, Ishii T, Nishiyama N, Kataoka K. ACS Nano. 2012;6:6693. doi: 10.1021/nn301164a. [DOI] [PubMed] [Google Scholar]
- 29.Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. Cell. 1990;61:1303. doi: 10.1016/0092-8674(90)90694-a. [DOI] [PubMed] [Google Scholar]
- 30.Zhang S, Balch C, Chan MW, Lai HC, Matei D, Schilder JM, Yan PS, Huang TH, Nephew KP. Cancer Res. 2008;68:4311. doi: 10.1158/0008-5472.CAN-08-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricardo S, Vieira AF, Gerhard R, Leitao D, Pinto R, Cameselle-Teijeiro JF, Milanezi F, Schmitt F, Paredes J. J Clin Pathol. 2011;64:937. doi: 10.1136/jcp.2011.090456. [DOI] [PubMed] [Google Scholar]
- 32.Oh EJ, Park K, Kim KS, Kim J, Yang JA, Kong JH, Lee MY, Hoffman AS, Hahn SK. J Control Release. 2010;141:2. doi: 10.1016/j.jconrel.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 33.Kim E, Yang J, Park J, Kim S, Kim NH, Yook JI, Suh JS, Haam S, Huh YM. ACS Nano. 2012;6:8525. doi: 10.1021/nn300289u. [DOI] [PubMed] [Google Scholar]
- 34.Dreaden EC, Morton SW, Shopsowitz KE, Choi JH, Deng ZJ, Cho NJ, Hammond PT. ACS Nano. 2014;8:8374. doi: 10.1021/nn502861t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel MC, Astruc D. Chem Rev. 2004;104:293. doi: 10.1021/cr030698+. [DOI] [PubMed] [Google Scholar]
- 36.Brewer SH, Glomm WR, Johnson MC, Knag MK, Franzen S. Langmuir. 2005;21:9303. doi: 10.1021/la050588t. [DOI] [PubMed] [Google Scholar]
- 37.Chang K, Elledge SJ, Hannon GJ. Nat Methods. 2006;3:707. doi: 10.1038/nmeth923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.