Abstract
The goal of the present study was to develop a fully three-dimensional (3D) coculture system that would allow for systematic evaluation of the interplay between activated macrophages (AMs) and chondrocytes in osteoarthritic disease progression and treatment. Toward this end, our coculture system was first validated against existing in vitro osteoarthritis models, which have generally cultured healthy normal chondrocytes (NCs)—in two-dimensional (2D) or 3D—with proinflammatory AMs in 2D. In this work, NCs and AMs were both encapsulated within poly(ethylene glycol) diacrylate hydrogels to mimic the native 3D environments of both cell types within the osteoarthritic joint. As with previous studies, increases in matrix metalloproteinases (MMPs) and proinflammatory cytokines associated with the early stages of osteoarthritis were observed during NC-AM coculture, as were decreases in protein-level Aggrecan and collagen II. Thereafter, the coculture system was extended to osteoarthritic chondrocytes (OACs) and AMs to evaluate the potential effects of AMs on pre-existing osteoarthritic phenotypes. OACs in coculture with AMs expressed significantly higher levels of MMP-1, MMP-3, MMP-9, MMP-13, IL-1β, TNF-α, IL-6, IL-8, and IFN-γ compared to OACs in mono-culture, indicating that proinflammatory macrophages may intensify the abnormal matrix degradation and cytokine secretion already associated with OACs. Likewise, AMs cocultured with OACs expressed significantly more IL-1β and VEGF-A compared to AM mono-culture controls, suggesting that OACs may intensify abnormal macrophage activation. Finally, OACs cultured in the presence of nonactivated macrophages produced lower levels of MMP-9 and proinflammatory cytokines IL-1β, TNF-α, and IFN-γ compared to OACs in the OAC-AM system, results that are consistent with anti-inflammatory agents temporarily reducing certain OA symptoms. In summary, the 3D coculture system developed herein captures several key features of inflammatory OA and may prove useful in future screening of therapeutic agents and/or assessment of disease progression mechanisms.
Keywords: : osteoarthritis, inflammation, 3D coculture, chondrocytes, macrophages
Introduction
Osteoarthritis (OA) is a degenerative disease of certain joints that is primarily characterized by cartilage degradation and bony outgrowths at joint margins.1 Worldwide, OA is one of the leading causes of chronic disability,2,3 affecting close to 50% of the population over the age of 75.4 Although much attention has been focused on the treatment and management of OA symptoms, the factors governing disease onset and progression are still unclear. Over the last several years, the role of synovial inflammation in contributing to knee OA has been increasingly recognized.5 This inflammation is believed to result in part from macrophage activation in the synovium. Activated synovial macrophages secrete proinflammatory factors, such as IL-1β, TNF-α, and nitric oxide (NO)6,7 that act upon chondrocytes in a paracrine manner. In turn, chondrocytes produce excess matrix metalloproteinases (e.g., MMP-3 and MMP-13) and aggrecanases (e.g., ADAMTS-4 and ADAMTS-5), which result in the degradation of extracellular matrix (ECM) components such as collagen II (Col-2) and Aggrecan.8–11 Diseased chondrocytes also secrete inflammatory cytokines like IL-1β, TNF-α, IL-6, and IL-8 that contribute to the inflammatory environment within the knee.12–14 The products of ECM degradation and inflammatory cytokines are believed to stimulate increased synovial inflammation, resulting in a repeating cycle of inflammation and cartilage degradation.
Thus, current pharmacological treatment options for OA include the administration of analgesics and nonsteroidal anti-inflammatory drugs, which mitigate pain and temporarily reduce inflammation.15 However, these positive effects diminish with time, and the medications are also not tolerated well by certain patient groups. In such cases, visco-supplementation with intra-articular hyaluronic acid (HA) injections has been used to promote joint lubrication. HA is also known to exert an anti-inflammatory effect by suppressing MMPs/ADAMTSs in addition to counteracting the degenerative effects caused by IL-1β.16 However, HA supplementation often requires additional follow-up procedures and may not provide any significant benefits for patients over the age of 65 or those with advanced radiographic OA.17,18 Thus, a deeper understanding of inflammation in OA and insights into the interactions between cartilage and synovial macrophages may allow for improved development and screening of OA therapeutic agents.
The goal of the current work was to build and validate a fully 3D coculture system that can be used to study paracrine interactions between chondrocytes and macrophages in the context of OA inflammation. While several previous OA models have employed 2D monolayers,19–23 the literature reveals that cell behavior in 2D is often not representative of cell responses observed in physiologically relevant 3D contexts.24–27 Indeed, an increasing number of studies recently reporting in vitro OA models have cultured chondrocytes within 3D microenvironments.28–32 However, important auxiliary cell types such as macrophages have continued to be cultured in 2D,28,31,33 despite the primary location of the macrophages in the knee joint being within the synovial membrane. Moreover, phenotypic changes in macrophages cocultured with chondrocytes have largely been unexplored in previous in vitro studies.
Poly(ethylene glycol) diacrylate (PEGDA)-based hydrogels were used in this study for the establishment of the 3D coculture system, due to their widespread use in cartilage tissue engineering,34–37 their easily tunable mechanical properties,38,39 and their ability to significantly resist cell adhesion even in serum-containing culture environments relative to many biomaterials.34,40,41 This latter property permits desired cell adhesion to be “programmed” into the PEGDA network through conjugation of bioactive moieties.42,43 In the present work, we have tethered the peptide RGD to the PEGDA network to enable uniform levels of initial cell-matrix interactions across experimental groups.
In this study, healthy normal chondrocytes (NCs) and classically activated, proinflammatory (M1) macrophages (AMs) were first encapsulated within separate PEGDA hydrogels and placed in coculture, permitting comparisons with previously reported in vitro OA models. Thereafter, the culture system was extended to osteoarthritic chondrocytes (OACs) and AMs to evaluate the potential effects of AMs in exacerbating existing osteoarthritic phenotypes. Concurrently, OACs were cocultured with nonactivated macrophages (NMs) to determine whether the presence of NMs could influence the diseased phenotype of OACs.
Materials and Methods
Polymer synthesis and functionalization of cell adhesive peptides
PEGDA, possessing >95% acrylation, was synthesized from PEG-diol (3.4 kDa, Sigma Aldrich) as reported previously.43 NH2-Arg-Gly-Asp-Ser-COOH (RGDS; American Peptide Company) and 3.4 kDa acryoyl-PEG succinimidyl valerate (ACRL-PEG-SVA; Laysan Bio, Inc.) were reacted a 1:1 molar ratio for 2 h in 50 mM sodium bicarbonate buffer (pH 8.5). The product (ACRL-PEG-RGDS) was purified by dialysis, lyophilized, and stored at −80°C until further use.
Culture of chondrocytes and activation of macrophages
Cryopreserved primary human articular NCs (passage 3; Lonza) and human OACs (passage 2; Cell Applications, Inc.) were thawed and expanded for 5–7 days in Chondrogenic Growth Medium (Lonza) within a 37°C—5% CO2 jacketed incubator, after which the cells were transitioned to regular growth medium: Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (Hyclone; Fisher Scientific). Concurrently, mouse RAW 264.7 macrophage cells (ATCC) were expanded in regular growth medium in a 37°C—5% CO2 jacketed incubator and activated before encapsulation. The RAW 264.7 murine macrophage cell line is well characterized and is known to not only respond to stimulation with LPS, IFN-γ, and rh IL-10,44 but also express markers of both M1 and M2 macrophage phenotypes following appropriate stimulation.45
For activation, macrophages were harvested using a cell scraper and subsequently seeded within fresh T75 flasks at 6500 cells/cm2. After 24 h in growth medium, the cells were switched to activation medium (regular growth medium supplemented with 1 μg/mL lipopolysaccharide (LPS; Salmonella enterica serotype enteritidis; Sigma Aldrich)), and were maintained in this medium for 4 days as per established protocols.46 Cells thus activated with LPS were designated activated macrophages (AM), whereas cells that continued to receive regular growth medium were designated nonactivated macrophages (NM). Morphologically, AMs were larger and well spread, while also extending several lamellopodia not observed in the NMs. These differences between AM and NM are consistent with previous literature reporting macrophage polarization upon stimulation with LPS.47
Encapsulation of chondrocytes and macrophages
Before cell encapsulation, hydrogel precursor solutions were prepared by adding 4 mg of ACRL-PEG-RGDS and 10 μL of a photoinitiator solution (262 mg Irgacure 2959 dissolved in 1 mL of 70% ethanol) to 1 mL of a 10% (w/v) PEGDA solution in phosphate-buffered saline (PBS). The precursor solution was then filtered to remove endotoxins using an Acrodisc Unit with a Mustang E membrane (Fisher Scientific). NCs (between passages 5 and 6) or OACs (between passages 4 and 5) were harvested and resuspended at a concentration of 4 million cells/mL in separate aliquots of the hydrogel precursor solution. Two hundred microliter aliquots of the cell suspension were photopolymerized within a 48-well plate by exposure to long-wave ultraviolet light (∼10 mW/cm2; 365 nm) for 6 min, resulting in cell-laden hydrogel discs ∼1 mm in thickness and 10 mm in diameter. This polymerization process has previously been demonstrated to be cytocompatible and to result in gels with homogenously distributed cells.48–50 The hydrogel discs were then rinsed in PBS and transferred to 12-well plates containing regular growth medium supplemented with 1% antibiotic/antimycotic (10,000 U/mL penicillin, 10,000 μg/mL streptomycin, and 25 μg/mL amphotericin; Life Technologies).
Macrophages were encapsulated within PEGDA hydrogels in a similar manner. Briefly, NMs or AMs were separately suspended in the hydrogel precursor solutions at a concentration of 5 million cells/mL. Two hundred microliters of cell suspension was dispensed into Falcon 12-well cell culture inserts (pore size: 8 μm) and photopolymerized by exposure to long-wave ultraviolet light. Postfabrication, discs were washed in PBS and cultured for 24 h in regular growth medium supplemented with 1% antibiotic/antimycotic.
Culture of cell-laden hydrogels
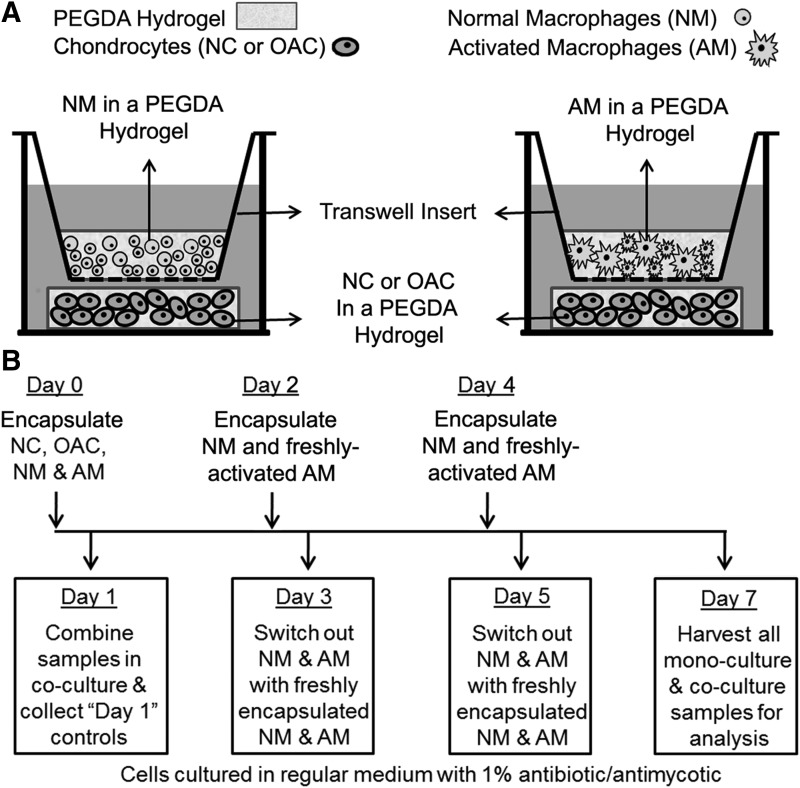
Twenty-four hours postencapsulation, randomly selected chondrocyte and macrophage discs were flash-frozen in liquid nitrogen and stored at −80°C to allow for characterization of initial cell phenotype before coculture. The remaining chondrocyte and macrophage hydrogels were combined in coculture as depicted in Figure 1A. Individually cultured chondrocyte and macrophage hydrogels served as mono-culture controls. All constructs were maintained in regular growth medium supplemented with 1% antibiotic/antimycotic for the entire duration of the study. To maintain AMs in a state of activation throughout the coculture period, AM hydrogels in coculture were replaced with fresh hydrogels containing newly activated AMs every 2 days following the first 24 h of culture. Correspondingly, NM 3D hydrogels were also replaced with hydrogels containing freshly encapsulated NMs every 2 days. After 6 days of coculture, samples were washed in PBS for 10 min, harvested by flash-freezing in liquid nitrogen, and stored at −80°C until further analysis. The time course of the study is depicted in Figure 1B. An additional set of samples were formalin-fixed for 10 min followed by embedding in OCT Compound (Tissue-Tek).
FIG. 1.
(A) Schematic representation of the trans-well coculture system used to investigate paracrine interactions between chondrocytes and macrophages encapsulated within separate PEGDA hydrogels. (B) Timeline describing the macrophage activation schedule, the encapsulation of chondrocytes and macrophages within PEGDA hydrogels at day 0, and the coculture of hydrogel-encapsulated chondrocytes and macrophages for 6 days thereafter. Note that LPS was utilized to induce macrophages to take on a classically activated phenotype before encapsulation within PEGDA hydrogels. After macrophage encapsulation, LPS supplementation was removed, as LPS can have detrimental effects on chondrocytes and prolonged LPS stimulation can desensitize macrophages. However, following LPS removal, AMs are known to slowly lose their proinflammatory phenotype over the course of 7 days. In the present work, AM discs in coculture were therefore replaced with discs containing freshly activated AMs every 2 days. This process ensured that chondrocytes were exposed to relatively consistent dosing of proinflammatory factors throughout the 6-day coculture period. NC, normal chondrocyte; OAC, osteoarthritic chondrocyte; PEGDA, poly(ethylene glycol) diacrylate.
Extraction of mRNA and protein
Chondrocytes or macrophages encapsulated within the hydrogel discs were used for the extraction of mRNA and protein. Here, we did not measure proteins released into the culture medium by the encapsulated cells, because we wanted to separately assess the contributions of the chondrocytes and the macrophages, which would have been challenging had we evaluated proteins released into the media. Accordingly, hydrogel discs with cells were transferred to 1.7 mL RNase-free microcentrifuge tubes containing 300 μL of the lysis buffer provided with the Dynabeads mRNA Direct kit (Life Technologies). The samples were homogenized using plastic RNase-free pestles (Kimble Chase) and incubated in lysis buffer at room temperature for 10 min. Thereafter, the samples were centrifuged for 5 min at 10,000 rpm and the polyA-mRNA in the supernatant was extracted using 20 μL of Dynabeads oligo(dT) magnetic beads according to the manufacturer's protocol. The supernatant was then stored at −20°C until further DNA and protein analyses. The mRNA-laden Dynabeads were rinsed and transferred to 100 μL of a 10 mM Tris-HCl buffer, into which the bound polyA-mRNA was released by heating the beads to 80°C for 2 min. The mRNA solution was then stored at −80°C until further analysis.
Quantitative reverse transcription polymerase chain reaction
To compare gene expression across the various experimental groups, quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using a StepOne Real-Time PCR system (Life Technologies) and the SuperScript III Platinum One-Step qRT-PCR kit (Life Technologies) according to the manufacturer's instructions. Validated qRT-PCR human primers for genes related to chondrocyte phenotype (Table 1) and validated qRT-PCR mouse primers for genes related to macrophage polarization (Table 2) were obtained from Origene or Qiagen. Approximately 3–6 ng of polyA-mRNA was mixed with 5 μL of 1 μM primer solution, 12 μL of 2 × SYBR Green master mix and 0.5 μL of Superscript III-Taq polymerase mix in a total reaction volume of 25 μL. Each reaction was monitored over 40 cycles by measuring the change in SYBR Green fluorescence, with ROX dye serving as a passive reference. The threshold fluorescence value in the exponential phase of amplification was internally set, and a threshold cycle (Ct) was determined for each sample. Gene expression was normalized to a reference gene (RPL-32) and the ΔΔCt method51 was used for the estimation of sample gene expression relative to a selected reference control. Melting temperature analysis was performed for each reaction to verify the amplification product.
Table 1.
Genes Assessed in Evaluating Shifts in Human Chondrocyte Phenotype
| Primer sequence | ||
|---|---|---|
| Function | Gene marker (supplier) | Forward 5′-3 (F), Reverse 5′-3′ (R) |
| Reference gene | RPL-32 (Origene) | F: ACAAAGCACATGCTGCCCAGTG |
| R: TTCCACGATGGCTTTGCGGTTC | ||
| Chondrogenic transcription factor | Sox-9 (Origene) | F: AGGAAGCTCGCGGACCAGTAC |
| R: GGTGGTCCTTCTTGTGCTGCAC | ||
| Matrix-degrading enzymes | MMP-1 (Origene) | F: ATGAAGCAGCCCAGATGTGGAG |
| R: TGGTCCACATCTGCTCTTGGCA | ||
| MMP-3 (Origene) | F: CACTCACAGACCTGACTCGGTT | |
| R: AAGCAGGATCACAGTTGGCTGG | ||
| MMP-13 (Origene) | F: CCTTGATGCCATTACCAGTCTCC | |
| R: AAACAGCTCCGCATCAACCTGC | ||
| ADAMTS-5 (Origene) | F: CCTGGTCCAAATGCACTTCAGC | |
| R: TCGTAGGTCTGTCCTGGGAGTT | ||
| Matrix building components | Col2A1 (Col-2) (Qiagen) | Proprietary sequence |
| Aggrecan (Qiagen) | Proprietary sequence | |
| Modulators of inflammation | PTGES-2 (Origene) | F: CCTCTATGAGGCTGCTGACAAG |
| R: ATCACACGCAGCACGCCATACA | ||
| IL-6 (Origene) | F: AGACAGCCACTCACCTCTTCAG | |
| R: TTCTGCCAGTGCCTCTTTGCTG |
Table 2.
Genes Assessed in Evaluating Shifts in Mouse Macrophage Phenotype
| Primer sequence | ||
|---|---|---|
| Association | Gene marker (supplier) | Forward 5′-3 (F), Reverse 5′-3′ (R) |
| Reference gene | RPL-32 (Origene) | F: ATCAGGCACCAGTCAGACCGAT |
| R: GTTGCTCCCATAACCGATGTTGG | ||
| Classical activation markers | IL-1β (Origene) | F: TGGACCTTCCAGGATGAGGACA |
| R: GTTCATCTCGGAGCCTGTAGTG | ||
| IL-12β (Origene) | F: TTGAACTGGCGTTGGAAGCACG | |
| R: CCACCTGTGAGTTCTTCAAAGGC | ||
| NOS-2 (Origene) | F: GAGACAGGGAAGTCTGAAGCAC | |
| R: CCAGCAGTAGTTGCTCCTCTTC | ||
| “Wound healing” marker | Arginase-1 (Operon) | F: TGTGTCATTTGGGTGGATGCT |
| R: TGGTACATCTGGGAACTTTCC | ||
| Proangiogenic marker | VEGF-A (Operon) | F: CCTGTGTGCCGCTGATG |
| R: CGCATGATCTGCATGGTGAT | ||
| Anti-inflammatory marker | IL-10 (Origene) | F: CGGGAAGACAATAACTGCACCC |
| R: CGGTTAGCAGTATGTTGTCCAGC |
Western blotting
Supernatant from the mRNA extraction process was utilized for western blot analysis to semi-quantitatively compare protein expression among the different experimental groups. Briefly, DNA levels in each sample homogenate were measured using the PicoGreen assay kit (Life Technologies). Thereafter, sample homogenate volumes representing 8000 ng DNA (for chondrocytes) or 2500 ng DNA (for macrophages) were concentrated using 3000 MWCO Amicon filter units (Millipore), followed by the addition of β-mercaptoethanol and heat treatment at 95°C for 10 min. The concentrated protein lysates were loaded into separate wells of either an 8–15% gradient (for chondrocytes) or a 12% (for macrophages) sodium dodecyl sulfate polyacrylamide gel and separated using electrophoresis. Subsequently, the proteins were transferred to nitrocellulose membranes and the membranes were blocked with a 5% bovine serum albumin (BSA; Fisher Scientific) solution in TBST/NaN3 (25 mM Tris-HCl, pH 7.5, 137 mM NaCl, 0.1% Tween 20, and 0.05% NaN3) for 1 h at room temperature.
Primary antibodies for Sox-9 (Clone E-9; Santa Cruz Biotechnology), MMP-1 (Clone N-17-R; Santa Cruz Biotechnology), and PTGES-2 (ab96189; Abcam) were diluted in 5% BSA solution in TBST/NaN3. The diluted antibodies were applied to the membranes overnight at 4°C with constant shaking. Bound primary antibodies were detected by the application of appropriate horseradish peroxidase-conjugated or alkaline phosphatase-conjugated secondary antibodies (Jackson ImmunoResearch) for 1 h at room temperature, followed by the application of Luminol (Santa Cruz Biotechnology) or Novex chemiluminescent substrate (Life Technologies), respectively, for the development of signal. Chemiluminescence was detected using a ChemiDoc™ XRS+ System equipped with Image Lab™ Software (Bio-Rad Laboratories), with exposure time controlled to avoid signal saturation. Band optical density was quantified using Adobe Photoshop and normalized to the corresponding amount of loaded DNA based on a DNA content estimation from the PicoGreen assay.
Multiplex protein assays
The levels of TNF-α, IL-1β, IFN-γ, IL-6, IL-8, MCP-1, MMP-3, MMP-9, and MMP-13 produced by chondrocytes were analyzed using Luminex xMAP technology (Luminex). Protein expression was assessed by multiplex assay kits (R&D Systems and EMD Millipore) using the previously extracted supernatants and as per manufacturer instructions. Briefly, samples were diluted 1:1 in the kit-provided buffer and dispensed into a 96-well plate, after which magnetic bead suspensions, detection antibodies, and streptavidin–phycoerythrin were added to sample wells. Sample concentrations for each analyte were obtained on the basis of a median fluorescence intensity relative to standards. Subsequently, resulting measures were normalized to DNA content in each sample.
Similarly, the levels of IL-1β, IL-12β, TNF-α, IL-6, VEGF, and IL-10 produced by macrophages were analyzed using the mouse cytokine array multiplex kit (EMD Millipore) according to manufacturer's protocol and normalized to sample DNA content.
Immunohistochemical analyses
Hyaline cartilage ECM proteins collagen II (Col-2) and Aggrecan were also analyzed using a standard immunohistochemical technique. Briefly, 30 μm sections were cut from each embedded histological sample (n = 3–4 per group) using a cryomicrotome (Zeiss). Rehydrated sections were blocked with peroxidase for 10 min, followed by 10 min exposure to Terminator (Biocare Medical). Primary antibodies for Col-2 (Clone M-2139; Santa Cruz Biotechnology) and Aggrecan (Clone D-20; Santa Cruz Biotechnology) were diluted in PBS containing 3% BSA and then applied to the sections for 1 h. Bound primary antibody was detected using appropriate alkaline phosphatase-conjugated secondary antibodies (Jackson ImmunoResearch) followed by application of chromogen Ferangi Blue (Biocare Medical). Stained sections were imaged using a Zeiss Axiovert microscope, and cell counts were carried out on sections from each sample to semi-quantitatively evaluate immunostaining results. These counting assessments were conducted according to established methods.42,52,53 For each cell, i, in a given section, a staining intensity, di, was recorded on a scale of 0–3 (with 0 = “no staining” and 3 = “highest intensity among all formulations for that antibody”) by a single observer blinded to outcome. The cumulative staining intensity, d, for a given antibody in a particular section was calculated using the following equation: d = (∑ di)/(total cell number).
Statistical analysis
Data are reported as mean ± standard deviation for n = 3–4 samples per experimental group. Sample means were compared using a two-tailed, independent sample Student's t-test (SPSS; IBM), with a p-value ≤0.05 considered statistically significant.
Results
Initial chondrocyte and macrophage phenotype before coculture
To evaluate the initial phenotypic state of hydrogel-encapsulated cells, cell-laden 3D hydrogel discs were analyzed 1 day after encapsulation but before the initiation of coculture.
Phenotypic assessment of hydrogel-encapsulated chondrocytes before coculture
Although the gene expression levels of Aggrecan, Sox-9, and MMP-1 were not significantly different between NCs and OACs, the diseased chondrocytes expressed significantly higher levels of MMP-3 (p = 0.003), MMP-13 (p = 0.012), and PTGES-2 (p = 0.032) mRNA compared to NCs (Supplementary Fig. S1A; Supplementary Data are available online at www.liebertpub.com/tea), consistent with previous reports of OAC expression profiles.8,54,55 At the protein level, when compared to NCs, OACs produced significantly higher levels of MMP-9 and certain inflammatory cytokines (IL-1β, IL-8, MCP-1, VEGF, and IL-13) (Supplementary Fig. S1B). Each of these enzymes and inflammatory cytokines has been shown to be increased in OA.5,12,56,57 These results demonstrate significant initial phenotypic differences between the NCs and OACs before coculture with macrophages.
Phenotypic assessment of hydrogel-encapsulated macrophages before coculture
AMs initially expressed higher levels of NOS-2 mRNA (∼4-fold; p = 0.019) and TNF-α mRNA (∼2.7-fold; p = 0.007) compared to NMs, consistent with classical activation58 (Supplementary Fig. S2A). AMs also initially produced significantly higher amounts of VEGF-A mRNA (p = 0.001) relative to NMs, in agreement with previous literature describing the production of VEGF by RAW 264.7 cells upon LPS stimulation.59 At the protein level, AMs produced significantly greater amounts of TNF-α (∼5.3-fold; p < 0.001) and IL-6 (∼8.6-fold; p < 0.001) and significantly less IL-10 (∼2-fold; p = 0.020) relative to NMs (Supplementary Fig. S2B). Collectively, these results indicate that stimulation with LPS in our model system is generating proinflammatory macrophages, as anticipated.
Validation of the 3D coculture system: NC-AM coculture
In the first set of studies, we cocultured NCs and AMs to allow for comparisons of our 3D coculture system with established in vitro OA-inflammation models, which have generally used NCs.
Effect of coculture on NCs
Before assessing NC gene and protein expression, chondrocyte viability within the PEGDA hydrogels was evaluated. Measures of DNA were used to assess viability, as they are considered standard indicators of net cell proliferation and death within hydrogel networks.60 Chondrocyte and macrophage DNA measures at the endpoint were ∼73% of their initial values at the time of encapsulation (Supplementary Fig. S3), consistent with PEG hydrogel literature.41,52,61–64
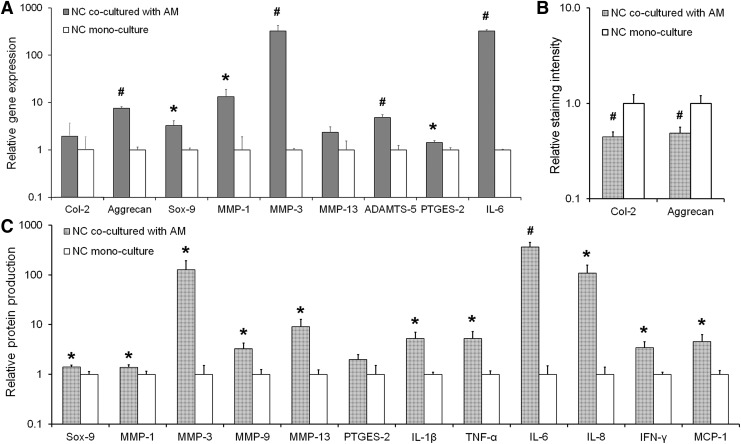
Following 7 days of culture, a variety of chondrocytic anabolic, catabolic, and inflammatory markers were analyzed by qRT-PCR, western blot, multiplex protein assays, and/or immunostaining. Representative western blot and immunostaining images are shown in Supplementary Figures S4–S6. Expression of matrix-building Aggrecan mRNA (∼7.5-fold; p < 0.001) and the chondrogenic transcription factor Sox-9 mRNA (∼3.2-fold; p = 0.013) by NCs in coculture was significantly higher compared to NCs in mono-culture (Fig. 2A). At the protein level, while Sox-9 expression by NCs in the two groups (Fig. 2C) (∼1.4-fold difference; p = 0.019) reflected the trend observed at the mRNA level, NCs in the NC-AM group produced significantly lower levels of Col-2 (∼2.5-fold; p < 0.001) and Aggrecan (∼2-fold; p < 0.001) relative to NC mono-culture (Fig. 2B).
FIG. 2.
(A) Relative gene expression by NCs cocultured with AMs as assessed by qRT-PCR. RPL-32 served as the reference gene, and the data are normalized to the NC mono-culture control. (B) Protein level production of Col-2 and aggrecan by NCs cocultured with AMs relative to NCs in mono-culture as assessed by semi-quantitative immunostaining. (C) Relative protein expression of the transcription factor Sox-9 and various MMPs and cytokines as assessed by western blot or multiplex assays. Data are normalized to the DNA content in each sample and further normalized to the NC mono-culture control. In each panel, significant differences between the coculture and mono-culture groups are indicated either by an asterisk (p < 0.05) or a pound symbol (p < 0.01). At least three samples per culture group were analyzed per assessment technique. qRT-PCR, quantitative reverse transcription polymerase chain reaction.
Gene expression levels of catabolic matrix-degrading enzymes such as MMP-1 (∼13-fold; p = 0.023), MMP-3 (∼323-fold; p = 0.005), and ADAMTS-5 (∼4.8-fold; p = 0.001) were markedly increased upon coculturing NCs with AMs (Fig. 2A), trends that were also reflected in protein production of MMP-1 (∼1.4-fold; p = 0.040) and MMP-3 (∼127-fold; p = 0.027) (Fig. 2C). While the increase in the expression of MMP-13 mRNA by NCs in the coculture group was not significantly higher than in the mono-culture control (p = 0.057) (Fig. 2A), the increase at the protein level was statistically significant (∼9.2-fold; p = 0.018) (Fig. 2C). When cocultured with AMs, NCs also expressed significantly higher amounts of PTGES-2 mRNA (∼1.4-fold; p = 0.014) and IL-6 mRNA (∼322-fold; p < 0.001) (Fig. 2A), both of which are associated with OA inflammation.65,66 At the protein level, however, only the increase in IL-6 (∼366-fold; p = 0.002) expression was statistically significant (Fig. 2C). Lastly, NCs in coculture demonstrated significantly increased production of proinflammatory cytokines IL-1β (∼5.3-fold; p = 0.013), TNF-α (∼5.3-fold; p = 0.020), IL-8 (∼108-fold; p = 0.018), IFN-γ (∼3.4-fold; p = 0.017), and MCP-1 (∼4.6-fold; p = 0.023) relative to NCs in the control group (Fig. 2C).
It is important to note here that the differences observed between the present gene and protein expression profiles are not unusual,67 and could be related to a host of complex factors including post-transcriptional and post-translational modifications and the fact that both the gene and protein expression data were measured at the same time point. Nevertheless, the NC protein production trends largely reflect the trends noted for gene expression. In addition, our results from the NC-AM system generally agree with cell responses observed in prior in vitro studies of NCs cultured under inflammatory conditions. For example, the expression trends for Aggrecan, Sox-9, and the MMPs are similar to previous reports by Sun et al.,28 who treated 3D encapsulated human chondrocytes with conditioned media obtained from differentiated THP-1 cells, and Peck et al.,33 who cocultured scaffold-free 3D cartilage with 2D RAW 264.7 macrophages in a trans-well setup. Similarly, the cytokine expression data are also similar to two previous reports—one in which healthy human chondrocytes were treated with the synovial fluid of OA patients,68 and another wherein healthy cartilage explants were cocultured with synovium from OA patients.69
Effect of coculture on AMs
In addition to NC response, we also investigated the phenotypic shift of AMs cocultured with NCs, which has generally remained unexplored in the literature. As AM hydrogel discs in coculture were replaced with fresh hydrogels containing newly activated AMs every 2 days following the first 24 h of culture (Fig. 1), we chose to conduct analyses of AMs cocultured with NCs from days 5 to 7 of the 7 day culture period. Thus, the AM data reported herein reflect shifts in AM polarization after 2 days of coculture with NCs that had previously been conditioned by AM paracrine signals.
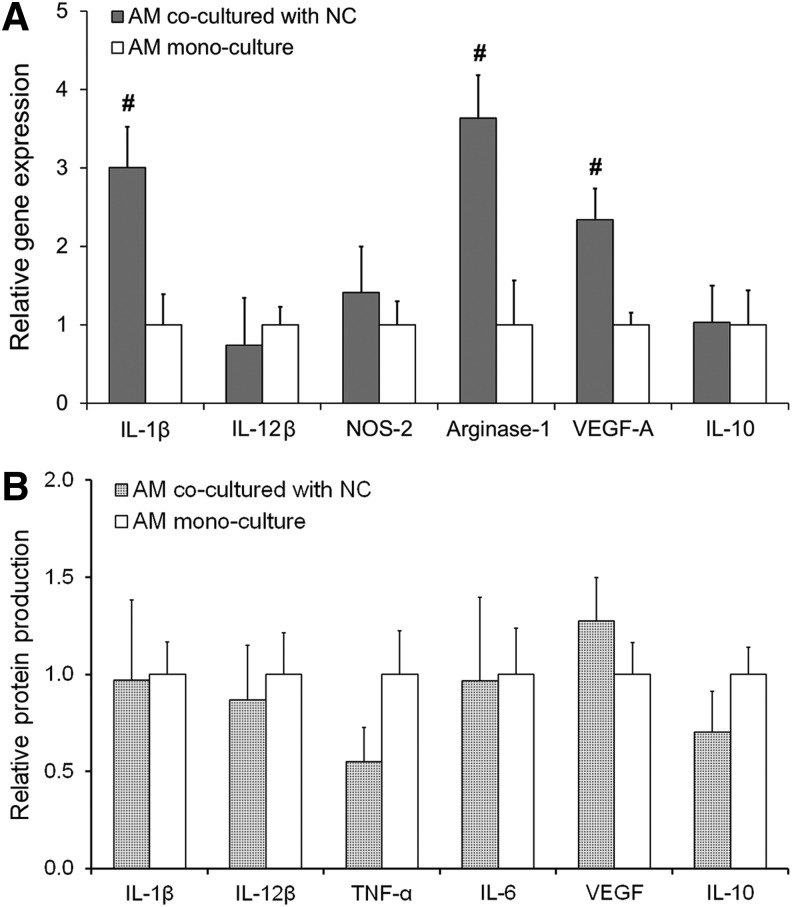
The gene expression analyses revealed that mRNA for IL-1β—a marker strongly associated with OA inflammation—was significantly increased (∼3-fold; p = 0.006) in AMs cocultured with NCs previously been stimulated with AMs (Fig. 3A). Interestingly, Arginase-1 (∼3.6-fold; p = 0.004) and VEGF-A mRNA (∼2.3-fold; p = 0.006) were also significantly elevated in NC-AM coculture compared to AM mono-culture controls. Increases in VEGF-A or Arginase-1 levels are generally associated with an M2 macrophage phenotype.46,59 Overall, these gene expression results are indicative of the emergence of mixed M1/M2 phenotypes in AMs exposed to previously stimulated NCs. However protein level data showed no change in any cytokine investigated (Fig. 3B), indicating that an M1 macrophage phenotype was being maintained despite shifts in the macrophage transcriptional profile.
FIG. 3.
(A) Relative gene expression by AMs cocultured with NCs as assessed by qRT-PCR. RPL-32 served as the reference gene, and the data are normalized to the AM mono-culture control. (B) Protein expression of various cytokines produced by AMs cocultured with NCs relative to mono-culture controls as assessed by multiplex assays. Significant differences between the coculture and mono-culture groups are indicated either by an asterisk (p < 0.05) or a pound symbol (p < 0.01). At least three samples per culture group were analyzed.
Collectively, these results indicate that our coculture system allows the investigation of paracrine interactions between the two cell types. The presence of AMs appears to drive the NCs toward overexpressing matrix-degrading enzymes and proinflammatory cytokines both at the gene and protein level, while the presence of the phenotypically evolving NCs appears to trigger the evolution of a mixed phenotype in macrophages at the transcriptional level but not at the protein level.
Establishment of an in vitro 3D OA model: Coculture of OAC and AM
Having confirmed the sensitivity of our system using NCs and the effects of conditioned NCs on AMs, we next examined the impact of AM paracrine factors on chondrocytes derived from osteoarthritic cartilage (OACs). Toward this end, OACs and activated AMs were cocultured and the expression of various markers was examined at the gene and protein levels.
Effect of coculture on OACs
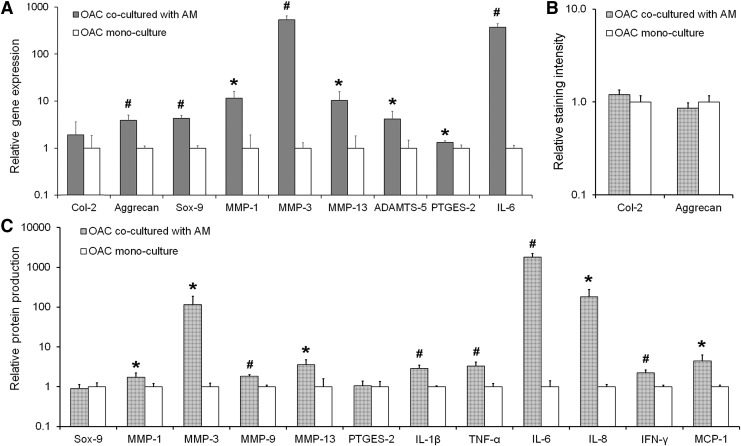
Both at the gene and protein levels, OACs cultured in the presence of AMs secreted higher levels of matrix-degrading enzymes and proinflammatory mediators (Fig. 4A, C). In terms of anabolic chondrogenic makers, OACs cocultured with AMs demonstrated significantly higher mRNA expression of Aggrecan (∼3.9-fold; p = 0.010) and Sox-9 (∼4.2-fold; p = 0.001) compared to OAC mono-culture controls (Fig. 4A). However, at the protein level, no significant differences in Sox-9 or Aggrecan were noted between the two experimental groups (Fig. 4B, C). Similarly, no significant differences in Col 2 levels were observed at the mRNA or protein level.
FIG. 4.
(A) Relative gene expression by OACs cocultured with AMs as assessed by qRT-PCR. RPL-32 served as the reference gene, and the data are normalized to the NC mono-culture control. (B) Protein level production of Col-2 and aggrecan by OACs cocultured with AMs relative to OACs in mono-culture as assessed by semi-quantitative immunostaining. (C) Relative protein expression of the transcription factor Sox-9 and various MMPs and cytokines as assessed by western blot or multiplex assays. Data are normalized to the DNA content in each sample and further normalized to the OAC mono-culture control. In each panel, significant differences between the coculture and mono-culture groups are indicated either by an asterisk (p < 0.05) or a pound symbol (p < 0.01). At least three samples per culture group were analyzed per assessment technique. OAC, osteoarthritic chondrocyte.
The mRNA levels of four key matrix-degrading enzymes, namely MMP-1 (∼11.5-fold; p = 0.018), MMP-3 (∼534-fold; p = 0.001), MMP-13 (∼10-fold; p = 0.040), and ADAMTS-5 (∼4.1-fold; p = 0.050) were significantly elevated in OAC-AM coculture relative to OAC mono-culture (Fig. 4A), and these increases were also reflected at the protein level for MMP-1 (∼1.7-fold; p = 0.035), MMP-3 (∼115-fold; p = 0.022) and MMP-13 (∼3.6-fold; p = 0.011) (Fig. 4C). In addition, MMP-9 protein expression by OACs in the coculture group was significantly higher than in the mono-culture group (1.8-fold; p < 0.001) (Fig. 4C).
Similarly, OACs in coculture expressed significantly higher levels of IL-6 (∼370-fold, p < 0.001) and PTGES-2 (∼1.3-fold, p = 0.042) mRNA compared to OAC controls (Fig. 4A). At the protein level, OACs cocultured with AMs produced significantly higher levels of the inflammatory cytokines IL-6 (∼1800-fold; p < 0.001), IL-1β (∼2.9-fold; p = 0.001), TNF-α (∼3.3-fold; p = 0.003), IL-8 (∼181-fold; p = 0.011), IFN-γ (∼2.2-fold; p = 0.001), and MCP-1 (∼4.5-fold; p = 0.011) relative to the OAC mono-culture control (Fig. 4C). As for the NC-AM system, the protein production trends largely reflected the trends noted for gene expression, and indicate that AMs can exacerbate the diseased condition of OACs.
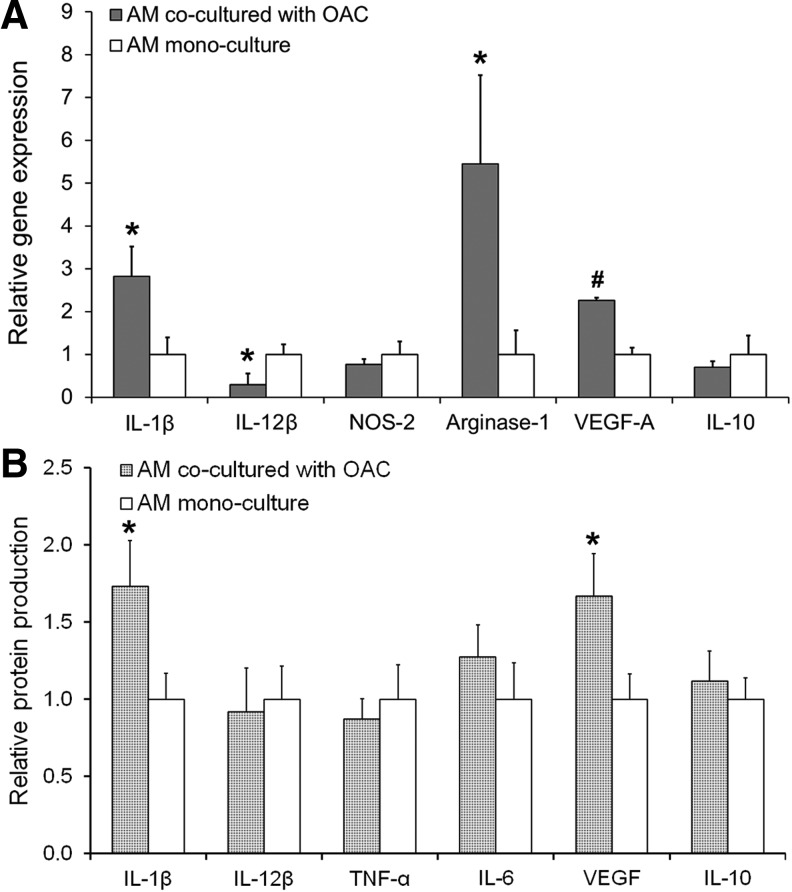
Effect of coculture on AMs
In assessing AM responses to OACs, we analyzed AM hydrogel discs cocultured with OACs from days 5 to 7 of the 7-day culture. AMs in coculture were significantly influenced by the presence of OACs compared to the AM mono-culture controls (Fig. 5). While the expression of IL-1β mRNA by AMs was increased in coculture (∼2.8-fold; p = 0.017), the expression of IL-12β—another marker associated with inflammation70—was reduced (∼3.3-fold; p = 0.024) upon coculture with OACs (Fig. 5A). Additionally, Arginase-1 (∼5.4-fold; p = 0.023) and VEGF-A (∼2.2-fold; p < 0.001) mRNA were elevated in the OAC-AM coculture system (Fig. 5A). Furthermore, at the protein level, AMs cocultured with OACs produced significantly elevated levels of the proinflammatory cytokine IL-1β (∼1.7-fold; p = 0.038) and VEGF (∼1.6-fold; p = 0.045) (Fig. 5B). These results suggest the emergence of mixed M1/M2 phenotypes at both the transcriptional and protein levels in AMs exposed to OACs, which is consistent with previously reported mixed macrophage subsets isolated from the synovium of OA patients.71
FIG. 5.
(A) Relative gene expression by AMs cocultured with OACs as assessed by qRT-PCR. RPL-32 served as the reference gene, and the data are normalized to the AM mono-culture control. (B) Protein expression of various cytokines produced by AMs cocultured with NCs relative to mono-culture controls as assessed by multiplex assays. Significant differences between the coculture and mono-culture groups are indicated either by an asterisk (p < 0.05) or a pound symbol (p < 0.01). At least three samples per culture group were analyzed.
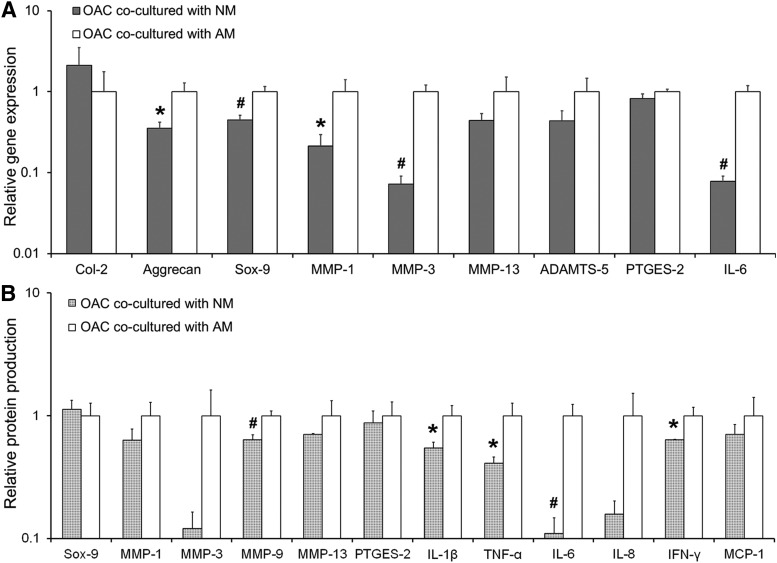
Toward modulating inflammation: Comparison of OAC-AM and OAC-NM
In the final set of experiments, we evaluated the effect of culturing OACs in the presence of NMs to determine whether this coculture mode resulted in a general decrease in the expression of matrix-degrading enzymes and proinflammatory cytokines compared to the OAC-AM system. A consistent decrease in the mRNA expression of almost all the analyzed markers was observed in the OAC-NM group relative to the OAC-AM group at both the gene and protein levels (Fig. 6). Specifically, the expression of Aggrecan (∼2.8-fold; p = 0.018) and Sox-9 (∼2.2-fold; p = 0.005) mRNA by OACs was significantly lower in the OAC-NM group compared to the OAC-AM group (Fig. 6A). Likewise, MMP-1 (∼4.6-fold, p = 0.03), MMP-3 (∼13.8-fold, p = 0.001), and IL-6 (∼12.8-fold, p = 0.001) mRNA were also markedly lower in the OAC-NM group (Fig. 6A). At the protein level, OACs cocultured with NM produced significantly lower levels of MMP-9 (∼1.6-fold; p = 0.031), IL-1β (∼1.8-fold; p = 0.024), TNF-α (∼2.4-fold; p = 0.019), IL-6 (∼9.1-fold; p = 0.012), and IFN-γ (∼1.6-fold; p = 0.021) relative to OACs cocultured with AM (Fig. 6B). Although MMP-3 and IL-8 appeared to be lower in the OAC-NM group relative to OAC-AM, these differences fell below statistical significance with p values of 0.072 and 0.051, respectively.
FIG. 6.
(A) Comparison of relative gene expression by OACs cocultured with AMs and OACs cocultured with NMs as assessed by qRT-PCR. RPL-32 served as the reference gene and the data are normalized to the OAC-AM coculture group. (B) Protein production by OACs cocultured with AMs as assessed by western blotting relative to OACs cocultured with NMs. Data are normalized to the DNA content in each sample and further normalized to the OAC-AM group. In both the panels, significant differences between the coculture and mono-culture groups are indicated either by an asterisk (p < 0.05) or a pound symbol (p < 0.01). At least three samples per culture group were analyzed per assessment technique. NM, normal macrophage.
Overall, the qRT-PCR and protein data suggest that OACs cocultured with NMs express significantly lower amounts of matrix-degrading enzymes and certain proinflammatory cytokines. These results are in line with certain anti-inflammatory agents moderately reducing certain OA symptoms.15,72 However, more detailed studies over longer time periods will be required to understand the mechanisms through which OAC-NM interactions differ from OAC-AM interactions.
Discussion
Recent literature indicates that inflammation plays a critical role in the onset and progression of knee OA.5 In vitro models that recapitulate key alterations associated with OA may enable the controlled investigation of cell–cell interactions and signaling mechanisms involved in OA, potentially allowing for improved understanding of OA disease progression at the cellular level and for the development of new therapeutics.73 Toward this eventual goal, we developed a fully 3D in vitro hydrogel-based system (Fig. 1A) that permits the independent evaluation of chondrocyte and macrophage phenotype in coculture, while concurrently allowing us to probe paracrine signaling between the two cell types. The culture of both chondrocytes and macrophages in 3D represents a significant difference between the present study and much of the previous in vitro work on OA and inflammation. As the primary location of the macrophages in the OA joint is within the synovial membrane, it is anticipated that 3D culture of macrophages and chondrocytes will more accurately reflect the in vivo environment.
First, we cultured NCs in the presence of proinflammatory AMs, which allowed us to compare our results with existing in vitro models of OA inflammation. The increased gene expression of Aggrecan and Sox-9 by NCs in coculture (Fig. 2A) is consistent with previously reported gene expression trends for stage 1 or early OA.30,74,75 This increased expression of matrix-building genes may also be related to the NCs trying to compensate for the loss in normal matrix proteins76 (Fig. 2B) due to the elevated expression of matrix-degrading enzymes (Fig. 2A, C). Our NC-AM ECM formation and degradation (i.e., Aggrecan MMPs) gene expression data are also consistent with previous in vitro results from several groups who reported increased expression of Aggrecan, MMP-1, and MMP-3 mRNA by 3D cartilage explants following either coculture with mouse macrophages or media conditioned by THP-1 cells.28,29,33 Similarly, our inflammatory cytokine data showing significantly increased production of IL-1β, TNF-α, IL-8, IFN-γ, and MCP-1 also matches well with other groups who focused on chondrocyte-produced inflammatory markers. These studies have demonstrated the enhanced production of key inflammatory mediators from chondrocytes exposed to OA synovial fluid or the synovium itself, which may act to further stimulate macrophage inflammation.12,56,57,68,69 Cumulatively, our results agree well with changes seen in early OA and with similar in vitro model systems using NCs.
The use of diseased rather than normal chondrocytes may enable modeling of the later stages of OA progression, and therefore may be more appropriate for testing potential therapeutics than OA models employing NCs. Thus, we next extended our coculture system to examine the impact of proinflammatory macrophages on OACs to evaluate the potential effects of AMs in exacerbating existing osteoarthritic phenotypes (Fig. 4). When comparing the chondrocyte response in the NC-AM coculture and OAC-AM coculture systems, many of the fold increases in gene and protein expression appeared to be similar (Fig. 2 vs. Fig. 4). However, two major differences between these culture systems were revealed. First, in the NC-AM system, significant decreases in Aggrecan and Col-2 protein levels were noticed upon coculture with AM (in contrast to the gene expression data, Fig. 2B), but no differences in these two proteins were apparent in the OAC-AM system (Fig. 4B). Second, the increases in IL-6 and IL-8 proteins were notably greater in the OAC-AM system compared to the NC-AM system (Fig. 2C vs. Fig. 4C).
In terms of the stages of human disease, Aggrecan and Col-2 protein levels have previously been demonstrated to decrease in early OA, whereas they may slightly increase or decrease at the final stages of disease.77,78 It is therefore possible that, at some point, the chondrocyte response switches from catabolism to anabolism, and we are capturing this transition with our OAC-AM system. Although we did not observe significant differences between the OAC-AM and the NC-AM groups for most of the major cytokines and MMPs examined, several reports have suggested that MMP profiles and the “major” cytokines such as TNF-α and IL-1β are not reliable delineators of various stages of OA.79–81 In contrast, the two cytokines for which we observed differences between the OAC-AM and NC-AM group - namely IL-6 and IL-8 - have been demonstrated to increase in late versus early shoulder OA.82 Combined, these points of comparison of NC-AM to OAC-AM to human disease suggest that our OAC-AM system may be serving as a disease mimic for later stage OA than the NC-AM system.
In addition to evaluating the chondrocyte response, we investigated the phenotypic evolution of macrophages in the final stages of coculture to assess the effect of NCs and OACs on AMs (Figs. 3 and 5). The macrophage response to either normal or diseased chondrocytes has remained largely uncharacterized in previous in vitro OA models.28,33 Interestingly, our gene expression data revealed that both NCs and OACs induce similar gene expression profiles in AMs and indicate the emergence mixed M1/M2 macrophage phenotypes (Fig. 3A and Fig. 5A), in line with the presence of mixed M1/M2 phenotypes in the synovium of OA patients.71 In particular, the proinflammatory marker IL-1β and wound healing markers Arginase-1 and VEGF-A mRNA were all significantly elevated in cocultured AMs regardless of chondrocyte disease state (i.e., cocultured with NCs or OACs). Interestingly, however, protein level analyses suggested that only OACs resulted in actual shifts in macrophage activation (Fig. 3B vs. Fig. 5B). In particular, IL-1β and VEGF protein levels were significantly elevated in AMs cocultured with OACs relative to AM mono-culture controls, a phenomenon that did not occur in the presence of NCs. The increased IL-1β protein production by AMs in the coculture group is potentially significant due to the known involvement of IL-1β in promoting inflammation in OA.6 Similarly, the increase in VEGF protein is potentially significant because it has been previously been shown to be enhanced in OA,83 with angiogenesis tightly linked to inflammation.84
OA progression represents a complex set of processes that are mediated by a number of factors. While the current work examined only a limited subset of the interactions believed to contribute to OA, examining the contribution of other cell types, the contribution of the various native ECM components, and the precise mechanisms aiding OA progression are beyond the scope of this study. In addition, the use of cells from two separate species (human chondrocytes vs. mouse macrophages) likely reduced the efficacy of chondrocyte-macrophage crosstalk. However, the cumulative ECM and phenotypic data indicate that proinflammatory macrophage paracrine signaling promotes abnormal chondrocyte phenotypes. It is also important to note that a number of studies have analyzed the response of human cells to murine RAW 264.7 cells, with results that frequently replicate key features of disease pathology.85–93 Future studies will use primary human macrophages to further develop this coculture system. Eventual testing of new therapeutics, particularly those targeting macrophage activation, using our developed coculture system will also be a key focus of future work.
Conclusions
In this study, the progression of inflammation in OA was studied by coculturing normal and diseased chondrocytes in the presence of classically activated, proinflammatory macrophages. Chondrocyte and macrophage responses to coculture were assessed by analyzing the expression of markers associated with ECM anabolism, matrix degradation, inflammation, and angiogenesis. The current results indicate that this coculture system recapitulates several key features of OA at the cellular level (in terms of expression profiles) and can potentially be used for studying the interplay between macrophages and chondrocytes in OA disease progression.
Supplementary Material
Acknowledgments
The authors would like to acknowledge an NSF CAREER award (Award No. 1346807) and NIH R01 DC013508 for funding. We would also like to thank Sean Quinn and Drew Kopicki for assistance with RNA extraction and western blotting, and Dr. Juan Felipe Diaz-Quiroz and Jonathan Kulwatno for assistance with immunostaining and immunostaining assessment.
Disclosure Statement
No competing financial interests exist.
References
- 1.Martel-Pelletier J., Boileau C., Pelletier J.P., and Roughley P.J. Cartilage in normal and osteoarthritis conditions. Best Pract Res Clin Rheumatol 22, 351, 2008 [DOI] [PubMed] [Google Scholar]
- 2.Heidari B. Knee osteoarthritis prevalence, risk factors, pathogenesis and features: part I. Caspian J Intern Med 2, 205, 2011 [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper C., Dennison E., Edwards M., and Litwic A. Epidemiology of osteoarthritis. Medicographia 35, 145, 2013 [Google Scholar]
- 4.Jordan J.M., Helmick C.G., Renner J.B., Luta G., Dragomir A.D., Woodard J., Fang F., Schwartz T.A., Abbate L.M., Callahan L.F., Kalsbeek W.D., and Hochberg M.C. Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians: the Johnston County Osteoarthritis Project. J Rheumatol 34, 172, 2007 [PubMed] [Google Scholar]
- 5.de Lange-Brokaar B.J., Ioan-Facsinay A., van Osch G.J., Zuurmond A.M., Schoones J., Toes R.E., Huizinga T.W., and Kloppenburg M. Synovial inflammation, immune cells and their cytokines in osteoarthritis: a review. Osteoarthritis Cartilage 20, 1484, 2012 [DOI] [PubMed] [Google Scholar]
- 6.Bondeson J., Wainwright S.D., Lauder S., Amos N., and Hughes C.E. The role of synovial macrophages and macrophage-produced cytokines in driving aggrecanases, matrix metalloproteinases, and other destructive and inflammatory responses in osteoarthritis. Arthritis Res Ther 8, R187, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pham T.N., Rahman P., Tobin Y.M., Khraishi M.M., Hamilton S.F., Alderdice C., and Richardson V.J. Elevated serum nitric oxide levels in patients with inflammatory arthritis associated with co-expression of inducible nitric oxide synthase and protein kinase C-eta in peripheral blood monocyte-derived macrophages. J Rheumatol 30, 2529, 2003 [PubMed] [Google Scholar]
- 8.Li W., Du C., Wang H., and Zhang C. Increased serum ADAMTS-4 in knee osteoarthritis: a potential indicator for the diagnosis of osteoarthritis in early stages. Genet Mol Res 13, 9642, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Jalba B.A., Jalba C.S., Vladoi A.D., Gherghina F., Stefan E., and Cruce M. Alterations in expression of cartilage-specific genes for aggrecan and collagen type II in osteoarthritis. Rom J Morphol Embryol 52, 587, 2011 [PubMed] [Google Scholar]
- 10.Li N.G., Shi Z.H., Tang Y.P., Wang Z.J., Song S.L., Qian L.H., Qian D.W., and Duan J.A. New hope for the treatment of osteoarthritis through selective inhibition of MMP-13. Curr Med Chem 18, 977, 2011 [DOI] [PubMed] [Google Scholar]
- 11.Baragi V.M., Becher G., Bendele A.M., Biesinger R., Bluhm H., Boer J., Deng H., Dodd R., Essers M., Feuerstein T., Gallagher B.M., Gege C., Hochguertel M., Hofmann M., Jaworski A., Jin L.X., Kiely A., Korniski B., Kroth H., Nix D., Nolte B., Piecha D., Powers T.S., Richter F., Schneider M., Steeneck C., Sucholeiki I., Taveras A., Timmermann A., Van Veldhuizen J., Weik J., Wu X.Y., and Xia B. A new class of potent matrix metalloproteinase 13 inhibitors for potential treatment of osteoarthritis evidence of histologic and clinical efficacy without musculoskeletal toxicity in rat models. Arthritis Rheum 60, 2008, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Wojdasiewicz P., Poniatowski L.A., and Szukiewicz D. The role of inflammatory and anti-inflammatory cytokines in the pathogenesis of osteoarthritis. Mediators Inflamm 2014, 561459, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnson C.I., Argyle D.J., and Clements D.N. In vitro models for the study of osteoarthritis. Vet J 209, 40, 2016 [DOI] [PubMed] [Google Scholar]
- 14.Takahashi A., de Andres M.C., Hashimoto K., Itoi E., and Oreffo R.O. Epigenetic regulation of interleukin-8, an inflammatory chemokine, in osteoarthritis. Osteoarthritis Cartilage 23, 1946, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roubille C., Martel-Pelletier J., and Pelletier J. Osteoarthritis treatments: where do we stand at the moment. Medicographia 35, 172, 2013 [Google Scholar]
- 16.Masuko K., Murata M., Yudoh K., Kato T., and Nakamura H. Anti-inflammatory effects of hyaluronan in arthritis therapy: not just for viscosity. Int J Gen Med 2, 77, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evanich J.D., Evanich C.J., Wright M.B., and Rydlewicz J.A. Efficacy of intraarticular hyaluronic acid injections in knee osteoarthritis. Clin Orthop Relat Res 173, 2001 [DOI] [PubMed] [Google Scholar]
- 18.Wang C.T., Lin J., Chang C.J., Lin Y.T., and Hou S.M. Therapeutic effects of hyaluronic acid on osteoarthritis of the knee. A meta-analysis of randomized controlled trials. J Bone Joint Surg Am 86-A, 538, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Fioravanti A., Tinti L., Pascarelli N.A., Di Capua A., Lamboglia A., Cappelli A., Biava M., Giordani A., Niccolini S., Galeazzi M., and Anzini M. In vitro effects of VA441, a new selective cyclooxygenase-2 inhibitor, on human osteoarthritic chondrocytes exposed to IL-1 beta. J Pharmacol Sci 120, 6, 2012 [DOI] [PubMed] [Google Scholar]
- 20.Zhou X.D., Li W.J., Jiang L.F., Bao J.P., Tao L.J., Li J., and Wu L.D. Tetrandrine inhibits the Wnt/beta-catenin signalling pathway and alleviates osteoarthritis: an in vitro and in vivo study. Evid Based Compl Altern Med 2013. Article id: 809579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bondeson J., Wainwright S., Hughes C., and Caterson B. The regulation of the ADAMTS4 and ADAMTS5 aggrecanases in osteoarthritis: a review. Clin Exp Rheumatol 26, 139, 2008 [PubMed] [Google Scholar]
- 22.Flannery C.R., Little C.B., Caterson B., and Hughes C.E. Effects of culture conditions and exposure to catabolic stimulators (IL-1 and retinoic acid) on the expression of matrix metalloproteinases (MMPs) and disintegrin metalloproteinases (ADAMs) by articular cartilage chondrocytes. Matrix Biol 18, 225, 1999 [DOI] [PubMed] [Google Scholar]
- 23.Reginato A.M., Sanz-Rodriguez C., Diaz A., Dharmavaram R.M., and Jimenez S.A. Transcriptional modulation of cartilage-specific collagen gene expression by interferon γ and tumour necrosis factor α in cultured human chondrocytes. Biochem J 294, 761, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caron M.M., Emans P.J., Coolsen M.M., Voss L., Surtel D.A., Cremers A., van Rhijn L.W., and Welting T.J. Redifferentiation of dedifferentiated human articular chondrocytes: comparison of 2D and 3D cultures. Osteoarthritis Cartilage 20, 1170, 2012 [DOI] [PubMed] [Google Scholar]
- 25.Schulze-Tanzil G. Activation and dedifferentiation of chondrocytes: implications in cartilage injury and repair. Ann Anat 191, 325, 2009 [DOI] [PubMed] [Google Scholar]
- 26.Bartneck M., Heffels K.H., Pan Y., Bovi M., Zwadlo-Klarwasser G., and Groll J. Inducing healing-like human primary macrophage phenotypes by 3D hydrogel coated nanofibres. Biomaterials 33, 4136, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharjee M., Schultz-Thater E., Trella E., Miot S., Das S., Loparic M., Ray A.R., Martin I., Spagnoli G.C., and Ghosh S. The role of 3D structure and protein conformation on the innate and adaptive immune responses to silk-based biomaterials. Biomaterials 34, 8161, 2013 [DOI] [PubMed] [Google Scholar]
- 28.Sun L., Wang X., and Kaplan D.L. A 3D cartilage—inflammatory cell culture system for the modeling of human osteoarthritis. Biomaterials 32, 5581, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murab S., Chameettachal S., Bhattacharjee M., Das S., Kaplan D.L., and Ghosh S. Matrix-embedded cytokines to simulate osteoarthritis-like cartilage microenvironments. Tissue Eng Part A 19, 1733, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bau B., Gebhard P.M., Haag J., Knorr T., Bartnik E., and Aigner T. Relative messenger RNA expression profiling of collagenases and aggrecanases in human articular chondrocytes in vivo and in vitro. Arthritis Rheum 46, 2648, 2002 [DOI] [PubMed] [Google Scholar]
- 31.Blasioli D.J., Matthews G.L., and Kaplan D.L. The degradation of chondrogenic pellets using cocultures of synovial fibroblasts and U937 cells. Biomaterials 35, 1185, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dreier R., Wallace S., Fuchs S., Bruckner P., and Grassel S. Paracrine interactions of chondrocytes and macrophages in cartilage degradation: articular chondrocytes provide factors that activate macrophage-derived pro-gelatinase B (pro-MMP-9). J Cell Sci 114, 3813, 2001 [DOI] [PubMed] [Google Scholar]
- 33.Peck Y., Ng L.Y., Goh J.Y., Gao C., and Wang D.A. A three-dimensionally engineered biomimetic cartilaginous tissue model for osteoarthritic drug evaluation. Mol Pharm 11, 1997, 2014 [DOI] [PubMed] [Google Scholar]
- 34.Williams C.G., Kim T.K., Taboas A., Malik A., Manson P., and Elisseeff J. In vitro chondrogenesis of bone marrow-derived mesenchymal stem cells in a photopolymerizing hydrogel. Tissue Eng 9, 679, 2003 [DOI] [PubMed] [Google Scholar]
- 35.Bahney C.S., Hsu C.W., Yoo J.U., West J.L., and Johnstone B. A bioresponsive hydrogel tuned to chondrogenesis of human mesenchymal stem cells. FASEB J 25, 1486, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nguyen Q.T., Hwang Y., Chen A.C., Varghese S., and Sah R.L. Cartilage-like mechanical properties of poly (ethylene glycol)-diacrylate hydrogels. Biomaterials 33, 6682, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin S., Sangaj N., Razafiarison T., Zhang C., and Varghese S. Influence of physical properties of biomaterials on cellular behavior. Pharm Res 28, 1422, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Munoz-Pinto D.J., Jimenez-Vergara A.C., Gharat T.P., and Hahn M.S. Characterization of sequential collagen-poly(ethylene glycol) diacrylate interpenetrating networks and initial assessment of their potential for vascular tissue engineering. Biomaterials 40, 32, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Munoz-Pinto D.J., Samavedi S., Grigoryan B., and Hahn M.S. Impact of secondary reactive species on the apparent decoupling of poly(ethylene glycol) diacrylate hydrogel average mesh size and modulus. Polymer 77, 227, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gombotz W.R., Guanghui W., Horbett T.A., and Hoffman A.S. Protein adsorption to poly(ethylene oxide) surfaces. J Biomed Mater Res 25, 1547, 1991 [DOI] [PubMed] [Google Scholar]
- 41.Nuttelman C.R., Tripodi M.C., and Anseth K.S. Synthetic hydrogel niches that promote hMSC viability. Matrix Biol 24, 208, 2005 [DOI] [PubMed] [Google Scholar]
- 42.Benoit D.S.W., Schwartz M.P., Durney A.R., and Anseth K.S. Small functional groups for controlled differentiation of hydrogel-encapsulated human mesenchymal stem cells. Nat Mater 7, 816, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Becerra-Bayona S., Guiza-Arguello V., Qu X., Munoz-Pinto D.J., and Hahn M.S. Influence of select extracellular matrix proteins on mesenchymal stem cell osteogenic commitment in three-dimensional contexts. Acta Biomater 8, 4397, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahim S.S., Khan N., Boddupalli C.S., Hasnain S.E., and Mukhopadhyay S. Interleukin-10 (IL-10) mediated suppression of IL-12 production in RAW 264.7 cells also involves c-rel transcription factor. Immunology 114, 313, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barth K.A., Waterfield J.D., and Brunette D.M. The effect of surface roughness on RAW 264.7 macrophage phenotype. J Biomed Mater Res A 101, 2679, 2013 [DOI] [PubMed] [Google Scholar]
- 46.Lynn A.D., and Bryant S.J. Phenotypic changes in bone marrow-derived murine macrophages cultured on PEG-based hydrogels activated or not by lipopolysaccharide. Acta Biomater 7, 123, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams L.M., and Ridley A.J. Lipopolysaccharide induces actin reorganization and tyrosine phosphorylation of Pyk2 and paxillin in monocytes and macrophages. J Immunol 164, 2028, 2000 [DOI] [PubMed] [Google Scholar]
- 48.Bryant S.J., Nuttelman C.R., and Anseth K.S. Cytocompatibility of UV and visible light photoinitiating systems on cultured NIH/3T3 fibroblasts in vitro. J Biomater Sci Polym Ed 11, 439, 2000 [DOI] [PubMed] [Google Scholar]
- 49.Bryant S.J., and Anseth K.S. Hydrogel properties influence ECM production by chondrocytes photoencapsulated in poly(ethylene glycol) hydrogels. J Biomed Mater Res 59, 63, 2002 [DOI] [PubMed] [Google Scholar]
- 50.Williams C.G., Malik A.N., Kim T.K., Manson P.N., and Elisseeff J.H. Variable cytocompatibility of six cell lines with photoinitiators used for polymerizing hydrogels and cell encapsulation. Biomaterials 26, 1211, 2005 [DOI] [PubMed] [Google Scholar]
- 51.Livak K.J., and Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402, 2001 [DOI] [PubMed] [Google Scholar]
- 52.Salinas C.N., and Anseth K.S. The influence of the RGD peptide motif and its contextual presentation in PEG gels on human mesenchymal stem cell viability. J Tissue Eng Regen Med 2, 296, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Salinas C.N., and Anseth K.S. The enhancement of chondrogenic differentiation of human mesenchymal stem cells by enzymatically regulated RGD functionalities. Biomaterials 29, 2370, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Attur M., Al-Mussawir H.E., Patel J., Kitay A., Dave M., Palmer G., Pillinger M.H., and Abramson S.B. Prostaglandin E2 exerts catabolic effects in osteoarthritis cartilage: evidence for signaling via the EP4 receptor. J Immunol 181, 5082, 2008 [DOI] [PubMed] [Google Scholar]
- 55.Aigner T., Zien A., Gehrsitz A., Gebhard P.M., and McKenna L. Anabolic and catabolic gene expression pattern analysis in normal versus osteoarthritic cartilage using complementary DNA-array technology. Arthritis Rheum 44, 2777, 2001 [DOI] [PubMed] [Google Scholar]
- 56.Kapoor M., Martel-Pelletier J., Lajeunesse D., Pelletier J.P., and Fahmi H. Role of proinflammatory cytokines in the pathophysiology of osteoarthritis. Nat Rev Rheumatol 7, 33, 2011 [DOI] [PubMed] [Google Scholar]
- 57.Zeng G.Q., Chen A.B., Li W., Song J.H., and Gao C.Y. High MMP-1, MMP-2, and MMP-9 protein levels in osteoarthritis. Genet Mol Res 14, 14811, 2015 [DOI] [PubMed] [Google Scholar]
- 58.Mosser D.M., and Zhang X. Activation of murine macrophages. Curr Protoc Immunol Chapter 14, Unit 14.2, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Du M., Roy K.M., Zhong L.H., Shen Z., Meyers H.E., and Nichols R.C. VEGF gene expression is regulated post-transcriptionally in macrophages. FEBS J 273, 732, 2006 [DOI] [PubMed] [Google Scholar]
- 60.Munoz-Pinto D.J., Jimenez-Vergara A.C., Gelves L.M., McMahon R.E., Guiza-Arguello V., and Hahn M.S. Probing vocal fold fibroblast response to hyaluronan in 3D contexts. Biotechnol Bioeng 104, 821, 2009 [DOI] [PubMed] [Google Scholar]
- 61.Jimenez-Vergara A.C., Munoz-Pinto D.J., Becerra-Bayona S., Wang B., Iacob A., and Hahn M.S. Influence of glycosaminoglycan identity on vocal fold fibroblast behavior. Acta Biomater 7, 3964, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinmetz N.J., and Bryant S.J. The effects of intermittent dynamic loading on chondrogenic and osteogenic differentiation of human marrow stromal cells encapsulated in RGD-modified poly(ethylene glycol) hydrogels. Acta Biomater 7, 3829, 2011 [DOI] [PubMed] [Google Scholar]
- 63.Benoit D.S.W., Durney A.R., and Anseth K.S. The effect of heparin-functionalized PEG hydrogels on three-dimensional human mesenchymal stem cell osteogenic differentiation. Biomaterials 28, 66, 2007 [DOI] [PubMed] [Google Scholar]
- 64.Burdick J.A., and Anseth K.S. Photoencapsulation of osteoblasts in injectable RGD-modified PEG hydrogels for bone tissue engineering. Biomaterials 23, 4315, 2002 [DOI] [PubMed] [Google Scholar]
- 65.Li X., Ellman M., Muddasani P., Wang J.H., Cs-Szabo G., van Wijnen A.J., and Im H.J. Prostaglandin E2 and its cognate EP receptors control human adult articular cartilage homeostasis and are linked to the pathophysiology of osteoarthritis. Arthritis Rheum 60, 513, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Livshits G., Zhai G., Hart D.J., Kato B.S., Wang H.Z., Williams F.M.K., and Spector T.D. Interleukin-6 is a significant predictor of radiographic knee osteoarthritis the Chingford Study. Arthritis Rheum 60, 2037, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogel C., and Marcotte E.M. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13, 227, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoff P., Buttgereit F., Burmester G.R., Jakstadt M., Gaber T., Andreas K., Matziolis G., Perka C., and Rohner E. Osteoarthritis synovial fluid activates pro-inflammatory cytokines in primary human chondrocytes. Int Orthop 37, 145, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beekhuizen M., Bastiaansen-Jenniskens Y.M., Koevoet W., Saris D.B., Dhert W.J., Creemers L.B., and van Osch G.J. Osteoarthritic synovial tissue inhibition of proteoglycan production in human osteoarthritic knee cartilage: establishment and characterization of a long-term cartilage-synovium coculture. Arthritis Rheum 63, 1918, 2011 [DOI] [PubMed] [Google Scholar]
- 70.Ma X., Sun J., Papasavvas E., Riemann H., Robertson S., Marshall J., Bailer R.T., Moore A., Donnelly R.P., Trinchieri G., and Montaner L.J. Inhibition of IL-12 production in human monocyte-derived macrophages by TNF. J Immunol 164, 1722, 2000 [DOI] [PubMed] [Google Scholar]
- 71.Fahy N., de Vries-van Melle M.L., Lehmann J., Wei W., Grotenhuis N., Farrell E., van der Kraan P.M., Murphy J.M., Bastiaansen-Jenniskens Y.M., and van Osch G.J.V.M. Human osteoarthritic synovium impacts chondrogenic differentiation of mesenchymal stem cells via macrophage polarisation state. Osteoarthr Cartilage 22, 1167, 2014 [DOI] [PubMed] [Google Scholar]
- 72.Lapane K.L., Yang S.B., Driban J.B., Liu S.H., Dube C.E., McAlindon T.E., and Eaton C.B. Effects of prescription nonsteroidal antiinflammatory drugs on symptoms and disease progression among patients with knee osteoarthritis. Arthritis Rheumatol 67, 724, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thysen S., Luyten F.P., and Lories R.J.U. Targets, models and challenges in osteoarthritis research. Dis Models Mech 8, 17, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lorenz H., Wenz W., Ivancic M., Steck E., and Richter W. Early and stable upregulation of collagen type II, collagen type I and YKL40 expression levels in cartilage during early experimental osteoarthritis occurs independent of joint location and histological grading. Arthritis Res Ther 7, R156, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang Q., Ji Q., Wang X., Kang L., Fu Y., Yin Y., Li Z., Liu Y., Xu X., and Wang Y. SOX9 is a regulator of ADAMTSs-induced cartilage degeneration at the early stage of human osteoarthritis. Osteoarthr Cartilage 23, 2259, 2015 [DOI] [PubMed] [Google Scholar]
- 76.Fukui N., Purple C.R., and Sandell L.J. Cell biology of osteoarthritis: the chondrocyte's response to injury. Curr Rheumatol Rep 3, 496, 2001 [DOI] [PubMed] [Google Scholar]
- 77.Mankin H.J., Dorfman H., Lippiell L., and Zarins A. Biochemical and metabolic abnormalities in articular cartilage from osteo-arthritic human hips. II. Correlation of morphology with biochemical and metabolic data. J Bone Joint Surg Am 53, 523, 1971 [PubMed] [Google Scholar]
- 78.Miosge N., Hartmann M., Maelicke C., and Herken R. Expression of collagen type I and type II in consecutive stages of human osteoarthritis. Histochem Cell Biol 122, 229, 2004 [DOI] [PubMed] [Google Scholar]
- 79.Heard B.J., Martin L., Rattner J.B., Frank C.B., Hart D.A., and Krawetz R. Matrix metalloproteinase protein expression profiles cannot distinguish between normal and early osteoarthritic synovial fluid. BMC Musculoskeletal Disord 13, 126, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Scanzello C.R., Umoh E., Pessler F., Diaz-Torne C., Miles T., DiCarlo E., Potter H.G., Mand L., Marx R., Rodeo S., Goldring S.R., and Crow M.K. Local cytokine profiles in knee osteoarthritis: elevated synovial fluid interleukin-15 differentiates early from end-stage disease. Osteoarthr Cartilage 17, 1040, 2009 [DOI] [PubMed] [Google Scholar]
- 81.Sandy J.D., Chan D.D., Trevino R.L., Wimmer M.A., and Plaas A. Human genome-wide expression analysis reorients the study of inflammatory mediators and biomechanics in osteoarthritis. Osteoarthr Cartilage 23, 1939, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wanner J.P., Subbaiah R., Skomorovska-Prokvolit Y., Shishani Y., Boilard E., Mohan S., Gillespie R., Miyagi M., and Gobezie R. Proteomic profiling and functional characterization of early and late shoulder osteoarthritis. Arthritis Res Ther 15, R180, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Murata M., Yudoh K., and Masuko K. The potential role of vascular endothelial growth factor (VEGF) in cartilage: how the angiogenic factor could be involved in the pathogenesis of osteoarthritis? Osteoarthr Cartilage 16, 279, 2008 [DOI] [PubMed] [Google Scholar]
- 84.Lambert C., Mathy-Hartert M., Dubuc J.E., Montell E., Verges J., Munaut C., Noel A., and Henrotin Y. Characterization of synovial angiogenesis in osteoarthritis patients and its modulation by chondroitin sulfate. Arthritis Res Ther 14, R58, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ghosh A., Koziol-White C.J., Asosingh K., Cheng G., Ruple L., Groneberg D., Friebe A., Comhair S.A.A., Stasch J.P., Panettieri R.A., Aronica M.A., Erzurum S.C., and Stuehr D.J. Soluble guanylate cyclase as an alternative target for bronchodilator therapy in asthma. Proc Natl Acad Sci USA 113, E2355, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawata M., Koinuma D., Ogami T., Umezawa K., Iwata C., Watabe T., and Miyazono K. TGF-beta-induced epithelial-mesenchymal transition of A549 lung adenocarcinoma cells is enhanced by pro-inflammatory cytokines derived from RAW 264.7 macrophage cells. J Biochem 151, 205, 2012 [DOI] [PubMed] [Google Scholar]
- 87.Li W., Zhang Q., Wang M., Wu H., Mao F., Zhang B., Ji R., Gao S., Sun Z., Zhu W., Qian H., Chen Y., and Xu W. Macrophages are involved in the protective role of human umbilical cord-derived stromal cells in renal ischemia-reperfusion injury. Stem Cell Res 10, 405, 2013 [DOI] [PubMed] [Google Scholar]
- 88.Li X., and Tai H.H. Activation of thromboxane A2 receptor (TP) increases the expression of monocyte chemoattractant protein −1 (MCP-1)/chemokine (C-C motif) ligand 2 (CCL2) and recruits macrophages to promote invasion of lung cancer cells. PLoS One 8, e54073, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lin C.Y., Lin C.J., Chen K.H., Wu J.C., Huang S.H., and Wang S.M. Macrophage activation increases the invasive properties of hepatoma cells by destabilization of the adherens junction. FEBS Lett 580, 3042, 2006 [DOI] [PubMed] [Google Scholar]
- 90.Liu C.Y., Xu J.Y., Shi X.Y., Huang W., Ruan T.Y., Xie P., and Ding J.L. M2-polarized tumor-associated macrophages promoted epithelial-mesenchymal transition in pancreatic cancer cells, partially through TLR4/IL-10 signaling pathway. Lab Invest 93, 844, 2013 [DOI] [PubMed] [Google Scholar]
- 91.Lossdorfer S., Gotz W., and Jager A. PTH(1-34)-induced changes in RANKL and OPG expression by human PDL cells modify osteoclast biology in a co-culture model with RAW 264.7 cells. Clin Oral Invest 15, 941, 2011 [DOI] [PubMed] [Google Scholar]
- 92.Schroder H.C., Wang X.H., Wiens M., Diehl-Seifert B., Kropf K., Schlossmacher U., and Muller W.E.G. Silicate modulates the cross-talk between osteoblasts (SaOS-2) and osteoclasts (RAW 264.7 Cells): inhibition of osteoclast growth and differentiation. J Cell Biochem 113, 3197, 2012 [DOI] [PubMed] [Google Scholar]
- 93.Virgili F., Kim D., and Packer L. Procyanidins extracted from pine bark protect alpha-tocopherol in ECV 304 endothelial cells challenged by activated RAW 264.7 macrophages: role of nitric oxide and peroxynitrite. FEBS Lett 431, 315, 1998 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.