Abstract
Viral internal ribosome entry sites (IRESs) are unique RNA elements, which use stable and dynamic RNA structures to recruit ribosomes and drive protein synthesis. IRESs overcome the high complexity of the canonical eukaryotic translation initiation pathway, often functioning with a limited set of eukaryotic initiation factors. The simplest types of IRESs are typified by the cricket paralysis virus intergenic region (CrPV IGR) and hepatitis C virus (HCV) IRESs, both of which independently form high-affinity complexes with the small (40S) ribosomal subunit and bypass the molecular processes of cap-binding and scanning. Owing to their simplicity and ribosomal affinity, the CrPV and HCV IRES have been important models for structural and functional studies of the eukaryotic ribosome during initiation, serving as excellent targets for recent technological breakthroughs in cryogenic electron microscopy (cryo-EM) and single-molecule analysis. High-resolution structural models of ribosome : IRES complexes, coupled with dynamics studies, have clarified decades of biochemical research and provided an outline of the conformational and compositional trajectory of the ribosome during initiation. Here we review recent progress in the study of HCV- and CrPV-type IRESs, highlighting important structural and dynamics insights and the synergy between cryo-EM and single-molecule studies.
This article is part of the themed issue ‘Perspectives on the ribosome’.
Keywords: eukaryotic translation, ribosome, internal ribosome entry site, cryogenic electron microscopy, single-molecule biophysics
1. Introduction
Translation of messenger RNA (mRNA) is a fundamental stage of gene expression, entailing the stochastic assembly, dynamic structural rearrangement and subsequent catalysis of protein synthesis by ribosomal complexes. The basic process of translation by the ribosome is conserved across all kingdoms of life, yet the molecular details can differ among species for any of the four stages of the translation cycle: initiation, elongation, termination and recycling [1]. This is particularly the case for eukaryotic initiation, which is probably the most complex phase of eukaryotic translation [2–4]. Eukaryotic initiation differs substantially from the relatively simple process in prokaryotes; in particular, ribosomal (r)RNA : mRNA base-pairing by the Shine–Dalgarno (SD)/anti-Shine–Dalgarno (SD/anti-SD) interaction mediates small (30S) ribosomal subunit recruitment to most prokaryotic mRNA, while no general mechanism of rRNA : mRNA base-pairing is used for ribosome recruitment in eukaryotes. Additionally, at least 12 eukaryotic initiation factors (eIFs), composed of many more polypeptides, are needed for efficient eukaryotic initiation, and only a set of three protein initiation factors (IF1-3) are used in prokaryotic initiation.
(a). The canonical model of eukaryotic translation initiation
Irrespective of mechanism, there are three general molecular events in initiation: (i) small ribosomal subunit and initiator tRNA recruitment to an mRNA, (ii) start codon recognition to establish the correct reading frame, and (iii) large ribosomal subunit joining to form an elongation-competent ribosome. Each eukaryotic mRNA has a 5′ end-modified 7-methylguanylate (m7G) cap structure (5′ cap), long 5′ untranslated regions (UTRs) and even longer 3′ UTRs with polyadenylated (PolyA) tails [5]. In canonical eukaryotic initiation, the small (40S) ribosomal subunit is first recruited to an mRNA at the 5′ cap as a pre-formed initiator tRNA- and protein factor–containing 43S pre-initiation complex (PIC), by the eIF4F ‘cap-binding’ complex (composed of eIF4A, 4E and 4G). The 43S complex then inspects sequences along the 5′ UTR in a 5′ to 3′ directionality (scanning) in an ATP-dependent manner [6], requiring auxiliary factors such as DHX29 to proceed through structured RNAs within the UTR [7]. Upon locating the start codon (typically AUG) via base-pairing of Met-tRNAiMet with mRNA in the P site, a series of conformational and compositional rearrangements take place to trigger the formation of the 48S PIC, followed by large (60S) subunit joining and eIF departure to configure the elongation-competent 80S ribosome. While there are structural and functional analogues of prokaryotic initiation factors IF1-3 in eukaryotes (eIF1A, eIF5B and eIF1, respectively), numerous additional protein factors are required to carry out the processes of cap-binding and scanning (figure 1), and endow the pathway with many regulatory inputs for control by cell signalling [8]. Eukaryotic translation initiation has been extensively reviewed elsewhere [2–4,9].
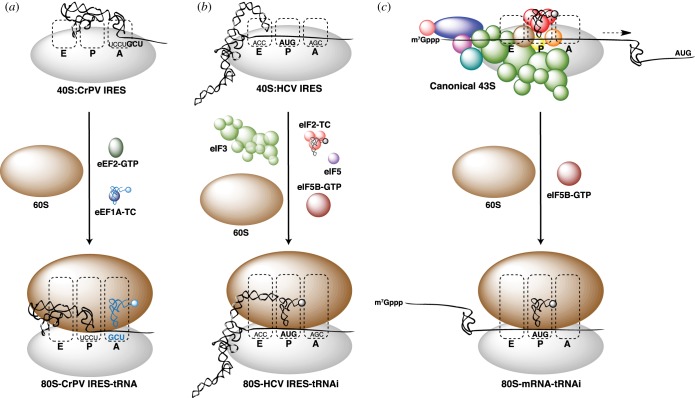
Figure 1.
Canonical and viral IRES-mediated eukaryotic translation initiation. All pathways are depicted from the first 40S : mRNA binding event to the formation of an 80S ribosome with a tRNA paired to the codon of the first amino acid. (a) Initiation on the type 4 CrPV-like IGR IRES requires the factors eEF1A and eEF2. The first GCU (Ala) codon is paired with an elongator tRNA in the A site. (b) Initiation on the type 3 HCV-like IRES requires the factors eIF2, eIF3, eIF5 and eIF5B. The AUG (Met) start codon is paired with an initiator tRNA in the P site. (c) Canonical initiation on cellular mRNAs requires numerous additional factors to promote 43S PIC recruitment to the m7Gppp cap and subsequent scanning to the AUG start codon (dashed arrow). As with the HCV IRES, the AUG start codon is paired with an initiator tRNA in the P site.
(b). IRES-mediated translation initiation
To overcome the inherent complexity of eukaryotic translation, certain viruses employ specialized RNA elements, termed internal ribosome entry sites (IRESs), to recruit ribosomal complexes to their mRNA in a manner independent of the 5′ cap [10]. These RNA elements were first discovered within the poliovirus and encephalomyocarditis virus genomes in the late 1980s [11,12], and have subsequently been identified in other viruses and potentially within cellular mRNAs [13]. IRESs have been classified into four types that are related to the complexity of their initiation mechanism [2,14,15] (figure 1). The simplest IRES families (types 3 and 4), or using an alternate classification—type IV and type V [16]—are exemplified by the HCV IRES and CrPV IRES, respectively. Both of these IRESs are composed of multi-domain RNAs, which form high-affinity complexes with ribosomal subunits, and dynamically manipulate their conformation to promote protein synthesis. The ability of these IRESs to form defined, high-affinity ribosomal complexes has enabled the production of structural and biochemical models for certain steps of initiation, and provided important insights into the structural dynamics of the eukaryotic ribosome. In particular, the IRES-bound ribosomes have been speculated to undergo similar conformation changes to those that occur in the 43S PIC during canonical initiation [17,18], and IRES RNA structures may functionally substitute for canonical eIFs and tRNA [14]. Thus, IRESs are not only a significant area of study for revealing the biology of major human pathogens, but also contribute to fundamental knowledge of human translation, which is often misregulated in cancer [19].
Technological advances in imaging have revolutionized structural biology and biophysics. Single-molecule imaging of biological systems in vitro and in vivo has allowed analysis of biological dynamics and pathways [20,21]. By harnessing the intrinsic sensitivity of fluorophores, single biological systems can be monitored without averaging from large numbers of molecules. This allows the composition and conformation (using energy transfer from dipolar coupling of two fluorescent dyes) of, for example, a single translating ribosome in real time [22]. Similar methods have been modified to image biological systems at resolutions below the optical diffraction limit [23]. In parallel, cryogenic electron microscopy (cryo-EM) has been transformed from a low-resolution (5–8 Å) method for large (greater than 500 kDa) complexes to a molecular-resolution (2.5–4 Å) method on both large and small (greater than 200 kDa) complexes [24–26]. Improvements in detector technology, in particular, direct electron detectors that can correct for specimen movement, plus better computational methods have driven this revolution. Translation has been a central focus of these improved approaches.
Recent high-resolution structures from cryo-EM of ribosome : IRES complexes have provided a mechanistic foundation for IRES-mediated translation. Coupled with these structures, we have developed single-molecule methods to probe the dynamic evolution of IRES complexes, and provide a time axis for the static structures from cryo-EM. Here we review this recent progress in the study of the CrPV and HCV-like IRESs with a focus on the compositional and conformational dynamics of ribosome : IRES initiation complexes.
2. The type 4 IGR IRES
Viruses within the Dicistroviridae family have approximately 8 500–10 000 nt, positive-sense RNA genomes. An IRES in the 5′ UTR directs polyprotein synthesis of non-structural proteins, while an intergenic region (IGR) IRES directs polyprotein synthesis of structural proteins (figure 2a). The IGR IRES used by viruses within the Dicistroviridae family has the simplest known mechanism of IRES translation initiation: it recruits the ribosome in the absence of host eIFs, and initiates translation from a non-AUG codon without using an initiator tRNA [27,30,31]. IGR IRESs from viruses within the Dicistroviridae family, which include the Plautia stali intestine virus, Taura syndrome virus, Israeli acute paralysis virus and the most well-studied cricket paralysis virus (CrPV), have a conserved secondary and tertiary structure [27,32–34]. The IGR IRESs are universally active in eukaryotes including yeasts [35], insects [36], rabbit and human [37], suggesting a mechanism involving evolutionarily conserved ribosomal regions to capture and manipulate eukaryotic ribosomes. Dicistroviridae IRESs are also active in prokaryotes, but the mechanism of initiation is different, as the 70S : IGR IRES complex forms on a distinct translation start site [38].
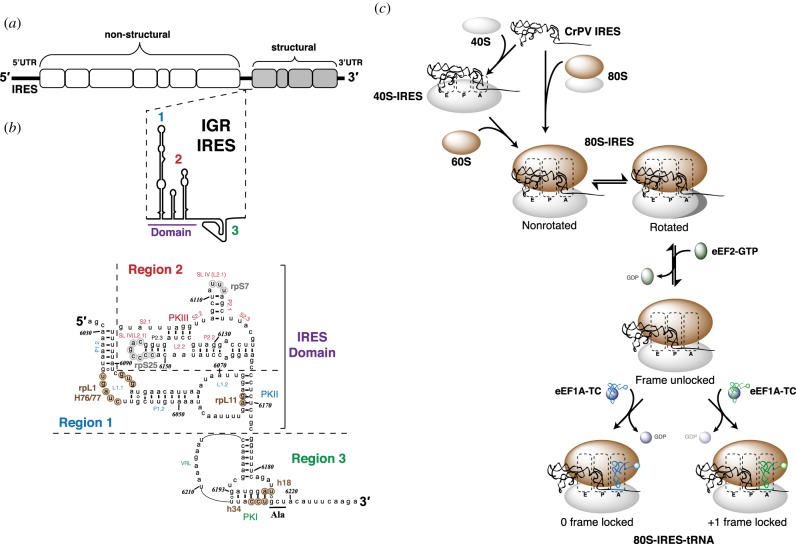
Figure 2.
CrPV IRES secondary structure and initiation pathway. (a) Schematic of the CrPV positive-sense RNA genome. (b) CrPV IRES secondary structure [27,28]. IRES regions 1 (red), 2 (blue) and 3 (green) are delineated by dotted lines. Structural elements (pseudoknot (PK); stem loop (SL); loop (L); single strand (S); paired helix (P)) are labelled and IRES nucleotides that interact with 40S (grey circles) and 60S (brown circles) ribosomal elements are identified. (c) Model of the CrPV IRES initiation pathway: 80S : IRES assembly occurs via sequential recruitment of the 40S and 60S ribosomal subunits and by direct recruitment of the 80S ribosome. The 80S : IRES complexes are in equilibrium between the non-rotated and rotated state. eEF2-mediated pseudotranslocation moves IRES PKIII from the A site to the P site of the ribosome to facilitate tRNA arrival. The translocated complexes are unstable and lack a defined reading frame. tRNA binding captures and stabilizes the translocated state of the ribosome. Reading frame selection depends on the relative rates of 0 and +1 frame tRNA arrival to the A site [29].
Biochemical and structural research has outlined the mechanisms by which the type 4 IRESs manipulate the ribosome to initiate viral protein synthesis, revealing ribosome dynamics that are conserved during both IRES-mediated and canonical translation. The following section provides an overview of IGR IRES structure and function, highlighting recent advancements in our understanding of IGR IRES-mediated translation initiation, with specific focus on the CrPV IGR IRES (herein, referred to as the CrPV IRES).
The CrPV IRES is a 190 nt segment of structured RNA upstream of the first Ala codon of the second CrPV cistron (figure 2a). The IRES has three structural regions, each of which contain a pseudoknot (PK) essential for IRES translational activity (figure 2b) [27,32]. Region 1, which begins at the 5′ end of the IRES, contains a large stem loop (SL) with an internal loop (L1.1) and an apical loop (L1.2). L1.1 segments the stem structure into separate paired helices (P), P1.1 and P1.2. Nucleotides within L1.2 form PKII by base-pairing with nucleotides downstream of single-stranded (S) segment S2.4. Region 2 of the IRES contains PKIII, SLIV and SLV. PKIII is formed by base-pairing between the internal loop (L2.2) of SLV and nucleotides between S2.1 and S2.2 upstream of SLIV. Regions 1 and 2 form a compact, globular IRES domain (figure 2b) [33]. Portions of PKII and PKIII make up the tightly packed RNA core of the IRES domain. Regions 1 and 2 accessory sequences, SLIV and SLV, and L1.1, are positioned on the surface of the IRES domain for interactions with the 40S and 60S ribosomal subunits, respectively [27,28,33,39,40]. Region 3 of the IRES, which is connected to the IRES domain by a short linker segment, contains PKI and the A-rich variable loop (VRL) [32]. The apical loop, L3.1, base pairs with nucleotides adjacent to the first codon to form PKI. Region 3 folds independently and does not make any contacts with the IRES domain in ribosome-bound or unbound states [28,33,41]. Region 3 is dispensable for ribosome binding [27], but is essential for initiation events that occur downstream of 80S ribosome assembly on the IRES [42,43].
(a). Mechanism of CrPV IRES initiation
The IRES binds the host 40S ribosome with high affinity, which is irreversible on the time scale of initiation [27,29]. The pre-folded, compact structure of type 4 IGR IRESs is essential for high-affinity 40S–IRES interaction [28,33]. The 40S : CrPV IRES complex recruits the 60S ribosomal subunit, resulting in 80S ribosome assembly on the IRES (figure 2c). These 80S ribosomes become elongation-competent on the IRES in the absence of host eIFs and initiator tRNA [30,31,35]. The first tRNA is recruited to the ribosome as a part of a ternary complex with eukaryotic elongation factor (eEF) 1A and GTP (eEF1A-TC). Toeprint analysis of initiation complexes [31,37,44] and tRNA binding assays [45,46] established the requirement of eEF1A and eEF2 (eukaryotic EF-G homologue) for tRNA recruitment. However, these experiments did not establish the precise position of the IRES in the 80S : IRES complex, or the order of translocation and tRNA arrival.
Early biochemical and structural data suggested that PKI is positioned in the P site of 80S : IRES initiation where it adopts a conformation that mimics P-site tRNA, while the Ala codon is subsequently positioned in the decoding centre (of the A site) of the ribosome [14,28,30,31,41,47]. However, recent studies have revealed that PKI of the IRES initially occupies the ribosomal A site, blocking tRNA binding [46,48]. Consistent with the requirements mentioned above, eEF2-mediated pseudotranslocation, which is defined as translocation in the absence of peptide bond formation, moves PKI from the A to the P site prior to accommodation of the first tRNA [45,46,48].
(b). Low-resolution cryo-EM structures ribosome : IRES complexes
In early low-resolution cryo-EM reconstructions of the CrPV IRES bound to the human 40S ribosome (20.3 Å), the IRES adopts an elongated shape with the majority of its density originating from the IRES domain [41]. Density from PKI, though not well resolved, extends from the E to the P site [41]. In the 80S : IRES structure (17.3 Å), the IRES is positioned within the inter-subunit space in a similar manner to that observed in 40S-IRES particles [41] (figure 3a). However, local changes in the IRES result in a slight retraction of the PKI region and overall shift of the IRES domain towards the E site compared with the 40S : IRES complex. The ribosome also undergoes conformational changes upon binding to the IRES. In the 80S : IRES complexes, the 40S ribosome head position shifts, facilitating widening of the mRNA entry channel. The electron density from the L1 stalk of the 60S subunit is well resolved in the 80S : IRES binary complex, while L1 stalk density is weak in empty 80S ribosome particles, suggesting that IRES binding induces ordering of the stalk.
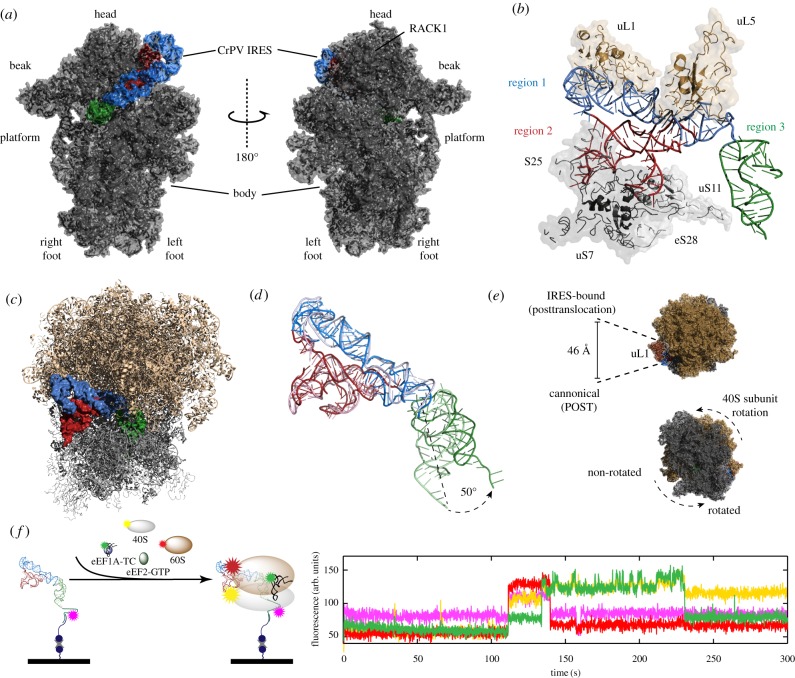
Figure 3.
Structure and dynamics of the ribosome-bound CrPV IRES. (a) 40S : IRES structure [46] (PDB entry 4V92). The 40S subunit inter-subunit face (left) and solvent face (right) structures are annotated with features of the small subunit. The IRES is coloured by region: region 1 (blue); region 2 (red); region 3 (green). (b) Structure of the 40S-bound CrPV IRES and IRES-interacting 40S (grey) and 60S (brown) ribosomal proteins [46] (PBD entry 4V92). (c) Structure of the 80S : IRES complex [46] (PBD entry 4V92). (d) Alignment of the pre- [28] (PBD entry 2NOQ) and post- [49] (PBD entry 4D5N) pseudotranslocated IRES. The post-pseudotranslocated IRES (dark colours) adopts a more extended conformation compared with the pre-pseudotranslocated IRES (light colours). (e) 60S subunit solvent face view of the 80S : IRES complex structure (top) highlighting L1 stalk displacement of the IRES-bound, post-pseudotranslocated state [49]. The 40S subunit solvent face view of the 80S : IRES complex structure showing 40S subunit rotation. (f) Observation of CrPV IRES-mediated initiation using single-molecule fluorescence. A schematic representation (left) of a ZMW single-molecule fluorescence delivery experiment where fluorescently labelled 40S (yellow) and 60S (red) subunits, eEF1A-TC tRNAPhe-Phe (green) and eEF2-GTP are co-delivered to surface-immobilized CrPV IRES-Cy5.5 (magenta). In the example trace from the experiment (right), simultaneous arrival of the 40S and 60S subunits (burst of red and yellow fluorescence at 110 s) to the immobilized IRES is followed by tRNA binding to the 80S : CrPV IRES complex (burst of green fluorescence at 130 s). Panel (f) has been reprinted with permission from Petrov et al. [29].
Using improved technologies, the yeast 80S : CrPV IGR structure was subsequently resolved to 7.3 Å, yielding detailed molecular insights into key ribosome : IRES interactions [28] (figure 3b). The exposed SLIV and SLV in region 2 of the IRES contact the 40S subunit proteins uS7 and a protein later identified as eS25 [28,50]. PKII contacts uL5, while the internal loop L1.1 interacts with uL1 and 25S rRNA. The density of region 3 is weak in this structure, consistent with the conformational flexibility of PKI. The head and shoulder of the 40S subunit, which interact with the anticodon stem loop of A-site tRNA during canonical translation, contact the PKI region of the CrPV IRES near the first codon [28].
(c). High-resolution 80S : IRES cryo-EM structures
(i). Pretranslocated 80S : CrPV IRES complexes
Using the recently improved cryo-EM methods, Fernández et al. [46] applied maximum-likelihood particle sorting to cryo-EM images of yeast 80S : CrPV IRES particles (figure 3c), and identified two major 80S : IRES subpopulations. Complete atomic models of the two major subpopulations of 80S : IRES complexes revealed ribosomes in both non-rotated (3.8 Å) and rotated (3.7 Å) states (anticlockwise rotation of 40S), similar to those observed in canonical prokaryotic translation (figure 3e) [51]. L1.1-mediated stabilization of the L1 stalk enabled its complete structural modelling. During canonical prokaryotic translation, the L1 stalk adopts closed, open and intermediate positions to facilitate interactions with P/E or E/E tRNA [52]. When bound to the CrPV IRES, the L1 stalk is oriented in an atypical outward position [46]. In the rotated state 80S : CrPV IRES complexes, the outward position is more pronounced than in the non-rotated complexes, resulting in contacts between the stalk and 60S ribosomal protein eL36 in the body of the 60S subunit. Similar ordering of the L1 stalk occurs during GTPase-ligand binding and activation during canonical translation in prokaryotes and eukaryotes [53,54], suggesting that the IRES-induced extension of the stalk may facilitate tRNA arrival and eEF2-mediated translation [41,46].
(ii). eEF2 translocation is required for binding of the first tRNA
A key point of mechanistic ambiguity in CrPV IRES-driven translation is the location of PKI within the 80S : IRES complex. Early biochemical analysis suggested that 80S : IRES assembly resulted in the positioning of PKI within the ribosomal P site [30,31]. The addition of eEF2 and eIF1A-TC to the 80S : IRES complex resulted in a 6 nt shift of the ribosome on the mRNA, as seen through toeprint analyses. This finding suggested the occurrence of two translocation events, although the precise placement of the ribosome on the IRES and the order of translocation and tRNA arrival were not determined. X-ray crystal structures of an isolated PKI fragment of the CrPV IRES suggested that PKI positions itself in the P site of the ribosome [47,55]. As mentioned above, the low-resolution cryo-EM structure of the 80S-bound CrPV IRES placed PKI near the P site [28]. Biochemical analysis with a similar IGR IRES indicated that PKI is initially positioned within the A site [45]. These ambiguities were eliminated by the higher resolution EM structures.
In the atomic models of the 80S : IRES complexes, PKI unequivocally mimics codon anticodon interactions in the A site [46,48]. Placement of PKI in the A site upon initial 80S complex assembly indicates that pseudotranslocation is required to move the IRES element from the A to the P site, positioning the first codon in the A site. A second round of pseudotranslocation moves the first tRNA to the P site and PK1 to the E site. Thus, CrPV IRES initiation requires two pseudotranslocation events prior to formation of the first peptide bond.
(iii). Architecture of the eEF2 : 80S : CrPV IRES complex
To explore how eEF2 drives this pseudotranslocation step, the cryo-EM structure of eEF2 : 80S : CrPV IRES complex was determined at high resolution (3.6 Å) [56]. eEF2•GDPCP and eEF2-mediated ribosome and IRES conformational changes were observed [56]. eEF2 binding and GTPase activation occur via a conserved mechanism previously described for prokaryotic GTPases EF-G and EF-Tu [57]. Domains II and V of eEF2 anchor the GTPase to the ribosome through contacts with uS12, resulting in approximately 3° hyper-rotation of the 40S subunit and further outward displacement of the L1 stalk compared with the rotated state of the 80S : CrPV IRES [46,56]. PKI is displaced 10 Å towards the P site adopting an A/P hybrid-like conformation [56,58]. Domain IV of eEF2 rests near the major groove of the ASL-like element of PKI, while eEF2 residues H694 and H699 interact directly with PKI. The diphthamide modification on H699 stabilizes the eEF2–PKI interaction to sterically block ribosome : IRES interactions in the A site. How the IRES–ribosome interactions are disrupted during pseudotranslocation remains an open question.
(iv). Post-translocated 80S : CrPV IRES complexes
The post-translocated state of the 80S : CrPV IRES complex was captured by replacing the first Ala codon with a stop codon [49]. Following the first round of eEF2 pseudotranslocation, the stop codon is positioned in the A site of the ribosome and recognized by eukaryotic release factor 1 (eRF1), whose binding stabilizes the translocated state of the ribosome, preventing spontaneous back-translocation [49,59]. The cryo-EM structure of the 80S : CrPV Stop : eRF1 complex was resolved to 8.7 Å [49]. In the post-translocated complexes, the IRES adopts an extended conformation within the ribosomal inter-subunit space (figure 3d). The IRES domain and PKI shift laterally by 25 Å and 22 Å, respectively, moving the IRES domain towards the E site and PKI towards the P site. The apical portion of PKI mimics an anticodon stem loop (ASL)-codon duplex in the P site. PKI is also rotated 50° about the linker element between the IRES domain. L3.2 of PKI contacts rpS5, while contacts between L2.1 and rpS5 and L2.3 and rpS25 are no longer apparent. IRES contacts with the L1 stalk were preserved, although the position of the stalk is extended outward. The 13-Å displacement of the L1 stalk is in the opposite direction, though similar in magnitude, to the mammalian elongating ribosome (figure 3e) [60]. The molecular details of the pretranslocated [46] and post-translocated [49] 80S : CrPV IRES structures have demonstrated how ribosome and IRES conformational dynamics work in concert during CrPV IRES-driven translation initiation.
(d). The CrPV IRES variable loop dynamics
Prior to elongation, reading frame must be established. For the CrPV IRES, and other IGR IRESs, both the 0 and +1 frames are translated, with the 0 frame being disproportionally favoured [43,61,62]. The dynamics of region 3 of the IRES are critical for positioning PKI in productive conformations within the ribosome, frame selection and translocation [43,63,64]. PKI can adopt conformations that mimic classical and hybrid tRNA states during IRES initiation [45,55]; however, the molecular mimicry is imperfect. In particular, during canonical translation, both the acceptor stem and peptidyl moiety are needed for frame maintenance during translocation [65,66], and their absence from PKI suggests that additional IRES elements are probably involved in frame selection during pseudotranslocation. In the cryo-EM reconstruction of 40S : CrPV IRES particles (3.8 Å), the VRL density is fragmented, suggesting a partial ordering of the IRES element that is not apparent in 80S : CrPV IRES reconstructions [56]. One possibility is that transient interactions of the VRL with the 40S subunit facilitate events downstream of 80S recruitment [56]. The IRES elements that functionally complete the tRNA mimicry to facilitate translocation and frame selection remain to be determined.
CrPV IRES translational activity is dependent on both the length and nucleotide sequence of the VLR [64]. Toeprint analysis of initiation by IRES mutants with a shortened VRL indicates that the loss of translational activity is due to an inability to pseudotranslocate, while toeprints for the sequence mutants are consistent with several rounds of translocation. However, the toeprints of the translocated complexes with the VRL sequence mutant IRES had lower band intensity compared with wild-type IRES translocated complexes, suggesting that subtle changes in translocation efficiency are responsible for the decreased translational activity of the VRL sequence mutants [64].
Co-sedimentation and fluorescent anisotropy experiments, which monitor P-site and A-site tRNA incorporation, were used to parse the two eEF2-mediated pseudotranslocation steps. Decreasing the length of the VRL inhibited both eEF2-mediated pseudotranslocation events, while changes in the sequence of the VRL inhibited only the second round of pseudotranslocation. The results indicate that the VRL facilitates both the first and second eEF2-mediated pseudotranslocation; however, the structural basis of this process remains to be explored.
(e). Applying single-molecule approaches to CrPV IRES-mediated translation
The structural studies discussed here provide a plausible collection of states during CrPV IRES-mediated initiation, yet to delineate the mechanism, real-time data are needed. Even in the simplest systems, translation initiation is a nonlinear process, wherein initiation occurs via several kinetically accessible pathways. Single-molecule experiments provide the tools to observe the time evolution of biochemical systems directly, allowing the sorting of kinetic pathways [67,68].
The entire process of CrPV IRES translation initiation and transition to elongation has now been observed in real time [29] using single-molecule (sm) fluorescence in zero-mode waveguides (ZMWs), nanostructures that allow single-molecule fluorescence detection in the presence of high (100–1000 nM) concentrations of free fluorescent ligand [69]. Direct tracking of the IRES, yeast ribosomal subunits and tRNA-enabled pathways of CrPV IRES initiation to be sorted and unified within the context of previous structural and biochemical research [29] (figure 3).
Formation of the 80S : CrPV IRES initiation complex was observed by co-delivering fluorescently labelled 40S and 60S ribosomal subunits to surface-immobilized CrPV IRES mRNA within ZMWs (figure 3f). 80S : IRES complexes form through two distinct pathways: sequential recruitment of the 40S subunit (kon = 70 nM−1 s−1) and the 60S subunit (kon = 700 nM−1 s−1), or direct recruitment of the 80S ribosome (kon < 30 nM−1 s−1) (figure 2c). Under the conditions tested, the sequential recruitment pathway was dominant, and yet both pathways result in 80S : IRES complexes with equivalent tRNA binding activity. Direct recruitment of the 80S ribosome by the CrPV IRES has been hypothesized as an alternative recruitment pathway that allows the virus to evade initiation inhibition by host translation initiation factors [37]. Direct 80S ribosome recruitment may also be important for viral protein synthesis under particular cellular conditions, as evidenced by an increase in the proportion of direct 80S recruitment by the CrPV IRES at low Mg2+ concentrations [29].
Following 80S : CrPV IRES complex formation, eEF2 catalyses movement of PKI from the A to the P site of the ribosome [45,46]. This initial pseudotranslocation event results in an unstable intermediate that readily back-translocates in the absence of an A-site tRNA [49,59]. In delivery experiments to surface-immobilized 80S : IRES complexes, efficient tRNA binding requires the presence of eEF2 both during the assembly of the 80S : CrPV IRES complexes and during the delivery of the tRNA [29]. The results indicate that initiation complexes adopt multiple conformational states following eEF2-mediated pseudotranslocation, which differ in their ability to bind tRNA. eEF2 drives formation of an unstable, pseudotranslocated intermediate that is captured by the arrival of tRNA, and tRNA binding serves as an essentially irreversible step during initiation.
Prior to tRNA arrival, the 80S : IRES complexes are in exchange between 0 and +1 frame states [29]. Incomplete tRNA mimicry by the IRES may decouple frame selection and pseudotranslocation in 80S : CrPV IRES initiation complexes, given that both the acceptor stem and peptidyl moiety are required for frame maintenance during canonical translocation [43,62,65,66]. Arrival of the first tRNA to the 80S : IRES complexes is the commitment step that establishes the reading frame (figure 2c). In sm-fluorescence experiments in which the 60S subunit, eEF2 and both 0-frame and +1-frame tRNAs, as eEF1A-TCs, were co-delivered to surface-immobilized 40S : IRES complexes, fast 0-frame tRNA binding biased against +1 frame selection [29]. The frame selection ratio of 0 frame to +1 frame (13.4 : 1) is nearly identical to the ratio of 0 frame to +1 frame apparent arrival rates (13.2 : 1), indicating that reading frame selection is defined by the kinetics of the first tRNA binding event.
The CrPV IRES uses stable and dynamic RNA structures to manipulate the eukaryotic ribosome. The defined structure of the IRES domain is essential for capturing and independently assembling active 80S ribosomes, while dynamic elements guide eEF2-mediated pseudotranslocation and accommodation of elongator tRNA. Ribosome and IRES conformational dynamics work in concert to channel ribosome : IRES complexes through multiple kinetically controlled pathways. Kinetic partitioning is probably a general characteristic of IRES-mediated and canonical eukaryotic translation initiation.
3. The type 3 HCV-like IRES
HCV uses a single IRES in the 5′ UTR of its approximately 9600 nt positive-sense RNA genome to initiate translation via a more complex mechanism than that of the IGR IRESs. The HCV IRES forms initiation complexes at an AUG start codon, initiating the continuous translation a polyprotein ORF, whose product is co- and post-translationally cleaved into 10 polypeptides [70] (figure 4a). By harnessing the IRES, HCV bypasses the canonical cap-binding and scanning process, such that many of the factors required in those processes are dispensable for IRES-mediated translation [74]. Given that translation is an essential early event in the HCV life cycle [75], and HCV chronically affects 130–150 million people worldwide [76], the IRES is also an attractive drug target [77].
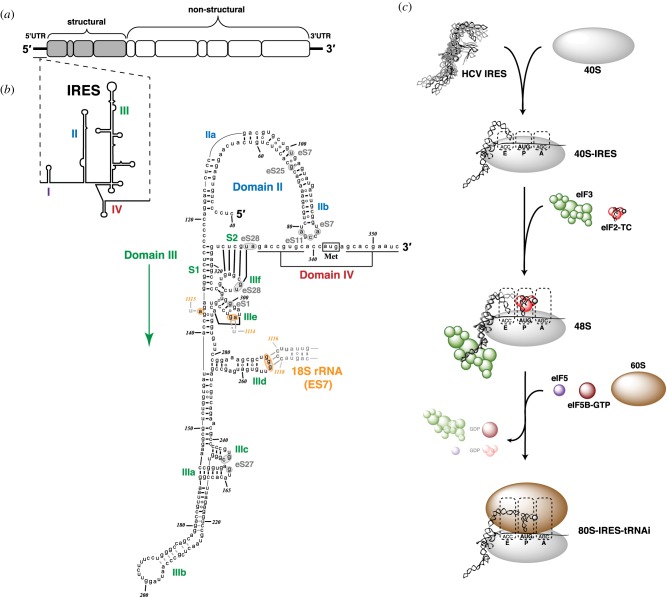
Figure 4.
HCV IRES secondary structure and mechanism. (a) Schematic of the HCV positive-sense RNA genome. (b) Secondary structure, domain architecture and ribosomal interactions of HCV IRES RNA. Intra-IRES and IRES : rRNA base-pairing is based on conformation of the ribosome-bound IRES [71,72]. Base-pairing of nt 178–221 of domain IIIb are drawn in grey as they are not resolved in the 40S : IRES structure, and are thus hypothetical. The short hairpin of domain IV is shown unfolded, encompassing the ribosomal toeprint-defined region of the 40S : IRES complex [73]. (c) Model of HCV IRES-mediated initiation: 40S subunit recruitment to form the binary complex, initiator tRNA recruitment during 48S PIC formation and 60S subunit joining to form the elongation-competent 80S ribosome.
Mechanistic investigations of HCV IRES initiation have revealed general principles of mammalian ribosome structure and function, and demonstrated how RNA structure may substitute for canonical protein-based initiation factors. Although skipping the scanning and cap-binding processes, the HCV IRES initiation mechanism encompasses the steps of start codon positioning, initiator tRNA recruitment and ribosomal subunit joining. Unlike the IGR IRESs, the HCV IRES uses certain eIFs, and is an important model for studying the large (approx. 800 kDa) eIF3 mulitprotein complex. The following section describes functional aspects of HCV IRES initiation, specifically highlighting recent advances resulting from the production of high-resolution cryo-EM structural models.
(a). The pathways of HCV IRES-mediated initiation
HCV IRES initiation can be divided into three main steps defined by the formation of stable PICs (figure 4c) [78–81]. The most common model begins with formation of the high-affinity 40S : IRES binary complex. In the second step, the binary complex recruits the multisubunit eIF3 complex and the Met-tRNAiMet-eIF2-GTP ternary complex (eIF2-TC) to form the 48S PIC. Initiation is completed by joining of the large (60S) subunit facilitated by the combined GTPase activation of eIF2-GTP by eIF5, GTP hydrolysis by eIF5B and dissociation of factors. As in canonical initiation, the elongation-competent 80S : IRES ribosome contains an initiator tRNA base paired with the start codon in the P site, using initiator tRNA at an AUG codon, rather than elongator tRNA used by IGR IRESs.
Initiation on the HCV IRES requires transient association of protein factors, dynamic structural rearrangement of ribosomal initiation complexes, and energy expenditure in the form of GTP. Although the three-step model accounts for many observed phenomena, the ordered release of factors from the 48S PIC has been observed [82], and multiple alternative pathways do exist. In particular, translation by the HCV IRES proceeds under stress conditions when eIF2 has been inactivated. Factor requirements may be circumvented through elevated Mg2+ concentration and alternative or minimal factor pathways by eIF3/eIF5B, eIF2A or eIF2D [36,83–86]. Although the favourability of these pathways remains unresolved, these finding indicate that HCV IRES forms compositionally heterogeneous complexes, perhaps explaining the adaptability of the virus to the multiple conditions encountered in an infected cell.
(b). The HCV IRES is a flexible ensemble of structured RNA modules
The HCV IRES encompasses approximately 340 nt in the 5′ UTR of the HCV genome [87] (figure 4a). It is composed of several independently folding domains (II–IV) and subdomains, and specifically binds the 40S subunit and the eIF3 complex (figure 4b). A short stretch of RNA upstream of the IRES, which comprises domain I with two binding sites for miR-122, is not critical for IRES activity and plays a predominant role in mRNA stability [87]. Domain II is a long, irregular stem loop that adopts a distorted L-shape structure [88], and contains two subdomains: IIa and IIb. The basal domain IIa harbours a multinucleotide bulge hinge, while IIb contains a loop E motif internal loop and an apical hairpin. A short six-nucleotide linker and the nine–base-pair stem 1 (S1) connects the base of domain II to domain III, which forms an elongated branched structure and consists of six subdomains (a–f). A four-way junction of the stem loop domains IIIabc protrudes at the apical end, while the irregular stem loop domain IIId projects out of a three-way junction below. At the base of domain III sits an irregular four-way junction, bridging the small step loop of IIIe to IIIf, which engages in base-pairing with stem 2 (S2) to form the pseudoknot structure. The start codon resides in the domain IV stem loop, which must be unfolded in the context of the mRNA-binding channel [89].
Biochemical studies have mapped regions of the IRES that are crucial for translational activity and binding partner affinity [78,79,90–93,74]. Factor and ribosome affinities for the IRES are not always correlated with translational activity. For instance, deletion of domain II, which strongly reduces translation activity, does not significantly alter 40S : IRES affinity. Similarly, mutations and deletions of the eIF3-interacting region (the apical portion of domain III) do not drastically alter 40S : IRES affinity, but strongly reduce eIF3 : IRES affinity and translation activity. Interestingly, while deletion of domain IIIb strongly reduces eIF3 : IRES affinity in isolation, the factor still remains in 48S complexes assembled on the mutant IRES, indicating that specific IRES : eIF3 interactions are required for translational activity [79]. Together, these findings suggest that the HCV IRES functions not only analogous to the prokaryotic SD sequence by increasing the local concentration of the mRNA start codon on the small subunit, but manipulates ribosomal complexes through its RNA structural elements to promote initiation.
The structure of the free IRES in solution is inherently flexible [14,94]. Early structural work applied a ‘divide and conquer’ approach to determine three-dimensional structures of isolated IRES domains using NMR and X-ray crystallography [14,81]. By incorporating structures of known fragments, a recent study using SAXS and molecular dynamics simulations found that the free IRES is best fit by an ensemble of at least five conformations as a result of rigid domains moving with respect to one another by flexible linkers [95]. Many of the HCV IRES structural elements are conserved in related flaviviruses, as well as certain picornavirus genera, with variability especially in domain II and apical domain III regions [16].
(c). The HCV IRES forms an extended interaction network on the 40S subunit
The HCV IRES forms a high-affinity (KD = 2–4 nM) binary complex with the 40S ribosomal subunit, whose formation is dictated by a fast bimolecular association rate constant (kon = 1.1 µM−1 s−1) and a slow off-rate (koff = 0.002 s−1, complex lifetime > 500 s), indicating that binary-complex formation is irreversible on the timescale of initiation [96]. The binary complex was first visualized at low resolution (20 Å) by cryo-EM, showing that the IRES binds the rabbit 40S subunit in a single conformation mediated largely by interactions of domain III at the solvent side [97]. Domain II was shown to loop around and interact with a region of the head and platform edge, where it partially overlaps with the E site. By comparing a wild-type IRES with a full domain II deletion mutant, this study demonstrated that the HCV IRES RNA induces a domain II-dependent rotation of the 40S subunit head relative to the body and a concomitant closure of the mRNA binding channel [97]. These conformational changes are similar to some of those seen in the 40S : CrPV IRES binary complex [41], and during cap-dependent translation [18], suggesting that similar ribosome conformational dynamics are induced during several unrelated mechanisms of initiation.
The recent breakthrough in cryo-EM has led to two subnanometer structural models of the mammalian 40S : IRES complexes (figure 5a) [71,72]. These two models are remarkably similar with an RMSD value of 2.0 Å in the ordered regions of the IRES, although the complexes were prepared and modelled using distinct methods. The Ban group directly assembled 80S : IRES complexes by incubating human 80S ribosomes with the HCV IRES, and used focused refinement by applying a mask around the 40S : IRES to reach a 3.9 Å resolution structure [71]. By contrast, the Spahn study formed binary complexes of the 40S : IRES with rabbit 40S subunits to determine their 6.7 Å resolution structure, which were subsequently used to form 80S : IRES complexes by the addition of excess 60S ribosomal subunits [72]. The structural models show slight differences in IRES resolution at the 5′ end, in the apical region of domain III, and the domain IV single-stranded region within the mRNA binding channel; however, the overall similarity is striking. Thus, in the absence of initiation factors and tRNA, 40S : IRES complexes appear to form a homogeneous complex independent of their assembly.
Figure 5.
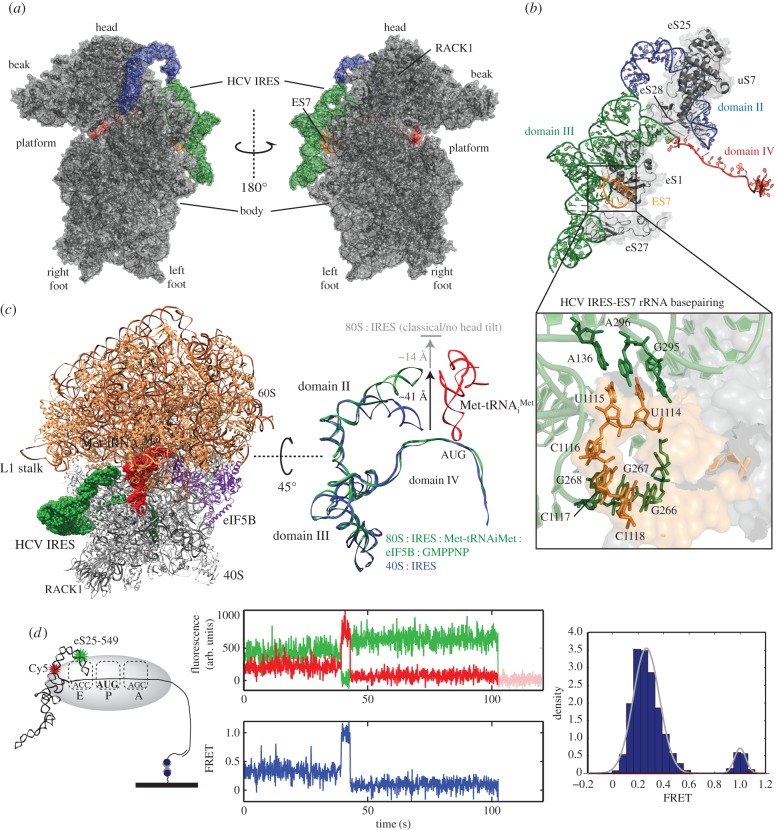
Structure and dynamics of the ribosome-bound HCV IRES. (a) 40S : IRES structure [72] (PDB entry 5FLX). Structure is annotated with features of the small subunit. (b) Structure of the 40S-bound HCV IRES and IRES-interacting ribosomal proteins and rRNA. Inset shows HCV IRES domain IIId interaction with ES7 of 18S rRNA. (c) Structure of the HCV IRES bound to the 80S ribosome with eIF5B and initiator tRNA [98] (PDB entry 4UJD). The IRES and tRNA are shown at the right, following alignment against the 40S-bound IRES (PDB entry 5FLX). (d) Conformational changes of 40S : IRES [96]. Structural basis of the FRET signal is shown at the far left, with labels on eS25 and at the base of HCV IRES domain II. In the centre, an example single-molecule fluorescence trace shows the transition between the low- and high-FRET states of the 40S : IRES binary complex. The FRET intensity distribution of 146 molecules is shown at the far right. The fluorescence trace and histogram of D were modified from Fuchs et al. [96] with permission.
These new structures explain prior functional data through interactions of the HCV IRES with ribosomal proteins and 18S rRNA [99–105] (figure 4b). The kissing-loop interaction formed between the apical loop of domain IIId and expansion segment 7 (ES7) of 18S rRNA, where Watson–Crick base-pairing between the highly conserved 266GGG268 of the IRES domain IIId and 1116CCC1118 of ES7, serves as a crucial anchor for the binary complex [104,105] (figure 5c). Mutations of 266GGG268 reduce binary-complex affinity and IRES activity, which can be restored by compensatory mutations in 1116CCC1118 of ES7 [78,79,93,94,104,105]. Consistent with ES7 serving as an anchor, mutations of 266GGG268 that maintain 40S affinity, but fail to induce the necessary conformational changes in the binary complex, result in non-functional ribosome : IRES complexes [106]. Although the mRNA : rRNA kissing loop is reminiscent of the SD/anti-SD interaction of prokaryotes, the binding takes place distal to the AUG codon of domain IV and extends only 3 bp. Instead, domain IIIf of the pseudoknot engages in a stretch of intra-domain base-pairing that helps present the AUG within the mRNA channel (figure 5d) [71,72]. Start codon positioning by the HCV IRES is therefore factor-independent, instead relying on the affinity and orientation provided by the IRES RNA structures.
The HCV IRES makes an array of additional contacts with the 40S subunit, guided by the overall fold of the RNA (figure 4b). Basal regions of IRES domain III (IIIe) interact with 1114UU1115 of ES7 rRNA, and numerous small ribosomal proteins make contacts across the IRES (figure 4b). Along the four-way junction, which adopts an L-shaped cL-type fold, domains IIIa and IIIc interact with eS27 while stacked in an anti-parallel configuration [107]. Interestingly, the conformation of the IIIabc four-way junction in the binary complex is not in agreement with the cH-type orientation seen in the crystal structure of the isolated junction [108], where the IIIa and IIIc stem loops are arranged in a parallel conformation, demonstrating the role of the full RNA and interactions with the 40S subunit in structuring the IRES.
(d). The structural and functional basis of HCV IRES : eIF3 interactions
The 13-subunit mammalian eIF3 complex is a central mediator of canonical and IRES-mediated translation. During canonical initiation, eIF3 is stoichiometrically bound to the solvent-exposed surface of the 40S subunit and engages in a network of interactions with mRNA and several other eIFs: 1, 1A, 4B, 4G and 5 [109,110]. These interactions enable eIF3 to stimulate several important steps during the initiation pathway [110]. eIF3 also serves as a hub for the cell signalling proteins mTOR and S6K1 [111], and dysregulation of eIF3 subunits appears in multiple cancers [112]. Consistent with its central role in translation, multiple viruses co-opt and modulate eIF3 function, including HIV, which uses a viral protease to specifically cleave the eIF3d subunit [113]. The high-affinity binding of eIF3 to the HCV IRES RNA is another case of the factor's importance in virology, wherein eIF3 may orient 48S PICs on the IRES for proper 80S formation [74].
In canonical initiation, eIF3 interacts with eS27 at the solvent-exposed surface of the 40S subunit. The interaction between the HCV IRES and eS27, as seen in the high-resolution binary-complex structures, suggest that the IRES binds eIF3 and excludes the factor from its canonical binding site on the ribosome by a mutually exclusive interaction. Structural studies of a HCV-like CSFV IRES support this hypothesis, as the IRES excludes eIF3 from its normal binding site on the 40S subunit (figure 5e) [114–116]. This contrasts with earlier structural studies that predicted a more overlapping conformation of eIF3 on the 40S : IRES : eIF3 complex [117], and suggested that the function of eIF3 in HCV IRES-mediated translation could be analogous to the eIF4F : eIF3 interaction (via eIF4G) used in canonical initiation to regulate mRNA attachment and scanning. As HCV IRES-mediated translation does not use scanning and eIF3 only slightly increases the affinity of the 40S : IRES complex [93], the IRES : eIF3 interaction is probably functionally distinct from that of eIF3 : eIF4F.
(e). The interplay of multiple eIF3 subunits with the HCV IRES during initiation
The multiple subunits of eIF3 are orchestrated to drive productive initiation. IRES : eIF3 interactions take place via the apical section of HCV IRES domain III, involving the subdomains IIIa-c and several eIF3 subunits. Approximately 50 nt of subdomain IIIb are largely unresolved in the high-resolution ribosome : IRES structures [71,72] (figure 5a,b), probably a result of conformational flexibility that is stabilized by eIF3 binding [116]. The eIF3a, b, d and f subunits were shown in early studies to cross-link with HCV and CSFV IRES RNA [118], while a later mass spectrometry study demonstrated that a subset of subunits (g, f and h) are stabilized by IRES binding, while others (i, l and k) remain loosely bound [119]. Intriguingly, eIF3 is still capable of binding the IRES after being subjected to limited proteolysis [120], and a reconstituted octamer of eIF3 (a, c, e, f, h, k, l, m) is capable of high-affinity binding to an RNA fragment containing domain IIIa–c [121]. Although a full eIF3 complex appears to be important for efficiently forming 48S complexes [121], this suggests that IRES : eIF3 affinity does not require the full complex. IRES : eIF3 interactions are mediated predominantly through helix-loop-helix (HLH) RNA binding motifs in subunits eIF3a and c, and may play different functions over the course of initiation [122]. The eIF3a and c subunits are also the most-proximal subunits to the 40S-subunit mRNA-binding cleft within 43S complexes, further showing the subunit-selective means by which the IRES excludes eIF3 from its canonical position. While mutations of the HLH motif of eIF3a greatly weaken IRES : eIF3 affinity, mutations of the eIF3c HLH motif only modestly alter affinity and cause slight defects in the assembly of functional 80S : IRES complexes containing eIF5B [122].
Peripheral subunits of eIF3, including subunits b, d, g, i and j, probably play important functions during initiation that are not related to IRES affinity per se. For instance, the loosely bound eIF3j subunit must be removed from its entry channel location prior to stable accommodation of the HCV IRES mRNA start codon within the P site, possibly promoted by recruitment of the eIF2-TC [123]. Additionally, the eIF3d subunit resides on the rear side of the 40S subunit head (in canonical 43S complexes) near ribosomal protein RACK1, which is intriguingly required for IRES-mediated, but not canonical, translation [124]. Rather than a direct physical interaction between the HCV IRES and RACK1, one possibility is that RACK1 makes remote interactions to the IRES mediated by eIF3. These interactions could transiently influence a productive conformation of the ribosome during initiation that promotes elongation, as it has been shown that approximately 50% of natively assembled 80S : IRES complexes are lacking RACK1 density [125]. A recent structural study of the canonical 43S PIC suggested that peripheral subunits of eIF3 (g and i) may regulate the binding of eIF5B, as they overlap at the GTPase-binding site, such that the eIF3 g and i subunits must be removed prior to full initiator tRNA accommodation and 60S subunit joining [126]. Thus, while not required for IRES affinity, the eIF3 g and i subunits could also play an important role in IRES-mediated translation by controlling eIF5B-mediated 60S subunit joining. Taken together, current structural and functional data indicate that the IRES functionally hijacks eIF3 from its canonical 43S PIC position, and actively engages multiple eIF3 subunits to promote steps along the initiation pathway. eIF3 may exist as heterogeneous complexes within the cell [127], potentially as a result of dysregulation of individual subunits as seen in cancer [112]. As with the IRES binding, unique eIF3 assemblies might target specific mRNAs, and result in altered translational profiles [128].
(f). Domain II engages within the E site to aid in start codon positioning
Domain II of the IRES engages the core of the 40S subunit adjacent to the E site. The apical loop of domain II interacts with uS7 and uS11 adjacent to the E site, where it also appears to make tertiary interactions near the AUG codon of domain IV (figure 4b). Mutations within the stem loop IIb alter the configuration of the dIV RNA in the mRNA-binding groove and the placement of domain II [129]. Surprisingly, a domain IIb mutated IRES is still semi-competent for translation, and 48S complexes can be formed even with full domain II deletion (dIIΔ) IRES [78,82], indicating that domain II is not required for IRES recruitment, but instead is needed for later steps of initiation. Notably, the dIIΔ HCV IRES is defective in promoting eIF5-induced GTP hydrolysis of eIF2 and factor release from 48S complexes [82], and a dIIΔ of the CSFV IRES fails to promote efficiently subunit joining of 48S complexes assembled both with or without eIF2 [84]. These findings suggest that domain II function is at least partially agnostic to 48S PIC composition, and might operate by mediating the dII-dependent conformational rearrangement of the 40S subunit or later initiation events.
(g). Dynamic rearrangements within the 80S : IRES complex
Structures of the native mammalian 80S : IRES complexes have been solved at both low (15 Å) [125] and recently high (3.9 Å) resolution [71,72]. Although the initial structural model proposed interactions between the IRES and the both ribosomal subunits, recent models lack connections between the IRES and the 60S subunit (at the L1 stalk). The density connecting the L1 stalk to the IRES could result from the extra RNA of IRES domain I or an RNA aptamer used in the earlier study [71]. Alternatively, as the low-resolution study was performed on 80S : IRES complexes purified from native human extracts stalled by cycloheximide, while the high-resolution complexes were assembled from rabbit ribosomal subunits in vitro, the density could potentially be related to the methods of 80S : IRES complex assembly.
Recent high-resolution 80S : IRES complexes were classified into three subpopulations differentiated by the rotational position of the 40S : IRES relative to the 60S subunit [72]. The major subpopulation exists in a classical conformation and is related to the structure of the elongating ribosome in the post-translocational (POST) state. A second tier of classification of the major subpopulation resulted in the identification of one additional class with P site–bound tRNA, which probably resulted from co-purification within the E site of 80S ribosomes, as observed previously [130]. The other two subpopulations represent conformation of the 80S : IRES in a pre-translocation (PRE) state, where the complex is in either a classical (rolled) or rotated state. The rolled state is defined as a rotation of the small subunit around its long axis, orthogonal to the previously described inter-subunit rotation [60]. In the case of the 80S : IRES : eIF5B : Met-tRNAiMet : GMPNP complex, the ribosome was classified into two major subpopulations of PRE and POST states, where the PRE state ribosomes were observed as rolled rather than rotated relative to the POST state ribosomes [98]. Thus, the IRES-bound initiating ribosome is conformationally flexible and adopts conformations that are reminiscent of an elongating ribosome, even in the presence of numerous factors and P site–bound initiator tRNA.
(h). HCV IRES domain II serves as a ‘wedge’ to ensure proper mRNA: tRNAi orientation
The high-resolution structures have begun to reveal the structural dynamics of the 80S : IRES complex during the recruitment and positioning of initiator tRNA. During the transition from the 40S : IRES to 80S : IRES complexes, the 40S subunit head reorients relative to the body with a 17° 40S head tilt upon association with the 60S subunit, which is reversed upon association of P site–bound tRNA [72]. This reversal coincides with a large-scale movement of domain II, where the apical end moves approximately 55 Å from its position adjacent to the mRNA within the E site to a location adjacent to helix 68 (H68) of the 60S subunit's 28S rRNA. This domain II rearrangement is also present in eIF5B-containing complexes, albeit at an intermediate degree (approx. 41 Å), where the apical loop positions itself adjacent to the elbow of the initiator tRNA [98] (figure 5c). Domain II was proposed to serve as a ‘wedge’ by positioning itself to prop open the mRNA channel and widen the P site for tRNAi binding. Consequently, tRNAi binding reverses the head tilt, narrowing the P site and locking in the tRNA.
These observed positions of domain II are probably only a subset sampled during initiation, as a complete 48S complex of the 40S : IRES : eIF3 : eIF2-TC has yet to be solved. It has been proposed that the IRES must undergo a multi-step rearrangement during binary-complex formation to position domain II [95]; and clearly, for 80S ribosomes to transition into elongation, domain II must be entirely vacated from its E site position to make room for deacyclated initiator tRNA moving from the P site. The structural observations help explain the multipurpose role of domain II during tRNA recruitment and positioning, where it participates in both the formation of productive 48S complexes and the progression of 80S initiation complexes into elongation [82,129,131].
Intriguingly, the domain II of the CSFV IRES prevents tRNA binding in the absence of factors, makes 48S complexes sensitive to eIF1-induced destabilization, and is required for efficient subunit joining [84]. Given that eIF1 binds near the P site and induces a similar conformational change of the 40S ribosome to that of the HCV IRES [17], these results suggest potential competition between domain II and eIF1. As eIF1, together with eIF1A, aids in start codon recognition during scanning, and eIF1 is responsible for dissociating incorrectly associated 48S complexes during canonical initiation [132], one possibility is that domain II functionally replaces the role of eIF1 and/or eIF1A, albeit not in the context of a scanning ribosome. Regardless of factor substitution, domain II motions are crucial, and their inhibition by a small molecule ligand, which binds within the hinge bulge of subdomain IIa and locks domain II in an elongated conformation, suppresses translation of an HCV replicon [72,77]. Such RNA conformational switches may be general in a range of IRESs [133], with potential as a drug target, but also as a simple means for manipulating initiation complexes to replace the functions of canonical factors.
(i). Single-molecule FRET reveals domain II and 40S head conformational dynamics
Within the binary complex, the medial region of domain II makes contacts with eS25 near the 40S head region (figures 4b and 5b). Intriguingly, eS25 is non-essential for canonical translation, but required for both CrPV and HCV IRES-mediated initiation [50,134], suggesting that the eS25 interactions may be crucial for proper domain II function. As eS25 is non-essential for cells cultured in vitro, the fluorescently labelling of human 40S ribosomes has now been accomplished, via the genetic complementation of a SNAP-tag RPS25 gene fusion to a CRISPR/Cas9-mediate knockout cell line of RPS25 [96]. Investigation of the fluorescently labelled 40S subunit as a complex and the HCV IRES, labelled upstream of domain II, enabled single-molecule FRET (smFRET) analysis of the binary complex (figure 5d).
All cryo-EM structural models to date have suggested that the 40S : IRES complex exists in a single conformation. By contrast, the smFRET experiments observed domain II sampling at least two conformations with respect to the 40S subunit head [96] (figure 5d). These motions may reflect functionally relevant conformational changes, such as rotation of the 40S subunit head or domain II occupancy of the E site, similar to conformations adopted in 80S : IRES complexes. Still, these results must be further clarified by cryo-EM and smFRET analyses, as the FRET state changes could reflect motions of IRES RNA, the ribosome and/or the SNAP-tag label on eS25. Curiously, however, both the addition of translation extract and reduced Mg2+ concentration (2.5 versus 5 mM) funneled the 40S : IRES complexes into a single conformation, suggesting that the 40S : IRES conformational dynamics are sensitive to their chemical environment [135].
Further single-molecule studies should clarify these conformational dynamics and may reveal the 40S : IRES conformational substrates that are accessible during tRNA recruitment and 60S subunits joining. Given that no structures are currently available of the full HCV IRES 48S PIC intermediate, single-molecule studies have the potential to reveal conformational and compositional states of this relatively unstudied species. Improvements in cryo-EM and particle classification of the 40S : IRES complexes, as well as high-resolution structures of mutant 40S ribosomal subunits and/or IRES, will further shed light on the rare and dynamic conformations adopted by the ribosome : IRES complexes during initiation.
4. Outlook
The confluence of structures and dynamics reviewed above represents the future of molecular biophysics. Cryo-EM structures have provided multiple snapshots of IRES-mediated translation, and the single-particle method encompasses conformational and compositional heterogeneity. Using these structures as guides, single-molecule fluorescence reagents have been created for the study of eukaryotic translation, allowing real-time analysis of conformational and compositional dynamics during IRES-mediated initiation. The combined data from structure and dynamics explain decades of biochemical study in molecular detail.
The cryo-EM methods will continue to evolve, improving resolution and classification of distinct conformers. Regions of poorly resolved density in ribosome : IRES models often demarcate structurally disordered regions, such as the variable loop of the CrPV IRES. These regions will be targets for real-time dynamics studies, and are likely to be stabilized and/or manipulated by eIFs, as evidenced by the apical region of CSFV IRES domain III in the 40S : IRES : eIF3 structure [116]. The future cryo-EM studies of IRES-mediated translation may include assembly of unique eIF-containing complexes, improvements in particle classification, and application of time-resolved methods [136]. These will undoubtedly be complemented by classical structure–function studies and biochemistry of reconstituted translation components.
Even the simplest processes of translation initiation by the CrPV IRES are highly complex and undergo a number of parallel pathways distinguished by favourability at kinetic branch-points determined by factor concentrations [29]. Translation initiation from more structurally dynamic mRNAs, such as the HCV IRES, appear to undergo a number of alternative pathways within their cellular context, and these too are probably dictated by the kinetic favourability of molecular interactions. These features make IRES-mediated translation extremely well suited for single-molecule studies, which can reveal the compositional and conformational dynamics of macromolecule assemblies [69]. Single-molecule FRET can measure the time evolution of distance changes during translation, such as ribosomal rotation during bacterial elongation [22] and 40S head motions during initiation [96]. However, FRET provides only a single distance constraint on a biochemical system requiring synergistic integration of detailed static structural data from cryo-EM [137,138].
Dynamic tools for investigation of human translation in vitro will continue to improve [96,139], and will be integrated with in vivo genome wide profiling studies [140] of eukaryotic translation. The study of IRES-mediated translation through structure and dynamics will be a springboard to extending these approaches to the general mechanisms of translation initiation and its regulation.
5. Molecular graphics
Molecular graphics and analyses were performed with Pymol [141] and the UCSF Chimera package [142], using structure entries deposited on PDB [143]. Chimera is developed by the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Acknowledgements
We thank Peter Sarnow for collaboration, friendship and many useful discussions over the past decade.
Authors' contributions
All the authors wrote, edited and approved the manuscript.
Competing interests
We have no competing interests.
Funding
A.G.J. is supported by an NSF Graduate Research Fellowship (DGE-114747). R.G. is supported by NIH grant no. AI099245. Research on eukaryotic translation is supported by NIH grants nos. GM099687, AI047365 and AI099506 to J.D.P.
References
- 1.Melnikov S, Ben-Shem A, Garreau de Loubresse N, Jenner L, Yusupova G, Yusupov M. 2012. One core, two shells: bacterial and eukaryotic ribosomes. Nat. Struct. Mol. Biol. 19, 560–567. ( 10.1038/nsmb.2313) [DOI] [PubMed] [Google Scholar]
- 2.Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 11, 113–127. ( 10.1038/nrm2838) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aitken CE, Lorsch JR. 2012. A mechanistic overview of translation initiation in eukaryotes. Nat. Struct. Mol. Biol. 19, 568–576. ( 10.1038/nsmb.2303) [DOI] [PubMed] [Google Scholar]
- 4.Fraser CS. 2015. Quantitative studies of mRNA recruitment to the eukaryotic ribosome. Biochimie 114, 58–71. ( 10.1016/j.biochi.2015.02.017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mignone F, Gissi C, Liuni S, Pesole G. 2002. Untranslated regions of mRNAs. Genome Biol. 3, pREVIEWS0004 ( 10.1186/gb-2002-3-3-reviews0004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 83, 779–812. ( 10.1146/annurev-biochem-060713-035802) [DOI] [PubMed] [Google Scholar]
- 7.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. 2008. Translation initiation on mammalian mRNAs with structured 5'UTRs requires DExH-box protein DHX29. Cell 135, 1237–1250. ( 10.1016/j.cell.2008.10.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell 136, 731–745. ( 10.1016/j.cell.2009.01.042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Valasek LS. 2012. 'Ribozoomin'--translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr. Protein Pept. Sci. 13, 305–330. ( 10.2174/138920312801619385) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellen CU, Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 15, 1593–1612. ( 10.1101/gad.891101) [DOI] [PubMed] [Google Scholar]
- 11.Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334, 320–325. ( 10.1038/334320a0) [DOI] [PubMed] [Google Scholar]
- 12.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. 1988. A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62, 2636–2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson RJ. 2013. The current status of vertebrate cellular mRNA IRESs. Cold Spring Harb. Perspect. Biol. 5, a011569 ( 10.1101/cshperspect.a011569) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieft JS. 2008. Viral IRES RNA structures and ribosome interactions. Trends Biochem. Sci. 33, 274–283. ( 10.1016/j.tibs.2008.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balvay L, Soto Rifo R, Ricci EP, Decimo D, Ohlmann T. 2009. Structural and functional diversity of viral IRESes. Biochim. Biophys. Acta 1789, 542–557. ( 10.1016/j.bbagrm.2009.07.005) [DOI] [PubMed] [Google Scholar]
- 16.Asnani M, Kumar P, Hellen CU. 2015. Widespread distribution and structural diversity of type IV IRESs in members of Picornaviridae. Virology 478, 61–74. ( 10.1016/j.virol.2015.02.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell 26, 41–50. ( 10.1016/j.molcel.2007.03.018) [DOI] [PubMed] [Google Scholar]
- 18.Hellen CU. 2009. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim. Biophys. Acta 1789, 558–570. ( 10.1016/j.bbagrm.2009.06.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. 2015. Targeting the translation machinery in cancer. Nat. Rev. Drug Discov. 14, 261–278. ( 10.1038/nrd4505) [DOI] [PubMed] [Google Scholar]
- 20.Li GW, Xie XS. 2011. Central dogma at the single-molecule level in living cells. Nature 475, 308–315. ( 10.1038/nature10315) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deniz AA, Mukhopadhyay S, Lemke EA. 2008. Single-molecule biophysics: at the interface of biology, physics and chemistry. J. R. Soc. Interface 5, 15–45. ( 10.1098/rsif.2007.1021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. 2013. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat. Struct. Mol. Biol. 20, 718–727. ( 10.1038/nsmb.2567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang B, Bates M, Zhuang X. 2009. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 78, 993–1016. ( 10.1146/annurev.biochem.77.061906.092014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nogales E, Scheres SH. 2015. Cryo-EM: a unique tool for the visualization of macromolecular complexity. Mol. Cell 58, 677–689. ( 10.1016/j.molcel.2015.02.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng Y. 2015. Single-particle cryo_EM at crystallographic resolution. Cell 161, 450–457. ( 10.1016/j.cell.2015.03.049) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Skiniotis G, Southworth DR. 2016. Single-particle cryo-electron microscopy of macromolecular complexes. Microscopy (Oxf.) 65, 9–22. ( 10.1093/jmicro/dfv366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jan E, Sarnow P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324, 889–902. ( 10.1016/S0022-2836(02)01099-9) [DOI] [PubMed] [Google Scholar]
- 28.Schüler M, et al. 2006. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 13, 1092–1096. ( 10.1038/nsmb1177) [DOI] [PubMed] [Google Scholar]
- 29.Petrov A, Grosely R, Chen J, O'Leary SE, Puglisi JD. 2016. Multiple parallel pathways of translation initiation on the CrPV IRES. Mol. Cell 62, 92–103. ( 10.1016/j.molcel.2016.03.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson JE, Pestova TV, Hellen CU, Sarnow P. 2000. Initiation of protein synthesis from the A site of the ribosome. Cell 102, 511–520. ( 10.1016/S0092-8674(00)00055-6) [DOI] [PubMed] [Google Scholar]
- 31.Pestova TV, Hellen CU. 2003. Translation elongation after assembly of ribosomes on the Cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 17, 181–186. ( 10.1101/gad.1040803) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanamori Y, Nakashima N. 2001. A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7, 266–274. ( 10.1017/S1355838201001741) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Costantino D, Kieft JS. 2005. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA 11, 332–343. ( 10.1261/rna.7184705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pfingsten JS, Costantino DA, Kieft JS. 2007. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J. Mol. Biol. 370, 856–869. ( 10.1016/j.jmb.2007.04.076) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson SR, Gulyas KD, Sarnow P. 2001. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl Acad. Sci. USA 98, 12 972–12 977. ( 10.1073/pnas.241286698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lancaster AM, Jan E, Sarnow P. 2006. Initiation factor-independent translation mediated by the hepatitis C virus internal ribosome entry site. RNA 12, 894–902. ( 10.1261/rna.2342306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pestova TV, Lomakin IB, Hellen CU. 2004. Position of the CrPV IRES on the 40S subunit and factor dependence of IRES/80S ribosome assembly. EMBO Rep. 5, 906–913. ( 10.1038/sj.embor.7400240) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, Kieft JS. 2014. The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature 511, 366–369. ( 10.1038/nature13378) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. 2003. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 31, 2434–2442. ( 10.1093/nar/gkg336) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pfingsten JS, Costantino DA, Kieft JS. 2006. Structural basis for ribosome recruitment and manipulation by a viral IRES RNA. Science 314, 1450–1454. ( 10.1126/science.1133281) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. 2004. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell 118, 465–475. ( 10.1016/j.cell.2004.08.001) [DOI] [PubMed] [Google Scholar]
- 42.Jang CJ, Lo MC, Jan E. 2009. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J. Mol. Biol. 387, 42–58. ( 10.1016/j.jmb.2009.01.042) [DOI] [PubMed] [Google Scholar]
- 43.Au HH, Cornilescu G, Mouzakis KD, Ren Q, Burke JE, Lee S, Butcher SE, Jan E. 2015. Global shape mimicry of tRNA within a viral internal ribosome entry site mediates translational reading frame selection. Proc. Natl Acad. Sci. USA 112, E6446–E6455. ( 10.1073/pnas.1512088112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jan E, Kinzy TG, Sarnow P. 2003. Divergent tRNA-like element supports initiation, elongation, and termination of protein biosynthesis. Proc. Natl Acad. Sci. USA 100, 15 410–15 415. ( 10.1073/pnas.2535183100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto H, Nakashima N, Ikeda Y, Uchiumi T. 2007. Binding mode of the first aminoacyl-tRNA in translation initiation mediated by Plautia stali intestine virus internal ribosome entry site. J. Biol. Chem. 282, 7770–7776. ( 10.1074/jbc.M610887200) [DOI] [PubMed] [Google Scholar]
- 46.Fernández IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157, 823–831. ( 10.1016/j.cell.2014.04.015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu J, Korostelev A, Costantino DA, Donohue JP, Noller HF, Kieft JS. 2011. Crystal structures of complexes containing domains from two viral internal ribosome entry site (IRES) RNAs bound to the 70S ribosome. Proc. Natl Acad. Sci. USA 108, 1839–1844. ( 10.1073/pnas.1018582108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koh CS, Brilot AF, Grigorieff N, Korostelev AA. 2014. Taura syndrome virus IRES initiates translation by binding its tRNA-mRNA–like structural element in the ribosomal decoding center. Proc. Natl Acad. Sci. USA 111, 9139–9144. ( 10.1073/pnas.1406335111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muhs M, Hilal T, Mielke T, Skabkin MA, Sanbonmatsu KY, Pestova TV, Spahn CM. 2015. Cryo-EM of ribosomal 80S complexes with termination factors reveals the translocated cricket paralysis virus IRES. Mol. Cell 57, 422–432. ( 10.1016/j.molcel.2014.12.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landry DM, Hertz MI, Thompson SR. 2009. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 23, 2753–2764. ( 10.1101/gad.1832209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. 2013. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340, 1235490 ( 10.1126/science.1235490) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cornish PV, Ermolenko DN, Staple DW, Hoang L, Hickerson RP, Noller HF, Ha T. 2009. Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl Acad. Sci. USA 106, 2571–2576. ( 10.1073/pnas.0813180106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. 1999. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat. Struct. Biol. 6, 643–647. ( 10.1038/10695) [DOI] [PubMed] [Google Scholar]
- 54.Gomez-Lorenzo MG, et al. 2000. Three-dimensional cryo-electron microscopy localization of EF2 in the Saccharomyces cerevisiae 80S ribosome at 17.5 A resolution. EMBO J. 19, 2710–2718. ( 10.1093/emboj/19.11.2710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. 2008. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 15, 57–64. ( 10.1038/nsmb1351) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Murray J, Savva CG, Shin BS, Dever TE, Ramakrishnan V, Fernández IS. 2016. Structural characterization of ribosome recruitment and translocation by type IV IRES. Elife 5, e13567 ( 10.7554/eLife.13567) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voorhees RM, Ramakrishnan V. 2013. Structural basis of the translational elongation cycle. Annu. Rev. Biochem. 82, 203–236. ( 10.1146/annurev-biochem-113009-092313) [DOI] [PubMed] [Google Scholar]
- 58.Ratje AH, et al. 2010. Head swivel on the ribosome facilitates translocation by means of intra-subunit tRNA hybrid sites. Nature 468, 713–716. ( 10.1038/nature09547) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, Ng MY, Chen Y, Cooperman BS. 2016. Kinetics of initiating polypeptide elongation in an IRES-dependent system. Elife 5, e13429 ( 10.7554/eLife.13429) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Budkevich TV, et al. 2014. Regulation of the mammalian elongation cycle by subunit rolling: a eukaryotic-specific ribosome rearrangement. Cell 158, 121–131. ( 10.1016/j.cell.2014.04.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ren Q, Wang QS, Firth AE, Chan MM, Gouw JW, Guarna MM, Foster LJ, Atkins JF, Jan E. 2012. Alternative reading frame selection mediated by a tRNA-like domain of an internal ribosome entry site. Proc. Natl Acad. Sci. USA 109, E630–E639. ( 10.1073/pnas.1111303109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ren Q, Au HH, Wang QS, Lee S, Jan E. 2014. Structural determinants of an internal ribosome entry site that direct translational reading frame selection. Nucleic Acids Res. 42, 9366–9382. ( 10.1093/nar/gku622) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Banerjee PR, Deniz AA. 2014. Shedding light on protein folding landscapes by single-molecule fluorescence. Chem. Soc. Rev. 43, 1172–1188. ( 10.1039/c3cs60311c) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruehle MD, Zhang H, Sheridan RM, Mitra S, Chen Y, Gonzalez RL, Cooperman BS, Kieft JS. 2015. A dynamic RNA loop in an IRES affects multiple steps of elongation factor-mediated translation initiation. Elife 4, e08146 ( 10.7554/eLife.08146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fredrick K, Noller HF. 2002. Accurate translocation of mRNA by the ribosome requires a peptidyl group or its analog on the tRNA moving into the 30S P site. Mol. Cell 9, 1125–1131. ( 10.1016/S1097-2765(02)00523-3) [DOI] [PubMed] [Google Scholar]
- 66.Joseph S, Noller HF. 1998. EF-G-catalyzed translocation of anticodon stem-loop analogs of transfer RNA in the ribosome. EMBO J. 17, 3478–3483. ( 10.1093/emboj/17.12.3478) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tsai A, Petrov A, Marshall RA, Korlach J, Uemura S, Puglisi JD. 2012. Heterogeneous pathways and timing of factor departure during translation initiation. Nature 487, 390–393. ( 10.1038/nature11172) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O'Leary SE, Petrov A, Chen J, Puglisi JD. 2013. Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure 21, 2197–2207. ( 10.1016/j.str.2013.09.016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen J, et al. 2014. High-throughput platform for real-time monitoring of biological processes by multicolor single-molecule fluorescence. Proc. Natl Acad. Sci. USA 111, 664–669. ( 10.1073/pnas.1315735111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lindenbach BD, Rice CM. 2005. Unravelling hepatitis C virus replication from genome to function. Nature 436, 933–938. ( 10.1038/nature04077) [DOI] [PubMed] [Google Scholar]
- 71.Quade N, Boehringer D, Leibundgut M, van den Heuvel J, Ban N. 2015. Cryo-EM structure of Hepatitis C virus IRES bound to the human ribosome at 3.9-A resolution. Nat. Commun. 6, 7646 ( 10.1038/ncomms8646) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamamoto H, et al. 2015. Molecular architecture of the ribosome-bound hepatitis C virus internal ribosomal entry site RNA. EMBO J. 34, 3042–3058. ( 10.15252/embj.201592469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl Acad. Sci. USA 98, 7029–7036. ( 10.1073/pnas.111145798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. 1998. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 12, 67–83. ( 10.1101/gad.12.1.67) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Niepmann M. 2013. Hepatitis C virus RNA translation. Curr. Top. Microbiol. Immunol. 369, 143–166. ( 10.1007/978-3-642-27340-7_6) [DOI] [PubMed] [Google Scholar]
- 76.World Heath Organization. 2016. Hepatitis C fact sheet. http://www.who.int/mediacentre/factsheets/fs164/en/.
- 77.Dibrov SM, et al. 2014. Hepatitis C virus translation inhibitors targeting the internal ribosomal entry site. J. Med. Chem. 57, 1694–1707. ( 10.1021/jm401312n) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Otto GA, Puglisi JD. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119, 369–380. ( 10.1016/j.cell.2004.09.038) [DOI] [PubMed] [Google Scholar]
- 79.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. 2004. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl Acad. Sci. USA 101, 16 990–16 995. ( 10.1073/pnas.0407402101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fraser CS, Doudna JA. 2007. Structural and mechanistic insights into hepatitis C viral translation initiation. Nat. Rev. Microbiol. 5, 29–38. ( 10.1038/nrmicro1558) [DOI] [PubMed] [Google Scholar]
- 81.Lukavsky PJ. 2009. Structure and function of HCV IRES domains. Virus Res. 139, 166–171. ( 10.1016/j.virusres.2008.06.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Locker N, Easton LE, Lukavsky PJ. 2007. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 26, 795–805. ( 10.1038/sj.emboj.7601549) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. 2008. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 15, 836–841. ( 10.1038/nsmb.1445) [DOI] [PubMed] [Google Scholar]
- 84.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 27, 1060–1072. ( 10.1038/emboj.2008.49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J. Biol. Chem. 285, 26 779–26 787. ( 10.1074/jbc.M110.119693) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kim JH, Park SM, Park JH, Keum SJ, Jang SK. 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 30, 2454–2464. ( 10.1038/emboj.2011.146) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sagan SM, Chahal J, Sarnow P. 2015. cis-Acting RNA elements in the hepatitis C virus RNA genome. Virus Res. 206, 90–98. ( 10.1016/j.virusres.2014.12.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. 2003. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 10, 1033–1038. ( 10.1038/nsb1004) [DOI] [PubMed] [Google Scholar]
- 89.Honda M, Brown EA, Lemon SM. 1996. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA 2, 955–968. [PMC free article] [PubMed] [Google Scholar]
- 90.Brown EA, Zhang H, Ping LH, Lemon SM. 1992. Secondary structure of the 5' nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 20, 5041–5045. ( 10.1093/nar/20.19.5041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. 1992. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 66, 1476–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang C, Sarnow P, Siddiqui A. 1993. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J. Virol. 67, 3338–3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kieft JS, Zhou K, Jubin R, Doudna JA. 2001. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA 7, 194–206. ( 10.1017/S1355838201001790) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292, 513–529. ( 10.1006/jmbi.1999.3095) [DOI] [PubMed] [Google Scholar]
- 95.Perard J, Leyrat C, Baudin F, Drouet E, Jamin M. 2013. Structure of the full-length HCV IRES in solution. Nat. Commun. 4, 1612 ( 10.1038/ncomms2611) [DOI] [PubMed] [Google Scholar]
- 96.Fuchs G, et al. 2015. Kinetic pathway of 40S ribosomal subunit recruitment to hepatitis C virus internal ribosome entry site. Proc. Natl Acad. Sci. USA 112, 319–325. ( 10.1073/pnas.1421328111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. 2001. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40 s ribosomal subunit. Science 291, 1959–1962. ( 10.1126/science.1058409) [DOI] [PubMed] [Google Scholar]
- 98.Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Burger J, Mielke T, Spahn CM. 2014. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat. Struct. Mol. Biol. 21, 721–727. ( 10.1038/nsmb.2859) [DOI] [PubMed] [Google Scholar]
- 99.Fukushi S, Okada M, Kageyama T, Hoshino FB, Katayama K. 1999. Specific interaction of a 25-kilodalton cellular protein, a 40S ribosomal subunit protein, with the internal ribosome entry site of hepatitis C virus genome. Virus Genes 19, 153–161. ( 10.1023/A:1008131325056) [DOI] [PubMed] [Google Scholar]
- 100.Fukushi S, Okada M, Stahl J, Kageyama T, Hoshino FB, Katayama K. 2001. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 276, 20 824–20 826. ( 10.1074/jbc.C100206200) [DOI] [PubMed] [Google Scholar]
- 101.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. 2002. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA 8, 913–923. ( 10.1017/S1355838202022057) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lu H, Li W, Noble WS, Payan D, Anderson DC. 2004. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 3, 949–957. ( 10.1021/pr0499592) [DOI] [PubMed] [Google Scholar]
- 103.Joseph AP, Bhat P, Das S, Srinivasan N. 2014. Re-analysis of cryoEM data on HCV IRES bound to 40S subunit of human ribosome integrated with recent structural information suggests new contact regions between ribosomal proteins and HCV RNA. RNA Biol. 11, 891–905. ( 10.4161/rna.29545) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Malygin AA, Kossinova OA, Shatsky IN, Karpova GG. 2013. HCV IRES interacts with the 18S rRNA to activate the 40S ribosome for subsequent steps of translation initiation. Nucleic Acids Res. 41, 8706–8714. ( 10.1093/nar/gkt632) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Matsuda D, Mauro VP. 2014. Base pairing between hepatitis C virus RNA and 18S rRNA is required for IRES-dependent translation initiation in vivo. Proc. Natl Acad. Sci. USA 111, 15 385–15 389. ( 10.1073/pnas.1413472111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Angulo J, Ulryck N, Deforges J, Chamond N, Lopez-Lastra M, Masquida B, Sargueil B. 2016. LOOP IIId of the HCV IRES is essential for the structural rearrangement of the 40S-HCV IRES complex. Nucleic Acids Res. 44, 1309–1325. ( 10.1093/nar/gkv1325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Laing C, Schlick T. 2009. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 390, 547–559. ( 10.1016/j.jmb.2009.04.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kieft JS, Zhou K, Grech A, Jubin R, Doudna JA. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9, 370–374. ( 10.1038/nsb781) [DOI] [PubMed] [Google Scholar]
- 109.Sundkvist IC, Staehelin T. 1975. Structure and function of free 40S ribosome subunits: Characterization of initiation factors. J. Mol. Biol. 99, 401–418. ( 10.1016/S0022-2836(75)80135-5) [DOI] [PubMed] [Google Scholar]
- 110.Hinnebusch AG. 2006. eIF3: a versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 31, 553–562. ( 10.1016/j.tibs.2006.08.005) [DOI] [PubMed] [Google Scholar]
- 111.Holz MK, Ballif BA, Gygi SP, Blenis J. 2005. mTOR and S6K1 mediate assembly of the translation preinitiation complex through dynamic protein interchange and ordered phosphorylation events. Cell 123, 569–580. ( 10.1016/j.cell.2005.10.024) [DOI] [PubMed] [Google Scholar]
- 112.Hershey JW. 2015. The role of eIF3 and its individual subunits in cancer. Biochim. Biophys. Acta 1849, 792–800. ( 10.1016/j.bbagrm.2014.10.005) [DOI] [PubMed] [Google Scholar]
- 113.Jager S, et al. 2012. Global landscape of HIV-human protein complexes. Nature 481, 365–370. ( 10.1038/nature10719) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. 2015. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525, 491–495. ( 10.1038/nature14891) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. 2013. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153, 1108–1119. ( 10.1016/j.cell.2013.04.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. 2013. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503, 539–543. ( 10.1038/nature12658) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. 2005. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science 310, 1513–1515. ( 10.1126/science.1118977) [DOI] [PubMed] [Google Scholar]
- 118.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. 1998. Specific interaction of eukaryotic translation initiation factor 3 with the 5' nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 72, 4775–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhou M, et al. 2008. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc. Natl Acad. Sci. USA 105, 18 139–18 144. ( 10.1073/pnas.0801313105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cai Q, Todorovic A, Andaya A, Gao J, Leary JA, Cate JH. 2010. Distinct regions of human eIF3 are sufficient for binding to the HCV IRES and the 40S ribosomal subunit. J. Mol. Biol. 403, 185–196. ( 10.1016/j.jmb.2010.07.054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sun C, et al. 2011. Functional reconstitution of human eukaryotic translation initiation factor 3 (eIF3). Proc. Natl Acad. Sci. USA 108, 20 473–20 478. ( 10.1073/pnas.1116821108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sun C, Querol-Audi J, Mortimer SA, Arias-Palomo E, Doudna JA, Nogales E, Cate JH. 2013. Two RNA-binding motifs in eIF3 direct HCV IRES-dependent translation. Nucleic Acids Res. 41, 7512–7521. ( 10.1093/nar/gkt510) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fraser CS, Hershey JW, Doudna JA. 2009. The pathway of hepatitis C virus mRNA recruitment to the human ribosome. Nat. Struct. Mol. Biol. 16, 397–404. ( 10.1038/nsmb.1572) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Majzoub K, et al. 2014. RACK1 controls IRES-mediated translation of viruses. Cell 159, 1086–1095. ( 10.1016/j.cell.2014.10.041) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Boehringer D, Thermann R, Ostareck-Lederer A, Lewis JD, Stark H. 2005. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure 13, 1695–1706. ( 10.1016/j.str.2005.08.008) [DOI] [PubMed] [Google Scholar]
- 126.Simonetti A, Brito Querido J, Myasnikov AG, Mancera-Martinez E, Renaud A, Kuhn L, Hashem Y. 2016. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol. Cell 63, 206–217. ( 10.1016/j.molcel.2016.05.033) [DOI] [PubMed] [Google Scholar]
- 127.Smith MD, Arake-Tacca L, Nitido A, Montabana E, Park A, Cate JH. 2016. Assembly of eIF3 mediated by mutually dependent subunit insertion. Structure 24, 886–896. ( 10.1016/j.str.2016.02.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lee AS, Kranzusch PJ, Cate JH. 2015. eIF3 targets cell-proliferation messenger RNAs for translational activation or repression. Nature 522, 111–114. ( 10.1038/nature14267) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Filbin ME, Kieft JS. 2011. HCV IRES domain IIb affects the configuration of coding RNA in the 40S subunit's decoding groove. RNA 17, 1258–1273. ( 10.1261/rna.2594011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP. 2015. Structure of the human 80S ribosome. Nature 520, 640–645. ( 10.1038/nature14427) [DOI] [PubMed] [Google Scholar]
- 131.Filbin ME, Vollmar BS, Shi D, Gonen T, Kieft JS. 2013. HCV IRES manipulates the ribosome to promote the switch from translation initiation to elongation. Nat. Struct. Mol. Biol. 20, 150–158. ( 10.1038/nsmb.2465) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Pestova TV, Borukhov SI, Hellen CU. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394, 854–859. ( 10.1038/29703) [DOI] [PubMed] [Google Scholar]
- 133.Boerneke MA, Dibrov SM, Gu J, Wyles DL, Hermann T. 2014. Functional conservation despite structural divergence in ligand-responsive RNA switches. Proc. Natl Acad. Sci. USA 111, 15 952–15 957. ( 10.1073/pnas.1414678111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hertz MI, Landry DM, Willis AE, Luo G, Thompson SR. 2013. Ribosomal protein S25 dependency reveals a common mechanism for diverse internal ribosome entry sites and ribosome shunting. Mol. Cell. Biol. 33, 1016–1026. ( 10.1128/MCB.00879-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Shenvi CL, Dong KC, Friedman EM, Hanson JA, Cate JH. 2005. Accessibility of 18S rRNA in human 40S subunits and 80S ribosomes at physiological magnesium ion concentrations—implications for the study of ribosome dynamics. RNA 11, 1898–1908. ( 10.1261/rna.2192805) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chen B, Kaledhonkar S, Sun M, Shen B, Lu Z, Barnard D, Lu TM, Gonzalez RL Jr, Frank J. 2015. Structural dynamics of ribosome subunit association studied by mixing-spraying time-resolved cryogenic electron microscopy. Structure 23, 1097–1105. ( 10.1016/j.str.2015.04.007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen J, Tsai A, O'Leary SE, Petrov A, Puglisi JD. 2012. Unraveling the dynamics of ribosome translocation. Curr. Opin. Struct. Biol. 22, 804–814. ( 10.1016/j.sbi.2012.09.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Frank J. 2016. Whither ribosome structure and dynamics research? (A perspective). J. Mol. Biol. 428, 3565–3569. ( 10.1016/j.jmb.2016.04.034) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ferguson A, et al. 2015. Functional dynamics within the human ribosome regulate the rate of active protein synthesis. Mol. Cell 60, 475–486. ( 10.1016/j.molcel.2015.09.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Archer SK, Shirokikh NE, Beilharz TH, Preiss T. 2016. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535, 570–574. ( 10.1038/nature18647) [DOI] [PubMed] [Google Scholar]
- 141.LLCS. The PyMOL molecular graphics system, version 1.8 Schrödinger, LLC.
- 142.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. ( 10.1002/jcc.20084) [DOI] [PubMed] [Google Scholar]
- 143.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res. 28, 235–242. ( 10.1093/nar/28.1.235) [DOI] [PMC free article] [PubMed] [Google Scholar]