Abstract
Recently, it was determined that representatives of the riboswitch candidates called ykkC and mini-ykkC directly bind free guanidine. These riboswitches regulate the expression of genes whose protein products are implicated in overcoming the toxic effects of this ligand. Thus, the relevant ykkC motif and mini-ykkC motif RNAs have been classified as guanidine-I and guanidine-II riboswitch RNAs, respectively. Moreover, we had previously noted that a third candidate riboswitch class, called ykkC-III, was associated with a distribution of genes similar to those of the other two motifs. Therefore, it was predicted that ykkC-III motif RNAs would sense and respond to the same ligand. In the current report, we present biochemical data supporting the hypothesis that ykkC-III RNAs represent a third class of guanidine-sensing RNAs called guanidine-III riboswitches. Members of the guanidine-III riboswitch class bind ligand with an affinity similar to that observed for members of the other two classes. Notably, there are some sequence similarities between guanidine-II and guanidine-III riboswitches. However, the characteristics of ligand discrimination by guanidine-III RNAs are different from those of the other guanidine-binding motifs, suggesting that the binding pockets have distinct features among the three riboswitch classes.
Keywords: aptamer, guanidine-III riboswitch, in-line probing, noncoding RNA, ykkC-III motif
INTRODUCTION
Orphan riboswitch candidates are well-conserved RNA motifs that have been proposed to be riboswitches, but whose ligands remain to be identified.1,2 The experimental validation of several orphan riboswitches recently has led to the discovery of novel riboswitch ligands, and has revealed new and unexpected areas of bacterial metabolism,3 cell signaling,4 metal ion homeostasis,5,6 and toxicity response mechanisms.7 Given the potential for novel biological insight to be gained, we became particularly interested in experimentally validating riboswitch candidates whose ligands have long remained undiscovered.
Recently, we determined that certain members of the ykkC orphan riboswitch class2,8 selectively recognize free guanidine and regulate various genes whose protein products overcome the toxic effects of this compound.9 As the number of distinct riboswitch classes has grown, it has become helpful to name riboswitch classes for the ligand they sense. Therefore members of this newly-validated RNA class have been renamed ‘guanidine-I’ riboswitches. Moreover, we had previously speculated that two additional RNA motifs, initially called mini-ykkC10 and ykkC-III,11 might also function as riboswitches that respond to the same ligand as guanidine-I RNAs.2,11 Indeed, we have evidence that that mini-ykkC motif RNAs also function as guanidine-sensing RNAs, hereafter called ‘guanidine-II’ riboswitches.12
Although ykkC-III RNAs share no sequence or structural characteristics with the ligand-binding aptamer domain of guanidine-I riboswitches, ykkC-III RNAs are commonly associated with the same types of genes. For example, the genes commonly associated with guanidine-I riboswitches and ykkC-III RNAs encode ‘small multidrug resistance’ (SMR) transporters, including those annotated as EmrE13 and SugE,14 that have been implicated as guanidine exporters.9 Furthermore, there are some similarities in the conserved sequences of ykkC-III motif RNAs and guanidine-II riboswitches. Specifically, many guanidine-II riboswitches are formed by two near-identical hairpins with conserved ‘ACGR’ loops. This same sequence also occurs once in the most highly-conserved portion of ykkC-III motif RNAs, although the sequence is not present in a hairpin loop.
Given the recent advances in guanidine riboswitch validation, and given the similarities noted above, we chose to further investigate the hypothesis that ykkC-III motif RNAs function as guanidine-sensing riboswitches. In the current report, we present our findings that confirm ykkC-III motif RNAs selectively bind guanidine, which indicates that they function as a distinct riboswitch class called ‘guanidine-III’. In addition, the biochemical attributes of guanidine-III riboswitches are compared and contrasted with representatives of guanidine-I and guanidine-II riboswitches.
MATERIALS AND METHODS
Chemicals and Oligonucleotides
All chemicals and chemically synthesized oligonucleotides were purchased from Sigma-Aldrich with the exception of guanidinosuccinic acid (Alfa Chemistry). [γ-32P]-ATP was purchased from PerkinElmer. Enzymes were purchased from New England BioLabs, unless otherwise noted. A complete list of DNA and RNA oligonucleotides used in this study can be found in Table S1.
Bioinformatics Analyses
Additional examples of ykkC-III RNAs were identified using Infernal 1.115 to search the RefSeq version 63 plus additional environmental microbial databases.16 Iterative searches for new sequences were performed based on the previously published alignment of the ykkC-III motif,11 revealing over 400 new examples. The consensus sequence and secondary structure model was constructed using R2R.17
RNA Oligonucleotide Preparation
5′ 32P-labeled RNA oligonucleotides were prepared as previously described.12 The only exception is the truncated 45 emrE RNA, for which synthetic RNA was chemically synthesized and subsequently purified and 5′ 32P labeled as previously described.
RNA In-line Probing Analyses
In-line probing assays were performed largely as previously described.18,19 Additional details can be found in the accompanying manuscript.12
RESULTS AND DISCUSSION
Updated Consensus Model and Expanded Gene Associations for ykkC-III Motif RNAs
Infernal, a homology search method,15 was used to identify examples of ykkC-III motif RNAs within fully-sequenced genomes and environmental sequences made available since the initial report of its discovery.11 This search revealed over 400 additional unique examples, with representatives mainly residing within Actinobacteria as previously reported. However, some representatives were also found in Proteobacteria, Deinococcus-Thermus, Planctomycetes, and Firmicutes.
A revised consensus sequence and structural model for the ykkC-III motif (Figure 1A), which is very similar to the model from the original report, was created by using the updated collection of representatives. As observed previously,11 the variable length pairing element (called P0) at the 5′ terminus of the motif lacks sequence conservation, and we identified numerous examples that appear to lack this substructure. Therefore, this variable hairpin is unlikely to be critical for the function of these RNAs. The majority of the nucleotides initially observed to be present in 97% of the representatives remain highly conserved in the updated model. Specifically, 18 very highly conserved nucleotides reside in three separate segments of the RNA, and 10 of these conserved nucleotides are predicted to reside in bulged regions. Most notably, the highly conserved ACGA sequence that is similar to the ACGR hairpin loop sequence in guanidine-II riboswitches resides in an unpaired region that joins stems P1 and P2.
Figure 1.
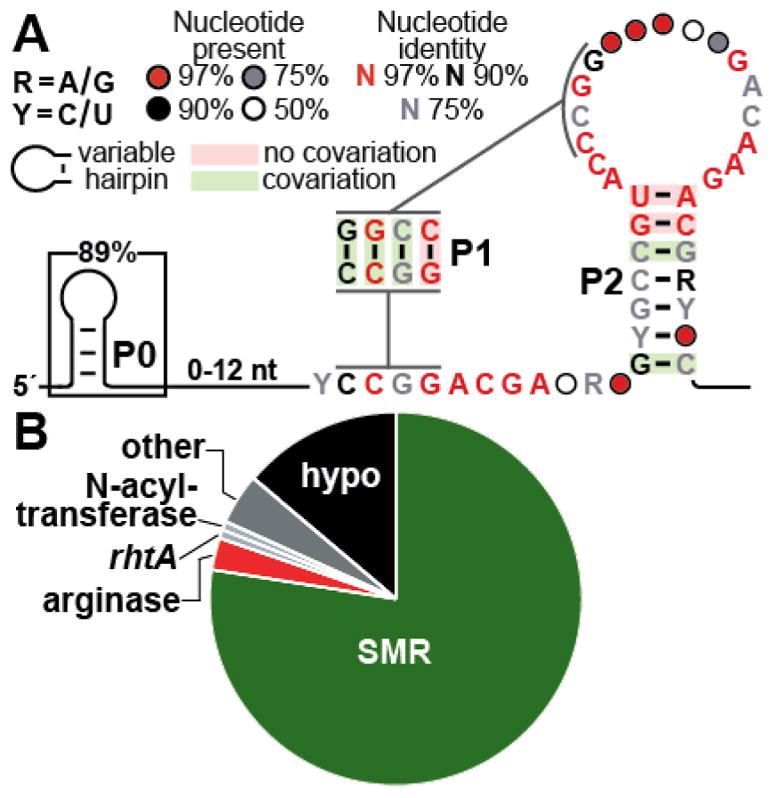
Structure and function of guanidine-III riboswitches. (A) The consensus sequence and structural model for the ~460 examples of ykkC-III motif (guanidine-III) RNAs. Covarying base pairs (indicated by green shading) are found in the P1 and P2 stems which form a pseudoknot as indicated by the gray lines. Many highly conserved (>97%) nucleotides are found in the loop regions between the stems. (B) Genes controlled by guanidine-III riboswitches, including a large number of predicted SMR transporters (green), arginases (red), and other (gray) or hypothetical (hypo, black) genes.
Similar to guanidine-II riboswitches, the conserved ykkC-III motif usually resides in the 5′-untranslated region (UTR) in close proximity to the ribosome binding site (RBS) and start codon of the adjacent open reading frame (ORF). Specifically, the purine-rich RBS is typically found just a few nucleotides downstream of the right shoulder of the P2 stem. Consequently, both guanidine-II riboswitches and the ykkC-III motif are predicted to function as genetic ‘ON’ switches that control the translation of the ORF by allowing ribosomes to access the RBS in the presence of ligand.
Genes encoding annotated EmrE and SugE transporters are most commonly associated with ykkC-III motif RNAs (Figure 1B). These transporters resemble those controlled by both guanidine-I and guanidine-II riboswitches, and fall into the category of SugE-like heterodimers (Randy Stockbridge, personal communication). Curiously, ykkC-III motif RNAs never associate with genes encoding the guanidine carboxylase protein, which was previously annotated as a urea carboxylase but was demonstrated to prefer guanidine as a substrate.9 The genes for guanidine carboxylase and its associated proteins are the most common types of protein-coding regions controlled by guanidine-I riboswitches, and are also frequently regulated by guanidine-II riboswitches.
Although ykkC-III motif RNAs lack associations with guanidine carboxylase genes, all three RNA classes share other gene associations that are consistent with the hypothesis that they sense the same ligand. One example is arginase, which is predicted to convert arginine to urea and ornithine. The role played by this enzyme in guanidine metabolism is not clear, but there are multiple possibilities.9 Finally, both guanidine-I riboswitches and ykkC-III motif RNAs are occasionally associated with a class of genes belonging to the superfamily of N-acyltransferases, again suggesting that the ligand for these RNA classes might be identical.
Guanidine is Bound by ykkC-III Motif RNAs With Affinities Similar to Guanidine-I and -II Riboswitch Aptamers
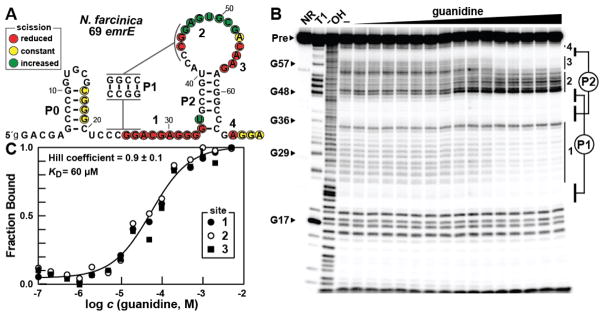
To assess the ability of ykkC-III motif RNAs to bind guanidine, we created an RNA construct carrying 69 nucleotides encompassing the representative located upstream of the emrE gene from Nocardia farcinica (Figure 2A). Although this and other ykkC-III motif RNAs are associated with genes annotated as emrE, the genes actually code for SugE-like members of the SMR protein class. This RNA construct, called 69 emrE, was subjected to in-line probing18,19 to identify any changes in structure upon incubation with guanidine. Robust structural modulation is observed (Figure 2B) primarily within the regions that also exhibit high sequence conservation, whereas regions that lack sequence or structure conservation remain largely unstructured, or even become less structured, in the presence of guanidine.
Figure 2.
Guanidine is bound by a guanidine-III RNA in vitro. (A) The sequence and observed secondary structure of the 69 emrE RNA from N. farcinica. Lowercase 5′ nucleotides indicate G residues added to enable transcription by T7 RNA Polymerase. Red, yellow, and green circles indicate nucleotides that undergo decreased, constant, or increased scission, respectively, upon addition of the ligand as determined by their relative band intensities in B. Regions of guanidine-mediated structural modulation numbered to correspond to those in B and C. (B) Polyacrylamide gel electrophoresis (PAGE) analysis of in-line probing reactions of 69 emrE RNA in the absence (−) and presence of guanidine ranging from 100 nM to 5 mM. NR, T1, and −OH indicate no reaction, RNA partially digested with T1 ribonuclease (cleavage after each G), and RNA partially digested under alkaline conditions (cleavage after every nucleotide). Precursor RNA (Pre) as well as certain G residues within the RNA sequence are indicated and regions of modulation are numbered and indicated with vertical bars. Base pairing of the pseudoknotted P1 and P2 stems is depicted. (C) Plot of the fraction of RNA bound to ligand versus the logarithm of the guanidine concentration as determined by quantification of B (see reference 12 for details). The KD and Hill coefficient were determined by the sigmoidal fit and the reported error represents the standard deviation reported in the fitting parameters.
A plot of the relative increase or decrease in spontaneous RNA scission at positions in three of the areas of modulation reveals a curve that is consistent with a 1:1 binding interaction and a dissociation constant (KD) of 60 μM (Figure 2C). These are characteristics similar to those observed for guanidine-I riboswitches, and are consistent with our hypothesis that ykkC-III motif RNAs represent a third guanidine riboswitch class.
Interestingly, nucleotides spanning position 8 to 20, which have the potential to form part of the P0 hairpin, exhibit an RNA cleavage pattern that is not consistent with hairpin formation as presented in our secondary structure model. This result, along with bioinformatics data indicating that the P0 stem is sometimes absent, suggests that this stem is not necessary for ligand binding. To investigate this possibility, a truncated version of the 69 emrE RNA, called 45 emrE (Figure S1A) was examined. The 45 emrE construct, which begins just one nucleotide prior to the P1 stem, is able to bind guanidine and modulates in similar regions as the full length construct (Figure S1B). P0 deletion only slightly diminishes the binding affinity, with a dissociation constant of 200 μM (Figure S1C). Thus, it appears that the P0 stem of this guanidine-III aptamer only modestly enhances the affinity for the ligand, which is similar to that observed when a poorly conserved P0 stem of the pre-Q1-I riboswitch is removed.20 However, this stem might be more important to other guanidine-III riboswitch representatives, or might serve other biological or biochemical roles in its natural setting.
To further validate the guanidine-III riboswitch class, three additional representative sequences were evaluated for guanidine binding. Aptamers associated with the arginase gene from Mycobacterium kansasii (65 arg, Figure S2), the smr gene from Streptomyces bingchenggensis (64 smr, Figure S3), and the emrE gene from Legionella pneumophila (71 emrE, Figure S4) all exhibit structural modulation in similar regions with 1:1 binding characteristics and dissociation constants of 100, 200, and 20 μM, respectively. Again, the binding affinities of these guanidine-III riboswitches are similar to those of guanidine-I9 and guanidine-II RNAs.12
Mutation of Conserved Nucleotides in Guanidine-III Riboswitch Aptamers Abolishes Ligand Binding
Single mutations were introduced separately at nine highly-conserved positions in the 69 emrE RNA (Figure S5A) to investigate the importance of these nucleotides for guanidine recognition. Each mutation causes the loss of the ability of the RNA to recognize guanidine at concentrations as high as 1 mM (Figure S5B). Some mutations, such as M1 and M9, appear to vastly alter the RNA structure, which explains the loss of ligand binding function. However, other mutations such as M2 through M6 yield folded RNAs that appear to have the same overall architecture, but these RNAs are unable to recognize guanidine at concentrations as much as 20-fold above the KD of the unaltered aptamer.
These results suggest that guanidine binding is due to specific interactions, rather than nonspecific interactions between the negatively-charged RNA and positively-charged guanidinium ions. Similarly, the conserved nucleotides present in guanidine-I riboswitch aptamers appear to serve important roles in forming the guanidine binding pocket, either by establishing the global architecture of the RNA or by directly interacting with the cognate ligand. Therefore, we predict that the highly-conserved nucleotides of guanidine-III riboswitches will be engaged in directly binding the ligand, or in establishing the global architecture necessary to form the guanidine-binding pocket.
Selective Ligand Binding by Guanidine-III Aptamers
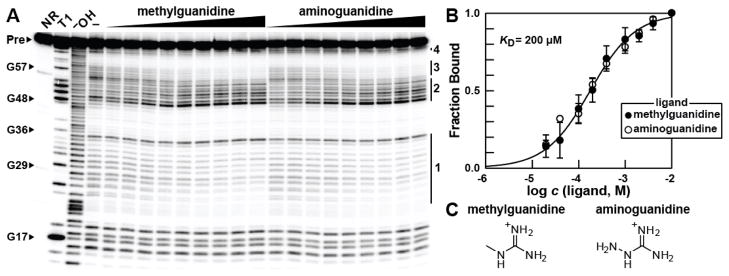
The ability of guanidine-III RNAs to discriminate against chemical analogs of guanidine was also evaluated by using in-line probing (Figure 3, Figure S6). Small substituents on a single nitrogen atom of guanidine are reasonably well tolerated, as is evident by examining the binding characteristics of methylguanidine and aminoguanidine (Figure 3). These two analogs modulate RNA structure in a manner that is identical to that observed for guanidine. Moreover, both analogs exhibit a KD of 200 μM, which is only ~3-fold poorer than the value measured for guanidine. Larger substituents or modifications to multiple nitrogen atoms are strongly rejected. These results suggest that the RNA forms a binding pocket and exploits a steric strategy to limit the size of any substituent attached to the guanidine moiety. Most notably, urea (KD poorer than 10 mM) is rejected by the ligand binding pocket with a discrimination factor of greater than 100 fold. This finding suggests that the presence of all three nitrogen atoms and their associated hydrogens, and perhaps also the positive charge of guanidinium, are key ligand recognition determinants whose absence in other natural guanidine analogs such as urea is deleterious to binding.
Figure 3.
Guanidine-III aptamers can discriminate against even close analogs of guanidine. (A) PAGE analysis of in-line probing reactions of 69 emrE RNA from N. farcinica in the absence (−) or presence of methylguanidine or aminoguanidine ranging from 1 μM to 40 mM. Additional annotations are as described in the legend to Figure 2B. (B) Plot of the fraction of RNA bound to ligand versus the logarithm of the methyl- or aminoguanidine concentration as determined by quantification of the data in A (see reference 12 for details). Each point represents the average and standard deviation of the fraction bound by assessing three sites of structure modulation (regions 1, 2, and 3, as indicated in Figure 2A). Only the fit calculated for methylguanidine binding is shown for clarity, although both fits yield an approximately identical KD. The resulting curves are consistent with a 1:1 binding interaction. (C) Chemical structures of methylguanidine and aminoguanidine.
These and other characteristics of the 69 emrE guanidine-III RNA are somewhat different than those observed for a guanidine-II riboswitch RNA. Although similarities in nucleotide sequences exist between guanidine-II and -III riboswitches, namely the conserved ACGR segments, high-resolution structural models will be needed to determine if the two classes form guanidine-binding pockets that are similar in structure. In contrast, ligand binding by guanidine-I riboswitches is much more selective than these other two classes. A representative of guanidine-I riboswitches was shown to more strongly discriminate against every guanidine analog tested.9 These differences in ligand selectivity between class I versus class II and III guanidine riboswitches might explain why guanidine-I riboswitches are more common in bacteria.
Concluding Remarks
The guanidine-III riboswitch class, frequently found in species of Actinobacteria, is abundant but less common than the guanidine-I and -II riboswitch classes. The genes most commonly controlled by guanidine-III RNAs mostly overlap with those of guanidine-I RNAs. However, some rarer and many hypothetical genes are uniquely associated with guanidine-III riboswitches. This fact prompts interesting questions for researchers who seek a deeper understanding of proteins that affect the metabolism of free guanidine. For example, one gene type associated with guanidine-III riboswitches is rhtA, which is annotated as encoding a threonine/homoserine efflux transporter. However, it seems possible that transporter genes associated with this riboswitch might have altered their original ligand specific to now recognize guanidine, as was the case for some transporters associated with fluoride riboswitches that converted from chloride to fluoride selectivity.7 To expand insight regarding guanidine metabolism, one could further examine the biochemical functions of the genes associated with all guanidine riboswitches.
Experimental validation of the ykkC-III motif as a third class of guanidine riboswitches again indicates that free guanidine is an important metabolite in bacteria, but its role is almost completely unknown. Every cognate ligand for the known riboswitches to date represents a fundamentally important molecule or ion for bacteria and often also for organisms from all three domains of life. A major hurdle in identifying the cognate ligand for all three ykkC motif RNAs was the lack of known roles for free guanidine in biology. Thus, the discovery and validation of three distinct classes of guanidine-responsive riboswitches has also revealed free guanidine as an important, but largely unexplored, compound in many species of bacteria.
Acknowledgments
We thank Adam Roth, Zasha Weinberg, Ruben Atilho, James Nelson and other members of the Breaker laboratory as well as Randy Stockbridge of the University of Michigan for helpful discussions. M.E.S. was supported by an NIH Cellular and Molecular Biology Training Grant (T32GM007223). This work was also supported by NIH grants (GM022778, DE022340) as well as funds from Howard Hughes Medical Institute to R.R.B.
Footnotes
ASSOCIATED CONTENT
Supporting Information (.doc)
References
- 1.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer MM, Hammond MC, Salinas Y, Roth A, Sudarsan N, Breaker RR. Challenges of ligand identification for riboswitch candidates. RNA Biol. 2011;8:5–10. doi: 10.4161/rna.8.1.13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim PB, Nelson JW, Breaker RR. An ancient riboswitch class in bacteria regulates purine biosynthesis and one-carbon metabolism. Mol Cell. 2015;57:317–328. doi: 10.1016/j.molcel.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson JW, Sudarsan N, Furukawa K, Weinberg Z, Wang JX, Breaker RR. Riboswitches in eubacteria sense the second messenger c-di-AMP. Nat Chem Biol. 2013;9:834–839. doi: 10.1038/nchembio.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dambach M, Sandoval M, Updegrove TB, Anantharaman V, Aravind L, Waters LS, Storz G. The ubiquitous yybP-ykoY riboswitch is a manganese-responsive regulatory element. Mol Cell. 2015;57:1099–1109. doi: 10.1016/j.molcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Price IR, Gaballa A, Ding F, Helmann JD, Ke A. Mn2+-sensing mechanisms of yybP-ykoY orphan riboswitches. Mol Cell. 2015;57:1110–1123. doi: 10.1016/j.molcel.2015.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker JL, Sudarsan N, Weinberg Z, Roth A, Stockbridge RB, Breaker RR. Widespread genetic switches and toxicity resistance proteins for fluoride. Science. 2012;335:233–235. doi: 10.1126/science.1215063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barrick JE, Corbino KA, Winkler WC, Nahvi A, Mandal M, Collins J, Lee M, Roth A, Sudarsan N, Jona I. New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc Natl Acad Sci USA. 2004;101:6421–6426. doi: 10.1073/pnas.0308014101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nelson JW, Atilho RM, Sherlock ME, Stockbridge RB, Breaker RR. Metabolism of free guanidine in bacteria is regulated by a widespread riboswitch class. Mol Cell. 2017 doi: 10.1016/j.molcel.2016.11.019. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weinberg Z, Barrick JE, Yao Z, Roth A, Kim JN, Gore J, et al. Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 2007;35:4809–4819. doi: 10.1093/nar/gkm487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy RH, Breaker RR. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaea, and their metagenomes. Genome Biol. 2010;11:31. doi: 10.1186/gb-2010-11-3-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sherlock ME, Malkowski SN, Breaker RR. Biochemical validation of a second guanidine riboswitch class in bacteria. Biochemistry. 2017 doi: 10.1021/acs.biochem.6b01270. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schuldiner SS. EmrE, a model for studying evolution and mechanism of ion-coupled transporters. Biochim Biophys Acta. 2009;1794:748–762. doi: 10.1016/j.bbapap.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 14.Jack DL, Storms ML, Tchieu JH, Paulsen IT, Saier MH., Jr A broad-specificity multidrug efflux pump requiring a pair of homologous SMR-type proteins. J Bacteriol. 2000;182:2311–2313. doi: 10.1128/jb.182.8.2311-2313.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nawrocki EP, Eddy SR. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lünse CE, Breaker RR. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol. 2015;11:606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinberg Z, Breaker RR. R2R-software to speed the depiction of aesthetic consensus RNA secondary structures. BMC Bioinformatics. 2011;12:1. doi: 10.1186/1471-2105-12-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regulski EE, Breaker RR. In-line probing analysis of riboswitches. Methods Mol Biol. 2008;419:53–67. doi: 10.1007/978-1-59745-033-1_4. [DOI] [PubMed] [Google Scholar]
- 19.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwik A, Kim JN, Welz R, et al. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol. 2007;14:308–317. doi: 10.1038/nsmb1224. [DOI] [PubMed] [Google Scholar]