Abstract
Exposure to early-life stress (ELS) may heighten the risk for psychopathology at adulthood. Here, in order to identify common genes that may keep the memory of ELS through changes in their methylation status, we intersected methylome analyses performed in different tissues and time points in rats, non-human primates and humans, all characterized by ELS. We identified Ankyrin-3 (Ank3), a scaffolding protein with a strong genetic association for psychiatric disorders, as a gene persistently affected by stress exposure. In rats, Ank3 methylation and mRNA changes displayed a specific temporal profile during the postnatal development. Moreover, exposure to prenatal stress altered the interaction of ankyrin-G, the protein encoded by Ank3 enriched in the post-synaptic compartment, with PSD95. Notably, to model in humans a gene by early stress interplay on brain phenotypes during cognitive performance, we demonstrated an interaction between functional variation in Ank3 gene and obstetric complications on working memory in healthy adult subjects. Our data suggest that alterations of Ank3 expression and function may contribute to the effects of ELS on the development of psychiatric disorders.
Introduction
Adverse events during prenatal and early postnatal periods have a pivotal role in the later susceptibility to neuropsychiatric disorders1, 2, 3 by interfering with the developmental trajectories of different systems, thus leading to long-lasting reprogramming consequences.4 A growing body of evidence in the last decade has ascribed a key role to the epigenome, an array of chemical modifications to the DNA and histone proteins that affect gene expression without altering the DNA sequence, in bridging the experience of early insults with the appearance of a pathological phenotype at adulthood.5, 6, 7 In particular, several rodent and human studies have proven that among epigenetic mechanisms, DNA methylation is dynamically sensitive to external cues particularly in the perinatal period, when most of the DNA methylation patterns are arranged to shape and define the cellular destiny and thus are also highly responsive and susceptible to environmental stressors that could alter such programming, thus increasing the vulnerability to later psychopathology.8, 9, 10, 11, 12, 13 In order to investigate the underlying causative mechanisms responsible of the long-term effects, several animal models have been developed that replicate the exposure to different types of stressors, of diverse intensities, in specific time windows during early perinatal life.14, 15, 16, 17 Although these animal models bear the advantages of studying, in few months, the effects of neonatal stress on adult behavior, both in the brain as well as at peripheral level, they have the limitation of lacking a direct relevance to the human condition. Here, in order to circumvent these problems, we used a cross-species genome-wide approach to identify shared DNA methylation patterns that are associated with early-life stress (ELS). In detail, we analyzed: (1) in humans the methylome signature characterizing CD34+ cells derived from the cord blood of newborns whose mothers have been characterized for stressful experiences during the last trimester of gestation;18 (2) in rhesus monkeys (Macaca mulatta) the DNA methylation patterns in the peripheral blood and in the prefrontal cortex (PFC) of monkeys that were exposed to different early-life social and rearing conditions;19, 20 (3) in rodents (Rattus norvegicus) we addressed the PFC of early-adult rats exposed to prenatal stress (PNS), a well-established model of vulnerability to altered adult behavior.18, 21, 22, 23, 24
This combined analysis pointed to Ankyrin-3 (Ank3), a genetic risk factor for bipolar disorder and schizophrenia 25, 26, 27 that plays a pivotal role as scaffolding protein in specific membrane domains, such as the node of Ranvier and the axon initial segment.28, 29, 30 Moreover, a role for ankyrin-G (ANKG) protein in regulating the post-synaptic compartment organization and function has recently emerged,31, 32 suggesting its potential for mediating the effects of stress exposure on dysfunctions associated with several psychiatric conditions.33, 34, 35, 36
Finally, we demonstrated, in healthy adult subjects, that variation in the Ank3 gene affecting gene expression interacted with an early stress factor (obstetric complications), in modulating prefrontostriatal connectivity and behavioral correlates of working memory, a cognitive phenotype tightly linked with different psychiatric disorders.37 Overall, our results suggest that alterations of Ank3 expression may contribute to the effects of perinatal stress on the vulnerability to psychiatric conditions.
Materials and Methods
Human cohort for MeDIP-chip analysis
Data were obtained from a cohort of mothers and their infants (n=180) recruited during the third trimester of pregnancy in the Rhine-Neckar Region of Germany. For inclusion and exclusion criteria see Supplementary Information. The study protocol was approved by the Ethics Committee of the Medical Faculty Mannheim of the University of Heidelberg. The study was conducted in accordance with the Declaration of Helsinki. All mothers provided written informed consent before participation. Investigator was blinded for the laboratory analyses.
Animals and experimental paradigms
Rhesus monkeys were reared as previously described.18, 19 Venous blood samples were obtained from 30 days old and 2-year-old monkeys, whole blood and buccal epithelial cell samples were taken at 2-years or older age, while PFCs were obtained from 7-year-old male monkeys. The Institutional Animal Care and Use Committee of the NICHD approved protocols for the use of experimental animals.
Pregnant rats were randomly assigned to control (Ctrl, n=7) or PNS (n=9) conditions. The stress paradigm was carried out as previously described.18, 38, 39 Male offspring PFC was dissected at different postnatal days (PND: 7 (Ctrl, n=7; PNS, n=9), 21 (Ctrl, n=6; PNS, n=6) and 62 (Ctrl, n=7; PNS, n=9)). Rat handling and experimental procedures were performed in accordance with the EC guidelines (EC Council Directive 86/609 1987) and with the Italian legislation on animal experimentation (D.L. 116/92), in accordance with the National Institute of Health Guide for the Care and Use of Laboratory Animals.
All efforts were made to minimize animal suffering and to reduce the number of animals used, which has been set, using G*Power 3 software,40 to take into account mortality and to allow n=7–9 in each final group, also based upon our own previous data.39 No pre-established inclusion/exclusion criteria were used for subsequent analyses. All samples were processed and analyzed by experimenters blind to rearing or prenatal stress conditions.
Extraction of DNA
Genomic DNA was extracted using Qiagen (Hilden, Germany) or Promega (Madison, WI, USA) systems, sheared by sonication and quantified using the Qubit system (Life Technologies, Burlington, ON, Canada).
Analysis of genome-wide promoter DNA methylation
MeDIP analysis was adapted from previously published protocols.18, 19 In detail, the final sample size for the methylome analysis was the following: rat PFC, Ctrl N=4, PNS N=4; human cohort, Ctrl N=8, ELS N=10; monkey PFC, Ctrl N=4, ELS N=4; monkey whole blood, Ctrl N=5, ELS N=5; monkey buccal samples, Ctrl N=3, ELS N=3; monkey CD3+ (30 days old), Ctrl N=10, ELS N=10; monkey CD3+ (2 years old), Ctrl N=6, ELS N=4 (the monkey CD3+ samples were pooled and subjected to 3 parallel MeDIP analyses, that is 3 Ctrl and 3 ELS pools per time point). Briefly, 2 μg of DNA were sonicated, and methylated DNA was immunoprecipitated using anti-5-methyl-cytosine (Cat. No. BI-MECY-0100, Eurogentec, Fremont, CA, USA). The DNA–antibody complex was immunoprecipitated with protein G, and the methylated DNA was re-suspended in digestion buffer (50 mm TrisHCl pH8; 10 mm EDTA; 0.5 % SDS) and treated with proteinase K overnight at 55 °C. The input and bound fractions were purified, amplified using the Whole Genome Amplification Kit (Sigma-Aldrich, St. Louis, MO, USA), and labeled for microarray hybridization with Cy3-dUTP and Cy5-dUTP, respectively, using the CGH Enzymatic Labeling Kit (Agilent Technologies, Mississauga, ON, Canada) in accordance with the manufacturer's instructions. Custom designed tiling arrays were used (Agilent Technologies). All steps of the hybridization, washing, scanning and feature extraction procedures were performed in accordance with the Agilent Technologies protocol for chip-on-chip analysis. Extracted microarray intensities were processed and analyzed using the R software environment for statistical computing (http://www.r-project.org/). The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus41 and are accessible through GEO series accession numbers GSE84028 (subseries accession numbers: GSE84018, GSE84020, GSE84021 and GSE84024).
Validation using qPCR
Gene-specific validation of MeDIP data was performed applying quantitative-real-time PCR (qPCR) (see Supplementary Table 2) using the 2−ΔΔCt method. Data are expressed as group means±s.e.m. To test for statistical significance, the Student's t-test was used (two-tailed), and the alpha level was set at 0.05.
Gene expression analysis in rats
RNA was isolated from rat PFC using AllPrep DNA/RNA Mini kit (Qiagen). RNA was analyzed by TaqMan qRT-PCR instrument (CFX384 real-time system, Bio-Rad Laboratories, Segrate, Italy) using the iScriptTM one-step RT-PCR kit for probes (Bio-Rad Laboratories) in triplicate as multiplexed reactions with a normalizing internal control (36b4).
Protein analysis
Protein subcellular fractionating was obtained homogenizing the tissues in a Teflon-glass potter in ice-cold 0.32 m sucrose buffer containing 1 mm HEPES, 1 mm MgCl2, 1 mm NaHCO3 and 0.1 mm phenylmethylsulfonyl fluoride, pH=7.4, in the presence of a complete set of protease (Roche, Monza, Italy) and phosphatase (Sigma-Aldrich) inhibitors. Total cell homogenate was next processed to obtain the crude membrane fraction (P2) and the Triton-Insoluble post-synaptic fraction (TIF) as described in detail in the Supplementary S1 Materials and Methods.
For western blot analysis, samples were run on SDS-polyacrylamide gel electrophoresis (PAGE) and analyzed using the Chemidoc MP Imaging System after normalization on β-actin levels.
Co-immunoprecipitation assays were performed as in Vastagh et al.42 with the introduction of some methodological modifications. Briefly, total cell homogenates from the rat PFC were immunoprecipitated with 3 μg of antibody overnight at 4 °C, followed by a 2 h incubation with protein A/G beads. Beads were then washed extensively and bound complexes were analyzed by SDS-PAGE and western blotting.
For all the above-mentioned molecular and biochemical analyses, all data met the assumptions of normal distribution and equality of variance.
Interaction between functional variation in Ank3 and obstetric complications on working memory processing
Association of rs9804190 with human post-mortem prefrontal Ank3 mRNA expression
Earlier results have indicated association of an intronic single-nucleotide polymorphism in the Ank3 gene, that is rs9804190, with gene expression levels in the superior temporal gyrus of patients with schizophrenia.28 We therefore tested such association in a large group of samples from PFC (BA46) of 268 non-psychiatric individuals using Braincloud (http://braincloud.jhmi.edu/) (Supplementary Information and Supplementary Table 3). In particular, analysis of covariance was performed, with rs9804190 genotype as the independent variable, Ank3 mRNA expression as the dependent variable, and non-matched variables between groups (that is, age, sex, RNA integrity number) and ethnicity as covariates of no interest. A statistical threshold of P<0.05 was used for this analysis.
In vivo fMRI and behavioral study
Three hundred six healthy adults (Supplementary Information and Supplementary Table 4) were enrolled in a behavioral study. One hundred seventy four of these individuals (supplementary Table 4) also participated in a functional magnetic resonance imaging (fMRI) study. In both studies, all individuals performed the 1- and 2-back versions of the N-back task, eliciting two loads of working memory (WM) processing and were genotyped for rs9804190 (Supplementary S1 Materials and Methods). Furthermore, mothers of all individuals completed the McNeil-Sjöström Scale,43 which allowed to split individuals based on the Obstetric Complications (OC) score (Supplementary S1 Materials and Methods).44 Sample sizes were: (a) in the fMRI sample, 57 CC with OC, 48 CC without OC, 38 T carriers with OC, 31 T carriers without OC; (b) in the behavioral study sample, 115 CC with OC, 66 CC without OC, 86 T carriers with OC, 39 T carriers without OC.
fMRI data acquisition and analysis
fMRI data were acquired with a 3T GE scanner and processed with SPM8 (see Supplementary S1 Materials and Methods). Second-level random effects multiple regression were performed to investigate Ank3 × OC interaction on prefrontal activity during performance of the 1- and 2-back WM tasks, using task load as the repeated-measures factor, and Ank3 rs9804190 genotype as well as OC (absence/presence) as the between-subjects factor. We used a statistical threshold of P<0.05, family-wise error small volume corrected for the left dorsolateral PFC (DLPFC), whose role is crucial in WM processing45 and the greater involvement of the left portion of DLPFC during WM tasks eliciting both verbal and visuospatial processing.46, 47 Then, we explored the interaction between ANK3 and OC on prefrontostriatal connectivity. With this aim, psychophysiological interaction analysis 48 was performed for each subject. In particular, we used a 5 mm region of interest centered on the peak activity (x=−48, y=38, z=30) in left DLPFC as seed region (Supplementary S1 Materials and Methods). Individual psychophysiological interaction contrasts were then entered in second-level random effects multiple regressions using task load as the repeated-measures factor, and Ank3 rs9804190 genotype as well as OC (absence/presence) as the between-subjects factors. We used a statistical threshold of P<0.05, family-wise error small-volume corrected within the left striatum (see Supplementary S1 Materials and Methods).
Analysis of behavioral data
A repeated measure factorial analysis of variance was performed, with genotype and OC presence/absence as the between-subjects factors, load (1- and 2-back) as the repeated-measures factor and behavioral accuracy (% of correct responses) or reaction time as the dependent variable. Tukey's test was used for post hoc analyses.
Results
Genome-wide DNA methylation analyses in different paradigms of ELS exposure
MeDIP-chip analysis at PND62 in the PFC of male rats uncovered 7660 probes associated with 3475 distinct genes whose methylation status was different between control (Ctrl) and animals exposed to PNS (false discovery rate (FDR)<0.2). In particular, 3362 probes, related to 1508 genes, were hypomethylated in PNS animals, whereas 4298 probes, corresponding to 1773 genes, were hypermethylated in PNS animals. Furthermore, the remaining 194 genes showed mixed methylation patterns. Using Ingenuity Pathway Analysis, we found enrichment for several neuronal functions, including ‘psychological disorders' (Supplementary Table 5), among other important biological mechanisms.
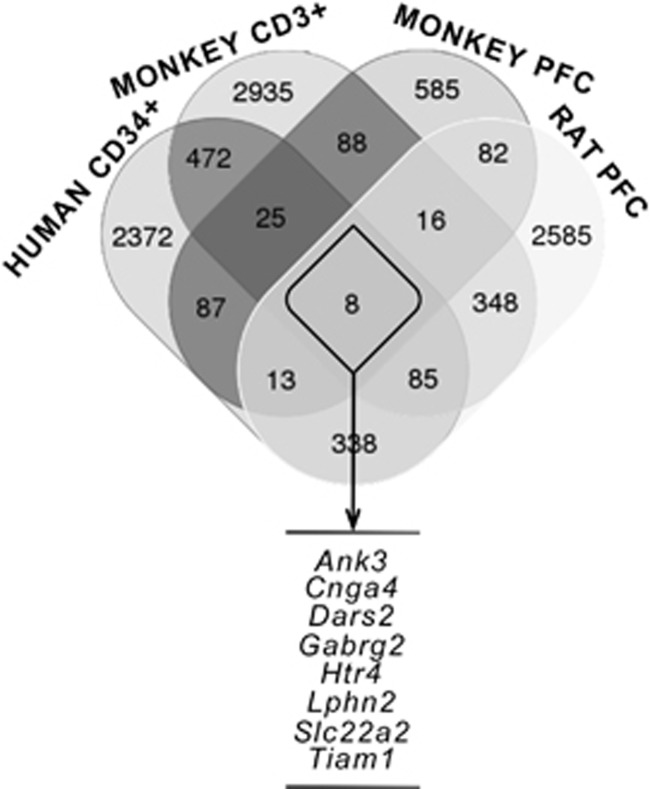
In order to prioritize the genes that may be important in keeping the memory of the adverse perinatal experience through the regulation of their methylation status, we performed a cross-species analysis overlapping the list of genes differentially methylated in the PFC of rats exposed to PNS with the results of MeDIP-chip genome-wide analyses in two different species, humans and non-human primates, which were also characterized by exposure to adversities during the perinatal life. In the human cohort, we analyzed the methylation status of genes in CD34+ hematopoietic stem cells collected from the umbilical cord of newborns from mothers with different levels of stress during gestation.18 In addition, we used two groups of monkeys characterized by different early-life social and rearing conditions, in which we performed genome-wide analyses in the PFC of 7-year-old animals and in the peripheral T cells (CD3+) at ages 30 days and 2 years, in order to look for stable peripheral DNA signatures in response to different early environmental conditions.49, 50 The analyses in monkeys, in particular, at a difference from our previously published work,18 fill a gap as the addition of the data from the monkey PFC further integrates the results obtained at brain level (in adult rats and monkeys) with data obtained at peripheral level (in monkeys and human newborns), providing more reliability and authenticity to our trans-species approach. We identified eight genes commonly affected—regardless the direction of the methylation changes—after exposure to ELS in all the conditions described above: Ankyrin-3 (Ank3), Cyclic nucleotide gated channel alpha 4 (Cnga4), Aspartyl-tRNA synthetase 2, mitochondrial (Dars2), GABA A receptor, gamma 2 (Gabrg2), 5-hydroxytryptamine receptor 4 (Htr4), Latrophilin 2 (Lphn2), Solute carrier family 22, member 2 (Slc22a2) and T-cell lymphoma invasion and metastasis 1 (Tiam1; Figure 1). Almost all these genes showed a mixed direction of the methylation changes, with the exception of Ank3 and Gabrg2, which showed a consistently higher methylation in the stressed group across all the conditions (Supplementary Table 6). Ank3 has previously emerged as a common genetic risk factor for neurodevelopmental and psychiatric disorders, such as bipolar disorder26, 27 and schizophrenia,28 whereas a strong dysfunction of the GABAergic system has also been demonstrated in anxiety and depression.51, 52 We focused our subsequent analyses on Ank3 since its methylation status was affected in the same direction also in the whole blood and buccal cells from 2-year-old monkeys exposed to ELS, whereas Gabrg2 did not (Supplementary Table 7). The extent of the methylation differences pertaining all the Ank3 probes hypermethylated in the stressed groups is listed in Supplementary Table 8 using similar significance (P-values) and effect size estimates (as differential expressed in log2) as previously reported.18, 19
Figure 1.
Venn diagram showing the overlap of the number of differentially methylated genes regulated by exposure to early-life stress in different species (see Materials and Methods for details). The eight genes listed below emerged as differentially methylated after early-life stress in all conditions. PFC, prefrontal cortex.
Validation of MeDIP-chip
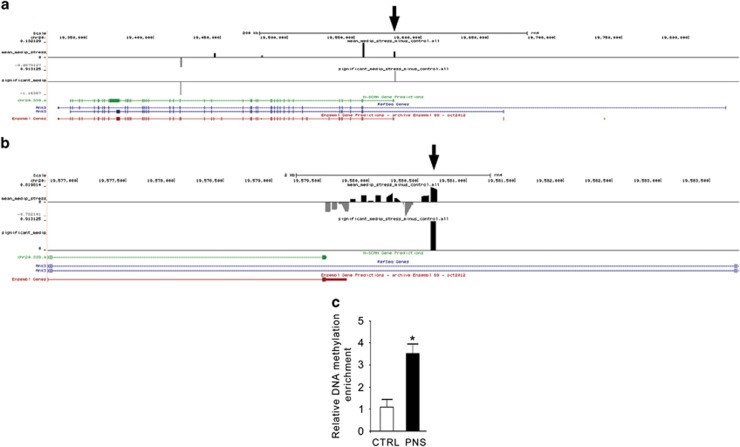
We next characterized in more detail the modulation of Ank3 in the rat model of PNS. Three genomic locations showed a different methylation status between Ctrl and PNS rats in the PFC at PND62 (Figure 2a and Supplementary Table 6). In detail, the probes located at chr20:19420344-19420403 and at chr20:19420705-19420764 were hypomethylated in PNS rats, whereas the probe located at chr20:19580638-19580697 was hypermethylated in PNS rats. qPCR analysis was used to validate the array data indicated by the arrow in Figures 2a and b, and confirmed the result obtained with MeDIP-chip (Figure 2c). We focused on this probe, as it was the only showing an increased methylation status in the stressed group, according with all the other conditions (Supplementary Tables 6 and 7).
Figure 2.
Expanded views from the UCSC genome browser at the rat Ank3 gene location are depicted. (a) The first track shows average methylation probe fold differences (Log2), whereas the second track shows the regions whose methylation status is significantly different as a consequence of PNS. The three tracks at the bottom show exons and introns boundaries taken from the rat N-SCAN Gene Predictions, NCBI Reference Sequence Database (RefSeq) and Ensembl Gene Predictions, respectively. Arrow indicates the location of DNA amplification for qPCR validation. (b) Zoomed view of a showing every single probe in the region selected for the validation in the rat prenatal stress model. (c) Bar graph of the qPCR validation of Ank3 relative DNA methylation enrichment between PNS and Ctrl groups, shown as relative bound fraction concentrations. The data represent the mean±s.e.m. of 3–4 independent determinations. *P<0.05 vs Ctrl (Student's t-test). Ctrl, control; PNS, prenatal stress.
Postnatal developmental expression of Ankyrin-3 in the PFC of male rats
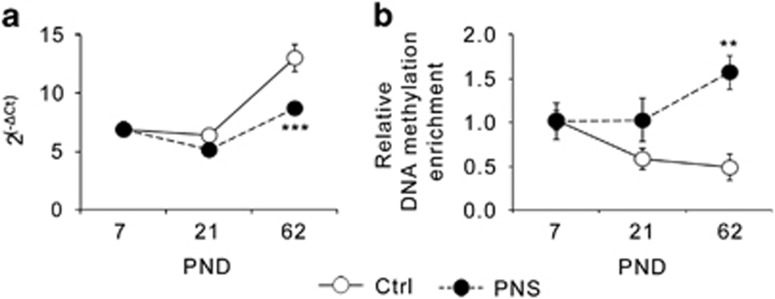
Using qRT-PCR, we next examined if the altered methylation status of Ank3 was associated with changes in its transcriptional regulation during the postnatal development until early adulthood. We found a significant effect of PNS exposure (F1,41=14.795, P<0.001), of AGE (F2,41=39.885, P<0.001), and a significant PNS × AGE interaction (F2,41=7.303, P<0.01). Indeed, as depicted in Figure 3a, Ctrl rats showed a relative stable expression of Ank3 from PND7 to PND21, followed by a steady increase at PND62, whereas PNS rats showed statistically significant lower Ank3 mRNA levels at PND62 (P<0.001), but not earlier (Figure 3a). Next, in order to establish a correlation between these changes and the epigenetic modifications at the Ank3 gene, we investigated DNA methylation at the same gene location found to be affected using MeDIP-chip and validated with qPCR at PND62 in PNS rats. As depicted in Figure 3b, Ctrl rats showed a progressive decrease in the methylation levels of Ank3 from PND7 to PND62, whereas PNS rats showed a stable methylation between PND7 and PND21, with a steady increase at early adulthood. Statistical analysis revealed a significant effect of PNS (F1,29=10.315, P<0.01) and a significant PNS × AGE interaction (F2,29=3.897, P<0.05), supporting the fact that PNS and Ctrl rats showed comparable methylation levels of Ank3 at PND7 that progressively diverge, leading to a statistical difference at PND62 (P<0.01).
Figure 3.
Exposure to prenatal stress in rats alters the DNA methylation status and the expression levels of Ank3 with a specific time-profile. (a) mRNA expression levels of Ankyrin-3 during the postnatal development in the prefrontal cortex of PNS male rats as compared with control animals (Ctrl). The data, expressed as fold change (where ΔCt is the difference between the threshold cycle of the target gene and the housekeeping gene) are the mean±s.e.m. of 6–9 independent determinations. (b) Relative DNA methylation enrichment of Ankyrin-3 during the postnatal development in the prefrontal cortex of PNS male rats as compared with Ctrl. The data, expressed as relative bound fraction concentration, are the mean±s.e.m. of 3–7 independent determinations. **P<0.01 and ***P<0.001 vs Ctrl at the same postnatal age (2-way analysis of variance (ANOVA) followed by Fisher's LSD post hoc comparison). PND, postnatal day; PNS, prenatal stress.
Analysis of ANKG protein modulation following PNS exposure in the PFC of male rats at PND62
Ankyrin-3 encodes for a protein, ANKG, which exists in different isoforms that are expressed in nearly all tissues, although with some peculiarities. It has been recently demonstrated, for example, that the 190 kDa isoform of ANKG localizes at post-synaptic level, where it interacts with both PSD95 and GLUR1.32 Since it has been amply demonstrated that neuropsychiatric disorders can be characterized by synaptic dysfunction,33, 34 we decided to investigate whether the long-term changes in Ank3 expression observed in PNS animals may also involve alterations in the synaptic compartment.
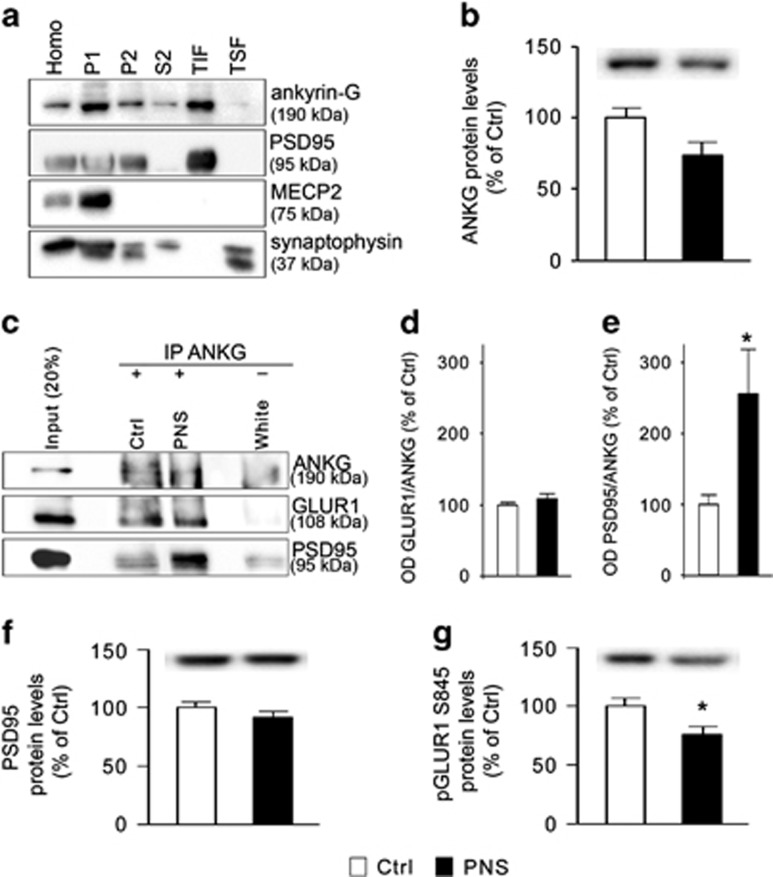
Using western blot analysis, we first established the expression of ANKG 190 kDa in different subcellular fractions from the rat brain. As shown in Figure 4a, ANKG 190 kDa is enriched in the crude membrane fraction (P2) and in the TIF, which represents the post-synaptic compartment, as also occurred for the post-synaptic density marker PSD95, but not for MECP2, a marker of the nuclear compartment, or synaptophysin, a presynaptic marker (Figure 4a). Moreover, we found that ANKG 190 kDa levels were reduced by PNS in the P2 (−26% vs Ctrl, P=0.057, Figure 4b).
Figure 4.
ANKG interaction with PSD95 and GLUR1: modulation following prenatal stress exposure in rats. (a) Western blot analysis of ANKG enrichment in different subcellular fractions, as compared with PSD95, MECP2 and synaptophysin. (b) Bar graph of ANKG protein levels in the crude membrane fraction from the prefrontal cortex of adult Ctrl and PNS male rats. (c) Western blot analysis of co-immunoprecipitation experiments of ANKG with GLUR1 or PSD95 from the prefrontal cortex of male rats at PND62. (d and e) Bar graph of GLUR1/ANKG (d) and PSD95/ANKG (e) interaction in the prefrontal cortex of Ctrl and PNS male rats at PND62. (f, g). Bar graphs of PSD95 (f) and pGLUR1 S845 (g) protein levels in the crude membrane fraction from the prefrontal cortex of adult Ctrl and PNS male rats. Data in bar graphs are presented as mean±s.e.m. of 7–8 independent determinations. *P<0.05 vs Ctrl (Student's t-test). ANKG, ankyrin-G; Homo, whole homogenate; P1, nuclear fraction; P2, crude membrane fraction; PNS, prenatal stress; S2, cytosolic fraction; TIF, triton insoluble fraction; TSF, triton soluble fraction.
Seen ANKG 190 kDa enrichment in the post-synaptic compartment (Figure 4a) and its trend toward a decrease in the P2 fraction (Figure 4b), we investigated, by co-immunoprecipitation assay, whether ANKG 190 kDa interacts with key molecules localized at the post-synaptic site and if PNS exposure may alter these interactions. As shown in Figure 4c, ANKG 190 kDa interacted with PSD95 as well as with GLUR1. Interestingly, we found that while GLUR1/ANKG interaction was not affected by PNS (+9% vs Ctrl, P>0.05, Figure 4d), PSD95/ANKG interaction was significantly increased in the PFC of adult rats that were exposed to PNS (+155% vs Ctrl, P<0.05, Figure 4e), possibly leading to alterations in the functional integrity of the post-synaptic compartment. Indeed, although we did not detect changes of PSD95 protein levels in the P2 (−13% vs Ctrl, P>0.05, Figure 4f), we found a significant decrease of the phosphorylated (pS845) form of the obligatory AMPA GLUR1 subunit (−24% vs Ctrl, P<0.05, Figure 4g), a proxy for receptor activation.53
Interaction between the Ank3 single-nucleotide polymorphism rs9804190 and OC on prefrontostriatal functional connectivity and behavior during working memory
Analysis of human post-mortem data, using the Braincloud data set,54 indicated that the Ank3 single-nucleotide polymorphism rs9804190 was associated with Ank3 mRNA expression in the DLPFC (F2, 261=6.66; P=0.001), with greater expression in the TT genotype as compared with CT (P<0.0001) and CC (P<0.0001).
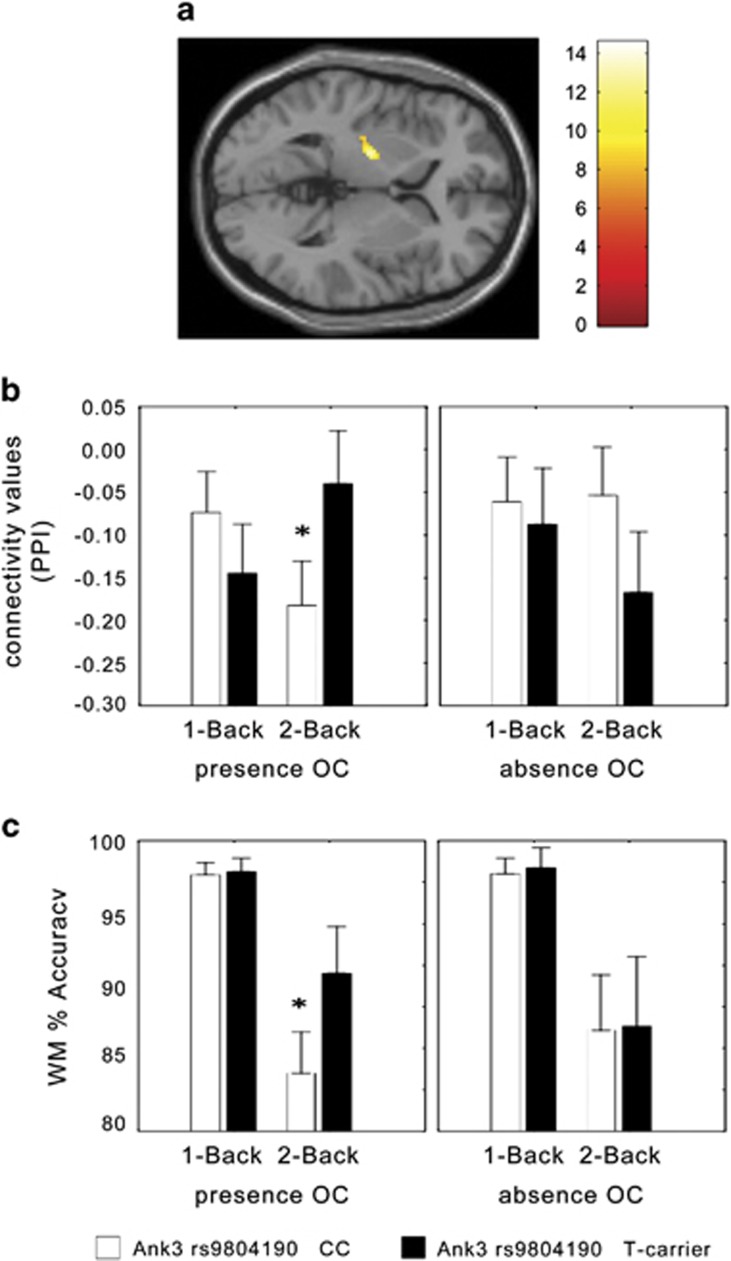
We next investigated the influence of rs9804190 on brain function and the possible modulation following exposure to early-life adversities, in the form of OC. fMRI results did not indicate significant main effects of OC, of rs9804190, or their interaction on brain activity. There was a rs9804190 genotype by WM load interaction in bilateral DLPFC (Right BA9, x, y, z=28, 40, 36; K=68; Z score=3.28; uncorrected P=0.001. Left BA9/10, x, y, z=−40, 46, 20; K=60; Z score=3.31; uncorrected P=0.001). Consistent with previous studies,28, 55 CC subjects exhibited greater recruitment of bilateral PFC during 2-back compared with T carriers. On the other hand, psychophysiological interaction analysis revealed a genotype × OC × WM load interaction on functional connectivity between left DLPFC and left striatum (x,y,z=−26, −16, 9; K=111; Z score=3.59; family-wise error corrected P=0.037). Post hoc analysis on values of connectivity extracted from the significant striatal cluster revealed, in presence of OC, a greater connectivity strength in T carriers as compared with CC individuals during 2-Back (P<0.007; Figures 5a and b). No other statistically significant difference was found in between genotype group comparisons within each load (Tukey's HSD post hoc test, all P>0.1).
Figure 5.
Healthy humans with a history of obstetric complications combined with functional variation in Ank3 gene, show altered prefrontostriatal connectivity and working memory performance. (a) Section of the brain depicting the left striatal cluster whose functional connection with the left DLPFC is associated with an Ank3 rs9804190 × OC interaction during WM processing. (b) Graph with functional connectivity values (arbitrary units) extracted from the cluster depicted in a. T carrier healthy subjects had greater connectivity strength compared with CC healthy individuals in the presence of OC. See text for statistics. (c) Graph showing an Ank3 rs9804190 × OC interaction on behavioral accuracy (% correct responses) during WM in healthy subjects. T carriers were associated with greater accuracy compared with subjects with the CC genotype (see text for statistics). DLPFC, dorsolateral prefrontal cortex; OC, Obstetric Complications; WM, working memory.
Analysis of WM behavioral data indicated a main effect of genotype reaching significance (F1.287=3.2; P=0.07), with greater mean values of percent correct responses in T carriers compared with CC individuals. Furthermore, there was a trend for an interaction between genotype and load (F1.287=3.5; P=0.06], with T carrier subjects having greater mean accuracy than CC at 2-Back. No significant main effect of OC or of its interaction with WM load or rs9804190 was found (all P>0.1). Notably, and similarly to the psychophysiological interaction analysis of fMRI data, there was a genotype × OC × WM load interaction on percent correct responses (F1,287=3.88; P=0.049). More specifically, post hoc analysis demonstrated greater accuracy in T carriers compared with CC subjects at 2-Back in presence of OC (Tukey's HSD post hoc test P=0.0005). No other statistically significant difference was found between genotypic group comparisons within each load (Tukey's test, all P>0.9; Figure 5c). No statistically significant main effects or interaction was present on reaction time data (all P>0.05).
Discussion
In the present study, using a ‘converging evolutionary' approach, we identified an association between the exposure to ELS and the methylation status of Ank3 gene, which may support its role in the vulnerability to psychiatric disorders. Ank3 methylation, mRNA and protein levels were indeed significantly altered in rats exposed to PNS with a specific temporal profile.33 Furthermore, in humans we found an interaction between a polymorphism affecting Ank3 expression and OC, modulating prefrontostriatal functional connectivity and behavior during WM, whose deficits are manifested in several psychiatric conditions.37
First, in line with previous studies showing the impact of ELS on the epigenome,49, 50, 56 we found that PNS in rats persistently affected a large set of genes involved in specific biological functions. Subsequently, our cross-species, cross-tissues and longitudinal approach pointed to the identification of eight genes whose methylation status was affected in all the investigated cohorts, which may be relevant for the long-lasting nature of the effects brought about by ELS. Although the heterogeneity of all the conditions in this study has to be taken into account (tissues, species and timing), we strongly believe that this assortment represents the strength of our experimental procedure, seeing that we, and others, already demonstrated the high validity of these convergent approaches.18, 22, 23, 24
We found that some of the overlapping genes have not yet been previously associated with stress or psychopathology (Cnga4, Dars2, Lphn2 and Tiam1). Instead, other groups have previously demonstrated an involvement of Slc22a2 or Htr4 in the efficacy of antidepressant medication,57, 58 whereas Gabrg2 holds high relevance seen the involvement of GABAergic transmission in several psychiatric disorders.51, 59
Seen the concordance of the methylation changes across all the conditions and species investigated in the present study, we decided to focus our attention on Ank3, which is among the most consistently replicated and statistically significant genetic risk factor for neuropsychiatric diseases.25, 26, 27, 28, 30
In the adult PFC from PNS rats, we detected an increase in DNA methylation in the first intron of Ank3, although we also found two differentially methylated regions downstream, within intron 24, which were more methylated in Ctrl rats. Interestingly, this same location was also found to be hypermethylated in the hippocampus of PNS male rats compared with Ctrl (chr20.19580650-19580709, FDR=0.006), an effect that we validated also at mRNA level (Ctrl: 100±8 and PNS: 77±7, P<0.05), as in the PFC. Although hypermethylation within a promoter is frequently correlated with transcriptional silencing,60, 61, 62 it has been also shown that hypermethylated promoters could be activated in a transient way through chromatin remodeling, without demethylation63 and, on the contrary, that the promoter hypomethylation does not necessarily correlate with active transcription, as other epigenetic mechanisms could act in concert to repress gene expression.64 Moreover, although DNA methylation of CpG islands associated to promoter regions has been amply investigated, several studies, including our own, have shown that also intragenic and intronic regions are widely modulated after environmental stressors through methylation, thus affecting transcriptional regulation using different mechanisms, such as alternative splicing rather than alternative promoters.62, 65 Indeed, we showed that PNS rats have decreased total mRNA levels of Ank3 at PND62 as a function of different methylation levels at this genomic location. Several studies, at least in mice, have shown the complexity of Ank3 gene organization and the existence of different mRNA isoforms with a specific tissue and cellular localization and function,66 and based on NCBI Genbank, rats seems to share a similar transcriptional organization to mice and humans. However, in this work, using qRT-PCR, we evaluated the total mRNA levels of Ank3, as the differentially methylated region is within a locus included in all Ank3 rat mRNA transcript variants. We found that Ank3 mRNA levels are relatively stable before weaning, with a steep increase thereafter to reach adult expression, suggesting a role for Ank3 across the postnatal neurodevelopment and in the impending maturational steps, as already hypothesized.67, 68, 69 Conversely, the shift between weaning and adulthood is affected in PNS rats, leading to a significant reduction of Ank3 expression at adulthood. Interestingly, we have recently shown that the expression of Bdnf, a neurotrophin involved in psychiatric disorders, is significantly downregulated in PNS rats only after adolescence,39 thus supporting the idea that exposure to ELS may influence life-long genome adaptation thus resulting in behavioral disorders, such as bipolar disorder, depression and schizophrenia, that frequently become manifest during adolescence.13, 70, 71, 72
Indeed, ELS can affect the epigenome in a persistent manner.18, 73, 74, 75 However, it is not clear whether DNA methylation changes represent a direct consequence of the early-life experiences or a result of the psychopathological phenotype associated with perinatal adversities. We demonstrated that PNS animals not only failed to show a progressive decrease in the DNA methylation levels of Ank3 as occurring in Ctrl, but they displayed a gradual increase after weaning. It may be inferred that two concomitant events may lead to a reduction of Ank3 expression from weaning to adulthood: a failure of the mechanisms that reduce gene methylation (as occurring in Ctrl rats), together with an activation of de novo methylation as long-term consequence of stress exposure. Similar to these findings, Weaver and colleagues have previously demonstrated that maternal behavior alters the methylation status of the glucocorticoid receptor during the first week of life, thus affecting its expression in the hippocampus until adulthood.76
We also showed that exposure to PNS led to functional alterations in the protein encoded by Ank3 gene, namely ANKG. We found that ANKG is enriched in the membrane compartment (P2 and TIF), as previously demonstrated using primary cortical neurons,32 and that PNS may affect its subcellular distribution reducing its protein levels selectively in this fraction (compartment). Furthermore, we showed that the interaction of ANKG with PSD95 was significantly increased in PNS animals, which may alter the functional integrity of the post-synaptic domain. As an example, Yaka et al.77 demonstrated that a mild form of PNS determined an increase of synaptic PSD95 levels in the hippocampus in juvenile rats, thus altering glutamate receptor clustering and leading to impairments in LTP and memory tasks. Even if we failed to detect changes of PSD95 protein levels in PNS animals, the abnormal interaction of PSD95 with ANKG 190 kDa could contribute to a disrupted AMPAR clustering and function, as also demonstrated by the reduction of the phosphorylation levels of GLUA1 S845 in PNS rats, a proxy for receptor activation.53 As the glutamatergic synapse represents a key player in the pathogenesis and development of neuropsychiatric disorders,33, 34 our data suggest that ANKG may mediate some of the detrimental effects of PNS exposure on the glutamatergic architecture.
Furthermore, we provided evidence for a functional outcome of reduced Ank3 expression in humans. Indeed, by investigating an Ank3 single-nucleotide polymorphism, rs9804190, which has previously been associated with heightened risk for different psychiatric disorders,27, 28, 78, 79, 80 we found that the C allele predicts lower post-mortem prefrontal Ank3 expression in non-psychiatric subjects. Interestingly, CC subjects with OC had lower prefrontostriatal functional connectivity strength and lower behavioral accuracy at the higher WM demand, as compared with T carriers with OC. These results suggest that a genetic context leading to lower Ank3 expression interacts with the presence of OC, a relevant factor of early stress, in determining sub-optimal patterns of brain functional connections and behavior during WM. Some limitations of our study need to be considered. First, the relatively small sample size in the MeDIP studies, which is due to technical issues. Second, for the animal cohorts (both rats and monkeys) only males have been used, which could limit the conclusions to a sex-specific effect. Seen the existence of sex bias in the prevalence and severity of many neurodevelopmental and psychiatric disorders, and evidence suggesting that environment has a sex-dependent effect on DNA methylation itself,81, 82 future studies are needed to investigate whether such alterations may characterize also the female counterpart. It is also important to mention that following ELS the methylation differences across species occur in different regions, whose functional impact on gene function remains to be established. Last, although we limited the analysis to the PFC for performing cross-species investigation, we cannot rule out the possibility that methylation changes in other key brain regions, such as the hippocampus and the basal ganglia, may mediate the long-term effect of ELS on the susceptibility to psychopathology.
The results in humans at the imaging and behavioral level, together with those obtained by our methylation analysis, suggest that Ank3 may represent an important link between ELS and the development of psychopathology, through alterations in neuronal circuits relevant for psychiatric diseases. Moreover, given the possibility that Ank3 expression can be modulated by pharmacological intervention 31 and that the behavioral phenotype induced by Ank3 suppression in the mouse brain may be reverted by chronic lithium administration,83 we believe that Ank3 may represent a novel molecular marker as well as a target for drug intervention in stress-related disorders with a neurodevelopmental origin.
Acknowledgments
This work was supported by an ERANET Neuron grant to MD, MS, MAR and PG, as well as by grants from the Italian Ministry of University and Research to MAR (PRIN – grant number 20107MSMA4_002) and from Fondazione CARIPLO (grant number 2012-0503) to MAR, FC and A Bertolino. In addition, characterization of the human cohort was supported by a grant of the Dietmar Hopp Foundation. VN received support from the Olympia-Morata-Programme of the University of Heidelberg and the Boehringer-Ingelheim Fonds. AL was supported by a Fondazione Umberto Veronesi Fellowship. ZN was supported by Marie Curie International Outgoing Fellowship within the 7th European Community Framework Programme. Work in MS's laboratory was supported by Canadian Institute of Health Research Grant MOP-42411. We are grateful to Elisa Zianni and Dr Jennifer Stanic for the help with co-immunoprecipitation.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
MAR reports having received compensation as speaker/consultant from Lundbeck, Otzuka, Sumitomo Dainippon Pharma, Sunovion and Takeda. A Bertolino disclosed consulting fees from Hoffman-La Roche. The remaining authors declare no conflict of interest.
Supplementary Material
References
- Barker DJ. Intrauterine programming of adult disease. Mol Med Today 1995; 1: 418–423. [DOI] [PubMed] [Google Scholar]
- Weinstock M. The long-term behavioural consequences of prenatal stress. Neurosci Biobehav Rev 2008; 32: 1073–1086. [DOI] [PubMed] [Google Scholar]
- Lupien SJ, McEwen BS, Gunnar MR, Heim C. Effects of stress throughout the lifespan on the brain, behaviour and cognition. Nat Rev Neurosci 2009; 10: 434–445. [DOI] [PubMed] [Google Scholar]
- Kolb B, Mychasiuk R, Muhammad A, Li Y, Frost DO, Gibb R. Experience and the developing prefrontal cortex. Proc Natl Acad Sci USA 2012; 109(Suppl 2): 17186–17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Binder EB. The neurobiological effects of stress as contributors to psychiatric disorders: focus on epigenetics. Curr Opin Neurobiol 2015; 30: 31–37. [DOI] [PubMed] [Google Scholar]
- Gudsnuk K, Champagne FA. Epigenetic influence of stress and the social environment. ILAR J 2012; 53: 279–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TY, Meaney MJ. Epigenetics and the environmental regulation of the genome and its function. Ann Rev Psychol 2010; 61: c431–c433. [DOI] [PubMed] [Google Scholar]
- Klengel T, Pape J, Binder EB, Mehta D. The role of DNA methylation in stress-related psychiatric disorders. Neuropharmacology 2014; 80: 115–132. [DOI] [PubMed] [Google Scholar]
- Szyf M. The genome- and system-wide response of DNA methylation to early life adversity and its implication on mental health. Can J Psychiatry 2013; 58: 697–704. [DOI] [PubMed] [Google Scholar]
- Babenko O, Kovalchuk I, Metz GA. Stress-induced perinatal and transgenerational epigenetic programming of brain development and mental health. Neurosci Biobehav Rev 2015; 48: 70–91. [DOI] [PubMed] [Google Scholar]
- Maccari S, Krugers HJ, Morley-Fletcher S, Szyf M, Brunton PJ. The consequences of early-life adversity: neurobiological, behavioural and epigenetic adaptations. J Neuroendocrinol 2014; 26: 707–723. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Maternal care as a model for experience-dependent chromatin plasticity? Trends Neurosci 2005; 28: 456–463. [DOI] [PubMed] [Google Scholar]
- Provencal N, Binder EB. The effects of early life stress on the epigenome: From the womb to adulthood and even before. Exp Neurol 2015; 268: 10–20. [DOI] [PubMed] [Google Scholar]
- Worlein JM. Nonhuman primate models of depression: effects of early experience and stress. ILAR J 2014; 55: 259–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi M, Horii-Hayashi N, Sasagawa T. Effects of early life adverse experiences on the brain: implications from maternal separation models in rodents. Front Neurosci 2014; 8: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidotti A, Dong E, Tueting P, Grayson DR. Modeling the molecular epigenetic profile of psychosis in prenatally stressed mice. Prog Mol Biol Transl Sci 2014; 128: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnaudery M, Maccari S. Epigenetic programming of the stress response in male and female rats by prenatal restraint stress. Brain Res Rev 2008; 57: 571–585. [DOI] [PubMed] [Google Scholar]
- Nieratschker V, Massart R, Gilles M, Luoni A, Suderman MJ, Krumm B et al. MORC1 exhibits cross-species differential methylation in association with early life stress as well as genome-wide association with MDD. Transl Psychiatry 2014; 4: e429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provencal N, Suderman MJ, Guillemin C, Massart R, Ruggiero A, Wang D et al. The signature of maternal rearing in the methylome in rhesus macaque prefrontal cortex and T cells. J Neurosci 2012; 32: 15626–15642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman IC, Rosenblum LA. Effects of separation from mother on the emotional behavior of infant monkeys. Ann N Y Acad Sci 1969; 159: 681–695. [DOI] [PubMed] [Google Scholar]
- Bock J, Wainstock T, Braun K, Segal M. Stress in utero: prenatal programming of brain plasticity and cognition. Biol Psychiatry 2015; 6: 00162–00166. [DOI] [PubMed] [Google Scholar]
- Bertsch B, Ogden CA, Sidhu K, Le-Niculescu H, Kuczenski R, Niculescu AB. Convergent functional genomics: a Bayesian candidate gene identification approach for complex disorders. Methods 2005; 37: 274–279. [DOI] [PubMed] [Google Scholar]
- Le-Niculescu H, Balaraman Y, Patel SD, Ayalew M, Gupta J, Kuczenski R et al. Convergent functional genomics of anxiety disorders: translational identification of genes, biomarkers, pathways and mechanisms. Transl Psychiatry 2011; 1: e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M, Brandwein C, Luoni A, Sandrini P, Calzoni T, Deuschle M et al. Morc1 knockout evokes a depression-like phenotype in mice. Behav Brain Res 2015; 296: 7–14. [DOI] [PubMed] [Google Scholar]
- Iqbal Z, Vandeweyer G, van der Voet M, Waryah AM, Zahoor MY, Besseling JA et al. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum Mol Genet 2013; 22: 1960–1970. [DOI] [PubMed] [Google Scholar]
- Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet 2008; 40: 1056–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze TG, Detera-Wadleigh SD, Akula N, Gupta A, Kassem L, Steele J et al. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol Psychiatry 2009; 14: 487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos P, Katsel P, Davis KL, Bitsios P, Giakoumaki SG, Jogia J et al. Molecular and genetic evidence for abnormalities in the nodes of Ranvier in schizophrenia. Arch Gen Psychiatry 2012; 69: 7–15. [DOI] [PubMed] [Google Scholar]
- Komada M, Soriano P. [Beta]IV-spectrin regulates sodium channel clustering through ankyrin-G at axon initial segments and nodes of Ranvier. J Cell Biol 2002; 156: 337–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar PR S, Scott, L J, Andreassen, O A, Cichon S, Craddock N et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet 2011; 43: 977–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanavati D, Austin DR, Catapano LA, Luckenbaugh DA, Dosemeci A, Manji HK et al. The effects of chronic treatment with mood stabilizers on the rat hippocampal post-synaptic density proteome. J Neurochem 2011; 119: 617–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KR, Kopeikina KJ, Fawcett-Patel JM, Leaderbrand K, Gao R, Schurmann B et al. Psychiatric risk factor ANK3/ankyrin-G nanodomains regulate the structure and function of glutamatergic synapses. Neuron 2014; 84: 399–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bartolomeis A, Latte G, Tomasetti C, Iasevoli F. Glutamatergic postsynaptic density protein dysfunctions in synaptic plasticity and dendritic spines morphology: relevance to schizophrenia and other behavioral disorders pathophysiology, and implications for novel therapeutic approaches. Mol Neurobiol 2014; 49: 484–511. [DOI] [PubMed] [Google Scholar]
- Duman RS, Aghajanian GK. Synaptic dysfunction in depression: potential therapeutic targets. Science 2012; 338: 68–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Focking M, Lopez LM, English JA, Dicker P, Wolff A, Brindley E et al. Proteomic and genomic evidence implicates the postsynaptic density in schizophrenia. Mol Psychiatry 2015; 20: 424–432. [DOI] [PubMed] [Google Scholar]
- Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM. Dendritic spine pathology in neuropsychiatric disorders. Nat Neurosci 2011; 14: 285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertolino A, Blasi G. The genetics of schizophrenia. Neuroscience 2009; 164: 288–299. [DOI] [PubMed] [Google Scholar]
- Maccari S, Piazza PV, Kabbaj M, Barbazanges A, Simon H, Le Moal M. Adoption reverses the long-term impairment in glucocorticoid feedback induced by prenatal stress. J Neurosci 1995; 15(1 Pt 1): 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luoni A, Berry A, Calabrese F, Capoccia S, Bellisario V, Gass P et al. Delayed BDNF alterations in the prefrontal cortex of rats exposed to prenatal stress: preventive effect of lurasidone treatment during adolescence. Eur Neuropsychopharmacol 2014; 24: 986–995. [DOI] [PubMed] [Google Scholar]
- Faul F, Erdfelder E, Lang AG, Buchner A. G*Power 3: a flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007; 39: 175–191. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastagh C, Gardoni F, Bagetta V, Stanic J, Zianni E, Giampa C et al. N-methyl-D-aspartate (NMDA) receptor composition modulates dendritic spine morphology in striatal medium spiny neurons. J Biol Chem 2012; 287: 18103–18114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeil TF, Cantor-Graae E, Sjostrom K. Obstetric complications as antecedents of schizophrenia: empirical effects of using different obstetric complication scales. J Psychiatr Res 1994; 28: 519–530. [DOI] [PubMed] [Google Scholar]
- Verdoux H, Sutter AL. Perinatal risk factors for schizophrenia: diagnostic specificity and relationships with maternal psychopathology. Am J Med Genet 2002; 114: 898–905. [DOI] [PubMed] [Google Scholar]
- Callicott JH, Mattay VS, Bertolino A, Finn K, Coppola R, Frank JA et al. Physiological characteristics of capacity constraints in working memory as revealed by functional MRI. Cereb Cortex 1999; 9: 20–26. [DOI] [PubMed] [Google Scholar]
- Rottschy C, Langner R, Dogan I, Reetz K, Laird AR, Schulz JB et al. Modelling neural correlates of working memory: a coordinate-based meta-analysis. Neuroimage 2012; 60: 830–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager TD, Smith EE. Neuroimaging studies of working memory: a meta-analysis. Cogn Affect Behav Neurosci 2003; 3: 255–274. [DOI] [PubMed] [Google Scholar]
- Friston KJ, Buechel C, Fink GR, Morris J, Rolls E, Dolan RJ. Psychophysiological and modulatory interactions in neuroimaging. Neuroimage 1997; 6: 218–229. [DOI] [PubMed] [Google Scholar]
- Massart R, Suderman M, Provencal N, Yi C, Bennett AJ, Suomi S et al. Hydroxymethylation and DNA methylation profiles in the prefrontal cortex of the non-human primate rhesus macaque and the impact of maternal deprivation on hydroxymethylation. Neuroscience 2014; 268: 139–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoda Z, Massart R, Suderman M, Hallett M, Li T, Coote M et al. Maternal depression is associated with DNA methylation changes in cord blood T lymphocytes and adult hippocampi. Transl Psychiatry 2015; 5: e545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Shen Q, Sahir N. The GABAergic deficit hypothesis of major depressive disorder. Molecular psychiatry 2011; 16: 383–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KS, Rudolph U. Anxiety and depression: mouse genetics and pharmacological approaches to the role of GABA(A) receptor subtypes. Neuropharmacology 2012; 62: 54–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JQ, Arora A, Yang L, Parelkar NK, Zhang G, Liu X et al. Phosphorylation of AMPA receptors: mechanisms and synaptic plasticity. Molecular neurobiology 2005; 32: 237–249. [DOI] [PubMed] [Google Scholar]
- Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature 2011; 478: 519–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delvecchio G, Dima D, Frangou S. The effect of ANK3 bipolar-risk polymorphisms on the working memory circuitry differs between loci and according to risk-status for bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 2015; 168b: 188–196. [DOI] [PubMed] [Google Scholar]
- Cao-Lei L, Massart R, Suderman MJ, Machnes Z, Elgbeili G, Laplante DP et al. DNA methylation signatures triggered by prenatal maternal stress exposure to a natural disaster: Project Ice Storm. PLoS One 2014; 9: e107653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacq A, Balasse L, Biala G, Guiard B, Gardier AM, Schinkel A et al. Organic cation transporter 2 controls brain norepinephrine and serotonin clearance and antidepressant response. Mol Psychiatry 2012; 17: 926–939. [DOI] [PubMed] [Google Scholar]
- Lucas G, Rymar VV, Du J, Mnie-Filali O, Bisgaard C, Manta S et al. Serotonin(4) (5-HT(4)) receptor agonists are putative antidepressants with a rapid onset of action. Neuron 2007; 55: 712–725. [DOI] [PubMed] [Google Scholar]
- Schmidt MJ, Mirnics K. Neurodevelopment, GABA system dysfunction, and schizophrenia. Neuropsychopharmacology 2015; 40: 190–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R, Razin A, Cedar H. In vitro methylation of the hamster adenine phosphoribosyltransferase gene inhibits its expression in mouse L cells. Proc Natl Acad Sci USA 1982; 79: 3418–3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP. CpG islands—'a rough guide'. FEBS Lett 2009; 583: 1713–1720. [DOI] [PubMed] [Google Scholar]
- Kulis M, Queiros AC, Beekman R, Martin-Subero JI, Intragenic DNA. methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta 2013; 1829: 1161–1174. [DOI] [PubMed] [Google Scholar]
- Raynal NJ, Si J, Taby RF, Gharibyan V, Ahmed S, Jelinek J et al. DNA methylation does not stably lock gene expression but instead serves as a molecular mark for gene silencing memory. Cancer Res 2012; 72: 1170–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 2002; 298: 1039–1043. [DOI] [PubMed] [Google Scholar]
- Nagy C, Suderman M, Yang J, Szyf M, Mechawar N, Ernst C et al. Astrocytic abnormalities and global DNA methylation patterns in depression and suicide. Mol Psychiatry 2015; 20: 320–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordeli E, Lambert S, Bennett V. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem 1995; 270: 2352–2359. [DOI] [PubMed] [Google Scholar]
- Birnbaum R, Jaffe AE, Hyde TM, Kleinman JE, Weinberger DR. Prenatal expression patterns of genes associated with neuropsychiatric disorders. Am J Psychiatry 2014; 171: 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durak O, de Anda FC, Singh KK, Leussis MP, Petryshen TL, Sklar P et al. Ankyrin-G regulates neurogenesis and Wnt signaling by altering the subcellular localization of beta-catenin. Mol Psychiatry 2015; 20: 388–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell 2012; 149: 525–537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paus T, Keshavan M, Giedd JN. Why do many psychiatric disorders emerge during adolescence? Nat Rev Neurosci 2008; 9: 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs D. Schizophrenia: The making of a troubled mind. Nature 2010; 468: 154–156. [DOI] [PubMed] [Google Scholar]
- Szyf M. Implications of a life-long dynamic epigenome. Epigenomics 2009; 1: 9–12. [DOI] [PubMed] [Google Scholar]
- Klengel T, Mehta D, Anacker C, Rex-Haffner M, Pruessner JC, Pariante CM et al. Allele-specific FKBP5 DNA demethylation mediates gene-childhood trauma interactions. Nat Neurosci 2013; 16: 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan PO, Sasaki A, D'Alessio AC, Dymov S, Labonte B, Szyf M et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci 2009; 12: 342–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bale TL. Epigenetic and transgenerational reprogramming of brain development. Nat Rev Neurosci 2015; 16: 332–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA, D'Alessio AC, Sharma S, Seckl JR et al. Epigenetic programming by maternal behavior. Nature neuroscience 2004; 7: 847–854. [DOI] [PubMed] [Google Scholar]
- Yaka R, Salomon S, Matzner H, Weinstock M. Effect of varied gestational stress on acquisition of spatial memory, hippocampal LTP and synaptic proteins in juvenile male rats. Behav Brain Res 2007; 179: 126–132. [DOI] [PubMed] [Google Scholar]
- Logue MW, Solovieff N, Leussis MP, Wolf EJ, Melista E, Baldwin C et al. The ankyrin-3 gene is associated with posttraumatic stress disorder and externalizing comorbidity. Psychoneuroendocrinology 2013; 38: 2249–2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke J, Witt SH, King AV, Nieratschker V, Poupon C, Gass A et al. Genome-wide supported risk variant for bipolar disorder alters anatomical connectivity in the human brain. Neuroimage 2012; 59: 3288–3296. [DOI] [PubMed] [Google Scholar]
- Tesli M, Koefoed P, Athanasiu L, Mattingsdal M, Gustafsson O, Agartz I et al. Association analysis of ANK3 gene variants in nordic bipolar disorder and schizophrenia case-control samples. Am J Med Genet B Neuropsychiatr Genet 2011; 156B: 969–974. [DOI] [PubMed] [Google Scholar]
- Liu J, Morgan M, Hutchison K, Calhoun VD. A study of the influence of sex on genome wide methylation. PloS one 2010; 5: e10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen J, Nivard MG, Willemsen G, Hottenga JJ, Helmer Q, Dolan CV et al. Genetic and environmental influences interact with age and sex in shaping the human methylome. Nat Commun 2016; 7: 11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leussis MP, Berry-Scott EM, Saito M, Jhuang H, de Haan G, Alkan O et al. The ANK3 bipolar disorder gene regulates psychiatric-related behaviors that are modulated by lithium and stress. Biol Psychiatry 2013; 73: 683–690. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.