Abstract
Melanoma differentiation associated gene-7/Interleukin-24 (mda-7/IL-24) displays broad-spectrum anti-cancer activity in vitro, in vivo in preclinical animal models, and in a phase I/II clinical trial in patients with advanced cancers without harming normal cells or tissues. Here we demonstrate that mda-7/IL-24 regulates a specific subset of miRNAs, including cancer-associated miR-221. Either ectopic expression of mda-7/IL-24 or treatment with recombinant His-MDA-7 protein resulted in downregulation of miR-221 and upregulation of p27 and PUMA in a panel of cancer cells, culminating in cell death. Mda-7/IL-24-induced cancer cell death was dependent on reactive oxygen species induction and was rescued by overexpression of miR-221. Beclin-1 was identified as a new transcriptional target of miR-221, and mda-7/IL-24 regulated autophagy through a miR-221/beclin-1 feedback loop. In a human breast cancer xenograft model, miR-221 overexpressing MDA-MB-231 clones were more aggressive and resistant to mda-7/IL-24-mediated cell death than parental clones. This is the first demonstration that mda-7/IL-24 directly regulates microRNA expression in cancer cells and highlights the novelty of the mda-7/IL-24-miR-221-beclin-1 loop in mediating cancer cell-specific death.
Keywords: Breast cancer, MDA-7/IL-24, miR-221, Beclin-1, Autophagy
Introduction
mda-7/IL-24 (1) has potent anti-tumor activity in almost all types of cancers (2–6). mda-7/IL-24 is a member of the IL-10-related cytokine gene family, which was cloned using subtraction hybridization and induction of terminal cancer cell differentiation in melanoma cells (1). Extensive research has confirmed the ubiquitous anti-tumor properties of mda-7/IL-24 both in in vitro cell cultures and animal models (6). mda-7/IL-24 displayed safety and efficacy in a Phase I/II clinical trial in patients with several advanced cancers (7, 8). Forced over expression of mda-7/IL-24 inhibits angiogenesis (9, 10), sensitizes cancer cells to radiation or chemotherapy (3–6) and elicits potent ‘bystander’ antitumor activity (11). Physical interaction of MDA-7/IL-24 protein with the chaperone protein BiP/GRP78 initiates an unfolded protein response (UPR) in cancer cells that leads to apoptosis (12). Gene expression studies have illustrated a number of apoptotic and cell cycle molecules regulated by mda-7/IL-24 (13).
MicroRNAs are small noncoding RNAs, which degrade RNAs, negatively affect the stability of RNAs or block the translation of mRNAs (14–16). MicroRNAs are aberrantly expressed in many diseases including cancer (17, 18). MicroRNA-221 is an important regulator, whose up regulation has been described in several types of cancers and several reports suggest that miR-221 can be used as a therapeutic target for cancer. Many tumor suppressors have been reported to be targets of miR-221. miR-221 regulates cell cycle through p27 (19) and apoptosis through PUMA (20). Additionally, by targeting the estrogen receptor (ER) it blocks the action of tamoxifen and hence targeting miR-221 can promote susceptibility to tamoxifen-mediated cell death in ER positive breast cancers (21). Other tumor suppressor targets of miR-221 include PTEN (22), p57 (23), FOXO3A (24), and TIMP3 (22). By regulating these targets miR-221 plays a critical role in cancer progression. Felicetti et al. reported that the promyelocytic leukemia zinc finger (PLZF) transcription factor functions as a transcriptional repressor of miR-221 (25), however, the mechanism(s) regulating miR-221 requires further elucidation.
Beclin-1, the mammalian homologue of Atg6 of yeast is a promoter of autophagy. Expression of beclin-1 is altered in different disease states including cancer. In several types of cancer aberrant mRNA/protein expression of beclin-1 has been observed (26). The underlying mechanism of this altered expression of beclin-1 is largely unknown. In the present study, we document crosstalk between tumor suppressors and oncogenes, i.e., mda-7/IL-24, miR-221, and beclin-1. Ad.mda-7 infection down regulates miR-221, which in turn up regulates beclin-1 and promotes toxic autophagy that switches to apoptosis. Our findings suggest that miR-221 is a downstream participant in mda-7/IL-24-mediated cell death and cells overexpressing miR-221 are resistant to mda-7/IL-24-mediated cell death. Finally, we show that ROS plays a key role in this pathway and a novel mda-7/IL-24-miR-221-beclin-1 axis is critical in mda-7/IL-24-mediated cell death.
Materials and methods
Plasmids, cell lines and stable clones
The miR-221 and anti-miR-221 constructs were from GeneCopoeia (Rockville, MD). Beclin-1 3′UTR construct was from Origene (Rockville, MD). Beclin-1 construct was from Addgene (Cambridge, MA). The Beclin-1-UTR mutant was cloned from the wild type Beclin-1-UTR by standard site-directed mutagenesis (27). Cell lines used in this study included DU-145, MCF-7, T-47D, MDA-MB-231, ZR-751, SK-BR-3, RPMI-7951, NB-1691, SK-N-SH, IM-PHFA, RWPE-1, HMEC, and A549. These cells were obtained from the American type culture collection (ATCC) (Manassas, VA), with the exception of IM-PHFA, which was established in our laboratory (28), and were maintained as described by the ATCC. All cell lines were obtained from ATCC between 2012–2016 and authenticated by ATCC using short tandem repeat (STR) analysis. All the cell lines were expanded and frozen immediately after receipt. The cumulative culture length of the cells was less than 6 months after recovery. Early passage cells were used for all experiments. Human mammary epithelial cells (HMEC) were purchased from Lonza, Basel, Switzerland. NB-1691 cells were a kind gift from Dr. Alan Houghton from St Jude children’s research hospital (Memphis, TN). All the cell lines were frequently tested for mycoplasma contamination using a mycoplasma detection kit from Sigma. Stable clones expressing miR-221 and beclin-1 were established in MDA-MB-231 cells as described previously (28).
Western blotting Analysis
Western blotting was done as described (29). The primary antibodies used in this study were MDA-7/IL-24 (Genhunter Corporation, Nashville, TN), EF1α (Upstate biotechnology, Lake Placid, NY), p27 and PUMA (Cell Signaling Technology, Danvers, MA), and beclin-1 (Abcam, Cambridge, MA). Secondary antibodies used in this study were from Sigma, (St. Louis, MO).
Real time PCR
Total RNA and microRNA-enriched fractions of RNA were isolated from cells using the RNA and microRNA isolation kits, respectively, from Qiagen (Hilden, Germany). Real time PCR was performed with the taqman master mix and probe were from Applied Biosystems, Foster City, CA. Data were analyzed by Graphpad prism software.
Transient transfection and reporter gene assay
Transient transfection used the lipofectamine reagent from Invitrogen, Carlsbad, CA. For luciferase assay, cells were transfected with the 3′UTR construct of beclin-1 with or without miR-221 with the pRLTK luc construct encoding renilla luciferase control. Cells were incubated for 24 hours and then luciferase assays were done using the dual-luciferase assay kit from Promega, Madison, WI.
Cell proliferation assay
Cell proliferation was measured by standard MTT (3-(4, 5-di methyl thiazol-2-yl)-2, 5 diphenyl tetrazolium bromide) assay as described earlier (11). Colony formation assays were done as described previously (29).
Tumor xenograft studies
Tumor xenografts were established subcutaneously in both flanks of 6-week old female athymic mice (Charles River Laboratories, Wilmington, MA) by injecting 0.5 x 106 MDA-MB-231 or MDA-MB-231 cells overexpressing miR-221 or beclin-1 mixed with Matrigel in a 1:1 ratio. Once tumors reached a measurable size of approximately 100 mm3, the mice were divided into different groups and treated as described in the figure and figure legend. When the tumors in the control group reached the maximum allowable limit, mice were sacrificed and tumor weight was measured. Tumor size was also measured and plotted. Animals were maintained under the guidelines of the National Institute of Health and under evaluation and approval of the Institutional Animal Care and Use Committee (Virginia Commonwealth University). Food and water were provided ad libitum.
Reactive Oxygen species (ROS) measurement
The amount of reactive oxygen species that is produced was quantified by staining cells with carboxy-2′, 7′-dichloro dihydro fluorescein diacetate (Life technologies, Molecular probes, Grand Island, NY) in 1X phosphate buffered saline. Fluorescence was measured using a green filter after 30 minutes. Experimental conditions are described in the figures and figure legends.
Live-dead assay
The number of live and dead cells was observed by confocal laser microscope (Zeiss, Germany) after staining with live/dead staining reagent (Invitrogen, Carlsbad, CA) as per the manufacturer’s protocol. The images were analyzed by Zeiss software.
Apoptosis assay
MDA-MB-231 cells were treated as indicated in the figure. After 72 hours, cells were analyzed for apoptosis using the Annexin-V-FITC/propidium iodide apoptosis detection kit (BD Biosciences, San Jose, CA) and subjected to flow cytometry analysis using BDFACS CantoII and BDFACS DIVA software (BD Biosciences, San Jose, CA).
Autophagy assay
The cellular acidic compartment was observed as a marker of autophagy and quantified by staining with acridine orange as described previously (30). Briefly, cells were stained with 1 μg/ml acridine orange (Sigma, St. Louis, MO) for 10 minutes. Cells were washed with 1X PBS and then the numbers of cells, which have increased acidic vacuoles, were measured using flow cytometry (BDFACS CantoII) and analyzed with BDFACS DIVA software. Approximately, 10000-gated cells were analyzed.
Statistical analysis
The data are presented as the mean +/− S.D. of the values from 3 to 5 independent experiments and statistical analysis was performed using either student’s t-test or one-way anova. P-value <0.05 was considered to be significant. This was done using the graph pad prism software.
Results
MDA-7 regulates miR-221
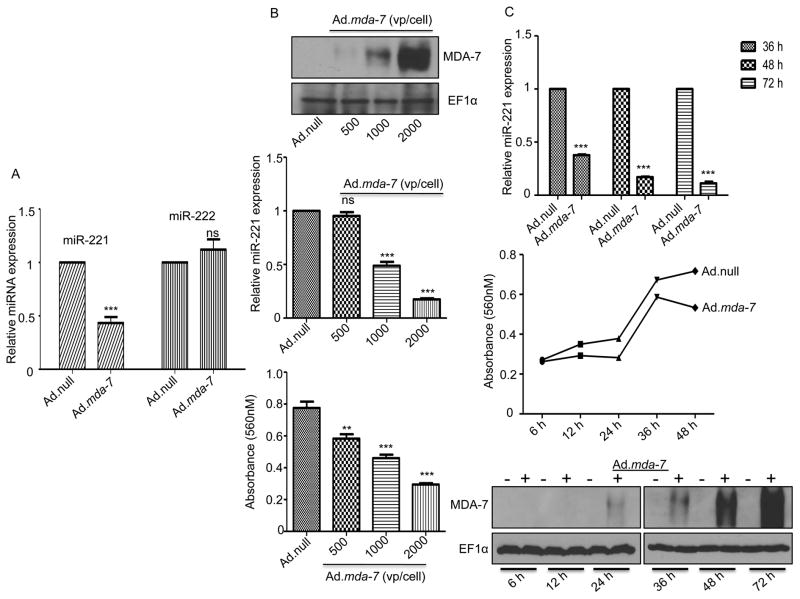
mda-7/IL-24 is recognized for its specific and selective tumor cell-killing effects without harming normal cells. To examine the role of mda-7/IL-24 on the regulation of different microRNAs that are potentially involved in cell death or apoptosis, we overexpressed mda-7/IL-24 using a viral vector expressing mda-7/IL-24 (Ad.mda-7) in MDA-MB-231 cells, an aggressive triple negative breast cancer cell line. The microRNA-enriched fraction was isolated and real time PCR was done for a series of microRNAs related to cell death/apoptosis. A number of microRNAs including miR-200c, let7c, and miR-320 were found to be deregulated after treatment with mda-7/IL-24 (Supplementary Fig. 1). miR-200c which regulates tumor metastasis and epithelial-mesenchymal transition, was found to be down regulated by mda-7/IL-24. Another microRNA, miR-17, reported in G1-S transition of cell cycle, was found to be up regulated by mda-7/IL-24. microRNA-185, a tumor suppressor, reported in many cancers was found to be up regulated in mda-7/IL-24-infected cells. The members of let-7 microRNA family were also deregulated in mda-7/IL-24-infected cells. While let-7e showed no change, let-7c was up regulated in mda-7/IL-24-infected cells. Among these miRNAs, miR-221 is one of the microRNAs reported in a number of cancers and it exhibits an expansive role in different pathways deregulated in cancer. miR-221 targets p27, a key modulator of cell cycle (19) and PUMA, a proapoptotic gene that is degraded by miR-221 (20). PTEN, a potent tumor suppressor is also down regulated by miR-221 (22). As shown in Fig. 1A, we found miR-221 was down regulated in mda-7/IL-24-treated MDA-MB-231 cells, while no alteration was observed in the level of miR-222. mda-7/IL-24-mediated down regulation of miR-221 occurred in a dose-dependent manner, which correlated with exogenous protein expression (MDA-7/IL-24) and inhibition of cell growth (Fig. 1B). The down regulation of miR-221 by mda-7/IL-24 also occurred in a temporal manner in a time point kinetic study (Fig. 1C).
Figure 1. MDA-7/IL-24 regulates miR-221.
A. MDA-MB-231 cells were infected with either Ad.null or Ad.mda-7. Seventy-two hours after infection miRNA fractions were isolated and real time PCR was done using different taqman probes, i.e., miR-221 or miR-222. RNU44 was used as an endogenous control. B. MDA-MB-231 cells were infected with increasing doses of Ad.mda-7 (500 vp, 1000 vp and 2000 vp per cell). Protein lysates were prepared at 72 hours after infection and Western blotting was done to check the levels of MDA-7/IL-24 and EF1α (loading control) (upper panel). miRNA fractions were also isolated at 72 hours after infection and real time PCR was done to check the level of miR-221 (middle panel). MTT assays were done to verify the inhibition of proliferation by mda-7/IL-24 (bottom panel). C. The down regulation of miR-221 by mda-7/IL-24 was temporal as confirmed by a time point kinetics study. MTT assays were done to check the effect of mda-7/IL-24 on the proliferation of cells. The level of MDA-7/IL-24 protein was checked by western blotting.
mda-7/IL-24 down regulates miR-221 in diverse cancer cell lines
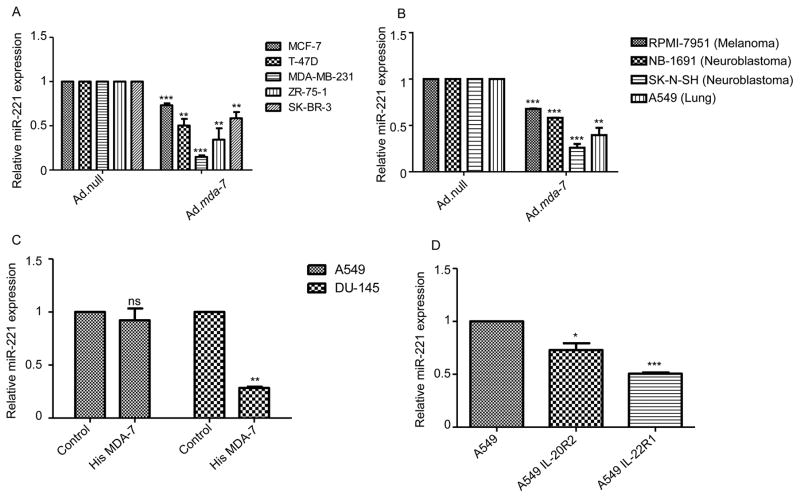
Breast cancer is classified on the basis of hormone receptor expression [estrogen receptor (ER) and progesterone receptor (PR)] and also HER2/Neu status. Triple negative breast cancers express higher levels of miR-221 than ER/PR/HER2 positive breast cancers (31). Our initial observation confirmed that miR-221 was down regulated by mda-7/IL-24 in MDA-MB-231 cells, a triple negative breast cancer cell line. Next we checked regulation of miR-221 after mda-7/IL-24 expression in a panel of breast cancer cell lines with variable ER/PR/HER2 status. We infected MCF-7, T-47D, ZR-75-1 (triple positive) and SK-BR-3 (ER −ve, PR −ve, HER2 +ve) with Ad.null and Ad.mda-7 and collected miRNA enriched fractions and checked the level of miR-221. Interestingly, we found that miR-221 levels decreased with mda-7/IL-24 over expression irrespective of the breast cancer cells receptor status (Fig. 2A). Additionally, we assayed other cancer cell lines, i.e., melanoma, neuroblastoma, and lung cancer, and found a similar downregulation of miR-221 following overexpression of mda-7/IL-24 (Fig. 2B). This endorses the hypothesis that miR-221 may be a potential target for mda-7/IL-24 in a diverse array of cancers and suggests a new pathway of mda-7/IL-24-mediated gene regulation.
Figure 2. MDA-7/IL-24 down regulates miR-221 in diverse cancer cell lines.
A. Different breast cancer cells were infected with Ad.null or Ad.mda-7 (2000 vp/cell) for 72 hours. RQ-PCR was performed to check the level of miR-221. B. Indicated cells were infected with Ad.null or Ad.mda-7 (2000 vp/cell) for 72 hours. RQ-PCR was performed to check the level of miR-221. C. A549 and DU-145 cells were treated with His-MDA-7. RQ-PCR was performed to check the level of miR-221. D. A549 cells were transfected with IL-20R2 or IL-22R1 and treated with His-MDA-7. RQ-PCR was performed to check the level of miR-221.
As a cytokine and a member of the IL-10 cytokine gene family, MDA-7/IL-24 signals through receptor dimers consisting of an R1 type receptor and an R2 type receptor (IL-20R1 and IL-20R2; IL-22R1 and IL-20R2; or a unique receptor pair IL-20R1 and IL-22R1) in order to activate downstream signaling events (5, 6). We used purified recombinant MDA-7/IL-24 protein (11) to confirm further the regulation of miR-221 by MDA-7/IL-24. We treated A549 cells (lung cancer cells which lack a full set of R1 and R2, IL-20/IL-22, receptors) and DU-145 cells (prostate cancer cells containing both receptor types) with His tagged MDA-7 and measured the level of miR-221. miR-221 expression decreased in DU-145 cells following treatment with His-MDA-7, while the level remained unchanged in A549 cells, which lacks the cognate receptor pairs (Fig. 2C). Overexpression of the IL-20R2 or IL-22R1 receptors in A549 cells rendered these cells sensitive to miR-221 down regulation after treatment with MDA-7/IL-24 recombinant protein (Fig. 2D).
To determine if the ability of mda-7/IL-24 to regulate miR-221 was a general phenomenon in both cancer and normal cells we also checked the level of miR-221 following Ad.mda-7 infection in a series of normal immortal human cell lines (IM-PHFA, RWPE-1 and HMEC). No substantial changes in miR-221 levels were evident in any of these normal cells following infection with Ad.mda-7 further supporting the cancer specificity of this cytokine (Supplementary Fig. 2).
Over expression of miR-221 rescues cells from mda-7/IL-24-mediated cell death
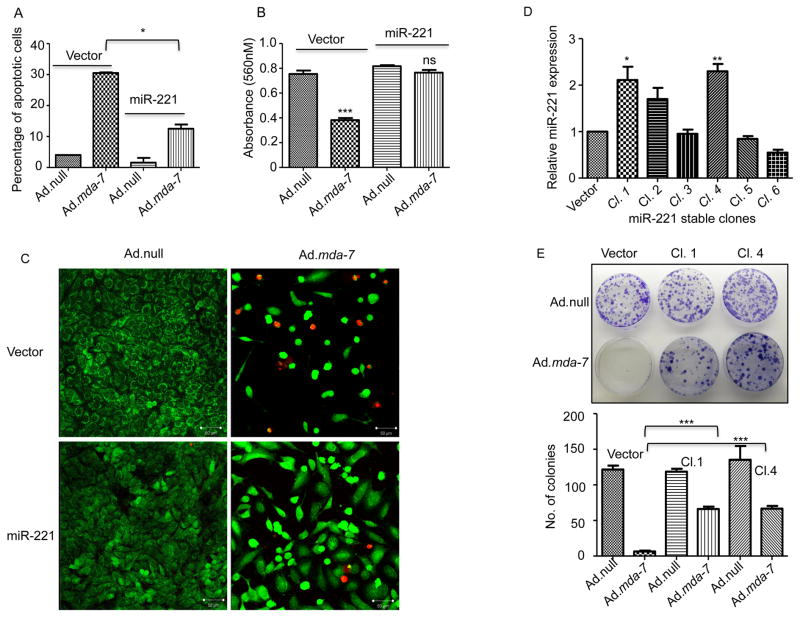
To study the effect of miR-221 on mda-7/IL-24-mediated cell death, MDA-MB-231 cells were transfected with a pCDNA3.1 or miR-221 vector and infected with Ad.null or Ad.mda-7 (2000vp/cells). After 72 hours cells were analyzed for cell death using the Annexin-V binding assay and flow cytometry. As shown in Fig. 3A, MDA-MB-231 cells showed increased apoptosis following infection with Ad.mda-7 vs. Ad.null. Cells that overexpress miR-221 had significantly less cell death suggesting a protective role of miR-221 in mda-7/IL-24-induced apoptotic death. This observation was also supported using a cell proliferation assay. Cells were transfected with miR-221 and infected with Ad.null or Ad.mda-7 and cell proliferation was analyzed with a MTT assay at 72 hours post-infection. As predicted, overexpression of mda-7/IL-24 decreased cell growth, which was rescued in miR-221 overexpressing cells (Fig. 3B). A live dead staining assay further confirmed this data (Fig. 3C). Next, we generated stable cell lines overexpressing miR-221 in MDA-MB-231 cells (Fig. 3D) and did a clonogenic assay following infection with Ad.null or Ad.mda-7 in pCDNA3.1 control vector or miR-221 vector overexpressing stable clones. Infection with Ad.mda-7 resulted in a complete inhibition of colony formation in pCDNA3.1-transfected cells, however there were viable colonies observed in miR-221 overexpressing stable cell clones Cl. 1 and Cl. 4 (Fig. 3E). A direct correlation between enhanced colony formation and levels of miR-221 expression was evident when plating Ad.mda-7 infected cells at low density (50 as opposed to 200 cells/6-cm plate) (data not shown).
Figure 3. Over expression of miR-221 can rescue cells from mda-7/IL-24-mediated cell death.
A. MDA-MB-231 cells were transfected with pCDNA3.1 (vector) or miR-221 and then treated with Ad.null or Ad.mda-7. After 72 hours cells were stained with Annexin-V and then analyzed by flow cytometer. B. Cells were treated as in A and MTT assays were done to check the effect of miR-221 overexpression on mda-7/IL-24-mediated cell growth inhibition. C. Cells were treated as in A and stained with live dead staining kit 72 hours after treatment. Images were obtained using confocal microscopy. D. MDA-MB-231 cells were stably transfected with pCDNA3.1 (vector) or miR-221. After selection, clones were checked for miR-221 expression. E. MDA-MB-231 cells stably overexpressing either pCDNA3.1 (vector) or miR-221 were treated with Ad.null or Ad.mda-7. Two thousand cells were plated and after two weeks they were stained with crystal violet. Number of colonies was counted and the data were plotted in the graph.
mda-7/IL-24 regulation of miR-221 expression is ROS-dependent
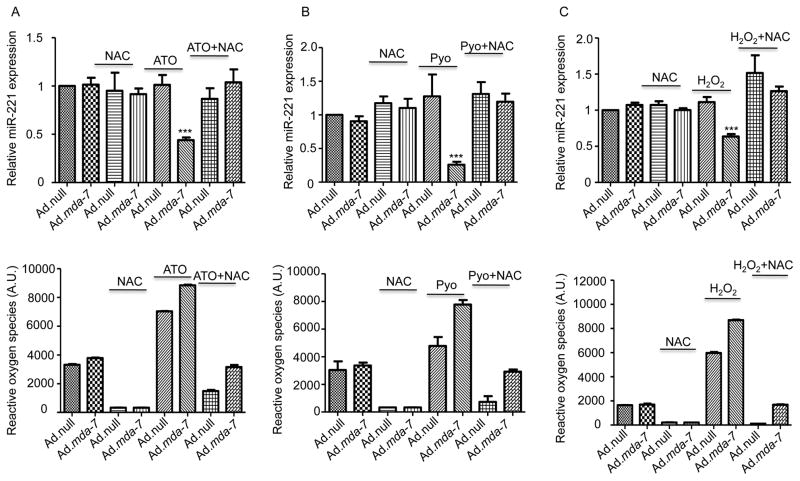
Ectopic expression of MDA-7/IL-24 in glioblastoma multiforme cells increases thioredoxin and manganese super oxide dismutase (SOD2) levels, without altering SOD1 protein levels (32). MDA-7/IL-24-mediated cell killing relies on reactive oxygen species (ROS) generation, which is one of the key mediators of MDA-7/IL-24 toxicity in cancer cells (33). In these contexts, we determined if ROS inducers could enhance mda-7/IL-24 down regulation of miR-221. Infection of MDA-MB-231 breast cancer cells with 500 vp/cell of Ad.mda-7 did not significantly alter miR-221 levels. Similarly, treatment with low doses of ROS inducers, Arsenic trioxide (ATO, 1μM), hydrogen peroxide (10 μM) or pyocyanin (50μM) did not alter miR-221 levels in MDA-MB-231 cells. In contrast, a combination of Ad.mda-7 plus ATO (Fig. 4A), pyocyanin (Fig. 4B) or hydrogen peroxide (Fig. 4C) at the low doses indicated above significantly decreased the level of miR-221. These observations confirm a role of reactive oxygen species in mda-7/IL-24-mediated down regulation of miR-221. While ATO, hydrogen peroxide or pyocyanin down regulated the miR-221 level with Ad.mda-7, treatment with NAC, a known anti-oxidant abrogated down regulation of miR-221. This also supports earlier published studies, which have shown the role of ROS in mda-7/IL-24-mediated cell death (33). As shown in supplementary Fig. 3, elevated levels of ROS inducers decreased the level of miR-221 further supporting a ROS-mediated regulation of miR-221.
Figure 4. mda-7/IL-24 regulates miR-221 expression in a ROS-dependent manner.
A. MDA-MB-231 cells were infected with Ad.null or Ad.mda-7 (500 vp/cell) for 72 hours, N-acetyl cysteine pretreatment was for 12 hours. Arsenic trioxide (ATO) was added to cells for 12 hours as indicated. RQ-PCR was performed to check the level of miR-221. The level of ROS was measured and presented below. B. MDA-MB-231 cells were treated as above in A. Cells were exposed to Pyocyanin for 12 hours as indicated. RQ-PCR was performed to check the level of miR-221. The level of ROS was measured and is represented below. C. MDA-MB-231 cells were treated as above in A. Hydrogen peroxide was added to cells for 4 hours as indicated. RQ-PCR was performed to determine the level of miR-221 expression. The graphs shown below represent the amount of ROS produced.
Beclin-1 is a direct target of miR-221
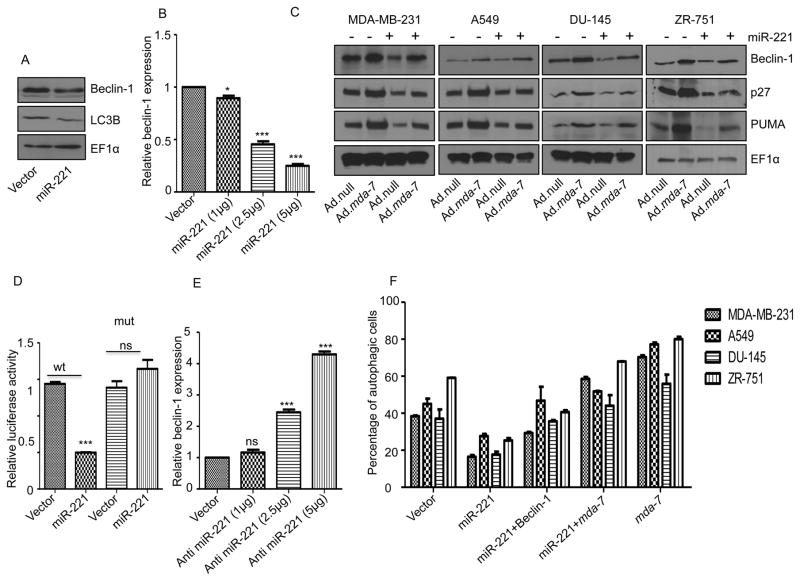
miR-221 inhibits autophagy induction, which leads to heart failure by deregulating the p27/CDK2/mTOR pathway (34). A regulatory link between miR-221 and Beclin-1 has been suggested, since Beclin-1 is regulated by HDAC6 (35) and HDAC6 is regulated by miR-221 (36). Additionally, a regulatory role of mda-7/IL-24 in toxic autophagy has been described (37). These observations prompted us to investigate the regulatory role of mda-7/IL-24-miR-221 axis in the autophagy process. First, we checked the expression pattern of Beclin-1 following overexpression of miR-221. MDA-MB-231 cells were transfected with a pCDNA3.1 vector or a miR-221 expressing construct. Overexpression of miR-221 resulted in beclin-1 down regulation (Fig. 5A) and increasing doses of miR-221 significantly down regulated beclin-1 at the transcript and protein level in a dose-dependent manner in MDA-MB-231 cells (Figs. 5B, 5C and Supplementary Fig. 4). To validate the role of miR-221 on the transcriptional regulation of beclin-1, we performed a luciferase reporter gene assay using a 3′ UTR beclin-1 construct that covers 600-bp downstream of the beclin-1 stop codon. The miR-221 transfected HeLa cells showed a significantly lower luciferase activity than the vector-transfected cells (Fig. 5D) suggesting that beclin-1 is a potential target of miR-221. The assay was validated with a mutated 3′ UTR construct of beclin-1. miR-221 failed to down regulate the mutant construct, which has no binding site for miR-221 (Fig. 5D). It was reported earlier that mda-7/IL-24 over expression led to enhanced beclin-1 expression (35). These data confirm that beclin-1 is a potential target of miR-221 and suggest a mechanism of mda-7/IL-24-mediated autophagy regulation through a miR-221 and beclin-1 pathway. To further confirm miR-221 regulation of beclin-1 we blocked miR-221 by a specific anti-miR-221 in MDA-MB-231 cells and measured the expression of beclin-1. The addition of anti-miR-221 prevented the degradation of beclin-1 in basal (Fig. 5E and Supplementary Fig. 5) and Ad.mda-7-infected cells (Supplementary Fig. 5). Expression of p27 and PUMA were also checked to validate the experimental controls (Supplementary Fig. 5). The same trend was observed in two other cell lines derived from other cancers, i.e., lung (A549) and prostate (DU-145) and also in an additional breast cancer cell line, i.e. ZR-751 (Fig. 5C, Supplementary Figs. 4 and 5). These data confirm that beclin-1 is a potential target of miR-221.
Figure 5. Beclin-1 is a direct target of miR-221.
A. MDA-MB-231 cells were transfected with control pCDNA3.1 (vector) or miR-221. Western blotting analysis was performed to show the expression of Beclin-1/LC3B/EF1α. B. MDA-MB-231 cells were transfected with increasing concentrations of miR-221, RNA was isolated 48-hours post-transfection and real time PCR was done to check the level of Beclin-1. C. Cells were transfected with a miR-221 construct and then infected with either Ad.null or Ad.mda-7 virus (2000 vp/cell) for 72 hours. Cell lysates were probed with Beclin-1, p27, and PUMA antibodies. EF1α was used as a loading control. D. Reporter gene assays were done in HeLa cells using the 3′UTR Beclin-1 construct; miR-221 over expression significantly decreased the luciferase activity of the wt Beclin-1 UTR. E. MDA-MB-231 cells were transfected with increasing concentrations of anti-miR-221, RNA was isolated after 48 hours and real time PCR was done to check the level of Beclin-1. F. Cells were transfected/treated with the indicated constructs and after 24 hours of transfection they were serum-starved by growth in serum-free medium for 24 hours. Cells were stained with acridine orange and then analyzed by flow cytometry.
To investigate further the potential regulatory axis of mda-7/IL-24-miR-221-beclin-1, we transfected cells with a miR-221 construct and infected cells with Ad.mda-7. As observed earlier, mda-7/IL-24 up regulated beclin-1 and simultaneous over expression of mda-7/IL-24 with miR-221 diminished the mda-7/IL-24-mediated enhanced expression of beclin-1 (Fig. 5C). Additionally, we evaluated autophagy induction using an acridine orange-based staining method. As shown in Fig 5F, ectopic expression of miR-221 diminished autophagy induction and overexpression of beclin-1 or mda-7/IL-24 rescued the autophagy process in these cells confirming further a role of the mda-7/IL-24-miR-221-beclin-1 regulatory loop in autophagy induction.
Rapamycin, another autophagy inducer, up regulates beclin-1 (38). To study the role of Rapamycin and miR-221 on beclin-1 levels, different cancer cell lines were transfected with miR-221 and treated with Rapamycin. While Rapamycin up regulated beclin-1 protein levels, ectopic expression of miR-221 decreased this upregulation (Supplementary Fig. 6). This result confirms miR-221-mediated beclin-1 regulation and also explains yet another mechanism showing miR-221 can deregulate Rapamycin-induced autophagy.
mda-7/IL-24 regulates miR-221expression in vivo
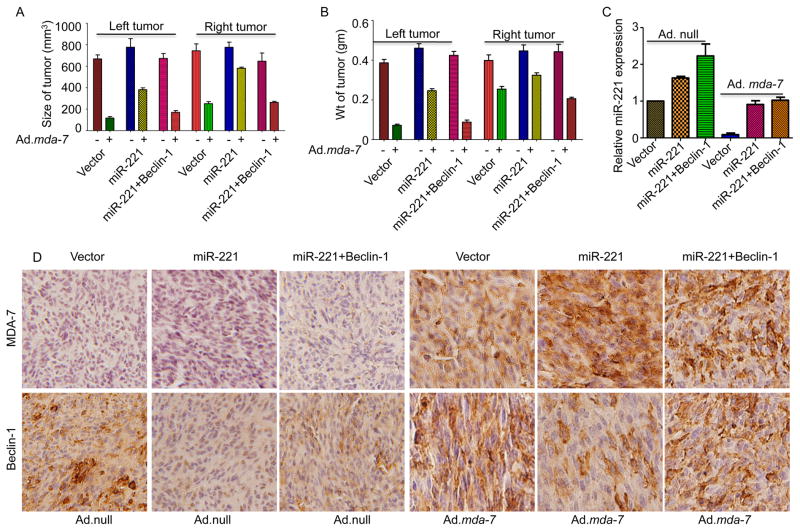
To investigate the role of mda-7/IL-24 on miR-221 in vivo, MDA-MB-231 cells over expressing miR-221 or miR-221 plus beclin-1 were injected subcutaneously to establish tumor xenografts in female athymic nude mice. After a palpable tumor (100 mm3) developed in approximately 10 days, the tumors were injected with 8 intramural injections over a 3-week period with 1 X 108 viral particles of Ad.null or Ad.mda-7. In control vector-transfected cells a significant growth inhibitory effect was evident, but in miR-221 over expressing cells the effect of Ad.mda-7 was less apparent both in the injected left tumor, and in the uninjected right tumor, as previously observed when infecting these cells in vitro (Fig. 6A and B). Interestingly, overexpressing beclin-1 in miR-221-transfected cells sensitized these cells to mda-7/IL-24-induced cell death. The expression of miR-221 was confirmed by RQ-PCR (Fig. 6C) and the expression of MDA-7/IL-24 and beclin-1 was validated and quantified in the tumor sections by immunohistochemistry (Fig. 6D, Supplementary Fig. 7). A modest increase in p27 and PUMA expression was also evident in tumor sections from Ad.mda-7-treated tumors vs. Ad.null-treated tumors (Supplementary Fig. 8). Taken together these results demonstrate the significance of miR-221 and beclin-1 in triggering mda-7/IL-24-mediated cell death in cancer cells.
Figure 6. Intratumoral injections of mda-7/IL-24 induces miR-221-mediated cell death.
A. MDA-MB-231 human breast cancer cells, stably expressing a control pCDNA3.1 vector, miR-221 or miR-221 and Beclin-1, were subcutaneously implanted in both flanks of nude mice. Left sided tumors were treated with 8 intratumoral injections of Ad.mda-7. Ad.null was used as control. A total of 5 mice were studied in each group. Once the control tumors reached maximum allowable limit, tumors were isolated from both flanks. A. Tumor volumes on both flanks were measured and results are presented in a graphical manner. B. Graphical representation of the weight of the tumors on both flanks. C. RNA was isolated from the tumor sections (injected tumors) and real time PCR was done to validate the level of miR-221. D. Immunohistochemical analysis of MDA-7/IL-24 and Beclin-1 in tumor sections (injected tumors).
Discussion
Reprogramming of cancerous cells to undergo toxic autophagy (39) or apoptosis (40) is a viable strategy for treatment of cancer. The discovery of mda-7/IL-24 using subtraction hybridization in melanoma has further advanced this opportunity (1, 2). The multiple distinctive functions of mda-7/IL-24 in cancer therapy include tumor-specific killing through combined effects of apoptosis and toxic autophagy, potent “bystander” anti-cancer activity, immunomodulation, inhibition of cell proliferation, and suppression of angiogenesis (3–6). We now demonstrate that miR-221, an oncogenic miRNA, is down regulated by over expression of mda-7/IL-24 in a cancer cell-specific manner. Using a panel of breast, lung, prostate, and neuroblastoma cell lines we document a significant decrease in the level of miR-221 following adenoviral-mediated delivery of mda-7/IL-24. This down regulation of miR-221 correlates with mda-7/IL-24-mediated cell death and over expression of miR-221 blocks cell death induced by mda-7/IL-24. Production and secretion of MDA-7/IL-24 following treatment with purified recombinant cytokine or infection with Ad.mda-7 decreases cell growth and induces apoptosis in cancer cells, but not in normal cells. Additionally, secreted MDA-7/IL-24 also induces apoptosis in surrounding cells as well as distant tumor cells through “bystander” antitumor effects (28). Furthermore, MDA-7/IL-24 protein induces production of endogenous MDA-7/IL-24 through an autocrine/paracrine loop (11). Using recombinant His-MDA-7 we found that MDA-7/IL-24 also down regulates miR-221, uniquely in IL-20/IL-22 receptor positive cancer cells. Reconstruction of cognate receptors in receptor complex negative cells renders them vulnerable to His MDA-7 treatment and consequently results in down regulation of miR-221. These results further demonstrate the relevance of the MDA-7/IL-24-miR-221 axis in promoting cancer-specific cell death. In these contexts, miR-221 represents a novel downstream target of mda-7/IL-24 specific to cancer cells that mediates its biological anti-cancer functions both in vitro and in vivo.
The profound anticancer action of mda-7/IL-24 in cell culture and pre-clinical animal models led to its entry into the clinic and has culminated in a successful phase-I/II clinical trial (3, 4, 7, 8, 41). These observations reinforce the relevance of defining the mechanism of anti-cancer activity of mda-7/IL-24. Additionally, many combinatorial approaches have been shown to further enhance mda-7/IL-24’s antitumor activities. Defining ways of making this therapeutic even better is of significant import and very relevant for the treatment of primary and advanced cancers.
Reactive oxygen species play a prominent role in mda-7/IL-24-restricted antitumor functions (33). Multiple cellular and physiological processes impacted on by mda-7/IL-24 are regulated by ROS. In the context of pancreatic cancer, where there is a ‘translational block’ of mda-7/IL-24 mRNA into protein, ROS can reverse this inhibition resulting in enhanced association of mda-7/IL-24 mRNA with polyribosomes and translation into protein thereby resulting in pancreatic cancer cell death (42–44). We now show that mda-7/IL-24-mediated down regulation of miR-221 is ROS-dependent and treatment with anti-oxidants can reverse this process. These results accentuate a path for the development of rational combinatorial approaches for the treatment of aggressive tumors by combining mda-7/IL-24 with other in-clinic ROS-inducing chemotherapeutic agents.
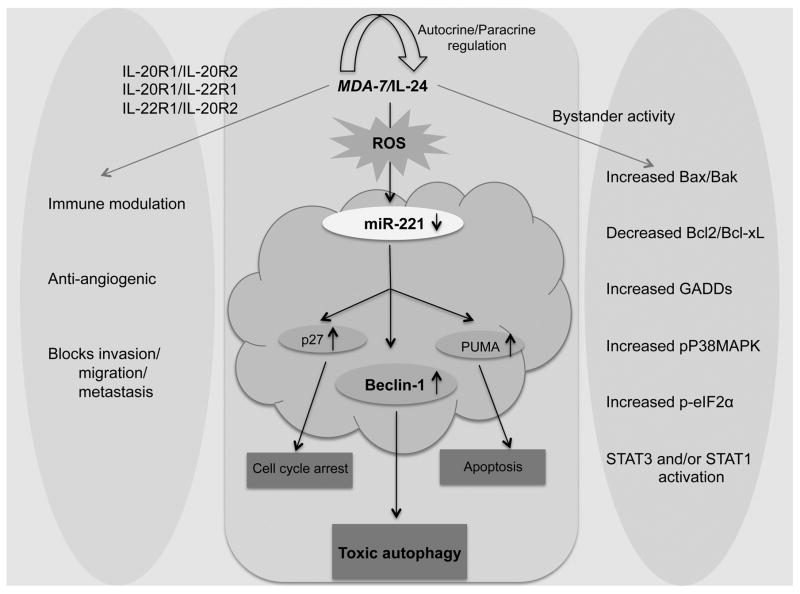
MicroRNAs (miRNAs) play a central role in regulating different normal and pathological pathways, including development and cancer, respectively. Different microRNAs instigate diverse effects in a cell and tissue context-dependent manner depending on the target gene they regulate (45). Prior studies indicate that miR-221 is significantly upregulated in different cancers (17). To identify potential new targets of miR-221 we investigated the expression of some of the major proteins involved in autophagy, apoptosis and cell cycle. The regulation of beclin-1 expression by miR-221 was demonstrated by transfection of tumor cells with a miR-221 mimic, which resulted in a decrease in beclin-1 expression at both the mRNA and protein level. This relationship was confirmed further by transfection with anti-miR-221, which resulted in up regulation of beclin-1 expression. Beclin-1-mediated protective/toxic autophagy, depending on cellular context, plays a decisive role in cell survival/death and aberrant expression of beclin-1 has been reported in different diseases including cancer (26, 46–49). Rapamycin is a well described autophagy inducer (50) and it also up regulates beclin-1 (36). Our results confirm that miR-221 not only degrades basal but also Rapamycin- and mda-7/IL-24-induced beclin-1 expression. Although it has been shown that mda-7/IL-24 promotes toxic autophagy, the detailed mechanism is not well understood. We now show that beclin-1, a key player in autophagy, is a new target of miR-221 and mda-7/IL-24 can promote toxic autophagy through a miR-221/beclin-1 axis. In addition to other regulators, i.e., p27, BAX, GADDs, Stat3, p38MAPKs, Bcl2/Bcl-xL and PUMA, Beclin-1 is a new target of mda-7/IL-24 (summarized in figure 7). This study of beclin-1 regulation by miR-221 and miR-221 regulation by mda-7/IL-24 warrants further investigation and these studies have potential to yield new insights into the regulation of autophagy and the association of this phenomenon with various disease states.
Figure 7. Schematic representation of Ad. mda-7-induced cell death in cancer cells.
MDA-7/IL-24 down regulates miR-221 which in turn up regulates Beclin-1 to induce toxic autophagy and cell death in cancer cells. Additionally, the pathways that are regulated by mda-7/IL-24 are depicted here schematically.
Supplementary Material
Acknowledgments
Grant Support
The present study was supported in part by NIH, NCI SPORE P50 CA058236 and the National Foundation for Cancer Research (NFCR). Microscopy was performed at the VCU - Dept. of Anatomy & Neurobiology Microscopy Facility, supported, in part, by funding from NIH-NINDS Center Core Grant 5 P30 NS047463 and, in part, by funding from NIH-NCI Cancer Center Support Grant P30 CA016059. The VCU Massey Cancer Center Flow Cytometry Shared Resource, supported, in part, with funding from NIH-NCI Cancer Center Support Grant P30 CA016059, generated services and products in support of the research project. D.S. is the Harrison Foundation Distinguished Professor in Cancer Research. P.B.F. holds the Thelma Newmeyer Corman Chair in Cancer Research.
Footnotes
Conflict of Interest: PBF is co-founder of serves as a consultant to and has ownership interest in Cancer Targeting Systems (CTS), Inc. Virginia Commonwealth University, Johns Hopkins University and Columbia University have ownership interest in CTS, Inc. Competing financial interests had no role in study design, data collection and interpretation, or the decision to submit the work for publication. The other authors declare that no competing interests exist.
Author’s Contributions: Conception and design: A.K. Pradhan, L. Emdad, S.K. Das, P.B. Fisher; Development of methodology: A.K. Pradhan, S. Talukdar, P. Bhoopathi; Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): A.K. Pradhan, S. Talukdar, P. Bhoopathi, X.-N. Shen; Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): A.K. Pradhan, L. Emdad, S.K. Das, D. Sarkar, P.B. Fisher; Administrative, technical or material support (i.e., reporting or organizing data, constructing databases): A.K. Pradhan, S.K. Das, D. Sarkar, L. Emdad, P.B. Fisher; Study Supervision: S.K. Das, D. Sarkar, L. Emdad, P.B. Fisher; Wrote Paper: A.K. Pradhan and P.B. Fisher.
References
- 1.Jiang H, Lin JJ, Su ZZ, Goldstein NI, Fisher PB. Subtraction hybridization identifies a novel melanoma differentiation associated gene, mda-7, modulated during human melanoma differentiation, growth and progression. Oncogene. 1995;11:2477–86. [PubMed] [Google Scholar]
- 2.Jiang H, Su ZZ, Lin JJ, Goldstein NI, Young CS, Fisher PB. The melanoma differentiation associated gene mda-7 suppresses cancer cell growth. Proceedings of the National Academy of Sciences of the United States of America. 1996;93:9160–5. doi: 10.1073/pnas.93.17.9160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emdad L, Lebedeva IV, Su ZZ, Gupta P, Sauane M, Dash R, et al. Historical perspective and recent insights into our understanding of the molecular and biochemical basis of the antitumor properties of mda-7/IL-24. Cancer biology & therapy. 2009;8:391–400. doi: 10.4161/cbt.8.5.7581. [DOI] [PubMed] [Google Scholar]
- 4.Lebedeva IV, Sauane M, Gopalkrishnan RV, Sarkar D, Su ZZ, Gupta P, et al. mda-7/IL-24: exploiting cancer’s Achilles’ heel. Molecular therapy : the journal of the American Society of Gene Therapy. 2005;11:4–18. doi: 10.1016/j.ymthe.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Dash R, Bhutia SK, Azab B, Su ZZ, Quinn BA, Kegelmen TP, et al. mda-7/IL-24: a unique member of the IL-10 gene family promoting cancer-targeted toxicity. Cytokine Growth Factor Rev. 2010;21:381–91. doi: 10.1016/j.cytogfr.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menezes ME, Bhatia S, Bhoopathi P, Das SK, Emdad L, Dasgupta S, et al. MDA-7/IL-24: multifunctional cancer killing cytokine. Advances exper med biol. 2014;818:127–53. doi: 10.1007/978-1-4471-6458-6_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisher PB, Gopalkrishnan RV, Chada S, Ramesh R, Grimm EA, Rosenfeld MR, et al. mda-7/IL-24, a novel cancer selective apoptosis inducing cytokine gene: from the laboratory into the clinic. Cancer Biol Ther. 2003;2:S23–37. [PubMed] [Google Scholar]
- 8.Cunningham CC, Chada S, Merritt JA, Tong A, Senzer N, Zhang Y, et al. Clinical and local biological effects of an intratumoral injection of mda-7 (IL24; INGN 241) in patients with advanced carcinoma: a phase I study. Molecular therapy : the journal of the American Society of Gene Therapy. 2005;11:149–59. doi: 10.1016/j.ymthe.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 9.Nishikawa T, Ramesh R, Munshi A, Chada S, Meyn RE. Adenovirus-mediated mda-7 (IL24) gene therapy suppresses angiogenesis and sensitizes NSCLC xenograft tumors to radiation. Molecular therapy : the journal of the American Society of Gene Therapy. 2004;9:818–28. doi: 10.1016/j.ymthe.2004.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Ramesh R, Mhashilkar AM, Tanaka F, Saito Y, Branch CD, Sieger K, et al. Melanoma differentiation-associated gene 7/interleukin (IL)-24 is a novel ligand that regulates angiogenesis via the IL-22 receptor. Cancer research. 2003;63:5105–13. [PubMed] [Google Scholar]
- 11.Sauane M, Su ZZ, Gupta P, Lebedeva IV, Dent P, Sarkar D, et al. Autocrine regulation of mda-7/IL-24 mediates cancer-specific apoptosis. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9763–8. doi: 10.1073/pnas.0804089105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gupta P, Walter MR, Su ZZ, Lebedeva IV, Emdad L, Randolph A, et al. BiP/GRP78 is an intracellular target for MDA-7/IL-24 induction of cancer-specific apoptosis. Cancer research. 2006;66:8182–91. doi: 10.1158/0008-5472.CAN-06-0577. [DOI] [PubMed] [Google Scholar]
- 13.Fisher PB. Is mda-7/IL-24 a “magic bullet” for cancer? Cancer research. 2005;65:10128–38. doi: 10.1158/0008-5472.CAN-05-3127. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;129:303–17. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 15.Gu S, Jin L, Zhang F, Sarnow P, Kay MA. Biological basis for restriction of microRNA targets to the 3′ untranslated region in mammalian mRNAs. Nature structural & molecular biology. 2009;16:144–50. doi: 10.1038/nsmb.1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:9667–72. doi: 10.1073/pnas.0703820104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blower PE, Verducci JS, Lin S, Zhou J, Chung JH, Dai Z, et al. MicroRNA expression profiles for the NCI-60 cancer cell panel. Molecular cancer therapeutics. 2007;6:1483–91. doi: 10.1158/1535-7163.MCT-07-0009. [DOI] [PubMed] [Google Scholar]
- 18.Guo L, Xu J, Qi J, Zhang L, Wang J, Liang J, et al. MicroRNA-17-92a upregulation by estrogen leads to Bim targeting and inhibition of osteoblast apoptosis. Journal of cell science. 2013;126:978–88. doi: 10.1242/jcs.117515. [DOI] [PubMed] [Google Scholar]
- 19.le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. The EMBO journal. 2007;26:3699–708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Molecular cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. The Journal of biological chemistry. 2008;283:31079–86. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 22.Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer cell. 2009;16:498–509. doi: 10.1016/j.ccr.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–61. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 24.Hamada N, Fujita Y, Kojima T, Kitamoto A, Akao Y, Nozawa Y, et al. MicroRNA expression profiling of NGF-treated PC12 cells revealed a critical role for miR-221 in neuronal differentiation. Neurochemistry international. 2012;60:743–50. doi: 10.1016/j.neuint.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 25.Felicetti F, Errico MC, Bottero L, Segnalini P, Stoppacciaro A, Biffoni M, et al. The promyelocytic leukemia zinc finger-microRNA-221/-222 pathway controls melanoma progression through multiple oncogenic mechanisms. Cancer research. 2008;68:2745–54. doi: 10.1158/0008-5472.CAN-07-2538. [DOI] [PubMed] [Google Scholar]
- 26.Miracco C, Cosci E, Oliveri G, Luzi P, Pacenti L, Monciatti I, et al. Protein and mRNA expression of autophagy gene Beclin 1 in human brain tumours. International journal of oncology. 2007;30:429–36. [PubMed] [Google Scholar]
- 27.Su Z, Shi Y, Friedman R, Qiao L, McKinstry R, Hinman D, et al. PEA3 sites within the progression elevated gene-3 (PEG-3) promoter and mitogen-activated protein kinase contribute to differential PEG-3 expression in Ha-ras and v-raf oncogene transformed rat embryo cells. Nucleic Acids Res. 2001;29:1661–71. doi: 10.1093/nar/29.8.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Su Z, Emdad L, Sauane M, Lebedeva IV, Sarkar D, Gupta P, et al. Unique aspects of mda-7/IL-24 antitumor bystander activity: establishing a role for secretion of MDA-7/IL-24 protein by normal cells. Oncogene. 2005;24:7552–66. doi: 10.1038/sj.onc.1208911. [DOI] [PubMed] [Google Scholar]
- 29.Dash R, Bhoopathi P, Das SK, Sarkar S, Emdad L, Dasgupta S, et al. Novel mechanism of MDA-7/IL-24 cancer-specific apoptosis through SARI induction. Cancer research. 2014;74:563–74. doi: 10.1158/0008-5472.CAN-13-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson EN, Bristol ML, Di X, Maltese WA, Koterba K, Beckman MJ, et al. A switch between cytoprotective and cytotoxic autophagy in the radiosensitization of breast tumor cells by chloroquine and vitamin D. Hormones & cancer. 2011;2:272–85. doi: 10.1007/s12672-011-0081-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nassirpour R, Mehta PP, Baxi SM, Yin MJ. miR-221 promotes tumorigenesis in human triple negative breast cancer cells. PloS one. 2013;8:e62170. doi: 10.1371/journal.pone.0062170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Yacoub A, Hamed HA, Allegood J, Mitchell C, Spiegel S, Lesniak MS, et al. PERK-dependent regulation of ceramide synthase 6 and thioredoxin play a key role in mda-7/IL-24-induced killing of primary human glioblastoma multiforme cells. Cancer research. 2010;70:1120–9. doi: 10.1158/0008-5472.CAN-09-4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lebedeva IV, Su ZZ, Sarkar D, Kitada S, Dent P, Waxman S, et al. Melanoma differentiation associated gene-7, mda-7/interleukin-24, induces apoptosis in prostate cancer cells by promoting mitochondrial dysfunction and inducing reactive oxygen species. Cancer research. 2003;63:8138–44. [PubMed] [Google Scholar]
- 34.Su M, Wang J, Wang C, Wang X, Dong W, Qiu W, et al. MicroRNA-221 inhibits autophagy and promotes heart failure by modulating the p27/CDK2/mTOR axis. Cell death and differentiation. 2015;22:986–99. doi: 10.1038/cdd.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jung KH, Noh JH, Kim JK, Eun JW, Bae HJ, Chang YG, et al. Histone deacetylase 6 functions as a tumor suppressor by activating c-Jun NH2-terminal kinase-mediated beclin 1-dependent autophagic cell death in liver cancer. Hepatology. 2012;56:644–57. doi: 10.1002/hep.25699. [DOI] [PubMed] [Google Scholar]
- 36.Bae HJ, Jung KH, Eun JW, Shen Q, Kim HS, Park SJ, et al. MicroRNA-221 governs tumor suppressor HDAC6 to potentiate malignant progression of liver cancer. J Hepatol. 2015;63:408–19. doi: 10.1016/j.jhep.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 37.Bhutia SK, Dash R, Das SK, Azab B, Su ZZ, Lee SG, et al. Mechanism of autophagy to apoptosis switch triggered in prostate cancer cells by antitumor cytokine melanoma differentiation-associated gene 7/interleukin-24. Cancer research. 2010;70:3667–76. doi: 10.1158/0008-5472.CAN-09-3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie ZG, Xie Y, Dong QR. Inhibition of the mammalian target of rapamycin leads to autophagy activation and cell death of MG63 osteosarcoma cells. Oncology letters. 2013;6:1465–69. doi: 10.3892/ol.2013.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thorburn A, Thamm DH, Gustafson DL. Autophagy and cancer therapy. Molecular pharmacology. 2014;85:830–8. doi: 10.1124/mol.114.091850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reed JC. Apoptosis-based therapies. Nature reviews Drug discovery. 2002;1:111–21. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- 41.Tong AW, Nemunaitis J, Su D, Zhang Y, Cunningham C, Senzer N, et al. Intratumoral injection of INGN 241, a nonreplicating adenovector expressing the melanoma-differentiation associated gene-7 (mda-7/IL24): biologic outcome in advanced cancer patients. Molecular therapy. 2005;11:160–72. doi: 10.1016/j.ymthe.2004.09.021. [DOI] [PubMed] [Google Scholar]
- 42.Lebedeva IV, Su ZZ, Sarkar D, Gopalkrishnan RV, Waxman S, Yacoub A, et al. Induction of reactive oxygen species renders mutant and wild-type K-ras pancreatic carcinoma cells susceptible to Ad. mda-7-induced apoptosis. Oncogene. 2005;24:585–96. doi: 10.1038/sj.onc.1208183. [DOI] [PubMed] [Google Scholar]
- 43.Lebedeva IV, Su ZZ, Emdad L, Kolomeyer A, Sarkar D, Kitada S, et al. Targeting inhibition of K-ras enhances Ad. mda-7-induced growth suppression and apoptosis in mutant K-ras colorectal cancer cells. Oncogene. 2007;26:733–44. doi: 10.1038/sj.onc.1209813. [DOI] [PubMed] [Google Scholar]
- 44.Sarkar S, Azab BM, Das SK, Quinn BA, Shen X, Dash R, et al. Chemoprevention gene therapy (CGT): novel combinatorial approach for preventing and treating pancreatic cancer. Current molecular medicine. 2013;13:1140–59. doi: 10.2174/1566524011313070008. [DOI] [PubMed] [Google Scholar]
- 45.Roldo C, Missiaglia E, Hagan JP, Falconi M, Capelli P, Bersani S, et al. MicroRNA expression abnormalities in pancreatic endocrine and acinar tumors are associated with distinctive pathologic features and clinical behavior. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2006;24:4677–84. doi: 10.1200/JCO.2005.05.5194. [DOI] [PubMed] [Google Scholar]
- 46.Pickford F, Masliah E, Britschgi M, Lucin K, Narasimhan R, Jaeger PA, et al. The autophagy-related protein beclin 1 shows reduced expression in early Alzheimer disease and regulates amyloid beta accumulation in mice. The Journal of clinical investigation. 2008;118:2190–9. doi: 10.1172/JCI33585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diskin T, Tal-Or P, Erlich S, Mizrachy L, Alexandrovich A, Shohami E, et al. Closed head injury induces upregulation of Beclin 1 at the cortical site of injury. J Neurotrauma. 2005;22:750–62. doi: 10.1089/neu.2005.22.750. [DOI] [PubMed] [Google Scholar]
- 48.Li DD, Wang LL, Deng R, Tang J, Shen Y, Guo JF, et al. The pivotal role of c-Jun NH2-terminal kinase-mediated Beclin 1 expression during anticancer agents-induced autophagy in cancer cells. Oncogene. 2009;28:886–98. doi: 10.1038/onc.2008.441. [DOI] [PubMed] [Google Scholar]
- 49.Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z, et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer research. 2008;68:9167–75. doi: 10.1158/0008-5472.CAN-08-1573. [DOI] [PubMed] [Google Scholar]
- 50.Sarkar S, Ravikumar B, Floto RA, Rubinsztein DC. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell death and differentiation. 2009;16:46–56. doi: 10.1038/cdd.2008.110. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.