Abstract
To explore the origins and consequences of tetraploidy in the African clawed frog, we sequenced the Xenopus laevis genome and compared it to the related diploid X. tropicalis genome. We demonstrate the allotetraploid origin of X. laevis by partitioning its genome into two homeologous subgenomes, marked by distinct families of “fossil” transposable elements. Based on the activity of these elements and the age of hundreds of unitary pseudogenes, we estimate that the two diploid progenitor species diverged ~34 million years ago (Mya) and combined to form an allotetraploid ~17–18 Mya. 56% of all genes are retained in two homeologous copies. Protein function, gene expression, and the amount of flanking conserved sequence all correlate with retention rates. The subgenomes have evolved asymmetrically, with one chromosome set more often preserving the ancestral state and the other experiencing more gene loss, deletion, rearrangement, and reduced gene expression.
Ancient polyploidization events have shaped diverse eukaryotic genomes1, including two rounds of whole genome duplication at the base of the vertebrate radiation2. While such polyploidy is rare in amniotes, presumably due to constraints on sex chromosome dosage3,4, it is common in fish5 and amphibian lineages6,7, and in plants8. Polyploidy provides raw material for evolutionary diversification, since gene duplicates can support new functions and networks9. However, the component subgenomes of a polyploid must cooperate to mediate potential incompatibilities of dosage, regulatory controls, protein-protein interactions, and transposable element activity.
The African clawed frog Xenopus laevis is one of a polyploid series that ranges from diploid to dodecaploid, and thus is ideal for studying the impact of genome duplication10, especially given its status as a premier model for cell and developmental biology11. X. laevis has a chromosome number (2N=36) nearly double that of the Western clawed frog Xenopus (formerly Silurana) tropicalis (2N=20) and most other diploid frogs12, and is proposed to be an allotetraploid that arose via the interspecific hybridization of diploid progenitors with 2N=18, followed by subsequent genome doubling to restore meiotic pairing and disomic inheritance 10,13 (See Supplementary Note 1, Extended Data Fig. 1 for discussion of the Xenopus allotetraploidy hypothesis).
Here we prove the allotetraploid hypothesis by tracing the origins of the X. laevis genome from its extinct progenitor diploids. The two subgenomes are distinct and maintain separate recombinational identities. Despite sharing the same nucleus, we find that the subgenomes have evolved asymmetrically: one of the two subgenomes has experienced more intrachromosomal rearrangement, gene loss by deletion and pseudogenization, changes in levels of gene expression, and in histone and DNA methylation. Superimposed on these global trends are local gene family expansions and alteration of gene expression patterns.
Results
Assembly, annotation, and karyotype
We sequenced the genome of the X. laevis inbred “J” strain by whole genome shotgun methods in combination with long-insert clone-based end sequencing, (Supplementary Note 2) and organized the assembled sequences into chromosomes using fluorescence in situ hybridization (FISH) of 798 bacterial artificial chromosome clones (BACs) and in vivo and in vitro chromatin conformation capture analysis (Supplementary Note 3; Online Methods). These complementary methods produced a high quality chromosome-scale draft that includes all previously known X. laevis genes and assigns >91% of the assembled sequence (and 90% of the predicted protein-coding genes) to a chromosomal location.
We annotated 45,099 protein-coding genes and 342 microRNAs using RNAseq from 14 oocyte/developmental stages and 14 adult tissues and organs (Supplementary Note 4), analysis of histone marks associated with transcription, and homology with X. tropicalis and other tetrapods (Supplementary Note 5; Online Methods). 24,419 X. laevis protein-coding genes can be placed in 2:1 or 1:1 correspondence with 15,613 X. tropicalis genes, defining 8,806 homeologous pairs of X. laevis genes with X. tropicalis orthologs, and 6,807 single copy orthologs. The remaining genes are members of larger gene families (olfactory receptor genes, etc.) whose X. tropicalis orthology is more complex.
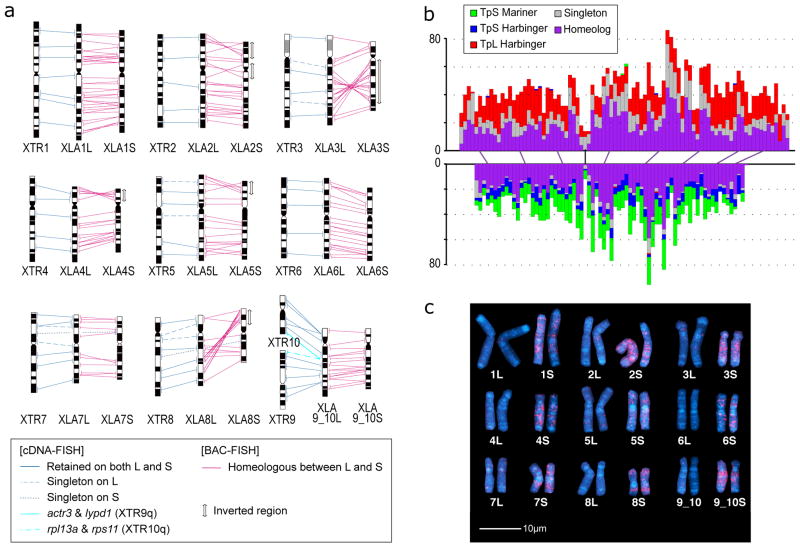
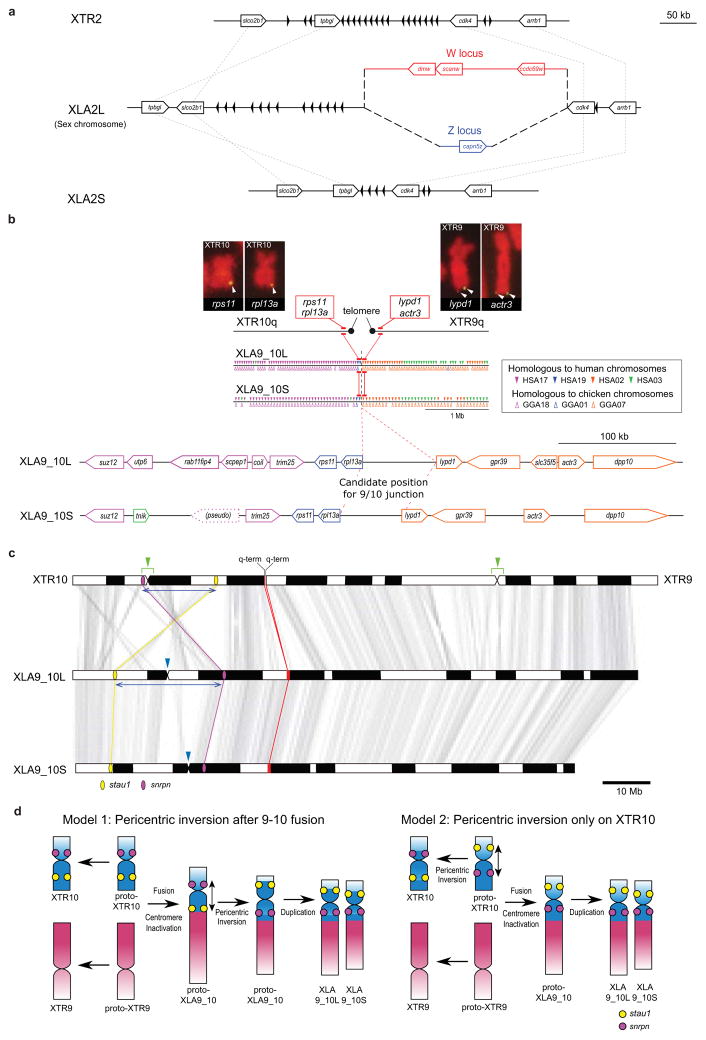
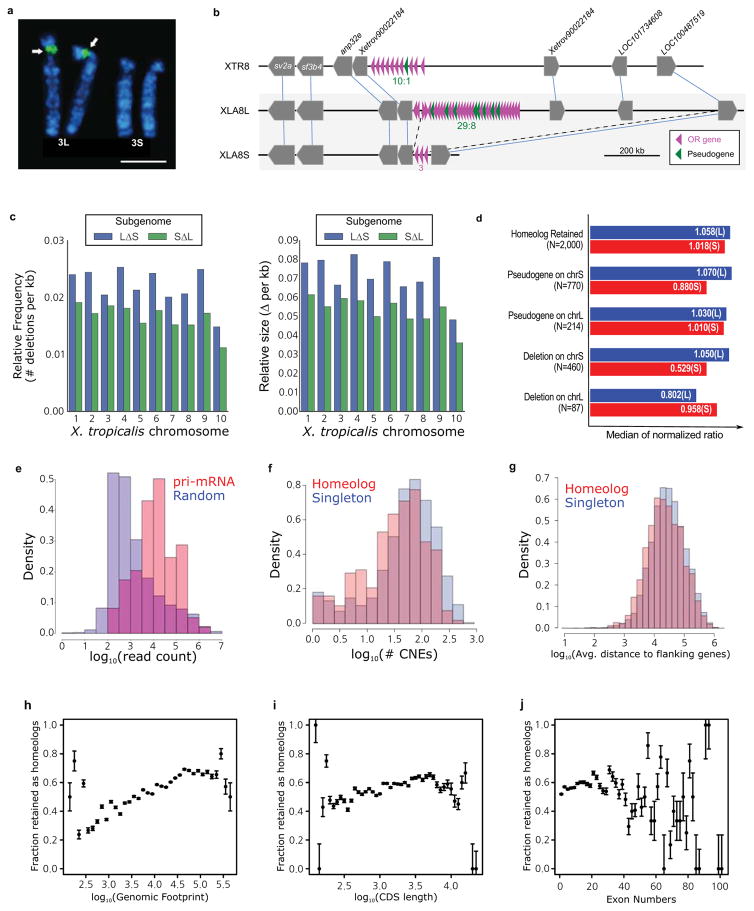
The X. laevis karyotype (Fig. 1a) reveals nine pairs of homeologous chromosomes 1,14,15. Each of the first eight pairs is co-orthologous to and named for a corresponding X. tropicalis chromosome, appending an “L” and “S” for the longer and shorter homeologs, respectively16. XLA2L is the Z/W sex chromosome17, for which we determined a W-specific sequence in the q-subtelomeric region that includes the sex-determining gene dmw17, and a corresponding Z-specific haplotype. The homeologous XLA2Sq, by contrast, has no such locus, and neither does XTR2 (Extended Data Fig. 2a, Supplemental Note 6). The ninth pair of homeologs is a q-q fusion of proto-chromosomes homologous to XTR9 and XTR10, which likely occurred prior to allotetraploidization (Extended Data Fig. 2b–d; Supplementary Note 6). The S chromosomes are on average 13.2% shorter karyotypically16 and 17.3% shorter in assembled sequence than their L counterparts. The single nucleotide polymorphism rate in X. laevis is ~0.4%, far less than the ~6% divergence between homeologous genes (Extended Data Fig. 1c; Supplementary Note 8.8).
Figure 1. Chromosome evolution in Xenopus.
a. Comparative cytogenetic map of XLA and XTR chromosomes. Magenta lines show relationships of chromosomal locations of 198 homeologous gene pairs between XLAL and S chromosomes, identified by FISH mapping using BAC clones (Supplemental Table 1, see Supplementary Note 3.1). Blue lines show relationships of chromosomal locations of orthologous genes between XTR chromosomes and (i) both XLA L and S chromosomes (solid line) (lines between XLAL and S are omitted), (ii) only XLAL (dashed), or (iii) only XLAS (dotted), which were taken from our previous studies14,15. Light blue lines indicate positional relationships of actr3 and lypd1 on XTR9q and rpl13a and rps11 on XTR10q with those on XLA9_10LS chromosomes (see Supplementary Note 6.2). Double-headed arrows on the right of XLAS chromosomes indicate the chromosomal regions in which inversions occurred. Ideograms of XTR and XLA chromosomes were taken from our previous reports15,16.
b. Distribution of homeologous genes (purple), singletons (grey), and subgenome-specific repeats across XLA1L (top) and XLA1S (bottom). Xl-TpL_Harb is red, Xl-TpS_Harb is blue, and Xl-TpS_Mar is green. Purple lines mark homeologous genes present in both L and S chromosomes, the black line marks the approximate centromere location on each chromosome. The homeologous gene pairs, from left to right: rnf4, spcs3, intsl2, foxa1, sds, ap3s1, lifr, aqp7. Each bin is 3 MB in size, with 0.5 MB overlap with the previous bin.
c. Chromosomal localization of the Xl-TpS_Mar sequence. Hybridization signals were only observed on the S chromosomes. Scale bar shows 10 μm.
Subgenome identity and timing of allotetraploidization
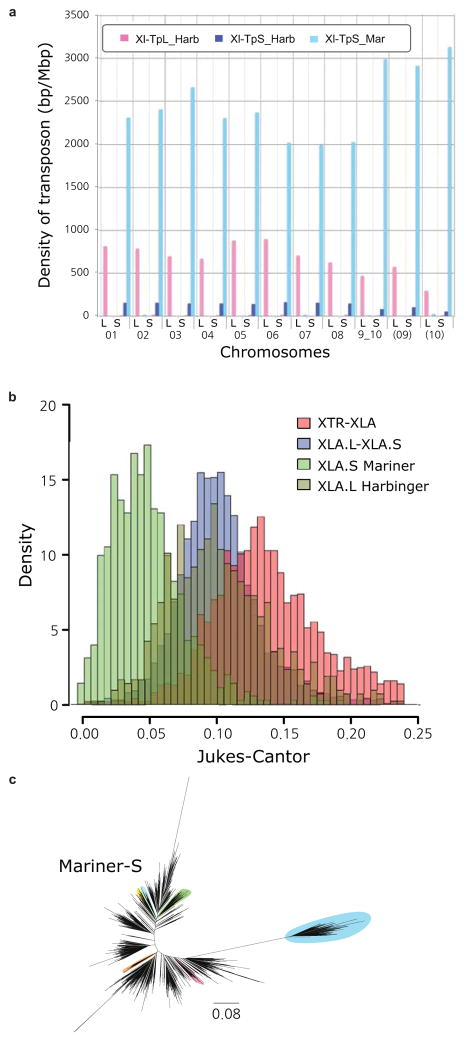
We reasoned that dispersed relicts of transposable elements specific to each progenitor would mark the descendent subgenomes in an allotetraploid (Fig. 2c, Extended Data Fig. 1). Three classes of DNA transposon relicts appear almost exclusively on either the L or S chromosomes (Supplementary Note 7). Xl-TpL_Harb and Xl-TpS_Harb are novel subfamilies of miniature inverted-repeat transposable elements (MITE) of the PIF/Harbinger superfamily18,19 whose relicts are almost completely restricted to L or S chromosomes, respectively (Fig. 1b, Extended Data Fig. 3a). Similarly, sequence relicts of the Tc1/mariner superfamily member Xl-TpS_Mar (closely related to the fish MMTS subfamily20) are found almost exclusively on the S chromosomes (Fig. 1b), as confirmed by FISH analysis using Xl-TpS_Mar as a probe (Fig. 1c, Supplemental Note 7.4; see Supplemental Note 7.3 for details on the rare elements that map to the opposite subgenome).
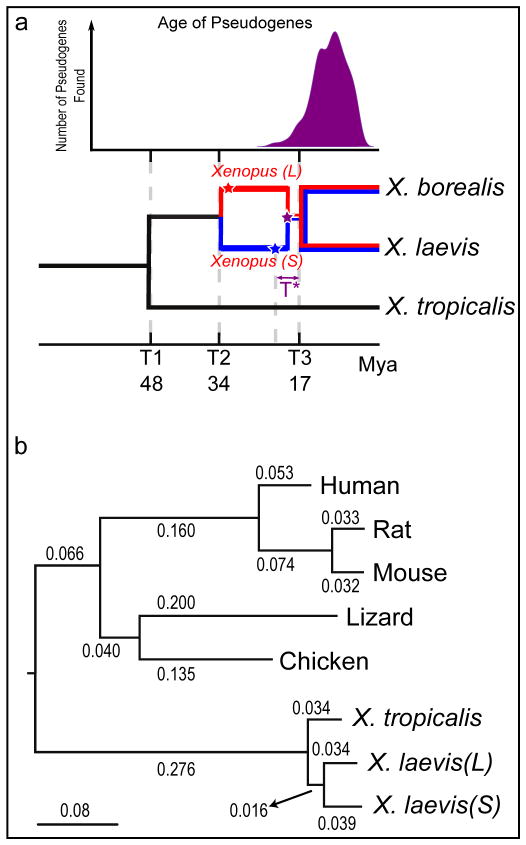
Figure 2. Molecular evolution and allotetraploidy.
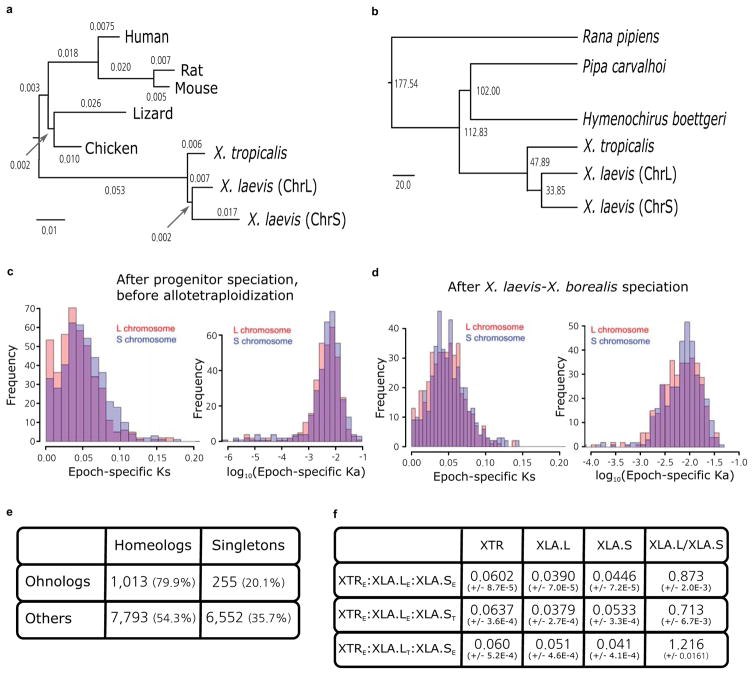
a. (top) The distribution of pseudogene ages, as described in Supplemental Note 9. (bottom) Phylogenetic tree illustrating the different epochs in Xenopus, with times based on protein-coding gene phylogeny of pipids, including Xenopus, Pipa carvalhoi, Hymenochirus boettgeri, and Rana pipiens (only Xenopus depicted). We date the speciation of X. tropicalis and the X. laevis ancestor at 48 Mya, the L and S polyploid progenitors at 34 Mya, and the divergence of the polyploid Xenopus radiation at 17 Mya. Using these times as calibration points, we estimate bursts of transposon activity at 18 Mya (mariner, blue star) and 33–34 Mya (harbinger, red star). The purple star is the time of hybridization, around 17–18 Mya.
b. Phylogenetic tree based on protein-coding genes of tetrapods, rooted by elephant shark (not shown). Alignments were done by MACSE, the maximum-likelihood tree was built by PhyML. Branch length scale shown on bottom. The difference in branch length between Xenopus laevis-L and Xenopus laevis-S is similar to that seen between mouse and rat. Both subgenomes of X. laevis have longer branch lengths than X. tropicalis.
The L and S chromosome sets therefore represent the descendants of two distinct diploid progenitors, confirming the allotetraploid hypothesis even in the absence of extant progenitor species. Based on analysis of synonymous divergence of protein-coding genes, the L and S subgenomes diverged from each other ~34 Mya (T2) and from X. tropicalis ~48 Mya (T1)(Fig. 2a), consistent with prior gene-by-gene estimates from transcriptomes 21–24 (Supplementary Note 8, Extended Data Fig. 4; Online Methods). L- and S-specific transposable elements were active ~18–34 Mya, indicating that the two progenitors were independently evolving diploids during that period (Fig. 2a; Supplementary Note 7.5; Extended Data Fig. 3). More recent transposon activity is more uniformly distributed across the L and S chromosomes (not shown). Finally, consistent with a common origin for tetraploid Xenopus species, we can clearly identify orthologs of L and S genes in whole genome sequences of another allotetraploid frog, X. borealis, and estimate the X. laevis-X. borealis divergence to be ~17 Mya (T3). These considerations constrain the allotetraploid event to ~17–18 Mya (T*). This timing is consistent with other estimates of the radiation of tetraploid Xenopus species, which are presumed to emerge from the bottleneck of a shared allotetraploid founder population23,24.
Karyotype stability
Remarkably, with the exception of the chromosome 9/10 fusion, X. laevis and X. tropicalis chromosomes have maintained conserved synteny since their divergence ~48 Mya (Fig. 1a,b). The absence of inter-chromosomal rearrangements is consistent with the relative stability of amphibian and avian karyotypes compared to mammals25, which typically show dozens of inter-chromosome rearrangements26. It also contrasts with many plant polyploids, which can show considerable inter-subgenome rearrangement27. The distribution of L- and S-specific repeats along entire chromosomes implies the absence of crossover recombination between homeologs since allotetraploidization, presumably because the two progenitors were sufficiently diverged to avoid meiotic pairing between homeologous chromosomes, though we cannot rule out very limited localized inter-homeolog exchanges (Supplementary Note 7).
The extensive collinearity between homologous X. laevis L and X. tropicalis chromosomes (Fig. 1a) implies that they represent the ancestral chromosome organization. In contrast, the S subgenome shows extensive intra-chromosomal rearrangements, evident in the large inversions of XLA2S, XLA3S, XLA4S, XLA5S and XLA8S, as well as shorter rearrangements (Fig. 1a). The S subgenome has also experienced more deletions. For example, the 45S pre-ribosomal RNA gene cluster is found on X. laevis XLA3Lp, but its homeologous locus on XLA3Sp is absent (Extended Data Fig. 5a). Extensive small-scale deletions (Extended Data Fig. 5b) reduce the length of S chromosomes relative to the L and X. tropicalis counterparts (see below).
Response of subgenomes to allotetraploidy
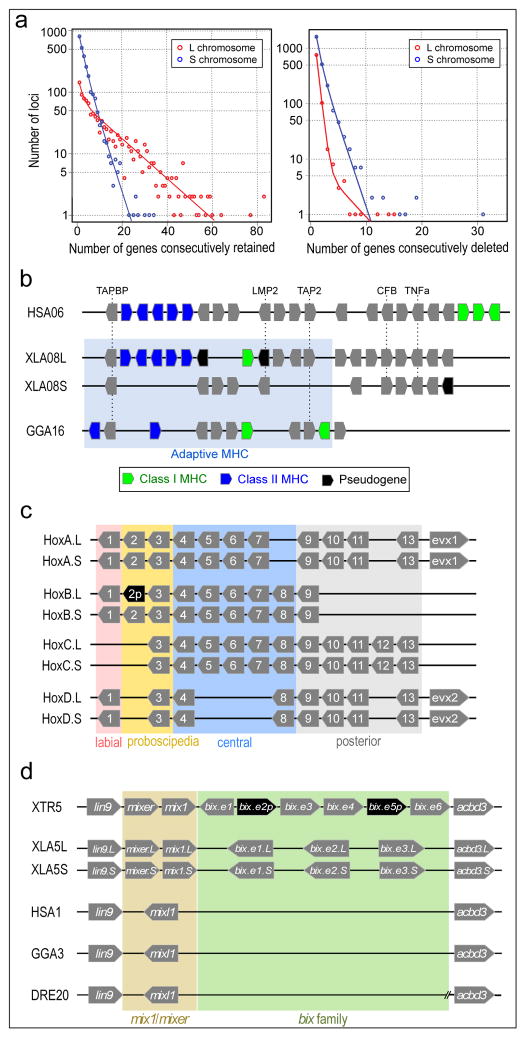
Redundant functional elements in a polyploid are expected to rapidly revert to single copy through the fixation of disabling mutations and/or loss28 unless prevented by neofunctionalization8, subfunctionalization26, or selection for gene dosage29. Differential gene loss between homeologous chromosomes is sometimes referred to as “genome fractionation”30–32 (see Supplementary Note 1) At least 56.4% of the protein-coding genes duplicated by allotetraploidization have been retained in the X. laevis genome (Supplementary Note 10; 60.2% if genes on unassigned short scaffolds are included). Previous studies that rely on cDNA21 and EST surveys22,33,34 have observed far lower rates of retention, probably due to sampling biases from gene expression (Supplementary Note 8.2).
Even higher retention rates are found for homeologous microRNAs (156 of 180, 86.7%), as also found in the salmonid-specific duplication5, and both primary copies are expressed for intergenic homeologous microRNAs (Supplementary Note 8.6; Extended Data Fig. 5e). Pan-vertebrate putatively cis-regulatory conserved non-coding elements35 are also highly retained (541 of 550, 98.4%; Supplementary Note 8.7; Table 1). CNEs conserved between X. laevis and X. tropicalis, however, are retained at a significantly lower rate (49%; Table 1). Longer genes (by genomic span, exon number, or coding length) are more likely to be retained (Wilcoxon p-value <= 1E-5; Supplementary Note 10.5; Extended Data Fig. 5 h–j), broadly consistent with the idea that longer genes have more independently mutable functions and are therefore more susceptible to subfunctionalization and subsequent retention36.
Table 1.
Summary of retention of different genomic elements, in comparison to the diploid X. tropicalis genome. More detailed information is available in Supplementary Tables 2 and 3.
| Sequence element | XTR | XLA-L | XLA-S | Retention |
|---|---|---|---|---|
| Protein Coding Genes | 15,613 | 13,781 | 10,241 | 56.4% |
| Genomic DNA (MB) | 1,227 | 1,222 | 1,010 | N/A |
| miRNAs | 180 | 166 | 168 | 86.7% |
| Pan Vertebrate Conserved Noncoding Elements | 550 | 542 | 536 | 96.6% |
| H3K4me3 Peaks | 7,473 | 6,927 | 5,833 | 70.6% |
| p300 Peaks | 4,321 | 3,457 | 2,702 | 42.5% |
| CACTUS | 1,294,342 | 1,026,204 | 888,899 | 49.0% |
| MitoCarta | 917 | 717 | 501 | 46.0% |
| GermPlasm | 15 | 15 | 6 | 40.0% |
Genes have been lost asymmetrically between the two subgenomes of X. laevis. Similar results have been reported for some plant polyploids30 but not in rainbow trout5. For X. laevis protein-coding genes with clear 1:1 or 2:1 orthologs in X. tropicalis, we find that significantly more genes are lost on the S subgenome (31.5%) vs. the L subgenome (8.3%; χ2 test p-value=2.23E-50, Supplemental Table 2), with the same trend for other types of functional elements, such as H3K4me3-enriched promoters and p300-bound enhancers (Table 1). Across most of the genome, genes appear to be lost independently of their neighbors, as the distribution of runs of gene losses are nearly geometrically distributed (Fig. 3a, right). We do observe some large block deletions (e.g., several olfactory clusters (Extended Data Fig. 5b) and a few unusually long blocks of functionally unrelated genes that are retained in two copies without loss (Fig 3a, left).
Figure 3. Structural response to allotetraploidy.
a. Distributions of consecutive retentions (left) and deletions (right) in the L (red) and S (blue) subgenomes. The distributions were fit using the equation y= a*(ebx) + c*(edx). The y-axis is shown on a log scale. Significant differences were seen between L and S subgenomes in both distributions (Student’s t-test, retention p=3.6E-22, deletion p=4.5E-84).
b. Evolutionary conservation of the Xenopus MHC and differential MHC silencing on the two X. laevis subgenomes. Selected gene names shown above. The ‘Adaptive MHC’ encodes tightly-linked essential genes involved in antigen presentation to T cells; this group of genes is the primordial linkage group and has been preserved in most non-mammalian vertebrates, including Xenopus. Differential gene silencing is particularly pronounced as four genes around the class I gene are functional on S chromosome but absent (dma, dmb) or pseudogenes (ring3, lmp2) on L chromosome. The gene map is not to scale; pseudogenes (p) are noted as indicated. HSA, XLA, GGA: human, Xenopus, and chicken MHC, respectively. Refer to the Supplemental Table 8 for a more detailed MHC map.
c. Hox gene clusters. X. laevis retains eight Hox clusters, consisting of pairs of HoxA, B, C and D clusters, on L and S chromosomes. even-skipped genes (evx1 or evx2) are positioned flanking Hoxa and Hoxd clusters. hox genes are classified into four, labial, proboscipedia, central, and posterior groups. Note that hoxb2.L (black) is a pseudogene.
d. Syntenies around the mix gene family. Abbreviations for species and chromosome numbers: human (H. sapiens; HSA1), chicken (G. gallus; GGA3), X. tropicalis (XTR5), X. laevis (XLA5L and XLA5S), zebrafish (D. rerio; DRE20). Each Xenopus (sub)genome experienced its own independent expansion of the family (see Extended Data Fig. 5 for details).
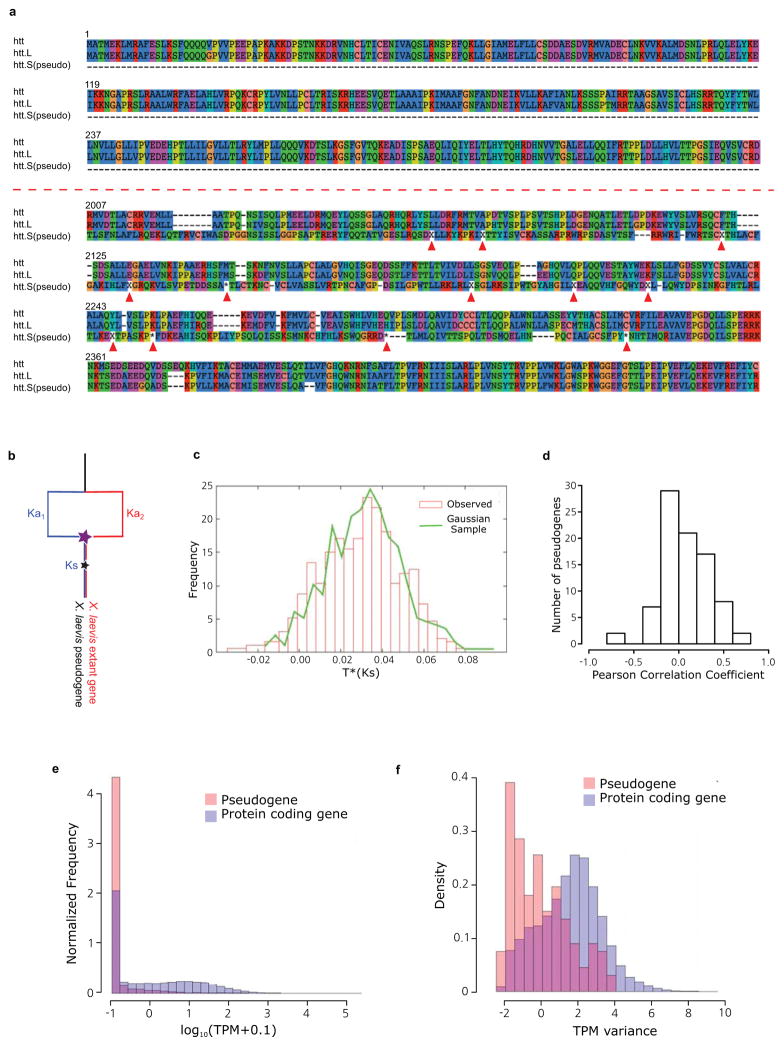
Many lost genes are simply deleted, as demonstrated by significantly shorter distances between conserved flanking genes. Both the size and number of deletions are greater on the S subgenome (Extended Data Fig. 5c). We identified 985 “unitary” (i.e., non-retrotransposed) pseudogenes out of 1,531 loci examined in detail. This 64% detection rate is similar between subgenomes in X. laevis and comparable to that reported in trout5. Based on the accumulation of non-synonymous mutations37 we estimate that most of these pseudogenes escaped evolutionary constraint ~15 Mya (Fig. 2a, Extended Data Fig. 6), consistent with the onset of extensive redundancy in the allotetraploid, though the precision of our pseudogene age estimates is low (Supplementary Note 9). Most pseudogenes show no evidence of expression, but of 769 pseudogenes longer than 100 bp, 133 (17.2%) showed residual expression (Extended Data Fig. 6). Conversely, among homeologous gene pairs, we found 760 for which one member had little to no expression across our 28 sampled conditions. Although these retained some gene structure (start and stop codon, no frame shifts, good splice signals) they showed increased rates of amino acid change and appear to be under relaxed selection (Extended Data Fig. 5f). We call these nominally dying genes “thanagenes” (Supplementary Note 12.5). Reduced expression may be due to mutated cis-regulatory elements, exemplified by the six6 gene pair (Fig. 4e; Extended Data Fig. 8 g–i; Supplementary Note 13.1).
Figure 4. Retention and functional differentiation.
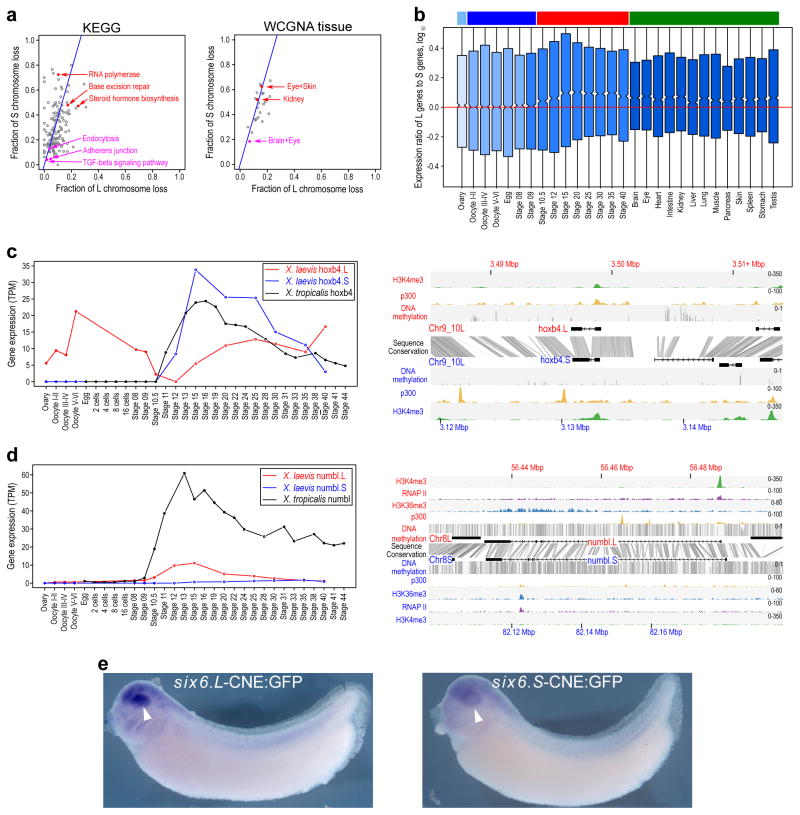
a. (left) Comparison of L and S gene loss by KEGG categories. X-axis is fraction loss of L genes, and the y-axis is fraction loss of S genes. Blue line is expected L/S loss based on genome-wide average (56.4%). Red points are functional categories that show a high degree of loss (χ2 test p<0.01). Magenta points are functional categories that show a high degree of retention (p<0.01). (right) Similar scatterplot for tissue WGCNA categories. See Supplemental Note 10.1 for a more detailed discussion.
b. Boxplot of log10(Ltpm/Stpm) for homeologous gene pairs, zoomed in to show medians. Ovary and maternally-controlled developmental time points are on the left (light blue and dark blue bars respectively), zygotically-controlled developmental time points and adult tissues are on the right (red and green bars respectively). The red line shows the equal ratio log10(1). On average maternal data sets express the L gene of a homeologous pair 12% higher than as S (median = 0%), while the zygotic tissues and time points express the L gene of a homeologous pair 25% higher than S (median = 1.8%). The difference between the mean and medians is explained by many genes with large differences between homeologs, illustrated by the full distribution in Extended Data Fig. 8c. Here, to illustrate the difference in median of zygotic expression, we zoom in on the center of the boxplot.
c. (left) Developmental expression plot and (right) epigenetic landscape surrounding hoxb4. L expression is red, S expression is blue, tropicalis expression is shown in black. The right panel shows the genomic profiles of H3K4me3 (green) and p300 (yellow) ChIP-seq tracks, as well as DNA methylation levels determined by whole-genome bisulfite sequencing (grey). The gene annotation track shows the hoxb4 gene on L (top) and S. The conservation between the L and S genomic sequence is shown in grey between the gene annotation tracks.
d. (left) Developmental expression plot and (right) epigenetic landscape surrounding numbl. L expression is red, S expression is blue, tropicalis expression is shown in black. The small amount of expression seen in maternal numbl and numbl.L is consistent between replicates. In addition to the tracks described for c), the right panel shows RNA Polymerase II (RNAPII; purple) and H3K36me3 (blue) ChIP-seq profiles.
e. Representative embryos with GFP expression driven by either six6.L-CNE or six6.S-CNE linked to a basal promoter-GFP cassette (six6.L-CNE:GFP and six6.S-CNE:GFP, respectively). GFP expression was detected by in situ hybridization. Semi-quantitative image analysis revealed a statistically significant difference in their average expression level (p < 0.01); the expression driven by six6.S-CNE (n = 27) was 0.6-fold weaker than that by six6.L-CNE in the eye region (n = 32). Given eye-specific patterns of their endogenous expression, the six6 genes likely have additional silencers for restricting enhancer activity of the CNEs in the eye.
Although tetraploidy created two “copies” of nearly every gene, additional gene copies are continually produced by tandem duplication (Fig. 3d; Extended Data Fig. 7). The number of tandem clusters is greater in X. tropicalis than in the X. laevis L subgenome, which in turn is greater than in S (Supplementary Note 11.1). Although tandem duplication is faster in X. tropicalis than in X. laevis, there is also a higher rate of loss. Since tandem duplications and deletions occur by unequal crossing over during meiosis, these differing rates are consistent with a shorter generation time of X. tropicalis (Extended Data Fig. 7 f, g). The mean time to loss of an old tandem duplicate is ~40 Mya in X. laevis (on either subgenome) compared with ~16 Mya in X. tropicalis. Homeologous gene loss and tandem duplication can combine to yield complex histories for some gene families. We discuss how these families contribute to the literature on whole genome duplication evolution in Supplemental Notes 10 and 13.
Functional patterns of gene retention and loss
We find preferential retention or loss of many functional categories (Fig 4a; Extended Data Figs. 4e, 9, 10; Supplemental Note 13). DNA binding proteins and components of developmentally-regulated signaling pathways (TGFβ, Wnt, Hh and Hippo) and cell cycle regulation are retained at a significantly higher rate (> 90%) than average (Extended Data Fig. 10). Genes retained in multiple copies after the ancient vertebrate genome duplications are also more likely to be retained as homeologs in X. laevis (Supplemental Note 10.4), as found for the teleost and trout genome duplications5. A notable example is the nearly complete retention of 37/38 duplicated genes in the four pairs of homeologous Hox clusters, with a single pseudogene (Fig. 3c). High rates of homeolog retention in most genes in these categories suggest that stoichiometrically controlled expression levels may be needed or subfunctionalization of homeologs may have occurred, either in their expression domain or target specificity.
Conversely, homeologous genes in other functional categories have been lost at a higher rate, presumably because of a corresponding lack of selection for dosage. For example, genes involved in DNA repair are lost at a high rate (79%) (Supplementary Note 10.1), consistent with reduced selection for repair in the immediate aftermath of allotetraploidy, when all genes were present in four copies per somatic cell5. Other metabolic categories are also prone to loss, presumably because single loci encoding enzymes are sufficient38. Genomic regions with notable loss include the major histocompatibility complex genes on the S subgenome (Fig. 3b) and several olfactory receptor clusters (Extended Data Fig. 5b). We hypothesize that homeologous genes may be functionally incompatible in these cases, leading to en bloc deletion in response to this selection pressure. Specific case studies of duplicate gene retention and loss are detailed in Extended Data Figures 9,10 and Supplemental Note 13.
Evolution of gene expression
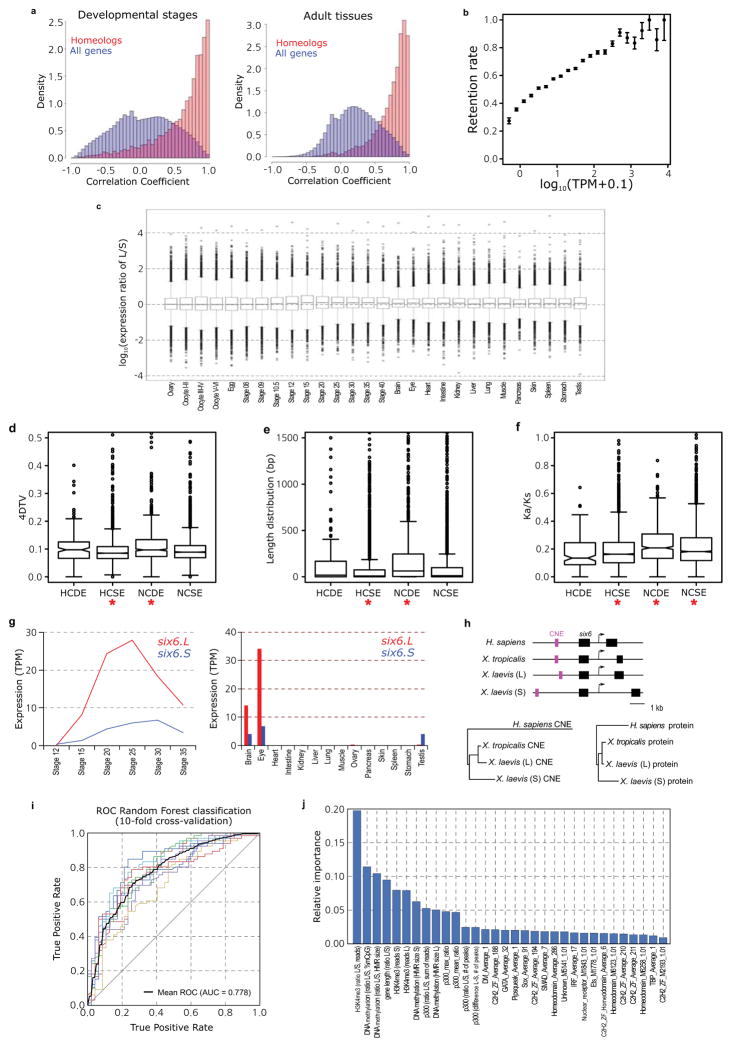
Gene expression is also a predictor of retention, with more highly expressed genes more likely to be retained (Extended Data Fig. 8b), similar to results seen in Paramecium 39,40. Developmentally regulated genes whose expression levels peak at the maternal-zygotic transition (MZT) or during neural differentiation are retained at higher levels (p < 0.01), based on gene expression networks constructed from developmental and adult tissue expression (Online Methods; Fig. 4a (right); Extended Data Fig. 10e; Supplementary Note 12.3). We speculate that the exceptional retention of developmentally regulated genes is due to selection for stoichiometric dosage of these factors, and in some cases higher expression in the physically larger allotetraploid cells and embryos relative to those of diploid frogs, although a propensity36 of these genes for sub- or neo-functionalization could also contribute. In the adult, genes whose expression peaks in the brain and eye are also retained at higher levels (Figure 4b).
In X. laevis, the expression of homeologs is highly correlated (Extended Data Fig. 8a), showing that the overall expression of homeologs diverges similarly to orthologs between Xenopus species41. Many homeologous pairs, however, are differentially expressed throughout development or across adult tissues, either in spatiotemporal pattern (a form of sub-functionalization36; Supplemental Note 12.4; Extended Data Fig. 8d–f) or in the same pattern but with differing expression levels. When homeologous gene pairs are both expressed the average L copy expression level is ~25% higher than the S copy consistently across adult tissues, and after the MZT42 (Fig. 4b; Supplemental Note 12.2). Excess L expression, however, averages only ~12% in oocyte and early pre-MZT stages, suggesting that the two subgenomes are more evenly expressed as maternal transcripts but develop an increased asymmetry after MZT. Strikingly, we found 391 cases in which one homeolog had no detectable maternal mRNA (oocytes, egg and stage 8; Fig. 4c,d; Extended Data Fig. 8c). Comparing with similar transcript data from X. tropicalis, we found cases of apparent loss of expression (“maternal subfunctionalization”: that is, X. tropicalis and one X. laevis gene expressed, the other X. laevis gene silenced pre-MZT; 238 genes, e.g., numbl.S) as well as a surprising gain (“maternal neofunctionalization”: that is, X. tropicalis gene not expressed maternally, but one X. laevis gene expressed; 153 genes, e.g., hoxb4.L). We do not see such a large divergence in other expression domains (Supplemental Note 12.2; Extended Data Fig. 8c), suggesting a high level of plasticity of maternal mRNA regulation between X. laevis homeologs, similar to the trend seen between Xenopus species41.
Overall, thousands of homeolog pairs have either divergent spatiotemporal patterns or similar patterns with differing expression levels. Such homeolog pairs differ in substitution rate, and CDS length difference, more than those that are similar in expression (Supplementary Note 12.4; Extended Data Fig. 8Fig. 8d–f), a pattern also found in trout homeologous pairs5. These expression differences can largely be attributed to changes in epigenetic regulation (Random Forest classification; ROC AUC 0.78), with changes in H3K4me3 and DNA methylation contributing the most explanatory power among our epigenetic variables (Supplementary Note 14). Detailed comparison of the two subgenomes will facilitate identification of specific sequences that control cis-regulatory differences between homeologs.
Conclusion
The two subgenomes of Xenopus laevis have evolved asymmetrically, with the L-subgenome more consistently resembling the ancestral condition and the S-subgenome more disrupted by deletion and rearrangement. Asymmetric gene loss has been observed in allopolyploid plants30 and yeast43 at the segmental level, but it has not been shown directly that similarly “fractionated” segments derive from the same progenitor (Fig. 1c). Our results are consistent with the model that optimized gene expression levels are an important force affecting gene retention following polyploidy39,40. The asymmetry between L and S could have been the result of an intrinsic difference between their diploid progenitors. Alternately, the remodeling of the S genome could have been a response to the L-S merger itself, a “genomic shock,”44 resulting from the activation of transposable elements (Fig 2a; Supplemental Note 8.5). Xenopus’ position as a premier model for the study of vertebrate development, cell biology, and immunology, and the existence of a number of related polyploids, will continue to provide rich material for the study of vertebrate polyploidy.
Online Methods
Notation and terminology
“Homeologous” chromosomes are anciently orthologous chromosomes that diverged by speciation but were reunited in the same nucleus by a polyploidization event. They are a special case of paralogs. Homeologous genes are sometimes called “alloalleles” to emphasize their role as alternate forms of a gene, but since homeologs are unlinked and assort independently, we do not use this terminology. Similarly, loss of homeologous genes is sometimes referred to as “diploidization.” We prefer the simpler and more descriptive term “gene loss.” Note that an allotetraploid like Xenopus laevis has two related subgenomes, but these subgenomes are each transmitted to progeny via conventional disomic inheritance. So immediately after allotetraploidization, the new species is already genetically diploid. This is clearly the case for X. laevis, since we find no evidence for recombination between homeologous chromosomes, which would create new sequences with mixed “L” and “S” type transposable elements.
Sequencing and assembly
DNA was extracted from the blood of a single female from the inbred J-strain for whole genome shotgun sequencing. We generated 4.6 billion paired-end Illumina reads from a range of inserts, and used Sanger dideoxy sequencing to obtain fosmid- and bacterial artificial chromosome (BAC)-end pairs and full BAC sequences. We used meraculous45 as the primary genome assembler. See supplementary notes for more detailed information.
Chromosome scale organization
We identified 798 bacterial artificial chromosomes (BACs) containing genes of interest distributed across the Xenopus genome, and performed fluorescence in situ hybridization (FISH) to assign these BACs to specific chromosomes based on Hoechst 33258-stained late-replication banding patterns (Supplemental Table 1). “HiC” chromatin capture from animal caps was performed as previously reported46 and assembled with HiRise47.
Characterization of sex locus
Sex determination in X. laevis follows a female heterogametic ZZ/ZW system48. We fully sequenced BAC clones representing both W and Z haplotypes, and identified both W- and Z-specific sequences (Extended Data Fig. 2a). The existence of the Z-specific sequence was unexpected and therefore verified by PCR analysis using specific primer sets and DNA from gynogenetic frogs having either W or Z loci.
Gene annotation
We made use of extensive previously generated transcriptome data for X. laevis and X. tropicalis, including 697,015 X. laevis EST sequences (see a review49). In addition, more than 1 billion RNAseq reads were generated for this project from 14 oocyte/developmental stages and 14 adult tissues from J-strain X. laevis (see Supplementary Note 4). These data were combined with homology and ab initio predictions using the Joint Genome Institute’s Integrated Gene Call pipeline (See Supplementary Note 4 and 8 for more details).
Characterization of subgenome-specific transposable elements
We found subgenome specific repeats using a RepeatMasker50 result. The repeats were used to reconstruct full-length subgenome specific transposon sequences. The specific transposons, Xl-TpL_Harb, Xl-TpS_Harb, and Xl-TpS_Mar, were classified based on their target site sequence and terminal inverted repeat (TIR) sequences. The coverage lengths of the transposons on each chromosome were calculated from the results of BLASTN search (E-value < 1E-5) using the consensus sequences of the transposons as queries. The chromosomal distribution of the Xl-TpS_Mar was revealed by a FISH analysis (See Supplemental Note 7.4).
Phylogeny, divergence time, and evolutionary rates
We used Hymenochirus boettgeri, Pipa carvalhoi, and Rana pipiens sequences as outgroups to estimate the evolutionary rate of duplicated genes in X. laevis and their relationship to X. tropicalis. See Supplementary Note 7 and 8 for more detail.
Deletions and pseudogenes
Pseudogene sequences contain various defects including premature stop codons, frameshifts, disrupted splicing, and/or partial coding deletions. 985 pseudogenes were identified among 1,531 “2-1-2 regions”, with the others deleted or rendered unidentifiable by mutation. 368/985 could be timed, based on the accumulation of non-synonymous and synonymous substitution between a pseudogene, its homeolog, and its ortholog in X. tropicalis, providing a time since the loss of constraint for each pseudogene37.
Functional annotation of genes
We used several bioinformatic methods and high throughput datasets to assign functional annotations to Xenopus genes. Protein domains were assigned using InterPro (including PFAM and Panther)51 and KEGG52. Gene Ontology was assigned using InterPro2Go51. We identified genes that encode mitochondrial proteins by mapping the MitoCarta53 database from mouse to the most recent X. tropicalis proteome. Xenopus genes associated with germ plasm were manually curated using the extensive Xenopus literature (See Supplement Note 13).
Gene expression
We analyzed transcriptome data generated for 14 oocyte/developmental stages and 14 adult tissues in duplicate except for oocyte stages (see Supplementary Note 4). Expression levels were measured by mapping paired-end RNA-seq reads to predicted full length cDNA and reporting transcripts per one million mapped reads (TPM). We consider the limit of detectable expression to be TPM > 0.5 Co-expression modules were defined by Weighted Gene Co-expression Network Analysis (WGCNA) clustering54 (See Supplementary Note 12).
Epigenetic analysis
We determined DNA methylation levels (DNAme) by whole genome bisulfite sequencing, and used ChIP seq to generate profiles of the promoter mark histone H3 lysine 4 trimethylation (H3K4me3), the transcription elongation mark H3K36me3, as well as RNA polymerase II (RNAPII) and the enhancer-associated co-activator p300. To test which regulatory features would contribute most to the L versus S expression differences, we applied a Random Forest machine learning algorithm to analyze differential expression between the L and S homeologs (See Supplementary Note 14).
Extended Data
EDF 1. ALLOTETRAPLOIDY AND ASSEMBLY.
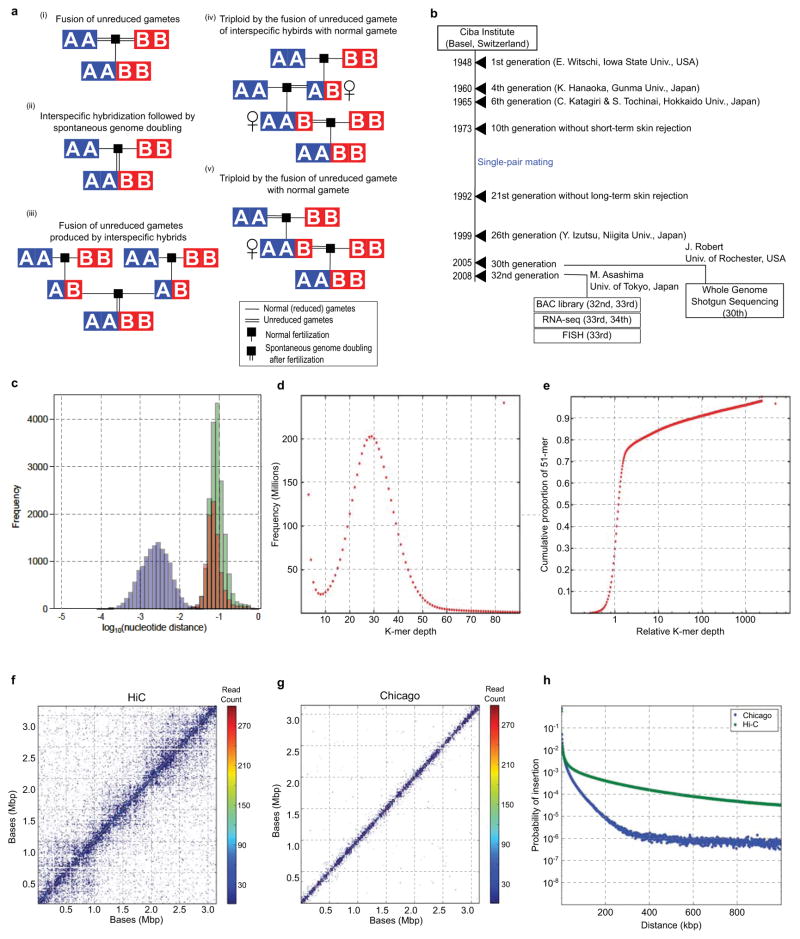
- (i) Fusion of unreduced gametes from species A and B. (ii) Interspecific hybridization followed by spontaneous doubling. (iii) Fusion of unreduced gametes produced by interspecific hybrids. (iv) Interspecific hybrids produce unreduced gametes, which fuse with normal gametes from species A. The resulting triploid again produces unreduced gametes, which fuse with normal gametes from species B (v) Unreduced gamete from species A fuses with normal gamete from species B. The resulting AAB triploid produces unreduced gametes that are fertilized by normal gametes species B. See Supplemental Note 1.1 for a more detailed discussion.
- History of the J strain. See Supplementary Note 2.1 for details. The years of events and generation numbers (e.g., frog transfer to another institute, establishment of homozygosity, construction of materials) are indicated in the scheme. Generation numbers are estimates due to loss of old breeding records.
- The nucleotide distance of orthologs (green), homeologs (red), and alleles (blue) is discussed in Supplemental Note 8.7. The distances are shown on a log scale to differentiate between the distributions.
- 51-mer frequency histogram showing the number of 51-mers with specified count in the shotgun dataset. The prominent peak implies that each genomic locus is sampled 29x in 51-mers. Note the absence of a feature at twice this depth, indicating that homeologous features with high identity are rare.
- Cumulative proportion of 51-mers as a function of relative depth (i.e., depth/29). Relative depth provides an estimate of genomic copy number. The rapid rise at relative depth 1 implies that 70–75% the X. laevis genome is single copy with respect to 51-mers. The remainder of the genome is primarily concentrated in repetitive sequences with copy number ≫100. Note logarithmic scale.
- The contact map of 85,260 Chicago read pairs for JGIv72.000090484.chr4S, a 3.1Mb scaffold in the XENLA_JGI_v72 assembly.
- The contact map of 85,260 HiC read pairs for JGIv72.000090484.chr4S. Read pairs were binned at 10kb intervals. For each read pair, the forward and reverse reads map with at least 20 map quality score.
- The insert distribution of HiC and Chicago read pairs that map to the same scaffold of XENLA_JGI_v72 with at least 20 map quality score. The x-axis is the read pair separation distance. The y-axis is the counts for that bin divided by the total number of reads. The bins are 1kb.
EDF 2. CHROMOSOME STRUCTURE.
- Structure of the sex chromosome of X. laevis (XLA2L) and comparison with XLA2S and XTR2. The W version of XLA2L harbors W-specific sequence containing the female sex-determining gene, dmw (red), while Z has a different Z-specific sequence (blue). Pentagon arrows and black triangles indicate genes and olfactory receptor genes, respectively. Their tips correspond to their 3’-ends.
- Alignment of the q-terminal regions of XTR9 and 10 with corresponding regions of XLA9_10L and XLA9_10S. Genes near the q terminal regions of XTR 9 and XTR10 were missing in the X. tropicalis genome assembly v9, but rps11, rpl13a, lypd1, and actr3 were expected to be located there based on the synteny with human chromosomes, and then verified by cDNA FISH (upper panels). Small triangles on XLA9_10L and S indicate the distribution of gene models showing both identity and coverage greater than 30%, against the human and chicken peptide sequences from Ensembl, in the region between ±2 Mb from the prospective 9/10 junction. HSA: human chromosome. GGA: chicken chromosome. The magnified view represents syntenic genes to scale with colors corresponding to human genes.
- The orders of orthologous genes across XTR9, XTR10, XLA9_10L and XLA9_10S. Green arrowheads: positions of centromeres in XTR9 and 10 predicted by examination of the cytogenetic chromosome length ratio of p versus q arms15. Blue arrowheads: positions of centromere repeats, frog centromeric repeat-162, in XLA9_10L and S. Magenta and yellow ellipses: chromosomal locations of snrpn (magenta) and stau1 (yellow) from X. tropicalis v9 and X. laevis v9.1 assemblies. Red ellipses: chromosomal locations of four genes, rps11, rpl13a, lypd1, and actr3. XTR9 is flipped to facilitate comparison. Blue bidirectional arrows indicate the homologous regions where pericentric inversions may have occurred on proto-chromosomes (see Extended Data Fig. 2d).
- Schematic representation for the two hypothetical processes of chromosomal rearrangements (fusion and inversion) that occurred between the hypothetical proto-XTR9 and 10 to produce proto-XLA9_10, and eventually XLA9_10L and S. The process of chromosome rearrangements is explained parsimoniously in two different ways (left and right panels), starting from proto-XTR9 and 10. Actual and hypothetical ancestral chromosomal locations of snrpn and stau1 are shown by magenta and yellow circles, respectively. Note that the chromosomal locations of these genes on the proto-XTR10 differ between the two models. Chromosome segments homologous to XTR9 and XTR10 are shown in red and blue, respectively. XTR9 is inverted to facilitate comparison. Bidirectional arrows indicate the regions where pericentric inversions may have occurred. Black arrows indicate the direction of chromosomal evolution.
EDF 3. TRANSPOSONS.
- Density of the subgenome specific transposons on each chromosome (coverage length of transposable element [bp]/chromosome length [Mbp]). The coverage lengths of transposons were calculated from the results of BLASTN search (E-value cutoff 1E-5) using the consensus sequences as queries.
- Jukes-Cantor distances across non-CpG sites, corrected as in Supplemental Note 7.5. Distances between X. tropicalis and X. laevis transposons consensus sequences are shown. The X. laevis-specific transposon differences are each individual transposon sequence against the consensus sequence for that subfamily.
- Phylogenetic tree of Xl-TpS_Mar transposon expansions in the X. laevis genome, built using Jukes-Cantor corrected distances (Supplemental Note 7.5). Sub-clusters with enough members to determine accurate timings are highlighted. The scale bar represents the corrected Jukes-Cantor distance.
EDF 4. PHYLOGENY.
- Phylogenetic tree of pan-vertebrate conserved non-coding elements (pvCNEs), rooted by elephant shark. Alignments were done by MUSCLE, and the maximum-likelihood tree was built by PhyML. Branch length scale shown on bottom. The difference in branch lengths of tetrapods follows the same topology as the protein-coding tree (Fig. 2b).
- Complete phylogenetic tree from Fig. 2a, with divergence times computed by r8s.
- Distribution of Ks and Ka on specific subgenomes during the time between L and S speciation, before X. laevis and X. borealis speciation. We find accelerated mutations rates between T2 and T3 in Ks and Ka (p=1.4e-5 (left), 8.6e-3 (right)).
- Distribution of Ks and Ka on specific subgenomes during the time after laevis and borealis speciation. We do not find significantly accelerated substitution rates. (p= 0.10 (left) and 0.03 (right)).
- Table showing the number of homeologs and singletons identified as homeologs from the ancient vertebrate duplication (or ohnologs as they are historically called)63. 79.9% of ohnologs retain both copies in X. laevis today, significantly more than the 54.3% of the rest of the genome after excluding ohnologs (χ2 test p-value= 4.44E-69).
- Table showing the branch lengths of bootstrapped maximum likelihood trees described in Supplemental Note 12.5. The columns refer to the X. tropicalis (XTR), L chromosome of X. laevis (XLA.L), S chromosome of X. laevis (XLA.S), and XLA.L/XLA.S branch lengths respectively. The first row is triplets where all genes show expression, the second row is triplets where L is a thanagene, and the third row is triplets where S is a thanagene. The L branch length is significantly smaller when all genes are expressed, or when S is a thanagene (Wilcoxon p-value=1.7E-216 and 6.4E-212 respectively). The S branch length is smaller when L is a thanagene (p=2.4E-223). The ratio of branch lengths (L/S) is significantly different for either L or S thanagene datasets compared to when all genes are expressed (p=3.55E-214 and 7.48E-220 respectively). The ratio is different between the two thanagene datasets as well (p=1.79E-217).
EDF 5. STRUCTURAL EVOLUTION.
- Chromosomal locations of the 45S pre-ribosomal RNA gene (rna45s), which encodes a precursor RNA for 18S, 5.8S, and 28S rRNAs, was determined using pHr21Ab (5.8-kb for the 5′ portion) and pHr14E3 (7.3-kb for the 3′ portion) fragments as FISH probes. DNA fragments used for the probes were provided by National Institutes of Biomedical Innovation, Health and Nutrition, Osaka, and labeled with biotin-16-dUTP (Roche Diagnostics) by nick translation. After hybridization, the slides were incubated with FITC-avidin (Vector Laboratories). Hybridization signals (arrows) were detected to the short arm of XLA3L, but not XLA3S. Scale bar represents 5 μm.
- A large deletion including an olfactory receptor gene (or) cluster. Schematic structures of or gene clusters and adjacent genes on the 8th chromosomes of X. tropicalis (XTR8) and X. laevis (XLA8L and XLA8S). Chromosomal locations: XTR8:107,524,547-108,927,581; XLA8L:105,062,063–106,610,199; XLA8S:91,630,596–92,060,451. Horizontal bars, genomic DNA sequences; triangles, genes. Outside of or gene cluster, only representative genes are shown. The length of triangle is to scale. The orientation of triangles indicates 5′ to 3′ direction of genes. Thin lines connect orthologous/homeologous genes. Magenta triangles, or genes; green triangles, pseudogenes (point-mutated or truncated or genes). The number of or genes is shown underneath gene clusters. Dotted lines, a deleted region in XLA8S in comparison with XLA8L. The centromere is located on the left side and the telomere is on the right.
- The relative frequency (left panel) and size (right panel) of genomic regions deleted in the S (blue) and L (green) chromosomes respectively. Both subgenomes experienced sequence loss through deletions, however, the deletions on the S subgenome are larger and have been more frequent. Deletions were called based on the progressive Cactus sequence alignment between the X.laevis L and S subgenomes and the X. tropicalis genome. Chromosome 9_10 of laevis was split into 9 and 10 on basis of alignment with the X. tropicalis chromosomes. Sequences from L that were not present on S, but could at least partially be identified in X. tropicalis, and consisted of gaps for no more than 25% of their length were called as deleted regions in S. The same procedure was followed for deleted regions in L.
- Identification of triplet loci is described in Supplemental Note 8.1. Loci were classified into groups based on the presence of gene 2 in both X. laevis subgenomes (homeolog retained), versus those that had a pseudogene in the middle (pseudogene) or no remnant of the middle gene as assessed by Exonerate (deletion). To normalize the intergenic lengths we divided the nucleotide distance between genes 1 and 3 in either X. laevis subgenome by the orthologous distance in X. tropicalis. The median of the normalized ratio distribution is plotted on the bar chart. On average S deletions appear to be larger than L deletions (52.9% length vs 80.2% the size of the orthologous X. tropicalis region respectively).
- The number of RNA-seq reads aligning +/− 1kb of precursor miRNA loci (red) was compared to the read count for 10,000 random unannotated 2.1 kb regions of the genome (blue). All 83 homeologous, intergenic miRNA pairs showed alignment within their regions, as opposed to 4,127/10,000 (41.27%) of the randomly chosen intergenic sequences. The putative primary-miRNA loci have a higher read count than the expressed randomly chosen regions as well (Wilcoxon p=1.4E-38).
- The CACTUS alignment was parsed to identify flanking CNE around each X. tropicalis gene. The number of CNEs > 50bp in length for singletons is shown in red, homeologs in blue. Komologrov-Smirnov test p-value is 1E-11.
- The average distance to the nearest gene was computed for each chromosomal locus in X. tropicalis. The average intergenic distance for those with a single X. laevis gene is shown in red, those with two shown in blue. Wilcoxon p-value= 9.8E-24.
- The distribution of gene retention by genomic footprint of the X. tropicalis ortholog. We define genomic footprint as the genomic distance from the start signal of the CDS to the stop signal, including introns. The x axis shows log10(genomic footprint), the y-axis is the retention rate of each bin. The error bars are the standard deviation of the total divided by the number of genes in each bin. We tested for significant differences in length between homeologs and singletons by a Wilcoxon test (p-value = 2.4E-96).
- The distribution of gene retention by CDS length of the X. tropicalis ortholog. The x axis shows log10(CDS length), the y-axis is the retention rate of each bin. The error bars are the standard deviation of the total divided by the number of genes in each bin. We tested for significant differences in length between homeologs and singletons by a Wilcoxon test (p-value= 1.7E-21).
- The distribution of gene retention by exon number of the X. tropicalis ortholog. The x axis shows number of exons; the y-axis is the retention rate of each bin. The error bars are the standard deviation of the total divided by the number of genes in each bin. We tested for significant differences in length between homeologs and singletons by a Wilcoxon test (p-value= 3.2E-8).
EDF 6. PSEUDOGENES.
- Illustration of htt.S pseudogene alignment to X. tropicalis htt, and the extant X. laevis htt.L, translated to amino acids. The amino acid position is shown at the beginning of each line. Missing codons are marked by ‘−’. Frameshifts and premature stops are marked by ‘X’ and ‘*’ respectively (and pointed to with red arrows). (top) the first exon of the pseudogene is completely missing from the S chromosome. The characteristic poly-Q region is maintained by both htt and htt.L. (bottom) An exon with conservation in the pseudogene, illustrating that despite many frameshifts, premature stops, and the lack of a proper start, and insertions of new sequence, we identify many codons in the pseudogene that occur in large conserved blocks.
- Illustration of our model to compute pseudogene ages. The star represents the point of nonfunctionalization for a currently pseudogenized locus. We assume the expected rate of nonsynonymous changes can be estimated by the Ka of the extant gene and X. tropicalis. We then compare the Ks and Ka of the pseudogene sequence to estimate the time of nonfunctionalization. See Supplemental Note 9 for a more detailed discussion.
- Estimated epochs of pseudogenization for 430 genes are indistinguishable from a burst of pseudogenization > 10 Mya (Ks > 0.03). See Supplemental Note 9 for a more detailed discussion.
- Correlation of pseudogene expression with its extant homeolog. The little expression seen in pseudogenes tends to be uncorrelated with the extant homeolog.
- Histogram of pseudogene expression values across all 28 tissues and developmental stages (red) compared to all extant genes (blue). The pseudogenes are rarely expressed, and tend to be expressed at lower levels than extant protein-coding genes.
- Histograms of expression variance of pseudogenes (red) compared to extant genes (blue). The small amount of pseudogene expression observed does not tend to vary across tissues and developmental stages in the same way that extant genes do.
EDF 7. TANDEM DUPLICATIONS.
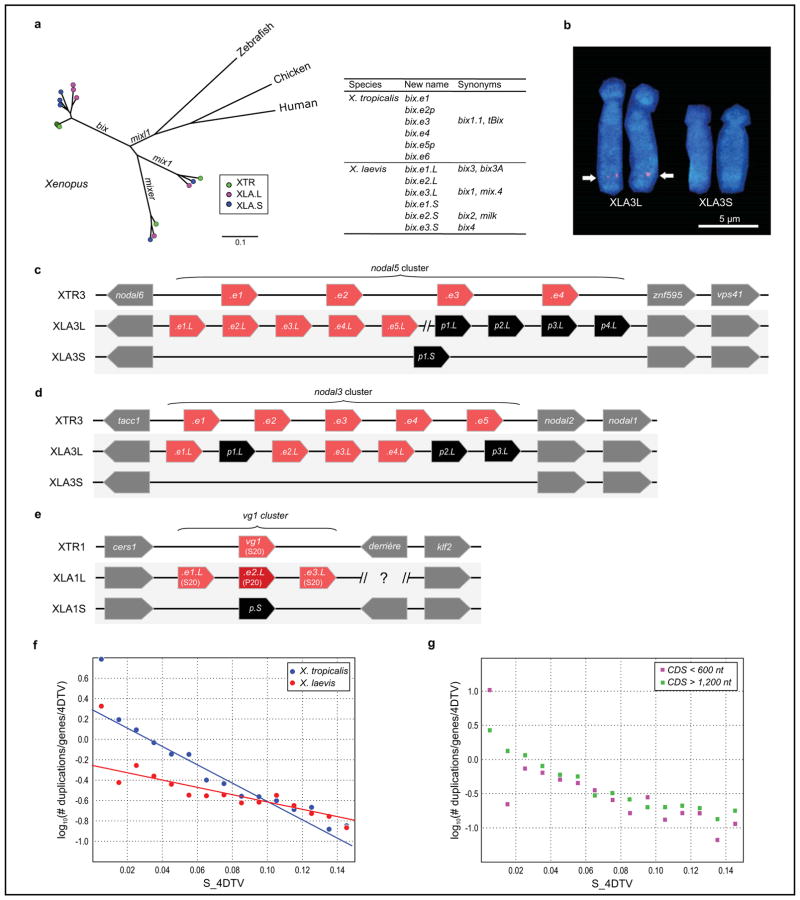
- Phylogenetic trees of the mix/bix cluster. Nucleotide sequences were aligned using MUSCLE, and a phylogenetic diagram was generated by the ML method with 1,000 bootstraps (MEGA6). Circles with different colors represent X. laevis L genes (magenta), X. laevis S genes (blue), and X. tropicalis genes (green). The table shows the correspondence of bix gene names proposed in this study and previously used (synonyms).
- FISH analysis showing XLA3S-specific deletion of the nodal5 gene cluster. One unit of the nodal5 gene region, including exons, introns, and an intergenic region was used as a probe for FISH (counterstained with Hoechst). Arrows indicate the hybridization signals of nodal5s. Scale bar indicates 5 um.
- Comparison of the nodal5 gene cluster. Genome sequencing revealed that nodal5.e1.L~.e5.L (in pink) and nodal6.L are clustered. Amplification of nodal5 gene in XLA3L and loss of this cluster in XLA3S were confirmed. Pseudogenes (nodal5p1.L~p4.L and nodal5p1.S) are indicated in black. The nodal5 cluster of X. tropicails does not contain any pseudogene.
- X. laevis L chromosome has four complete copies of nodal3 (nodal3.e1.L~.e4.L), whereas the gene cluster is lost from the X. laevis S chromosome. A truncated nodal3 gene (nodal3p1.L) is likely to be a pseudogene, and highly degenerate pseudogenes (nodal3p2.L and nodal3p3.L) also exist on the L chromosome.
- Like nodal3, vg1 is lost from the S chromosome although there is a pseudogene (vg1p.S). vg1 is specifically amplified on the X. laevis L chromosome (vg1.e1.L~.e3.L) in comparison with X. tropicalis. An amino acid change (Ser20 to Pro20) in Vg1 protein has been shown to result in functional differences (Supplementary Note 13.9). vg1 and derrière are orthologous to mammalian gdf1.
- Fraction of all genes duplicated and retained to present epoch per 1 expected 4DTV(four-fold degenerate transversions) at different epochs (semi-log scale). Shown also are linear fits, which would be consistent with constant birth- and death rate models (first epoch is omitted from both fitted data sets, as is second epoch from X. laevis). See Supplemental Note 11 for a more detailed discussion.
- Same, but for “short genes” (CDS < 600 bp) and “long genes” (CDS > 1200 bp) separately. The loss rate of new duplicates appears to be similar. If the extra copy of a newly duplicated gene were lost when the first 100% disabling mutation occurred, we would expect, on average, the longer genes to be lost.
EDF 8. GENE EXPRESSION ANALYSIS.
- Pairwise Pearson correlation distributions between homeologous genes (red) and all genes (blue). The left histogram is for stage data; right is for adult data. The x-axis is the correlation; the y-axis is the percent of data. The homeologous genes have a correlation distribution closer to one due to their being the same locus recently. X. laevis TPM values 0.5 were lowered to 0. Any gene with no TPM > 0 was removed from analysis. We then added 0.1 to all TPM values and log transformed (log10).
- Scatterplot comparing binned genes by their median X. tropicalis expression64 to the retention rate of their X. laevis (co)-orthologs. Error bars are the standard deviation for the whole data set divided by the square root of the number of genes analyzed in a bin. We assessed significance by a Wilcoxon test of the homeologous and singleton distributions, p-value = 6.31E-113.
- Complete boxplot shown in Fig. 4c. The difference between subgenomes is difficult to see at this magnification, illustrating that many loci deviate from the whole genome median of preferring the L homeolog. There are some L outliers expressed 104 as much as their S homeologs, whereas no S genes shows such a strong trend. These differences are discussed in more detail in Supplemental Note 12.
- Boxplot of 4DTv (four-fold degenerate transversions) by homeolog class defined in Supplemental Note 12.4. Significant differences are marked by a red asterisk (Wilcoxon p<1E-5). HCSE group shows lower sequence change than others (p=3.7E-12) and the NCDE group shows high rates of sequence change (p=5.6E-14).
- Boxplot of CDS length difference between X. laevis homeologs by homeolog class defined in Supplemental Note 12.4. Significant differences are marked by a red asterisk (Wilcoxon p<1E-5). HCSE group shows smaller CDS length differences than others (p=2.4E-13) and the NCDE group shows large differences in homeolog CDS length (p=2.1E-32).
- Boxplot of Ka/Ks between X. laevis homeologs by homeolog class defined in Supplemental Note 12.4. Significant differences are marked by a red asterisk (t-test p<1E-5). HCSE group shows lower non-synonymous sequence change than others (p=8.2E-19) and the NCDE and NCSE groups shows higher rates of non-synonymous sequence change (p=2.0E-12 and p=7.0E-9 respectively).
- RNA-seq analysis of six6.L (red) and six6.S (blue) during X. laevis development (left panel) and in the adult tissues (right panel). Expression levels of six6.S were lower than those of six6.L at most developmental stages and in adult tissues.
- Diagram of Homo sapiens, X. tropicalis and X. laevis six6 loci (upper panel). Magenta and black boxes indicate CNEs and exons, respectively. The phylogenetic tree analyses of H. sapiens, X. tropicalis and X. laevis six6 CNEs (lower left panel), and Six6 proteins (lower right panel). Notably, six6.S is more diverged from X. tropicalis six6 than six6.L, both in the encoded protein sequences and in conserved non-coding elements (CNEs) within 3 kb from the transcription start sites. Materials, methods and the CNE locations on genome assemblies are described in Supplementary Materials (Supplementary Note 13.1).
- On the basis of chromatin state properties, a Random Forest machine-learning algorithm can accurately predict L versus S expression bias. The classification is based on all genes with greater than 3-fold expression difference at NF stage 10.5 (a set of 1,129 genes). The mean (dotted black line) of the ROC area under the curve is 0.778 (10-fold cross-validation). Features were selected using Linear Support Vector Classification and are shown in Extended Data Fig. 8j.
- Relative importance (based on Gini impurity) of selected features used in the Random Forest classification. All features used in the classification are shown. Among various variables, the ratios of H3K4me3 and DNA methylation at the promoter contributed most to the decision tree model. A difference in p300 binding in the genomic region surrounding the gene also contributed to the Random Forest classification, as did the presence or absence of a number of specific transcription factor motifs in the promoter.
EDF 9. EXAMPLES OF PATHWAY RESPONSES.
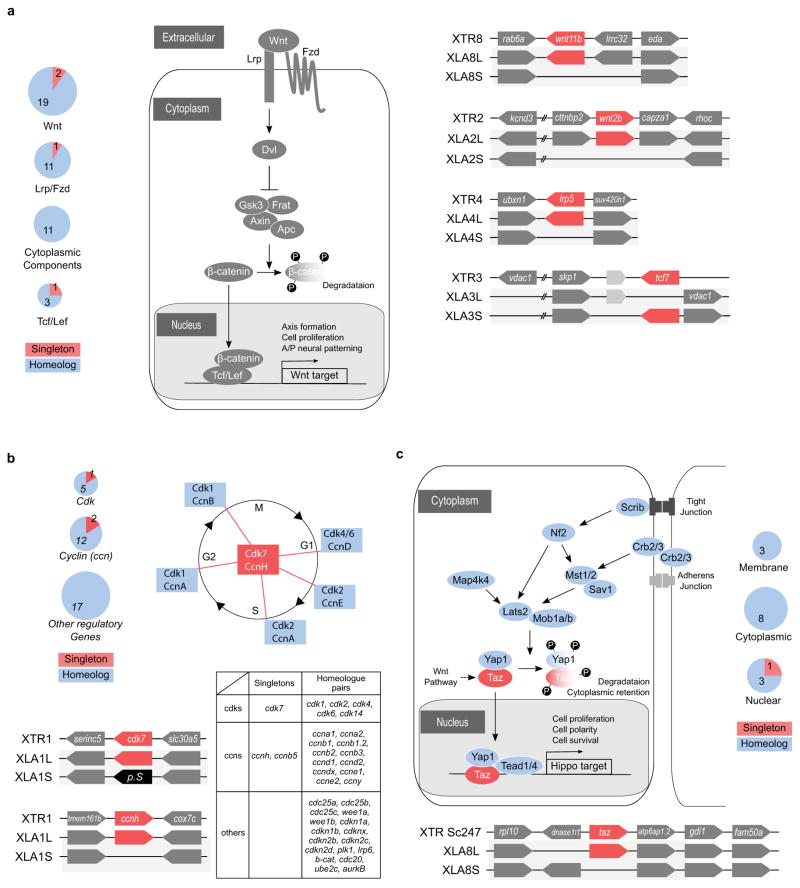
- Wnt pathway. Left panel: Several key components of the canonical Wnt pathway in the X. laevis genome. The numbers in brackets show the number of paralogs. Components that have homeologous pair of genes or singleton are shown in blue and red, respectively. Each (Wnt:21 genes, LRP:2 genes, Fzd:10 genes, Dvl:3 genes, Frat(GBP):1 gene, GSK3:2 genes, Axin:2 genes, bcatenin: 1 gene, APC: 2 genes, TCF/LEF:4 genes) were classified into 4 groups according to subcellular localization, and the number of singleton and homeolog retained genes is shown by pie charts. Right panel: Syntenies around four singleton genes.
- Cell cycle. Upper right panel: Diagram of the cell cycle and regulatory proteins critical to each phase. Cyclin H (CcnH) and Cdk7 constitute Cdk-activating kinase (CAK), a key factor required for activation of all Cdks. Genes encoding Cyclin H and Cdk7 (red), but not other regulators (blue), became singletons. Upper left panel: Pie charts show the numbers of homeologous pairs (blue) and singletons (red) in each functional category as indicated. Lower left panel: Syntenies of ccnh and cdk7 loci in X. tropicalis and X. laevis. Abbreviations for species and chromosome numbers: X. tropicalis (XTR1), X. laevis (XLA1L and XLA1S). Lower right table: Individual genes used for drawing the pie charts are shown in the table.
- Hippo pathway. Upper panel: Hippo pathway components and retention of their homeologous gene pairs. All genes for Hippo pathway components as indicated were identified in the whole genome of X. laevis. Blue icons indicate that both of the homeologous genes are expressed in normal development and adult organs. The red icon, Taz, indicates a singleton. Yap is interchangeable with Taz in most cases, but TAZ, but not YAP, serves as a mediator of Wnt signaling (broken line). Pie charts show the numbers of homeolog pairs (blue) and singleton (red) in each category of Hippo pathway components classified according to subcellular localization. Lower panel: Comparative analysis of syntenies around the taz gene. X. tropicalis scaffold247 is not incorporated into the chromosome-scale assembly (v9) and hence its chromosomal location is not known yet. The p arm termini of XLA8L and XLA8S are on the left.
EDF 10. PATHWAYS PART 2.
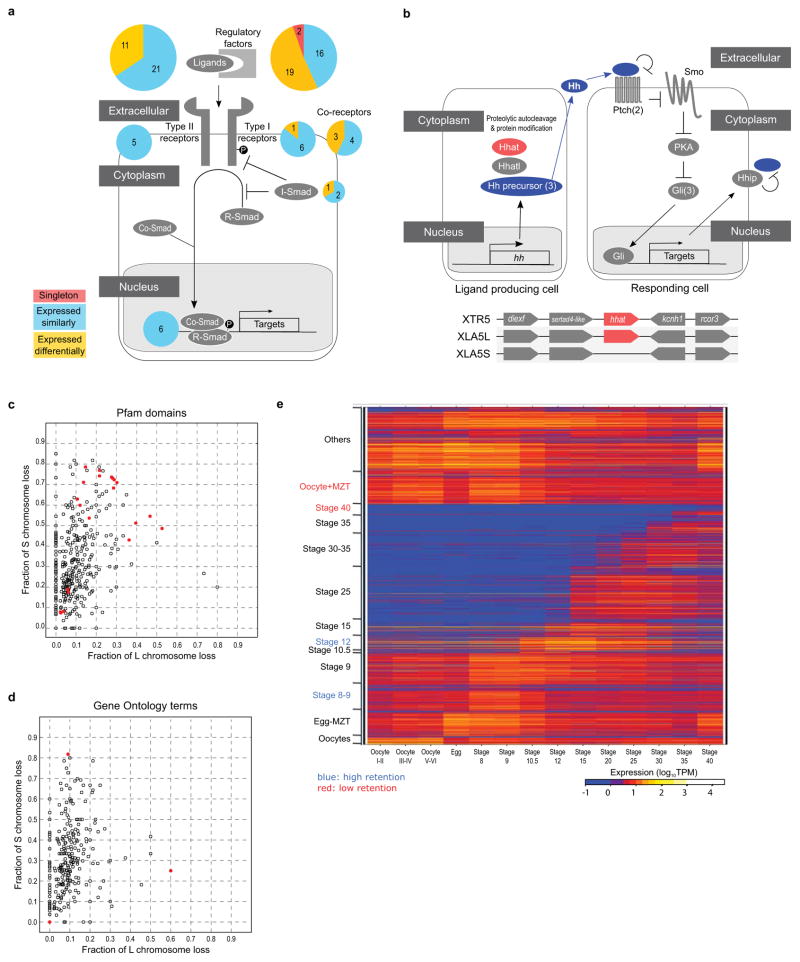
- TGF-beta pathway. Pie charts indicate the ratio of differentially expressed homeologous pairs (orange) and singleton (red). A large portion of the extracellular regulatory factors is either differentially regulated or became singleton. Genes for a type I receptor, co-receptors, an inhibitory Smad are also differentially regulated. Multicopy genes like nodal3, nodal5, and vg1 are not counted as singletons, though those S genes are deleted. Instead, these and duplicated chordin genes are categorized into differentially regulated genes.
- Hedgehog pathway. Upper panel: The simplified Hedgehog pathway known in Shh signalling is schematically shown. Most signalling components are encoded by both homeologous genes, whereas Hhat (shown in red) is encoded by a singleton gene. In case paralogs exist, the numbers of paralogs are shown in parenthesis. In the left cell, the Shh precursor (Hh precursor) is matured through the process involving Hhat and Hhatl, and secreted. In the right cell, the binding of Shh (Hh) to Ptch1 (Ptch) receptor inhibits Ptch1-mediated repression of Smo, leading to Smo activation and subsequent inhibition of PKA; otherwise PKA converts Gli activators to truncated repressors. As a consequence, Gli proteins activate target genes, such as Ptch1 and Hhip. The transmembrane protein Hhip binds Shh and suppresses Shh activity. Lower panel: Schematic comparison of syntenies around hhat genes of X. tropicalis chromosome 5 (top) and X. laevis 5L chromosome (middle), and the corresponding region of X. laevis 5S chromosome (bottom). The diagram is not drawn to scale.
- Deletions rates on L (x-axis), vs S (y-axis) for different Pfam groups. For Pfam groups we computed the number of X. laevis single-copy genes (singletons) vs homeolog pairs and computes a fraction retained. The line is expected L/S loss based on genome-wide average (56.4%). Red points show groups with high or low rates of loss (p<.01). See Supplemental Table 5 for more information.
- Deletions rates on L (x-axis), vs S (y-axis) for different stage WGCNA groups (visualized as a heatmap in Fig. 4a). For stage WGCNA groups we computed the number of X. laevis single-copy genes (singletons) vs homeolog pairs and computes a fraction retained. The line is expected L/S loss based on genome-wide average (56.4%). Red points show groups with high or low rates of loss (p<.01).
- Deletion rates on L (x-axis), vs S (y-axis) for different GO groups. For GO groups we computed the number of X. laevis single-copy genes (singletons) vs homeolog pairs and computes a fraction retained. The line is expected L/S loss based on genome-wide average (56.4%). Red points show groups with high or low rates of loss (p<0.01). See Supplemental Table 5 for more information.
Supplementary Material
Acknowledgments
Author Contributions
RMH, MT, DSR, GJcV, AF, AS, AS, TK, YU, AF, MK, and HO provided project leadership, with additional project management from YM, MA, YI, NU, JS, JW, EM, JS, AMZ, PDV, and MI. YI and JR inbred J strain frogs. AT, CH, AF, JG, JC, JL, JS, TM, and JL generated genome sequence data. JC, AS, TK, JJ, and JS performed genome assembly and validation. ST, TK, AS, US, TT, AT, AS, and MT generated and analyzed the transcriptome data. AS, TK, SvH, and SS generated the annotations. Manual validation of annotation was done by HO, ST, AF, AS, MK, HO, TT, TM, MW, TK, YO, SM, YH, TN, YY, JF, KB, VL, and KK. KM, AS, and RH generated the Hymenochirus transcriptome data. AS performed the phylogenetic analysis, with help from SM and UH. MW, AF, SM, YU, YM, and MT performed the chromosome structure analysis. AS, AH, OS, JC, and YU studied the transposable elements. BAC-FISH was performed by YU, AF, MK, AT, ST, HO, HO, YK, TT, TM, MW, TK, YO, YH, TY, CT, TN, AS, YM, NU, MA, YI, AF, and MT. IQ, SH, NP, and JS generated and analyzed the chromatin-libraries and their use in long-range scaffolding. HO and HO performed the transgenic enhancer analysis. SvH, GG, SP, IvK, OB, RL, and GJcV generated and analyzed the epigenetic data. AS, AS, TK, MK, MT, YO, TT, AF, MW, TM, TN, and LD conducted the gene and pathway analysis. DSR, AS, TK, RMH, MT, AS, YU, GV, MK, UH, SvH, AF, AH, OS, HO TTm IQ, JK, YO, ST, MW, TM, AT, HO, TK, SM, YS, TN, YI, and MFF wrote the paper and supplementary notes, with input from all authors.
Competing financial interest
Dovetail Genomics LLC is a commercial entity developing genome assembly methods. Nicholas Putnam and Jonathan Stites are employees of Dovetail Genomics, and Daniel Rokhsar is a scientific advisor to and minor investor in Dovetail.
Footnotes
Supplementary Information is linked to the online version of the paper. Please see Supplemental Note 15 for funding information and data deposition information.
References
- 1.Van de Peer Y, Maere S, Meyer A. The evolutionary significance of ancient genome duplications. Nat Rev Genet. 2009;10:725–32. doi: 10.1038/nrg2600. [DOI] [PubMed] [Google Scholar]
- 2.Holland PW, Garcia-Fernàndez J, Williams NA, Sidow A. Gene duplications and the origins of vertebrate development. Development. 1994:125–33. [PubMed] [Google Scholar]
- 3.Muller HJ. Why Polyploidy is Rarer in Animals Than in Plants. Am Nat. 1925;59:346–353. [Google Scholar]
- 4.Orr HA. ‘Why Polyploidy is Rarer in Animals Than in Plants’ Revisited. Am Nat. 1990;136:759–770. [Google Scholar]
- 5.Berthelot C, et al. The rainbow trout genome provides novel insights into evolution after whole-genome duplication in vertebrates. Nat Commun. 2014;5:3657. doi: 10.1038/ncomms4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods IG, et al. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–14. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Glasauer SMK, Neuhauss SCF. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol Genet Genomics. 2014;289:1045–60. doi: 10.1007/s00438-014-0889-2. [DOI] [PubMed] [Google Scholar]
- 8.Otto SP. The evolutionary consequences of polyploidy. Cell. 2007;131:452–62. doi: 10.1016/j.cell.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 9.Ohno S. Evolution by Gene Duplication. Springer; Berlin Heidelberg: 1970. [DOI] [Google Scholar]
- 10.Kobel HR, Du Pasquier L. Genetics of polyploid Xenopus. Trends Genet. 1986;2:310–315. [Google Scholar]
- 11.Harland RM, Grainger RM. Xenopus research: metamorphosed by genetics and genomics. Trends Genet. 2011;27:507–15. doi: 10.1016/j.tig.2011.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kuramoto M. A list of chromosome numbers of anuran amphibians. Bull Fukuoka Univ Educ. 1990;39:83–127. [Google Scholar]
- 13.Bisbee CA, Baker MA, Wilson AC, Haji-Azimi I, Fischberg M. Albumin phylogeny for clawed frogs (Xenopus) Science. 1977;195:785–7. doi: 10.1126/science.65013. [DOI] [PubMed] [Google Scholar]
- 14.Uno Y, Nishida C, Takagi C, Ueno N, Matsuda Y. Homoeologous chromosomes of Xenopus laevis are highly conserved after whole-genome duplication. Heredity (Edinb) 2013;111:430–6. doi: 10.1038/hdy.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Uno Y, et al. Inference of the protokaryotypes of amniotes and tetrapods and the evolutionary processes of microchromosomes from comparative gene mapping. PLoS One. 2012;7:e53027. doi: 10.1371/journal.pone.0053027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsuda Y, et al. A New Nomenclature of Xenopus laevis Chromosomes Based on the Phylogenetic Relationship to Silurana/Xenopus tropicalis. Cytogenet Genome Res. 2015;145:187–91. doi: 10.1159/000381292. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimoto S, et al. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105:2469–74. doi: 10.1073/pnas.0712244105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X, et al. P instability factor: an active maize transposon system associated with the amplification of Tourist-like MITEs and a new superfamily of transposases. Proc Natl Acad Sci U S A. 2001;98:12572–7. doi: 10.1073/pnas.211442198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jurka J, Kapitonov VV. PIFs meet Tourists and Harbingers: a superfamily reunion. Proc Natl Acad Sci U S A. 2001;98:12315–6. doi: 10.1073/pnas.231490598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn SJ, Kim MS, Jang JH, Lim SU, Lee HH. MMTS, a new subfamily of Tc1-like transposons. Mol Cells. 2008;26:387–95. [PubMed] [Google Scholar]
- 21.Morin RD, et al. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 2006;16:796–803. doi: 10.1101/gr.4871006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellsten U, et al. Accelerated gene evolution and subfunctionalization in the pseudotetraploid frog Xenopus laevis. BMC Biol. 2007;5:31. doi: 10.1186/1741-7007-5-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bewick AJ, Chain FJJ, Heled J, Evans BJ. The pipid root. Syst Biol. 2012;61:913–26. doi: 10.1093/sysbio/sys039. [DOI] [PubMed] [Google Scholar]
- 24.Cannatella D. Xenopus in Space and Time: Fossils, Node Calibrations, Tip-Dating, and Paleobiogeography. Cytogenet Genome Res. 2015;145:283–301. doi: 10.1159/000438910. [DOI] [PubMed] [Google Scholar]
- 25.Voss SR, et al. Origin of amphibian and avian chromosomes by fission, fusion, and retention of ancestral chromosomes. Genome Res. 2011;21:1306–12. doi: 10.1101/gr.116491.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson-Smith MA, Trifonov V. Mammalian karyotype evolution. Nat Rev Genet. 2007;8:950–62. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
- 27.Langham RJ, et al. Genomic duplication, fractionation and the origin of regulatory novelty. Genetics. 2004;166:935–45. doi: 10.1534/genetics.166.2.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haldane JBS. The Part Played by Recurrent Mutation in Evolution. Am Nat. 1933;67:5–19. [Google Scholar]
- 29.Birchler JA, Veitia RA. Gene balance hypothesis: connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci U S A. 2012;109:14746–53. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schnable JC, Springer NM, Freeling M. Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci U S A. 2011;108:4069–74. doi: 10.1073/pnas.1101368108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sankoff D, Zheng C, Wang B. A model for biased fractionation after whole genome duplication. BMC Genomics. 2012;13(Suppl 1):S8. doi: 10.1186/1471-2164-13-S1-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garsmeur O, et al. Two evolutionarily distinct classes of paleopolyploidy. Mol Biol Evol. 2014;31:448–54. doi: 10.1093/molbev/mst230. [DOI] [PubMed] [Google Scholar]
- 33.Sémon M, Wolfe KH. Preferential subfunctionalization of slow-evolving genes after allopolyploidization in Xenopus laevis. Proc Natl Acad Sci U S A. 2008;105:8333–8. doi: 10.1073/pnas.0708705105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chain FJJ, Dushoff J, Evans BJ. The odds of duplicate gene persistence after polyploidization. BMC Genomics. 2011;12:599. doi: 10.1186/1471-2164-12-599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol. 2011;28:1205–15. doi: 10.1093/molbev/msq304. [DOI] [PubMed] [Google Scholar]
- 36.Force A, et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–45. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meredith RW, Gatesy J, Murphy WJ, Ryder OA, Springer MS. Molecular Decay of the Tooth Gene Enamelin (ENAM) Mirrors the Loss of Enamel in the Fossil Record of Placental Mammals. PLoS Genet. 2009;5:e1000634. doi: 10.1371/journal.pgen.1000634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–90. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Aury JM, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–8. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 40.Gout JF, Kahn D, Duret L Paramecium Post-Genomics Consortium. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 2010;6:e1000944. doi: 10.1371/journal.pgen.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yanai I, Peshkin L, Jorgensen P, Kirschner MW. Mapping gene expression in two Xenopus species: evolutionary constraints and developmental flexibility. Dev Cell. 2011;20:483–96. doi: 10.1016/j.devcel.2011.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Langley AR, Smith JC, Stemple DL, Harvey SA. New insights into the maternal to zygotic transition. Development. 2014;141:3834–41. doi: 10.1242/dev.102368. [DOI] [PubMed] [Google Scholar]
- 43.Marcet-Houben M, Gabaldón T. Beyond the Whole-Genome Duplication: Phylogenetic Evidence for an Ancient Interspecies Hybridization in the Baker’s Yeast Lineage. PLoS Biol. 2015;13:e1002220. doi: 10.1371/journal.pbio.1002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 45.Chapman JA, et al. Meraculous: de novo genome assembly with short paired-end reads. PLoS One. 2011;6:e23501. doi: 10.1371/journal.pone.0023501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Burton JN, et al. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31:1119–25. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putnam NH, et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2015 doi: 10.1101/gr.193474.115. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chang CY, Witschi E. Genic control and hormonal reversal of sex differentiation in Xenopus. Proc Soc Exp Biol Med. 1956;93:140–4. doi: 10.3181/00379727-93-22688. [DOI] [PubMed] [Google Scholar]
- 49.Gilchrist MJ. From expression cloning to gene modeling: the development of Xenopus gene sequence resources. Genesis. 2012;50:143–54. doi: 10.1002/dvg.22008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smit AFA, Hubley R, Green P. RepeatMasker Open-4.0. http://www.repeatmasker.org.
- 51.Mitchell A, et al. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 2015;43:D213–D221. doi: 10.1093/nar/gku1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehisa M, et al. Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res. 2014;42:D199–205. doi: 10.1093/nar/gkt1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.