Summary
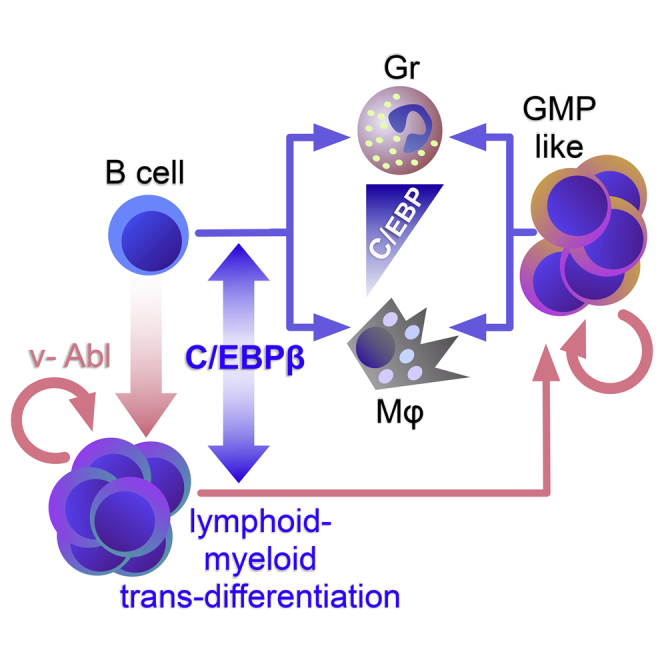
The lymphoid-myeloid transdifferentiation potentials of members of the C/EBP family (C/EBPα, β, δ, and ε) were compared in v-Abl-immortalized primary B cells. Conversion of B cells to macrophages was readily induced by the ectopic expression of any C/EBP, and enhanced by endogenous C/EBPα and β activation. High transgene expression of C/EBPβ or C/EBPε, but not of C/EBPα or C/EBPδ, also induced the formation of granulocytes. Granulocytes and macrophages emerged in a mutually exclusive manner. C/EBPβ-expressing B cells produced granulocyte-macrophage progenitor (GMP)-like progenitors when subjected to selective pressure to eliminate lymphoid cells. The GMP-like progenitors remained self-renewing and cytokine-independent, and continuously produced macrophages and granulocytes. In addition to their suitability to study myelomonocytic lineage bifurcation, lineage-switched GMP-like progenitors could reflect the features of the lympho-myeloid lineage switch observed in leukemic progression.
Keywords: hematopoiesis, leukemia, differentiation, stem cell, progenitor, lineage switch
Graphical Abstract
Highlights
-
•
Transactivating C/EBP family members transdifferentiate B cells to myeloid cells
-
•
C/EBPβ or C/EBPε transdifferentiate B cells to macrophages and granulocytes
-
•
Transgene dosage determines granulocyte versus macrophage cell-type outcome
-
•
C/EBP-mediated B cell conversion elicits GMP-like potential
In this article, Leutz and colleagues assessed the transdifferentiation potential of four C/EBP transcription factors in B cells. C/EBPβ and C/EBPε stimulated the formation of macrophage and granulocyte-like cells. Transdifferentiation involved endogenous C/EBP activation. High transgene dosage promoted granulocyte versus macrophage outcome. C/EBPβ also elicited a stable GMP-like phenotype that could serve as a model to study cell-fate decision.
Introduction
Hematopoiesis is thought to begin from stem cells that progress through consecutive precursor cell stages in a hierarchical fashion whereby lineage commitment precludes alternative cell differentiation. However, there has been increasing evidence of hematopoietic plasticity and cell lineage conversion, particularly during leukemogenesis (Cobaleda and Busslinger, 2008, Graf, 2008, Greaves et al., 1986, Regalo and Leutz, 2013). The transcription factors C/EBPα and C/EBPβ are potent inducers of myelomonocytic genes in heterologous cell types (Ness et al., 1993), and the experimental conversion of lymphoid cells to myeloid cells modulated by both C/EBPs has highlighted their lympho-myeloid transdifferentiation potential (Graf and Enver, 2009).
The C/EBP family members C/EBPα, C/EBPβ, C/EBPδ, and C/EBPε are expressed in myeloid cells (Cloutier et al., 2009, Scott et al., 1992). Loss-of-function studies in genetically modified mice suggested combinatorial and partially redundant functions of C/EBPs in myelopoiesis (Tsukada et al., 2011). Knockout studies showed that deletion of Cebpa has the strongest impact on myelopoiesis, resulting in an almost complete loss of neutrophils and impaired development of granulocyte-macrophage progenitor (GMP) cells (Zhang et al., 1997, Zhang et al., 2004). However, cytokines could compensate for the lack of Cebpa by the concomitant activation of Cebpb, and genetic replacement of Cebpa with Cebpb in the Cebpa locus compensates for the Cebpa requirement in hematopoiesis and liver functions (Chen et al., 2000, Hirai et al., 2006, Jones et al., 2002). Individual deletions of C/EBPβ, δ, and ε evoke milder and gene-specific phenotypes, such as susceptibility to infections, failure of emergency granulopoiesis, impaired cytokine production, and partial granulocyte deficiency that is intensified by compound C/EBP gene deletions. For example, compound Cebpb/Cebpe deletion mutants display impaired granulopoiesis, defective macrophage functions, and a disrupted innate immune regulatory gene expression network, confirming the compensatory and redundant functions of the C/EBPs (Akagi et al., 2010, Hirai et al., 2006, Litvak et al., 2009, Tanaka et al., 1995, Yamanaka et al., 1997).
C/EBPα can stimulate the transdifferentiation of B and T cells and, together with PU.1, even fibroblasts into macrophages (Bussmann et al., 2009, Feng et al., 2008, Ness et al., 1993, Xie et al., 2004). Conversion of B cells into inflammatory-type macrophages occurs rapidly after C/EBP expression, with high efficiency and through a direct route (Bussmann et al., 2009, Di Tullio et al., 2011, Xie et al., 2004). An experimental transdifferentiation system based on an estrogen-responsive, conditional C/EBPα protein in the v-H-ras-transformed pre-B cell line HAFTL1 (Holmes et al., 1986) has served as a tool to examine the mechanistic aspects of lympho-myeloid lineage conversion, including alterations of chromatin occupancy, gene expression kinetics, non-coding RNA expression, and DNA methylation (Barneda-Zahonero et al., 2013, Di Tullio et al., 2011, Kallin et al., 2012, Krijger et al., 2016, Rodriguez-Ubreva et al., 2012, Rodriguez-Ubreva et al., 2014, van Oevelen et al., 2015).
We recently found that structural alterations and post-translational modification sites of C/EBPβ may determine the path of transdifferentiation of primary progenitor B cells toward distinct myeloid cell fates (including granulocytes and dendritic cells), suggesting that epigenetic instructions beyond the inflammatory macrophage cell fate are encoded in the C/EBP structure and could account for cell-type specification (Stoilova et al., 2013). This observation has prompted us to compare the lineage conversion capacity of all transactivator C/EBP family members and to develop a lympho-myeloid transdifferentiation system that is amenable to targeted mouse genetics and cell-culture manipulation.
In this study, we generated murine v-Abl-immortalized B cells from wild-type and genetically altered mice to compare the lympho-myeloid transdifferentiation potential of the C/EBP family members C/EBPα, C/EBPβ, C/EBPδ, and C/EBPε. Our data showed that C/EBPβ and C/EBPε readily induce a granulocytic fate in addition to macrophage formation. Granulocytic conversion largely depended on transgene dosage. In addition, efficient transdifferentiation required endogenous Cebpa/Cebpb. Importantly, applying selective pressure on immortalized B cells expressing C/EBPβ by depriving β-mercaptoethanol resulted in the rapid extinction of B cells and massive expansion of stable myeloid cells. These myeloid progenitors displayed bipotential GMP-like properties and continuously produced macrophages and granulocytes. This process could suggest a link between C/EBP-induced lympho-myeloid lineage switch and a B cell-derived leukemic myelomonocytic GMP-like phenotype (Slamova et al., 2013).
Results
Transdifferentiation Potential of Various C/EBP Family Members
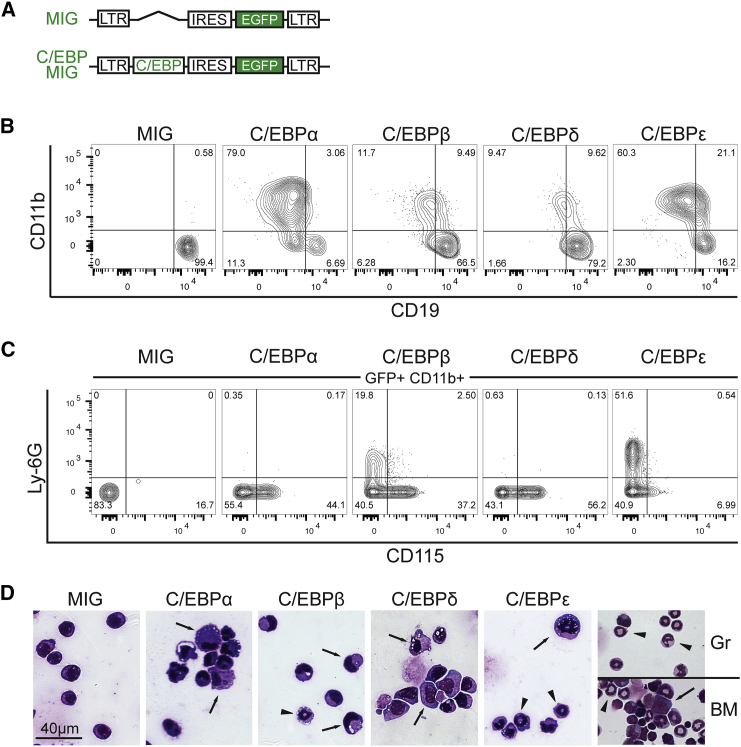
v-Abl-immortalized pre-B cells derived from primary mouse bone marrow cells (hereafter termed ‘‘B cells’’) were used to assess the ability of individual C/EBP family members to induce B-lymphoid to myeloid transdifferentiation (Rosenberg et al., 1975, Shore et al., 2002). pMSCV-based retroviral expression vectors (MIG) encoding full-length C/EBPα-, C/EBPβ-, C/EBPδ-, or C/EBPε-IRES-EGFP were constructed (Figure 1A). Transgene expression was confirmed in the retrovirus-packaging cell line (Figure S1A). As shown in Figure 1B, each C/EBP family member could induce expression of the myeloid marker CD11b (Mac-1) and caused the transdifferentiation of B cells into CD11b+CD19dim myeloid cells. Additional myeloid surface markers indicative of dendritic cells (CD11c+), macrophages (CD115+), or granulocytes (Ly-6G+), were included to assess myeloid subsets in the CD11b+ population (Figure S1B). While no CD11c+ cells were found, a fraction of transdifferentiated cells showed upregulated CD115 (MCSF-R), a macrophage marker and C/EBP target gene. A CD11b+Ly-6G+ population, characteristic of granulocytes, was detected with C/EBPε expression, consistent with observations of strong mRNA expression of C/EBPε in bone marrow-derived granulocytes (Figures S1B and S1C). C/EBPβ expression also caused the formation of a CD11b+Ly-6G+ population after prolonged transdifferentiation for 6 days, whereas C/EBPα and C/EBPδ expression did not (Figures 1C and S1B). Interestingly, the expression of Ly-6G and CD115 appeared to be mutually exclusive, as no double-positive cells could be detected, indicating a stringent mechanism of cell-fate decision (Figure 1C). Cytospin preparations of GFP+ cells confirmed that all C/EBPs stimulated the formation of myeloid cells with monocyte/macrophage features (Figure 1D). The C/EBPβ- and C/EBPε-transdifferentiated cell population contained small cells with segmented or indented nuclei, strongly resembling bone marrow-derived granulocytes and suggesting that C/EBPε or C/EBPβ may induce granulocyte/macrophage (G/M) cell fates. Transdifferentiation into granulocytes by C/EBPβ and C/EBPε was not restricted to the v-Abl B cell system, but also occurred in the fetal liver-derived H-ras-transformed HAFTL1 pre-B cell line (Figure S1D) (Bussmann et al., 2009). These findings suggest that all the tested C/EBP transcription factors could cause the conversion of B cells to myeloid cells and that C/EBPβ and C/EBPε could stimulate macrophage and granulocyte transdifferentiation of B cells.
Figure 1.
C/EBPα, β, δ, and ε Transdifferentiate B Cells to Myeloid Cells
(A) pMSCV-based vectors containing an internal ribosomal entry site (IRES) and EGFP (MIG) marker used for retroviral transduction. Full-length open reading frames of Cebpa, Cebpb, Cebpd, or Cebpe were inserted upstream of the IRES element. Empty vector (MIG) served as control.
(B) Flow cytometric analysis of GFP+-infected B cells 4 days after transduction with individual C/EBPs.
(C) Analysis of granulocyte (Ly-6G) and macrophage (CD115) cell-surface markers in GFP+CD11b+ transdifferentiated cells 6 days after transduction.
(D) May-Grünwald staining of GFP+-sorted B cells 4 days after transduction. Arrows indicate cells with typical macrophage morphology and arrowheads mark granulocyte-like cells. Primary bone marrow (BM) and sorted granulocytes (Gr) were used as references (far right).
See also Figure S1.
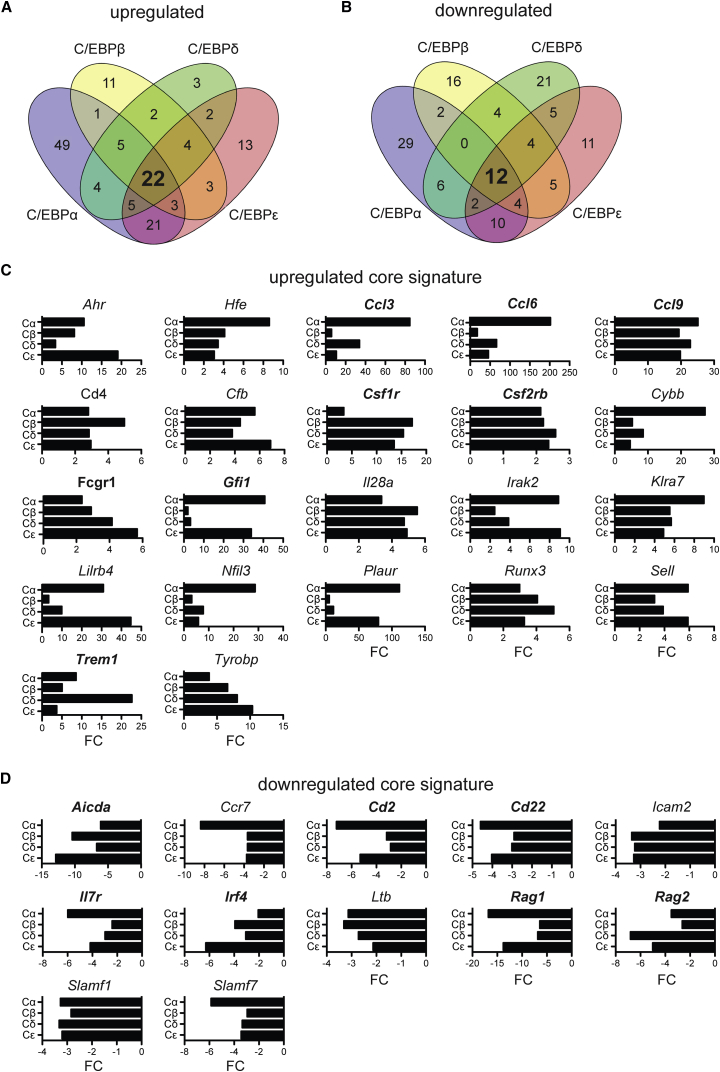
Quantitative gene expression analysis was performed by NanoString hybridization 1 day after transduction with individual C/EBPs, at the earliest emergence of GFP-positive cells (Figures 2A and 2B; Table S1). A predefined mouse immunology code set covering 547 probes, including key transcription factors, was used. Genes with 2-fold altered expression levels, compared with empty vector-transduced B cell controls, were considered. As shown in Figures 2A and 2B, all C/EBPs upregulated and downregulated a core set of 22 and 12 genes, respectively. Each C/EBP family member also displayed additional and partial overlapping regulatory specificity. The core transdifferentiation signature of 22 upregulated genes included myeloid factors, such as Csf1r, Csf2rb, Fcgr1, and Gfi1, and several chemokine CC-family members (Figure 2C). The 12 downregulated genes included lymphocyte genes, such as Il7r, Cd2, and the Rag genes (Figure 2D). These data suggest that C/EBPα, β, δ, and ε likely suppress the B cell program and induce lympho-myeloid conversion.
Figure 2.
Transdifferentiation Core Gene Signatures of B Cells Induced with C/EBP Family Members
Quantification of mRNA from GFP+-sorted B cells 24 hr after transduction with individual C/EBPs. Venn diagram of (A) upregulated and (B) downregulated genes, compared with empty vector (MIG) control. Overlapping core signatures of (C) 22 upregulated or (D) 12 downregulated genes. Key myeloid (in C) and lymphoid (in D) genes are shown in bold. The fold change (FC) relative to empty vector (MIG)-transduced B cells is shown. See also Table S1.
Deletion of Endogenous Cebpa and Cebpb Impairs Transdifferentiation but Has No Impact on Cell-Type Outcome
As previously reported by Bussmann et al. (2009) and shown in Figure S2A, activation of conditional C/EBPα-ER induced endogenous Cebpb and Cebpd gene expression in HAFTL1 B cells. We induced myeloid transdifferentiation via C/EBPε expression in HAFTL1 cells and analyzed endogenous C/EBPα and C/EBPβ protein expression. Interestingly, the expression of C/EBPε also led to a marked upregulation of endogenous C/EBPα after 16 hr, while only low levels of C/EBPβ were detected. However, after 24 hr, C/EBPβ protein expression increased while C/EBPα expression was diminished. A surge of C/EBPα and C/EBPβ expression was observed after 120 hr, indicating crosstalk between transgenic and endogenous C/EBPs during transdifferentiation (Figure S2B).
Synergistic collaboration between C/EBP family members was previously described as a key element to specify myeloid differentiation (Akagi et al., 2010). To examine the contribution of endogenous C/EBPs to lympho-myeloid transdifferentiation, we treated B cells generated from a homozygous Cebpaf/f;Cebpbf/f mouse with cell-permeable Cre-recombinase to generate double-knockout derivatives (Figure S2C). Verified CebpaΔ/Δ;CebpbΔ/Δ double-knockout B cell clones were designated as “B-DKO” (Figure S2D).
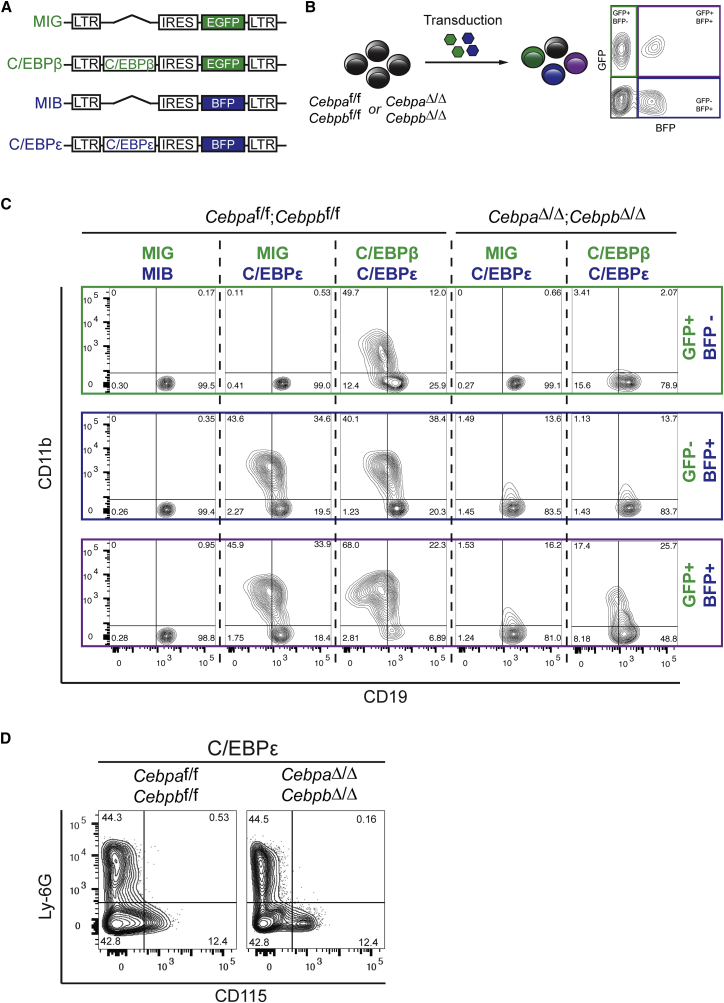
B-DKO cells or isogenic controls (not treated with Cre-recombinase) were infected with the G/M-transdifferentiation proficient retroviral constructs that co-expressed C/EBPβ and GFP (MIG) or C/EBPε and BFP (MIB) (Figures 3A and 3B). GFP+BFP+ gated cell populations, indicative of C/EBPβ and C/EBPε expression, were analyzed (Figure 3C). Deletion of endogenous Cebpa and Cebpb strongly impaired transdifferentiation induced by C/EBPε or C/EBPβ after 4 days, in comparison with isogenic controls (Figures 3C and S2E). Transdifferentiation efficacy in C/EBPε-expressing B-DKO cells could be partially rescued by retroviral co-expression of C/EBPβ. These data suggest synergistic effects between endogenous and exogenous C/EBPs during transdifferentiation (Figures 3C and S2E).
Figure 3.
Endogenous Cebpa and Cebpb Promote Transdifferentiation but Do Not Affect Lineage Outcome
(A) pMSCV-based vectors for the expression of C/EBPβ linked to EGFP marker or C/EBPε linked to BFP. Empty vectors (MIG/MIB) were used as controls.
(B) CebpaΔ/Δ;CebpbΔ/Δ or isogenic control B cells (Cebpaf/f;Cebpbf/f) were transduced with combinations of the vectors shown in (A).
(C) The GFP+BFP− fraction (top panels), GFP−BFP+ fraction (middle panels), and GFP+BFP+ fraction (bottom panels) were individually analyzed for myeloid (CD11b) and B cell (CD19) markers by flow cytometry after 4 days.
(D) CebpaΔ/Δ;CebpbΔ/Δ B cells and isogenic controls were transduced with C/EBPε, GFP+-sorted after 24 hr, and recultured. Flow cytometric analysis of granulocyte (Ly-6G+) and macrophage (CD115+) cell fractions in GFP+CD11b+-gated populations 7 days after transduction.
See also Figure S2.
Comparison of gene expression in C/EBPε-transduced wild-type or B-DKO cells revealed genes that were dependent on endogenous Cebpa and Cebpb (Figure S2F and Table S1). The list of refractory genes included Csf1r, Fcgr1, Itgam (CD11b), and the transcription factor Gfi1, known to promote granulocyte differentiation. These genes represent important myeloid cell factors, and failure of induction may explain the impaired transdifferentiation (Figures 3C and S2E). In addition, Pax5 or Pou2f2 (Oct-2) were refractory to downregulation, which could contribute to the persistence of the B cell phenotype (Figure S2G and Table S1).
To determine whether the G/M cell-type outcome was affected by deletion of endogenous Cebpa and Cebpb, we analyzed C/EBPε-transdifferentiated GFP+CD11b+ cells for Ly-6G or CD115 surface marker expression after 7 days of culture. Comparison of transdifferentiated B-DKO and control cells did not reveal differences in the frequency of Ly-6G+ versus CD115+ cells in the CD11b+-gated cell population (Figure 3D), suggesting that the continuously expressed transgene product but not endogenous Cebpa and Cebpb predominantly affected myeloid cell lineage choice.
C/EBP Dosage and Granulocyte-Macrophage Lineage Choice
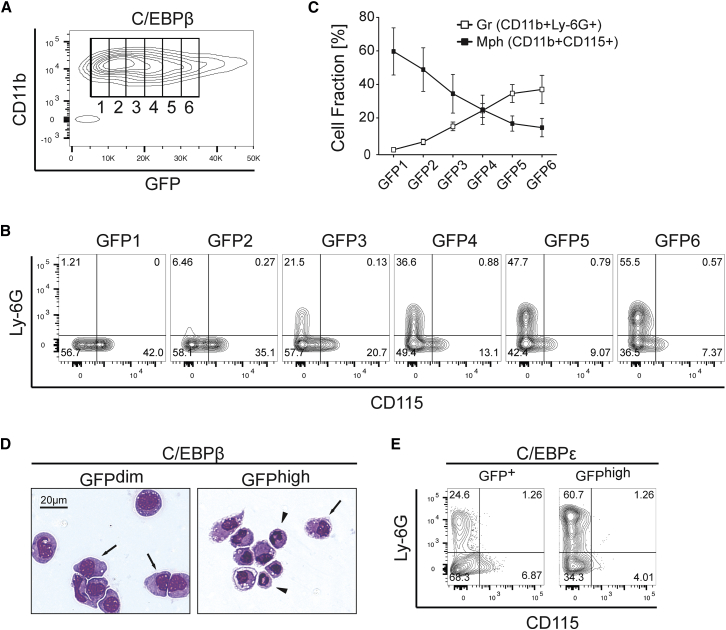
Transcription factor dosage is an important determinant in hematopoietic lineage choice (Dahl et al., 2003, DeKoter et al., 2007, DeKoter and Singh, 2000, Di Tullio and Graf, 2012, Ma et al., 2014, Rosenbauer et al., 2004, Simmons et al., 2012). We therefore examined how the level of C/EBP transgene expression would alter lympho-myeloid cell fate. As shown in Figures S3A and S3B, GFP intensity directly correlated with retroviral C/EBP protein expression and enabled us to examine transcription factor dosage effects on lympho-myeloid transdifferentiation. Based on increasing GFP intensity, C/EBPβ-transduced B cells were divided into six fractions, GFP1–6 (Figure 4A). Macrophage and granulocyte surface marker analysis of the CD11b+ cells revealed a bipartite profile when all GFP+ cells were included in the analysis (as in Figure 1C). As shown in Figures 4B and 4C, the distribution of CD11b+Ly-6G+ versus CD11b+CD115+ shifted according to the GFP intensity. GFPdim fractions were preferentially CD11b+CD115−/+ and Ly-6G−. The CD11b+Ly-6G+ cell population expanded with increasing GFP intensity, while the CD11b+CD115+ cell fraction diminished. The fraction with the highest GFP intensity (GFP6) consisted mostly of CD11b+Ly-6G+ cells (Figures 4B and 4C). Cytospin preparations of GFPdim- and GFPhigh-sorted cells showed that the GFPdim fraction consisted of large cells, characteristic of monocyte/macrophages, while the GFPhigh fraction contained cells with indented and segmented nuclei—typical features of mature granulocytes (Figure 4D). Finally, B cells transduced with C/EBPε were separated by flow cytometry according to their GFP intensity 24 hr post infection. Six days later, cell-surface marker expression analysis indicated that the GFPhigh cells were enriched for Ly-6G+ versus CD115+ cells when compared with the GFP+ culture. These data indicate a prospective cell-type outcome according to the initial C/EBP dosage (Figure 4E). To further exclude artifacts produced by the bicistronic C/EBP-IRES-EGFP MIG constructs, we examined the dosage effect using retroviral vectors with direct C/EBPε-GFP fusion. As shown in Figure S3C, higher GFP intensity correlated with the G/M surface marker profile. Taken together, these findings show that the dosage of C/EBPβ or C/EBPε transcription factors determines granulocyte versus macrophage cell-fate outcome during lympho-myeloid transdifferentiation.
Figure 4.
C/EBPβ Transgene Dosage Affects Macrophage versus Granulocyte Lineage Outcome
(A) Data binning of C/EBPβ-transduced B cells according to GFP intensity (GFP1–6).
(B) Flow cytometric analysis of GFP1–6 bins (CD11b + gated) 6 days after transduction.
(C) Quantification from five independent experiments. Data are presented as mean ± SEM.
(D) Cytospin preparation and May-Grünwald staining of GFPdim- and GFPhigh-sorted C/EBPβ-transduced B cells at day 7. Arrows indicate macrophage and arrowheads granulocyte-like cells.
(E) B cells were transduced with C/EBPε, sorted after 24 hr as GFP+ or GFPhigh, and recultured. Flow cytometric analysis of granulocyte (Ly-6G+) versus macrophage (CD115+) markers in GFP+CD11b+-gated populations 6 days post sorting.
See also Figure S3.
Long-Term Proliferating C/EBPβ-Lympho-Myeloid Progeny Display a GMP-like State and Spontaneously Differentiate into Granulocytes and Macrophages
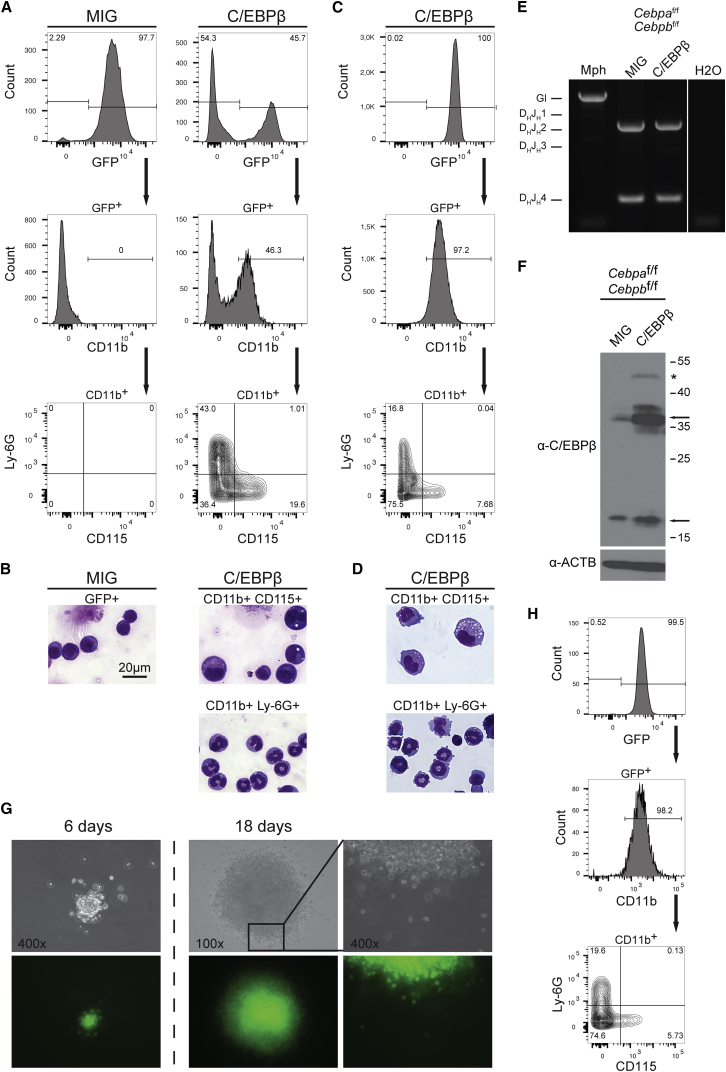
To assess the proliferation and lineage-forming capacity of C/EBPβ-transdifferentiated B cells, we aimed to establish a stably converted B cell line. After the removal of β-mercaptoethanol, an essential culture supplement for B cell maintenance, all B cells perished within a few days in C/EBPβ-transduced and empty vector-transduced cells. In contrast to the controls, however, proliferating GFP+ cells emerged from C/EBPβ-transduced B cells within 2 weeks after infection. Comparison of surface marker expression after 8 days and 6 weeks of cell culture showed that the long-term proliferating cells maintained both macrophage and granulocyte differentiation potential (Figures 5A and 5C). Cytospin preparations from CD11b+Ly-6G+ and CD11b+CD115+ sorted cells resembled typical granulocytes and macrophages, respectively (Figures 5B and 5D). Genotyping of rearranged immunoglobulin heavy chain (IgH) locus of C/EBPβ-transduced cells ruled out the possibility of a contamination with myeloid remnants from initiating cultures (Figure 5E). Immunoblot analysis showed that transgenic C/EBPβ protein is present and that endogenous C/EBPβ protein isoforms are also upregulated in transdifferentiated cells (Figure 5F). In addition, C/EBPβ-transduced cells grew in a semi-adherent fashion and showed irregular morphological features typical of myeloid progenitor cells, in contrast to round, refractile B cells. Upon treatment with recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF) the proliferation rate decreased and cells became adherent, large, and motile, resembling activated macrophages and suggesting that the transdifferentiated cells respond to myeloid signaling pathway activation (Figures S4A and S4B).
Figure 5.
Long-Term Proliferating GMP-like Cells Derived from C/EBPβ-Transduced B Cells
(A) Cebpaf/f;Cebpbf/f B cells transduced with C/EBPβ or vector control (MIG), sorted after 24 hr, and recultured. Flow cytometric analysis of myeloid surface markers was performed after 6 days. Gating strategy is indicated by arrows.
(B) May-Grünwald staining of macrophage (CD11b+CD115+) or granulocyte (CD11b+Ly-6G+) marker-sorted C/EBPβ-transduced cells from (A) and GFP+-purified empty vector (MIG)-transduced cells as control.
(C and D) Flow cytometry analysis (C) and May-Grünwald staining (D) according to (A) and (B) of non-purified C/EBPβ-transduced cells 6 weeks after removal of β-mercaptoethanol.
(E) Analysis of immunoglobulin heavy chain (IgH) rearrangement of long-term C/EBPβ-transduced cells or MIG-transduced controls. Monocytic Raw264.7 (Mph) cells served as a control for IgH germ line (Gl) configuration.
(F) Immunoblot analysis of C/EBPβ from protein lysates of long-term C/EBPβ- or empty vector (MIG)-transduced B cells. Arrows indicate C/EBP isoforms; asterisk indicates the size of transgene.
(G) Colony formation from single cells. Phase contrast (top) and fluorescent signal (bottom) micrographs of a C/EBPβ-transdifferentiated B cell colony. Clonogenic growth is shown at days 6 and 18. Magnification of inset area focuses on the border of the colony.
(H) Flow cytometric characterization of the recultured cell clone derived from (G) using myeloid markers.
See also Figure S5.
The G/M bilineage phenotype of the transdifferentiated cells was maintained during more than 6 months of continuous cell culture. Nevertheless, C/EBPβ may stimulate B cells to transdifferentiate into distinct G and M progenitors either directly or through a common GMP cell type. To distinguish between these possibilities, we seeded 4,500 of the stably transdifferentiated cells into semi-solid medium to derive colonies from single cells. Approximately 40–70 colonies were obtained in a single experiment. Individual GFP+ colonies were inspected from day 6 to day 18 and displayed a compact core and a periphery of larger motile, oval cells, reminiscent of colony-forming unit GM colonies (Figure 5G). Two colonies were isolated and propagated in liquid culture and both displayed a Ly-6G+ and CD115+ phenotype indicative of GMP-like potential (Figure 5H, result for one clone is shown). Subsequent recloning of one of the isolates in semi-solid medium resulted in stably growing secondary subclones (cloning efficiency between ∼4% and 6%) and persistence of the GMP-like phenotype of all 17 subclones examined, as shown in Figure S5.
We conclude that C/EBPβ-induced lympho-myeloid conversion confers a selective advantage by abrogating the dependence on B cell survival conditions. These C/EBPβ-converted cells acquire cytokine-independent long-term proliferation capacity and a robust GMP-like potential.
Discussion
Transcription factor-mediated conversion of one cell type into another is an important avenue by which we can advance our understanding of instructive mechanisms of cellular lineage commitment and differentiation. The primary B cells immortalized by v-Abl provide an experimental system to combine B cell mouse genetics and ectopic gene expression for the exploration of progenitor biology, lympho-myeloid lineage plasticity, maintenance of cell identity, and regulatory mechanisms of myeloid lineage commitment.
Experimental transdifferentiation of B cells by ectopic expression of conditional C/EBPα has shed light on the molecular dynamics of the conversion of lymphoid cells into inflammatory macrophages (Bussmann et al., 2009, Di Tullio et al., 2011, Krijger et al., 2016, Laiosa et al., 2006, Rodriguez-Ubreva et al., 2012, Rodriguez-Ubreva et al., 2014, van Oevelen et al., 2015, Xie et al., 2004). However, lympho-myeloid transdifferentiation may not be restricted to activated macrophages but may yield other myeloid cell types depending on the C/EBP structure and post-translational modification pattern (Stoilova et al., 2013). Here, we assessed the lympho-myeloid transdifferentiation potential of C/EBP α, β, δ, and ε in primary v-Abl-immortalized B cells. Our data show that v-Abl B cells represent a robust in vitro system for experimental cell lineage switch and may also be used for targeted murine genetics and in vitro genetic manipulation at the B cell stage.
Lineage conversion could be achieved using any of the four C/EBP members, revealing a common gene signature, and their common potential to extinguish B cell identity and induce the myeloid cell fate. C/EBPβ and C/EBPε induced granulocytic differentiation in addition to monocytic transdifferentiation. Granulopoietic capacity of C/EBPβ has previously been documented in emergency granulopoiesis (Hirai et al., 2006) and by restoring Cebpa deficiency (Jones et al., 2002). The lack of granulopoietic transdifferentiation potential of C/EBPα was surprising and has been discussed previously (Xie et al., 2004), because C/EBPα induced granulocyte differentiation in a myeloid progenitor cell line (Radomska et al., 1998, Suh et al., 2006). Although we do not know why C/EBPα and C/EBPδ fail to induce granulocyte formation, the strong proliferation arrest elicited by C/EBPα could account for the failure of granulocytic conversion. On the other hand, our data may also raise the possibility that some of the functions of C/EBPα reported in established progenitor cell lines may depend on the activation of endogenous C/EBPβ. Nevertheless, the v-Abl B cell system could be useful to resolve quantitative and qualitative C/EBP functions and help uncover mechanisms of lineage specification, e.g., by C/EBP domain-swap experiments and mutational analysis.
Our findings support the concept that subtle variations in transcription factor levels have an impact on hematopoietic cell lineage decisions. In addition to transcription factor dosage, specificity and functional redundancy play major roles; high levels of C/EBPβ or C/EBPε consistently increased the granulocyte over macrophage ratio. The exact interplay between C/EBP quantity and qualitative specificity remains to be explored, but is likely similar to graded PU.1 expression that has an impact on lympho-myeloid specification by occupancy of low-affinity binding sites only at high transcription factor concentrations (DeKoter and Singh, 2000, Pham et al., 2013). Relative expression levels of PU.1 and C/EBPα are also key determinants in granulocyte versus macrophage specification. Higher PU.1 levels promote macrophage differentiation, while higher C/EBPα levels lead to granulopoiesis (Dahl et al., 2003, Ma et al., 2014). Therefore, less accessible “granulocytic genes” may require higher C/EBP concentrations in addition to other features of C/EBPβ and C/EBPε proteins.
Activation of endogenous C/EBP genes and impaired lineage conversion after their removal suggests that autoregulatory loops are involved in the kinetics of lineage conversion (Bussmann et al., 2009, Lu et al., 2009). However, deletion of endogenous C/EBPα and C/EBPβ appears to affect the conversion efficiency but not the lineage outcome, suggesting a dominant and distinguished qualitative role for the constitutively expressed C/EBP transgene.
Leukemia with indifferent or promiscuous lineage association is frequently resistant to standard chemotherapy and may show changed lineages during relapse (Dorantes-Acosta and Pelayo, 2012, Golemovic et al., 2006, Matutes et al., 1997). Inter-myeloid and lympho-myeloid lineage ambiguity therefore emerges as an important pathological entity (Greaves et al., 1986, Smith et al., 1983). It is therefore conceivable that switching lineages reflect an epigenetic selection process in leukemogenesis associated with gain-of-function survival and proliferation capacity (Greaves et al., 1986, Janz et al., 2006, Regalo and Leutz, 2013). Our experimental data provide direct evidence that enhancing the selective pressure on B cells by removal of β-mercaptoethanol strongly promotes C/EBPβ-induced lympho-myeloid converts that remained immortalized and highly proliferative. We therefore conclude that cancerous B cells may escape from hostile conditions by lineage conversion through C/EBPβ activation. Furthermore, B cells could profit from lineage-foreign survival signals by aberrant expression of growth factor receptors, such as CSF1-R, a target gene of C/EBPβ (Lamprecht et al., 2010). Various C/EBP translocations have been associated with B cell-derived leukemia, implying their cancer-supporting function. However, the role played by transdifferentiation remains unknown (Akasaka et al., 2007, Chapiro et al., 2006, Slamova et al., 2013, Wiemels et al., 2015). Strikingly, DNA demethylation and upregulation of C/EBPα was suspected to be involved in lineage switching during leukemia progression (Slamova et al., 2013). In addition to pre-B cell lymphoma, v-Abl retroviral infection of murine bone marrow cells also evoked a chronic myeloid leukemia-like disease that involved immunoglobulin rearrangements (Kelliher et al., 1990).
In a recent study, pulsed C/EBPα expression was shown to induce a GMP-like state according to gene expression and chromatin accessibility data. However, during prolonged exposure only a macrophage fate was consolidated (Di Stefano et al., 2016). Remarkably, the stable C/EBPβ B cell converts reported here resembled self-renewing bipotential GMP progenitors that were responsive to GM-CSF but not dependent on cytokine signaling, and stably retained G/M differentiation potential. In the absence of cytokines, expression of macrophage- and granulocyte-specific markers emerged in a mutually exclusive fashion. These data suggest the involvement of C/EBPβ in maintenance, commitment, or both at the GMP stage.
The cytokine-independent, self-renewing, spontaneously differentiating GMP-like cells present a valuable experimental tool for the study of determinants of granulocyte versus macrophage specification by genetic manipulation and screening approaches. In addition, maintenance of the transdifferentiation capacity of the v-Abl B cells establishes a sound basis for using targeted genetic mouse models to examine signaling pathways and other instructive cues of cell specification. The fact that v-Abl B cells are amenable to DNA transfection permits experimental alteration at the B cell stage, such as stable transfection, gene deletion, or chromosomal engineering, and is an important technical advance in overcoming the hurdle of difficult genetic manipulation of myelomonocytic cells.
In summary, we present a robust in vitro lympho-myeloid transdifferentiation system to study determinants of lineage promiscuity, GMP progenitor biology, and macrophage versus granulocyte cell-fate decisions. In addition, our findings lend experimental evidence to lympho-myeloid transdifferentiation processes as a trait of lineage ambiguity in leukemia progression.
Experimental Procedures
Expression Constructs
C/EBP genes were inserted into pMSCV-IRES-EGFP (MIG) or pMSCV-IRES-BFP (MIB) retroviral vectors using XhoI/EcoRI restriction sites. C/EBPs lacking stop codons were cloned in-frame with the EGFP open reading frame and inserted into pMSCV for C/EBP-GFP fusion construct. Expression vectors encoding C/EBPα p42 isoform and C/EBPβ LAP∗ isoform were described previously (Stoilova et al., 2013). Murine C/EBPδ was isolated from pMEX-mC/EBPδ (a gift from Dr. E Sterneck). Codon-optimized human C/EBPε p32 isoform was commercially synthesized (MWG-Biotech) and subcloned from pEX-A2 into MIG or MIB using EcoRI sites flanking C/EBPε. The v-Abl-expressing pMSCV vector was a gift from Dr. F Melchers (Ohnishi and Melchers, 2003). For production of infectious supernatants the retroviral packaging cell line Plat-E was used. Infectious supernatants were collected 48 hr and 72 hr after transfection.
Generation of v-Abl-Transformed B Cells
Bone marrow cells were isolated from the femur and tibia of 8- to 9-week-old Cebpaf/f-crossed (Zhang et al., 2004) and Cebpbf/f-crossed (Sterneck et al., 2006) Cebpaf/f;Cebpbf/f mice and C57BL/6J wild-type controls. These mice were maintained and handled in compliance with protocols approved by the institutional Animal Care and Use Committee. After erythrolysis, bone marrow cells were transduced with v-Abl-expressing retroviral supernatants and 8 μg/mL hexadimethrine bromide in complete DMEM (10% fetal calf serum (FCS), 10 mM HEPES, and penicillin/streptomycin (Gibco)) supplemented with 50 μM β-mercaptoethanol. Bone marrow cells were washed on the following day and medium was changed every other day. A stable cell line with a pre-B cell-like phenotype (CD19+, c-kit−, CD25+, IgM−) emerged after 4 weeks of culture. For loxP site recombination, 5 × 105 B cells were washed three times in serum-free DMEM and incubated with purified TAT-Cre protein (50 μg/mL; a gift from Dr. K Rajewsky) in serum-free DMEM at 37°C for 45 min (Peitz et al., 2002). The cells were then washed and cultured for 24 hr and seeded subsequently as single-cell clones (by fluorescence-activated cell sorting (FACS)) into 96-well plates. Deletion of the Cebpa and Cebpb genes was checked by PCR to identify double-knockout clones. Primer sequences used are shown in Table S2.
Cell Lines
The monocytic/macrophage Raw264.7 cell line was cultured in complete DMEM. The fetal liver-derived pre-B cell line HAFTL1 was cultured in complete RPMI-1640 (10% FCS, 10 mM HEPES, penicillin/streptomycin (Gibco)) supplemented with 50 μM β-mercaptoethanol (Pierce and Aaronson, 1982). The HAFTL1-derived C10 clone (a gift from Dr. T Graf) containing a C/EBPα-ER_IRES_GFP transgene was cultured in complete phenol red-free RPMI-1640 (10% charcoal-stripped FCS (HyClone), 10 mM HEPES, penicillin/streptomycin (Gibco)) supplemented with 50 μM β-mercaptoethanol (Bussmann et al., 2009). Transdifferentiation of C10 cells was induced with 200 nM hydroxytamoxifen (Sigma). 293T-derived Plat-E line (Cell Biolabs) was used for packaging.
Retroviral Transduction
v-Abl-generated stable B cells or HAFTL1 cells (2 × 105/12-well or 1 × 106/6-well) were mixed with freshly prepared retroviral supernatants (1/3 of total volume) in complete medium supplemented with 8 μg/mL hexadimethrine bromide and 50 μM β-mercaptoethanol. Cells were centrifuged at 1,000 rpm without brakes for 10 min at room temperature and incubated overnight. Infected cells were washed and processed for flow cytometry or replated in 6-well plates (BD Falcon). For the generation of the long-term proliferating transdifferentiated cells, β-mercaptoethanol supplementation was omitted after 24 hr. Half of the medium was changed every other day.
Flow Cytometry
For flow cytometry or cell sorting, cells were transferred to 15-mL tubes and washed in cold buffer (2% FCS, 2 mM EDTA in PBS). Before antibody labeling, cells were incubated with Fc-Block (1:200, anti-CD16/32 antibody; 2.4G2, BD Pharmingen) for 10 min at 4°C and washed. Cells were stained in buffer with (1:100) fluorescently labeled antibodies against CD11b (clone M1/70, BD Pharmingen), CD11c (clone N418, Biolegend), CD115 (clone AFS98, eBioscience), CD19 (clone 1D3, BD Pharmingen), and Ly-6G (1A8, Biolegend) for 30 min at 4°C in the dark. 7-AAD (BD Pharmingen) was added before measurement to exclude dead cells. Analysis was performed on LSRFortessa and sorting on FACSAria II or III (BD Pharmingen). Unstained, empty vector-transduced cells or fluorescence-minus-one staining setups served as controls.
PCR
Total RNA was prepared using TriPure (Roche) following the manufacturer's instructions. DNA contamination was removed by DNase I treatment (Roche). For standard cDNA synthesis the RevertAid First Strand Synthesis Kit (Thermo Scientific) was used. Ten nanograms of cDNA was applied to PCR according to the MangoTaq kit protocol (Bioline). PCR reaction was subjected to 94°C for 3 min and 27 cycles of 94°C for 30 s, 56°C for 30 s, 72°C for 30 s, and a final extension step at 72°C for 5 min on a Mastercycler Pro (Eppendorf). For detection of DH-JH IgH rearrangements, DNA was prepared as described by Truett et al. (2000) and subjected to PCR conditions as reported by Ehlich et al. (1994), using the combination of primers DQ52 FW1, DFS FW2, and JH4A RV. Cebpa and Cebpb genotypes were assessed using “-flox” or “-Δ” primer pairs, respectively.
Immunoblotting
Total protein lysates were prepared by incubation of snap-frozen cell pellets with lysis buffer (50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% Nonidet P-40, 1 mM EDTA, 10% glycerol, and proteinase inhibitor cocktail [Roche] in H2O) for 30 min on ice. After centrifugation protein concentration was measured in supernatants using Pierce Reagent (Thermo Scientific). Protein lysates were subjected to electrophoresis and transferred to a nitrocellulose membrane on the Trans-Blot Turbo System (Bio-Rad). Specific protein signals were detected by incubation with antibodies against C/EBPα (14AA, Santa Cruz Biotechnology), C/EBPβ (C-19, Santa Cruz), C/EBPδ (C-22, Santa Cruz), C/EBPε (C-22, Santa Cruz), GFP (7.1/13.1, Roche), and ACTB (AC-15, Sigma).
NanoString Analysis
Total RNA from freshly FACS-enriched cells or cell cultures was prepared and quality checked on a 2100 Bioanalyzer (Agilent Technologies). Detection of mouse protein-coding transcripts was performed using a predefined panel (GXA-MIM1-12, nCounter), containing 547 probes against immunology-related mRNAs and 14 probes against housekeeping genes (G6pdx, Hprt, Gapdh, Alas1, Oaz1, Sdha, Rpl19, Eef1g, Tbp, Ppia, Polr2a, Gusb, Tubb5, and Polr1b) for normalization. Primary data were processed using default settings of the nSolver software. Genes were defined as “upregulated” or “downregulated” if normalized expression values were increased or decreased at least 2-fold compared with empty vector-transduced controls.
Statistical Analysis
Processed data were visualized using GraphPad Prism software (Version 5.0a). Error bars represent the SEM. Hypothesis testing was performed from at least three independent experiments using the t-test function (two-tailed) in Rstudio (R version 3.2.1, R Foundation). p Values of <0.05 were defined as significant and p < 0.01 highly significant.
Author Contributions
B.C. conceived the methodology, performed the experiments, analyzed the data, and wrote the draft. V.B. provided Cebpaf/f;Cebpbf/f mouse resources and experimental advice. E.K.-L. generated CEBP-EGFP fusion construct resources, and J.S. and C.K. validated experiments. V.B., J.I., N.P., J.S., and E.K.-L. were involved in reviewing the manuscript. B.C. and A.L. conceptualized the work and wrote the manuscript. A.L. was responsible for supervision, project administration, and funding acquisition.
Acknowledgments
We thank Dr. B Niesler and R. Roeth of the nCounter Core Facility, Heidelberg, for providing services; Dr. H.-P. Rahn and K. Rautenberg (MDC, Berlin, Germany) for flow cytometry support; Dr. T. Graf (CRG, Barcelona, Spain) for the C10 cell line; M. Hofstätter and J. Bergemann (MDC, Berlin, Germany) for technical assistance; and Dr. E. Sterneck (NCI, Frederick, MD), Dr. F. Melchers (MPI for Infection Biology, Berlin, Germany), and Dr. K. Rajewsky (MDC, Berlin, Germany) for reagents and mice.
Published: January 19, 2017
Footnotes
Supplemental Information includes five figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.12.015.
Supplemental Information
References
- Akagi T., Thoennissen N.H., George A., Crooks G., Song J.H., Okamoto R., Nowak D., Gombart A.F., Koeffler H.P. In vivo deficiency of both C/EBPbeta and C/EBPepsilon results in highly defective myeloid differentiation and lack of cytokine response. PLoS One. 2010;5:e15419. doi: 10.1371/journal.pone.0015419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akasaka T., Balasas T., Russell L.J., Sugimoto K.J., Majid A., Walewska R., Karran E.L., Brown D.G., Cain K., Harder L. Five members of the CEBP transcription factor family are targeted by recurrent IGH translocations in B-cell precursor acute lymphoblastic leukemia (BCP-ALL) Blood. 2007;109:3451–3461. doi: 10.1182/blood-2006-08-041012. [DOI] [PubMed] [Google Scholar]
- Barneda-Zahonero B., Roman-Gonzalez L., Collazo O., Rafati H., Islam A.B., Bussmann L.H., di Tullio A., De Andres L., Graf T., Lopez-Bigas N. HDAC7 is a repressor of myeloid genes whose downregulation is required for transdifferentiation of pre-B cells into macrophages. PLoS Genet. 2013;9:e1003503. doi: 10.1371/journal.pgen.1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussmann L.H., Schubert A., Vu Manh T.P., De Andres L., Desbordes S.C., Parra M., Zimmermann T., Rapino F., Rodriguez-Ubreva J., Ballestar E. A robust and highly efficient immune cell reprogramming system. Cell Stem Cell. 2009;5:554–566. doi: 10.1016/j.stem.2009.10.004. [DOI] [PubMed] [Google Scholar]
- Chapiro E., Russell L., Radford-Weiss I., Bastard C., Lessard M., Struski S., Cave H., Fert-Ferrer S., Barin C., Maarek O. Overexpression of CEBPA resulting from the translocation t(14;19)(q32;q13) of human precursor B acute lymphoblastic leukemia. Blood. 2006;108:3560–3563. doi: 10.1182/blood-2006-03-010835. [DOI] [PubMed] [Google Scholar]
- Chen S.S., Chen J.F., Johnson P.F., Muppala V., Lee Y.H. C/EBPbeta, when expressed from the C/ebpalpha gene locus, can functionally replace C/EBPalpha in liver but not in adipose tissue. Mol. Cell. Biol. 2000;20:7292–7299. doi: 10.1128/mcb.20.19.7292-7299.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloutier A., Guindi C., Larivee P., Dubois C.M., Amrani A., McDonald P.P. Inflammatory cytokine production by human neutrophils involves C/EBP transcription factors. J. Immunol. 2009;182:563–571. doi: 10.4049/jimmunol.182.1.563. [DOI] [PubMed] [Google Scholar]
- Cobaleda C., Busslinger M. Developmental plasticity of lymphocytes. Curr. Opin. Immunol. 2008;20:139–148. doi: 10.1016/j.coi.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Dahl R., Walsh J.C., Lancki D., Laslo P., Iyer S.R., Singh H., Simon M.C. Regulation of macrophage and neutrophil cell fates by the PU.1:C/EBPalpha ratio and granulocyte colony-stimulating factor. Nat. Immunol. 2003;4:1029–1036. doi: 10.1038/ni973. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.1. Science. 2000;288:1439–1441. doi: 10.1126/science.288.5470.1439. [DOI] [PubMed] [Google Scholar]
- DeKoter R.P., Kamath M.B., Houston I.B. Analysis of concentration-dependent functions of PU.1 in hematopoiesis using mouse models. Blood Cells Mol. Dis. 2007;39:316–320. doi: 10.1016/j.bcmd.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano B., Collombet S., Jakobsen J.S., Wierer M., Sardina J.L., Lackner A., Stadhouders R., Segura-Morales C., Francesconi M., Limone F. C/EBPalpha creates elite cells for iPSC reprogramming by upregulating Klf4 and increasing the levels of Lsd1 and Brd4. Nat. Cell Biol. 2016;18:371–381. doi: 10.1038/ncb3326. [DOI] [PubMed] [Google Scholar]
- Di Tullio A., Graf T. C/EBPalpha bypasses cell cycle-dependency during immune cell transdifferentiation. Cell Cycle. 2012;11:2739–2746. doi: 10.4161/cc.21119. [DOI] [PubMed] [Google Scholar]
- Di Tullio A., Manh T.P., Schubert A., Mansson R., Graf T. CCAAT/enhancer binding protein alpha (C/EBP(alpha))-induced transdifferentiation of pre-B cells into macrophages involves no overt retrodifferentiation. Proc. Natl. Acad. Sci. USA. 2011;108:17016–17021. doi: 10.1073/pnas.1112169108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorantes-Acosta E., Pelayo R. Lineage switching in acute leukemias: a consequence of stem cell plasticity? Bone Marrow Res. 2012;2012:406796. doi: 10.1155/2012/406796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlich A., Martin V., Muller W., Rajewsky K. Analysis of the B-cell progenitor compartment at the level of single cells. Curr. Biol. 1994;4:573–583. doi: 10.1016/s0960-9822(00)00129-9. [DOI] [PubMed] [Google Scholar]
- Feng R., Desbordes S.C., Xie H., Tillo E.S., Pixley F., Stanley E.R., Graf T. PU.1 and C/EBPalpha/beta convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. USA. 2008;105:6057–6062. doi: 10.1073/pnas.0711961105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golemovic M., Sucic M., Zadro R., Mrsic S., Mikulic M., Labar B., Rajic L.J., Batinic D. IgH and TCRgamma gene rearrangements, cyclin A1 and HOXA9 gene expression in biphenotypic acute leukemias. Leuk. Res. 2006;30:211–221. doi: 10.1016/j.leukres.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Graf T. Immunology: blood lines redrawn. Nature. 2008;452:702–703. doi: 10.1038/452702a. [DOI] [PubMed] [Google Scholar]
- Graf T., Enver T. Forcing cells to change lineages. Nature. 2009;462:587–594. doi: 10.1038/nature08533. [DOI] [PubMed] [Google Scholar]
- Greaves M.F., Chan L.C., Furley A.J., Watt S.M., Molgaard H.V. Lineage promiscuity in hemopoietic differentiation and leukemia. Blood. 1986;67:1–11. [PubMed] [Google Scholar]
- Hirai H., Zhang P., Dayaram T., Hetherington C.J., Mizuno S., Imanishi J., Akashi K., Tenen D.G. C/EBPbeta is required for 'emergency' granulopoiesis. Nat. Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- Holmes K.L., Pierce J.H., Davidson W.F., Morse H.C., 3rd Murine hematopoietic cells with pre-B or pre-B/myeloid characteristics are generated by in vitro transformation with retroviruses containing fes, ras, abl, and src oncogenes. J. Exp. Med. 1986;164:443–457. doi: 10.1084/jem.164.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janz M., Dorken B., Mathas S. Reprogramming of B lymphoid cells in human lymphoma pathogenesis. Cell Cycle. 2006;5:1057–1061. doi: 10.4161/cc.5.10.2737. [DOI] [PubMed] [Google Scholar]
- Jones L.C., Lin M.L., Chen S.S., Krug U., Hofmann W.K., Lee S., Lee Y.H., Koeffler H.P. Expression of C/EBPbeta from the C/ebpalpha gene locus is sufficient for normal hematopoiesis in vivo. Blood. 2002;99:2032–2036. doi: 10.1182/blood.v99.6.2032. [DOI] [PubMed] [Google Scholar]
- Kallin E.M., Rodriguez-Ubreva J., Christensen J., Cimmino L., Aifantis I., Helin K., Ballestar E., Graf T. Tet2 facilitates the derepression of myeloid target genes during CEBPalpha-induced transdifferentiation of pre-B cells. Mol. Cell. 2012;48:266–276. doi: 10.1016/j.molcel.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelliher M.A., McLaughlin J., Witte O.N., Rosenberg N. Induction of a chronic myelogenous leukemia-like syndrome in mice with v-abl and BCR/ABL. Proc. Natl. Acad. Sci. USA. 1990;87:6649–6653. doi: 10.1073/pnas.87.17.6649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger P.H., Di Stefano B., de Wit E., Limone F., van Oevelen C., de Laat W., Graf T. Cell-of-origin-specific 3D genome structure acquired during somatic cell reprogramming. Cell Stem Cell. 2016;18:597–610. doi: 10.1016/j.stem.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laiosa C.V., Stadtfeld M., Xie H., de Andres-Aguayo L., Graf T. Reprogramming of committed T cell progenitors to macrophages and dendritic cells by C/EBP alpha and PU.1 transcription factors. Immunity. 2006;25:731–744. doi: 10.1016/j.immuni.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Lamprecht B., Walter K., Kreher S., Kumar R., Hummel M., Lenze D., Kochert K., Bouhlel M.A., Richter J., Soler E. Derepression of an endogenous long terminal repeat activates the CSF1R proto-oncogene in human lymphoma. Nat. Med. 2010;16:571–579. doi: 10.1038/nm.2129. 1p following 579. [DOI] [PubMed] [Google Scholar]
- Litvak V., Ramsey S.A., Rust A.G., Zak D.E., Kennedy K.A., Lampano A.E., Nykter M., Shmulevich I., Aderem A. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat. Immunol. 2009;10:437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y.C., Kim I., Lye E., Shen F., Suzuki N., Suzuki S., Gerondakis S., Akira S., Gaffen S.L., Yeh W.C., Ohashi P.S. Differential role for c-Rel and C/EBPbeta/delta in TLR-mediated induction of proinflammatory cytokines. J. Immunol. 2009;182:7212–7221. doi: 10.4049/jimmunol.0802971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma O., Hong S., Guo H., Ghiaur G., Friedman A.D. Granulopoiesis requires increased C/EBPalpha compared to monopoiesis, correlated with elevated cebpa in immature G-CSF receptor versus M-CSF receptor expressing cells. PLoS One. 2014;9:e95784. doi: 10.1371/journal.pone.0095784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matutes E., Morilla R., Farahat N., Carbonell F., Swansbury J., Dyer M., Catovsky D. Definition of acute biphenotypic leukemia. Haematologica. 1997;82:64–66. [PubMed] [Google Scholar]
- Ness S.A., Kowenz-Leutz E., Casini T., Graf T., Leutz A. Myb and NF-M: combinatorial activators of myeloid genes in heterologous cell types. Genes Dev. 1993;7:749–759. doi: 10.1101/gad.7.5.749. [DOI] [PubMed] [Google Scholar]
- Ohnishi K., Melchers F. The nonimmunoglobulin portion of lambda5 mediates cell-autonomous pre-B cell receptor signaling. Nat. Immunol. 2003;4:849–856. doi: 10.1038/ni959. [DOI] [PubMed] [Google Scholar]
- Peitz M., Pfannkuche K., Rajewsky K., Edenhofer F. Ability of the hydrophobic FGF and basic TAT peptides to promote cellular uptake of recombinant Cre recombinase: a tool for efficient genetic engineering of mammalian genomes. Proc. Natl. Acad. Sci. USA. 2002;99:4489–4494. doi: 10.1073/pnas.032068699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T.H., Minderjahn J., Schmidl C., Hoffmeister H., Schmidhofer S., Chen W., Langst G., Benner C., Rehli M. Mechanisms of in vivo binding site selection of the hematopoietic master transcription factor PU.1. Nucleic Acids Res. 2013;41:6391–6402. doi: 10.1093/nar/gkt355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce J.H., Aaronson S.A. Balb- and Harvey-murine sarcoma virus transformation of a novel lymphoid progenitor cell. J. Exp. Med. 1982;156:873–887. doi: 10.1084/jem.156.3.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radomska H.S., Huettner C.S., Zhang P., Cheng T., Scadden D.T., Tenen D.G. CCAAT/enhancer binding protein alpha is a regulatory switch sufficient for induction of granulocytic development from bipotential myeloid progenitors. Mol. Cell. Biol. 1998;18:4301–4314. doi: 10.1128/mcb.18.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regalo G., Leutz A. Hacking cell differentiation: transcriptional rerouting in reprogramming, lineage infidelity and metaplasia. EMBO Mol. Med. 2013;5:1154–1164. doi: 10.1002/emmm.201302834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ubreva J., Ciudad L., Gomez-Cabrero D., Parra M., Bussmann L.H., di Tullio A., Kallin E.M., Tegner J., Graf T., Ballestar E. Pre-B cell to macrophage transdifferentiation without significant promoter DNA methylation changes. Nucleic Acids Res. 2012;40:1954–1968. doi: 10.1093/nar/gkr1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ubreva J., Ciudad L., van Oevelen C., Parra M., Graf T., Ballestar E. C/EBPa-mediated activation of miR-34a and miR-223 inhibits Lef1 expression to achieve efficient reprogramming into macrophages. Mol. Cell. Biol. 2014;34:1145–1157. doi: 10.1128/MCB.01487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbauer F., Wagner K., Kutok J.L., Iwasaki H., Le Beau M.M., Okuno Y., Akashi K., Fiering S., Tenen D.G. Acute myeloid leukemia induced by graded reduction of a lineage-specific transcription factor, PU.1. Nat. Genet. 2004;36:624–630. doi: 10.1038/ng1361. [DOI] [PubMed] [Google Scholar]
- Rosenberg N., Baltimore D., Scher C.D. In vitro transformation of lymphoid cells by Abelson murine leukemia virus. Proc. Natl. Acad. Sci. USA. 1975;72:1932–1936. doi: 10.1073/pnas.72.5.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott L.M., Civin C.I., Rorth P., Friedman A.D. A novel temporal expression pattern of three C/EBP family members in differentiating myelomonocytic cells. Blood. 1992;80:1725–1735. [PubMed] [Google Scholar]
- Shore S.K., Tantravahi R.V., Reddy E.P. Transforming pathways activated by the v-Abl tyrosine kinase. Oncogene. 2002;21:8568–8576. doi: 10.1038/sj.onc.1206084. [DOI] [PubMed] [Google Scholar]
- Simmons S., Knoll M., Drewell C., Wolf I., Mollenkopf H.J., Bouquet C., Melchers F. Biphenotypic B-lymphoid/myeloid cells expressing low levels of Pax5: potential targets of BAL development. Blood. 2012;120:3688–3698. doi: 10.1182/blood-2012-03-414821. [DOI] [PubMed] [Google Scholar]
- Slamova L., Starkova J., Fronkova E., Zaliova M.K., Reznickova L.R., van Delft F.W., Vodickova E., Volejnikova J., Zemanova Z., Polgarova K. CD2-positive B-cell precursor acute lymphoblastic leukemia with an early switch to the monocytic lineage. Leukemia. 2013;28:609–620. doi: 10.1038/leu.2013.354. [DOI] [PubMed] [Google Scholar]
- Smith L.J., Curtis J.E., Messner H.A., Senn J.S., Furthmayr H., McCulloch E.A. Lineage infidelity in acute leukemia. Blood. 1983;61:1138–1145. [PubMed] [Google Scholar]
- Sterneck E., Zhu S., Ramirez A., Jorcano J.L., Smart R.C. Conditional ablation of C/EBP beta demonstrates its keratinocyte-specific requirement for cell survival and mouse skin tumorigenesis. Oncogene. 2006;25:1272–1276. doi: 10.1038/sj.onc.1209144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoilova B., Kowenz-Leutz E., Scheller M., Leutz A. Lymphoid to myeloid cell trans-differentiation is determined by C/EBPbeta structure and post-translational modifications. PLoS One. 2013;8:e65169. doi: 10.1371/journal.pone.0065169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh H.C., Gooya J., Renn K., Friedman A.D., Johnson P.F., Keller J.R. C/EBPalpha determines hematopoietic cell fate in multipotential progenitor cells by inhibiting erythroid differentiation and inducing myeloid differentiation. Blood. 2006;107:4308–4316. doi: 10.1182/blood-2005-06-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Akira S., Yoshida K., Umemoto M., Yoneda Y., Shirafuji N., Fujiwara H., Suematsu S., Yoshida N., Kishimoto T. Targeted disruption of the NF-IL6 gene discloses its essential role in bacteria killing and tumor cytotoxicity by macrophages. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- Truett G.E., Heeger P., Mynatt R.L., Truett A.A., Walker J.A., Warman M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT) Biotechniques. 2000;29:52–54. doi: 10.2144/00291bm09. [DOI] [PubMed] [Google Scholar]
- Tsukada J., Yoshida Y., Kominato Y., Auron P.E. The CCAAT/enhancer (C/EBP) family of basic-leucine zipper (bZIP) transcription factors is a multifaceted highly-regulated system for gene regulation. Cytokine. 2011;54:6–19. doi: 10.1016/j.cyto.2010.12.019. [DOI] [PubMed] [Google Scholar]
- van Oevelen C., Collombet S., Vicent G., Hoogenkamp M., Lepoivre C., Badeaux A., Bussmann L., Sardina J.L., Thieffry D., Beato M. C/EBPalpha activates pre-existing and de novo macrophage enhancers during induced pre-B cell transdifferentiation and myelopoiesis. Stem Cell Rep. 2015;5:232–247. doi: 10.1016/j.stemcr.2015.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemels J.L., de Smith A.J., Xiao J., Lee S.T., Muench M.O., Fomin M.E., Zhou M., Hansen H.M., Termuhlen A., Metayer C. A functional polymorphism in the CEBPE gene promoter influences acute lymphoblastic leukemia risk through interaction with the hematopoietic transcription factor Ikaros. Leukemia. 2015;30:1194–1197. doi: 10.1038/leu.2015.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Ye M., Feng R., Graf T. Stepwise reprogramming of B cells into macrophages. Cell. 2004;117:663–676. doi: 10.1016/s0092-8674(04)00419-2. [DOI] [PubMed] [Google Scholar]
- Yamanaka R., Barlow C., Lekstrom-Himes J., Castilla L.H., Liu P.P., Eckhaus M., Decker T., Wynshaw-Boris A., Xanthopoulos K.G. Impaired granulopoiesis, myelodysplasia, and early lethality in CCAAT/enhancer binding protein epsilon-deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:13187–13192. doi: 10.1073/pnas.94.24.13187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D.E., Zhang P., Wang N.D., Hetherington C.J., Darlington G.J., Tenen D.G. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc. Natl. Acad. Sci. USA. 1997;94:569–574. doi: 10.1073/pnas.94.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M.L., Dayaram T., Owens B.M., Shigematsu H., Levantini E., Huettner C.S., Lekstrom-Himes J.A. Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBP alpha. Immunity. 2004;21:853–863. doi: 10.1016/j.immuni.2004.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.