Abstract
The Cullin-RING ubiquitin ligase 4 (CRL4) is implicated in controlling cell cycle, DNA damage repair, and checkpoint response based on studies employing cell lines and mouse models. CRL4 proteins, including CUL4A and CUL4B, are often highly accumulated in human malignancies. Elevated CRL4 attenuates DNA damage repair and increases genome instability that is believed to facilitate tumorigenesis. However, this has yet to be evaluated in human patients with cancer. In our study, 352 lung cancer and 62 normal lung specimens of Asian origin were constructed into tissue microarrays of four distinct lung cancer subtypes. Expression of CUL4A, CUL4B, and their substrates was detected by immunohistochemistry and analyzed statistically for their prognostic value and association with DNA damage response and genomic instability. Our results show that both CUL4A and CUL4B are overexpressed in the majority of lung carcinomas (PCUL4A <0.001 and PCUL4B <0.001) and significantly associated with tumor size (PCUL4A <0.001 and PCUL4B = 0.002), lymphatic invasion (PCUL4A = 0.004 and PCUL4B <0.001), metastasis (PCUL4A = 0.019 and PCUL4B = 0.006), and advanced TNM stage (PCUL4A <0.001 and PCUL4B <0.001), which parallels gene amplification and abnormal activation of the canonical WNT signaling. Moreover, overexpression of CUL4A, but not CUL4B, is significantly associated with tobacco smoking (p = 0.01) and is inversely correlated with XPC and P21, both of which are substrates of CUL4A (PCUL4A = 0.019 and PCUL4B = 0.006). Higher levels of CUL4A or CUL4B are significantly associated with the overall survival of patients (PCUL4A <0.001 and PCUL4B <0.001) and progression-free survival (PCUL4A <0.001 and PCUL4B = 0.001). Our findings revealed that CUL4A and CUL4B are differentially associated with etiologic factors for pulmonary malignancies and are independent prognostic markers for the survival of distinct lung cancer subtypes.
Keywords: cell signaling, E3 ubiquitin ligase, immunohistochemistry, lung cancer, protein degradation, ubiquitin, ubiquitin ligase
Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide. The death rate of lung cancer has increased more than 800% in males and has more than doubled in females during the last 25 years (1). In recent years, intense investigations have led to an exponential increase in the understanding of the molecular mechanisms underlying lung cancer growth, metastasis, relapse, and drug resistance, resulting in the development of novel intervention strategies, such as EGF receptor mutation-targeted therapy by tarceva and irasa and immunotherapy by Nivolumab (Opdivo). However, these Food and Drug Administration-approved drugs are directed against small subgroups of non-small cell lung cancer. Targeting a specific trait for managing different lung cancer subtypes remains a daunting task.
The CRL4 ubiquitin ligase regulates a wide variety of cellular processes, including control of cell cycle, signal transduction, DNA repair, and embryonic development (2, 3), and it is frequently implicated in human pathologic conditions such as cancer and neuronal diseases. CRL4 contains four subunits, in which CUL4A or CUL4B serves as an elongated docking platform to assemble the ubiquitin ligase complex; the N-terminal region of CUL4 recruits substrates via the adaptor protein DDB1 and various DDB1 and CUL4-associated factor substrate receptors, whereas the C terminus of CRL4 binds to the RBX1/ROC1 adaptor that recruits ubiquitin-bearing E2 enzymes. As such, substrates and activated ubiquitin are brought into close proximity to facilitate ubiquitin transfer to the substrates (4, 5). There are two members in the CRL4 family, CUL4A and CUL4B, that are derived from the same CRL4 ancestor (6). CUL4A was initially identified as an amplified gene in primary human breast cancers (7). High levels of CUL4A were observed in squamous cell carcinomas (8), adrenocortical carcinomas (9), childhood medulloblastomas (10), hepatocellular carcinomas (11), and malignant pleural mesotheliomas (12). Overexpression of CUL4A was significantly associated with poor overall survival (OS)5 in patients with prostate cancer (13). Additionally, inhibition of CUL4A induced cell death in ovarian cancer cells (14). Mutations of the cul4b gene are causal in the X-linked mental retardation syndrome (15–17), as well as developmental abnormalities, including short stature, seizure, central obesity, mild macrocephaly, and other abnormalities in humans (18). High levels of CUL4B in colon cancer have been shown to correlate with tumor progression and pathogenesis (19). However, the association of CUL4A and CUL4B status with pulmonary tumorigenesis has yet to be investigated.
Human CUL4A and CUL4B share 82% sequence identity and 90% homology and are functionally redundant in supporting embryonic development, as mice deleted of either cul4a or cul4b are viable, healthy, and display no overt abnormalities except male infertility (15, 18, 20–22). Inactivation of both CUL4A and CUL4B led to rapid growth arrest and loss of survival in primary embryonic fibroblasts (15). However, more recent studies suggest distinct activities of CUL4A and CUL4B in regulating a variety of cellular processes (23–26). Mutations of cul4b cause X-linked mental retardation, suggesting a distinct role of CUL4B as it is not rescued by the intact CUL4A in these patients. Moreover, CUL4A is primarily responsible for controlling the degradation of DDB2 and XPC, the rate-limiting damage recognition factors for nucleotide excision repair, and P21, the effector of the G1/S DNA damage checkpoint pathways, respectively, whereas CUL4B has no obvious role (15). Strikingly, cul4a null mice are hyper-resistant to UV-induced skin cancer (15). CUL4A and CUL4B are also expressed at different stages of spermatogenesis, and germ line deletion of either cul4 gene results in male infertility (24, 27). Therefore, CUL4A and CUL4B share overlapping functions in growth and survival, while also playing distinct roles in diverse cellular processes.
In this study, we measured the expression of CUL4A and CUL4B in four different subtypes of lung carcinoma in patients of Asian ethnic group, including adenocarcinoma (ADC), large cell carcinoma (LCC), squamous cell carcinoma (SCC), and small cell lung carcinoma (SCLC), and we analyzed the relationship between their expression levels and the clinicopathologic factors, as well as the OS and PFS of these patients.
Results
High Level Expression of CUL4A and CUL4B in Lung Carcinoma
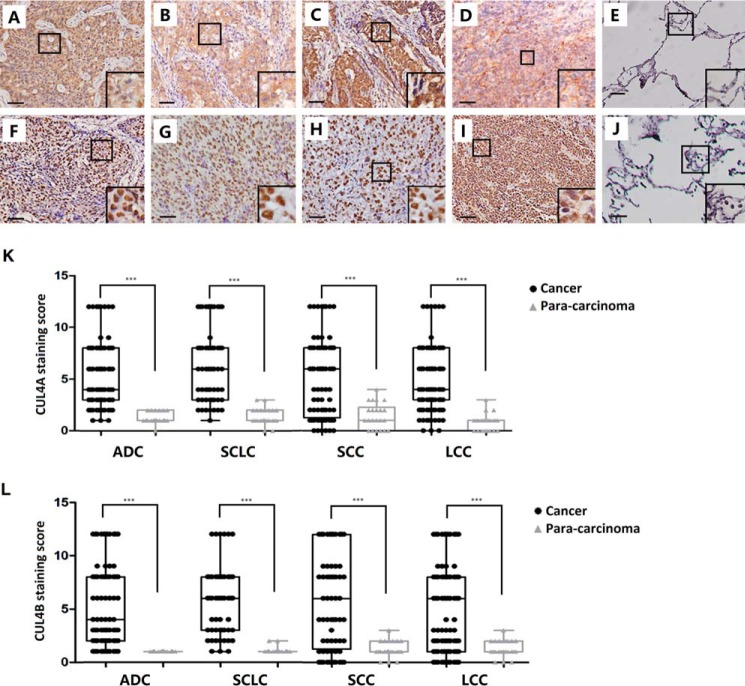
To determine the expression of CUL4A and CUL4B in lung cancers, subtype-specific lung cancer TMAs were constructed from 92 cases of ADC, 61 cases of LCC, 127 cases of SCC, 72 cases of SCLC, and 62 para-carcinoma tissues as control. Immunohistochemical staining was carried out with antibodies against CUL4A or CUL4B and scored by lung pathologists at TMUCIH using the two-way scoring system (28–30). Unlike the low/negative expression in normal lung, significantly higher expression of CUL4A was detected in the cytoplasm and nucleus of lung carcinoma (p < 0.001; Fig. 1, A–E and K; Table 1). Approximately 50% of cases in each subtype of lung carcinoma express high levels of CUL4A, with ADC (48%), LCC (61%), SCC (46%), and SCLC (51%). CUL4B was expressed exclusively in the nucleus, and about half of the lung carcinoma cases displayed abnormally higher expression levels of CUL4B compared with normal lung (p < 0.001; Fig. 1, F–J and L; Table 1). Of all four subtypes, overexpression of CUL4B was detected in 49% ADC, 64% LCC, 50% SCC, and 54% SCLC. Among all the 352 cases, 33.2% had high expression of both CUL4A and CUL4B, whereas 29.8% were low CUL4A and CUL4B expressers.
FIGURE 1.
Immunohistochemical staining showing high level expression of CUL4A and CUL4B in lung cancer. Representative staining of CUL4A (A–E) or CUL4B (F–J) in squamous cell carcinoma, adenocarcinoma, large cell lung carcinoma, small cell lung carcinoma, and para-carcinoma tissue is shown. K and L, CUL4A and CUL4B are both expressed at significantly higher levels in cancer tissue than that in the corresponding para-carcinoma of all subtypes of lung cancer (p < 0.001). Images in A–J are at 20 × 10 (=200) magnification, and insets are at 40 × 10 (=400) magnification, Bar, 50 μm. ***, significant p values (<0.001).
TABLE 1.
CUL4A and CUL4B expression in lung carcinoma patients
| Total no. of cases | CUL4A, case no. (%) |
CUL4B, case no. (%) |
|||
|---|---|---|---|---|---|
| Low | High | Low | High | ||
| Tissue types | |||||
| All lung carcinoma | 352 | 175 (50) | 177 (50) | 1650 (47) | 187 (53) |
| Para-carcinoma | 62 | 62 (100) | 0 (00) | 62 (100) | 0 (00) |
| Subtypes | |||||
| Adenocarcinoma | 92 | 48 (52) | 44 (48) | 47 (51) | 45 (49) |
| Large cell lung cancer | 61 | 24 (39) | 37 (61) | 22 (36) | 39 (64) |
| Squamous cell lung cancer | 127 | 68 (54) | 59 (46) | 63 (58) | 64 (50) |
| Small cell lung cancer | 72 | 35 (49) | 37 (51) | 33 (46) | 39 (54) |
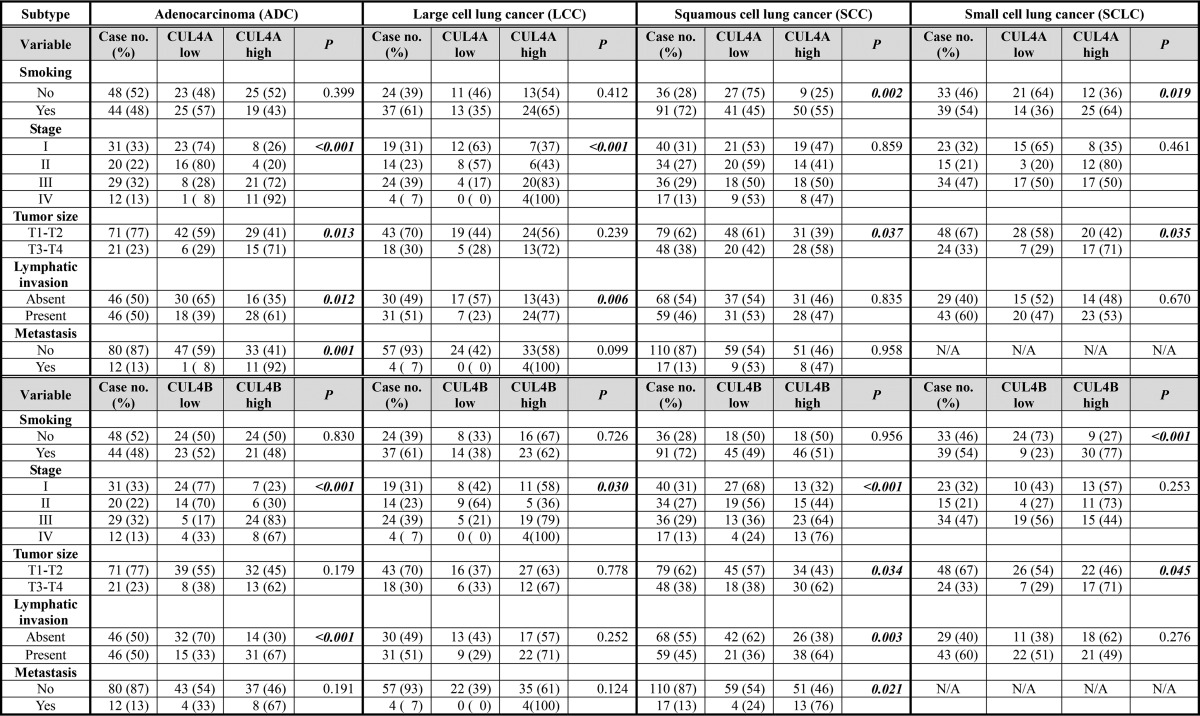
To identify the association of CUL4A and CUL4B expression with clinicopathologic parameters, univariable analysis was performed. High expression of CUL4A was significantly associated with tumor size (p < 0.001), presence of lymphatic invasion (p = 0.004), metastasis (p = 0.019), and advanced TNM stage (p < 0.001), suggesting the crucial role of CUL4A in lung carcinoma growth and progression (Table 2). CUL4A was recently shown to inhibit nucleotide excision repair (15), a key DNA repair pathway that removes alkylating DNA adducts induced by cigarette smoking or environmental pollutions. Indeed, high CUL4A expression is significantly associated with tobacco smoking (p = 0.010), suggesting that CUL4A could be a smoking-related risk factor for lung carcinogenesis among smokers. However, CUL4A is not related to the smoking score in smokers (Table 3). More interestingly, the association between CUL4A expression and tobacco smoking was found to be subtype-specific; in SCC and SCLC that are known as smoking-related subtypes (31, 32), the correlation between CUL4A expression and smoking is statistically significant (PSCC = 0.002 and PSCLC = 0.019) (Table 4). In ADC and LCC, which are not directly related to smoking (31, 32), there was no correlation between CUL4A and tobacco smoking (p > 0.05). Instead, CUL4A overexpression was found to significantly associate with TNM stage in ADC (p < 0.001) and LCC (p < 0.001) but not SCC or SCLC (p > 0.05). Moreover, different subtypes of lung cancer displayed certain level of divergences in clinicopathologic parameters in relation to CUL4A status; tumor size was related to CUL4A expression in ADC (p = 0.013), SCC (p = 0.037), and SCLC (p = 0.035) but not in LCC (p = 0.239); the presence of lymphatic invasion was associated with high expression of CUL4A in both ADC (p = 0.012) and LCC (p = 0.006), whereas distant metastasis showed positive correlation with CUL4A expression only in ADC (p = 0.001) (Table 4).
TABLE 2.
Association between clinicopathologic factors and CUL4A/CUL4B expression
The boldface italic P values indicate statistical significance.
| Variable | Case no. (%) | CUL4A, case no. (%) |
CUL4B, case no. (%) |
||||
|---|---|---|---|---|---|---|---|
| Low | High | P | Low | High | P | ||
| Age (years) | |||||||
| <60 | 164 (47) | 84 (51) | 80 (49) | 0.599 | 84 (51) | 80 (49) | 0.128 |
| >=60 | 188 (53) | 91 (48) | 97 (52) | 81 (43) | 107 (57) | ||
| Gender | |||||||
| Male | 233 (66) | 116 (49) | 117 (51) | 0.971 | 101 (43) | 132 (57) | 0.064 |
| Female | 119 (34) | 59 (49) | 60 (51) | 64 (54) | 55 (46) | ||
| Smoking | |||||||
| No | 141 (40) | 82 (58) | 59 (42) | 0.010 | 74 (52) | 67 (48) | 0.085 |
| Yes | 211 (60) | 93 (44) | 118(56) | 91 (43) | 120 (57) | ||
| Stage | |||||||
| I | 113 (32) | 71 (63) | 42 (37) | <0.001 | 69 (61) | 44 (39) | <0.001 |
| II | 83 (24) | 47 (57) | 36 (43) | 46 (55) | 37 (45) | ||
| III | 123 (35) | 47 (38) | 76 (62) | 42(34) | 81 (66) | ||
| IV | 33 (09) | 10 (30) | 23 (70) | 8 (24) | 25 (76) | ||
| Tumor size | |||||||
| T1-T2 | 242 (69) | 138 (57) | 104 (43) | <0.001 | 127 (52) | 115 (48) | 0.002 |
| T3-T4 | 110 (31) | 37 (34) | 73 (66) | 38 (35) | 72 (65) | ||
| Lymphatic invasion | |||||||
| Absent | 174 (49) | 100 (57) | 74 (43) | 0.004 | 99 (57) | 75 (43) | <0.001 |
| Present | 178 (51) | 75 (42) | 103 (58) | 66 (37) | 112 (63) | ||
| Metastasis | |||||||
| No | 319 (91) | 165 (52) | 154 (48) | 0.019 | 157 (49) | 162 (51) | 0.006 |
| Yes | 33 (09) | 10 (30) | 23 (70) | 8 (24) | 25 (76) | ||
TABLE 3.
Association between smoking index and CUL4A expression in different subtypes of lung cancer
| Smoking index | Case no. (%) | CUL4A, no. (%) |
||
|---|---|---|---|---|
| Low | High | P | ||
| All subtypes | ||||
| I | 40 (11) | 22 (55) | 18 (45) | I versus II 0.662 |
| II | 43 (12) | 21 (49) | 22 (51) | II versus III 0.484 |
| III | 140 (40) | 59 (42) | 81 (58) | I versus III 0.155 |
| Adenocarcinoma | ||||
| I | 6 (07) | 4 (67) | 2 (33) | I versus II 1.000 |
| II | 11 (12) | 8 (73) | 3 (27) | II versus III 0.282 |
| III | 27 (29) | 13 (48) | 14 (52) | I versus III 0.656 |
| Large cell lung cancer | ||||
| I | 2 (03) | 0 (00) | 2 (100) | I versus II 0.500 |
| II | 7 (11) | 3 (43) | 4 (57) | II versus III 1.000 |
| III | 29 (48) | 11 (38) | 18 (62) | I versus III 0.527 |
| Squamous cell lung cancer | ||||
| I | 30 (23) | 18 (60) | 12 (40) | I versus II 0.180 |
| II | 25 (20) | 10 (40) | 15 (60) | II versus III 0.805 |
| III | 47 (37) | 21 (45) | 26 (55) | I versus III 0.244 |
| Small cell lung cancer | ||||
| I | 2 (03) | 0 (00) | 2 (100) | I versus III 0.528 |
| III | 37 (51) | 14 (38) | 23 (62) | |
TABLE 4.
Association between clinicopathologic factors and CUL4A/B expression in different subtypes of lung cancer
CUL4B expression levels were found significantly associated with tumor size (p = 0.002), lymphatic invasion (p < 0.001), tumor metastasis (p = 0.006), and TNM stage (p < 0.001) (Table 2). Contrary to CUL4A, CUL4B expression is not significantly associated with tobacco smoking (p = 0.085) except in SCLC (p < 0.001) (Tables 2 and 4), consistent with the finding that CUL4B inactivation had no effect on NER activity in primary cells (15). Subtype-specific associations between CUL4B and clinicopathologic parameters were also observed (Table 4). In ADC, CUL4Bhigh cases had more advanced TNM stages (p < 0.001) and more prevalent lymphatic invasion (p = 0.001). In SCC, CUL4B was strongly associated with TNM stage (p < 0.001), tumor size (p = 0.034), lymphatic invasion (p = 0.003), and tumor metastasis (p = 0.021). CUL4B expression also correlated significantly with TNM stage in LCC (p = 0.030) and tumor size in SCLC (p = 0.045) (Table 4).
Prognostic Value of CUL4A and CUL4B
Because of the unavailable survival information of 25 cases, 327 cases were included in the survival analysis. 192 (58.7%) were seen with disease progression post-surgery, and 161 (49.2%) patients eventually died of tumor within the follow-up period of 2 months to 125 months. The median OS was 40 months, and the median PFS was 25 months.
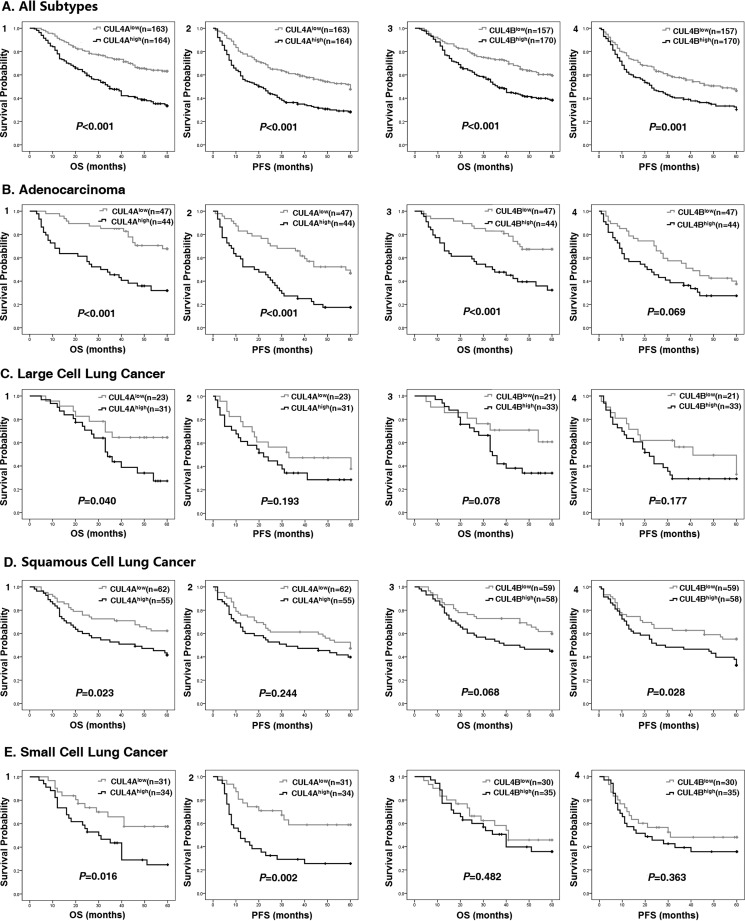
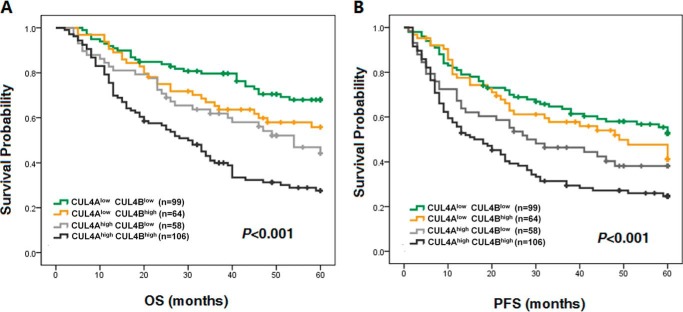
To investigate whether CUL4A or CUL4B overexpression correlates with the survival probability of patients with lung cancer, survival analysis was performed. As shown in Fig. 2, both CUL4Ahigh and CUL4Bhigh patients had significantly worse prognosis than those with low levels of CUL4A or CUL4B expression. For all lung cancer cases, both CUL4A and CUL4B are of prognostic value for OS (PCUL4A < 0.001 and PCUL4B < 0.001) and PFS (PCUL4A < 0.001 and PCUL4B = 0.001) (Table 5 and Fig. 2), indicating patients with low expression of CUL4A or CUL4B had significantly longer survival than the high expressers. Furthermore, combination of CUL4A and CUL4B (CUL4Ahigh CUL4Bhigh) could serve as a better prognostic marker for poor OS and PFS than that of CUL4A or CUL4B alone; meanwhile, CUL4Alow CUL4Blow predicted more favorable OS and PFS (POS < 0.001 and PPFS < 0.001, Table 6 and Fig. 3). Aside from the expression of CUL4A and CUL4B, TNM stage, tumor size, lymphatic invasion, and tumor metastasis were also shown to significantly associate with OS and PFS (Table 5).
FIGURE 2.
Poor OS and PFS in CUL4Ahigh and CUL4Bhigh lung cancer patients. Kaplan-Meier survival curves based on high versus low expression levels of CUL4A and CUL4B in all patients with lung cancers (A) or subtypes of ADC (B), LCC (C), SCC (D), and SCLC (E) are shown. OS and PFS of CUL4A groups (panels 1 and 2) or CUL4B groups (panels 3 and 4) are shown.
TABLE 5.
Association between clinicopathologic factors and OS and PFS in lung carcinoma
The boldface italic P values indicate statistical significance.
| Variable | Case no. (%) | OS |
PFS |
||
|---|---|---|---|---|---|
| 5-Year OS (%) | P | 5-Year PFS (%) | P | ||
| Age (years) | |||||
| <60 | 154 (47) | 48.0 | 0.505 | 40.2 | 0.627 |
| >=60 | 173 (53) | 48.1 | 36.4 | ||
| Gender | |||||
| Male | 218 (67) | 49.6 | 0.327 | 36.3 | 0.981 |
| Female | 109 (33) | 46.1 | 40.6 | ||
| Smoking | |||||
| No | 130 (40) | 45.4 | 0.278 | 39.2 | 0.422 |
| Yes | 197 (60) | 50.4 | 35.8 | ||
| Stage | |||||
| I-II | 194 (59) | 62.9 | <0.001 | 50.6 | <0.001 |
| III-IV | 133 (41) | 27.1 | 19.3 | ||
| Tumor size | |||||
| T1-T2 | 250 (76) | 52.0 | 0.016 | 42.9 | <0.001 |
| T3-T4 | 77 (24) | 37.1 | 22.0 | ||
| Lymphatic invasion | |||||
| Absent | 171 (52) | 64.5 | <0.001 | 49.1 | <0.001 |
| Present | 156 (48) | 30.8 | 25.8 | ||
| Metastasis | |||||
| No | 300 (92) | 49.9 | 0.004 | 39.4 | 0.007 |
| Yes | 27 (08) | 32.1 | 19.4 | ||
| CUL4A | |||||
| Low | 163 (49) | 63.2 | <0.001 | 47.6 | <0.001 |
| High | 164 (51) | 33.5 | 28.2 | ||
| CUL4B | |||||
| Low | 157 (48) | 59.4 | <0.001 | 46.3 | 0.001 |
| High | 170 (52) | 38.3 | 30.1 | ||
TABLE 6.
OS and PFS of four CUL4A and CUL4B combination groups in lung carcinoma
The boldface italic P values indicate statistical significance.
| Variable | Case no. (%) | OS |
PFS |
||
|---|---|---|---|---|---|
| 5-Year OS (%) | P | 5-Year PFS (%) | P | ||
| Combinations | <0.001 | <0.001 | |||
| CUL4Alow CUL4Blow | 99 (30) | 68.0 | 53.2 | ||
| CUL4Alow CUL4Bhigh | 64 (20) | 55.8 | 40.4 | ||
| CUL4Ahigh CUL4Blow | 58 (18) | 44.1 | 38.2 | ||
| CUL4Ahigh CUL4Bhigh | 106 (32) | 27.6 | 24.6 | ||
FIGURE 3.
Poor OS and PFS in CUL4Ahigh CUL4Bhigh lung cancer patients. Kaplan-Meier overall survival curve (A) and progression-free survival curve (B) based on four combinations of CUL4A and CUL4B expression pattern in all patients with lung cancers.
Multivariable analysis was also performed to determine the independent prognostic value of CUL4A and CUL4B expression. TNM stages, tumor size, presence of lymphatic invasion, and distant metastasis were also included based on their significance in univariable analysis. For OS, CUL4A expression (p = 0.001), CUL4B expression (p = 0.024), lymphatic invasion (p < 0.001), as well as tumor metastasis (p = 0.025) were independent prognostic factors. However, only CUL4A expression (p = 0.001) and lymphatic invasion (p < 0.001) were independent prognostic factors for PFS (Table 7).
TABLE 7.
Multivariable analyses of factors associated with overall survival and progression free survival
The following abbreviations are used: 95% CI is 95% confidence interval; HR, hazards ratio. Bold italic indicates statistical significance using multivariable analysis. P < 0.05 was considered statistically significant.
| Variable | OS |
PFS |
||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| CUL4Ahigh/CUL4Alow | 1.610 (1.204–2.152) | 0.001 | 1.739 (1.267–2.386) | 0.001 |
| CUL4Bhigh/CUL4Blow | 1.409 (1.047–1.896) | 0.024 | 1.200 (0.876–1.644) | 0.256 |
| Tumor size (III, IV)/tumor size (I, II) | 1.144 (0.967–1.354) | 0.117 | 1.195 (1.000–1.428) | 0.050 |
| Lymphatic invasion(+)/lymphatic invasion(−) | 2.234 (1.664–3.000) | <0.001 | 1.968 (1.442–2.685) | <0.001 |
| Distant metastasis(+)/distant metastasis(−) | 1.637 (1.063–2.552) | 0.025 | 1.383 (0.861–1.644) | 0.180 |
Mechanisms Underlying High Expression of CUL4A/B in Lung Cancer
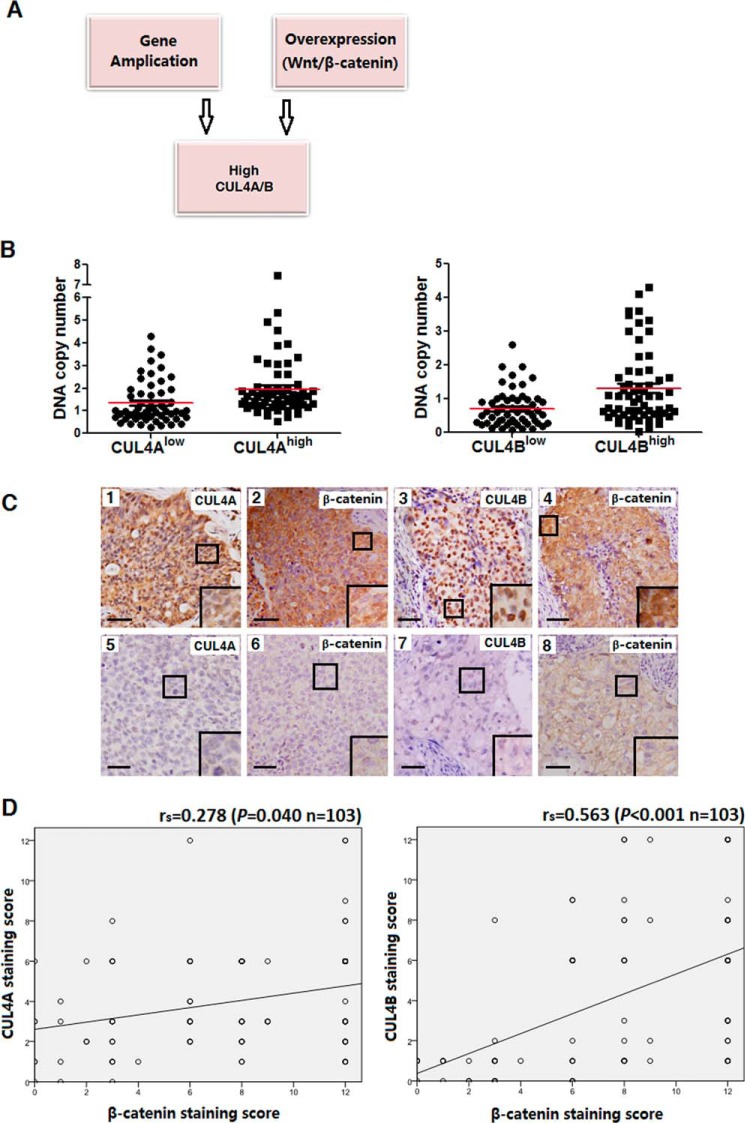
Increased accumulation of oncoproteins as a result of amplification of oncogenes is often seen in cancer cells as a means of conferring growth or selective advantage (7). To this end, 117 patients with lung cancer from the available 352 individuals were randomly selected for DNA copy number analysis by qPCR (33). As shown in Fig. 4B, CUL4Ahigh or CUL4Bhigh expression groups were found to contain significantly higher levels of cul4a and cul4b DNA than those of their low expression counterparts (PCUL4A < 0.001 and PCUL4B < 0.001), indicating that gene amplification accounts, at least in part, for the increased accumulation of CUL4A and CUL4B proteins in patients with lung cancer.
FIGURE 4.
Mechanisms underlying high level expression of CUL4A and CUL4B. A, gene amplification and activation of the canonical WNT/β-catenin signaling contribute to accumulation of CUL4A/B in patients with lung cancer. B, increased gene copy numbers in CUL4Ahigh or CUL4Bhigh expression groups compared with those in CUL4Alow or CUL4Blow groups, as determined by qPCR. C, association between CUL4A/B and β-catenin expression of lung cancer specimens. Immunohistochemical staining of lung cancer specimens used antibodies against CUL4A, CUL4B, and β-catenin, respectively. Representative images of CUL4Ahigh β-cateninhigh (panels 1 and 2), CUL4Bhigh β-cateninhigh (panels 3 and 4), CUL4Alow β-cateninlow (panels 5 and 6), and CUL4Blow β-cateninlow (panels 7 and 8) are shown. D, correlation between CUL4A or CUL4B and β-catenin expression. rs indicates the correlation ratio.
Elevated gene transcription is another mechanism by which cancer cells accumulate aberrant high levels of oncoproteins. Miranda-Carboni et al. (34) identified both CUL4A and CUL4B as target genes of the canonical WNT signaling in both human and mouse mammary cells, which are measured by the abundance of cytosolic and/or nuclear β-catenin. We determined cytoplasmic and nuclear β-catenin expression and the association with CUL4Ahigh and CUL4Bhigh expression in a total of 103 squamous cell lung cancer patients analyzed. As shown in Fig. 4, C and D, soluble β-catenin indeed correlated with both CUL4A (rs = 0.278, p = 0.040) and CUL4B expression (rs = 0.563, p < 0.001). More interestingly, the correlation between soluble β-catenin and CUL4A was only significant among smokers (p = 0.023) but not non-smokers (p = 0.133, Table 8). As such, activation of the canonical WNT/β-catenin pathway could also contribute to the increased accumulation of CUL4A and CUL4B in lung cancer, similar to what was observed in breast cancer (34).
TABLE 8.
Association between β-catenin and CUL4A expression in non-smoking and smoking patients of lung cancer
Bold italic P indicates statistical significance using multivariable analysis.
| Variable | Case no. (%) | CUL4A, case no. (%) |
||
|---|---|---|---|---|
| Low | High | P | ||
| Non-smokers | ||||
| β-Cateninlow | 5 (22) | 5 (100) | 0 (00) | 0.133 |
| β-Cateninhigh | 18 (78) | 14 (78) | 4 (22) | |
| Smokers | ||||
| β-Cateninlow | 22 (28) | 15 (68) | 7 (32) | 0.023 |
| β-Cateninhigh | 58 (72) | 26 (45) | 32 (55) | |
Inverse Correlation of CUL4A with XPC or P21 in Lung Cancer
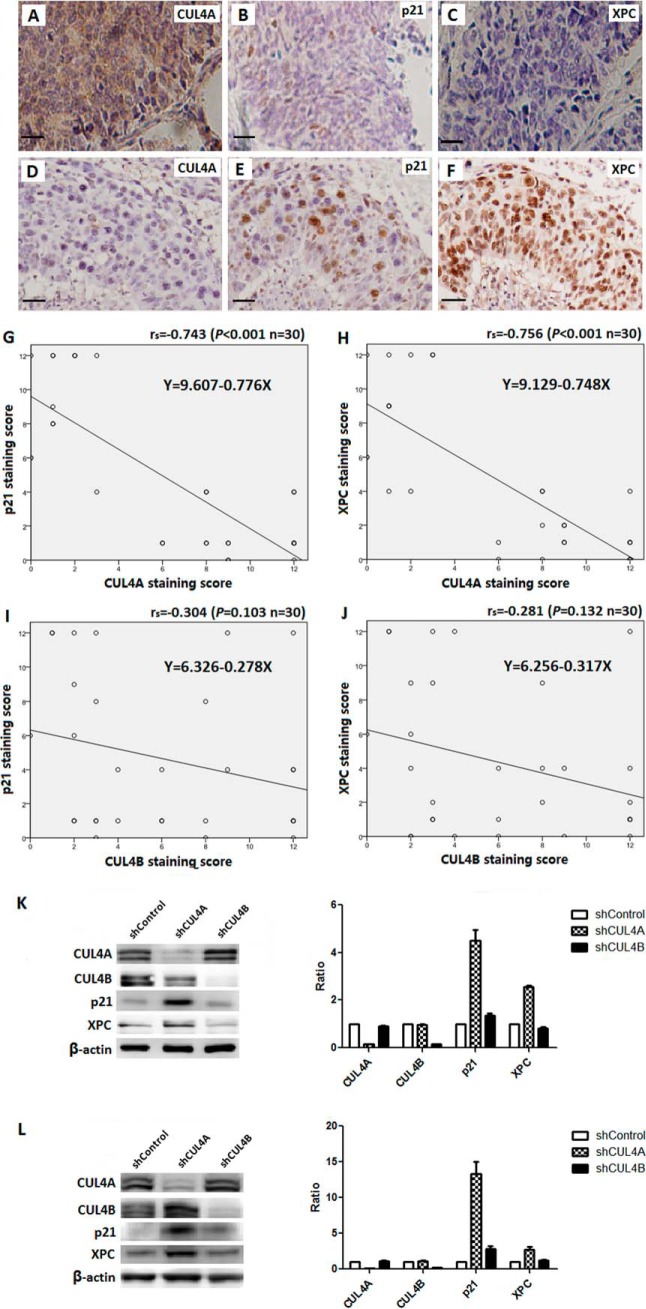
Tobacco smoking induces strand-distorting DNA damages that are repaired by the nucleotide excision repair (35). Moreover, the DNA damage checkpoint pathways are activated that halt the cell cycle and allow time for the NER machinery to identify and remove DNA lesions. Interestingly, both the NER and G1/S DNA damage checkpoint pathways are suppressed by CUL4A; the rate-limiting NER damage recognition factor XPC and the effector of the G1/S checkpoint protein P21 are both substrates of the CRL4A ubiquitin ligase (15). As such, overexpression of CUL4A is expected to accelerate the destruction of XPC and P21, compromising the NER and checkpoint activities, and lead to unrepaired DNA adducts and genomic instability. Whereas CUL4A abrogation indeed conferred marked protection against UV-induced skin carcinogenesis in mice (15), the association of CUL4A overexpression with NER and checkpoint factors has yet to be examined in patients with cancer.
Here, a randomly selected cohort of 30 lung carcinoma specimens representing different subtypes was evaluated for expression of CUL4A, XPC, and P21 by IHC. As seen in Fig. 5, A–H, inverse correlation was seen between protein levels of CUL4A and XPC (rs = 0.756, p < 0.001) and between CUL4A and P21 (rs = 0.743, p < 0.001). The expression of CUL4B was also assessed in parallel and showed no significant correlation with that of XPC or P21 (Fig. 5, I and J). Therefore, high CUL4A expression was significantly associated with reduced XPC and P21, consistent with the notion of excessive destruction of these key NER and G1/S checkpoint proteins in patients with lung cancer. By contrast, there was no correlation detected between the expression levels of CUL4B and XPC or CUL4B and P21. This is also consistent with the finding by Liu et al. (15) that CUL4B is not involved in XPC or P21 turnover.
FIGURE 5.
Inverse correlation of CUL4A and P21 or XPC expression in SCC patients. A–F, representative staining of CUL4A, P21, and XPC in SCC specimens. Regression line plots showed significant inverse correlation of P21 or XPC with CUL4A (G and H) but not CUL4B (I and J). K and L, depletion of CUL4A resulted in increased accumulation of XPC and P21 in NCI-H520 SCC cells (K) or NCI-H446 SCLC cells (L), as determined by immunoblotting using antibodies again CUL4A, CUL4B, and β-actin (internal loading control). The levels of CUL4A, CUL4B, XPC, and P21 (relative to that of β-actin) in the shCUL4A and shCUL4B cell lines were quantitatively determined using Odyssey and plotted against those of the shControl (right panels).
To further investigate the biochemical basis of the inverse expression of CUL4A and XPC/P21 in lung cancer cells, we transduced lentiviral shCUL4A or shCUL4B into the SCC cell line NCI-H520 and SCLC cell line NCI-H446, and we established stable CUL4A and CUL4B knockdown cell lines. As a negative control, NCI-H520 and NCI-H446 cells were infected with lentiviral shControl vector alone and were selected for stable cell lines. Cell extracts were prepared for immunoblotting to measure the steady-state levels of CUL4A, CUL4B, XPC, and P21. In both cell lines, XPC and P21 accumulated upon silencing of CUL4A but not CUL4B (Fig. 5, K and L). Taken together, these data revealed that CUL4Ahigh lung cancer cells are compromised in the NER and G1/S checkpoint functions as a result of excess destruction of XPC and P21.
Discussion
Amplification or overexpression of the cul4a or cul4b gene has been reported in a wide variety of cancers (8–12, 36). However, there was no systematic investigation of the prognostic significance of CUL4A and CUL4B in lung carcinoma. This study is the first to investigate the clinical implications of CUL4A and CUL4B expression in a large cohort of lung cancer patients of distinct subtypes with long term follow-up and to interrogate the underlying DNA damage response pathways that respond to elevated CUL4A and CUL4B in lung carcinomas. Here, we showed both gene amplification and WNT-β-catenin-induced transcriptional activation led to increased steady-state levels of CUL4A and CUL4B. OS and PFS are significantly reduced in both CUL4Ahigh and CUL4Bhigh patient groups. On multivariable analyses, both CUL4A and CUL4B are independent prognostic factors of OS, whereas CUL4A is also an independent prognostic factor of PFS. This study defines CUL4A and CUL4B as new prognostic markers for lung cancer.
Tobacco smoking and/or environmental exposure to polycyclic aromatic hydrocarbons and other chemical pollutants are main risk factors for lung tumorigenesis. These carcinogens induce alkylating DNA damage and genome instability (37, 38), which are repaired with by NER. In this regard, CUL4A is the first identified inhibitor of NER, and we observed a strong association of high CUL4A levels with tobacco smoking risk in SCLC and SCC, the two smoking-related lung cancer subtypes. Furthermore, CUL4A suppresses NER as well as G1/S DNA damage checkpoint by facilitating ubiquitin-dependent proteolysis of the rate-limiting NER factors, DDB2 and XPC, as well as the P21 G1/S checkpoint effector (15). Consistently, we showed inverse correlation of expression levels between CUL4A and XPC or P21 in patients with lung carcinoma (Fig. 5). Interestingly, CUL4B is not involved in XPC and P21 turnover, and inactivation of CUL4B in primary murine MEF cells had no effect on NER (15). Consistently, we detected no correlation between CUL4B expression and the cellular levels of XPC and P21, and CUL4B is not associated with the smoking.
High levels of CUL4A and CUL4B expression in lung carcinoma are significantly associated with worse clinicopathologic variables, including lymph node metastasis, higher TNM stage, distant metastasis, and larger tumor size in SCC, ADC, LCC, and SCLC. Thus, dysregulation of CUL4A not only alters DNA damage repair and genomic integrity but is also expected to affect the later stages of lung carcinogenesis through excess degradation of other yet-to-be-determined CUL4 substrates and pathways. Collectively, our data revealed the prognostic value of CRL4 ubiquitin ligases in distinct types of lung carcinoma and highlight a role of CUL4A dysregulation in suppressing cellular DNA damage response pathways in pulmonary carcinogenesis.
Experimental Procedures
Patient Samples and Tissue Microarray
352 cases of patients from Asian origin with lung cancer who underwent surgical resection or bronchoscopic biopsies at TMUCIH, China, between 2003 and 2010 were collected. The 62 samples of normal lung taken at 10 cm from the lung carcinoma were used as para-carcinoma controls. Tissues were formalin-fixed, paraffin-embedded, and constructed into TMA at the Shanghai Outdo Biotech Co. (Shanghai, China).
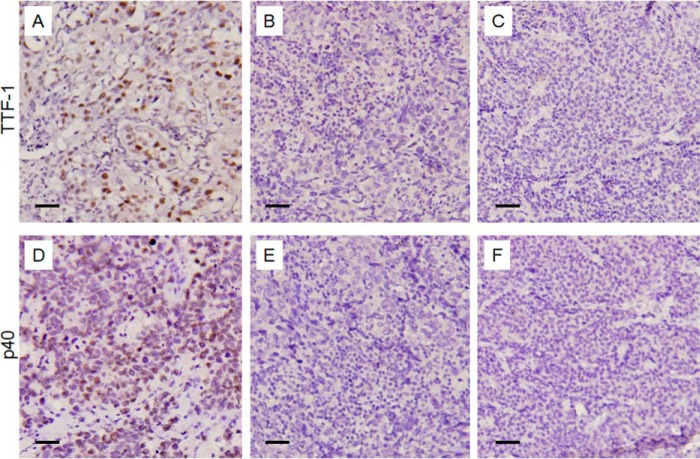
Clinical data were collected from a prospectively maintained database at the Department of Pathology, TMUCIH, China. Subtypes and TNM stages were defined based on the NCCN Guidelines Version 1.2014. Large cell lung cancer specimens were diagnosed based on morphologic features of paraffin-embedded tumor specimens as well as double negative staining of TTP-1 and P40 (Fig. 6).
FIGURE 6.
Negative IHC staining of nuclear TTF-1 and P40 for diagnosis of large cell lung carcinomas. A, representative staining of TTF-1 in adenocarcinoma as positive control. B and C, IHC staining of TTF-1 in two representative large cell lung carcinoma cases. D, representative staining of P40 in squamous cell lung carcinoma as positive control. E and F, IHC staining of P40 in the corresponding specimens as B and C.
The ages of the patients ranged from 30 to 83 years old, with a median age of 60. A total of 233 (66%) male and 119 (34%) female patients participated in this study. 211 (60%) patients were smokers. Patients were followed from the time of surgery and censored as the status at the time of the last follow-up. The average follow-up time was 46 months (median, 41 months; ranging from 2 to 125 months). Because of the unavailable survival information of 25 cases, 327 cases were included in the survival analysis.
DNA Samples and qPCR
Genomic DNA samples from 117 of 352 patients were deposited in the TMUCIH tumor tissue bank. qPCR was carried out (33). Cycle of threshold (Ct) values were determined by ABI software. If a sample showed a Ct of >34 or <27, a lower or a higher dilution of the DNA sample was measured again in the range of the standard curve (39).
Cell Lines, Antibodies, and Reagents
Human squamous cell carcinoma cell lines NCI-H520 and human small cell lung cancer cell lines NCI-H446 were obtained from American Type Culture Collection. Antibodies used for IHC of lung carcinoma tissues were purchased from commercial sources as follows: rabbit anti-CUL4A antibody (Bethyl Laboratories, Montgomery, TX; 1:100); rabbit anti-CUL4B antibody (Novus Biologicals, Littleton, CO; 1:50); β-catenin (D10A8; 1:100) and P21Waf1/Cip1 (12D1) rabbit mAbs (Cell Signaling Technology, Danvers, MA; 1:100); anti-XPC antibody(3.26) (Abcam, ab116211, Cambridge, MA; 1:100); and secondary antibody (Fuzhou New Biotechnology Development Co., China; 1:1000). Antibodies against CUL4A (Cell Signaling Technology, 1:1000), CUL4B (Proteintech, Rosemont, IL; 1:1000), XPC antibody (GeneTex, Irvine, CA; 1:1000), and P21Waf1/Cip1 (Cell Signaling Technology; 1:1000) were used for Western blotting. For DNA copy number assessment, genomic DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Valencia, CA). DNA quality was examined by 1% agarose gel electrophoresis, and DNA concentrations were measured using the PicoGreen dsDNA assay (Invitrogen). qPCR was carried out on genomic DNA using SYBR® Premix Ex TaqTM II (Takara Bio Inc, Shiga, Japan) universal master mix and Multiscript RT on a 7500 Fast System (Applied Biosystems, Foster City, CA).
Plasmid Construction, Transfection, and Lentiviral Infection
The lentiviral shRNA plasmids for knocking down CUL4A and CUL4B were described previously (40). To generate the recombinant shRNA lentiviruses, 293T cells were transfected with 12 μg of shRNA, 6 μg of VSV-G, and 9 μg of Δ8.9 plasmids using the Lipofectamine® 2000 transfection reagent (Invitrogen). 48 hours post-transfection, supernatants containing the indicated lentivirus were collected for infection of NCI-H520 and NCI-H446 cells. Infected cells were selected in RPMI 1640 medium (Invitrogen) containing 4 μg/ml puromycin dihydrochloride (Sigma, Buchs SG, Switzerland).
Immunohistochemical Scoring and Analysis
The 4-μm-thick sections from the tissue microarray blocks were processed for immunohistochemical staining. The two-way scoring system was used for staining quantification (28–30). The staining intensity was scored in four categories as follows: negative, 0; weak, 1; moderate, 2; and strong, 3. The proportion of positively stained cells of interest was determined as follows: 0–25%, 1; 26–50%, 2; 51–75%, 3; and 76–100%, 4. The final expression level of each protein in each sample was obtained by multiplying the proportion and the intensity. For CUL4A and CUL4B, a median score of 6 was used as the cutoff to divide the cases into two groups, the low expression group (scores of 0–4) and the high expression group (scores of 6–12) according to the published standard scoring method (41–44).
Statistical Analysis
The correlations between CUL4A/CUL4B expression and continuous clinicopathologic factors were assessed using Spearman's correlation coefficient. The associations between CUL4A/CUL4B expression and categorical clinicopathologic factors were assessed using χ2 or Fisher's exact test. The median OS and PFS and their 95% confidence intervals were determined by the Kaplan-Meier method and tested using log-rank test. Multivariable analysis of the independent factors associated with survival was performed using the Cox proportional hazard model (45). Two-tailed, two-sample Student's t test was used to compare the DNA copy number between CUL4A and CUL4B high and low expression groups. All differences were considered significant when the associated p value was less than 0.05. The software package of SPSS 16.0 for Windows was used for statistical analyses (46).
Author Contributions
L. J. and F. Y. carried out the experiments, analyzed the data, and wrote most of the paper. W. C. and Y. P. read the H&E and IHC slides and scored the expression levels. Z. C. contributed to the statistical analysis. H. Z. and Haixin Li provided frozen samples and patient records. N. N. led the pathologic analysis of lung cancer subtypes. X. R. and Hui Li analyzed the data and contributed to the manuscript preparation. P. Z. conceived the idea, designed the experiments, and wrote the manuscript with L. J. and F. Y.
This work was supported by National Key Technology R&D Program of China Grant 2015BAI12B12, National Sciences Foundation of China Grants 81272221 and 81472471 (to X. R.), Tianjin Medical University Cancer Institute and Hospital Research Grant B1309 (to F. Y.), National Institutes of Health Grants R01 CA159925 and CA098210, and translational grant from the V Foundation for Cancer Research (to P. Z.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- OS
- overall survival
- IHC
- immunohistochemistry
- PFS
- progression-free survival
- ADC
- adenocarcinoma
- SCC
- squamous cell carcinoma
- LCC
- large cell carcinoma
- SCLC
- small cell lung carcinoma
- TMA
- tissue microarrays
- qPCR
- quantitative PCR
- NER
- nucleotide excision repair
- TMUCIH
- Tianjin Medical University Cancer Institute and Hospital
- IHC
- immunohistochemistry.
References
- 1. Siegel R. L., Miller K. D., and Jemal A. (2016) Cancer statistics. CA Cancer J. Clin. 66, 7–30 [DOI] [PubMed] [Google Scholar]
- 2. Kerzendorfer C., Whibley A., Carpenter G., Outwin E., Chiang S. C., Turner G., Schwartz C., El-Khamisy S., Raymond F. L., and O'Driscoll M. (2010) Mutations in Cullin 4B result in a human syndrome associated with increased camptothecin-induced topoisomerase I-dependent DNA breaks. Hum. Mol. Genet. 19, 1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kerzendorfer C., Hart L., Colnaghi R., Carpenter G., Alcantara D., Outwin E., Carr A. M., and O'Driscoll M. (2011) CUL4B-deficiency in humans: understanding the clinical consequences of impaired Cullin 4-RING E3 ubiquitin ligase function. Mech. Ageing Dev. 132, 366–373 [DOI] [PubMed] [Google Scholar]
- 4. Lee J., and Zhou P. (2012) Pathogenic role of the CRL4 ubiquitin ligase in human disease. Front. Oncol. 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marín I. (2009) Diversification of the cullin family. BMC Evol. Biol. 9, 267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Higa L. A., and Zhang H. (2007) Stealing the spotlight: CUL4-DDB1 ubiquitin ligase docks WD40-repeat proteins to destroy. Cell Div. 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen L. C., Manjeshwar S., Lu Y., Moore D., Ljung B. M., Kuo W. L., Dairkee S. H., Wernick M., Collins C., and Smith H. S. (1998) The human homologue for the Caenorhabditis elegans cul-4 gene is amplified and overexpressed in primary breast cancers. Cancer Res. 58, 3677–3683 [PubMed] [Google Scholar]
- 8. Shiyanov P., Nag A., and Raychaudhuri P. (1999) Cullin 4A associates with the UV-damaged DNA-binding protein DDB. J. Biol. Chem. 274, 35309–35312 [DOI] [PubMed] [Google Scholar]
- 9. Dohna M., Reincke M., Mincheva A., Allolio B., Solinas-Toldo S., and Lichter P. (2000) Adrenocortical carcinoma is characterized by a high frequency of chromosomal gains and high-level amplifications. Genes Chromosomes Cancer 28, 145–152 [PubMed] [Google Scholar]
- 10. Michiels E. M., Weiss M. M., Hoovers J. M., Baak J. P., Voûte P. A., Baas F., and Hermsen M. A. (2002) Genetic alterations in childhood medulloblastoma analyzed by comparative genomic hybridization. J. Pediatr. Hematol. Oncol. 24, 205–210 [DOI] [PubMed] [Google Scholar]
- 11. Yasui K., Arii S., Zhao C., Imoto I., Ueda M., Nagai H., Emi M., and Inazawa J. (2002) TFDP1, CUL4A, and CDC16 identified as targets for amplification at 13q34 in hepatocellular carcinomas. Hepatology 35, 1476–1484 [DOI] [PubMed] [Google Scholar]
- 12. Hung M. S., Mao J. H., Xu Z., Yang C. T., Yu J. S., Harvard C., Lin Y. C., Bravo D. T., Jablons D. M., and You L. (2011) Cul4A is an oncogene in malignant pleural mesothelioma. J. Cell. Mol. Med. 15, 350–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ren S., Xu C., Cui Z., Yu Y., Xu W., Wang F., Lu J., Wei M., Lu X., Gao X., Liang Y., Mao J. H., and Sun Y. (2012) Oncogenic CUL4A determines the response to thalidomide treatment in prostate cancer. J. Mol. Med. 90, 1121–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pan W. W., Zhou J. J., Yu C., Xu Y., Guo L. J., Zhang H. Y., Zhou D., Song F. Z., and Fan H. Y. (2013) Ubiquitin E3 ligase CRL4(CDT2/DCAF2) as a potential chemotherapeutic target for ovarian surface epithelial cancer. J. Biol. Chem. 288, 29680–29691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu L., Lee S., Zhang J., Peters S. B., Hannah J., Zhang Y., Yin Y., Koff A., Ma L., and Zhou P. (2009) CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 34, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zou Y., Liu Q., Chen B., Zhang X., Guo C., Zhou H., Li J., Gao G., Guo Y., Yan C., Wei J., Shao C., and Gong Y. (2007) Mutation in CUL4B, which encodes a member of cullin-RING ubiquitin ligase complex, causes X-linked mental retardation. Am. J. Hum. Genet. 80, 561–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tarpey P. S., Raymond F. L., O'Meara S., Edkins S., Teague J., Butler A., Dicks E., Stevens C., Tofts C., Avis T., Barthorpe S., Buck G., Cole J., Gray K., Halliday K., et al. (2007) Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am. J. Hum. Genet. 80, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zou Y., Mi J., Cui J., Lu D., Zhang X., Guo C., Gao G., Liu Q., Chen B., Shao C., and Gong Y. (2009) Characterization of nuclear localization signal in the N terminus of CUL4B and its essential role in cyclin E degradation and cell cycle progression. J. Biol. Chem. 284, 33320–33332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jiang T., Tang H. M., Wu Z. H., Chen J., Lu S., Zhou C. Z., Yan D. W., and Peng Z. H. (2013) Cullin 4B is a novel prognostic marker that correlates with colon cancer progression and pathogenesis. Med. Oncol. 30, 534. [DOI] [PubMed] [Google Scholar]
- 20. Higa L. A., Yang X., Zheng J., Banks D., Wu M., Ghosh P., Sun H., and Zhang H. (2006) Involvement of CUL4 ubiquitin E3 ligases in regulating CDK inhibitors Dacapo/p27Kip1 and cyclin E degradation. Cell Cycle 5, 71–77 [DOI] [PubMed] [Google Scholar]
- 21. Cox B. J., Vollmer M., Tamplin O., Lu M., Biechele S., Gertsenstein M., van Campenhout C., Floss T., Kühn R., Wurst W., Lickert H., and Rossant J. (2010) Phenotypic annotation of the mouse X chromosome. Genome Res. 20, 1154–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin Y., Lin C., Kim S. T., Roig I., Chen H., Liu L., Veith G. M., Jin R. U., Keeney S., Jasin M., Moley K., Zhou P., and Ma L. (2011) The E3 ubiquitin ligase Cullin 4A regulates meiotic progression in mouse spermatogenesis. Dev. Biol. 356, 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson S., and Xiong Y. (2009) CRL4s: the CUL4-RING E3 ubiquitin ligases. Trends Biochem. Sci. 34, 562–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Liu L., Yin Y., Li Y., Prevedel L., Lacy E. H., Ma L., and Zhou P. (2012) Essential role of the CUL4B ubiquitin ligase in extra-embryonic tissue development during mouse embryogenesis. Cell Res. 22, 1258–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sarikas A., Hartmann T., and Pan Z. Q. (2011) The cullin protein family. Genome Biol. 12, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhong W., Feng H., Santiago F. E., and Kipreos E. T. (2003) CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423, 885–889 [DOI] [PubMed] [Google Scholar]
- 27. Yin Y., Liu L., Yang C., Lin C., Veith G. M., Wang C., Sutovsky P., Zhou P., and Ma L. (2016) Cell autonomous and nonautonomous function of CUL4B in mouse spermatogenesis. J. Biol. Chem. 291, 6923–6935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kyo S., Sakaguchi J., Ohno S., Mizumoto Y., Maida Y., Hashimoto M., Nakamura M., Takakura M., Nakajima M., Masutomi K., and Inoue M. (2006) High twist expression is involved in infiltrative endometrial cancer and affects patient survival. Hum. Pathol. 37, 431–438 [DOI] [PubMed] [Google Scholar]
- 29. Nenutil R., Smardova J., Pavlova S., Hanzelkova Z., Muller P., Fabian P., Hrstka R., Janotova P., Radina M., Lane D. P., Coates P. J., and Vojtesek B. (2005) Discriminating functional and non-functional p53 in human tumours by p53 and MDM2 immunohistochemistry. J. Pathol. 207, 251–259 [DOI] [PubMed] [Google Scholar]
- 30. Bremnes R. M., Veve R., Gabrielson E., Hirsch F. R., Baron A., Bemis L., Gemmill R. M., Drabkin H. A., and Franklin W. A. (2002) High-throughput tissue microarray analysis used to evaluate biology and prognostic significance of the E-cadherin pathway in non-small-cell lung cancer. J. Clin. Oncol. 20, 2417–2428 [DOI] [PubMed] [Google Scholar]
- 31. Morabia A., and Wynder E. L. (1991) Cigarette smoking and lung cancer cell types. Cancer 68, 2074–2078 [DOI] [PubMed] [Google Scholar]
- 32. Selamat S. A., Galler J. S., Joshi A. D., Fyfe M. N., Campan M., Siegmund K. D., Kerr K. M., and Laird-Offringa I. A. (2011) DNA methylation changes in atypical adenomatous hyperplasia, adenocarcinoma in situ, and lung adenocarcinoma. PLoS One 6, e21443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Salvi S., Casadio V., Conteduca V., Lolli C., Gurioli G., Martignano F., Schepisi G., Testoni S., Scarpi E., Amadori D., Calistri D., Attard G., and De Giorgi U. (2016) Circulating AR copy number and outcome to enzalutamide in docetaxel-treated metastatic castration-resistant prostate cancer. Oncotarget 7, 37839–37845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miranda-Carboni G. A., Krum S. A., Yee K., Nava M., Deng Q. E., Pervin S., Collado-Hidalgo A., Galic Z., Zack J. A., Nakayama K., Nakayama K. I., and Lane T. F. (2008) A functional link between Wnt signaling and SKP2-independent p27 turnover in mammary tumors. Genes Dev. 22, 3121–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sakoda L. C., Loomis M. M., Doherty J. A., Julianto L., Barnett M. J., Neuhouser M. L., Thornquist M. D., Weiss N. S., Goodman G. E., and Chen C. (2012) Germ line variation in nucleotide excision repair genes and lung cancer risk in smokers. Int. J. Mol. Epidemiol. Genet. 3, 1–17 [PMC free article] [PubMed] [Google Scholar]
- 36. Wang Y., Wen M., Kwon Y., Xu Y., Liu Y., Zhang P., He X., Wang Q., Huang Y., Jen K. Y., LaBarge M. A., You L., Kogan S. C., Gray J. W., Mao J. H., and Wei G. (2014) CUL4A induces epithelial-mesenchymal transition and promotes cancer metastasis by regulating ZEB1 expression. Cancer Res. 74, 520–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ishida M., Ishida T., Tashiro S., Uchida H., Sakai C., Hironobe N., Miura K., Hashimoto Y., Arihiro K., Chayama K., Kihara Y., and Yoshizumi M. (2014) Smoking cessation reverses DNA double-strand breaks in human mononuclear cells. PLoS One 9, e103993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meyers D. G., and Neuberger J. S. (2008) Cardiovascular effect of bans on smoking in public places. Am. J. Cardiol. 102, 1421–1424 [DOI] [PubMed] [Google Scholar]
- 39. Cawthon R. M. (2002) Telomere measurement by quantitative PCR. Nucleic Acids Res. 30, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee J., Shieh J. H., Zhang J., Liu L., Zhang Y., Eom J. Y., Morrone G., Moore M. A., and Zhou P. (2013) Improved ex vivo expansion of adult hematopoietic stem cells by overcoming CUL4-mediated degradation of HOXB4. Blood 121, 4082–4089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hotakainen K., Ljungberg B., Haglund C., Nordling S., Paju A., and Stenman U. H. (2003) Expression of the free β-subunit of human chorionic gonadotropin in renal cell carcinoma: prognostic study on tissue and serum. Int. J. Cancer 104, 631–635 [DOI] [PubMed] [Google Scholar]
- 42. Camerini A., Donati S., Viacava P., Siclari O., Puccetti C., Tartarelli G., Valsuani C., De Luca F., Martini L., Cavazzana A., and Amoroso D. (2011) Evaluation of HER2 and p53 expression in predicting response to docetaxel-based first-line chemotherapy in advanced breast cancer. J. Exp. Clin. Cancer Res. 30, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. van Cruijsen H., Ruiz M. G., van der Valk P., de Gruijl T. D., and Giaccone G. (2009) Tissue microarray analysis of ganglioside N-glycolyl GM3 expression and signal transducer and activator of transcription (STAT)-3 activation in relation to dendritic cell infiltration and microvessel density in non-small cell lung cancer. BMC Cancer 9, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Idikio H. A. (2011) Quantitative analysis of p53 expression in human normal and cancer tissue microarray with global normalization method. Int. J. Clin. Exp. Pathol. 4, 505–512 [PMC free article] [PubMed] [Google Scholar]
- 45. Fujikawa H., Tanaka K., Toiyama Y., Saigusa S., Inoue Y., Uchida K., and Kusunoki M. (2012) High TrkB expression levels are associated with poor prognosis and EMT induction in colorectal cancer cells. J. Gastroenterol. 47, 775–784 [DOI] [PubMed] [Google Scholar]
- 46. Dai W., Sun C., Huang S., and Zhou Q. (2016) Carvacrol suppresses proliferation and invasion in human oral squamous cell carcinoma. OncoTargets Therapy 9, 2297–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]