Abstract
Bacterial ribosome biogenesis has been an active area of research for more than 30 years and has served as a test-bed for the development of new biochemical, biophysical and structural techniques to understand macromolecular assembly generally. Recent work inspecting the process in vivo has advanced our understanding of the role of ribosome biogenesis factors, the co-transcriptional nature of assembly, the kinetics of the process under sub-optimal conditions, and the rRNA folding and ribosome protein binding pathways. Additionally, new structural work enabled by single-particle electron microscopy has helped to connect in vitro ribosomal protein binding maps to the underlying RNA. This review summarizes the state of these in vivo studies, provides a kinetic model for ribosome assembly under sub-optimal conditions, and describes a framework to compare newly emerging assembly intermediate structures.
This article is part of the themed issue ‘Perspectives on the ribosome’.
Keywords: ribosome assembly, macromolecular assembly, RNA folding, quantitative mass spectrometry, cryo-electron microscopy, assembly factors
1. Introduction
Prokaryotic ribosome synthesis is a complex, multistep process requiring the coordinated synthesis, cleavage, post-transcriptional modification and folding of ribosomal RNA (rRNA), and the translation, post-translational modification, folding and binding of approximately 50 ribosomal proteins (r-proteins). The fundamentals of the basic steps of this process have been reviewed extensively [1–5]. Ribosome biogenesis is energetically costly, with the majority of cellular transcription and translational capacity dedicated to the production of new ribosomes [6,7]. This process is both rapid, requiring ∼2 minutes for production of a single ribosome [8,9], and efficient, with the vast majority of assembly events resulting in mature, translationally active complexes. The assembly of ribosomes is tightly regulated in a growth-rate–dependent manner primarily at the level of rRNA synthesis [10], which, in turn, regulates r-protein synthesis through an elegant autoregulatory feedback translational control mechanism that prevents the r-proteins' levels from exceeding the availability of rRNA [11]. The net effect of this network of regulatory mechanisms is a linear relationship between cellular growth rate and ribosomal content that optimally allocates the proteome between catabolic enzymes that produce amino acids, and ribosomes that convert these precursors into new biomass [12].
(a). A co-transcriptional assembly process
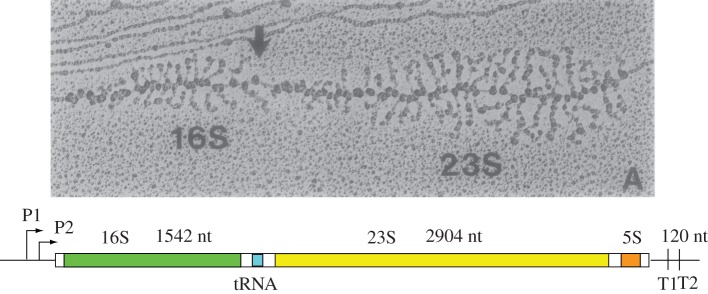
Despite the central cellular role of ribosome assembly in cell physiology, and decades of study, the details of ribosome biogenesis are only beginning to emerge. There has been a trade-off between the detailed studies that can be accomplished by studying assembly in vitro, and less precise but more biologically relevant studies that can be carried out in cells. The primary difference between the in vitro and in vivo studies is the co-transcriptional assembly of rRNA that occurs in cells. Direct evidence for co-transcriptional assembly arose from observation of ultrastructure in the rRNA operons, known as ‘Miller spreads’ [13–16]. Miller found that gently fixing the chromatin of rapidly growing cells cross-linked the RNA polymerase (RNAP) and nascent rRNA transcripts, and that these preparations could be visualized by negative-stain electron microscopy (EM), as shown in figure 1. These images revealed high packing density of RNAP on the rRNA gene with clear evidence for association of additional protein components to the nascent rRNA as the polymerase moved through the operon. Furthermore, a discrete transition from long to short nascent chains at the approximate position of the 3′-terminus of the 16S rRNA coding region provided strong evidence for co-transcriptional rRNA cleavage and release of a 30S precursor early during transcription of the pre-23S rRNA (figure 1) [17].
Figure 1.
Co-transcriptional assembly of ribosomes in the E. coli ribosomal RNA operon. The negative-stain electron micrograph of fixed chromatin shows the high transcriptional level of the ribosomal operon, where over a hundred RNAP molecules can be seen over the 5.5 kb segment [13]. The organization of the rRNA genes is shown below, where the primary rRNA precursor transcript is processed by a series of endonucleolytic cleavage reactions to produce the mature 16S, 23S and 5S rRNAs. The series of increasingly long nascent transcripts can be seen progressing from left to right, with evidence for co-transcriptional binding of ribosomal proteins. In addition, co-transcriptional rRNA processing is observed, indicated by the arrow, which liberates a pre-30S ribonucleoprotein complex prior to initiating the transcription of the 23S rRNA gene.
(b). Assembly guided by RNA folding
Elegant small-angle neutron-scattering measurements by Dr Ramakrishnan suggested that assembly of the small ribosomal subunit (SSU) required RNA folding and compaction, which occurred through both r-protein-dependent and -independent events [18]. These initial experiments further suggested the proteins and RNA were intermingled, a hypothesis that would later be validated when atomic structures of the SSU [19], large subunit (LSU) [20] and mature 70S [21] were solved. These structural studies, when combined with a variety of biochemical and biophysical experiments, led to the hypothesis that ribosome assembly is effectively an RNA-folding problem [22], wherein proteins are used to ‘lock in’ productive RNA folding and drive the structure towards its mature conformation [23,24]. RNA structure probing experiments by the Noller, Woodson and Weeks labs have provided evidence that much of the native rRNA secondary structure is formed in a protein-independent manner, whereas native tertiary rRNA contacts are often stabilized through protein-binding events [25–28].
(c). In vitro ribosomal protein assembly maps
Despite the highly complex and co-transcriptional nature of the assembly process, active subunits can be reconstituted in vitro using purified r-proteins and rRNA [29,30]. While these in vitro assembly reactions are generally less efficient than the process in vivo, the success of in vitro assembly implies that the folding determinants are primarily encoded in the ribosomal components themselves. Extensive in vitro reconstitution experiments by the Nomura and Nierhaus groups directly tested the interdependence of r-protein binding [31,32]. By withholding specific r-proteins, they could classify the r-proteins into groups that bound to rRNA independently (primary binders) and those whose binding was improved by the addition of other r-proteins (secondary binders). These distinct protein-binding classes were the first evidence that ribosome assembly consisted both of sequential and parallel processing steps, and direct structural evidence of these parallel pathways was later observed for both the SSU [33] and LSU [34].
This mixture of sequential and parallel elements provides for a rich landscape of potential assembly pathways. Under a particular growth condition, ribosome assembly flux will partition between the available parallel assembly pathways according to the rates of those pathways, with greater flux along the more rapid pathways. However, the availability of alternative and potentially slower parallel pathways provides an added degree of robustness to buffer the system against transient changes in r-protein or assembly factor availability, stochastic rRNA misfolding or environmental fluctuations (e.g. temperature).
Prokaryotic ribosome biogenesis remains an active and rich area of research, with particular interest in (i) understanding the role of assembly cofactors, (ii) determining the structure and composition of assembly intermediates, and (iii) understanding the kinetics of the process in vivo including the role of and extent to which parallel processing is used. Below, we discuss recent progress in the field.
2. The role of ribosome biogenesis factors
Dozens of accessory biogenesis factors help guide the assembly process, including GTPases [1], rRNA modification enzymes, helicases and other maturation factors [4]. Putative roles for these factors include aiding the maturing particle in avoiding kinetic traps that are thought to occur during RNA misfolding [35], facilitating r-protein association or serving to block premature or non-native r-protein binding [36]. Much recent research has focused on the role of these factors, with researchers testing their activity by adding the factors to in vitro reconstitutions and identifying r-protein binding rates that are enhanced upon factor addition [37], or withholding them in vivo using genetic deletions or depletions and monitoring ribosome assembly for defects [38–43]. Interestingly, many ribosome assembly factors are non-essential, indicating that the reactions they catalyse can be bypassed in a parallel pathway, that redundant factors are present or that they are only necessary under particular environmental conditions like cold-stress. Notably, recent ribosome profiling experiments [44] as well as direct quantitative mass spectrometry measures [45] have revealed that the assembly factors are present at low levels (less than 5% of total ribosomes) and are thus likely to act on the maturing subunits rapidly and catalytically and must have relatively low affinity for the mature subunits to ensure they are not titrated away from intermediates undergoing maturation.
To understand the role of assembly factors in ribosome biogenesis, a series of recent studies have used genetic or pharmacological assembly factor inhibitors [46]. Briefly, cells are grown under factor-restricted conditions, which results in the accumulation of apparent assembly intermediates whose composition and structure is then determined. Using this approach, researchers have suggested roles for assembly factors in aiding in docking of entire structural domains (e.g. RimP) [47], properly positioning r-proteins on the ribosome (e.g. RbgA) [36], or in blocking the SSU–LSU association until maturation is complete (e.g. YjeQ/RbfA) [48].
(a). Ribosome assembly under perturbed conditions
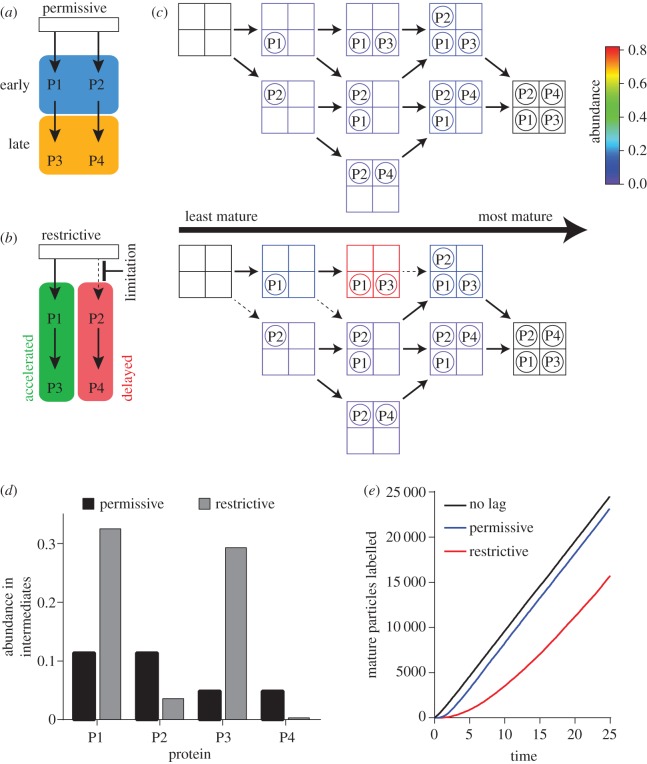
While much of our understanding of ribosome assembly in vivo has resulted from analysis of cells grown under non-optimal conditions (e.g. assembly factor depletions), it is worthwhile noting that interpreting the point of action for a particular factor can be difficult. In these experiments, the accumulating intermediates may have progressed significantly beyond the point of the initial perturbation due to the presence of alternative, parallel assembly pathways. As a result, the intermediates that accumulate during perturbation may not reflect the same intermediates that accumulate in the absence of perturbation. These effects can be illustrated using a simplified model system where four proteins (P1, P2, P3 and P4) bind using two parallel pathways. In this model, P1 and P2 binding are independent, but P3 binding depends on P1, and P4 binding depends on P2 (figure 2a). In this system, the flux through potential assembly pathways, the steady-state intermediate populations (figure 2b), and the total protein occupancy across all intermediates (figure 2c) can be calculated using a stochastic chemical simulation [49]. Assuming equal rates of protein binding at any given step, each of the eight possible intermediates are populated at similarly low levels. These balanced kinetic rates result in many parallel pathways that all converge on the fully assembled complex, and proteins that can be classified as either ‘early’ (P1, P2) or ‘late’ (P3, P4) based on their total abundance in the intermediate population (figure 2a,c).
Figure 2.
Modelling the effects of perturbations on the populations of ribosome assembly intermediates. (a) A simple assembly map involving two primary binding proteins, P1 and P2, and two secondary binding proteins, P3 and P4, in analogy to the Nomura and Nierhaus maps. Binding of P3 depends on prior binding of P1, and binding of P4 depends on prior binding of P2. This leads to a class of early binding proteins in blue, and a class of late-binding proteins in yellow. (b) Perturbation of assembly by limitation of P2 binding results in apparent changes in the observed binding order of assembly. Accelerated proteins shown in green and delayed proteins shown in red. (c) An explicit model enumerating all possible intermediates, based on the assembly maps in (a,b), can be used to simulate the flux through the assembly pathways and the populations of all of the intermediates using the Gillespie algorithm for stochastic dynamics [49]. The RNA is schematically shown as a grid of binding sites, and the proteins as circles. Icons are coloured by abundance according to the colour bar. In the permissive scheme, all protein-binding rate constants are the same. In the restrictive scheme, the rate constant for all steps involving P2 binding are reduced fivefold (dashed arrows). (d) The total population of each protein summed across all intermediates is shown for the permissive condition (black) and the restrictive condition (grey). These values correspond to the protein levels that would be measured by quantitative mass spectrometry. (e) Simulated pulse-labelling kinetics of the mature particle under permissive (blue) or restrictive (red) conditions. A no-intermediate reference is also shown (black).
The effects of a perturbation can easily be modelled by decreasing the rate constant for binding of a particular protein. For example, if P2 binding is limited either by deletion of a cofactor that facilitates that step, or by limiting the availability of P2, there is accumulation of a significant population of intermediates. If the overall rate of mature ribosome assembly is unchanged, then the steady-state pool of assembly intermediates will increase upon the limitation. Furthermore, the accumulated intermediates will bear an altered ribosomal protein composition, with strong enrichment for proteins that can bind in the unperturbed (left) pathway. Indeed, in the permissive condition, P1 and P2 binding precede P3 and P4 binding, as reflected in the assembly map in part (a). In the restrictive condition, P1 and P3 binding appear ‘accelerated’ based on their abundance in the intermediates, whereas and P2 and P4 binding appear ‘delayed’ (figure 2b,d). In reality, the availability of the parallel pathway allows for P2-independent P3 binding. These data show the evidence for the parallel pathways inherent in the assembly map, but also show the complex relationship between the observed order of binding during perturbation and the order of binding without perturbation. This simple example highlights a point of caution when interpreting such assembly inhibition experiments and the dangers of inferring r-protein binding order under wild-type conditions from the binding order observed upon perturbation. Notably, the limitation's effect on P2 and P4 binding becomes apparent upon comparing the composition of the ensemble of intermediates between restrictive and permissive conditions (figure 2c). There, one will note that P3 levels increase by accumulation of an intermediate that is not well-populated in the absence of the perturbation.
An additional concern when perturbing assembly is whether the accumulating particles are dead-end intermediates or whether they are competent to mature, and thus may provide information about the native pathway. One can distinguish between these possibilities using a pulse-chase experiment, which determines whether the rRNA in the intermediate can be radiolabelled and then chased into the mature particles [45,46,50]. Alternatively, if cells are pulse labelled with a stable isotope, quantitative mass spectrometry of r-proteins can be used to show that (i) the intermediate labels before the 70S particle, placing the intermediate upstream, and (ii) that the presence of the intermediate induces a lag in the labelling kinetics of the mature particle, demonstrating that the intermediate is competent to mature [8,34,41,47,51]. These behaviours are captured in the stochastic simulation as a lag introduced into the labelling kinetics of the fully assembled complex. Under the permissive conditions, there is a small population of assembly intermediates, resulting in a small but measureable lag in formation of the full complex. By contrast, owing to the significant population of intermediates awaiting the limiting P2 binding, there is a significant lag in labelling of the full complex (figure 2f). Notably, this lag can be used to estimate the abundance of the maturation-competent intermediates [41].
3. Structure and composition of assembly intermediates
The seminal Nierhaus and Nomura experiments demonstrated thermodynamic cooperativity in r-protein binding, suggesting the presence of underlying cooperatively folded domains in both the SSU and LSU. In the SSU, these folding domains were hypothesized to consist of the individual domains (5′, central, 3′ major and 3′ minor) predicted from the secondary structure as few tertiary contacts were identified linking those domains [52]. In general, r-protein binding in the SSU can be directly attributed to a single domain [52], with a small number of r-proteins linking distal domains in the final structure [53]. The structural observations are consistent with previous experiments demonstrating that individual domains can be folded and reconstituted with the appropriate subsets of ribosomal protein in vitro [54–56]. Indeed, these results indicate that tertiary contacts observed in the mature particle are dispensable for folding of the individual domains. These data were further validated in a series of structural studies of assembly intermediates generated from time-resolved in vitro reconstitutions [33], or from structural analysis of intermediates arising in vivo under various perturbed conditions [47]. For example, Sashital et al. used negative-stain electron microscopy reconstructions of particles arising in a rimP-deletion strain to identify intermediates bearing various combinations of the body, platform, head and terminal stems. Notably, these structural regions correspond to 16S rRNA arising from the 5′, central, 3′ major and 3′ minor secondary structure domains, respectively [47,52]. Later stage assembly intermediates were structurally characterized by single-particle cryo-electron microscopy using various assembly factor-depletion strains, and these structures suggested that the head was the last element to dock as each of the structures bore completely or nearly completely formed body and platform structures with much of the head region unresolved [47,57–59]. Additionally, these complexes only lacked tertiary binding proteins, consistent with nearly mature intermediates.
(a). Folding blocks in the small subunit
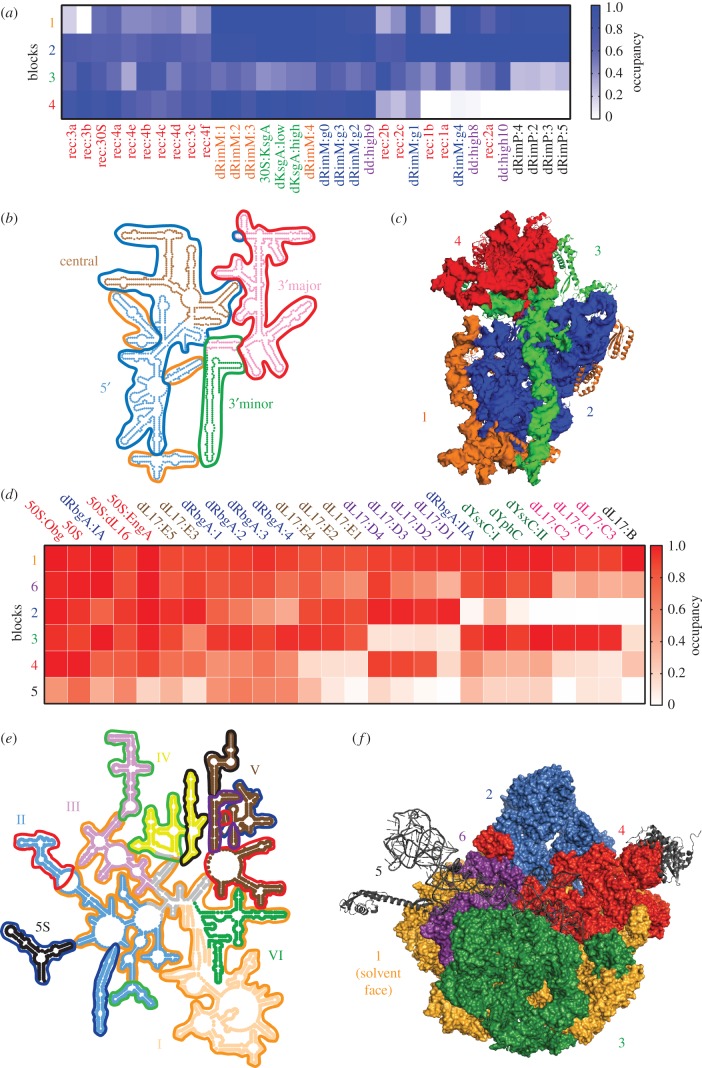
To determine how these SSU intermediates are related to one another and to understand how various SSU structural elements co-assemble, we have analysed the majority of the available SSU assembly intermediate structures on a protein-by-protein, and helix-by-helix basis following the approach described by Davis et al. [34] (figure 2a; electronic supplementary material, figure S1). For each structure and each protein or helix, we calculated the amount of density docked in a native conformation as defined by the assembled 70S subunit. We then clustered this data, revealing a set of four blocks whose ‘native occupancy’ differed between classes; and we name these blocks 1–4.
Consistent with the domain architecture described above, we found that blocks 3 and 4 directly corresponded to the 3′ minor and 3′ major domains in the 16S secondary structure map [60] (figure 3b and table 1). Block 4 was particularly distinguished by its low occupancy in ‘early’ intermediates found in in vitro reconstitution experiments, as well as intermediates from rimP−, rimM− and rbfA−/rsgA− strains. When mapped onto the crystal structure (figure 3c), it is clear that block 4 rRNA helices and proteins form the SSU ‘head’, consistent with reports of misdocking or incomplete docking of the head in these structures [33,47,57,62]. Block 3 exhibited the greatest degree of heterogeneity, but was generally under-occupied across the structures inspected. Notably, this group included the late-binding proteins uS2, uS3, and bS21 as well as helix 44, whose docking is hypothesized to be one of the last assembly events [25,65,66]. As with block 4, the block 3 elements nicely co-localize on both the secondary and tertiary structures, forming the 3′ minor domain (figure 3b,c).
Figure 3.
Cooperative folding blocks of the bacterial large (LSU) and small (SSU) ribosomal subunits. (a) Heatmap of median folding block occupancy in various published SSU assembly intermediate structures. Blocks 1–4 were derived from hierarchical clustering of the calculated occupancy for each r-protein or rRNA helix across the set of available SSU assembly intermediate structures (electronic supplementary material, figure S1). Structures are labelled according to table 1. (b) 16S secondary structure coloured and labelled by domain. Folding blocks as defined by hierarchical clustering in the electronic supplementary material, figure S1, are outlined and coloured according to (a). (c) SSU structure model (PDB: 4ybb) with blocks labelled and coloured according to (a). (d) Heatmap of median folding block occupancy in various published LSU assembly intermediate structures. Blocks 1–6 were derived from hierarchical clustering as in (a) using the set of available LSU assembly intermediate structures (electronic supplementary material, figure S2) and are labelled according to table 1. (e) 23S secondary structure coloured and labelled by domain and folding blocks as in (b). (f) LSU structure model (PDB: 4ybb) with blocks labelled and coloured according to (d).
Table 1.
Electron microscopy structures used in native occupancy analysis. Subunit, label used in figures 2 and 3, sample description, structure ID, nominal resolution, density threshold used and reference provided.
| subunit | label | sample source | EMDB ID | resolution (Å) | threshold | reference |
|---|---|---|---|---|---|---|
| SSU | rec:[1a-4f] | in vitro reconstituted subunits | 1783, 2453–2470 | 30 | 1.24 | [33] |
| dRimM:[1–4] | isolated from a rimM− strain | 5595–5598 | 17 | 2.6 | [59] | |
| dRimM:g[0–4] | isolated from a rimM− strain | 5500–5504 | 14 | −3.125 | [57] | |
| dKsgA:low | isolated from a ksgA− strain with low Mg++ | 2019–2020 | 15.5 | 60 | [61] | |
| dKsgA:high | isolated from a ksgA− strain with high Mg++ | 2019–2020 | 17.7 | 39 | [61] | |
| 30S:KsgA | 30S SSU bound to KsgA | 2017 | 13.5 | 65 | [61] | |
| dd:high[8–10] | isolated from a rsgA−/rbfA− strain with high salt | 5908–5910 | 21.2 | 2.95 | [62] | |
| dRimP:2–5 | pulled out of a rimP− strain with affinity oligo | 6142–6145 | 27.6 | 0.56 | [47] | |
| LSU | 50S:Obg | 50S subunit bound to Obg | 2605 | 5.5 | 0.045 | [63] |
| 50S | 50S subunit isolated from B. subtilis | 5787 | 13 | 1.38 | [41] | |
| dRbgA:[I/II]A | isolated from a rbgA depletion strain | 5642–5643 | 13.3 | 2.7 | [42] | |
| 50S:dL16 | 50S subunit lacking uL16 | 5788 | 13 | 1.38 | [41] | |
| 50S:EngA | 50S subunit bound to EngA | 6149 | 5 | 0.045 | [64] | |
| dL17[B-E5] | isolated from a bL17 depletion strain | 8440, 8442-8444, 8446-8449, 8451-8454, 8456 | 5 | 0.026 | [34] | |
| dRbgA:[1–4] | isolated from a rbgA depletion strain | 5789–5792 | 13 | 1.38 | [41] | |
| dYsxC:[I/II] | isolated from a ysxC depletion strain | 8274–8275 | 6.5 | 0.014 | [43] | |
| dYphC | isolated from a yphC depletion strain | 8276 | 6.5 | 0.014 | [43] |
By contrast, block 2, which was present in the vast majority of the structures, consisted of elements from both the 5′ and central domains (figure 3b), as well as helix 36, which forms extensive contacts with the central pseudoknot and links the 3′ major domain to the rest of the structure (electronic supplementary material, figure S3). Owing to the central pseudoknot's proximity to the 5′ and central domains, this ‘native occupancy’ analysis may overestimate the presence of helix 36, particularly in the lower resolution structures. Regardless, it is interesting to note that none of the isolated and characterized intermediates consisted exclusively of either the 5′ or central domains despite the fact that, according to in vitro studies, these isolated domains are competent for folding. This result suggests either that these domains co-fold very rapidly in vivo, or that the individually folded domains have gone unnoticed in current datasets due to their small size. As single-particle cryo-EM techniques mature, it will be interesting to search for evidence of independent folding of the 5′ or central domains in intermediates isolated in vivo. Finally, block 1 was composed of a subset of rRNA helices from the 5′ and central domains as well as the early binding proteins uS4, uS15 and bS20. The fact that these elements are located on the periphery of the structure (figure 2c) suggests that these elements may be incompletely or less stably docked in the structures analysed. Interestingly, the rRNA helices from block 1 are strongly connected through tertiary interactions (electronic supplementary material, figure S3), indicating that they may fold and dock in a cooperative manner.
(b). Folding blocks in the large subunit
Identifying cooperative folded domains in the LSU has been significantly more difficult because the rRNA domains defined by the secondary structure are highly intertwined and linked through tertiary contacts in the mature subunit [21,67]. Further complicating matters, numerous r-proteins contact link distal rRNA secondary structure domains [21,68]. As a result of this highly interlocked RNA tertiary structure, it is perhaps not surprising that in vitro assembly of the LSU is significantly less efficient than for the SSU, and proper maturation requires various temperature and buffer changes [29]. Moreover, it has not been possible to systematically reconstitute individual domains of the LSU in analogy to the SSU. Finally, unlike the SSU, cooperative protein binding events observed by Nierhaus et al. are generally difficult to reconcile with the LSU rRNA secondary structure domains.
In studying the role of various LSU assembly cofactors, the Gao and Ortega groups determined the structure of a variety of assembly intermediates that arise upon cofactor depletion or deletion [41–43]. Interestingly, these intermediates largely lack the central protuberance, consistent with late folding of this region, and suggesting that the remaining LSU structure can fold largely independently. Notably however, the peptidyl transferase centre is highly disordered in each of these structures suggesting its maturation occurs late in assembly. This result raises the possibility that the modern ribosome assembles along a significantly different assembly pathway than its PTC-only primordial ancestor, which is thought have lacked many of the modern domains now observed to assemble before the PTC.
More recently, we determined the structure of 13 assembly intermediates that arise upon depletion of the r-protein bL17 [34]. These structures are highly disparate and allow for the identification of five folding blocks that are differentially present across the various structures, consistent with the individual blocks folding cooperatively and largely independently. Notably, block 2 from these structures contains the central protuberance and we find a high degree of overlap in structural elements lacking in block 2 and those elements missing in the aforementioned cofactor depletion structures. This overlap could be quantified by calculating the native occupancy of each protein or helix and clustering the resulting data matrix as described for the SSU above (figure 3d). Additionally, the degree to which different structures are related can be quantified using their linkage distance, providing an unbiased approach to group various structures.
This approach revealed that the folding blocks observed in the bL17-depletion strain intermediates are largely conserved across the full swath of intermediate structures that could be analysed. Interestingly, including the additional factor-depletion structures did reveal a single new block 6, which was present at full occupancy in all non-bL17 depletion structures, but was largely absent in the bL17 depletion structures. Unsurprisingly, block 6 contained bL17 and bL17-dependent proteins, such as bL32 and bL28. This result suggests that many of the major groups we observed in the bL17 depletion are also present in the other structures, and it allows us to now segregate the structural effects that were specific to the bL17 perturbation.
Mapping these blocks onto the 23S secondary structure highlights why understanding the folding pathway of the LSU has been significantly more difficult than that of the SSU. The folding blocks span multiple domains, and, although they are connected through tertiary contacts, these connections only represent a small fraction of the total tertiary interaction connectivity map. In contrast to the secondary structure maps, painting the tertiary structure by folding blocks provides a coherent view of how the blocks are related. Block 1, which is present in all of the structures, forms the solvent face of the LSU. Block 2, which is distinguished by its absence in each of the GTPase assembly cofactor depletion strains (rbgA−, yphC− and ysxC−), forms the central protuberance. The remaining blocks 3, 4 and 6 form various regions of the subunit interface. Finally, block 5 is composed strictly of elements in flexible regions of the structure, and their low abundance in all structures is probably a simple reflection of this flexibility.
As noted, our helix/protein occupancy approach also allows for the identification of structurally related intermediates. Satisfyingly, each of the mature 50S structures analysed co-clustered, and they were found to be most closely related to two late-assembly intermediates present in the bL17-depletion strain intermediates (E3 and E5). Next, we find that the bL17-depletion structures lacking a properly docked central protuberance (B, C3, C1 and C2) co-clustered with the least mature intermediates isolated from GTPase assembly cofactor depletion strains, which also lack this structure. Finally, this approach revealed that the ‘D’ classes isolated from the bL17-depletion strains are completely unique among the structures analysed. Given the rapid advances in single-particle cryo-electron microscopy, we expect a large number of new LSU assembly intermediate structures will be reported in the coming years, which will allow for the determination of how unique these ‘D’ class structures truly are.
4. Summary
The field of ribosome assembly is rapidly advancing due to technical advances in mass spectrometry, electron microscopy and other biophysical techniques. While the combined sequential and parallel nature of the assembly process has been appreciated for decades, it has been difficult to understand the role of RNA folding that underlies the cooperative assembly inherent in the Nierhaus and Nomura maps. Furthermore, the substrates and mechanistic roles for the dozens of assembly cofactors remain relatively obscure. By comparing the structures of a wide range of assembly intermediates, we are getting the first molecular glimpses of how ribosome assembly proceeds. While a great deal remains to be understood, it is now clear that the tools are in hand that will ultimately decipher the intricate details of the key process of ribosome biogenesis.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
Competing interests
We declare we have no competing interests.
Funding
This study was funded by National Institute of General Medical Sciences (R37 GM535757) and National Institute on Aging (K99 AG050749).
References
- 1.Britton RA. 2009. Role of GTPases in bacterial ribosome assembly. Annu. Rev. Microbiol. 63, 155–176. ( 10.1146/annurev.micro.091208.073225) [DOI] [PubMed] [Google Scholar]
- 2.Connolly K, Culver G. 2009. Deconstructing ribosome construction. Trends Biochem. Sci. 34, 256–263. ( 10.1016/j.tibs.2009.01.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donhofer A, Sharma MR, Datta PP, Nierhaus KH, Agrawal RH, Wilson DN. 2009. Factor-mediated ribosome assembly in bacteria. In Encyclopedia of life sciences. Chichester, UK: John Wiley & Sons. [Google Scholar]
- 4.Shajani Z, Sykes MT, Williamson JR. 2011. Assembly of bacterial ribosomes. Annu. Rev. Biochem. 80, 501–526. ( 10.1146/annurev-biochem-062608-160432) [DOI] [PubMed] [Google Scholar]
- 5.Woodson SA. 2008. RNA folding and ribosome assembly. Curr. Opin Chem. Biol. 12, 667–673. ( 10.1016/j.cbpa.2008.09.024) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bremer H, Dennis P. 1996. Modulation of chemical composition and other parameters of the cell by growth rate. In Escherichia coli and Salmonella (ed. Neidhart FC.), pp. 1553–1569. Washington, DC: ASM Press. [Google Scholar]
- 7.Maitra A, Dill KA. 2015. Bacterial growth laws reflect the evolutionary importance of energy efficiency. Proc. Natl Acad. Sci. USA 112, 406–411. ( 10.1073/pnas.1421138111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen SS, Sperling E, Silverman JM, Davis JH, Williamson JR. 2012. Measuring the dynamics of E. coli ribosome biogenesis using pulse-labeling and quantitative mass spectrometry. Mol. Biosyst. 8, 3325–3334. ( 10.1039/c2mb25310k) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindahl L. 1975. Intermediates and time kinetics of the in vivo assembly of Escherichia coli ribosomes. J. Mol. Biol. 92, 15–37. ( 10.1016/0022-2836(75)90089-3) [DOI] [PubMed] [Google Scholar]
- 10.Paul BJ, Ross W, Gaal T, Gourse RL. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38, 749–770. ( 10.1146/annurev.genet.38.072902.091347) [DOI] [PubMed] [Google Scholar]
- 11.Zengel JM, Lindahl L. 1994. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acid Res. Mol. Biol. 47, 331–370. ( 10.1016/S0079-6603(08)60256-1) [DOI] [PubMed] [Google Scholar]
- 12.Scott M, Klumpp S, Mateescu EM, Hwa T. 2014. Emergence of robust growth laws from optimal regulation of ribosome synthesis. Mol. Syst. Biol. 10, 747 ( 10.15252/msb.20145379) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotta SL, Miller OL Jr, French SL. 1991. rRNA transcription rate in Escherichia coli. J. Bacteriol. 173, 6647–6649. ( 10.1128/jb.173.20.6647-6649.1991) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hofmann S, Miller OL Jr. 1977. Visualization of ribosomal ribonucleic acid synthesis in a ribonuclease III-Deficient strain of Escherichia coli. J. Bacteriol. 132, 718–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller OL Jr, Beatty BR. 1969. Visualization of nucleolar genes. Science 164, 955–957. ( 10.1126/science.164.3882.955) [DOI] [PubMed] [Google Scholar]
- 16.Miller OL Jr, Hamkalo BA, Thomas CA Jr. 1970. Visualization of bacterial genes in action. Science 169, 392–395. ( 10.1126/science.169.3943.392) [DOI] [PubMed] [Google Scholar]
- 17.Mougey EB, O'Reilly M, Osheim Y, Miller OL Jr, Beyer A, Sollner-Webb B. 1993. The terminal balls characteristic of eukaryotic rRNA transcription units in chromatin spreads are rRNA processing complexes. Genes Dev. 7, 1609–1619. ( 10.1101/gad.7.8.1609) [DOI] [PubMed] [Google Scholar]
- 18.Ramakrishnan V. 1986. Distribution of protein and RNA in the 30S ribosomal subunit. Science 231, 1562–1564. ( 10.1126/science.3513310) [DOI] [PubMed] [Google Scholar]
- 19.Clemons WM Jr, May JL, Wimberly BT, McCutcheon JP, Capel MS, Ramakrishnan V. 1999. Structure of a bacterial 30S ribosomal subunit at 5.5 Å resolution. Nature 400, 833–840. ( 10.1038/23631) [DOI] [PubMed] [Google Scholar]
- 20.Ban N, Nissen P, Hansen J, Capel M, Moore PB, Steitz TA. 1999. Placement of protein and RNA structures into a 5 Å-resolution map of the 50S ribosomal subunit. Nature 400, 841–847. ( 10.1038/23641) [DOI] [PubMed] [Google Scholar]
- 21.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289, 905–920. ( 10.1126/science.289.5481.905) [DOI] [PubMed] [Google Scholar]
- 22.Williamson JR. 2003. After the ribosome structures: How are the subunits assembled? RNA 9, 165–167. ( 10.1261/rna.2164903) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Agalarov SC, Sridhar Prasad G, Funke PM, Stout CD, Williamson JR. 2000. Structure of the S15,S6,S18-rRNA complex: assembly of the 30S ribosome central domain. Science 288, 107–113. ( 10.1126/science.288.5463.107) [DOI] [PubMed] [Google Scholar]
- 24.Culver GM. 2003. Assembly of the 30S ribosomal subunit. Biopolymers 68, 234–249. ( 10.1002/bip.10221) [DOI] [PubMed] [Google Scholar]
- 25.Adilakshmi T, Bellur DL, Woodson SA. 2008. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature 455, 1268–1272. ( 10.1038/nature07298) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA. 2013. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol. Cell. 52, 506–516. ( 10.1016/j.molcel.2013.09.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 11, 959–965. ( 10.1038/nmeth.3029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern S, Powers T, Changchien LM, Noller HF. 1989. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science 244, 783–790. ( 10.1126/science.2658053) [DOI] [PubMed] [Google Scholar]
- 29.Nierhaus KH, Dohme F. 1974. Total reconstitution of functionally active 50S ribosomal subunits from Escherichia coli. Proc. Natl Acad. Sci. USA 71, 4713–4717. ( 10.1073/pnas.71.12.4713) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Traub P, Nomura M. 1968. Structure and function of E. coli ribosomes. V. Reconstitution of functionally active 30S ribosomal particles from RNA and proteins. Proc. Natl Acad. Sci. USA 59, 777–784. ( 10.1073/pnas.59.3.777) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herold M, Nierhaus KH. 1987. Incorporation of six additional proteins to complete the assembly map of the 50 S subunit from Escherichia coli ribosomes. J. Biol. Chem. 262, 8826–8833. [PubMed] [Google Scholar]
- 32.Mizushima S, Nomura M. 1970. Assembly mapping of 30S ribosomal proteins from E. coli. Nature 226, 1214 ( 10.1038/2261214a0) [DOI] [PubMed] [Google Scholar]
- 33.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. 2010. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science 330, 673–677. ( 10.1126/science.1193220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Davis JH, Tan YZ, Carragher B, Potter CS, Lyumkis D, Williamson JR. 2016. Modular assembly of the bacterial large ribosomal subunit. Cell 167, 1610–1622. ( 10.1016/j.cell.2016.11.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Woodson SA. 2000. Recent insights on RNA folding mechanisms from catalytic RNA. Cell. Mol. Life Sci. 57, 796–808. ( 10.1007/s000180050042) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gulati M, Jain N, Davis JH, Williamson JR, Britton RA. 2014. Functional interaction between ribosomal protein L6 and RbgA during ribosome assembly. PLoS Genet. 10, e1004694 ( 10.1371/journal.pgen.1004694) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bunner AE, Nord S, Wikstrom PM, Williamson JR. 2010. The effect of ribosome assembly cofactors on in vitro 30S subunit reconstitution. J. Mol. Biol. 398, 1–7. ( 10.1016/j.jmb.2010.02.036) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Charollais J, Dreyfus M, Iost I. 2004. CsdA, a cold-shock RNA helicase from Escherichia coli, is involved in the biogenesis of 50S ribosomal subunit. Nucleic Acids Res. 32, 2751–2759. ( 10.1093/nar/gkh603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Charollais J, Pflieger D, Vinh J, Dreyfus M, Iost I. 2003. The DEAD-box RNA helicase SrmB is involved in the assembly of 50S ribosomal subunits in Escherichia coli. Mol. Microbiol. 48, 1253–1265. ( 10.1046/j.1365-2958.2003.03513.x) [DOI] [PubMed] [Google Scholar]
- 40.Jiang M, Datta K, Walker A, Strahler J, Bagamasbad P, Andrews PC, Maddock JR. 2006. The Escherichia coli GTPase CgtAE is involved in late steps of large ribosome assembly. J. Bacteriol. 188, 6757–6770. ( 10.1128/JB.00444-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jomaa A, Jain N, Davis JH, Williamson JR, Britton RA, Ortega J. 2013. Functional domains of the 50S subunit mature late in the assembly process. Nucleic Acids Res. 42, 3419–3435. ( 10.1093/nar/gkt1295) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li N, Chen Y, Guo Q, Zhang Y, Yuan Y, Ma C, Deng H, Lei J, Gao N. 2013. Cryo-EM structures of the late-stage assembly intermediates of the bacterial 50S ribosomal subunit. Nucleic Acids Res. 41, 7073–7083. ( 10.1093/nar/gkt423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ni X, et al. 2016. YphC and YsxC GTPases assist the maturation of the central protuberance, GTPase associated region and functional core of the 50S ribosomal subunit. Nucleic Acids Res. 44, 8442–8455. ( 10.1093/nar/gkw678) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li G-W, Burkhardt D, Gross C, Weissman JS. 2014. Quantifying absolute protein synthesis rates reveals principles underlying allocation of cellular resources. Cell 157, 624–635. ( 10.1016/j.cell.2014.02.033) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thurlow B, Davis JH, Leong V, Moraes TF, Williamson JR, Ortega J. 2016. Binding properties of YjeQ (RsgA), RbfA, RimM and Era to assembly intermediates of the 30S subunit. Nucleic Acids Res. 44, 9918–9932. ( 10.1093/nar/gkw613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stokes JM, Davis JH, Mangat CS, Williamson JR, Brown ED. 2014. Discovery of a small molecule that inhibits bacterial ribosome biogenesis. Elife 3, e03574 ( 10.7554/eLife.03574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR. 2014. A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli. Elife 3, e04491 ( 10.7554/eLife.04491) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jeganathan A, Razi A, Thurlow B, Ortega J. 2015. The C-terminal helix in the YjeQ zinc-finger domain catalyzes the release of RbfA during 30S ribosome subunit assembly. RNA 21, 1203–1216. ( 10.1261/rna.049171.114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillespie DT. 2007. Stochastic simulation of chemical kinetics. Annu. Rev. Phys. Chem. 58, 35–55. ( 10.1146/annurev.physchem.58.032806.104637) [DOI] [PubMed] [Google Scholar]
- 50.Al Refaii A, Alix J-H. 2009. Ribosome biogenesis is temperature-dependent and delayed in Escherichia coli lacking the chaperones DnaK or DnaJ. Mol. Microbiol. 71, 748–762. ( 10.1111/j.1365-2958.2008.06561.x) [DOI] [PubMed] [Google Scholar]
- 51.Chen SS, Williamson JR. 2013. Characterization of the ribosome biogenesis landscape in E. coli using quantitative mass spectrometry. J. Mol. Biol. 425, 767–779. ( 10.1016/j.jmb.2012.11.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wimberly BT, Brodersen DE, Clemons WM Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. 2000. Structure of the 30S ribosomal subunit. Nature 407, 327–339. ( 10.1038/35030006) [DOI] [PubMed] [Google Scholar]
- 53.Brodersen DE, Clemons WM Jr, Carter AP, Wimberly BT, Ramakrishnan V. 2002. Crystal structure of the 30 s ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 s RNA. J. Mol. Biol. 316, 725–768. ( 10.1006/jmbi.2001.5359) [DOI] [PubMed] [Google Scholar]
- 54.Agalarov SC, Zheleznyakova EN, Selivanova OM, Zheleznaya LA, Matvienko NI, Vasiliev VD, Spirin AS. 1998. In vitro assembly of a ribonucleoprotein particle corresponding to the platform domain of the 30S ribosomal subunit. Proc. Natl Acad. Sci. USA 95, 999–1003. ( 10.1073/pnas.95.3.999) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Samaha RR, O'Brien B, O'Brien TW, Noller HF. 1994. Independent in vitro assembly of a ribonucleoprotein particle containing the 3' domain of 16S rRNA. Proc. Natl Acad. Sci. USA 91, 7884–7888. ( 10.1073/pnas.91.17.7884) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J. 1993. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 7, 177–180. [DOI] [PubMed] [Google Scholar]
- 57.Guo Q, et al. 2013. Dissecting the in vivo assembly of the 30S ribosomal subunit reveals the role of RimM and general features of the assembly process. Nucleic Acids Res. 41, 2609–2620. ( 10.1093/nar/gks1256) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jomaa A, Stewart G, Martin-Benito J, Zielke R, Campbell TL, Maddock JR, Brown ED, Ortega J. 2011. Understanding ribosome assembly: the structure of in vivo assembled immature 30S subunits revealed by cryo-electron microscopy. RNA 17, 697–709. ( 10.1261/rna.2509811) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leong V, Kent M, Jomaa A, Ortega J. 2013. Escherichia coli rimM and yjeQ null strains accumulate immature 30S subunits of similar structure and protein complement. RNA 19, 789–802. ( 10.1261/rna.037523.112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Petrov AS, et al. 2014. Secondary structures of rRNAs from all three domains of life. PLoS ONE 9, e88222 ( 10.1371/journal.pone.0088222) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boehringer D, O'Farrell HC, Rife JP, Ban N. 2012. Structural insights into methyltransferase KsgA function in 30S ribosomal subunit biogenesis. J. Biol. Chem. 287, 10 453–10 459. ( 10.1074/jbc.M111.318121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang Z, et al. 2014. Structural insights into the assembly of the 30S ribosomal subunit in vivo: functional role of S5 and location of the 17S rRNA precursor sequence. Protein Cell 5, 394–407. ( 10.1007/s13238-014-0044-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feng B, et al. 2014. Structural and functional insights into the mode of action of a universally conserved Obg GTPase. PLoS Biol. 12, e1001866 ( 10.1371/journal.pbio.1001866) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang X, et al. 2014. Structural insights into the function of a unique tandem GTPase EngA in bacterial ribosome assembly. Nucleic Acids Res. 42, 13 430–13 439. ( 10.1093/nar/gku1135) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dutca LM, Culver GM. 2008. Assembly of the 5' and 3' minor domains of 16S ribosomal RNA as monitored by tethered probing from ribosomal protein S20. J. Mol. Biol. 376, 92–108. ( 10.1016/j.jmb.2007.10.083) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang ZJ, Chee CE, Huang S, Sinicrope FA. 2011. The role of autophagy in cancer: therapeutic implications. Mol. Cancer Ther. 10, 1533–1541. ( 10.1158/1535-7163.MCT-11-0047) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292, 883–896. ( 10.1126/science.1060089) [DOI] [PubMed] [Google Scholar]
- 68.Klein DJ, Moore PB, Steitz TA. 2004. The roles of ribosomal proteins in the structure assembly, evolution of the large ribosomal subunit. J. Mol. Biol. 340, 141–177. ( 10.1016/j.jmb.2004.03.076) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.