Abstract
Spermatogenesis is a classic model of cycling cell lineages that depend on a balance between stem cell self-renewal for continuity and the formation of progenitors as the initial step in the production of differentiated cells. The mechanisms that guide the continuum of spermatogonial stem cell (SSC) to progenitor spermatogonial transition and precise identifiers of subtypes in the process are undefined. Here we used an Id4-eGfp reporter mouse to discover that EGFP intensity is predictive of the subsets, with the ID4-EGFPBright population being mostly, if not purely, SSCs, whereas the ID4-EGFPDim population is in transition to the progenitor state. These subsets are also distinguishable by transcriptome signatures. Moreover, using a conditional overexpression mouse model, we found that transition from the stem cell to the immediate progenitor state requires downregulation of Id4 coincident with a major change in the transcriptome. Collectively, our results demonstrate that the level of ID4 is predictive of stem cell or progenitor capacity in spermatogonia and dictates the interface of transition between the different functional states.
KEY WORDS: ID4, Progenitor, Spermatogonia, Stem cell
Summary: The level of ID4 expression distinguishes stem cells and immediate progenitors in mammalian spermatogonial cells, while downregulation of ID4 is required for transition between the functional states.
INTRODUCTION
Spermatogenesis produces millions of genetically unique sperm every day in males from puberty until old age. The continuity and robustness of the process depend on the actions of spermatogonial stem cells (SSCs) and progenitor spermatogonia that comprise an undifferentiated population (de Rooij and Russell, 2000; Oatley and Brinster, 2008). In mammals, spermatogenesis occurs at periodic intervals, with the transition of progenitor spermatogonia from an undifferentiated type A to a differentiating type A1 state in response to a retinoic acid (RA) pulse (Hogarth et al., 2015). The differentiating spermatogonia then undergo a consistent series of mitotic divisions before initiating meiosis. The SSC pool serves as a self-renewing reservoir from which the next cohort of progenitors will arise. Thus, the balance between self-renewing divisions to sustain the stem cell pool and the production of daughter cells that will transition to a progenitor state is essential for continuity of the spermatogenic lineage.
In all mammals studied, SSC and progenitor spermatogonial subsets are components of a heterogeneous undifferentiated type A population (de Rooij, 1973; de Rooij and Russell, 2000). In mice and rats, the subsets can be classified as single cells (Asingle), a pair of cells (Apair), or chains of 4-16 aligned cells (Aaligned4-16). The Apair and Aaligned cells are linked by a persistent intercellular bridge due to incomplete cytokinesis during mitotic division. The traditional model posits that the SSC pool comprises Asingle, and divisions that produce Apair are the first step in transition to a transit amplifying progenitor state (Oakberg, 1971; Huckins and Oakberg, 1978). This paradigm has been challenged using mouse models with a variety of reporter transgenes that mark subsets of spermatogonia (Nakagawa et al., 2010; Hara et al., 2014). Outcomes based on time-lapse imaging suggest that fragmentation of chains occurs in steady-state conditions at low frequency, but more prevalently in conditions of regeneration to re-establish the SSC pool, which might suggest that the subsets of spermatogonia with stem cell potential are more diverse than predicted by the Asingle model. However, this premise is complicated by the fact that a majority of undifferentiated spermatogonia (64% of Apair, 94% of Aaligned4, and 100% of Aaligned8-16) transition to a differentiating state after every periodic pulse of RA (Tegelenbosch and de Rooij, 1993). By contrast, none of the Asingle spermatogonia are thought to undergo this transition. Thus, the persisting Asingle and rare Apair cells are tasked with re-establishing the progenitor population in preparation for a subsequent RA pulse.
At present, molecular markers that distinguish SSC and progenitor spermatogonia and mechanisms that control the interface of the transition are undefined. In previous studies, we discovered that expression of the helix-loop-helix protein ID4 (Riechmann et al., 1994) is selective for a subset of Asingle in mouse testes and plays a role in maintenance of the SSC pool (Oatley et al., 2011). To study the population in more detail, we generated an Id4-eGfp transgenic mouse line in which EGFP signal reflects ID4 protein levels, although the half-life of EGFP may extend beyond that of normal ID4, and discovered that ID4-EGFP+ spermatogonia are primarily Asingle, although some Apair cells can be observed (Chan et al., 2014). Notably, EGFP+ Apair cells could be false pairs that form transiently when Asingle divide to form new Asingle cells, for example because abscission is delayed and the cells may not have migrated away from each other. In addition, we utilized primary cultures of undifferentiated spermatogonia to compare the regenerative capacity of ID4-EGFP+ and ID4-EGFP− subsets. Outcomes of those experiments suggested that most, if not all, SSC activity resides in the ID4-EGFP+ population (Chan et al., 2014). Furthermore, lineage-tracing studies confirmed that at least some ID4-expressing spermatogonia are SSCs in testes during steady-state conditions (Sun et al., 2015). Although the stem cell purity of the population has not been determined, these findings suggested that the levels of ID4 influence the stem cell-to-progenitor transition.
In the current study, we utilized Id4-eGfp transgenic mice and transplantation analyses to discover that the levels of ID4 expression are associated with regenerative capacity. Importantly, the outcomes of limiting dilution transplantation analyses revealed that a population defined as being ID4-EGFPBright is mostly, if not purely, SSCs, and that most ID4-EGFPDim spermatogonia lack stem cell capacity and are therefore likely to be in transition to a progenitor state. In addition, we discovered that the spermatogonial subsets are distinguishable based on unique transcriptome signatures. Furthermore, we generated a novel mouse model for manipulating Id4 levels and found that induction of constitutive expression in prospermatogonia, which are precursors of SSCs, leads to the formation of an initial SSC pool, but development of the progenitor spermatogonial population is impaired and initiation of the transition to a differentiating state is blocked. Moreover, we discovered that constitutive expression of Id4 leads to dramatic alteration of the transcriptome. Taken together, these findings indicate that the level of ID4 expression is a key factor in the mechanism regulating the transition from a stem cell to progenitor state in mammalian spermatogonia.
RESULTS
Identification of ID4-EGFPBright and ID4-EGFPDim spermatogonial subsets
In the Id4-eGfp transgenic mouse line that we generated in a previous study, EGFP signal represents ID4 protein levels and bright cells appear to exist primarily as Asingle (Chan et al., 2014). Here, we sought to explore further whether subsets of undifferentiated spermatogonia could be distinguished based on intensity of the ID4-EGFP signal. We utilized mice at postnatal day (P) 8 of development because testes are enriched for undifferentiated spermatogonia at this age and the composition of the population is identical to that in adults (Drumond et al., 2011). Cells with different EGFP fluorescent intensity were clearly distinguishable in whole tubules by confocal microscopy (Fig. 1A, Fig. S1A). In confirmation of our previous observations, cells with the brightest EGFP intensity appeared to be Asingle, but some EGFPBright Apair cells were also observed. In addition, cells with a lower intensity of EGFP were observed as both Asingle and Apair. It is important to note that although it is likely that all Asingle are EGFP+ at some level, we could not determine this unequivocally, nor could we clearly determine whether intercellular bridges existed between the ID4-EGFP+ Apair cells, but they were in close enough proximity and appeared to possess an obvious cellular connection to suggest cohort identity. In addition, we could not definitively observe Aaligned cells with EGFP signal. To further define the observations, we measured relative EGFP intensity in images of cells with different identities, i.e. single or pair. Analysis of the dataset by linear regression revealed a significant (P<0.0001) association between EGFP intensity and single or pair status (Fig. 1B, Fig. S1B). The mean EGFP intensity of Asingle cells was overall significantly greater (by 1.6-fold, P<0.0001) than that for Apair cells. Using flow cytometric analysis (FCA), we also observed a gradient of EGFP intensity and could classify subsets of cells as being EGFPBright or EGFPDim (Fig. 1C), representing ∼20% and ∼41% of the population, respectively (Fig. 1D). The remaining ∼39% of the population could be classified as possessing intermediate levels of EGFP. Moreover, qRT-PCR analysis of FACS isolated cell subsets revealed that endogenous Id4 transcript levels correlate with EGFP intensity (Fig. S1C). Collectively, these findings suggested that reduction in the level of ID4 expression associates with transition from an Asingle to Apair state.
Fig. 1.
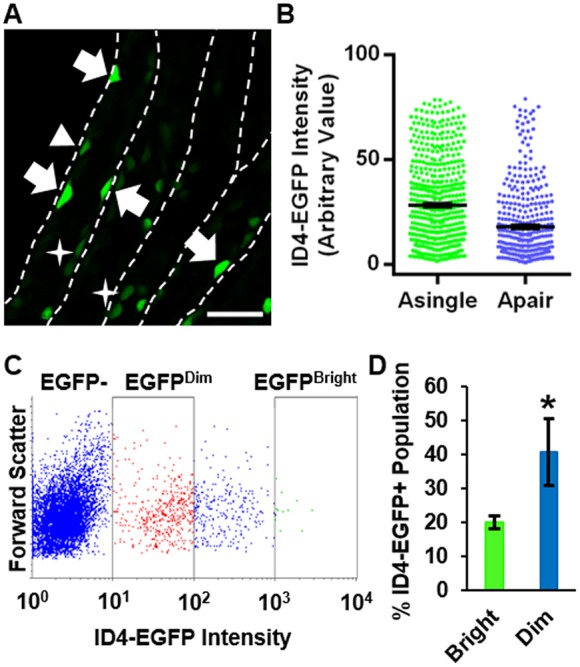
Distinction of undifferentiated spermatogonial subsets by ID4-EGFP expression in testes of mice. (A) Whole-mount confocal image of live seminiferous tubules from an Id4-eGfp transgenic mouse at P8. EGFP+ cells are ID4-expressing spermatogonia. Arrows indicate Asingle with bright EGFP intensity (ID4-EGFPBright). Arrowhead indicates Asingle with dim EGFP intensity (ID4-EGFPDim). Stars indicate Apair with dim EGFP intensity (ID4-EGFPDim). Scale bar: 50 µm. (B) Quantitative comparison of EGFP intensity in Asingle and Apair ID4+ spermatogonia. Each dot represents an individual cell (n=4 different animals, 13.2 mm of tubules, 944 total cells) and the mean±s.e.m. is indicated by black bars for each population. (C) Dot plot of flow cytometric analysis (FCA) for subsets of spermatogonia based on ID4-EGFP intensity. (D) Quantitative comparison of the percentage of the ID4-EGFP+ population that can be classified as ID4-EGFPBright or ID4-EGFPDim from FCA. Data are mean±s.e.m. for three independent experiments. *P<0.05.
Regenerative capacity of ID4-EGFP+ spermatogonial subsets
To explore whether differences in ID4 expression levels are linked with functional capacity to regenerate the spermatogenic lineage, we performed a series of transplantation experiments with ID4-EGFPBright and ID4-EGFPDim subsets isolated from testes of P8 mice (Fig. 2A). To conduct the experiments in a quantitative manner, we generated F1 hybrid males possessing Id4-eGfp and Rosa26-lacZ transgenes to serve as the source of donor germ cells. The Rosa26-lacZ transgene allows for colonies of donor-derived spermatogenesis within recipient testes to be clearly identified by blue staining and to be quantified visually (Brinster and Zimmermann, 1994; Oatley and Brinster, 2006; Helsel and Oatley, 2017). Because each colony is clonally derived from a single SSC (Nagano et al., 1999; Kanatsu-Shinohara et al., 2006), colony number is a direct measure of stem cell content in a transplanted cell population.
Fig. 2.
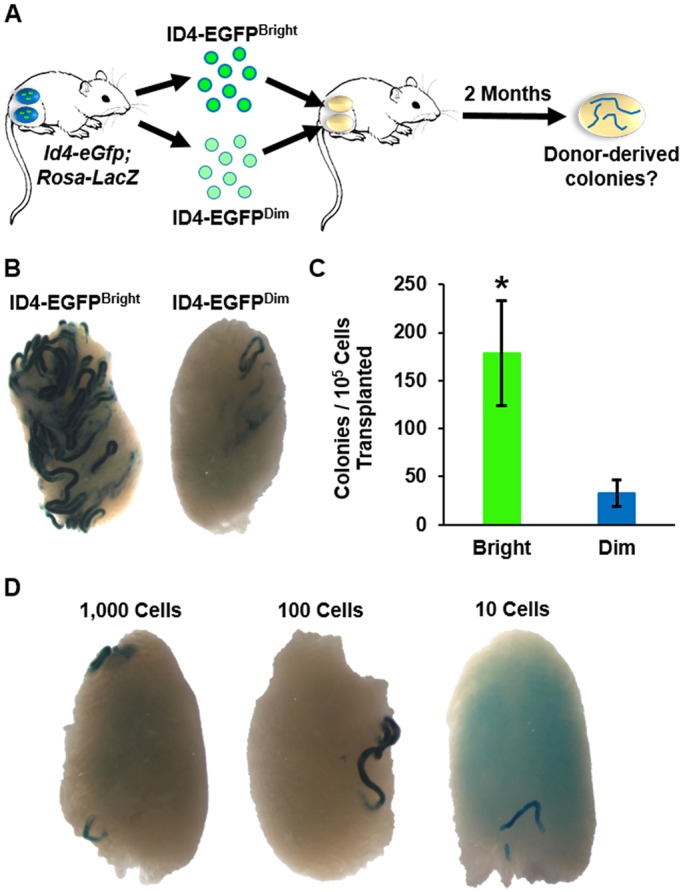
Functional distinction of ID4-EGFP+ spermatogonial subsets by transplantation. (A) Experimental scheme to assess regenerative capacity of ID4-EGFPBright and ID4-EGFPDim spermatogonial subsets by transplantation analyses. (B) Representative images of recipient testes transplanted with 1×104 ID4-EGFPBright or ID4-EGFPDim cells. Each blue segment is a colony of spermatogenesis derived from an individual transplanted donor SSC. (C) Quantitative comparison of donor-derived spermatogenic colonies derived from transplantation of ID4-EGFPBright or ID4-EGFPDim spermatogonia (1×104 cells transplanted/recipient testis). Data are mean±s.e.m. and n=3 different cell preparations and 29 recipient testes. *P<0.05. (D) Representative images of recipient testes transplanted with 1000, 100 or 10 ID4-EGFPBright spermatogonia.
In the first set of experiments, we made a direct comparison of regenerative capacity between ID4-EGFPBright and ID4-EGFPDim subsets. The cells were isolated by FACS and a standard number (1×104) transplanted into each testis of immunologically compatible recipient mice prepared by busulfan treatment. To remove variation in the recipient environment as a confounding factor of donor cell colonization, one testis of each recipient received ID4-EGFPBright cells and the contralateral testis received ID4-EGFPDim cells. Recipient testes were then analyzed for colonies of donor-derived spermatogenesis 2 months after transplantation. Outcomes revealed that ID4-EGFPBright cells generated a significantly greater (by 5.5-fold, P<0.05) number of colonies than ID4-EGFPDim cells (Fig. 2B,C). In total, ∼85% of the SSCs were captured in the ID4-EGFPBright population.
Next, we aimed to determine the relative stem cell potency of the ID4-EGFPBright population. We conducted transplantation analyses in a limiting dilution (LD) manner by microinjecting 1000, 100 or 10 ID4-EGFPBright cells into recipient testes to determine the lowest number that would result in at least one donor-derived colony of spermatogenesis. Two months after transplantation, we observed colonies in testes receiving all of the different cell numbers (Fig. 2D). With 10 cells being the limiting number to observe colonies, we conducted a larger scale experiment to determine the relative SSC content of the ID4-EGFPBright population. In total, transplantation of 110 donor cells into 11 recipient testes yielded 6 colonies of donor-derived spermatogenesis (Table S1), thus demonstrating that the ID4-EGFPBright population is highly enriched for SSCs. Based on these data, the relative regenerative potency of the population can be defined as LD10 with the relative SSC content being 5455 (6 colonies/105 cells transplanted). Furthermore, based on a colonization efficiency of 5% that was determined in previous transplantation studies to describe the efficiency of SSC engraftment in a recipient testis (Nagano et al., 1999; Ogawa et al., 2003), we estimated that 1 in 0.92 cells [110 cells transplanted/(6 colonies of donor-derived spermatogenesis/5% colonization efficiency)] is an SSC in the ID4-EGFPBright population. These findings suggest that the ID4-EGFPBright population is essentially pure stem cells.
ID4-EGFPBright and ID4-EGFPDim spermatogonia possess distinct transcriptomes
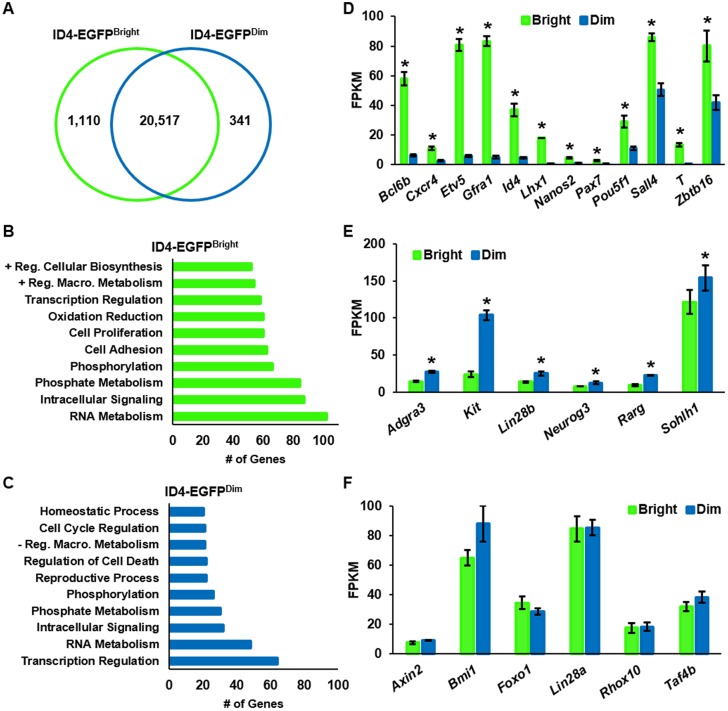
Considering that the ID4-EGFPBright and ID4-EGFPDim spermatogonial subsets were found to possess functional differences, we postulated that the subsets could also be distinguished based on unique transcriptome signatures. To explore this, RNA-seq analysis was conducted with both subsets that were isolated by FACS from testes of P8 mice. To define significant differences in gene expression, we chose a stringent cut-off of the mean FPKM being at least 1.0 for one of the subtypes and a false discovery rate of 1% (q≤0.01). Using these criteria, 1451 protein-coding genes were found to be differentially expressed between the two populations overall (Fig. 3A). Of these, 1110 genes were found to be upregulated in ID4-EGFPBright cells and 341 genes were upregulated in ID4-EGFPDim cells (Table S2). Mining of the dataset for the expression of genes that have been associated with undifferentiated spermatogonia via previous studies revealed these to make up only 1.8%. Thus, ∼98% of the differentially expressed genes are potential novel regulators of the stem cell and progenitor states in spermatogonia. Assessment of the differentially expressed genes by gene ontology (GO) analysis revealed common functional classifications between the ID4-EGFPBright and ID4-EGFPDim subsets (Fig. 3B,C). However, distinguishing GO classifications could also be made for upregulated genes of each spermatogonial subtype (Fig. 3B,C), including positive regulation of cellular and macromolecular biosynthesis, cell adhesion, and oxidation reduction for ID4-EGFPBright cells; and homeostatic processes, negative regulation of macromolecular biosynthesis, regulation of cell death, and reproductive processes for ID4-EGFPDim cells.
Fig. 3.
Transcriptome distinction of ID4-EGFP+ spermatogonial subsets by RNA-seq. (A) Venn diagram depicting the number of genes determined to be expressed at significantly greater levels in ID4-EGFPBright or ID4-EGFPDim spermatogonial populations at q≤0.01. Data are derived from FACS-isolated populations; n=3. (B,C) Functional classification of genes upregulated in ID4-EGFPBright (B) and ID4-EGFPDim (C) spermatogonia by GO analysis. Data are the number of genes binned into the top ten functional categories for each population. (D,E) Genes with significant (P≤0.05) upregulation in ID4-EGFPBright (D) or ID4-EGFPDim (E) spermatogonia. (F) Genes with no difference (P>0.05) in expression between ID4-EGFPBright and Id4-EGFPDim spermatogonia. For all graphs, data are mean±s.e.m. Fragments per kilobase of exon per million fragments mapped (FPKM) values from RNA-seq analyses of three matched samples from different pools of animals. *P<0.05.
Next, we mined the RNA-seq dataset for the expression of genes that have been associated previously with SSCs. A cut-off of P<0.05 was considered as a significant difference between the subtypes. As expected, expression of Id4 was significantly upregulated in the ID4-EGFPBright population, similar to the expression of Bcl6b, Etv5, Lhx1, T, Zbtb16 (Plzf), Gfra1, Pax7, Cxcr4, Sall4, Nanos2 and Pou5f1 (Fig. 3D), thus implicating the expression level of these molecules as distinguishing features of SSCs and confirming a role for some in SSC maintenance. By contrast, expression of Neurog3, Rarg, Kit, Taf4b, Lin28a, Lin28b, Sohlh1, Rhox10, Adgra3 (Gpr125), Axin2, Bmi1 and Foxo1 was significantly greater in ID4-EGFPDim cells or did not differ between the subtypes (Fig. 3E,F), thus indicating an association with progenitor status or an equally important role in SSC and progenitor maintenance.
Constitutive overexpression of Id4 in the germline impairs spermatogenesis
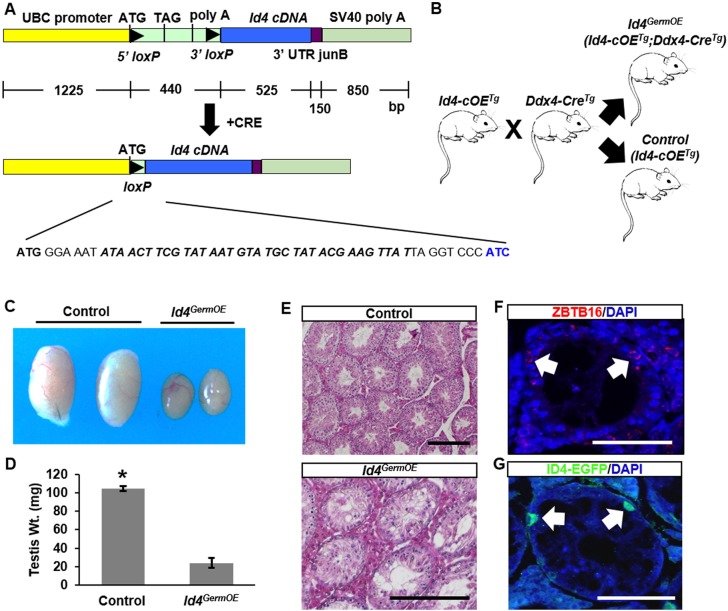
Our previous studies revealed an age-related increase in the percentage of seminiferous tubules that lack germline in mice deficient for Id4 expression, indicating impaired SSC maintenance (Oatley et al., 2011). In addition, the findings presented above from examining the ID4-EGFPBright and ID4-EGFPDim subsets suggested that the level of ID4 dictates the SSC-to-progenitor transition. To explore this further, a novel transgenic mouse model was generated for conditional overexpression of Id4. A transgene was assembled with a flox-stop sequence inserted between the constitutive human ubiquitin C (UBC) promoter and Id4 coding sequence (Fig. 4A). A founder line of FVB;129 genetic background was then generated using pronuclear microinjection (hereafter designated Id4cOE). In primary cultures of tail-tip fibroblasts from these mice, treatment with adenovirus-Cre resulted in induction of ID4 expression, confirming the conditional nature of the transgene (Fig. S2A). Next, we crossed Id4cOE and Ddx4Cre mice to produce animals with constitutive expression of Id4 in germ cells only (Fig. 4B; hereafter designated Id4GermOE). Expression of the Ddx4Cre transgene initiates at embryonic day 15.5 in prospermatogonia that are precursors of the SSC pool (Gallardo et al., 2007).
Fig. 4.
Impact of constitutive Id4 expression in the male germline. (A) Schematic of the transgene used for making a mouse model for conditional overexpression of Id4. The nucleotide sequence between the UBC promoter and Id4 cDNA remaining after LoxP recombination of the Stop cassette is indicated. (B) Experimental approach to produce male mice with constitutive expression of Id4 in the germline (Id4GermOE) and controls for comparison. (C) Images of testes from adult (2 months old) control and Id4GermOE mice. (D) Quantitative comparison of testis weight from control and Id4GermOE mice at 2 months of age. Data are mean±s.e.m. for three different mice of each genotype; *P<0.05. (E) Images of testis cross-sections from adult (2 months old) control and Id4GermOE mice stained with Hematoxylin and Eosin. (F) Immunofluorescence staining of a cross-section from an Id4GermOE mouse at 2 months of age for the pan-undifferentiated spermatogonial marker ZBTB16 (arrows). (G) Immunofluorescence staining of a cross-section from a multi-transgenic Id4eGfp;GermOE mouse at 2 months of age for ID4-EGFP+ spermatogonia (arrows). Scale bars: 50 µm.
First, we examined the fertility of Id4GermOE male mice at adulthood (beginning at 2 months of age) by pairing with at least three adult wild-type females for 4 months. Controls were Id4cOE littermates lacking the Ddx4Cre transgene. Outcomes of the breeding trials revealed that Id4GermOE mice (n=3) never sired a pup, even though mating occurred as evidenced by the presence of vaginal plugs. By contrast, control males sired ∼10 pups per litter [10.4±2.1 (s.d.), n=3-4 litters per male] over the same period of time. At 2 months of age, the testes of Id4GermOE mice were found to be reduced to only 23% of the size of testes from control mice, indicating impaired spermatogenesis (Fig. 4C,D). Assessment of cross-sections from Id4GermOE mice at 2 months of age revealed impaired spermatogenesis in a vast majority (∼98%) of the seminiferous tubules examined, whereas spermatogenesis was intact in all seminiferous tubules of control mice (Fig. 4E).
Although spermatogenesis was impaired in a majority of seminiferous tubules in testes of Id4GermOE mice at 2 months of age, every cross-section examined contained a few spermatogonia and some tubules contained pachytene spermatocytes, but a subsequent layer of differentiating spermatogonia or earlier stages of spermatocytes was not observable (Fig. 4E, Fig. S2B). However, at 6 months of age we observed tubules that contained spermatogonia and other tubules that appeared to completely lack germ cells, containing Sertoli cells only (Fig. 4E, Fig. S2B), and we confirmed these observations by immunostaining for the pan-germ cell marker TRA98 (Fig. S2C). Further immunostaining analyses revealed that all of the germ cells in Id4GermOE testes at 2 and 6 months of age expressed the undifferentiated spermatogonial marker ZBTB16 (Fig. 4F). Moreover, we produced triple-transgenic Id4eGfp;Id4cOE;Ddx4Cre mice by crossbreeding and found that the persisting spermatogonia were ID4-EGFP+ (Fig. 4G). Taken together, these results indicated that constitutive expression of Id4 beginning in prospermatogonia disrupts spermatogenesis by both impairing germ cell survival and inhibiting the differentiation process, but an SSC pool is initially established.
Postnatal development of the undifferentiated spermatogonial population in testes of mice with constitutive expression of Id4 in the germline
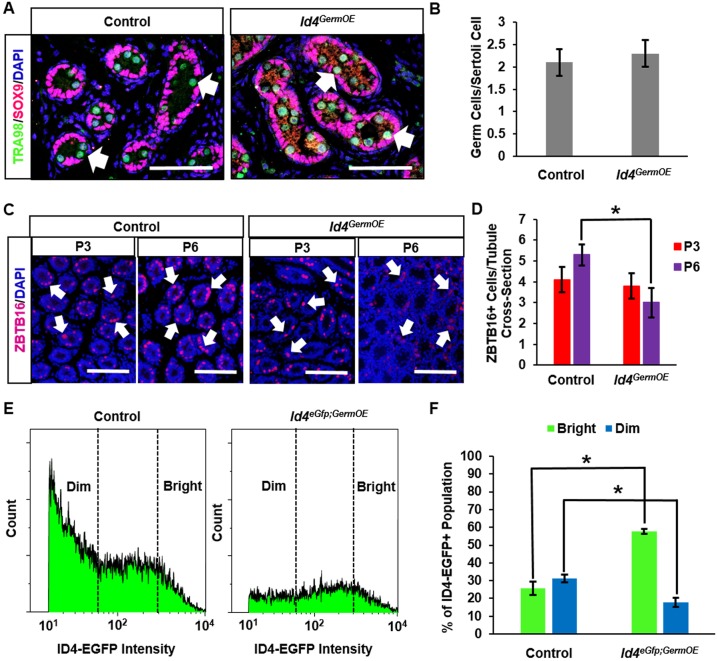
To further explore the reason for disrupted spermatogenesis in Id4GermOE mice, we examined whether formation of the different spermatogonial populations was compromised during neonatal development. A primary heterogeneous spermatogonial population is known to arise from precursor prospermatogonia during P0-6 in mice that includes formation of initial SSC and progenitor pools. By P8, the dynamics of the heterogeneous undifferentiated spermatogonial population are identical to that in adulthood (Drumond et al., 2011). At P0, the number of prospermatogonia in cross-sections of testes was found to be the same in Id4GermOE and control mice (Fig. 5A,B). Likewise, at P3, the number of undifferentiated spermatogonia (i.e. ZBTB16+ cells) in cross-sections was similar between Id4GermOE and control mice (Fig. 5C,D). However, at P6, the number of ZBTB16+ spermatogonia was significantly reduced in testes of Id4GermOE compared with control mice (Fig. 5C,D). The overall number of undifferentiated spermatogonia increased by ∼30% from P3-6 in control mice, but a comparable change was ablated in testes of Id4GermOE mice.
Fig. 5.
Impact of constitutive Id4 expression on postnatal development of the spermatogonial population. (A) Immunofluorescence staining of testis cross-sections from control and Id4GermOE mice at P0 for the pan-germ cell marker TRA98 (arrows and green staining) and the Sertoli cell marker SOX9 (pink). (B) Quantitative comparison of germ cell number in cross-sections of seminiferous cords from testes of control and Id4GermOE mice at P0. Data are mean±s.e.m. for three different mice of each genotype and normalized to the number of Sertoli cell nuclei in 30 different cross-sections. (C) Immunofluorescence staining of testis cross-sections from control and Id4GermOE mice at P3 and P6 for the pan-undifferentiated spermatogonial marker ZBTB16 (arrows and red staining). (D) Quantitative comparison of ZBTB16+ spermatogonial numbers in cross-sections of seminiferous tubules from testes of control and Id4GermOE mice at P3 and P6. Data are mean±s.e.m. for three different mice and 30 cross-sections of each genotype. *P≤0.05. (E) Histogram plots from flow cytometric analyses of the ID4-EGFP+ population in testes of control and Id4eGfp;GermOE mice at P8. (F) Quantitative comparison of the percentage of spermatogonia defined as ID4-EGFPBright and ID4-EGFPDim by FCA in testes of control and Id4eGfp;GermOE mice at P8. Data are mean±s.e.m. for three different mice of each genotype; *P≤0.05. Scale bars: 50 µm.
Next, we utilized the triple-transgenic model of Id4eGfp;Id4cOE;Ddx4Cre (Id4eGfp;GermOE) to assess development of the ID4-EGFPBright and ID4-EGFPDim subpopulations at P8 using FCA (Fig. 5E). The transplantation analyses described above demonstrated that ID4-EGFPBright cells represent the SSC pool, whereas most ID4-EGFPDim cells have lost this capacity. In testes of control mice (Id4eGfp;Id4cOE), the percentage of the ID4-EGFP+ population that could be classified as ID4-EGFPBright and ID4-EGFPDim was ∼25% and 32%, respectively. In comparison, these proportions were essentially opposite in testes of Id4eGfp;GermOE mice, with ∼58% and 18% of ID4-EGFP+ cells being classified as ID4-EGFPBright and ID4-EGFPDim, respectively (Fig. 5F). During normal development in mice, differentiating spermatogonia derived from the initial SSC and progenitor populations arise at P7-10. In cross-sections of testes from control mice at P10, differentiating spermatogonia were clearly present, as evidenced by expression of the marker STRA8, whereas only rare STRA8+ spermatogonia were observed in cross-sections of testes from Id4GermOE mice (Fig. S2D). Taken together, these findings indicate that normal expansion of the undifferentiated spermatogonial pool that occurs in postnatal development was hindered by constitutive expression of Id4 and development of the progenitor spermatogonial population is greatly impaired, and those that do develop are not capable of transitioning to a differentiating state in response to an RA pulse.
Overexpression of Id4 alters the transcriptome in spermatogonia
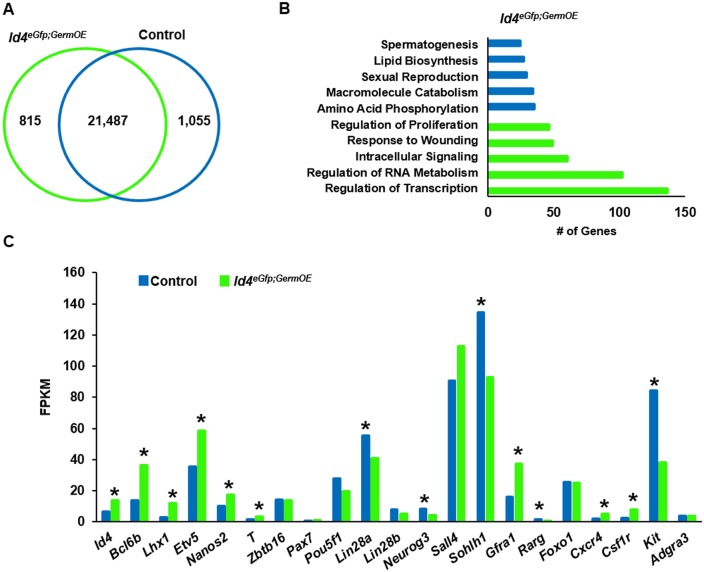
The dramatic phenotype of impaired spermatogenic lineage development in Id4GermOE mice suggested disruption in the expression of genes with a key role in the establishment of spermatogonial subtypes. To provide a molecular level assessment of the impact of Id4 overexpression on the dynamics of the developing SSC and progenitor pools, we compared the transcriptome profiles of the ID4-EGFP+ populations isolated by FACS from testes of triple-transgenic Id4eGfp;GermOE and control mice using RNA-seq analysis. Overall, the expression of 1870 protein-coding genes was found to be significantly (q≤0.01) different at a 2-fold or greater level between the genotypes (Fig. 6A). Of these, 815 genes were upregulated and 1055 genes downregulated in cells from Id4eGfp;GermOE mice compared with controls (Table S3). GO analysis revealed unique pathways for both upregulated and downregulated genes (Fig. 6B). Among the differentially expressed genes, several transcription factors shown previously to play an important role in SSC maintenance, including Bcl6b, Lhx1 and Etv5, as well as the key RNA-binding protein gene Nanos2, were upregulated in cells from Id4eGfp;GermOE mice (Fig. 6C). Also, the expression of genes encoding receptors to cytokines known to influence SSC self-renewal, including Gfra1, Csf1r and Cxcr4, were upregulated in Id4eGfp;GermOE cells (Fig. 6C). By contrast, the expression of multiple genes that influence progenitor spermatogonia development, including Lin28, Neurog3, Sall4 and Sohlh1, were downregulated in Id4eGfp;GermOE cells (Fig. 6C). Furthermore, the expression of several genes implicated to play a role in the regulation of SSC self-renewal, including Pou5f1, Foxo1, Zbtb16 and Rhox10, did not differ between Id4eGfp;GermOE and control cells, and expression of Pax7 was below the threshold of FPKM>1 to be considered as being expressed in ID4-EGFP+ cells from either control or Id4eGfp;GermOE mice (Fig. 6C). Collectively, these findings suggest that one or more subtle differences in developmental programming leads to spermatogonia that do or do not undergo self-renewal and these fates may be influenced by the level of ID4 expression.
Fig. 6.
Impact of constitutive Id4 expression on the transcriptome of SSCs and immediate progenitor spermatogonia. (A) Venn diagram depicting the number of genes expressed at significantly different levels (q≤0.05) in the ID4-EGFP+ spermatogonial population from testes of control and Id4eGfp;GermOE mice at P8. Data are derived from FACS-isolated cell populations; n=3. (B) Functional classification of genes differentially expressed (upregulated in green and downregulated in blue) in spermatogonia from Id4eGfp;GermOE mice by GO analysis. Data are the number of genes binned into the top five functional categories for upregulation or downregulation. (C) Comparison of expression for genes linked to SSC maintenance or progenitor development in the ID4-EGFP+ spermatogonial population from control and Id4eGfp;GermOE mice. Data are mean±s.e.m. for three different mice of each genotype; *P<0.05.
DISCUSSION
Stem cells are defined functionally as possessing the capacity to sustain a cycling cell lineage either during steady-state conditions or regeneration. For the male germline, the label SSC is reserved for cells with the capacity to regenerate the progenitor spermatogonial pool when it is depleted in situations such as successive rounds of RA-induced differentiation, cytotoxic insult or transplantation (Oatley and Brinster, 2008, 2012). The unequivocal identification of germ cells possessing regenerative capacity is important to clearly discern mechanisms driving the stem cell and progenitor states. To achieve this, knowledge of molecular markers that are expressed only in the SSC pool is invaluable. Although a multitude of identifiers for SSCs have been reported previously, the purity of the marked populations is undefined and most are expressed by all subsets of undifferentiated spermatogonia. These nuances have presented a barrier to progress in developing a deep understanding of the biology of SSCs and the molecular mechanisms underpinning their functions.
Outcomes of previous studies (Oatley et al., 2011; Chan et al., 2014; Sun et al., 2015) and those presented here demonstrate that the stem cell pool of the male germline is contained within the ID4+ spermatogonial population. Two major approaches for assigning stem cell function to specific populations are lineage tracing and transplantation. Although several cell populations, such as those marked by GFRA1 (Ebata et al., 2005; Grisanti et al., 2009; Hara et al., 2014), PAX7 (Aloisio et al., 2014), BMI1 (Komai et al., 2014) or NEUROG3 (Nakagawa et al., 2010) expression, have been identified as containing at least some SSCs using one of the approaches, the ID4 expressing population is the only one reported to date to be enriched for SSCs using both lineage tracing and transplantation (Chan et al., 2014; Sun et al., 2015). Moreover, in the current study we demonstrate through limiting dilution transplantation analysis that the ID4-EGFPBright population is mostly, if not purely, SSCs. In comparison, GFRA1 is often utilized as a marker of SSCs (Hara et al., 2014; Ikami et al., 2015), but the percentage of cells in the overall population that possess regenerative capacity, as defined by transplantation analysis (Ebata et al., 2005; Grisanti et al., 2009), is similar to that of the ID4-EGFPDim population that is reminiscent of cells transitioning to a progenitor state. In previous studies, we found that ID4-EGFP+ spermatogonia are also GFRA1+ (Chan et al., 2014). In the current study, our transcriptome analyses demonstrate that Gfra1 expression is significantly upregulated in ID4-EGFPBright SSCs; and a previous study found that levels of GFRA1 are positively associated with colonization activity in a transplantation assay (Takashima et al., 2015), although the potency is considerably less than that of the ID4-EGFPBright population. Thus, it is likely that spermatogonia expressing high levels of GFRA1 are the true SSCs and those with lower levels are immediate progenitors. This concept of a gradient of expression distinguishing SSC and progenitor populations is likely to be applicable to many molecules that have been considered SSC markers. In corroboration of this concept, a recent study utilized lineage tracing to show that SSCs are present in the BMI1 highly expressing population, whereas the population with lower expression is mostly progenitors (Komai et al., 2014).
Collective outcomes of the studies reported here with two different mouse models for examining the ID4-expressing spermatogonial population suggest that the levels of ID4 dictate the stem cell-to-progenitor transition in spermatogonia. Functional analyses from transplanting ID4-EGFPBright cells and the phenotype of Id4 overexpression mice support this notion. In addition, the expression of many genes reported previously to influence SSC maintenance was upregulated in both Id4 overexpression spermatogonia and ID4-EGFPBright spermatogonia, with the contrast holding true for the downregulation of several genes reported to regulate progenitor formation in Id4 overexpression spermatogonia and the ID4-EGFPDim spermatogonia. Taken together, these findings suggest that ID4 influences a core program for stem cell identity in mammalian spermatogonia and that downregulation in ID4 expression is required for transition to the progenitor state.
The spermatogenesis arrest phenotype in adult mice with constitutive Id4 overexpression is consistent with blocks in multiple aspects of the early differentiation continuum. At 2 months of age, many seminiferous tubules were found to contain only undifferentiated spermatogonia (evidenced by ZBTB16 expression) that were also ID4-EGFP+, and some tubules contained pachytene spermatocytes; however, underlying layers of differentiating germ cells (i.e. differentiating spermatogonia and earlier stage spermatocytes) were not observed. The few persisting spermatocytes could have been derivatives of the first round of spermatogenesis or rare differentiating spermatogonia that may have escaped the block caused by constitutive Id4 expression. By 6 months of age, tubules were found to contain rare ID4-EGFP+ undifferentiated spermatogonia or be completely devoid of germ cells, but meiotic cells were not observed in tubules with disrupted spermatogenesis. Taken together, these observations indicate a block in progenitor spermatogonial development when Id4 is constitutively expressed. Interestingly, round spermatids were not observed in cross-sections of mutant testes at 2 or 6 months of age. Although there are many possible explanations of this observation, it is conceivable that constitutive expression of Id4 disrupts meiotic progression, leading to an arrest in spermatocyte maturation.
Outcomes of RNA-seq analyses revealed that several genes previously associated with SSC maintenance were upregulated in spermatogonia with Id4 overexpression and in the ID4-EGFPBright population. Interestingly, the upregulated genes include Bcl6b, Etv5 and Lhx1, which we identified in previous studies as being induced by GDNF signaling in primary cultures of undifferentiated spermatogonia and playing an important role in maintenance of the SSC pool, a profile that is identical to that of Id4 (Oatley et al., 2006). These findings suggest that ID4 might be the key regulator of a core molecular circuit of GDNF-responsive genes that promote self-renewal of spermatogonia. It will be important in future studies to determine whether the influence of ID4 on the expression of these genes is direct at the level of promoter/enhancer regulation or indirect. Considering that ID4 lacks a DNA-binding domain, activity at promoters/enhancers would likely require binding with co-factors. Another possible mechanism of action is at the level of chromatin modification. Indeed, in a previous study we discovered an interaction between ID4 and retinoblastoma protein (RB) (Yang et al., 2013), and a multitude of evidence suggests a role for RB as a chromatin modifier (Osborne et al., 1997; Brehm et al., 1998; Sage, 2012).
For many years, attempts to examine gene expression in SSCs involved the analysis of isolated testis cell populations that were a heterogeneous mix of spermatogonial subtypes or a mix of somatic cells and spermatogonia (Hofmann et al., 2005; Oatley et al., 2006, 2007; Orwig et al., 2008; Hammoud et al., 2014). Although outcomes of those studies yielded a wealth of information, assigning a gene expression profile to SSCs specifically was challenging. The development of tools to isolate populations enriched or even purified for different spermatogonial subtypes, such as the Id4-eGfp transgenic mouse model, has refined the ability to explore SSCs specifically. Using these tools, a recent study profiled single ID4-EGFP+ spermatogonia from P6 mice to identify bimodal expression of a group of genes known to regulate spermatogonial functions, and identified clusters of cells expressing different levels of select cell surface markers including TSPAN8 (Hermann et al., 2015). In a follow-up study, the population of ID4-EGFP+ spermatogonia expressing a high level of TSPAN8 (TSPAN8-Hi) was found to have potent SSC activity by transplantation analyses, whereas the ID4-EGFP+ population expressing low levels of TSPAN8 (TSPAN8-Lo) possessed significantly less stem cell capacity (Mutoji et al., 2016). Transcriptome profiling of the ID4-EGFP+/TSPAN8-Hi and ID4-EGFP+/TSPAN8-Lo populations by RNA-seq uncovered distinct gene expression signatures (Mutoji et al., 2016). Included in those profiles was significant upregulation of previously defined core SSC maintenance genes (Id4, Bcl6b, Etv5, Lhx1, Nanos2 and T) and downregulation of known progenitor-promoting genes (Neurog3, Sohlh1 and Kit) in the TSPAN8-Hi population; a profile that is identical to the ID4-EGFPBright population measured in the current study. Taken together, these findings suggest that the ID4-EGFPBright and ID4-EGFP+/TSPAN8-Hi populations are similar and represent the SSC pool. Indeed, extrapolating the expression profile for Tspan8 from the transcriptome databases generated in the current study revealed significant upregulation in the ID4-EGFPBright population compared with the ID4-EGFPDim population (Table S2, Fig. S3).
Over the last few years, multiple studies have generated transcriptome profiles by RNA-seq for isolated subpopulations of spermatogonia. Comparison of the datasets is a solid approach for gaining insight into what might be the core molecular program that defines an SSC. As a first step, we compared the profiles of differentially expressed genes from three different studies: upregulated genes in ID4-EGFP+/TSPAN8-Hi versus ID4-EGFP+/TSPAN8-Lo spermatogonia (Mutoji et al., 2016); upregulated genes in THY1+ spermatogonia versus KIT+ (differentiating) spermatogonia (Hammoud et al., 2015); and upregulated genes in ID4-EGFPBright versus ID4-EGFPDim spermatogonia (Fig. S4). Outcomes revealed an overlap of 12 genes for all three profiles, which might include novel regulators of SSC functions (Table S4). In addition, comparing the upregulated gene expression profiles for ID4-EGFPBright cells and ID4-EGFP+/TSPAN8-Hi cells revealed an overlap of 123 genes, 111 of which are not represented in the profile for THY1+ cells (Fig. S4, Table S4). Although several expected genes are included in this list, such as Id4, Bcl6b and T, a multitude of potential novel regulators are also present. Considering that these populations are pure germ cells, whereas the THY1+ population contains some somatic cells, the core program of gene expression that influences the SSC state in spermatogonia is likely to be contained in this list of 123 genes. Although predicting a biological relevance for these genes in controlling SSC maintenance is tempting, most have not been explored in this capacity and future studies are needed to provide a functional assessment of their role.
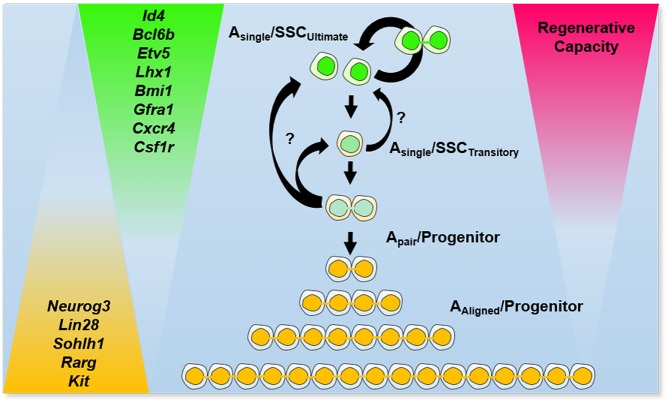
The traditional model for describing stem cell dynamics in the mammalian male germline proposes that all Asingle spermatogonia are stem cells and that transition to Apair represents initial commitment to differentiation (Huckins, 1971; Huckins and Oakberg, 1978). Collectively, outcomes of the current study support a variant of the Asingle model in which the interface of the stem cell-to-progenitor transition in spermatogonia involves alterations in the expression level of key genes concomitant with transition from an Asingle to Apair state (Fig. 7). We propose that a subset of the Asingle spermatogonia represent the ultimate stem cells (SSCultimate), which express a high level of core SSC genes (e.g. Id4, Bcl6b, Etv5, Lhx1, Cxcr4 and Gfra1), have suppressed expression of genes that drive the progenitor state (e.g. Neurog3, Lin28, Rarg and Kit), and possess potent regenerative capacity. In the course of self-renewing division, SSCultimate undergo a phase in which daughter cells are temporarily connected by an intercellular bridge and remain in close proximity after abscission, thereby existing as ‘false Apair’. The transition to a progenitor state is a continuum that initiates with downregulation of key SSC maintenance genes in accordance with increasing expression of progenitor genes, eventually yielding true Apair spermatogonia. At any given time, a portion of the Asingle population, possibly most of the cells, is in transition to progenitor status that includes initial downregulation of key SSC genes and reduction in the propensity for regenerative capacity and therefore can be considered transitory SSCs (SSCtransitory). As the cells progress in transition, a greater propensity to form true Apair upon the next division is gained. The true Apair cells possess a persistent intercellular bridge and divide to form Aaligned spermatogonia; both of these spermatogonial subtypes have low to no expression of core SSC genes, peak expression of progenitor genes and lack regenerative capacity, thereby being defined as progenitors. In response to damage, SSCtransitory may revert back to an SSCultimate state, and Apair in transition may revert to SSCtransitory or even SSCultimate to gain regenerative capacity. Conversely, true Apair and Aaligned always lack regenerative capacity and are fated for a pathway of terminal differentiation.
Fig. 7.
Model for the interface of the stem cell-to-progenitor transition in the mammalian male germline. The transition from a potent SSC to a transit amplifying progenitor state is a continuum that involves alterations in the expression of key genes concomitant with conversion from Asingle to Apair/Aaligned identity.
MATERIALS AND METHODS
Animals
All animal procedures were approved by the Washington State University Institutional Animal Care and Use Committee (IACUC). Id4-eGfp transgenic mice were as described previously (Chan et al., 2014). F1 hybrids to serve as donors for transplantation analyses were generated by crossing Id4-eGfp and Rosa26-lacZ mice (Jackson Laboratories, stock no. 002073). Recipient males for transplantation analyses were F1 hybrids of C57BL/6J (Jackson Laboratories, stock no. 000664) and 129S1/SvlmJ (Jackson Laboratories, stock no. 002448) treated with busulfan (Sigma, B2635) to eliminate the endogenous germline as described previously (Oatley and Brinster, 2006). Generation of the Id4 conditional overexpression mouse line is described in the supplementary Materials and Methods.
Whole-mount imaging
Testes from Id4-eGfp pups at P8 were excised and gently teased apart on a glass slide using fine forceps. Mounting media (VectaMount AQ, Vector Laboratories) and a coverslip were applied and slides visualized on a Leica TCS SP5 II confocal microscope. Whole-mount image optical slices were captured using LAS AF software (Leica) to classify ID4-EGFP+ cells as individuals (Asingle) or an interconnected cohort of two cells (Apair). Determination of cohort identity was based on criteria defined by de Rooij and Russell (2000). Specifically, we considered Apair as two EGFP+ cells being within half a cell length of one another and possessing a visible connection. Relative EGFP intensity of each Asingle and each cell in an Apair cohort was determined using Fiji software as described previously (Burgess et al., 2010; McCloy et al., 2014).
Testis histology and immunofluorescent staining
Testes were fixed and 5-7 μm cross-sections subject to Hematoxylin and Eosin staining or immunostaining for ZBTB16, STRA8, TRA98 or EGFP (primary antibodies and dilutions are listed in Table S5) and visualized by fluorescent microscopy as detailed in the supplementary Materials and Methods.
Fluorescence-activated cell sorting (FACS) and flow cytometric analysis (FCA)
Single-cell suspensions were generated from testes as described previously (Oatley and Brinster, 2006; Chan et al., 2014). FACS and FCA were performed using an SH800 machine (Sony Biotechnology) to isolate ID4-EGFPBright and ID4-EGFPDim populations. Selection of cells with different levels of ID4 was achieved by gating the fluorescence intensity scale for EGFP+ cells by thirds. Briefly, the EGFP+ gating area was based on the point of the fluorescence intensity axis where cells were considered as being EGFP+, set based on the background fluorescence intensity of a non-transgenic control testis cell population, and up to the point where cells with the greatest fluorescence intensity fell. This area was then subdivided into thirds to define the ID4-EGFP+ subsets as being EGFPDim (lower third) and EGFPBright (upper third).
Transplantation analyses
To compare the regenerative capacity of ID4-EGFPBright and ID4-EGFPDim spermatogonial populations, cells were suspended in mouse serum-free medium (Kubota et al., 2004) at 1×106 cells/ml and 10 µl (10,000 cells) was microinjected into each recipient testis as described previously (Oatley and Brinster, 2006; Chan et al., 2014). Recipient testes were evaluated for colonies of donor-derived spermatogenesis 2 months later as described previously (Oatley and Brinster, 2006; Chan et al., 2014). For limiting dilution transplantation analyses, single-cell suspensions of testes from adult W/Wv mice that lack endogenous germ cells were used as somatic support carriers via mixing with ID4-EGFPBright spermatogonia. For each replicate experiment, a single-cell suspension of FACS-isolated ID4-EGFPBright cells at 1×106 cell/ml was subjected to a series of 1:10 dilutions in mouse serum-free medium to yield suspensions of 1×105 cells/ml (1000 cells/10 µl), 1×104 cells/ml (100 cells/10 µl) and 1×103 cells/ml (10 cells/10 µl). Each diluted cell suspension was then mixed with W/Wv testis somatic cells to form a final concentration of 1×106 total cells/ml (10,000 total cells/10 µl) and 10 µl microinjected into each recipient testis, which translated to 1000, 100 and 10 ID4-EGFPBright cells being transplanted.
qRT-PCR
Total RNA from ID4-EGFPBright and ID4-EGFPDim spermatogonia isolated by FACS from P7 pups was subject to qRT-PCR as described in the supplementary Materials and Methods.
RNA-seq
Total RNA from ID4-EGFPBright and ID4-EGFPDim spermatogonia isolated by FACS from P8 mice was used to generate cDNA libraries for Illumina sequencing and mapping to the mouse genome (mm9 build). GO analysis was conducted using DAVID. For details, see the supplementary Materials and Methods.
Data presentation and statistical analysis
All quantitative data are presented as the mean±s.e.m. for at least three biological replicate experiments. Differences between means were determined statistically using the two-tailed t-test or linear regression analysis function of GraphPad Prism software and significance was set at P<0.05.
Acknowledgements
We thank members of the J.M.O. laboratory, including D. Miao, M. S. Waqas, N. Law and B. Hubbard, for valuable input and discussion, Y. Yamauchi for assistance with re-derivation of the Id4cOE mouse line, and E. Cinato for the Id4cOE construct.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
A.R.H.: experimental design, collection of data, analysis of data and manuscript writing. Q.-E.Y.: experimental design, collection of data, analysis of data and manuscript writing. M.J.O.: experimental design, collection and analysis of data. T.L.: collection and analysis of data. F.S.: generation of resources, financial support and manuscript writing. J.M.O.: experimental design, collection of data, analysis of data, financial support, manuscript writing and final approval of manuscript.
Funding
This research was supported by grant HD061665 awarded to J.M.O. from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Deposited in PMC for release after 12 months.
Data availability
RNA-seq data are available at Gene Expression Omnibus under accession number GSE93772.
Supplementary information
Supplementary information available online at http://dev.biologists.org/lookup/doi/10.1242/dev.146928.supplemental
References
- Aloisio G. M., Nakada Y., Saatcioglu H. D., Peña C. G., Baker M. D., Tarnawa E. D., Mukherjee J., Manjunath H., Bugde A., Sengupta A. L. et al. (2014). PAX7 expression defines germline stem cells in the adult testis. J. Clin. Invest. 124, 3929-3944. 10.1172/JCI75943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A., Miska E. A., McCance D. J., Reid J. L., Bannister A. J. and Kouzarides T. (1998). Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391, 597-601. 10.1038/35404 [DOI] [PubMed] [Google Scholar]
- Brinster R. L. and Zimmermann J. W. (1994). Spermatogenesis following male germ-cell transplantation. Proc. Natl. Acad. Sci. USA 91, 11298-11302. 10.1073/pnas.91.24.11298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A., Vigneron S., Brioudes E., Labbé J.-C., Lorca T. and Castro A. (2010). Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc. Natl. Acad. Sci. USA 107, 12564-12569. 10.1073/pnas.0914191107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F., Oatley M. J., Kaucher A. V., Yang Q.-E., Bieberich C. J., Shashikant C. S. and Oatley J. M. (2014). Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 28, 1351-1362. 10.1101/gad.240465.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rooij D. G. (1973). Spermatogonial stem cell renewal in the mouse. I. Normal situation. Cell Tissue Kinet. 6, 281-287. 10.1111/j.1365-2184.1973.tb01617.x [DOI] [PubMed] [Google Scholar]
- de Rooij D. G. and Russell L. D. (2000). All you wanted to know about spermatogonia but were afraid to ask. J. Androl. 21, 776-798. 10.1002/j.1939-4640.2000.tb03408.x [DOI] [PubMed] [Google Scholar]
- Drumond A. L., Meistrich M. L. and Chiarini-Garcia H. (2011). Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction 142, 145-155. 10.1530/REP-10-0431 [DOI] [PubMed] [Google Scholar]
- Ebata K. T., Zhang X. and Nagano M. C. (2005). Expression patterns of cell-surface molecules on male germ line stem cells during postnatal mouse development. Mol. Reprod. Dev. 72, 171-181. 10.1002/mrd.20324 [DOI] [PubMed] [Google Scholar]
- Gallardo T., Shirley L., John G. B. and Castrillon D. H. (2007). Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis 45, 413-417. 10.1002/dvg.20310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grisanti L., Falciatori I., Grasso M., Dovere L., Fera S., Muciaccia B., Fuso A., Berno V., Boitani C., Stefanini M.. et al. (2009). Identification of spermatogonial stem cell subsets by morphological analysis and prospective isolation. Stem Cells 27, 3043-3052. 10.1002/stem.206 [DOI] [PubMed] [Google Scholar]
- Hammoud S. S., Low D. H. P., Yi C., Carrell D. T., Guccione E. and Cairns B. R. (2014). Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell 15, 239-253. 10.1016/j.stem.2014.04.006 [DOI] [PubMed] [Google Scholar]
- Hammoud S. S., Low D. H. P., Yi C., Lee C. L., Oatley J. M., Payne C. J., Carrell D. T., Guccione E. and Cairns B. R. (2015). Transcription and imprinting dynamics in developing postnatal male germline stem cells. Genes Dev. 29, 2312-2324. 10.1101/gad.261925.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara K., Nakagawa T., Enomoto H., Suzuki M., Yamamoto M., Simons B. D. and Yoshida S. (2014). Mouse spermatogenic stem cells continually interconvert between equipotent singly isolated and syncytial states. Cell Stem Cell 14, 658-672. 10.1016/j.stem.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helsel A. R. and Oatley J. M. (2017). Transplantation as a quantitative assay to study mammalian male germline stem cells. Methods Mol. Biol. 1463, 155-172. 10.1007/978-1-4939-4017-2_12 [DOI] [PubMed] [Google Scholar]
- Hermann B. P., Mutoji K. N., Velte E. K., Ko D., Oatley J. M., Geyer C. B. and McCarrey J. R. (2015). Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol. Reprod. 92, 54 10.1095/biolreprod.114.125757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M.-C., Braydich-Stolle L. and Dym M. (2005). Isolation of male germ-line stem cells; influence of GDNF. Dev. Biol. 279, 114-124. 10.1016/j.ydbio.2004.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogarth C. A., Arnold S., Kent T., Mitchell D., Isoherranen N. and Griswold M. D. (2015). Processive pulses of retinoic acid propel asynchronous and continuous murine sperm production. Biol. Reprod. 92, 37 10.1095/biolreprod.114.126326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huckins C. (1971). The spermatogonial stem cell population in adult rats. I. Their morphology, proliferation and maturation. Anat. Rec. 169, 533-557. 10.1002/ar.1091690306 [DOI] [PubMed] [Google Scholar]
- Huckins C. and Oakberg E. F. (1978). Morphological and quantitative analysis of spermatogonia in mouse testes using whole mounted seminiferous tubules, I. The normal testes. Anat. Rec. 192, 519-528. 10.1002/ar.1091920406 [DOI] [PubMed] [Google Scholar]
- Ikami K., Tokue M., Sugimoto R., Noda C., Kobayashi S., Hara K. and Yoshida S. (2015). Hierarchical differentiation competence in response to retinoic acid ensures stem cell maintenance during mouse spermatogenesis. Development 142, 1582-1592. 10.1242/dev.118695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M., Inoue K., Miki H., Ogonuki N., Takehashi M., Morimoto T., Ogura A. and Shinohara T. (2006). Clonal origin of germ cell colonies after spermatogonial transplantation in mice. Biol. Reprod. 75, 68-74. 10.1095/biolreprod.106.051193 [DOI] [PubMed] [Google Scholar]
- Komai Y., Tanaka T., Tokuyama Y., Yanai H., Ohe S., Omachi T., Atsumi N., Yoshida N., Kumano K., Hisha H. et al. (2014). Bmi1 expression in long-term germ stem cells. Sci. Rep. 4, 6175 10.1038/srep06175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota H., Avarbock M. R. and Brinster R. L. (2004). Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA 101, 16489-16494. 10.1073/pnas.0407063101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCloy R. A., Rogers S., Caldon C. E., Lorca T., Castro A. and Burgess A. (2014). Partial inhibition of Cdk1 in G2 phase overrides the SAC and decouples mitotic events. Cell Cycle 13, 1400-1412. 10.4161/cc.28401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoji K., Singh A., Nguyen T., Gildersleeve H., Kaucher A. V., Oatley M. J., Oatley J. M., Velte E. K., Geyer C. B., Cheng K. et al. (2016). TSPAN8 expression distinguishes spermatogonial stem cells in the prepubertal mouse testis. Biol. Reprod. 95, 117 10.1095/biolreprod.116.144220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano M., Avarbock M. R. and Brinster R. L. (1999). Pattern and kinetics of mouse donor spermatogonial stem cell colonization in recipient testes. Biol. Reprod. 60, 1429-1436. 10.1095/biolreprod60.6.1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Sharma M., Nabeshima Y., Braun R. E. and Yoshida S. (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62-67. 10.1126/science.1182868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakberg E. F. (1971). A new concept of spermatogonial stem-cell renewal in the mouse and its relationship to genetic effects. Mutat. Res. 11, 1-7. 10.1016/0027-5107(71)90027-3 [DOI] [PubMed] [Google Scholar]
- Oatley J. M. and Brinster R. L. (2006). Spermatogonial stem cells. Methods Enzymol. 419, 259-282. 10.1016/S0076-6879(06)19011-4 [DOI] [PubMed] [Google Scholar]
- Oatley J. M. and Brinster R. L. (2008). Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 24, 263-286. 10.1146/annurev.cellbio.24.110707.175355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M. and Brinster R. L. (2012). The germline stem cell niche unit in mammalian testes. Physiol. Rev. 92, 577-595. 10.1152/physrev.00025.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Avarbock M. R., Telaranta A. I., Fearon D. T. and Brinster R. L. (2006). Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. USA 103, 9524-9529. 10.1073/pnas.0603332103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley J. M., Avarbock M. R. and Brinster R. L. (2007). Glial cell line-derived neurotrophic factor regulation of genes essential for self-renewal of mouse spermatogonial stem cells is dependent on Src family kinase signaling. J. Biol. Chem. 282, 25842-25851. 10.1074/jbc.M703474200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley M. J., Kaucher A. V., Racicot K. E. and Oatley J. M. (2011). Inhibitor of DNA binding 4 is expressed selectively by single spermatogonia in the male germline and regulates the self-renewal of spermatogonial stem cells in mice. Biol. Reprod. 85, 347-356. 10.1095/biolreprod.111.091330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T., Ohmura M., Yumura Y., Sawada H. and Kubota Y. (2003). Expansion of murine spermatogonial stem cells through serial transplantation. Biol. Reprod. 68, 316-322. 10.1095/biolreprod.102.004549 [DOI] [PubMed] [Google Scholar]
- Orwig K. E., Ryu B.-Y., Master S. R., Phillips B. T., Mack M., Avarbock M. R., Chodosh L. and Brinster R. L. (2008). Genes involved in post-transcriptional regulation are overrepresented in stem/progenitor spermatogonia of cryptorchid mouse testes. Stem Cells 26, 927-938. 10.1634/stemcells.2007-0893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne A., Tschickardt M. and Blanck G. (1997). Retinoblastoma protein expression facilitates chromatin remodeling at the HLA-DRA promoter. Nucleic Acids Res. 25, 5095-5102. 10.1093/nar/25.24.5095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann V., van Crüchten I. and Sablitzky F. (1994). The expression pattern of Id4, a novel dominant negative helix-loop-helix protein, is distinct from Id1, Id2 and Id3. Nucleic Acids Res. 22, 749-755. 10.1093/nar/22.5.749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage J. (2012). The retinoblastoma tumor suppressor and stem cell biology. Genes Dev. 26, 1409-1420. 10.1101/gad.193730.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F., Xu Q., Zhao D. and Degui Chen C. (2015). Id4 marks spermatogonial stem cells in the mouse testis. Sci. Rep. 5, 17594 10.1038/srep17594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S., Kanatsu-Shinohara M., Tanaka T., Morimoto H., Inoue K., Ogonuki N., Jijiwa M., Takahashi M., Ogura A. and Shinohara T. (2015). Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Reports 4, 489-502. 10.1016/j.stemcr.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegelenbosch R. A. J. and de Rooij D. G. (1993). A quantitative study of spermatogonial multiplication and stem cell renewal in the C3H/101 F1 hybrid mouse. Mutat. Res. 290, 193-200. 10.1016/0027-5107(93)90159-D [DOI] [PubMed] [Google Scholar]
- Yang Q.-E., Gwost I., Oatley M. J. and Oatley J. M. (2013). Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol. Reprod. 89, 113 10.1095/biolreprod.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]