C. difficile infects thousands of hospitalized patients every year, causing significant morbidity and mortality. C. difficile spores play a pivotal role in the transmission of the pathogen in the hospital environment. During infection, the spores germinate, and the vegetative bacterial cells produce toxins that damage host tissue. Thus, sporulation and toxin production are two important traits of C. difficile. In this study, we showed that a mutation in tcdR, the toxin gene regulator, affects both toxin production and sporulation in epidemic-type C. difficile strain R20291.
KEYWORDS: Clostridium difficile, sporulation, toxin gene regulation
ABSTRACT
Clostridium difficile is an important nosocomial pathogen and the leading cause of hospital-acquired diarrhea. Antibiotic use is the primary risk factor for the development of C. difficile-associated disease because it disrupts normally protective gut flora and enables C. difficile to colonize the colon. C. difficile damages host tissue by secreting toxins and disseminates by forming spores. The toxin-encoding genes, tcdA and tcdB, are part of a pathogenicity locus, which also includes the tcdR gene that codes for TcdR, an alternate sigma factor that initiates transcription of tcdA and tcdB genes. We created a tcdR mutant in epidemic-type C. difficile strain R20291 in an attempt to identify the global role of tcdR. A site-directed mutation in tcdR affected both toxin production and sporulation in C. difficile R20291. Spores of the tcdR mutant were more heat sensitive than the wild type (WT). Nearly 3-fold more taurocholate was needed to germinate spores from the tcdR mutant than to germinate the spores prepared from the WT strain. Transmission electron microscopic analysis of the spores also revealed a weakly assembled exosporium on the tcdR mutant spores. Accordingly, comparative transcriptome analysis showed many differentially expressed sporulation genes in the tcdR mutant compared to the WT strain. These data suggest that regulatory networks of toxin production and sporulation in C. difficile strain R20291 are linked with each other.
IMPORTANCE C. difficile infects thousands of hospitalized patients every year, causing significant morbidity and mortality. C. difficile spores play a pivotal role in the transmission of the pathogen in the hospital environment. During infection, the spores germinate, and the vegetative bacterial cells produce toxins that damage host tissue. Thus, sporulation and toxin production are two important traits of C. difficile. In this study, we showed that a mutation in tcdR, the toxin gene regulator, affects both toxin production and sporulation in epidemic-type C. difficile strain R20291.
INTRODUCTION
Clostridium difficile is a Gram-positive, spore-forming, anaerobic bacillus and is the leading cause of hospital-acquired diarrheal diseases (1, 2). Nearly 50% of all patients carry C. difficile asymptomatically after hospitalization (2, 3). Nearly 10% of all C. difficile-infected patients develop pseudomembranous colitis, and 3% develop severe, life-threatening complications such as fulminant colitis and toxic megacolon (4). C. difficile infection (CDI) is commonly acquired from C. difficile spores present in the hospital environment, and individuals become infected when the normal colonic microbiota is suppressed by antibiotic therapy (5). In the gut, C. difficile spores germinate to the toxin-producing vegetative form in response to certain bile acids, e.g., taurocholic acid (TA), and amino acids. C. difficile toxins A (TcdA) and B (TcdB) are then secreted from the vegetative cell and cause tissue damage, necrosis, and inflammation and are the main reasons for this disease outcome (6).
In C. difficile, the toxin genes, tcdA and tcdB, are located within a 19-kb pathogenicity locus (PaLoc) and the tcdR gene, located upstream of tcdB, is required for expression of the toxin genes. TcdR is an alternate sigma factor that directs transcription by recruiting RNA polymerase to the toxin gene promoters and its own promoter (7, 8). Previous studies have shown that other proteins can regulate toxin gene expression in response to different environmental stimuli by controlling the transcription of tcdR. The sigma factor SigD positively regulates toxin production by controlling the transcription of tcdR (9). CodY, a global transcriptional regulator, represses the toxin gene expression by binding with high affinity to the tcdR promoter region (10, 11). Finally, in response to sugar availability, CcpA, a major regulator of carbon catabolite repression, binds to the promoter region or the 5′ ends of several PaLoc genes, with the strongest affinity to the promoter region of tcdR (12, 13).
TcdR was the first member of the group V family of alternative sigma factors to be described (14). We recently determined that TcsR, a toxin gene regulator in Clostridium sordellii, is also a member of this family of sigma factors (15). Most of these alternative sigma factors are autoregulated (7, 16) and are induced by environmental stresses, such as nutritional limitation, DNA damage, or nonoptimal temperatures (8, 14, 17), suggesting that these sigma factors function under these suboptimal growth conditions.
In this study, we created and characterized a mutation in tcdR in the epidemic-type C. difficile R20291 strain to determine whether TcdR influenced cellular processes other than toxin production. We found that the tcdR mutant sporulated less efficiently than the wild-type (WT) strain. Moreover, spores prepared from the tcdR mutant were more heat sensitive and had lower germination efficiency than the wild-type parental strain. Electron microscopic (EM) analysis of the tcdR mutant spores also revealed a weakly assembled exosporium. In agreement with these findings, comparative transcriptome sequencing (RNA-seq) analyses of the WT and the tcdR mutant strains revealed several sporulation genes to be affected by the tcdR mutation. These results suggested that a mutation in tcdR not only affects toxin production but also influences the sporulation pathway in the C. difficile R20291 strain. Interestingly, however, mutating tcdR in the C. difficile 630Δerm strain did not result in this phenotype, suggesting that the TcdR regulon may be strain specific.
RESULTS
Mutation in tcdR affects both toxin production and sporulation in C. difficile strain R20291.
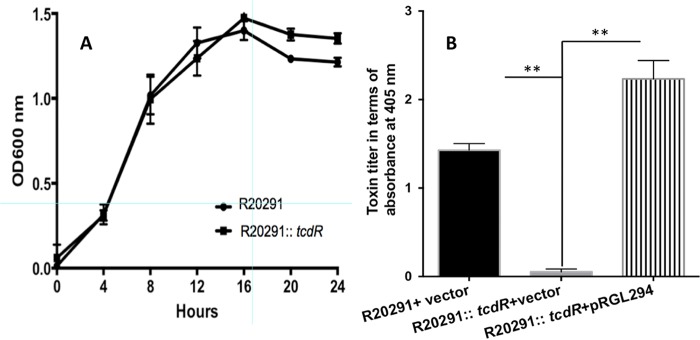
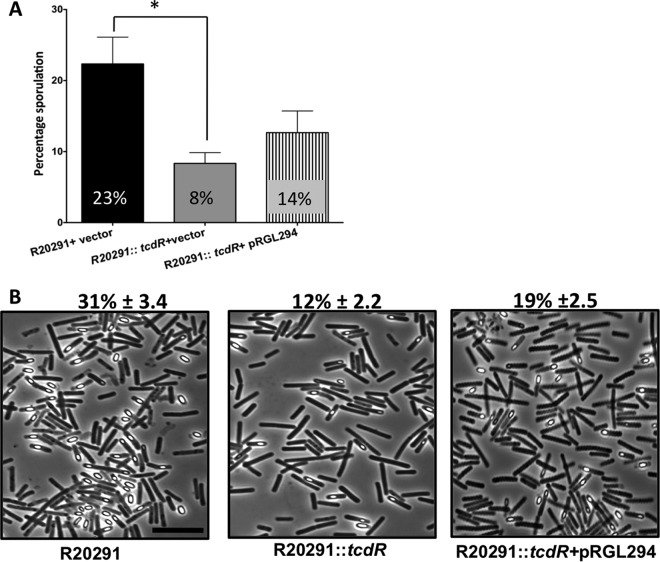
To analyze the global role of tcdR in C. difficile strain R20291, we used a Clostron system (18) to inactivate the tcdR gene. Insertion of the group II intron into the target gene (see Fig. S1A in the supplemental material) was verified by PCR using intron-specific primers and tcdR gene-specific primers (Fig. S1B and Table S1 in the supplemental material). Southern blotting confirmed the single chromosomal insertion of the intron in the tcdR gene (Fig. S1C). Growth kinetics analyses were performed and indicated that the inactivation of the tcdR gene did not affect the normal growth of the bacterium (Fig. 1A). A toxin enzyme-linked immunosorbent assay (ELISA) was performed with the cytosolic protein extracts of the tcdR mutant and the WT strain. We observed a dramatic reduction in toxin production (Fig. 1B) in the mutant compared to the WT, supporting the concept of the previously known function of TcdR as a positive regulator of the toxin genes (7, 8, 16). Further, we measured the sporulation efficiency of the tcdR mutant at the 24-h time point. A nearly 3-fold reduction in the level of ethanol-resistant spores was observed in the tcdR mutant compared to the WT strain (Fig. 2A). A similarly reduced sporulation rate (~2.6-fold) was observed when the number of sporulation cells in the population was counted microscopically (Fig. 2B). We then complemented the tcdR mutant by cloning and expressing tcdR from its own promoter. Toxin production in the complemented strain was fully recovered (Fig. 1B), whereas the effect on sporulation could be restored only partially (Fig. 2). Unlike toxin gene regulation (where TcdR directly regulates tcdA and tcdB transcription), sporulation is regulated by multiple transcription factors and alternative RNA polymerase sigma factors (19–21). Sporulation also involves finely tuned spatially and temporally regulated gene expression programs and may not be mimicked exactly in the complemented strain. All of these regulatory mechanisms could result in partial complementation of the sporulation. Another explanation could be that, when the TcdR sigma factor is overexpressed, the availability of RNA core polymerase for other sigma factors needed for sporulation could be limited and that limitation could result in partial complementation of the sporulation phenotype.
FIG 1 .
Effect of tcdR inactivation on bacterial growth kinetics and toxin production. (A) Growth curve of R20291 and R20291::tcdR in TY medium. (B) TcdA and TcdB levels in cytosolic fractions after 10 h of growth. C. difficile strains were grown in TY medium, and toxins were quantified using ELISA. The data represent the averages of the results of three independent assays. Error bars in both panel A and B correspond to the standard errors of the means. The asterisks (**) in panel B indicate statistical difference at a P value of <0.005.
FIG 2 .
Mutation in tcdR affects the sporulation efficiency in the R20291 strain. (A) Sporulation frequency (CFU per milliliter of ethanol-resistant spores) of R20291 plus pRPF185 (R20291+pRPF185), R20291::tcdR+pRPF185, and R20291::tcdR+pRGL294 (pRPF185 derivative plasmid carrying tcdR) strains grown for 24 h in 70:30 sporulation medium. The error bars correspond to standard errors of the means of results from 3 biological replicates. *, P < 0.05 (by two-tailed Student’s t test). At least three independent experiments were performed. (B) Phase-contrast microscopy of paraformaldehyde-fixed R20291, R20291::tcdR+pRPF185, and R20291::tcdR+pRGL294 strains grown for 24 h in 70:30 sporulation plate. Percent sporulation (± standard deviation) was calculated (using the number of spores divided by the total number of spores and vegetative cells) from results from at least three independent experiments. Bar, 10 µm.
Construction and characterization of tcdR mutant in C. difficile. (A) Schematic representation of insertional inactivation of tcdR by group II intron. (B) The intron insertion in the tcdR coding region was verified by PCR using the intron-specific primer EBS along with gene-specific primers ORG81 and ORG82 in the parent (R20291), in the tcdR mutant (R20291::tcdR), and in the tcdR complemented strain (R20291::tcdR+pRG294). The same strategy was followed to verify the tcdR mutation in the 630Δerm strain. (C) Southern blot analysis of genomic DNA from the WT and tcdR mutant strains with a tcdR-specific probe. The shift in the hybridization band indicates the integration of the intron within tcdR coding region. Download FIG S1, PDF file, 0.2 MB (180.9KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in the study. Download TABLE S1, DOCX file, 0.1 MB (128.7KB, docx) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Transcriptome analysis of tcdR mutant.
The global regulators ccpA and codY are known to influence both sporulation and toxin production in C. difficile (10, 11). We performed quantitative reverse transcription-PCR (qRT-PCR) analysis and found no significant change in their transcript levels in the tcdR mutant compared to the WT strain (Fig. S2). Since this initial analysis failed to explain the reasons behind the unexpected phenotype of the tcdR mutant, we decided to perform a transcriptome study using RNA-seq analysis. RNAs were prepared from stationary-phase cultures of the tcdR mutant (mutant R20291::tcdR) and the WT strain (strain R20291) and were subjected to RNA-seq analysis. The data observed for selected genes were confirmed by performing qRT-PCR analysis (Fig. S3 and S4). RNA-seq analysis of the tcdR mutant showed that most of the genes were underexpressed and revealed that two major classes of genes were particularly affected, i.e., the PaLoc genes and the sporulation-associated genes (see NCBI GEO accession number GSE85395). However, few genes were upregulated in the tcdR mutant. Among those that were overexpressed, we found the srlR gene encoding the regulator of glucitol/sorbitol-specific PTS system (CDR20291_0690 to CDR20291_0696). PaLoc genes (tcdA, tcdB, tcdR, and tcdE) were downregulated (33-fold, 12-fold, 5-fold, and 3-fold, respectively) in the tcdR mutant, as expected. Autoregulation of TcdR and its need for toxin gene transcription were well characterized previously (7, 8, 16). However, no report was available on the TcdR-mediated transcription of tcdE in the PaLoc. TcdE is a holin-like protein and was found to mediate toxin release from C. difficile cells (22, 23). Our data suggest that TcdR is also needed to initiate tcdE transcription in C. difficile.
Expression analysis of toxin genes and known toxin gene regulators in the tcdR mutant. RNA was prepared from R202091 and R20291::tcdR strains grown in TY medium for 16 h. qPCR analysis was performed for selected regulator-coding genes. Statistical analysis was performed using the t test, and the error bars indicate the standard errors of the means (s.e.m.). **, P value < 0.01. Download FIG S2, PDF file, 0.1 MB (55.9KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Many sporulation-associated genes were significantly repressed in the tcdR mutant.
In addition to the PaLoc genes, many genes in the sporulation pathways were repressed in the tcdR mutant compared to the WT (Table 1). Sporulation is a highly complex cellular process regulated by a cascade of events (20, 21, 24). Spo0A is the master regulator of sporulation, and its transcript levels were unchanged in the tcdR mutant as observed in both the RNA-seq and qRT-PCR analyses (Table 1 and Fig. 3). However, we saw that transcripts of specific sporulation sigma factor genes sigE, sigF, sigG, and sigK were underexpressed in the tcdR mutant (Table 1 and Fig. 3). Even though the levels of transcription of these genes were moderately (1.5-fold to 2-fold) reduced in the tcdR mutant compared to the WT strain in the RNA-seq analysis, we observed through qRT-PCR analyses that their transcription levels in the tcdR mutant were significantly reduced throughout the time of growth (Fig. 3). RNA-seq analysis also revealed several sporulation genes controlled by sigE, sigG, and sigK to be significantly affected in the tcdR mutant (Table 1) (19–21). SigE is a mother cell-specific sigma factor responsible for the transcription of early sporulation-specific genes, and the SigE-regulated genes identified to be affected in tcdR mutant included the following: spoIVA (stage IV sporulation protein A); spmBA (spore maturation proteins B and A); and sigK, the second mother cell-specific sigma factor. SigG is the forespore-specific factor that controls the final stages of sporulation. The SigG-regulated genes found to be repressed in tcdR mutant included the following: pdaA (spore specific deacetylase), sspA (small acid-soluble protein), and spoVAC and spoVAD (stage V sporulation proteins). SigG and SigE activities were previously found to be required for the production of heat-resistant spores (21). The sigK C. difficile mutant was able to make heat-resistant spores; however, the level of production was 3 log lower than that seen with the parent strain (21). SigK regulates many genes encoding spore structure proteins that participate in the synthesis of the spore coat and spore exosporium. In fact, we found that many of the SigK-regulated genes such as cotJBD, cotA, cotB, cotE, bclA3, and bclA2 as well as the sleC and cdeC genes were significantly underexpressed in the tcdR mutant compared to the WT strain. The downregulation of these genes was confirmed by qRT-PCR analysis (Fig. S3 and S4).
TABLE 1 .
| Gene ID | Gene name if assigned, known/predicted function | Fold downregulation in mutant (WT/tcdR mutant) | Known or predicted sigma factor needed for expression |
|---|---|---|---|
| CDR20291_0124 | Cell wall endopeptidase | 3.844 | SigF |
| CDR20291_2145 | Hypothetical protein | 5.993 | SigF |
| CDR20291_2363 | gpr, germination protease | 4.008 | SigF |
| CDR20291_3400 | Spore cortex-lytic enzyme | 5.652 | SigF |
| CDR20291_3401 | spoIIR, stage II sporulation protein | 4.228 | SigF |
| CDR20291_2530 | sigG | 2.14 | SigF |
| CDR20291_0125 | spoIIID, stage III sporulation protein D | 5.323 | SigE |
| CDR20291_0714 | Stage IV sporulation protein | 12.140 | SigE |
| CDR20291_1031 | spoIIIAB, stage III sporulation protein AB | 3.600 | SigE |
| CDR20291_1032 | spoIIIAC, stage III sporulation protein AC | 4.031 | SigE |
| CDR20291_1033 | spoIIIAD, stage III sporulation protein AD | 4.458 | SigE |
| CDR20291_1034 | spoIIIAE, stage III sporulation-related protein | 3.733 | SigE |
| CDR20291_2147 | cspBA, germination-specific protease | 4.346 | SigE |
| CDR20291_2513 | spoIVA, stage IV sporulation protein A | 4.773 | SigE |
| CDR20291_3376 | spmB, spore maturation protein B | 4.333 | SigE |
| CDR20291_3377 | spmA, spore maturation protein A | 5.447 | SigE |
| CDR20291_1073 | Hypothetical protein | 4.563 | SigE |
| CDR20291_0702 | spoVAC, stage V sporulation protein AC | 5.524 | SigG |
| CDR20291_0703 | spoVAD, stage V sporulation protein AD | 5.682 | SigG |
| CDR20291_1130 | Small acid-soluble spore protein | 4.816 | SigG |
| CDR20291_1131 | dacF, d-alanyl-d-alanine-carboxypeptidase | 5.891 | SigG |
| CDR20291_1529 | sodA, superoxide dismutase | 5.714 | SigG |
| CDR20291_2576 | sspA, small acid-soluble spore protein A | 4.500 | SigG |
| CDR20291_2802 | spoVFB, dipicolinate synthase subunit B | 3.914 | SigG |
| CDR20291_3080 | Small acid-soluble spore protein | 4.107 | SigG |
| CDR20291_3107 | sspB, small acid-soluble spore protein B | 4.690 | SigG |
| CDR20291_0212 | Spore coat protein | 6.600 | SigK |
| CDR20291_0316 | Spore coat assembly asparagine-rich protein | 6.101 | SigK |
| CDR20291_0337 | Fragment of putative exosporium glycoprotein | 12.666 | SigK |
| CDR20291_0522 | cotJB1, spore-coat protein | 8.666 | SigK |
| CDR20291_0523 | cotJC1, spore-coat protein | 6.842 | SigK |
| CDR20291_2290 | cotJB2, spore-coat protein | 5.679 | SigK |
| CDR20291_2291 | cotJC2, spore-coat protein | 5.165 | SigK |
| CDR20291_2803 | dpaA, dipicolinate synthase subunit A | 4.291 | SigK |
| CDR20291_3090 | bclA2, exosporium glycoprotein | 6.302 | SigK |
| CDR20291_3193 | bclA3, exosporium glycoprotein | 12.612 | SigK |
| CDR20291_3466 | Cell wall hydrolase | 4.631 | SigK |
| CDR20291_0476 | sleC, spore peptidoglycan hydrolase | 5.502 | Partly by SigF, SigK |
| CDR20291_2121 | sinR | 20.5 | Unknown |
| CDR20291_2122 | sinR like DNA binding protein | 27.25 | Unknown |
| CDR20291_0701 | sigF* | 1.23 | SigH |
| CDR20291_2531 | sigE* | 1.56 | SigH |
| CDR20291_1052 | spo0A* | 1.56 | SigH |
| CDR20291_1067B | sigK* | 1.78 | SigE |
Genes were considered differentially expressed if the fold change was ≥2.0 and their adjusted P value is ≤0.05. Expression levels of genes marked with (*) were not statistically significant. ID, identifier.
FIG 3 .
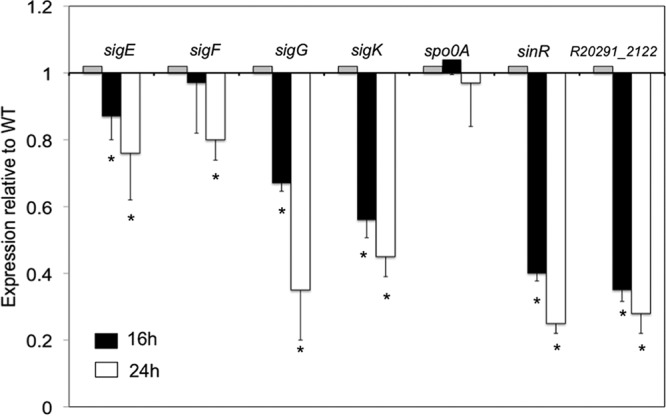
Decreased expression of key sporulation genes in the tcdR mutant. Data represent results of qRT-PCR analysis of sigE, sigF, sigG, sigK, spo0A, sinR, and R20291_2122 expression after 16 and 24 h of C. difficile growth in 70:30 sporulation medium. Error bars correspond to the standard errors of the means of results from at least three biological replicates. *, P < 0.05 (by two-tailed Student’s t test).
Expression analysis of selected sporulation genes during growth in 70:30 medium. RNA was prepared from R202091 and R20291::tcdR strains grown in 70:30 medium for 24 h. qRT-PCR analysis was performed for selected sporulation genes. Statistical analysis was performed using the t test, and the error bars indicate the standard errors of the means (s.e.m.). *, P value < 0.05. Download FIG S3, PDF file, 0.1 MB (70.4KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression analysis of selected sporulation genes during growth in TY medium. RNA was prepared from R202091 and R20291::tcdR strains that were grown in TY medium for 24 h. qRT-PCR analysis was performed for selected sporulation genes. Statistical analysis was performed using the t test, and the error bars indicate the standard errors of the means (s.e.m.). *, P value < 0.05. Download FIG S4, PDF file, 0.1 MB (57.1KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TEM analysis of spores from the R20291 and R20291::tcdR strains. The tcdR mutant spores (n = 60) were scored 100% for the presence of ruffled defective exosporium (marked with black arrow) and 98% for the presence of weakly stained core (marked as open triangle). Bar, 100 nm. Download FIG S5, PDF file, 1.5 MB (1.5MB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
CDR20291_2121 and CDR20291_2122 (sin operon) were repressed in the tcdR mutant.
Other than genes involved in sporulation morphology, we also found some regulatory genes potentially involved in sporulation to be affected in the tcdR mutant. In fact, the transcript levels of CDR20291_2121 (coding for a SinR-like protein of Bacillus subtilis) and CDR20291_2122 (coding for a DNA binding protein) genes were nearly 20-fold lower in the tcdR mutant than in the WT strain (Table 1). This result was confirmed by qRT-PCR (Fig. 3). In B. subtilis, SinR is encoded within the sin locus carrying both sinI and sinR genes. In B. subtilis, SinR forms tetramers, which repress spo0A transcription, although SinI is an inhibitor of SinR (25). If SinR functions similarly in C. difficile, a decrease in SinR activity should lead to an increase of sporulation. However, we observed decreased sporulation in the tcdR mutant (Fig. 2), suggesting that the products of the sin locus must function differently in C. difficile.
Spores derived from the tcdR mutant have increased heat sensitivity.
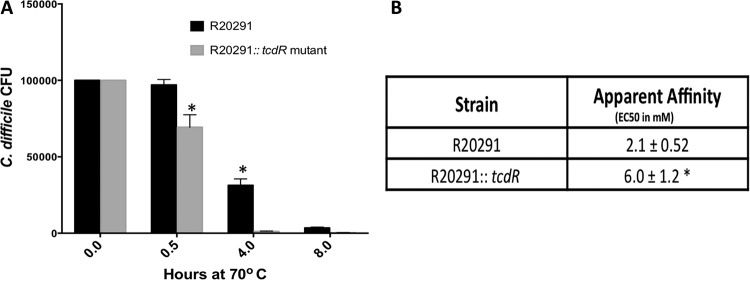
To compare the levels of heat sensitivity of spores between the WT and the tcdR mutant strains, we incubated purified spores at 70°C for 0.5 h, 4 h, and 8 h. When we monitored cell viability using the heat-treated spores, we found that spores from the tcdR mutant lost most of their viability upon 4 h of heat treatment and that they were nearly 10-fold more sensitive to heat than the WT spores (Fig. 4A). This could have been due to the decreased expression of both sigG and sigE in the tcdR mutant as observed in our transcriptional analysis; their activities are known to be involved in the formation of heat resistance of spores (21). In addition, the lower expression of many of the spore structure proteins (including cdeC) in the tcdR mutant can also explain the heat sensitivity of these spores.
FIG 4 .
(A) The tcdR mutant affects spore germination. Heat resistance of spores of C. difficile strain R20291 and its tcdR mutant derivatives was measured by heat-treating aliquots at 70°C for 0.5 h, 4 h, and 8 h. The surviving spores were enumerated as described in Materials and Methods. The data represent the averages of the results of three independent experiments, and error bars represent standard errors of the means. Asterisks (*) indicate statistical difference at a P value of <0.05. (B) Apparent affinity of taurocholate for C. difficile spores. EC50s were individually calculated from three independent germination experiments and are reported as averages with standard errors of the means. A Student’s t test was performed, and that asterisk indicates that the calculated P value is <0.05.
Increased taurocholate was required by tcdR mutant spores for germination.
To test if the lower transcription of sporulation-associated genes observed in the tcdR mutant (Table 1) affects the ability of C. difficile spores to germinate, we determined the apparent interaction of spores with taurocholic acid (Fig. 4B). C. difficile spores were suspended in rich medium alone or supplemented with increasing concentrations of the germinant taurocholate. The kinetics of spore germination were followed by measuring the rate of the decrease in optical density at 600 nm (OD600) as the spores germinated (see Materials and Methods). Though not traditional enzyme kinetics, this assay allows us to understand how spores interact with the taurocholate germinant. C. difficile R20291 spores display a 50% effective concentration (EC50) of 2.1 mM (similar to what has been previously reported for other strains) (26–28). However, the tcdR mutant spores display an EC50 of 6.0 mM, corresponding to a 3-fold reduction (<0.05 P value) in TA affinity. These results support the overall observation that spore-associated functions were affected when tcdR was inactivated in strain R20291.
Exosporium assembly was affected in the tcdR mutant.
Spores of the R20291::tcdR mutant were compared to those of the WT using electron microscopy to assess any effect on gross spore morphology. Samples were viewed as embedded thin sections, and the analysis revealed that tcdR mutant spores had a defect in their exosporium assembly (Fig. 5B and S5). The spore core of the tcdR mutant was stained weakly compared to the core of the WT spores, and darkly stained particulate materials were present over the spore coat and throughout these preparations. Weaker exosporium in the tcdR mutant spores could have made them susceptible to structural changes during chemical fixation procedures, resulting in these darker particles around the spores. In contrast, most of the R20291 WT spore had an intact exosporium that fully enclosed the spore coat (Fig. 5A) and was devoid of the darker debris observed in the tcdR mutant spores. This observation suggests that the tcdR mutation in R20291 affects the spore structure, with a profound effect on its exosporium assembly.
FIG 5 .
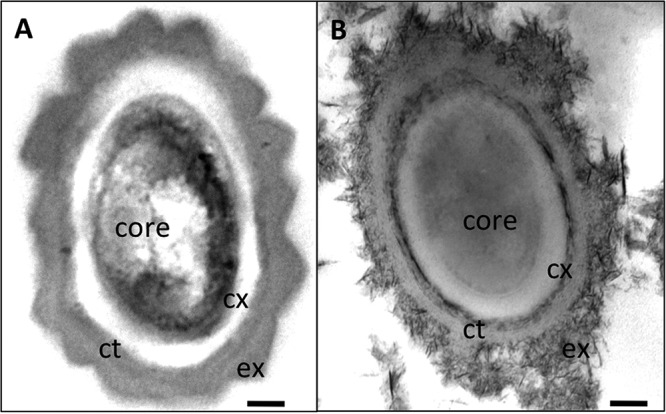
Transmission electron microscopic analysis of C. difficile spores. The images show thin sections of spores from the WT R20291 strain (A) and the R20291::tcdR mutant (B). Abbreviations: ex, exosporium; ct, coat; co, core; cx, cortex. Bar, 100 nm.
The effect of tcdR on sporulation is strain specific.
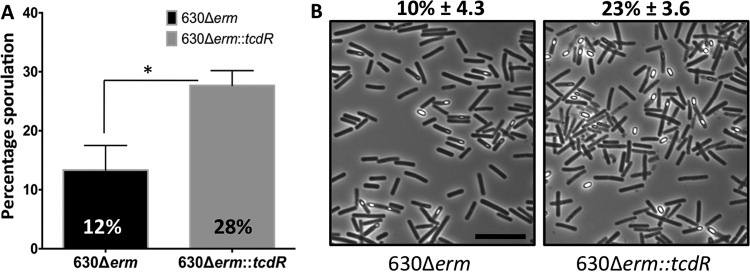
Previous studies have shown that mutations in a specific gene can result in different phenotypes in different C. difficile strain backgrounds (29, 30). To understand whether the effect of TcdR is strain dependent, we created a tcdR mutant in the 630Δerm strain using a ClosTron system. Toxin production in the 630Δerm::tcdR mutant was severely downregulated as observed in strain R20291::tcdR (Fig. S6). But unlike the results seen with the R20291::tcdR strain, the sporulation efficiency of the 630Δerm::tcdR strain was nearly 2-fold greater than that of its WT strain (Fig. 6). The similar opposing phenotype was previously reported for the spo0A mutants of strain R20291 versus the 630Δerm mutant, which also affects the toxin production (29, 30). Though the spo0A mutation resulted in increased toxin production in the R20291 strain, it resulted in reduced toxin production in the 630Δerm background. Even though the R20291 and 630 strains share 3,247 core genes, their genomes are significantly different from one another (31), whereas there are 47 coding sequences unique in R20291 compared to the 630 strain and 505 coding sequences unique in 630 compared to the R20291 strain (31). Therefore, the difference that we observed in these two strains concerning the impact of the tcdR mutation on sporulation might be related to the presence or absence of any of these unique genes. Even though we do not know the exact reason for these differences, these observations suggest that the C. difficile genome is dynamic and that its regulatory networks are fluid in nature.
FIG 6 .
Effect of tcdR on sporulation is strain specific. (A) Sporulation frequency (CFU per milliliter of ethanol-resistant spores) of 630Δerm and 630Δerm::tcdR strains grown for 24 h in 70:30 sporulation medium. The error bars correspond to standard deviations of results from at least three biological replicates. The asterisk (*) indicates a P value of <0.05 (by two-tailed Student’s t test). (B) Phase-contrast microscopy of paraformaldehyde-fixed 630Δerm and 630Δerm::tcdR strains grown for 48 h in a 70:30 sporulation plate. At least three independent experiments were performed to calculate percent sporulation (± standard errors of the means).
Toxin ELISA for 630Δerm and 630Δerm::tcdR strains. TcdA and TcdB expression levels in cytosolic fractions of C. difficile strains grown for 10 h in TY medium were quantified using ELISA. Data are from results of an experiment representative of three independent assays. Error bars correspond to the standard errors of the means of results from at least three biological replicates. The asterisks (***) in panel B indicate statistical difference at P < 0.001, estimated by Student’s t test. Download FIG S6, PDF file, 0.04 MB (44.2KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
DISCUSSION
TcdR-mediated toxin gene regulation is well studied in C. difficile (7, 8, 16). The aim of this study was to understand whether TcdR could influence cellular processes other than toxin production. To investigate this issue, we created a tcdR mutant in the R20291 strain and performed several phenotypic assays. As expected, the tcdR mutant strain either produced no toxins or produced toxins at levels that were not detectable. Surprisingly, we also observed that the level of spores produced by the mutant was significantly reduced compared to the level seen with wild type.
The link between toxin production and sporulation in C. difficile has always been suggested but has not been well studied. For example, in C. difficile R20291, a mutation in spo0A, the master regulator of sporulation, resulted in changes in toxin production (32). More recently, Edwards et al. reported that inactivation of CD3688 (rstA) in C. difficile strain 630 affects sporulation, toxin production, and motility (33). Moreover, it has been shown that the global regulators CodY and CcpA regulate toxin production along with sporulation (10, 12, 34). Thus, if the tcdR mutation affects codY, ccpA, or spo0A expression, both toxin production and sporulation could be influenced. When we measured transcript levels of these genes by qRT-PCR, we found no change in their levels in the tcdR mutant compared to the WT (see Fig. S2 in the supplemental material). However, the genome-wide transcriptome analyses of the tcdR mutant confirmed that many sporulation genes were affected.
Nearly 50% of the sporulation genes downregulated in the tcdR mutant are known (or predicted) to be under the control of SigE and SigK for their transcription (Table 1) (19–21, 35, 36). Among the downregulated SigE-dependent genes, we found sigK, whose presence could explain the transcriptional decrease in the levels of several SigK target genes in the tcdR mutant. The RNA-seq analyses of the tcdR mutant showed that transcription of spoIIR and spoIIID genes was reduced (Table 1). SpoIIR is essential for the activation of SigE (19, 35), and spoIIID, encoding a transcriptional regulator, is involved in the transcription of sigK (36). In C. difficile, as in B. subtilis, SigE is activated by proteolytic cleavage of the SigE precursor form (pro-SigE) (20). In B. subtilis, the enzyme SpoIIGA, which is responsible for pro-SigE processing, is coexpressed with sigE and is activated only when the mother cell and forespore compartments are formed (37–39). The trigger for SpoIIGA activation is the SpoIIR signal protein that is synthesized in the newly formed forespore and whose presence is communicated to the mother cell (40, 41). In B. subtilis, spoIIR is regulated by SigF, whereas in C. difficile, partial SigE processing is observed in sigF mutants, suggesting a lower level expression of spoIIR in the absence of SigF (19, 20). If the expression of spoIIR in a sigF mutant is influenced by TcdR, this could explain the partial processing of SigE in sigF mutants. Thus, a reduced abundance of spoIIR in the tcdR mutant could lead to low levels of activated SpoIIGA and part of pro-SigE would remain unprocessed and inactive. If so, this would also result in a decrease of spoIIID levels as observed in the transcriptome (Table 1); therefore, little or no transcription of sigK would occur, resulting in poor spore maturation.
Most of the genes identified as affected in the tcdR mutant code for proteins that are part of the spore proteome (20, 32, 42) and are involved in spore structure and germination. To determine whether tcdR mutant spore properties are different from those of the WT spores, we performed heat sensitivity and germination assays using purified spores. TcdR mutant spores were 10 times more heat-sensitive than WT spores (Fig. 4A). Accordingly, transcriptome analysis showed that several exosporium and coat protein coding genes were underexpressed in the tcdR mutant. A recent study on the C. difficile exosporium protein BclA3 demonstrated its role in spore heat resistance (43). The authors found that BclA3 is glycosylated by a glycosyltransferase encoded by the adjacent gene (CD3350) within the same operon whose mutation resulted in unglycosylated BclA3. They showed that spores from this mutant were highly susceptible to heat treatment compared to the WT spores (43). The same heat susceptibility was observed with the exosporium protein CdeC, which is present only in C. difficile and is needed for the assembly of exosporium (44). Also, C. difficile spoVAC and dpaAB mutants produced heat-sensitive spores (45). The dipicolinate synthase enzyme subunits (SpoVFB and DpaA) are responsible for the production of dipicolinic acid (DPA), which protects spores during heat treatment (46–48). Moreover, proteins encoded in the spoVA operon are responsible for transporting DPA from the mother cell to forespores during spore development (49).
All these results are consistent with the transcriptome analysis of the tcdR mutant, which showed decreased expression of bclA3, cdeC, spoVAC, and the DPA synthase coding operon. This probably results in the production of spores with weaker exosporium that must be more sensitive to heat treatment than the WT strain (Fig. 4A). Transmission electron microscopy (TEM) analysis of the tcdR mutant spores confirmed this speculation, where the exosporium was found to be defective and weakly assembled (Fig. 5).
Germination of bacterial spores is induced when the germinant receptors (GR) sense germinants and subsequently trigger the release of spore core DPA (46). The release of DPA from the spore core leads to the activation of cortex hydrolases that degrade the peptidoglycan (PG) cortex layer, which then allows core hydration. In C. difficile, CspC is the bile salt-sensing germinant receptor and is necessary for the release of DPA from spores (26). SleC is the spore cortex lytic enzyme, and its activation depends on CspC (through CspB-mediated cleavage of the prodomain to generate active SleC) (26, 50–52). A mutation in sleC was previously reported to affect germination in C. difficile (51, 53). Thus, lower transcription of sleC in tcdR mutant (Table 1) suggested that tcdR mutant spores could have inefficient germination. In agreement, we have shown that the TA affinity of C. difficile tcdR spores is low compared to that of the WT spores (Fig. 4B), indicating that germination is significantly reduced.
Several studies have previously identified the TTTACA sequence as the −35 region of the TcdR-dependent promoters (7, 8). To test whether some of the downregulated sporulation genes in the tcdR mutant can be directly controlled by TcdR, we looked for the presence of this consensus sequence in the promoter regions of these genes (Table 1). In fact, we found 9 genes/operons carrying the sequence in the −35 region of the TcdR-dependent promoters. These genes include bclA2, bclA3, cotJBD, spoVFB, cotA, cotB, cotE, dpaA, and sin. To test if any of these genes are directly controlled by TcdR, we constructed transcriptional fusions between the promoter of the bclA2 and bclA3 genes and the Escherichia coli β-glucuronidase (gusA) gene that we introduced in a gus-negative E. coli strain expressing or not expressing TcdR as we did previously (54). Compared to the control strains, we did not see any TcdR-mediated transcription of bclA2 or bclA3 promoters, indicating that TcdR is not a direct regulator of these genes (Fig. S7). However, we cannot exclude for these genes the possibility that TcdR may act together with a specific regulator present in the R20291 strain.
Beta-glucuronidase activity of bclA2 promoter-gusA and bclA3 promoter-gusA fusions in the presence or absence of TcdR. E. coli strains carrying promoter fusion plasmids (pbclA2-gusA and pbclA3-gusA) along with a TcdR-expressing plasmid (pRGL312) or the vector (pET16b) were grown for 6 h, and TcdR expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C. Bacterial cultures were harvested and were assayed for beta-glucuronidase activity (quantified in Miller units) as described previously (18). The values represent the means of the results from three independent experiments. Statistical analysis was performed using the t test, and the error bars indicate standard errors of the means. The asterisks (**) indicate statistical difference at P < 0.005, estimated by Student’s t test. Download FIG S7, PDF file, 0.1 MB (99.6KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Finally, the effect of TcdR on sporulation could be indirect. TcdR is an alternate sigma factor, and its presence or absence could influence the availability of the RNA polymerase core enzyme for other sigma factors in the cell, which in turn can influence the gene expression pattern. Thus, the absence of TcdR in the R20291::tcdR strain, increasing the availability of RNA polymerase core enzyme to other sigma factors, could indirectly affect those involved in the sporulation process. On the other hand, there may be common regulators that connect toxin gene regulation with the sporulation pathway in C. difficile that could be affected by the tcdR mutation. Previous studies have identified several regulators in C. difficile regulating toxin production along with sporulation, which strongly suggests that these two pathways were linked (10, 12, 33, 34, 55).
In the past decade, large C. difficile outbreaks, with higher relapse rates and increased mortality rates, were reported throughout the world and were attributed to C. difficile strains belonging to ribotype 027. Strain R20291 used in this study is a ribotype 027 isolate (56). Genetic and phenotypic features of this ribotype hint that the strains grouped as 027 ribotypes are different from other C. difficile strains (31). Recently, Lyon et al. reported that CdtR, a regulator in the binary toxin locus CdtLoc, could regulate toxin production only in 027 ribotypes and not in others (57). The authors of that study proposed that CdtR could be regulating toxin production by regulating the TcdR through a yet-to-be-identified intermediary regulator in the 027 ribotype. It has been previously proposed that the ability to regulate toxin production in response to various environmental cues with various regulatory responses may be different for 027 ribotypes in comparison to other C. difficile ribotypes (31). Results from subsequent studies are in agreement with this proposal. For example, a mutation in the highly conserved codY gene results in different phenotypes from 027 ribotypes and other ribotypes. The codY mutation results in a hypersporulation phenotype in a 027 ribotype (UK1 strain) and produces only a moderate effect on the sporulation in an 012 ribotype (630 strain) (34). It is also worth noting that sin locus expression levels were different in codY mutants in these two different C. difficile backgrounds (34). Similarly, a mutation in spo0A resulted in increased toxin production only in the 027 ribotype and not in the 012 ribotype (29, 30). In the current study, we observed the positive influence of TcdR on sporulation only in R20291 of the 027 ribotype and not in strain 630 of ribotype 012. Even though those previous studies, along with our observations, suggested that ribotype 027 has unique gene regulatory networks that differ from those of other C. difficile strains, variations may be present in strains within the 027 ribotype. Detailed study is needed to check whether the gene regulatory networks of the toxin synthesis and sporulation pathway are connected in all known ribotype 027 strains. In such a case, the ability to synchronize the toxin production and the sporulation can provide the selective advantage to ribotype 027 isolates to enable them to be more successful, with increased virulence and high transmission abilities. Deciphering the connections between toxins and the sporulation regulatory network could lead to the discovery of other novel regulators and pathways that can be targeted for the development of new therapeutics to manage C. difficile infections. Any treatment that leads to inhibition of toxin production and spore formation in patients with C. difficile infection can potentially lower the severity of the disease in addition to the transmission and recurrence of infection through dissemination of the spores.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Clostridium difficile strains (Table 2) were grown anaerobically in TY agar (tryptose, yeast extract) or 70:30 medium (58) as described previously (15, 54). Cefoxitin (Cef; 25 µg/ml), thiamphenicol (Thio; 15 µg/ml), and lincomycin (Lin; 15 µg/ml) were added to C. difficile cultures whenever necessary. Escherichia coli strains were grown in (LB) broth. E. coli strain S17-1 (59), used for conjugation, was supplemented with ampicillin (100 µg/ml) or chloramphenicol (25 µg/ml) when indicated and cultured aerobically in LB broth.
TABLE 2 .
Bacterial strains and plasmids used in this study
| Strain or plasmid used | Description | Reference or source |
|---|---|---|
| Strains | ||
| C. difficile R20291 | NAP1/027 ribotype | 31 |
| C. difficile R20291::tcdR | R20291with intron insertion in tcdR gene | This study |
| C. difficile 630Δerm | Erm′ derivative of strain 630 | 63 |
| C. difficile 630Δerm::tcdR | 630Δerm with intron insertion in tcdR gene | This study |
| E. coli DH5α | endA1 recA1 deoR hsdR17 (rK− mK+) | NEB laboratories |
| E. coli S17-1 | Strain with integrated RP4 conjugation transfer function for conjugation between E. coli and C. difficile | 59 |
| E. coli GM241(DE3) | gusA mutant lysogenized with DE3 phage and host for gusA reporter plasmids | 54 |
| Plasmids | ||
| pMTL007-CE5 | ClosTron plasmid | 18 |
| pMTL007-CE5::tcdR-141 | pMTL007-CE5 carrying tcdR-specific intron | This study |
| pRPF185 | C. difficile shuttle vector | 64 |
| pRGL294 | pRPF185 with tcdR expressed from its own promoter | This study |
| pACYC184 | E. coli cloning vector; compatible with pET16B | Neb |
| pACYC515 | pACYC184 vector carrying gusA gene under the control of the tcdR promoter | 54 |
| pET16b | E. coli expression vector | Novagen |
| pRGL312 | pET16B with tcdR | This study |
| pRGL320 | pACYC184 vector carrying gusA gene under the control of the bclA2 promoter | This study |
| pRGL321 | pACYC184 vector carrying gusA gene under the control of the bclA3 promoter | This study |
| C. difficile R20291::tcdR + pRGL294 | R20291::tcdR complemented with tcdR | This study |
| C. difficile R20291::tcdR + pRPF185 | R20291::tcdR with vector control | This study |
Construction of a tcdR mutant.
A tcdR mutation was constructed in a C. difficile strain using a ClosTron gene knockout system (18). The group II intron insertion site in the antisense orientation between nucleotides 141 and 142 of the tcdR ORF was selected using the Perutka algorithm, a Web-based design tool available at http://www.clostron.com. The designed retargeted intron was cloned into pMTL007-CE5, and the resulting plasmid, pMTL007-CE5::Cdi-tcdR-141a, was transferred into R20291 by conjugation as described previously (15, 22). The selection of thiamphenicol-resistant transconjugants in 15 µg·ml−1 lincomycin plates confers potential Lactococcus lactis ltrB (Ll.ltrB) insertions within the target tcdR gene in the chromosome of R20291. The presence of a putative tcdR mutant was identified by PCR using tcdR-specific primers (Table S1) in combination with the EBSu universal and ERM primers. Specific single integration of the group II intron into the genome was verified by Southern blotting using a (32P)dATP-radiolabeled probe specific for the tcdR gene as described previously (15, 22). Complementation of the C. difficile R20291::tcdR mutant is described in Text S1 in the supplemental materials.
Supplemental methods. I. Spore preparation. II. Toxin ELISA. III. Complementation of tcdR mutant. IV. RNA-seq analysis. V. Quantitative reverse transcriptase PCR. Download TEXT S1, DOCX file, 0.03 MB (29.1KB, docx) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Toxin assays.
Cultures of R20291 and the R20291::tcdR mutant were centrifuged after 10 h in TY medium, and toxin ELISAs were performed as described previously (15). Details are presented in Text S1.
Sporulation assay (microscopic analysis).
C. difficile cultures were grown overnight in TY medium supplemented with 0.1% taurocholate to induce germination of any spores that were present. Cells were then diluted in TY medium to an OD600 of 0.5, and then 100 µl was spread on 70:30 sporulation agar (58). Plates were incubated at 37°C and monitored for the production of spores. Cells were harvested from the plates after 24 h and were suspended in TY medium for phase-contrast microscopy as described previously (58). At least four fields per strain were obtained, and the numbers of spores and vegetative cells were counted to calculate the percentage of spores based on the total numbers of spores and vegetative cells. Experiments were performed at least three independent times.
Sporulation assay (ethanol resistance method).
C. difficile strains were inoculated into and grown on 70:30 sporulation agar as described above. After 24 h of growth, cells were scraped from the plates and suspended in 70:30 sporulation liquid medium to an OD600 of 1.0. Cells were immediately serially diluted and plated onto TY agar–0.1% taurocholate to enumerate viable vegetative cells and spores. A 0.5-ml aliquot of the culture was removed from the chamber, mixed with 0.5 ml of 95% ethanol, subjected to vortex mixing, and incubated at room temperature for 15 min. Ethanol-treated cells were serially diluted in 1× phosphate-buffered saline (PBS), returned to the anaerobic chamber, and plated onto TY agar–0.1% taurocholate plates to enumerate spores. After 24 h of growth, CFU were enumerated, and percent sporulation was calculated as the number of ethanol-resistant spores divided by the total number of viable cells (vegetative cells and spores).
Spore preparation.
Spores were generated and purified as previously described (26, 27). Details are presented in Text S1.
RNA-seq analysis and quantitative reverse transcription-PCR (qRT-PCR).
RNA-seq analysis was performed at the DNA Core Facility at the University of Missouri, and the data were analyzed using methods described previously (60–62). Details of the RNA-seq analysis and the qRT-PCR (19, 55) are provided in Text S1.
Germination.
Purified C. difficile spores were heat activated at 65°C for 30 min and then placed on ice. Ten microliters of the heat-activated spores was added to reach a final optical density at 600 nm (OD600) of 0.5 in 990 µl of BHIS medium (brain heart infusion [Difco] supplemented with 5 g/liter yeast extract and 0.1% l-cysteine) alone or supplemented with a 2, 5, 10, 20, or 50 mM concentration of taurocholic acid (TA). Germination was monitored at 600 nm for 30 min in a PerkinElmer (Waltham, MA) Lambda25 UV/Vis spectrophotometer. The data points at OD600 (Tx) were normalized to the starting OD600 value (T0). The germination rates and the 50% effective concentration (EC50) were calculated using the slopes of the linear portions of the germination plots as described previously (26, 28). The EC50 is the concentration of germinant needed to reach 50% of the maximum germination rate. EC50s were individually calculated from each germination experiment and are reported as averages with standard errors of the means.
Spore heat resistance.
Purified spores (nearly 1 × 105) prepared as described above were resuspended in 500 µl of water and incubated at 70°C. Samples were removed at 0.5 h, 4 h, and 8 h, serially diluted in PBS, plated onto TY agar plates with 0.1% taurocholate, and grown anaerobically for 48 h before counting was performed (44, 45). As a control for non-heat-treated spores, an aliquot was plated onto TY agar–0.1% taurocholate plates prior to the experiment and colonies were counted as described above.
Transmission electron microscopy.
All steps in sample preparation were performed at room temperature using pelleted spores in a 1.5-ml microcentifuge tube, and solutions were prepared in 1× PBS unless indicated otherwise. For transmission electron microscopy, spores (1010) were fixed for 2 h in a solution of 2% glutaraldehyde–2% paraformaldehyde. The spores were thoroughly rinsed three times in 1× PBS (for 5 min each time) and postfixed with 1% osmium tetroxide with constant rotation for 1 to 2 h. The samples were then washed thrice with 1× PBS (for 5 min each time) and stained en bloc with 2% aqueous uranyl acetate for 1 h under light-protected conditions and then washed three times (for 5 min each time) with distilled water. The spores were further dehydrated in a graded 50% (vol/vol)-to-95% (vol/vol) acetone series for 5 min and left in 100% acetone overnight. Infiltration was carried out in graded acetone/EMBED 812/araldite resin (Electron Microscopy Sciences) at ratios of 1:1 and 1:2 for 10 min each time at room temperature with constant rotation and incubated in 100% resin overnight. The resin was cured at 60°C for 24 to 48 h, and thin sections (silver to gold color) were cut and absorbed onto on 200-mesh copper grids. Sections were examined with a transmission electron microscope (CM100; FEI Company) at 100 kV, and images were captured using a side-mounted Hamamatsu digital camera (model C8484) with AMT image capture software version 602.591n.
Accession number(s).
Sequence data have been deposited in the NCBI GEO database under accession number GSE85395.
ACKNOWLEDGMENTS
We thank Nigel Minton, University of Nottingham, for sharing the plasmid pMTL007C-E5 and Robert Fagan for providing plasmid pRPF185. We also thank Jose E. Lopez for technical assistance throughout the study.
R.G. is supported by 1R15AI122173 from NIAID. Funds from the Johnson Cancer Center-KSU and a pilot project to R.G. from CBID-KU (1P20GM113117-01) also supported this work. J.A.S. is supported by award 5R01AI116895 from the National Institute of Allergy and Infectious Diseases.
The content is solely our responsibility and does not necessarily represent the official views of the funding agencies or the National Institutes of Health.
REFERENCES
- 1.CDC 2013. Antibiotic resistance threats in the United States, 2013. http://www.cdc.gov/drugresistance/threat-report-2013/pdf/ar-threats-2013-508.pdf.
- 2.McFarland LV. 1998. Epidemiology, risk factors and treatments for antibiotic-associated diarrhea. Dig Dis 16:292–307. doi: 10.1159/000016879. [DOI] [PubMed] [Google Scholar]
- 3.Cohen SH, Gerding DN, Johnson S, Kelly CP, Loo VG, McDonald LC, Pepin J, Wilcox MH, Society for Healthcare Epidemiology of America, Infectious Diseases Society of America . 2010. Clinical practice guidelines for Clostridium difficile infection in adults: 2010 update by the Society for Healthcare Epidemiology of America (SHEA) and the Infectious Diseases Society of America (IDSA). Infect Control Hosp Epidemiol 31:431–455. doi: 10.1086/651706. [DOI] [PubMed] [Google Scholar]
- 4.Kyne L, Hamel MB, Polavaram R, Kelly CP. 2002. Health care costs and mortality associated with nosocomial diarrhea due to Clostridium difficile. Clin Infect Dis 34:346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 5.Goudarzi M, Seyedjavadi SS, Goudarzi H, Mehdizadeh Aghdam E, Nazeri S. 2014. Clostridium difficile infection: epidemiology, pathogenesis, risk factors, and therapeutic options. Scientifica 2014:916826. doi: 10.1155/2014/916826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popoff MR, Bouvet P. 2009. Clostridial toxins. Future Microbiol 4:1021–1064. doi: 10.2217/fmb.09.72. [DOI] [PubMed] [Google Scholar]
- 7.Mani N, Dupuy B. 2001. Regulation of toxin synthesis in Clostridium difficile by an alternative RNA polymerase sigma factor. Proc Natl Acad Sci U S A 98:5844–5849. doi: 10.1073/pnas.101126598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mani N, Lyras D, Barroso L, Howarth P, Wilkins T, Rood JI, Sonenshein AL, Dupuy B. 2002. Environmental response and autoregulation of Clostridium difficile TxeR, a sigma factor for toxin gene expression. J Bacteriol 184:5971–5978. doi: 10.1128/JB.184.21.5971-5978.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.El Meouche I, Peltier J, Monot M, Soutourina O, Pestel-Caron M, Dupuy B, Pons JL. 2013. Characterization of the SigD regulon of C. difficile and its positive control of toxin production through the regulation of tcdR. PLoS One 8:e83748. doi: 10.1371/journal.pone.0083748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dineen SS, McBride SM, Sonenshein AL. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J Bacteriol 192:5350–5362. doi: 10.1128/JB.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dineen SS, Villapakkam AC, Nordman JT, Sonenshein AL. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol Microbiol 66:206–219. doi: 10.1111/j.1365-2958.2007.05906.x. [DOI] [PubMed] [Google Scholar]
- 12.Antunes A, Camiade E, Monot M, Courtois E, Barbut F, Sernova NV, Rodionov DA, Martin-Verstraete I, Dupuy B. 2012. Global transcriptional control by glucose and carbon regulator CcpA in Clostridium difficile. Nucleic Acids Res 40:10701–10718. doi: 10.1093/nar/gks864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antunes A, Martin-Verstraete I, Dupuy B. 2011. CcpA-mediated repression of Clostridium difficile toxin gene expression. Mol Microbiol 79:882–899. doi: 10.1111/j.1365-2958.2010.07495.x. [DOI] [PubMed] [Google Scholar]
- 14.Dupuy B, Matamouros S. 2006. Regulation of toxin and bacteriocin synthesis in Clostridium species by a new subgroup of RNA polymerase sigma-factors. Res Microbiol 157:201–205. doi: 10.1016/j.resmic.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Sirigi Reddy AR, Girinathan BP, Zapotocny R, Govind R. 2013. Identification and characterization of Clostridium sordellii toxin gene regulator. J Bacteriol 195:4246–4254. doi: 10.1128/JB.00711-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dupuy B, Sonenshein AL. 1998. Regulated transcription of Clostridium difficile toxin genes. Mol Microbiol 27:107–120. doi: 10.1046/j.1365-2958.1998.00663.x. [DOI] [PubMed] [Google Scholar]
- 17.Dupuy B, Mani N, Katayama S, Sonenshein AL. 2005. Transcription activation of a UV-inducible Clostridium perfringens bacteriocin gene by a novel sigma factor. Mol Microbiol 55:1196–1206. doi: 10.1111/j.1365-2958.2004.04456.x. [DOI] [PubMed] [Google Scholar]
- 18.Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, Scott JC, Minton NP. 2010. The ClosTron: mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80:49–55. doi: 10.1016/j.mimet.2009.10.018. [DOI] [PubMed] [Google Scholar]
- 19.Saujet L, Pereira FC, Serrano M, Soutourina O, Monot M, Shelyakin PV, Gelfand MS, Dupuy B, Henriques AO, Martin-Verstraete I. 2013. Genome-wide analysis of cell type-specific gene transcription during spore formation in Clostridium difficile. PLoS Genet 9:e1003756. doi: 10.1371/journal.pgen.1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fimlaid KA, Bond JP, Schutz KC, Putnam EE, Leung JM, Lawley TD, Shen A. 2013. Global analysis of the sporulation pathway of Clostridium difficile. PLoS Genet 9:e1003660. doi: 10.1371/journal.pgen.1003660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereira FC, Saujet L, Tomé AR, Serrano M, Monot M, Couture-Tosi E, Martin-Verstraete I, Dupuy B, Henriques AO. 2013. The spore differentiation pathway in the enteric pathogen Clostridium difficile. PLoS Genet 9:e1003782. doi: 10.1371/journal.pgen.1003782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Govind R, Dupuy B. 2012. Secretion of Clostridium difficile toxins A and B requires the holin-like protein TcdE. PLoS Pathog 8:e1002727. doi: 10.1371/journal.ppat.1002727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Govind R, Fitzwater L, Nichols R. 2015. Observations on the role of TcdE isoforms in Clostridium difficile toxin secretion. J Bacteriol 197:2600–2609. doi: 10.1128/JB.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Losick R, Stragier P. 1992. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature 355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 25.Bai U, Mandic-Mulec I, Smith I. 1993. SinI modulates the activity of SinR, a developmental switch protein of Bacillus subtilis, by protein-protein interaction. Genes Dev 7:139–148. doi: 10.1101/gad.7.1.139. [DOI] [PubMed] [Google Scholar]
- 26.Francis MB, Allen CA, Shrestha R, Sorg JA. 2013. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorg JA, Sonenshein AL. 2010. Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192:4983–4990. doi: 10.1128/JB.00610-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bhattacharjee D, Francis MB, Ding X, McAllister KN, Shrestha R, Sorg JA. 2015. Reexamining the germination phenotypes of several Clostridium difficile strains suggests another role for the CspC germinant receptor. J Bacteriol 198:777–786. doi: 10.1128/JB.00908-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackin KE, Carter GP, Howarth P, Rood JI, Lyras D. 2013. Spo0A differentially regulates toxin production in evolutionarily diverse strains of Clostridium difficile. PLoS One 8:e79666. doi: 10.1371/journal.pone.0079666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. 2012. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infect Immun 80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stabler RA, He M, Dawson L, Martin M, Valiente E, Corton C, Lawley TD, Sebaihia M, Quail MA, Rose G, Gerding DN, Gibert M, Popoff MR, Parkhill J, Dougan G, Wren BW. 2009. Comparative genome and phenotypic analysis of Clostridium difficile 027 strains provides insight into the evolution of a hypervirulent bacterium. Genome Biol 10:R102. doi: 10.1186/gb-2009-10-9-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pettit LJ, Browne HP, Yu L, Smits WK, Fagan RP, Barquist L, Martin MJ, Goulding D, Duncan SH, Flint HJ, Dougan G, Choudhary JS, Lawley TD. 2014. Functional genomics reveals that Clostridium difficile Spo0A coordinates sporulation, virulence and metabolism. BMC Genomics 15:160. doi: 10.1186/1471-2164-15-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards AN, Tamayo R, McBride SM. 2016. A novel regulator controls Clostridium difficile sporulation, motility and toxin production. Mol Microbiol 100:954–971. doi: 10.1111/mmi.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nawrocki KL, Edwards AN, Daou N, Bouillaut L, McBride SM. 2016. CodY-dependent regulation of sporulation in Clostridium difficile. J Bacteriol 198:2113–2130. doi: 10.1128/JB.00220-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Janoir C, Denève C, Bouttier S, Barbut F, Hoys S, Caleechum L, Chapetón-Montes D, Pereira FC, Henriques AO, Collignon A, Monot M, Dupuy B. 2013. Adaptive strategies and pathogenesis of Clostridium difficile from in vivo transcriptomics. Infect Immun 81:3757–3769. doi: 10.1128/IAI.00515-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pishdadian K, Fimlaid KA, Shen A. 2015. SpoIIID-mediated regulation of sigmaK function during Clostridium difficile sporulation. Mol Microbiol 95:189–208. doi: 10.1111/mmi.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fujita M, Losick R. 2002. An investigation into the compartmentalization of the sporulation transcription factor sigmaE in Bacillus subtilis. Mol Microbiol 43:27–38. doi: 10.1046/j.1365-2958.2002.02732.x. [DOI] [PubMed] [Google Scholar]
- 38.Jonas RM, Weaver EA, Kenney TJ, Moran CP Jr., Haldenwang WG. 1988. The Bacillus subtilis spoIIG operon encodes both sigma E and a gene necessary for sigma E activation. J Bacteriol 170:507–511. doi: 10.1128/jb.170.2.507-511.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaBell TL, Trempy JE, Haldenwang WG. 1987. Sporulation-specific sigma factor sigma 29 of Bacillus subtilis is synthesized from a precursor protein, P31. Proc Natl Acad Sci U S A 84:1784–1788. doi: 10.1073/pnas.84.7.1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofmeister AE, Londoño-Vallejo A, Harry E, Stragier P, Losick R. 1995. Extracellular signal protein triggering the proteolytic activation of a developmental transcription factor in B. subtilis. Cell 83:219–226. doi: 10.1016/0092-8674(95)90163-9. [DOI] [PubMed] [Google Scholar]
- 41.Karow ML, Glaser P, Piggot PJ. 1995. Identification of a gene, spoIIR, that links the activation of sigma E to the transcriptional activity of sigma F during sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A 92:2012–2016. doi: 10.1073/pnas.92.6.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawley TD, Croucher NJ, Yu L, Clare S, Sebaihia M, Goulding D, Pickard DJ, Parkhill J, Choudhary J, Dougan G. 2009. Proteomic and genomic characterization of highly infectious Clostridium difficile 630 spores. J Bacteriol 191:5377–5386. doi: 10.1128/JB.00597-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strong PC, Fulton KM, Aubry A, Foote S, Twine SM, Logan SM. 2014. Identification and characterization of glycoproteins on the spore surface of Clostridium difficile. J Bacteriol 196:2627–2637. doi: 10.1128/JB.01469-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barra-Carrasco J, Olguín-Araneda V, Plaza-Garrido A, Miranda-Cárdenas C, Cofré-Araneda G, Pizarro-Guajardo M, Sarker MR, Paredes-Sabja D. 2013. The Clostridium difficile exosporium cysteine (CdeC)-rich protein is required for exosporium morphogenesis and coat assembly. J Bacteriol 195:3863–3875. doi: 10.1128/JB.00369-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnelly ML, Fimlaid KA, Shen A. 2016. Characterization of Clostridium difficile spores lacking either SpoVAC or dipicolinic acid synthetase. J Bacteriol 198:1694–1707. doi: 10.1128/JB.00986-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Paredes-Sabja D, Setlow P, Sarker MR. 2011. Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19:85–94. doi: 10.1016/j.tim.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 47.Orsburn B, Melville SB, Popham DL. 2008. Factors contributing to heat resistance of Clostridium perfringens endospores. Appl Environ Microbiol 74:3328–3335. doi: 10.1128/AEM.02629-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen NY, Jiang SQ, Klein DA, Paulus H. 1993. Organization and nucleotide sequence of the Bacillus subtilis diaminopimelate operon, a cluster of genes encoding the first three enzymes of diaminopimelate synthesis and dipicolinate synthase. J Biol Chem 268:9448–9465. [PubMed] [Google Scholar]
- 49.Vepachedu VR, Setlow P. 2007. Role of SpoVA proteins in release of dipicolinic acid during germination of Bacillus subtilis spores triggered by dodecylamine or lysozyme. J Bacteriol 189:1565–1572. doi: 10.1128/JB.01613-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams CM, Eckenroth BE, Putnam EE, Doublié S, Shen A. 2013. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog 9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burns DA, Heap JT, Minton NP. 2010. SleC is essential for germination of Clostridium difficile spores in nutrient-rich medium supplemented with the bile salt taurocholate. J Bacteriol 192:657–664. doi: 10.1128/JB.01209-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gutelius D, Hokeness K, Logan SM, Reid CW. 2014. Functional analysis of SleC from Clostridium difficile: an essential lytic transglycosylase involved in spore germination. Microbiology 160:209–216. doi: 10.1099/mic.0.072454-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Francis MB, Allen CA, Sorg JA. 2015. Spore cortex hydrolysis precedes dipicolinic acid release during Clostridium difficile spore germination. J Bacteriol 197:2276–2283. doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Govind R, Vediyappan G, Rolfe RD, Dupuy B, Fralick JA. 2009. Bacteriophage-mediated toxin gene regulation in Clostridium difficile. J Virol 83:12037–12045. doi: 10.1128/JVI.01256-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saujet L, Monot M, Dupuy B, Soutourina O, Martin-Verstraete I. 2011. The key sigma factor of transition phase, SigH, controls sporulation, metabolism, and virulence factor expression in Clostridium difficile. J Bacteriol 193:3186–3196. doi: 10.1128/JB.00272-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Elliott B, Dingle KE, Didelot X, Crook DW, Riley TV. 2014. The complexity and diversity of the pathogenicity locus in Clostridium difficile clade 5. Genome Biol Evol 6:3159–3170. doi: 10.1093/gbe/evu248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lyon SA, Hutton ML, Rood JI, Cheung JK, Lyras D. 2016. CdtR regulates TcdA and TcdB production in Clostridium difficile. PLoS Pathog 12:e1005758. doi: 10.1371/journal.ppat.1005758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Putnam EE, Nock AM, Lawley TD, Shen A. 2013. SpoIVA and SipL are Clostridium difficile spore morphogenetic proteins. J Bacteriol 195:1214–1225. doi: 10.1128/JB.02181-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Teng F, Murray BE, Weinstock GM. 1998. Conjugal transfer of plasmid DNA from Escherichia coli to enterococci: a method to make insertion mutations. Plasmid 39:182–186. doi: 10.1006/plas.1998.1336. [DOI] [PubMed] [Google Scholar]
- 60.Criscuolo A, Brisse S. 2013. AlienTrimmer: a tool to quickly and accurately trim off multiple short contaminant sequences from high-throughput sequencing reads. Genomics 102:500–506. doi: 10.1016/j.ygeno.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 61.Monot M, Orgeur M, Camiade E, Brehier C, Dupuy B. 2014. COV2HTML: a visualization and analysis tool of bacterial next generation sequencing (NGS) data for postgenomics life scientists. OMICS 18:184–195. doi: 10.1089/omi.2013.0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée JY, Dupuy B, Martin-Verstraete I. 2013. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet 9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hussain HA, Roberts AP, Mullany P. 2005. Generation of an erythromycin-sensitive derivative of Clostridium difficile strain 630 (630Deltaerm) and demonstration that the conjugative transposon Tn916DeltaE enters the genome of this strain at multiple sites. J Med Microbiol 54:137–141. doi: 10.1099/jmm.0.45790-0. [DOI] [PubMed] [Google Scholar]
- 64.Fagan RP, Fairweather NF. 2011. Clostridium difficile has two parallel and essential Sec secretion systems. J Biol Chem 286:27483–27493. doi: 10.1074/jbc.M111.263889. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Construction and characterization of tcdR mutant in C. difficile. (A) Schematic representation of insertional inactivation of tcdR by group II intron. (B) The intron insertion in the tcdR coding region was verified by PCR using the intron-specific primer EBS along with gene-specific primers ORG81 and ORG82 in the parent (R20291), in the tcdR mutant (R20291::tcdR), and in the tcdR complemented strain (R20291::tcdR+pRG294). The same strategy was followed to verify the tcdR mutation in the 630Δerm strain. (C) Southern blot analysis of genomic DNA from the WT and tcdR mutant strains with a tcdR-specific probe. The shift in the hybridization band indicates the integration of the intron within tcdR coding region. Download FIG S1, PDF file, 0.2 MB (180.9KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Oligonucleotides used in the study. Download TABLE S1, DOCX file, 0.1 MB (128.7KB, docx) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression analysis of toxin genes and known toxin gene regulators in the tcdR mutant. RNA was prepared from R202091 and R20291::tcdR strains grown in TY medium for 16 h. qPCR analysis was performed for selected regulator-coding genes. Statistical analysis was performed using the t test, and the error bars indicate the standard errors of the means (s.e.m.). **, P value < 0.01. Download FIG S2, PDF file, 0.1 MB (55.9KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression analysis of selected sporulation genes during growth in 70:30 medium. RNA was prepared from R202091 and R20291::tcdR strains grown in 70:30 medium for 24 h. qRT-PCR analysis was performed for selected sporulation genes. Statistical analysis was performed using the t test, and the error bars indicate the standard errors of the means (s.e.m.). *, P value < 0.05. Download FIG S3, PDF file, 0.1 MB (70.4KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Expression analysis of selected sporulation genes during growth in TY medium. RNA was prepared from R202091 and R20291::tcdR strains that were grown in TY medium for 24 h. qRT-PCR analysis was performed for selected sporulation genes. Statistical analysis was performed using the t test, and the error bars indicate the standard errors of the means (s.e.m.). *, P value < 0.05. Download FIG S4, PDF file, 0.1 MB (57.1KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TEM analysis of spores from the R20291 and R20291::tcdR strains. The tcdR mutant spores (n = 60) were scored 100% for the presence of ruffled defective exosporium (marked with black arrow) and 98% for the presence of weakly stained core (marked as open triangle). Bar, 100 nm. Download FIG S5, PDF file, 1.5 MB (1.5MB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Toxin ELISA for 630Δerm and 630Δerm::tcdR strains. TcdA and TcdB expression levels in cytosolic fractions of C. difficile strains grown for 10 h in TY medium were quantified using ELISA. Data are from results of an experiment representative of three independent assays. Error bars correspond to the standard errors of the means of results from at least three biological replicates. The asterisks (***) in panel B indicate statistical difference at P < 0.001, estimated by Student’s t test. Download FIG S6, PDF file, 0.04 MB (44.2KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Beta-glucuronidase activity of bclA2 promoter-gusA and bclA3 promoter-gusA fusions in the presence or absence of TcdR. E. coli strains carrying promoter fusion plasmids (pbclA2-gusA and pbclA3-gusA) along with a TcdR-expressing plasmid (pRGL312) or the vector (pET16b) were grown for 6 h, and TcdR expression was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 4 h at 37°C. Bacterial cultures were harvested and were assayed for beta-glucuronidase activity (quantified in Miller units) as described previously (18). The values represent the means of the results from three independent experiments. Statistical analysis was performed using the t test, and the error bars indicate standard errors of the means. The asterisks (**) indicate statistical difference at P < 0.005, estimated by Student’s t test. Download FIG S7, PDF file, 0.1 MB (99.6KB, pdf) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Supplemental methods. I. Spore preparation. II. Toxin ELISA. III. Complementation of tcdR mutant. IV. RNA-seq analysis. V. Quantitative reverse transcriptase PCR. Download TEXT S1, DOCX file, 0.03 MB (29.1KB, docx) .
Copyright © 2017 Girinathan et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.