Abstract
Membrane-spanning 4-domains, subfamily A, member 6A (MS4A6A) has been identified as susceptibility loci of Alzheimer's disease (AD) by several recent genome-wide association studies (GWAS), whereas little is known about the potential roles of these variants in the brain structure and function of AD. In this study, we included a total of 812 individuals from the Alzheimer's disease Neuroimaging Initiative (ADNI) database. Using multiple linear regression models, we found MS4A6A genotypes were strongly related to atrophy rate of left middle temporal (rs610932: Pc = 0.017, rs7232: Pc = 0.022), precuneus (rs610932: Pc = 0.015) and entorhinal (rs610932, Pc = 0.022) on MRI in the entire group. In the subgroup analysis, MS4A6A SNPs were significantly accerlated the percentage of volume loss of middle temporal, precuneus and entorhinal, especially in the MCI subgroup. These findings reveal that MS4A6A genotypes affect AD specific brain structures which supported the possible role of MS4A6A polymorphisms in influencing AD-related neuroimaging phenotypes.
Keywords: Alzheimer's disease, MS4A6A, polymorphisms, phenotypes, brain structure, Gerotarget
INTRODUCTION
As the most common neurodegenerative disorder in the elderly, Alzheimer's disease (AD) is becoming a major challenge to the global health care system with the progressive increase burden on caregivers and society in most countries [1, 2]. Cognitive screening and detailed neuropsychological assessment still were the foundation of the diagnosis for AD dementia, but by the time the patient was diagnosed, the disease had been progressing for many years [3]. Recently, the latest guidelines on diagnosis of AD suggested that histological pathology (such as amyloid plaques, neurofibrillary tangles and so on) should be detected by specific abnormality on various biomarkers [4]. To date, multiple neuroimaging measures, including position emission tomography (PET), structural magnetic resonance imaging (MRI) and Pittsburgh Compound B position emission tomography (PiB-PET), had been used to detect the presence or absence of AD pathology [5-8], on account of they may infer the clinical progression in normal aging and MCI. Adult twin-studies showed these measures appeared to be explained by genetic factors with high heritability [9]. In addition, there is convincing evidence that these neuroimaging traits also be affected by genetic risk factors for AD which further confirm the important function for these genetic factors and suggest possible mechanisms through that they might be playing for AD.
Numbers of genome-wide association study (GWAS)-validated or GWAS-promising candidate loci were investigated and had been substantiated strongly related to neuroimaging or metabolic biomarkers in the AD process [10-14]. Membrane-spanning 4-domains, subfamily A, member 6A (MS4A6A), located in chromosome 11q12.1, has been identified as one of the most significantly associated risk locus with AD in serial recent large GWAS [15-24]. Although lots researchers found high levels of MS4A6A were associated with increased risk of AD, and in relation to AD-related neurofibrillary pathology and tau phosphorylation in vitro [25-28], whether the loci associate with the neuroimaging or metabolic biomarkers are still unclear.
In this article, we defined many brain regions as regions of interest (ROIs), including the main pathological change regions of AD and the atrophy of some regions in AD has been previously confirmed via MRI studies [29-33]. Then, we genotyped multi-loci in MS4A6A and searched the involvement of MS4A6A in the onset and progression of AD by investigating the influence of MS4A6A polymorphism on brain structure and function in the participants from the Alzheimer's Disease Neuroimaging Initiative (ADNI) dataset.
RESULTS
Characteristics of included subjects
Demographic features, cognitive status, number of APOE ε4 allele and neuroimaging phenotypes of AD, MCI and control subjects were summarized in Table 1. Finally, we recruited 281 cognitively normal (74.51±5.56 years), 483 MCI (72.28±7.45 years) and 48 AD patients (75.51±9.23 years) in our study. As we had expected, the frequency for the ε4 allele within ApoE gene was significantly higher in AD group than CN group. Based on various neuropsychological scales (including CDRSB, ADAS11, ADAS13, MMSE, RAVLT, ADAS-cog, etc.), AD dementia patients revealed the worst cognitive function than CN and MCI subjects. For the imaging endophenotypes, AD group had the most smallest cortical volume of hippocampus, middle temporal and entorhinal cortex as compared to MCI and NC group with MRI method (P < 0.01). Moreover, the lowest cerebral glucose metabolism rate and the highest Aβ tracer retention were found in AD patients than MCI and CN individuals.
Table 1. The characteristics of the ADNI subjects at baseline.
| Characteristics | CN | MCI | AD | Pa | |||
|---|---|---|---|---|---|---|---|
| Age (years) | 281 | 74.51±5.56 | 483 | 72.28±7.45 | 48 | 75.51±9.23 | - |
| Gender (male/female) | 281 | 136/145 | 483 | 282/201 | 48 | 30/18 | - |
| Education (years) | 281 | 16.41±2.66 | 483 | 15.98±2.82 | 48 | 15.73±2.62 | 0.08 |
| APOE ε4 (0/1/2) | 281 | 204/70/7 | 483 | 262/180/41 | 48 | 14/25/9 | <0.01 |
| CDR-SB | 207 | 0.03±0.13 | 406 | 1.44±0.87 | 47 | 4.44±1.69 | <0.01 |
| MMSE | 281 | 29.07±1.15 | 483 | 27.89±1.69 | 48 | 22.96±2.03 | <0.01 |
| ADAS-cog | 281 | 9.06±4.23 | 480 | 15.30±6.65 | 48 | 29.80±8.44 | <0.01 |
| RAVLT | 280 | 44.83±9.60 | 483 | 36.16±10.86 | 47 | 22.32±7.84 | <0.01 |
| FAQ | 281 | 0.17±0.66 | 481 | 2.85±3.99 | 48 | 12.6±7.14 | <0.01 |
| Hippocampus (mm3) | 257 | 7344±895 | 422 | 6996±1126 | 39 | 5757±948 | <0.01 |
| Middle Temporal (mm3) | 257 | 20298±2600 | 422 | 20186±2735 | 39 | 17776±3230 | <0.01 |
| Entorhinal (mm3) | 257 | 3803±650 | 422 | 3610±723 | 39 | 2919±705 | <0.01 |
| CMRgl | 207 | 6.55±0.55 | 406 | 6.32±0.64 | 47 | 5.30±0.72 | <0.01 |
| SUVR | 152 | 1.12±0.19 | 323 | 1.20±0.22 | 46 | 1.39±0.22 | <0.01 |
CN, cognitively normal; MCI, mild cognition impairment; AD, Alzheimer's disease; CDRSB, Clinical Dementia Rating scale sum of boxes; ADAS, Alzheimer's disease Assessment Scale; MMSE, Mini-Mental State Exam; RAVLT, Rey Auditory Verbal Learning Test; FAQ, Functional Activities Questionnaire; FDG, Cerebral Glucose Metabolism Rate measured with fluorodeoxyglucose-positron emission tomography(FDG-PET).
Pa values for continuous variables are from one-way analysis of variance (ANOVA). P values for categorical data are from chi square test. Data are given as mean ± standard deviation unless otherwise indicated
Brain structure and MS4A6A genotypes
In the follow-up study of one-year, we found that rs610932 were significantly associated with the percentage of decreases in the volume of left middle temporal, left precuneus and left entorhinal and these significant associations still survived after the FDR correction (Pc = 0.017, Pc = 0.015 and Pc = 0.022, respectively). In addition, rs610932 with minor A allele carriers decreased the atrophy rate of left middle temporal, left precuneus and left entorhinal in a dose-dependent manner (AA>CA>CC) (Figure 1A-1C). Moreover, the locus variation at rs7232 also significantly correlated the atrophy rate of left middle temporal (Pc = 0.022) and minor T allele carriers had less loss in the volume of left middle temporal than A allele homozygotes subjects (TT>TA>AA) (Figure 1D). SNPs also showed significant association with changes of volume in multiple ROIs, including left posterior cingulate (rs610932), left precuneus (rs610932), left parahippocampal (rs7232) and left entorhinal (rs7232) after two-year follow-up, but all associations failed to survive after the FDR correction. In our all group study, there was no evidence for an effect of rs12453 on structural MRI in these above ROIs (Supplementary Table 2).
Figure 1. The significant correlation between MS4A6A loci and morphological changes of AD specific structure on MRI in all group analysis.
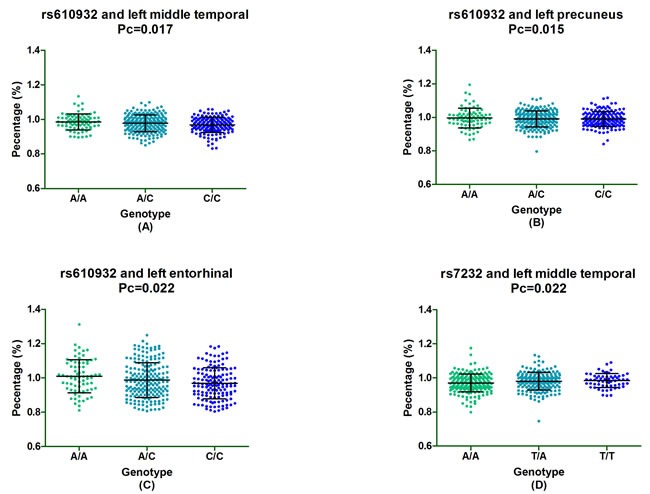
Figure 2A-2C depicted that rs610932 was associated with the atrophy rate of left middle temporal, left precuneus and left entorhinal in one year follow-up study. Figure 2D depicted that rs7232 was associated with the atrophy rate of left middle temporal after one-year follow-up.
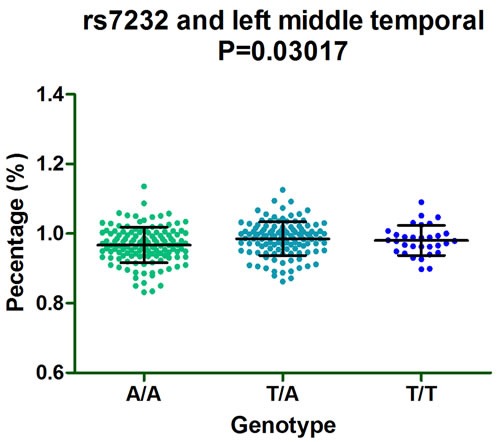
Furthermore, subgroup analysis discovered that rs610932 significantly correlated the left middle temporal, precuneus and entorhinal of MCI individuals (P = 0.03201, P = 0.01313 and P = 0.01298, respectively) (Figure 2A-2C) and also altered the left entorhinal volume (P = 0.007726) in CN subgroup in the follow-up of one year (Figure 2D). Moreover, rs7232 effected the atrophy rate of left middle temporal only in MCI group (P = 0.03017) in one year follow-up study (Figure 3).
Figure 2. The significant correlation between rs610932 and morphological changes of AD specific structure on MRI in subgroup analysis after one-year follow-up.
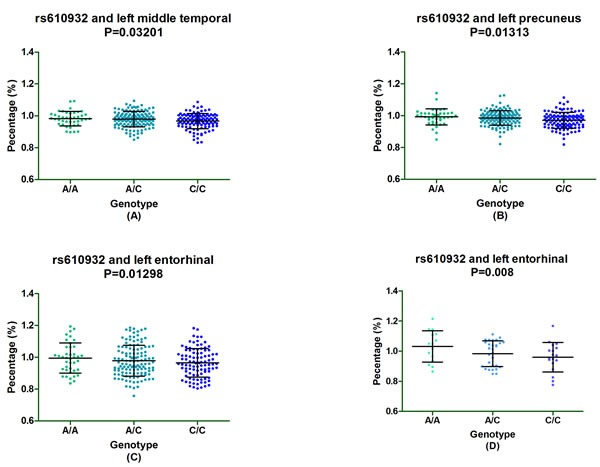
Figure 2A-2C depicted that rs610932 was associated with the atrophy rate of left middle temporal, left entorhinal and left precuneus in MCI group. Figure 4D depicted that rs610932 was associated with the atrophy rate of left entorhinal in NC group.
Figure 3. The significant correlation between rs7232 and morphological changes of AD specific structure on MRI in MCI group after one-year follow-up.
Rs610932 has been validated to be linked to AD in the many large GWAS, we also found that rs610932 and rs7232 were verified to associate with AD (P = 3.07×10-9 and P = 1.73×10-10, respectively) in meta-analysis of 74 046 participants.
Association of MS4A6A with cerebral glucose metabolism
Taken amygdala, posterior cingulate and temporal cortex as ROIs, we analyzed the influence of MS4A6A genotypes on cerebral metabolism rate of glucose (CMRgl) on FDG-PET imaging. Unfortunately, no significant association of three SNPs (rs610932, rs7232 and rs12453) with the CMRgl on FDG-PET was found in the follow up study (Supplementary Table 3).
Association of MS4A6A and Aβ deposition
Regarding the analysis of AV45-PET in frontal, parietal and temporal cortex and cingulate, we also not found the association of MS4A6A with Aβ deposition in the follow-up study (Supplementary Table 3).
DISCUSSION
Our study demonstrated that MS4A6A genotypes correlated the brain atrophic rate of some special brain regions, but not associated with cerebral glucose metabolism and Aβ deposition in the follow-up study. MS4A6A polymorphisms (rs610932 and rs7232) changed the atrophy rate of some brain structural in a dose-dependent manner. Rs610932 with minor A allele carriers decreased the atrophy rate of left middle temporal, left precuneus and left entorhinal and rs7232 with minor T allele carriers had less loss in the volume of left middle temporal than A allele homozygotes subjects. In all group analysis, we found rs610932 were decreased in the percentage of volume loss of left middle temporal, left entorhinal and left precuneus in a dose-dependent manner (AA>CA>CC). Moreover, rs7232 mainly correlated with atrophy rate of left middle temporal and minor T allele carriers had less loss in the volume of left middle temporal than A allele homozygotes subjects (TT>TA>AA). But no association was found in rs12453 with any above ROIs. In order to identify that at which stage these variants impacted these pathological markers in the pathogenesis of AD, the significant associations between these positive MS4A6A loci and suggestive phenotypes in total group will be further detected in subgroup (CN, MCI and AD). Subgroup analysis found those brain structures (left entorhinal, left middle temporal and left precuneus) atrophy rate were significantly correlated with MS4A6A variants.
Two large GWAS had identified the minor T allele of rs610932 as a protective allele in Europeans, and this result had been successfully validated by our group in Han Chinese population. It is well-known that the sequence and structural motifs of 3′-UTR regulated the process of messenger RNA stability, translation, and localization. The SNP rs610932 which located in the 3′-UTR of the MS4A6A gene may participate in the gene expression process and regulates the expression of the MS4A6A [34]. Previous studies have shown that rs610932 and rs7232 were strongly associated with the levels of MS4A6A expression in blood from MCI and AD patients (p < 0.05) [35]. Besides, rs610932, rs7232 and rs12453 were found to nominally significant in relation to MS4A6A expression in cerebellum and temporal cortex while increased minor T allele copies of rs610932 are accompanied with lower MS4A6A gene expression [15, 35]. Mounting evidences have suggested that high levels of MS4A6A are increased prevalence risk of AD and in relation to Braak tangle and Braak plaque scores [25-27]. Moreover, Martiskainen and his colleagues reported that highly MS4A6A expression was significantly correlated to AD-related neurofibrillary pathology and tau phosphorylation in the postmortem inferior temporal cortex in a sample containing 60 participants with different levels of AD-related neurofibrillary pathology [28]. Although the exact mechanisms underlying the effects of MS4A6A on the occurrence and development of AD are still largely unknown, several members of MS4A cluster are found to participate in the regulation of calcium signaling [36-40] and immune-system function [41-43]. All these above evidence, along with our finding, it could be linked to a hypothesis that the role of MS4A6A variants in AD might be mediated by modifying neurodegenerative changes, possibly by affecting calcium signaling and immune-system function in these regions and subsequently influencing memory function.
Endophenotype-based design would gain greater power and smaller sample size requirements to detect disease susceptibility loci than traditional case-control designs. Although lots of researches have identified the effect of genotype on AD-related neuroimaging phenotypes, to our knowledge, our investigation was the first study to explore the role of MS4A6A genotypes in neuroimaging phenotypes and detected that MS4A6A variants was significantly associated with the percentage change of volume of middle temporal, entorhinal and precuneus. But there are several potential limitations in this study. Firstly, the ADNI sample size was relatively small when considering follow-up imaging data of specific genotypes, and more studies are needed from a larger dataset to evaluate the association between MS4A6A and AD-related neuroimaging phenotypes. Secondly, only left hemisphere structures are associated with genetic variants, but whether the subjects are all right-handed or not is unknown. Thirdly, two years follow-up may be too short to observe the remark effect of MS4A6A on the changes of structural volumetric MRI, and an longer years of follow-up is need to replicate these findings. Finally, although GWAS identified several variants within MS4A6A (such as rs662196, rs583791 and rs983392), but we cannot extracted these SNPs from the GWAS of ADNI for all included participants [16, 35, 44]. So, more association studies with larger number of subjects and longer follow-up are still eager to sustain the present research findings.
In conclusion, our study provides evidence that supporting the possible role of MS4A6A genotypes in influencing AD-related neuroimaging phenotypes. These findings further confirmed the hypothesis that MS4A6A genetic variants may modulate the alteration of the biomarkers of neuronal degeneration or injury and then influence the risk of AD. Nonetheless, further work is essential to explain the effect of MS4S6A on AD in a larger sample with longer follow-up.
MATERIALS AND METHODS
ADNI dataset
The ADNI dataset is a consortium of the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, the Food and Drug Administration, private pharmaceutical companies, and nonprofit organizations to develop serial MRI, positron emission tomography (PET), other biological procedures, and neuropsychological and clinical assessment in cognitively normal older subjects, early or late MCI subjects, and early AD subjects (http://www.adni-info.org). The primary purpose of ADNI is to establish an accessible database that describes longitudinal changes in brain structure, function and metabolism according to parallel clinical, cognitive, and biochemical data. To date, the three protocols have recruited over 1,500 adults to participate in the research, ages from 55 to 90 [45]. Informed consent was acquired from all subjects or from authorized representatives and with approval from the institutional review boards of all participating centers.
Subjects
The subjects who classified as normal controls, MCI subjects, or AD subjects used in this research were downloaded from the ADNI database. The subjects with AD between the ages of 55-90, with an Mini-Mental State Examination (MMSE) score of 20-26 inclusive and meeting the NINCDS/ADRDA criteria for probable AD, and having a Clinical Dementia Rating (CDR) of 0.5 or 1. While the subjects with MCI have memory complaints, fulfilled the MMSE score between 24 and 30 and the CDR score was 0.5, essentially preserved activities of daily living. On the other hand, all individuals included in this study were free from any serious neurological disease except for possible AD, current or past history of brain lesions or head trauma, or psychoactive medication use [45]. Other detailed specification can be found on the ADNI cohort online (http://adni.loni.usc.edu/).
SNPs selection
In our study, MS4A6A genotypes data were downloaded from the ADNI GWAS using PLINK format. The quality control (QC) procedures were performed using PLINK software, and meeting the following criteria for the SNPs will be included in our research: Hardy-Weinberg equilibrium test P >0.001, minimum call rates >90%, minimum minor allele frequencies (MAF) >0.01.
We first selected four MS4A6A loci (rs610932, rs583791, rs662196 and rs138650483) which have been reported to be significantly correlated with AD in published GWAS for analysis [16, 46, 47]. In addition, we further searched seven promising candidate loci (rs7232, rs12453, rs646924, rs632185, rs983392, rs17602572 and rs2278867) from meta-analysis and replication studies [15, 17, 18, 44, 48, 49] (Supplementary Table 1). Although 11 SNPs were identified in the initial screening, 8 SNPs were further excluded due to their absence in ADNI. Therefore, we selected 3 loci (rs610932, rs7232 and rs12453), which meeting inclusion criteria for the SNP quality control, to research the association between MS4A6A genotypes and brain structure (Table 2).
Table 2. Characteristics of three SNPs included for our analysis.
| SNP | Chr | Position | Minor allele | SNP source | MAF | H-W (p value) | Ref. |
|---|---|---|---|---|---|---|---|
| rs610932 | 11 | 3′-untranslated region (3′-UTR) | A | GWAS & Meta-analysis & Replication | 0.406 | 0.9325 | [15-18],[30] |
| rs7232 | 11 | Nonsynonymous | T | Replication | 0.348 | 0.2644 | [20],[33] |
| rs12453 | 11 | Synonymous | C | Replication | 0.373 | 0.7207 | [33] |
Abbreviation: MAF= Minor Allele Frequency; SNP= Single Nucleotide Polymorphism; H-W= Hardy-Weinberg
Brain structures on MRI
All data of structural volumetric MRI were downloaded from the ADNI dataset for free. This technology provided by the University of California, San Francisco (UCSF) medical center. A detailed description of imaging data acquisition and processing can be acquired in other papers [50]. Previous studies have demonstrated that brain atrophy may begin to emerge many years before the clinical symptoms of mild MCI that associated to evidences of AD. We defined six cerebral areas, including hippocampus, parahippocampal, entorhinal, middle temporal, posterior cingulate and precuneus, as regions of interest as they are known to be affected by AD and mounting researches have been previously validated their atrophy in AD via MRI studies. In this study we calculated one and two years percent volumetric changes of the most discriminant ROIs for longitudinal analysis to analyze their associations with MS4A6A genotypes (Supplementary Table 2).
Glucose metabolism on imaging
The cerebral metabolic rate for glucose (CMRgl) on FDG-PET analysis data were provided by both UC Berkeley and Lawrence Berkeley National Laboratory [51]. We extracted the data from the site (http://adni.loni.usc.edu/data-samples/access-data/). Their researchers used five brain regions (right and left right temporal gyrus, right and left angular gyrus and bilateral posterior cingulate) as ROIs to analysis (Supplementary Table 3). We downloaded FDG-PET analysis data from LONI (http://loni.usc.edu/), and subsequently these images were spatially normalized in Statistical Parametric Mapping (SPM) to the Montreal Neurological Institute (MNI) PET template. At last, the mean counts of these five ROIs for each subject's FDG scans at each time point were used to compute the intensity values with SPM subroutines [52].
Aβ deposition on AV45-PET imaging
UC Berkeley-AV45 analysis dataset offered the PET imaging data with amyloid tracer, florbetapir (AV-45) at site (http://adni.loni.usc.edu/data-samples/access-data/). On this site, we can found the detailed description of PET image acquisition and processing. A native-space MRI scan for each subject was segmented with Freesurfer (version 4.5.0) to define cortical grey matter ROIs, including frontal, lateral parietal, lateral temporal anterior/posterior cingulate, which make up a summary cortical ROIs [53, 54] (Supplementary Table 3). The institute used cerebellum ROIs (grey matter only) as a reference region. Firstly, each florbetapir scan was applied to the corresponding MRI, and then they calculated the mean florbetapir uptake within the cortical and reference region. Finally, florbetapir standard uptake value ratios (SUVRs) were generated by averaging across the four ROIs and dividing this average by whole cerebellum.
Statistical analysis
Demographic and clinical characteristics of subjects (AD, MCI and control subjects) were described either in terms of means and standard deviations (SD) if quantitative or in terms of proportions. One-way analysis of variance (ANOVA) was used to compare the differences in continuous variables and chi-square test was used to examine the categorical data. Furthermore, taken age, gender, education and number of APOE ε4 allele as covariates, the multiple linear regression model was used to evaluate the possible associations between various phenotypes and MS4A6A genotypes. At first, we computed the influences of MS4A6A loci on the percentage change of the imaging phenotypes in follow-up study. Secondly, the significant associations between these positive MS4A6A loci and suggestive phenotypes in total group will be further detect in subgroup (CN, MCI and AD subjects) to distinguish at which stage these variants impacted these pathological markers in the pathogenesis of AD. Finally, we verify the associations between these positive loci in this study and AD patients from a large database which included 74 046 individuals of European descent [44].
Statistical analyses were performed using R 3.12. According to the method invented by Hochberg and Benjamini for the number of tests, we performed the false discovery rate (FDR) to account for multiple testing [55]. All tests were two-sided and FDR-corrected P < 0.05 was considered statistically significant.
SUPPLEMENTARY MATERIALS TABLES
Acknowledgments
Data collection and sharing for this project was funded by the Alzheimer's Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimer's Association; Alzheimer's Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare;; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer's disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuroimaging at the University of Southern California.
Footnotes
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
GRANTS SUPPORT
This work was also supported by grants from the National Natural Science Foundation of China (81471309, 81171209, 81371406, 81501103, 81571245), the Shandong Provincial Outstanding Medical Academic Professional Program, Qingdao Key Health Discipline Development Fund, Qingdao Outstanding Health Professional Development Fund, and Shandong Provincial Collaborative Innovation Center for Neurodegenerative Disorders.
REFERENCES
- 1.Jiang T, Yu JT, Tian Y, Tan L. Epidemiology and etiology of Alzheimer's disease: from genetic to non-genetic factors. Current Alzheimer research. 2013;10(8):852–867. doi: 10.2174/15672050113109990155. [DOI] [PubMed] [Google Scholar]
- 2.Takizawa C, Thompson PL, van Walsem A, Faure C, Maier WC. Epidemiological and economic burden of Alzheimer's disease: a systematic literature review of data across Europe and the United States of America. Journal of Alzheimer's disease. 2015;43(4):1271–1284. doi: 10.3233/JAD-141134. [DOI] [PubMed] [Google Scholar]
- 3.Salmon DP, Lange KL. Cognitive screening and neuropsychological assessment in early Alzheimer's disease. Clin Geriatr Med. 2001;17(2):229–254. doi: 10.1016/s0749-0690(05)70067-7. [DOI] [PubMed] [Google Scholar]
- 4.Holtzman DM, Morris JC, Goate AM. Alzheimer's disease: the challenge of the second century. 14414 Sci Transl Med. 2011;3(77):77sr71. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Santiago LM, Santos T, Miranda PR, Constantino L, Matias C, Rosendo I, Simoes AR, Neto MG, Francisco MP. Non steroidal anti-inflammatory drugs prescription in General Practice in the centre of Portugal from 2007 to 2009. Acta reumatologica portuguesa. 2010;35(5):447–454. [PubMed] [Google Scholar]
- 6.Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer's disease. Nature. 2009;461(7266):916–922. doi: 10.1038/nature08538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP, Bergstrom M, Savitcheva I, Huang GF, Estrada S, Ausen B, Debnath ML, Barletta J, et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Annals of neurology. 2004;55(3):306–319. doi: 10.1002/ana.20009. [DOI] [PubMed] [Google Scholar]
- 8.Jack CR, Jr, Lowe VJ, Senjem ML, Weigand SD, Kemp BJ, Shiung MM, Knopman DS, Boeve BF, Klunk WE, Mathis CA, Petersen RC. 11C PiB and structural MRI provide complementary information in imaging of Alzheimer's disease and amnestic mild cognitive impairment. Brain. 2008;131:665–680. doi: 10.1093/brain/awm336. (Pt 3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peper JS, Brouwer RM, Boomsma DI, Kahn RS, Hulshoff Pol HE. Genetic influences on human brain structure: a review of brain imaging studies in twins. 6924 Hum Brain Mapp. 2007;28(6):464–473. doi: 10.1002/hbm.20398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biffi A, Anderson CD, Desikan RS, Sabuncu M, Cortellini L, Schmansky N, Salat D, Rosand J. Genetic variation and neuroimaging measures in Alzheimer disease. Archives of neurology. 2010;67(6):677–685. doi: 10.1001/archneurol.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu XC, Tan L, Wang HF, Jiang T, Cao L, Wang C, Wang J, Tan CC, Meng XF, Yu JT. Rate of early onset Alzheimer's disease: a systematic review and meta-analysis. Annals of translational medicine. 2015;3(3):38. doi: 10.3978/j.issn.2305-5839.2015.01.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Yu JT, Wang HF, Hao XK, Yang YF, Jiang T, Zhu XC, Cao L, Zhang DQ, Tan L. Association between NME8 locus polymorphism and cognitive decline, cerebrospinal fluid and neuroimaging biomarkers in Alzheimer's disease. PloS one. 2014;9(12):e114777. doi: 10.1371/journal.pone.0114777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang C, Wang HF, Tan MS, Liu Y, Jiang T, Zhang DQ, Tan L, Yu JT. Impact of Common Variations in PLD3 on Neuroimaging Phenotypes in Non-demented Elders. Molecular neurobiology. 2015 doi: 10.1007/s12035-015-9370-4. [DOI] [PubMed] [Google Scholar]
- 14.Wang C, Tan L, Wang HF, Yu WJ, Liu Y, Jiang T, Tan MS, Hao XK, Zhang DQ, Yu JT. Common Variants in PLD3 and Correlation to Amyloid-Related Phenotypes in Alzheimer's Disease. Journal of Alzheimer's disease. 2015;46(2):491–495. doi: 10.3233/JAD-150110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, et al. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer's disease. Nature genetics. 2011;43(5):429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nature genetics. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tan L, Yu JT, Zhang W, Wu ZC, Zhang Q, Liu QY, Wang W, Wang HF, Ma XY, Cui WZ. Association of GWAS-linked loci with late-onset Alzheimer's disease in a northern Han Chinese population. Alzheimer's & dementia. 2013;9(5):546–553. doi: 10.1016/j.jalz.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Deng YL, Liu LH, Wang Y, Tang HD, Ren RJ, Xu W, Ma JF, Wang LL, Zhuang JP, Wang G, Chen SD. The prevalence of CD33 and MS4A6A variant in Chinese Han population with Alzheimer's disease. Human genetics. 2012;131(7):1245–1249. doi: 10.1007/s00439-012-1154-6. [DOI] [PubMed] [Google Scholar]
- 19.Cazzo E, Gestic MA, Utrini MP, Machado RR, Pareja JC, Chaim EA. Control of hypertension after roux-en-y gastric bypass among obese diabetic patients. Arquivos de gastroenterologia. 2014;51(1):21–24. doi: 10.1590/s0004-28032014000100005. [DOI] [PubMed] [Google Scholar]
- 20.Zhu XC, Wang HF, Jiang T, Lu H, Tan MS, Tan CC, Tan L, Tan L, Yu JT. Effect of CR1 Genetic Variants on Cerebrospinal Fluid and Neuroimaging Biomarkers in Healthy, Mild Cognitive Impairment and Alzheimer's Disease Cohorts. Molecular neurobiology. 2016 doi: 10.1007/s12035-015-9638-8. [DOI] [PubMed] [Google Scholar]
- 21.Wang ZX, Wang HF, Tan L, Liu J, Wan Y, Sun FR, Tan MS, Tan CC, Jiang T, Tan L, Yu JT. Effects of HLA-DRB1/DQB1 Genetic Variants on Neuroimaging in Healthy, Mild Cognitive Impairment, and Alzheimer's Disease Cohorts. Molecular neurobiology. 2016 doi: 10.1007/s12035-016-9890-6. [DOI] [PubMed] [Google Scholar]
- 22.Wang ZX, Wan Y, Tan L, Liu J, Wang HF, Sun FR, Tan MS, Tan CC, Jiang T, Tan L, Yu JT. Genetic Association of HLA Gene Variants with MRI Brain Structure in Alzheimer's Disease. Molecular neurobiology. 2016 doi: 10.1007/s12035-016-9889-z. [DOI] [PubMed] [Google Scholar]
- 23.Wang WY, Liu Y, Wang HF, Tan L, Sun FR, Tan MS, Tan CC, Jiang T, Tan L, Yu JT. Impacts of CD33 Genetic Variations on the Atrophy Rates of Hippocampus and Parahippocampal Gyrus in Normal Aging and Mild Cognitive Impairment. Molecular neurobiology. 2016 doi: 10.1007/s12035-016-9718-4. [DOI] [PubMed] [Google Scholar]
- 24.Li JQ, Wang HF, Zhu XC, Sun FR, Tan MS, Tan CC, Jiang T, Tan L, Yu JT. GWAS-Linked Loci and Neuroimaging Measures in Alzheimer's Disease. Molecular neurobiology. 2016 doi: 10.1007/s12035-015-9669-1. [DOI] [PubMed] [Google Scholar]
- 25.Proitsi P, Lee SH, Lunnon K, Keohane A, Powell J, Troakes C, Al-Sarraj S, Furney S, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, et al. Alzheimer's disease susceptibility variants in the MS4A6A gene are associated with altered levels of MS4A6A expression in blood. Neurobiology of aging. 2014;35(2):279–290. doi: 10.1016/j.neurobiolaging.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 26.Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer's disease risk genes in control and Alzheimer's disease brains. PloS one. 2012;7(11):e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen M, Zou F, Chai HS, Younkin CS, Crook J, Pankratz VS, Carrasquillo MM, Rowley CN, Nair AA, Middha S, Maharjan S, Nguyen T, Ma L, et al. Novel late-onset Alzheimer disease loci variants associate with brain gene expression. Neurology. 2012;79(3):221–228. doi: 10.1212/WNL.0b013e3182605801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martiskainen H, Viswanathan J, Nykanen NP, Kurki M, Helisalmi S, Natunen T, Sarajarvi T, Kurkinen KM, Pursiheimo JP, Rauramaa T, Alafuzoff I, Jaaskelainen JE, Leinonen V, et al. Transcriptomics and mechanistic elucidation of Alzheimer's disease risk genes in the brain and in vitro models. Neurobiology of aging. 2015;36(2):1221, e1215–1228. doi: 10.1016/j.neurobiolaging.2014.09.003. [DOI] [PubMed] [Google Scholar]
- 29.Simmons A, Westman E, Muehlboeck S, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Wahlund LO, Soininen H, Lovestone S, Evans A, Spenger C, AddNeuroMed C. MRI measures of Alzheimer's disease and the AddNeuroMed study. Annals of the New York Academy of Sciences. 2009;1180:47–55. doi: 10.1111/j.1749-6632.2009.05063.x. [DOI] [PubMed] [Google Scholar]
- 30.Kesslak JP, Nalcioglu O, Cotman CW. Quantification of magnetic resonance scans for hippocampal and parahippocampal atrophy in Alzheimer's disease. Neurology. 1991;41(1):51–54. doi: 10.1212/wnl.41.1.51. [DOI] [PubMed] [Google Scholar]
- 31.Convit A, De Leon MJ, Tarshish C, De Santi S, Tsui W, Rusinek H, George A. Specific hippocampal volume reductions in individuals at risk for Alzheimer's disease. Neurobiology of aging. 1997;18(2):131–138. doi: 10.1016/s0197-4580(97)00001-8. [DOI] [PubMed] [Google Scholar]
- 32.Jack CR, Jr, Petersen RC, Xu Y, O'Brien PC, Smith GE, Ivnik RJ, Tangalos EG, Kokmen E. Rate of medial temporal lobe atrophy in typical aging and Alzheimer's disease. Neurology. 1998;51(4):993–999. doi: 10.1212/wnl.51.4.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC Alzheimer's Disease Neuroimaging I. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Current Alzheimer research. 2009;6(4):347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yan H, Yuan W, Velculescu VE, Vogelstein B, Kinzler KW. Allelic variation in human gene expression. Science (New York, NY) 2002;297(5584):1143. doi: 10.1126/science.1072545. [DOI] [PubMed] [Google Scholar]
- 35.Proitsi P, Lee SH, Lunnon K, Keohane A, Powell J, Troakes C, Al-Sarraj S, Furney S, Soininen H, Kloszewska I, Mecocci P, Tsolaki M, Vellas B, et al. Alzheimer's disease susceptibility variants in the MS4A6A gene are associated with altered levels of MS4A6A expression in blood. Neurobiology of aging. 2014;35(2):279–290. doi: 10.1016/j.neurobiolaging.2013.08.002. [DOI] [PubMed] [Google Scholar]
- 36.Zundorf G, Reiser G. Calcium dysregulation and homeostasis of neural calcium in the molecular mechanisms of neurodegenerative diseases provide multiple targets for neuroprotection. Antioxidants & redox signaling. 2011;14(7):1275–1288. doi: 10.1089/ars.2010.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seaton G, Hogg EL, Jo J, Whitcomb DJ, Cho K. Sensing change: the emerging role of calcium sensors in neuronal disease. Seminars in cell & developmental biology. 2011;22(5):530–535. doi: 10.1016/j.semcdb.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 38.Hermes M, Eichhoff G, Garaschuk O. Intracellular calcium signalling in Alzheimer's disease. Journal of cellular and molecular medicine. 2010;14(1-2):30–41. doi: 10.1111/j.1582-4934.2009.00976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marambaud P, Dreses-Werringloer U, Vingtdeux V. Calcium signaling in neurodegeneration. Molecular neurodegeneration. 2009;4:20. doi: 10.1186/1750-1326-4-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LaFerla FM. Calcium dyshomeostasis and intracellular signalling in Alzheimer's disease. Nature reviews Neuroscience. 2002;3(11):862–872. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- 41.Zuccolo J, Deng L, Unruh TL, Sanyal R, Bau JA, Storek J, Demetrick DJ, Luider JM, Auer-Grzesiak IA, Mansoor A, Deans JP. Expression of MS4A and TMEM176 Genes in Human B Lymphocytes. Frontiers in immunology. 2013;4:195. doi: 10.3389/fimmu.2013.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuccolo J, Bau J, Childs SJ, Goss GG, Sensen CW, Deans JP. Phylogenetic analysis of the MS4A and TMEM176 gene families. PloS one. 2010;5(2):e9369. doi: 10.1371/journal.pone.0009369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ma J, Yu JT, Tan L. MS4A Cluster in Alzheimer's Disease. Molecular neurobiology. 2015;51(3):1240–1248. doi: 10.1007/s12035-014-8800-z. [DOI] [PubMed] [Google Scholar]
- 44.European Alzheimer's Disease I, Genetic, Environmental Risk in Alzheimer's D, Alzheimer's Disease Genetic C, Cohorts for H Aging Research in Genomic E. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nature genetics. 2013;45(12):1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, Jack CR, Jr, Jagust WJ, Shaw LM, Toga AW, Trojanowski JQ, et al. Alzheimer's Disease Neuroimaging Initiative (ADNI): clinical characterization. Neurology. 2010;74(3):201–209. doi: 10.1212/WNL.0b013e3181cb3e25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu X, Pickering EH, Hall SK, Naik S, Liu YC, Soares H, Katz E, Paciga SA, Liu W, Aisen PS, Bales KR, Samad TA, John SL. Genome-wide association study identifies multiple novel loci associated with disease progression in subjects with mild cognitive impairment. Translational psychiatry. 2011;1:e54. doi: 10.1038/tp.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vardarajan BN, Ghani M, Kahn A, Sheikh S, Sato C, Barral S, Lee JH, Cheng R, Reitz C, Lantigua R, Reyes-Dumeyer D, Medrano M, Jimenez-Velazquez IZ, et al. Rare coding mutations identified by sequencing of Alzheimer's disease GWAS loci. Annals of neurology. 2015 doi: 10.1002/ana.24466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung SJ, Lee JH, Kim SY, You S, Kim MJ, Lee JY, Koh J. Association of GWAS top hits with late-onset Alzheimer disease in Korean population. Alzheimer disease and associated disorders. 2013;27(3):250–257. doi: 10.1097/WAD.0b013e31826d7281. [DOI] [PubMed] [Google Scholar]
- 49.Antunez C, Boada M, Gonzalez-Perez A, Gayan J, Ramirez-Lorca R, Marin J, Hernandez I, Moreno-Rey C, Moron FJ, Lopez-Arrieta J, Mauleon A, Rosende-Roca M, Noguera-Perea F, et al. The membrane-spanning 4-domains, subfamily A (MS4A) gene cluster contains a common variant associated with Alzheimer's disease. Genome medicine. 2011;3(5):33. doi: 10.1186/gm249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang D, Wang Y, Zhou L, Yuan H, Shen D Alzheimer's Disease Neuroimaging I. Multimodal classification of Alzheimer's disease and mild cognitive impairment. NeuroImage. 2011;55(3):856–867. doi: 10.1016/j.neuroimage.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landau SM, Harvey D, Madison CM, Reiman EM, Foster NL, Aisen PS, Petersen RC, Shaw LM, Trojanowski JQ, Jack CR, Jr, Weiner MW, Jagust WJ Alzheimer's Disease Neuroimaging I. Comparing predictors of conversion and decline in mild cognitive impairment. Neurology. 2010;75(3):230–238. doi: 10.1212/WNL.0b013e3181e8e8b8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Landau SM, Harvey D, Madison CM, Koeppe RA, Reiman EM, Foster NL, Weiner MW, Jagust WJ Alzheimer's Disease Neuroimaging I. Associations between cognitive, functional, and FDG-PET measures of decline in AD and MCI. Neurobiology of aging. 2011;32(7):1207–1218. doi: 10.1016/j.neurobiolaging.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jagust WJ, Landau SM, Shaw LM, Trojanowski JQ, Koeppe RA, Reiman EM, Foster NL, Petersen RC, Weiner MW, Price JC, Mathis CA Alzheimer's Disease Neuroimaging I. Relationships between biomarkers in aging and dementia. Neurology. 2009;73(15):1193–1199. doi: 10.1212/WNL.0b013e3181bc010c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mormino EC, Kluth JT, Madison CM, Rabinovici GD, Baker SL, Miller BL, Koeppe RA, Mathis CA, Weiner MW, Jagust WJ Alzheimer's Disease Neuroimaging I. Episodic memory loss is related to hippocampal-mediated beta-amyloid deposition in elderly subjects. Brain. 2009;132(Pt 5):1310–1323. doi: 10.1093/brain/awn320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hochberg Y, Benjamini Y. More powerful procedures for multiple significance testing. Statistics in medicine. 1990;9(7):811–818. doi: 10.1002/sim.4780090710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


