ABSTRACT
The genomic basis of the response to dietary intervention of human gut beneficial bacteria remains elusive, which hinders precise manipulation of the microbiota for human health. After receiving a dietary intervention enriched with nondigestible carbohydrates for 105 days, a genetically obese child with Prader-Willi syndrome lost 18.4% of his body weight and showed significant improvement in his bioclinical parameters. We obtained five isolates (C1, C15, C55, C62, and C95) of one of the most abundantly promoted beneficial species, Bifidobacterium pseudocatenulatum, from a postintervention fecal sample. Intriguingly, these five B. pseudocatenulatum strains showed differential responses during the dietary intervention. Two strains were largely unaffected, while the other three were promoted to different extents by the changes in dietary carbohydrate resources. The differential responses of these strains were consistent with their functional clustering based on the COGs (Clusters of Orthologous Groups), including those involved with the ABC-type sugar transport systems, suggesting that the strain-specific genomic variations may have contributed to the niche adaption. Particularly, B. pseudocatenulatum C15, which had the most diverse types and highest gene copy numbers of carbohydrate-active enzymes targeting plant polysaccharides, had the highest abundance after the dietary intervention. These studies show the importance of understanding genomic diversity of specific members of the gut microbiota if precise nutrition approaches are to be realized.
IMPORTANCE
The manipulation of the gut microbiota via dietary approaches is a promising option for improving human health. Our findings showed differential responses of multiple B. pseudocatenulatum strains isolated from the same habitat to the dietary intervention, as well as strain-specific correlations with bioclinical parameters of the host. The comparative genomics revealed a genome-level microdiversity of related functional genes, which may have contributed to these differences. These results highlight the necessity of understanding strain-level differences if precise manipulation of gut microbiota through dietary approaches is to be realized.
INTRODUCTION
Recent evidence indicates that the dysbiosis of the gut microbiome plays a pivotal role in human diseases such as obesity and diabetes (1, 2). Specific members of the gut microbiota with a causative contribution to disease/health phenotypes can serve not only as a powerful tool for disease diagnosis (3, 4) but also as targets for disease alleviation/treatment through various methods such as drugs (5), fecal microbiota transplantation (6), and diet (7). However, due to the complexity and interindividual variation of the gut microbiota itself and its interactions with the host and diet (8), the precision manipulation of the gut microbiota for achieving optimum human health needs a more rigorous understanding at the genomic and molecular levels.
In one of our previous dietary intervention studies, we found that a diet enriched in nondigestible but fermentable carbohydrates, which was composed of whole grains, traditional Chinese medicinal foods, and prebiotics (the WTP diet), not only significantly changed the gut microbiota but also improved the bioclinical parameters and inflammatory conditions in genetically obese children with Prader-Willi syndrome (9). One particular beneficial bacterium, Bifidobacterium pseudocatenulatum was significantly enriched after the dietary intervention, negatively correlated with other potential detrimental species, and positively related to improvement of the clinical parameters of the host (9).
One child from that cohort finished the dietary intervention over 105 days. His bioclinical parameters were improved, with an initial body weight loss of more than 25.8 kg, together with a significant shift of the gut microbial community such as increases of Faecalibacterium, Lactobacillus, and Bifidobacterium spp. Moreover, we found that B. pseudocatenulatum was the most dominant Bifidobacterium species after the dietary intervention. Intriguingly, we had isolated five strains from his fecal sample on day 105 (10), and these strains had differential responses to the carbohydrate-enriched interventions. To understand the genetic traits involved in the bacterial niche adaption in the gut ecosystem and the interactions with the host and diet (11), we performed comparative genomics on B. pseudocatenulatum.
RESULTS AND DISCUSSION
Improved bioclinical parameters and shifted gut microbiota.
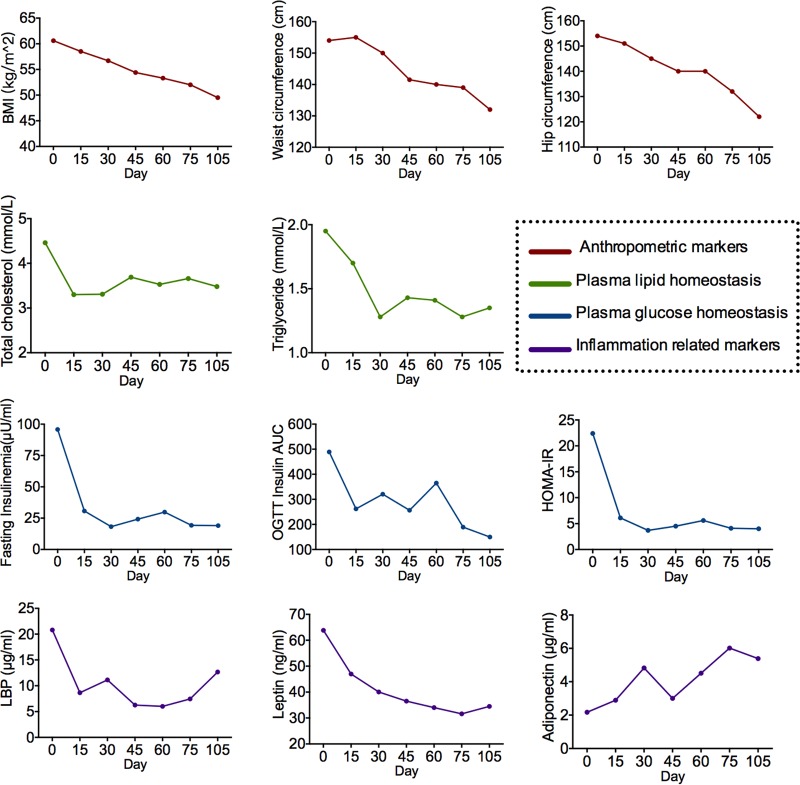
The bioclinical variables of the obese child improved during the dietary intervention (Fig. 1; see Fig. S1 in the supplemental material). The child’s body weight was reduced from 140.1 kg to 114.3 kg, and both the plasma glucose and lipid homeostasis were improved to the normal range. Two systemic inflammation markers, C-reactive protein (CRP) and serum amyloid A protein (SAA), were decreased after the dietary intervention. Adiponectin increased from 2.17 μg/ml to 5.39 μg/ml, and leptin decreased from 63.82 ng/ml to 34.47 ng/ml, indicating an alleviation of the “at-risk” phenotype (12). In addition, lipopolysaccharide-binding protein (LBP), a surrogate marker for bacterial antigen load in blood (13), decreased.
FIG 1 .
Improved bioclinical parameters and inflammatory conditions after the dietary intervention. Anthropometric markers, plasma lipid homeostasis, plasma glucose homeostasis, and inflammation-related markers are shown. Abbreviations: BMI, body mass index; OGTT, oral glucose tolerance test; LBP, lipopolysaccharide-binding protein; HOMA-IR, homeostatic model assessment of insulin resistance.
Bioclinical parameters that changed during the dietary intervention. Download FIG S1, PDF file, 0.2 MB (248.5KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
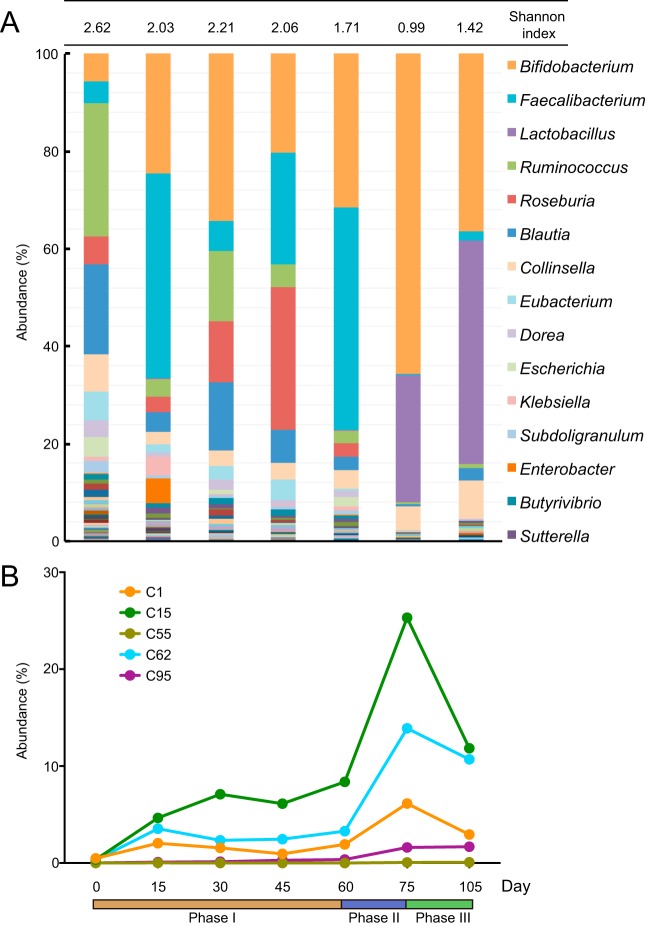
We obtained 25.2 × 106 ± 4.8 × 106 (mean ± standard deviation [SD]) high-quality paired-end reads per fecal sample at seven time points (0, 15, 30, 45, 60, 75, and 105 days) via metagenomic sequencing. The composition of the gut microbiota shifted during the dietary intervention (Fig. 2A) and showed different patterns responding to the three interventional phases (phase I, basic intervention from day 0 to day 60; phase II, basic intervention with 100 g more Formula No. 3 from day 60 to day 75; phase III, reduced Formula No. 1 with 100 g more Formula No. 3 from day 75 to day 105 [see Materials and Methods for details]). The defecation frequencies of the obese child in the different phases were similar (on average, three or four times per day), and no diarrhea occurred. The diversity of the gut bacterial community decreased during the dietary intervention, which is consistent with the changes in the cohort (9). At the baseline, Ruminococcus and Blautia were the two most abundant genera, accounting for 26.95% and 18.41%, respectively. Meanwhile, Bacteroides and Prevotella were present at low levels, suggesting that the community may belong to enterotype 3 (14). After the dietary intervention, Ruminococcus and Blautia were reduced to low abundance, while Bacteroides and Prevotella were almost unaffected by the intervention. Compared with the baseline, Faecalibacterium, which was reported to be an anti-inflammatory species (15, 16) and a beneficial (5) commensal bacterium, increased in phase I, with its abundance reaching 41.95% at day 15. The sharp increase in Faecalibacterium might contribute to the alleviation of inflammation in the first 15 days, as CRP and SAA decreased by 33.37% and 50.85%, respectively, during this period. Faecalibacterium decreased to a low abundance in phases II and III, when more oligosaccharides were provided. On the other hand, Lactobacillus, which is well equipped to metabolize oligosaccharides (17), had a low abundance at the baseline and during phase I but became one of the most dominant genera starting from phase II. Several studies have reported the beneficial effects of Lactobacillus strains on insulin resistance (18, 19). The dramatic increase of Lactobacillus may have contributed to the improvement of insulin sensitivity from phase II as indicated by the remarkable decrease of the oral glucose tolerance test (OGTT) insulin area under the curve (AUC) and steady the OGTT glucose AUC during this period. The abundance of Bifidobacterium microorganisms, which have the ability to metabolize a variety of carbohydrates via intra- and extracellular ways (20), was remarkable from day 15 and remained a predominant genus through the entire dietary intervention period: increasing from 27.47% when the child received the basic intervention to 65.53% by day 75, then decreasing to 36.41% when less Formula No. 1 containing complex dietary fibers was provided. These results show that the Bifidobacterium population responds to the shifts in carbohydrate resources provided in the three phases, and in each phase, it occupied a superior ecological niche. Given the known beneficial effects of Bifidobacterium strains on obesity (21, 22), their essential role in the interaction network of our previous cohort study (9) and their sustained high abundance during the dietary intervention, we speculate that they might have contributed to the continual decreases in body weight, body mass index (BMI), waist circumference, and hip circumference. Hence, we performed a deeper analysis on this genus.
FIG 2 .
Shifts of the gut microbiota during the dietary intervention. (A) Genus-level gut microbiota compositions at the seven time points. The Shannon index values are shown in panel A. According to the average abundance, the top 15 genera are labeled with their taxonomic names. (B) The differential abundances of the 5 B. pseudocatenulatum strains. Phase I, day 0 to day 60, basic dietary intervention; phase II, day 60 to day 75, basic intervention plus 100 g more Formula No. 3; phase III, less Formula No. 1 plus 100 g more Formula No. 3.
In total, nine Bifidobacterium species were identified in the postintervention samples (Fig. S2). Of the nine Bifidobacterium species, B. pseudocatenulatum was the most dominant, with an abundance of 29.36% in the whole-gut microbiota on day 105. Bifidobacterium longum, B. breve, and B. adolescentis represented 9.94%, 7.61%, and 3.75%, respectively, of the microbiota, and the other five species had abundances of less than 1%.
Abundance of the nine identified Bifidobacterium species in the gut microbial community on day 105. Download FIG S2, PDF file, 0.3 MB (316.9KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Differential responses of the B. pseudocatenulatum strains to the dietary intervention.
For a detailed study of the B. pseudocatenulatum population, we isolated and completely sequenced five B. pseudocatenulatum strains (strains C1, C15, C55, C62, and C95 [23]) from the fecal sample collected from our subject on day 105 (10). The abundance changes of these five strains at each time point were identified using the Sigma algorithm (24) by aligning the metagenomic data with the complete genomes (Fig. 2B). Before the dietary intervention, all of the strains had low abundances (maximum of 0.5% for B. pseudocatenulatum C1). The levels of B. pseudocatenulatum C55 and B. pseudocatenulatum C95 seemed unresponsive to the dietary interventions, as the former remained low throughout the trial and the latter showed a small increase only during phases II and III. In contrast, the B. pseudocatenulatum C1, B. pseudocatenulatum C62, and B. pseudocatenulatum C15 strains were responsive to the dietary interventions. Their changes in abundance were consistent with those observed for the Bifidobacterium genus, suggesting that these B. pseudocatenulatum strains primarily account for these changes. Indeed, after the dietary intervention, these three strains had increased significantly, especially B. pseudocatenulatum C15.
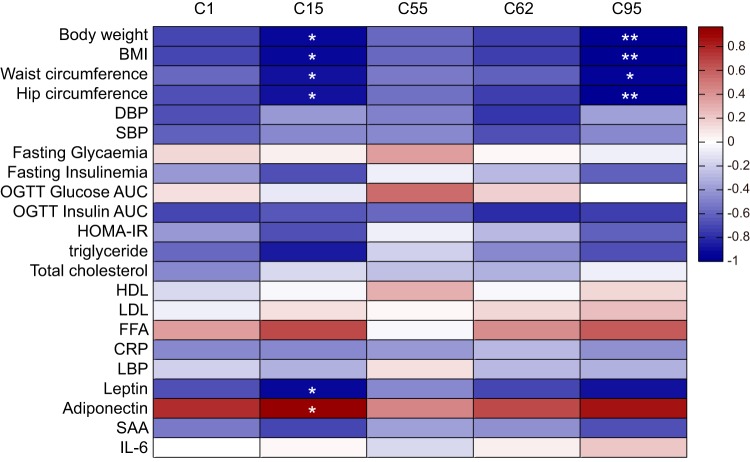
Consistent with the findings of the previous study (9), the improvements in the bioclinical variables were correlated with the increase in the B. pseudocatenulatum strains (Fig. 3). B. pseudocatenulatum C95 was negatively correlated with anthropometric markers, including body weight, BMI, waist circumference and hip circumference. Furthermore, B. pseudocatenulatum C15 was also correlated with the improvement of inflammatory markers, including the decrease in leptin and the increase in adiponectin.
FIG 3 .
Correlations between the changes in the clinical parameters with the abundance of the five B. pseudocatenulatum strains. Spearman correlation was performed. Correlations with an adjusted P value of <0.05 (*) or with an adjusted P value of <0.01 (**) are indicated by white asterisk(s) (60). Abbreviations: DBP, diastolic blood pressure; SBP, systolic blood pressure; HDL, high-density lipoprotein; LDL, low-density lipoprotein; FFA, free fatty acids; IL-6, interleukin 6.
Pan-genome analysis of B. pseudocatenulatum.
Based on our 5 complete B. pseudocatenulatum genomes and the available public data, including 5 draft B. pseudocatenulatum genomes and the complete genome of B. pseudocatenulatum JCM1200T (25), a total of 11 B. pseudocatenulatum genomes were included in the pan-genome analysis. The pan-genome curve showed an asymptotic trend with an average growing rate of 100 genes per genome in the first six iterations and then decreased to a much smaller rate (Fig. S3A). The curve finally arrived at 2,482 genes. This shows that the further incorporation of additional genomes would lead to only a minor increase in the pan-genome size. The core genome curve showed a more evidently asymptotic trend and a clear decrease in the first six iterations (Fig. S3B). The curve finally arrived at 1,427 genes. The pan-genome and core genome trends suggest that B. pseudocatenulatum displayed a closed pan-genome, and six genomes are almost sufficient to describe the gene properties of B. pseudocatenulatum. On the basis of these results and to avoid the illegibility caused by draft genomes, we used only the six complete genomes in our subsequent analysis to explore the genomic features of B. pseudocatenulatum.
Pan-genome and core genome curves of B. pseudocatenulatum. Download FIG S3, PDF file, 0.2 MB (218.8KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
General features of B. pseudocatenulatum.
The genomes of our five strains and the B. pseudocatenulatum JCM1200T displayed an average of 2,355,185 bp and 56.63 G+C%, which is consistent with the G+C% range of the Bifidobacterium genus (26). Five or six rRNA operon loci were identified in the genomes of B. pseudocatenulatum, and in each genome, there was one additional copy of the 5S rRNA gene (Table 1). In addition, the heterogeneity across the 16S rRNA genes existed in all the genomes except for that of B. pseudocatenulatum JCM1200T (Table S1). On average, 54 tRNA genes were found in each B. pseudocatenulatum genome.
TABLE 1 .
General features of the six complete genomes of B. pseudocatenulatum
| Feature | Value of feature for strain: |
|||||
|---|---|---|---|---|---|---|
| C15 | C1 | C55 | C62 | C95 | JCM1200T | |
| Genome length (bp) | 2,341,029 | 2,380,612 | 2,352,149 | 2,393,824 | 2,349,745 | 2,313,752 |
| No. of genes | 1,854 | 1,883 | 1,913 | 1,895 | 1,868 | 1,817 |
| No. of tRNA genes | 55 | 54 | 54 | 54 | 54 | 55 |
| No. of 16S rRNA genes | 6 | 5 | 6 | 5 | 5 | 6 |
| No. of 23S rRNA genes | 6 | 5 | 6 | 5 | 5 | 6 |
| No. of 5S rRNA genes | 7 | 6 | 7 | 6 | 6 | 7 |
| No. of hypothetical proteins | 372 | 381 | 366 | 380 | 363 | 372 |
| Hypothetical proteins (%) | 20 | 20 | 19 | 20 | 19 | 20 |
| Gene with assigned function (%) | 80 | 80 | 81 | 80 | 81 | 80 |
| G+C content (%) | 56.59 | 56.76 | 56.65 | 56.78 | 56.63 | 56.38 |
| No. of ORFs identified as CAZy | 106 | 104 | 104 | 106 | 106 | 111 |
Heterogeneity of the 16S rRNA genes. Download TABLE S1, DOCX file, 0.1 MB (103KB, docx) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
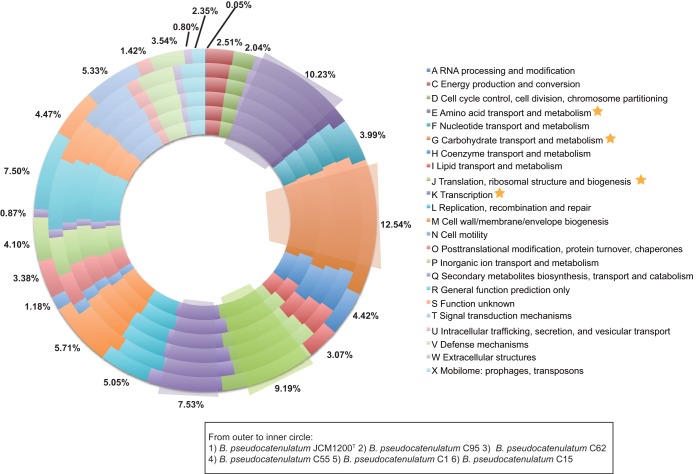
An average of 1,871 open reading frames (ORFs) per genome was predicted, with 80% of the detected ORFs with functional assignments via in silico prediction based on BLAST against the NCBI nr database, and the remaining 20% were predicted to be hypothetical proteins (Table 1). The identification of orthologous genes according to the COG (Clusters of Orthologous Groups) database (27) showed that the majority of the genes in the genome of B. pseudocatenulatum were involved in various housekeeping functions especially those for carbohydrate transport and metabolism (12.54%) as well as amino acid transport and metabolism (10.23%) (Fig. 4). These percentages were in agreement with those of other Bifidobacterium genomes (28, 29).
FIG 4 .
Cluster of Orthologous Groups (COG) classification of the orthologous genes of B. pseudocatenulatum. For each COG, the average percentage among the six complete B. pseudocatenulatum genomes has been indicated.
Based on the genomic analysis and supported by experimental evidence, several host colonization factors of bifidobacteria have already been identified, including functions involved in bile resistance and adhesins (11). Resistance to bile is important for the colonization of many intestinal bacteria, as bile acids can have antimicrobial activity at physiological concentrations (30). Bile salt hydrolase and/or bile acid transporter, which confer bile resistance, were identified in all six B. pseudocatenulatum strains. In the aspect of adhesion, all of the genomes harbored genes encoding enolase, and DnaK, which has been shown to be a plasminogen-binding related protein in Bifidobacterium animalis subsp. lactis BI07 (31, 32). Moreover, the genes encoding transaldolase, which is involved in the mucin binding found in four B. bifidum strains (33) and von Willebrand factor A, which has been reported to promote adhesion to extracellular matrices (34), existed in each genome as well. The existence of these functional genes suggests that, similar to other Bifidobacterium species, B. pseudocatenulatum has the genomic basis to colonize in the human intestine.
Genomic microdiversity of B. pseudocatenulatum.
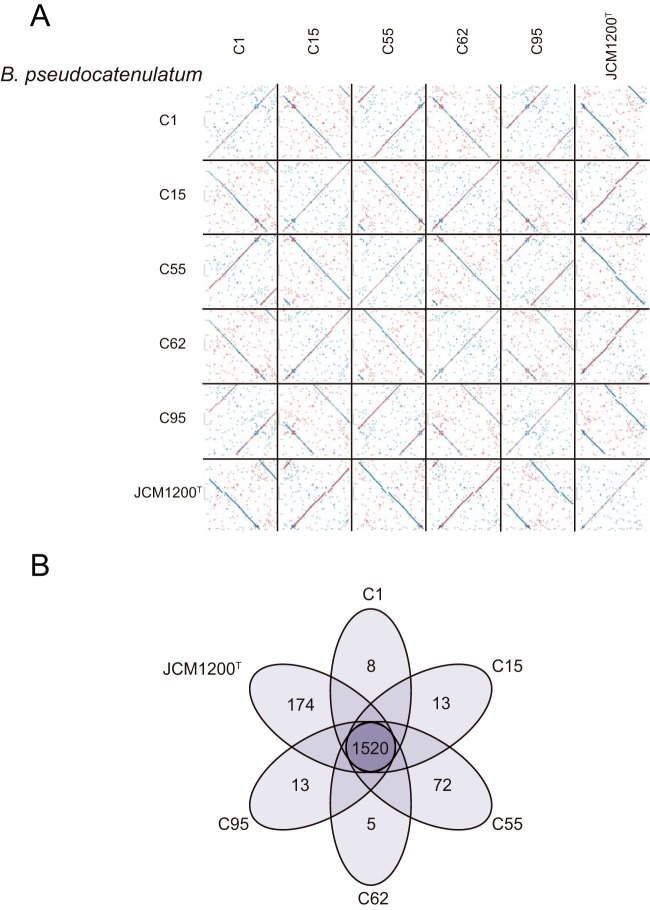
The average nucleotide identity (ANI) among the six complete B. pseudocatenulatum genomes met the threshold for species demarcation (35), as the minimum value was 97.76%. The similarity within the strains isolated from the same habitat (B. pseudocatenulatum C1, C15, C55, C62, and C95, minimum ANI of 99.88%) was slightly higher that the similarity of the five strains to B. pseudocatenulatum JCM1200T (maximum ANI of 97.80%), which was isolated from a different habitat. These results indicated the high degree of synteny across all these genomes, which is confirmed by the dot plot alignments for the genomes (Fig. 5A), although less colinearity and some differences (including indels) were apparent in the dot plots between B. pseudocatenulatum JCM1200T and the other five strains. A notable example of this variation was the eps gene cluster, which encodes exopolysaccharides (EPSs). EPS can form a slime layer that is attached to the cell and also can be released into the environment (36). Some EPSs produced by Bifidobacterium were considered to potentially contribute several beneficial activities to their hosts, including the modulation of the immune system, antagonism against pathogens, functions as scavenging agents, and modulation of the microbial community (37). One copy of the eps gene cluster was identified in each complete genome of B. pseudocatenulatum (Fig. S4), and these eps clusters were identical in our five B. pseudocatenulatum strains but quite different from that found in B. pseudocatenulatum JCM1200T.
FIG 5 .
Genomic variations among the B. pseudocatenulatum strains. (A) Pairwise dot plot comparisons based on genomic sequences alignment with MUMmer among the six complete B. pseudocatenulatum genomes. (B) The Venn diagram shows the number of core and unique gene groups in each strain.
Physical maps of the predicted eps clusters from the six complete B. pseudocatenulatum genomes. Download FIG S4, PDF file, 0.2 MB (177.1KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
All ORFs identified on the six complete genomes were compared with BLASTP and further clustered with the Markov cluster (MCL) algorithm, which revealed the presence of 2,115 gene groups (Fig. 5B). Of these gene groups, ~72% (1,520) were shared by all six B. pseudocatenulatum genomes, which represent the core genome of B. pseudocatenulatum. A total of 312 dispensable gene groups, which were present only in a subset of the examined B. pseudocatenulatum genomes, were identified. More than 61.48% of the unique groups were specific to B. pseudocatenulatum JCM1200T. Similar results were obtained based on COG assignments, with 1,101 COG families identified from the six complete B. pseudocatenulatum genomes. Of these COG families, 59 COG families were present only in a subset of the examined B. pseudocatenulatum genomes, and 37 additional COGs were unique to a single strain, with 35 of these identified only in B. pseudocatenulatum JCM1200T (Fig. S5). There were also 46 COG families that were found in our isolates that were absent from the B. pseudocatenulatum JCM1200T genome. Interestingly, at least some of these differences relate to the mobilome and DNA rearrangement, with our isolates possessing a larger number of unique COGs related to prophage functions, cellular processes, and signaling, while the B. pseudocatenulatum JCM1200T possessed a larger number of unique COGs, including those involved with the clustered regularly interspaced short palindromic repeat (CRISPR)/Cas system(s).
Distribution of dispensable and unique COGs in the six B. pseudocatenulatum genomes. Download FIG S5, PDF file, 0.6 MB (638.1KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Particularly, our isolates and B. pseudocatenulatum JCM1200T were exposed to distinct carbohydrate resources. The former were isolated from the fecal sample of the postintervention child, who received mixed materials from whole grains and traditional Chinese medicine (TCM) food plants that are rich in dietary fiber and powders, including fructo-oligosaccharides and oligoisomaltoses, while the latter was isolated from feces from an infant, who was speculated to receive relatively simpler carbohydrates. Correspondingly, genetic variations involved in carbohydrate transport and metabolism were found; for instance, B. pseudocatenulatum JCM1200T lacked COG0383 (alpha-mannosidase), COG3594 (fucose 4-O-acetylase or related acetyltransferase), COG4209 (ABC-type polysaccharide transport system, permease component), and COG4214 (ABC-type xylose transport system, permease component) but uniquely had COG1554 (trehalose and maltose hydrolase [possible phosphorylase]).
The different copy numbers of the core COGs also contributed to the microdiversity of B. pseudocatenulatum. Among the 1,005 core COGs that appeared in each complete B. pseudocatenulatum genome, 51 COGs involved in various functional categories had a disparity in the copy number with at least two copies (Fig. 6). According to the distribution of these COGs, B. pseudocatenulatum JCM1200T was the most different strain compared to our five strains.
FIG 6 .
Distribution of core COGs having a disparity in copy number with at least two copies. The heatmap shows the copy number of genes annotated as the particular COG function. The strains are clustered with the Euclidean distance and the Ward linkage method.
Previous studies have reported the microdiversity of different strains in the same species originating from the same habitat (38–40). Among our five B. pseudocatenulatum strains, the differences appeared to result from variations in the copy numbers of genes assigned to specific core COG families, instead of differences in terms of the presence/absence of unique and/or dispensable COGs. The clustering result (Fig. 6) was consistent with the differential responses of the strains to the carbohydrate interventions. In that context, B. pseudocatenulatum C95 and B. pseudocatenulatum C55, which were largely unaffected by the dietary intervention, were clearly separated from the three responsive strains. B. pseudocatenulatum C1 and B. pseudocatenulatum C62, which had moderate responses to the dietary intervention, grouped together. B. pseudocatenulatum C15, the strain that was the most responsive to the dietary intervention, was further separable from B. pseudocatenulatum C1 and B. pseudocatenulatum C62.
With respect to carbohydrate transport and metabolism, B. pseudocatenulatum C15 had the highest copy number of COG2814 (predicted arabinose efflux permease, major facilitator superfamily [MFS]) and COG3250 (beta-galactosidase/beta-glucuronidase). Moreover, among the five strains, 106 ± 2 (mean ± SD) ORFs were identified as carbohydrate-active enzyme (CAZy) genes, which accounted for 41 CAZy families. The B. pseudocatenulatum C15 genome harbored all of the identified CAZy families and had the greatest copy numbers of these genes. It also harbored the highest copy numbers of genes encoding carbohydrate esterases, which are reported to deacetylate plant polysaccharides to overcome the complexity and cooperate with glycoside hydrolases in plant polysaccharide degradation (41). These features are presumed to be the principal differences accounting for the responsiveness of B. pseudocatenulatum C15 to the dietary intervention, whereby it accounts for more than 50% of the total B. pseudocatenulatum, and a substantial proportion of the entire bifidobacterial population throughout the dietary intervention.
Conclusions.
In a previous study investigating how the WTP diet results in weight loss in genetically obese persons, B. pseudocatenulatum was identified as the most abundant Bifidobacterium species. Here, we show that specific strains of B. pseudocatenulatum show variations in their response to the dietary intervention, and we use comparative genomics to identify possible reasons underlying these dynamics. The five B. pseudocatenulatum strains isolated as part of this study showed some differences relative to B. pseudocatenulatum JCM1200T, which was isolated from feces from an infant, which indicates the effect of different environmental parameters on genome microdiversity (42). Much of the observed microdiversity among our five isolates are the variations in the copy numbers of core COG families, and these differences provide a plausible explanation for the variations in the populations of the five strains during the dietary intervention. In particular, B. pseudocatenulatum C15, which genetically had the more diverse and greater copy numbers of CAZy genes for plant polysaccharides, had the greatest abundance in response to the dietary intervention. The coexistence and distribution of multiple strains are intuitive because it would support the survival of the population as a whole over a broader range of environmental conditions than would be possible for a homogeneous population (43). Thus, the coexistence of the five strains with diverse responses to the dietary intervention may work as a mechanism to ensure the stability and restoration of important beneficial species such as B. pseudocatenulatum in human gut. Importantly though, the five B. pseudocatenulatum strains identified in this study were also found to have different correlations with bioclinical parameters of the host, suggesting that at least some of the beneficial functions ascribed to changes in the gut microbiota are strain specific (44). More studies of the type presented here will be necessary to ensure that the full potential of the health benefits sought from dietary manipulations of the gut microbiota are realized.
MATERIALS AND METHODS
Clinical investigation.
The study was performed with the approval of the Ethics Committee of the School of Life Sciences and Biotechnology, Shanghai Jiao Tong University (2012-2016). The clinical trial was registered at the Chinese Clinical Trial Registry (ChiCTR-ONC-12002646). Written informed consent was obtained from the guardians of the obese child.
In Guangdong Women and Children Hospital, Guangzhou, China, the obese child received a dietary intervention for 105 days in the hospital. The volunteer did not take part in any exercise program. A diet based on whole grains, traditional Chinese medicinal foods, and prebiotics (WTP diet) (7) was administered in combination with appropriate amounts of vegetables, fruits, and nuts according to a dietician’s advice. The three ready-to-consume preprepared foods, Formula No. 1, Formula No. 2, and Formula No. 3, in the diet were manufactured by Perfect (China) Co., Ltd. The intervention was divided into three phases. In phase I (day 0 to day 60), the child received the basic diet (9). In phase II (day 60 to day 75), he consumed 100 g more Formula No. 3. During phase III (day 75 to day 105), he still consumed 100 g more Formula No. 3, but less Formula No. 1 was provided.
Biological samples, anthropometric data, and clinical laboratory analysis were obtained at the seven time points (0, 15, 30, 45, 60, 75, and 105 days). The bioclinical parameters were measured as in our previous study (9).
Metagenomic sequencing and analysis.
DNA extraction from fecal samples for metagenomic sequencing was conducted as previously described (45). Seven samples were sequenced using the Illumina HiSeq 2000 platform at Shanghai Genergy Biotechnology Co., Ltd. DNA libraries were prepared following Illumina’s instructions. Cluster generation, template hybridization, isothermal amplification, linearization, blocking, and denaturing and hybridization of the sequencing primers were performed according to the workflow indicated by the provider. Libraries were constructed followed by high-throughput sequencing to obtain paired-end reads with 151 bp in the forward and reverse directions.
Flexbar (46) was used to trim the adapter from the reads. Prinseq (47) was employed (i) to trim the reads from the 3′ end until reaching the first nucleotide with a quality threshold of 20, (ii) to remove read pairs if either read was shorter than 75 bp or contained “N” bases, and (iii) to deduplicate the reads. Reads that could be aligned to the human genome (Homo sapiens, UCSC hg19) were removed (aligned with Bowtie 2 [48], using –reorder–no-hd–no-contain–dovetail). On average, 25.3 × 106 ± 4.1 × 106 (mean ± SD) paired-end reads for each sample were retained and used for further analysis.
MetaPhlan (49) (–bt2_ps very-sensitive-local) was used to calculate the abundance of Bifidobacterium species on day 105. Sigma (24) was used to calculate the abundance of our five B. pseudocatenulatum strains.
Whole-genome sequencing and data assembly.
The genomes of B. pseudocatenulatum C1, B. pseudocatenulatum C15, B. pseudocatenulatum C55, B. pseudocatenulatum C62, and B. pseudocatenulatum C95 were sequenced on a PacBio RS II sequencing instrument with approximately 245-, 415-, 285-, 459-, and 198-fold coverage, respectively (Nextomics Biosciences, Wuhan, China). HGAP/Quiver (50) was used to de novo assemble the subreads, followed by Minimus2 (51) and Quiver. The genome of each strain was assembled into one contig that corresponded to its chromosome.
General feature prediction.
Open reading frame (ORF) prediction was performed by the combination of Prodigal v2_60 (52) and BLAST alignment. The identified ORFs were then annotated using BLASTP against the NCBI nr database. rRNA genes were detected using RNAmmer v1.2 (53), and tRNA genes were identified with tRNAscan-SE v.14 (54). The identity matrix between all 16S rRNA gene was calculated with USEARCH v8.0.1517 (55). The assignment of COGs was done with COGtriangles (ftp://ftp.ncbi.nih.gov/pub/wolf/COGs/COGsoft/).
Pan-genome calculation.
PGAPv1.12 (56) was used to calculate the pan-genome of B. pseudocatenulatum. Functional gene clustering was performed by the GF (gene family) method, and the pan-genome profile was then built.
Genomic comparison.
Software package MUMmer v3.0 (57) was used to perform the whole-genome sequence alignments at the nucleotide level. For each genome pair, the average nucleotide identity (ANI) was calculated using the program JSpecies version 1.2.1 (35). At the protein level, all sequences were pairwisely compared using BLASTP (maximum E value of 1e−10, minimum alignment identity of 50%, and minimum alignment coverage of 50% for either protein) and then clustering into gene families using the Markov cluster (MCL) algorithm implemented in PGAP v1.12.
Identification of exopolysaccharide (EPS) gene cluster.
In silico analysis of the bifidobacterial eps cluster was performed by searching for the putative priming-GTF (p-gtf) gene, rfbP (GenBank accession no. NP_695444) and cpsD (accession no. NP_695447), in each genome (37), followed by manually checking the genes surrounding the p-gtf gene.
Identification of Carbohydrate-Activated Enzymes (CAZys).
A local-version database of dbCAN v3.0 (58) was downloaded. Genes in each genome were aligned to the database using HMMscan (59). The alignment was parsed with hmmscan-parser.sh provided by dbCAN, and the best hit was retained.
Availability of data and materials.
All the genomes generated in this study and the metagenomic data set have been deposited in the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena) under accession number PRJEB18557. All the other genomes of B. pseudocatenulatum used for our analysis were downloaded from the NCBI database with the following GenBank accession numbers: AP012330.1, CDPW00000000.1, ABXX02000001, JEOD01000001, and JGZF01000001. The B. pseudocatenulatum D2CA genome was downloaded from MetaHIT (http://www.sanger.ac.uk/resources/downloads/bacteria/metahit/).
ACKNOWLEDGMENTS
We thank Jingjing Wang for assistance with the DNA extraction for metagenomic sequencing. We thank the reviewers and appreciate their constructive comments on this study.
This work was supported by grants from the National Natural Science Foundation of China (31330005, 30730005, 81401141, and 20875061) and the Science and Technology Commission of Shanghai Municipality (14YF1402200). A computing facility award for the PI cluster at Shanghai Jiao Tong University is acknowledged.
Footnotes
Citation Wu G, Zhang C, Wu H, Wang R, Shen J, Wang L, Zhao Y, Pang X, Zhang X, Zhao L, Zhang M. 2017. Genomic microdiversity of Bifidobacterium pseudocatenulatum underlying differential strain-level responses to dietary carbohydrate intervention. mBio 8:e02348-16. https://doi.org/10.1128/mBio.02348-16.
REFERENCES
- 1.Zhao L. 2013. The gut microbiota and obesity: from correlation to causality. Nat Rev Microbiol 11:639–647. doi: 10.1038/nrmicro3089. [DOI] [PubMed] [Google Scholar]
- 2.Boulangé CL, Neves AL, Chilloux J, Nicholson JK, Dumas ME. 2016. Impact of the gut microbiota on inflammation, obesity, and metabolic disease. Genome Med 8:42. doi: 10.1186/s13073-016-0303-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin N, Yang F, Li A, Prifti E, Chen Y, Shao L, Guo J, Le Chatelier E, Yao J, Wu L, Zhou J, Ni S, Liu L, Pons N, Batto JM, Kennedy SP, Leonard P, Yuan C, Ding W, Chen Y, Hu X, Zheng B, Qian G, Xu W, Ehrlich SD, Zheng S, Li L. 2014. Alterations of the human gut microbiome in liver cirrhosis. Nature 513:59–64. doi: 10.1038/nature13568. [DOI] [PubMed] [Google Scholar]
- 4.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J. 2015. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 5.Xu J, Lian F, Zhao L, Zhao Y, Chen X, Zhang X, Guo Y, Zhang C, Zhou Q, Xue Z, Pang X, Zhao L, Tong X. 2015. Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J 9:552–562. doi: 10.1038/ismej.2014.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly CR, Kahn S, Kashyap P, Laine L, Rubin D, Atreja A, Moore T, Wu G. 2015. Update on fecal microbiota transplantation 2015: indications, methodologies, mechanisms, and outlook. Gastroenterology 149:223–237. doi: 10.1053/j.gastro.2015.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiao S, Fei N, Pang X, Shen J, Wang L, Zhang B, Zhang M, Zhang X, Zhang C, Li M, Sun L, Xue Z, Wang J, Feng J, Yan F, Zhao N, Liu J, Long W, Zhao L. 2014. A gut microbiota-targeted dietary intervention for amelioration of chronic inflammation underlying metabolic syndrome. FEMS Microbiol Ecol 87:357–367. doi: 10.1111/1574-6941.12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Human Microbiome Project Consortium 2012. Structure, function and diversity of the healthy human microbiome. Nature 486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang C, Yin A, Li H, Wang R, Wu G, Shen J, Zhang M, Wang L, Hou Y, Ouyang H, Zhang Y, Zheng Y, Wang J, Lv X, Wang Y, Zhang F, Zeng B, Li W, Yan F, Zhao Y, Pang X, Zhang X, Fu H, Chen F, Zhao N, Hamaker BR, Bridgewater LC, Weinkove D, Clement K, Dore J, Holmes E, Xiao H, Zhao G, Yang S, Bork P, Nicholson JK, Wei H, Tang H, Zhang X, Zhao L. 2015. Dietary modulation of gut microbiota contributes to alleviation of both genetic and simple obesity in children. EBioMedicine 2:968–984. doi: 10.1016/j.ebiom.2015.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu H, Wang R, Zhao Y, Pang X, Shen J, Zhang C. 2015. Exploring carbohydrate utilization capacity of Bifidobacterium pseudocatenulatum isolated from a morbidly obese child after dietary intervention. Genom Appl Biol 34:1384–1391. [Google Scholar]
- 11.Grimm V, Westermann C, Riedel CU. 2014. Bifidobacteria-host interactions—an update on colonisation factors. BioMed Res Int 2014:960826. doi: 10.1155/2014/960826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Labruna G, Pasanisi F, Nardelli C, Caso R, Vitale DF, Contaldo F, Sacchetti L. 2011. High leptin/adiponectin ratio and serum triglycerides are associated with an ‘at-risk’ phenotype in young severely obese patients. Obesity (Silver Spring) 19:1492–1496. doi: 10.1038/oby.2010.309. [DOI] [PubMed] [Google Scholar]
- 13.Zweigner J, Schumann RR, Weber JR. 2006. The role of lipopolysaccharide-binding protein in modulating the innate immune response. Microbes Infect 8:946–952. doi: 10.1016/j.micinf.2005.10.006. [DOI] [PubMed] [Google Scholar]
- 14.Arumugam M, Raes J, Pelletier E, Le Paslier D, Yamada T, Mende DR, Fernandes GR, Tap J, Bruls T, Batto JM, Bertalan M, Borruel N, Casellas F, Fernandez L, Gautier L, Hansen T, Hattori M, Hayashi T, Kleerebezem M, Kurokawa K, Leclerc M, Levenez F, Manichanh C, Nielsen HB, Nielsen T, Pons N, Poulain J, Qin J, Sicheritz-Ponten T, Tims S, Torrents D, Ugarte E, Zoetendal EG, Wang J, Guarner F, Pedersen O, de Vos WM, Brunak S, Doré J, MetaHIT Consortium, Antolin M, Artiguenave F, Blottiere HM, Almeida M, Brechot C, Cara C, Chervaux C, Cultrone A, Delorme C, Denariaz G, Dervyn R, et al. . 2011. Enterotypes of the human gut microbiome. Nature 473:174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fujimoto T, Imaeda H, Takahashi K, Kasumi E, Bamba S, Fujiyama Y, Andoh A. 2013. Decreased abundance of Faecalibacterium prausnitzii in the gut microbiota of Crohn’s disease. J Gastroenterol Hepatol 28:613–619. doi: 10.1111/jgh.12073. [DOI] [PubMed] [Google Scholar]
- 16.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermúdez-Humarán LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottière HM, Doré J, Marteau P, Seksik P, Langella P. 2008. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A 105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gänzle MG, Follador R. 2012. Metabolism of oligosaccharides and starch in lactobacilli: a review. Front Microbiol 3:340. doi: 10.3389/fmicb.2012.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreasen AS, Larsen N, Pedersen-Skovsgaard T, Berg RM, Møller K, Svendsen KD, Jakobsen M, Pedersen BK. 2010. Effects of Lactobacillus acidophilus NCFM on insulin sensitivity and the systemic inflammatory response in human subjects. Br J Nutr 104:1831–1838. doi: 10.1017/S0007114510002874. [DOI] [PubMed] [Google Scholar]
- 19.Hulston CJ, Churnside AA, Venables MC. 2015. Probiotic supplementation prevents high-fat, overfeeding-induced insulin resistance in human subjects. Br J Nutr 113:596–602. doi: 10.1017/S0007114514004097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charbonneau MR, Blanton LV, DiGiulio DB, Relman DA, Lebrilla CB, Mills DA, Gordon JI. 2016. A microbial perspective of human developmental biology. Nature 535:48–55. doi: 10.1038/nature18845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang J, Tang H, Zhang C, Zhao Y, Derrien M, Rocher E, van-Hylckama Vlieg JE, Strissel K, Zhao L, Obin M, Shen J. 2015. Modulation of gut microbiota during probiotic-mediated attenuation of metabolic syndrome in high fat diet-fed mice. ISME J 9:1–15. doi: 10.1038/ismej.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stenman LK, Waget A, Garret C, Klopp P, Burcelin R, Lahtinen S. 2014. Potential probiotic Bifidobacterium animalis ssp. lactis 420 prevents weight gain and glucose intolerance in diet-induced obese mice. Benef Microbes 5:437–445. doi: 10.3920/BM2014.0014. [DOI] [PubMed] [Google Scholar]
- 23.Staley J, Krieg N. 1984. Classification of prokaryotes organisms: an overview, p 1–4. In Krieg NR, Holt JG (ed), Bergey’s manual of systematic bacteriology, vol 1 Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 24.Ahn TH, Chai J, Pan C. 2015. Sigma: strain-level inference of genomes from metagenomic analysis for biosurveillance. Bioinformatics 31:170–177. doi: 10.1093/bioinformatics/btu641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morita H, Toh H, Oshima K, Nakano A, Arakawa K, Takayama Y, Kurokawa R, Takanashi K, Honda K, Hattori M. 2015. Complete genome sequence of Bifidobacterium pseudocatenulatum JCM 1200(T) isolated from infant feces. J Biotechnol 210:68–69. doi: 10.1016/j.jbiotec.2015.06.416. [DOI] [PubMed] [Google Scholar]
- 26.Bottacini F, Medini D, Pavesi A, Turroni F, Foroni E, Riley D, Giubellini V, Tettelin H, van Sinderen D, Ventura M. 2010. Comparative genomics of the genus Bifidobacterium. Microbiology 156:3243–3254. doi: 10.1099/mic.0.039545-0. [DOI] [PubMed] [Google Scholar]
- 27.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bottacini F, O’Connell Motherway M, Kuczynski J, O’Connell KJ, Serafini F, Duranti S, Milani C, Turroni F, Lugli GA, Zomer A, Zhurina D, Riedel C, Ventura M, van Sinderen D. 2014. Comparative genomics of the Bifidobacterium breve taxon. BMC Genomics 15:170. doi: 10.1186/1471-2164-15-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O’Callaghan A, Bottacini F, O’Connell Motherway M, van Sinderen D. 2015. Pangenome analysis of Bifidobacterium longum and site-directed mutagenesis through by-pass of restriction-modification systems. BMC Genomics 16:832. doi: 10.1186/s12864-015-1968-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Begley M, Gahan CGM, Hill C. 2005. The interaction between bacteria and bile. FEMS Microbiol Rev 29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Candela M, Biagi E, Centanni M, Turroni S, Vici M, Musiani F, Vitali B, Bergmann S, Hammerschmidt S, Brigidi P. 2009. Bifidobacterial enolase, a cell surface receptor for human plasminogen involved in the interaction with the host. Microbiology 155:3294–3303. doi: 10.1099/mic.0.028795-0. [DOI] [PubMed] [Google Scholar]
- 32.Candela M, Centanni M, Fiori J, Biagi E, Turroni S, Orrico C, Bergmann S, Hammerschmidt S, Brigidi P. 2010. DnaK from Bifidobacterium animalis subsp. lactis is a surface-exposed human plasminogen receptor upregulated in response to bile salts. Microbiology 156:1609–1618. doi: 10.1099/mic.0.038307-0. [DOI] [PubMed] [Google Scholar]
- 33.González-Rodríguez I, Sánchez B, Ruiz L, Turroni F, Ventura M, Ruas-Madiedo P, Gueimonde M, Margolles A. 2012. Role of extracellular transaldolase from Bifidobacterium bifidum in mucin adhesion and aggregation. Appl Environ Microbiol 78:3992–3998. doi: 10.1128/AEM.08024-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pareti FI, Fujimura Y, Dent JA, Holland LZ, Zimmerman TS, Ruggeri ZM. 1986. Isolation and characterization of a collagen binding domain in human von Willebrand factor. J Biol Chem 261:15310–15315. [PubMed] [Google Scholar]
- 35.Richter M, Rosselló-Móra R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci U S A 106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmid J, Sieber V, Rehm B. 2015. Bacterial exopolysaccharides: biosynthesis pathways and engineering strategies. Front Microbiol 6:496. doi: 10.3389/fmicb.2015.00496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hidalgo-Cantabrana C, Sánchez B, Milani C, Ventura M, Margolles A, Ruas-Madiedo P. 2014. Genomic overview and biological functions of exopolysaccharide biosynthesis in Bifidobacterium spp. Appl Environ Microbiol 80:9–18. doi: 10.1128/AEM.02977-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang XL, Gao PP, Chao QF, Wang LH, Senior E, Zhao LP. 2004. Microdiversity of phenol hydroxylase genes among phenol-degrading isolates of Alcaligenes sp. from an activated sludge system. FEMS Microbiol Lett 237:369–375. doi: 10.1016/j.femsle.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 39.Mao YJ, Zhang XJ, Xia X, Zhong HH, Zhao LP. 2010. Versatile aromatic compound-degrading capacity and microdiversity of Thauera strains isolated from a coking wastewater treatment bioreactor. J Ind Microbiol Biotechnol 37:927–934. doi: 10.1007/s10295-010-0740-7. [DOI] [PubMed] [Google Scholar]
- 40.Patra R, Chattopadhyay S, De R, Ghosh P, Ganguly M, Chowdhury A, Ramamurthy T, Nair GB, Mukhopadhyay AK. 2012. Multiple infection and microdiversity among Helicobacter pylori isolates in a single host in India. PLoS One 7:e43370. doi: 10.1371/journal.pone.0043370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Biely P. 2012. Microbial carbohydrate esterases deacetylating plant polysaccharides. Biotechnol Adv 30:1575–1588. doi: 10.1016/j.biotechadv.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 42.Jezbera J, Jezberová J, Kasalický V, Šimek K, Hahn MW. 2013. Patterns of Limnohabitans microdiversity across a large set of freshwater habitats as revealed by reverse line blot hybridization. PLoS One 8:e58527. doi: 10.1371/journal.pone.0058527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moore LR, Rocap G, Chisholm SW. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464–467. doi: 10.1038/30965. [DOI] [PubMed] [Google Scholar]
- 44.Zhang C, Zhao L. 2016. Strain-level dissection of the contribution of the gut microbiome to human metabolic disease. Genome Med 8:41. doi: 10.1186/s13073-016-0304-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fei N, Zhao L. 2013. An opportunistic pathogen isolated from the gut of an obese human causes obesity in germfree mice. ISME J 7:880–884. doi: 10.1038/ismej.2012.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dodt M, Roehr JT, Ahmed R, Dieterich C. 2012. FLEXBAR —flexible barcode and adapter processing for next-generation sequencing platforms. Biology (Basel) 1:895–905. doi: 10.3390/biology1030895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C. 2012. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 51.Sommer DD, Delcher AL, Salzberg SL, Pop M. 2007. Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8:64. doi: 10.1186/1471-2105-8-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Rognes T, Ussery DW. 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964. doi: 10.1093/nar/25.5.0955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 56.Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J. 2012. PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418. doi: 10.1093/bioinformatics/btr655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. 2004. Versatile and open software for comparing large genomes. Genome Biol 5:R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. 2012. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 40:W445–W451. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Finn RD, Clements J, Eddy SR. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res 39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B 57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bioclinical parameters that changed during the dietary intervention. Download FIG S1, PDF file, 0.2 MB (248.5KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Abundance of the nine identified Bifidobacterium species in the gut microbial community on day 105. Download FIG S2, PDF file, 0.3 MB (316.9KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Pan-genome and core genome curves of B. pseudocatenulatum. Download FIG S3, PDF file, 0.2 MB (218.8KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Heterogeneity of the 16S rRNA genes. Download TABLE S1, DOCX file, 0.1 MB (103KB, docx) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Physical maps of the predicted eps clusters from the six complete B. pseudocatenulatum genomes. Download FIG S4, PDF file, 0.2 MB (177.1KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Distribution of dispensable and unique COGs in the six B. pseudocatenulatum genomes. Download FIG S5, PDF file, 0.6 MB (638.1KB, pdf) .
Copyright © 2017 Wu et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
All the genomes generated in this study and the metagenomic data set have been deposited in the European Nucleotide Archive (ENA) (http://www.ebi.ac.uk/ena) under accession number PRJEB18557. All the other genomes of B. pseudocatenulatum used for our analysis were downloaded from the NCBI database with the following GenBank accession numbers: AP012330.1, CDPW00000000.1, ABXX02000001, JEOD01000001, and JGZF01000001. The B. pseudocatenulatum D2CA genome was downloaded from MetaHIT (http://www.sanger.ac.uk/resources/downloads/bacteria/metahit/).