Abstract
Background
Diabetic cardiomyopathy is a major risk factor in diabetic patients but its pathogenesis remains poorly understood. The ubiquitin-proteasome system (UPS) facilitates protein quality control by degrading unnecessary and damaged proteins in eukaryotic cells, and dysfunction of UPS is implicated in various cardiac diseases. However, the overall functional status of the UPS and its pathophysiological role in diabetic cardiomyopathy have not been determined.
Methods and results
Type I diabetes was induced in wild-type and transgenic mice expressing a UPS functional reporter (GFPdgn) by injections of streptozotocin (STZ). STZ-induced diabetes progressively impaired cardiac UPS function as evidenced by the accumulation of GFPdgn proteins beginning two weeks after diabetes induction, and by a buildup of total and lysine (K) 48-linked polyubiquitinated proteins in the heart. To examine the functional role of the UPS in diabetic cardiomyopathy, cardiac overexpression of PA28α (PA28αOE) was used to enhance proteasome function in diabetic mouse hearts. PA28αOE diabetic mice displayed exhibited restoration of cardiac UPS function, as demonstrated by the diminished accumulation of GFPdgn and polyubiquitinated proteins. Moreover, PA28αOE diabetic mice exhibited reduced myocardial collagen deposition, decreased cardiomyocyte apoptosis, and improved cardiac systolic and diastolic function.
Conclusion
Impairment of cardiac UPS function is an early event in STZ-induced diabetes. Overexpression of PA28α attenuates diabetes-induced proteotoxic stress and cardiomyopathy, suggesting a potential therapeutic role for enhancement of cardiac proteasome function in this disorder.
Keywords: PA28α, ubiquitin, proteasome, diabetic cardiomyopathy
1. Introduction
Diabetes mellitus remains a major health problem worldwide and is associated with a high rate of mortality and morbidity due cardiovascular complications [1]. Diabetic cardiomyopathy, defined as ventricular dysfunction in the absence of coronary artery disease and hypertension, is one major risk factor among others in diabetic patients [2, 3]. Manifestations of diabetic cardiomyopathy include myocyte hypertrophy, interstitial fibrosis, cardiomyocyte dropout, and decreased diastolic and systolic function [4, 5]. Research into the pathophysiology underlying the progression of diabetic cardiomyopathy to heart failure has identified increased oxidative stress, altered substrate metabolism, mitochondrial dysfunction, impaired calcium homeostasis and upregulation of the renin-angiotensin system as possible contributory factors [4–6]. However, the precise molecular and cellular mechanisms underpinning its pathogenesis remain to be elucidated.
The ubiquitin proteasome system (UPS) maintains protein homeostasis by timely degradation of unnecessary and/or damaged proteins such as terminally misfolded proteins and oxidized proteins. UPS-mediated protein degradation generally involves two steps, the ubiquitin-specific E1-E2-E3 enzyme-mediated covalent attachment of ubiquitin to protein targets (i.e., ubiquitination) and degradation of the modified targets by the proteasome [7, 8]. Eukaryotic cells have several types of proteasome complexes, which are generally comprised of a 20S core proteasome (20S) and different regulatory complexes such as the 19S proteasome (19S) and the 11S proteasome (11S), to carry out protein degradation. While the 19S-20S-19S complexes, known as the 26S proteasome, are responsible for selective degradation of most intracellular proteins in a ubiquitin- and ATP-dependent manner, the 11S-activated proteasome is generally believed to mediate protein degradation in a ubiquitin- and ATP-independent manner [7]. The 11S proteasome can be formed by either PA28α and PA28β heteroheptamers (α3β4 or α4β3) or by PA28γ in homoheptamers (γ7).
As a major player of intracellular protein quality control, the UPS is essential to the survival and function of most eukaryotic cells including cardiomyocytes. Dysfunction of the UPS disrupts protein homeostasis and has been implicated in human cardiomyopathies and various forms of murine cardiomyopathies including desmin-related, hypertrophic and ischemic cardiomyopathy[9–11]. UPS dysfunction in these diseased hearts is typically manifested by increased ubiquitinated proteins, decreased proteasomal peptidase activities, oxidative damage to proteasome subunits and aberrant protein aggregates, indicating a proteolytic demand-supply imbalance. Emerging evidence suggests that impairment of UPS function is sufficient to cause cardiomyopathy and heart failure or to expedite maladaptive cardiac remodeling[12–15]. Moreover, improvement of proteasome function by overexpression of 11S proteasome subunit PA28α was shown to protect the heart against pathological stresses in experimental models of myocardial ischemia reperfusion, desmin-related cardiomyopathy and right ventricle hypertrophy[16, 17]. These lines of compelling evidence suggest that UPS dysfunction may be a common mechanism in the pathogenesis of diverse cardiac diseases.
Circumstantial data links UPS dysfunction to diabetic cardiomyopathy. Alterations in the expression of UPS components, changes in proteasomal peptidase activities and increased ubiquitinated and oxidized proteins have been detected in the hearts of different diabetes models[18–20]. However, the effects of diabetes on the overall cardiac UPS function and its pathophysiological role in diabetic cardiomyopathy have not been examined. Using an in vivo UPS functional reporter in combination with other biochemical analyses, we have unveiled here that cardiac UPS function is impaired soon after the onset of type I diabetes and further deteriorates with diabetes progression. Moreover, enhancement of cardiac proteasome function by overexpression of PA28α alleviated the proteotoxic stress in diabetic hearts and attenuated cardiac dysfunction. Our findings suggest that insufficient proteasome function is a novel mechanism underlying the development of diabetic cardiomyopathy.
2. Methods
2.1 Animals
The transgenic mouse models expressing a tetracycline-controlled transactivator protein (tTA), PA28α or GFPdgn were previously described[17]. These mice were maintained in the FVB/N inbred background for our studies. Eight- to 10-week-old male mice were injected intraperitoneally with streptozotocin (STZ, 50 mg/Kg, Sigma-Aldrich) for five consecutive days to induce type I diabetes. Animals with fasting blood glucose levels greater than 250 mg/dl at day 7 post the first STZ injection were deemed diabetic and included in the studies. All animal experiments were approved by the Augusta University Institutional Animal Care and Use Committee.
2.2 Immunoblotting, fluorescence confocal microscopy and histology staining
Proteins were extracted from ventricular myocardium tissues or cultured cells and subjected to SDS-PAGE as previously described [21]. Membranes were incubated overnight at 4°C with antibodies against GFP (sc-9996, 1:5000, Santa Cruz), ubiquitin (#04-263, 1:1000, Sigma-Aldrich), K48-ubiquitin (#8081, 1:1000, Cell Signaling Technology), GAPDH (SAB1405848, 1:6000, Sigma-Aldrich), β-tubulin (E7, 1:5000, Developmental Studies Hybridoma Bank) and PA28α (customized[22]).
Fluorescence confocal microscopy was performed as described previously[21]. Direct GFP fluorescence on O.C.T.-embedded ventricular myocardium sections (5 μm) was visualized using a Zeiss LSM 510 upright confocal microscope (Zeiss). The images were captured with the same PMT voltage setting among groups and digitalized using the associated software.
Histological analyses were performed using 5-μm paraffin-embedded ventricular myocardium sections. Cardiac fibrosis was assessed by Masson’s Trichrome staining using the Trichrome Stain kit (American MasterTech). Data were quantified by Image-Pro Plus (Media Cybernetics).
2.3 Determination of proteasome peptidase activity
Proteasome peptidase activity was quantified as previously described [23]. Briefly, myocardial tissues were homogenized at 4°C in 10 volumes of HEPES buffer (50 mmol/L, pH 7.5). The supernatants of the homogenates were immediately used for protein concentration assay followed by determination of peptidase activities. The following synthetic fluorogenic peptides: Suc-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (AMC) (18 μmol/L, Enzo Life Sciences), Z-Leu-Leu-Glu-AMC (45 μmol/L, Enzo Life Sciences), and Ac-Arg-Leu-Arg-AMC (40μmol/L, Enzo Life Sciences) were used respectively for measuring chymotrypsin-like, caspase-like, and trypsin-like peptidase activities in the absence or presence of a proteasome inhibitor, MG-132 (20 μmol/L, for chymotrypsin-like and caspase-like activity) or Epoxomicin (5 μmol/L, for trypsin-like activity), and in the absence or presence of ATP (28 μmol/L), respectively. Measurements of each specimen were performed in triplicates. Twenty micrograms of crude protein extracts were added to 200 μl of the HEPES buffer containing the fluorogenic substrate to each well in 96-well plates, and incubated at 37°C. The fluorescence intensity was measured after 60 min of incubation using a Synergy H4 plate reader (Bio-Tek) with excitation wave length of 380 nm and emission wave length at 460 nm. The portion of peptidase activity inhibited by the proteasome inhibitor was attributed to the proteasome.
2.4 Pressure-volume loop analysis
For the invasive assessment of left ventricular (LV) pressure–volume (P–V) relationship, mice were anesthetized with isoflurane (2%) in medical grade oxygen, intubated and artificially ventilated. Body temperature was monitored with a rectal thermometer and maintained at 37°C. Retrograde catheterization of the LV was performed via the right carotid artery with a 1.2-F mouse P–V catheter (Millar). The instrumented animal was stabilized for 10min, data were recorded at a sampling rate of 1,500 Hz during steady-state conditions, and P–V loops were constructed using LabChart8 (ADInstruments). The raw conductance volumes were corrected for parallel conductance by the hypertonic saline bolus.
2.5 RNA preparation and quantitative real-time polymerase chain reaction (qRT-PCR)
Isolation of total RNA and reverse transcription into single-stranded cDNA were performed as previously described[21]. Gene expression levels were measured in triplicate per biological sample by real-time PCR (StepOnePlus Real-Time PCR system, Life Technologies) using the SYBR-Green assay with gene-specific primers at a final concentration of 200 nM. The following primers were used: GFP-forward: GGGCACAAGCTGGAGTACAACT, GFP-reverse: ATGTTGTGGCGGATCTTGAAG. Relative gene expression was calculated using the 2−ΔΔct method against a mouse house-keeping gene hypoxanthine guanine phosphoribosyl transferase (Hprt).
2.6 Terminal dUTP nick end-labeling (TUNEL) assays
Myocardial cryosections were fixed with 4% paraformaldehyde and rinsed with cold PBS. TUNEL staining was performed using the In Situ Cell Death Detection Kit (Sigma-Aldrich) according to the manufacturer’s instruction. To identify cardiomyocytes, the sections were counterstained with Alexa Fluor 568-conjugated phalloidin and DAPI. The number of TUNEL-positive cardiomyocyte nuclei and the total number of cardiomyocyte nuclei were counted by examining the entire section.
2.7 Statistical Analysis
Data were expressed as mean ± SD unless specified. Data between 2 groups were compared using unpaired t tests. A probability value of P<0.05 was considered to indicate statistical significance.
3. Results
3.1 Evaluation of a UPS functional reporter in STZ-induced diabetic mouse hearts
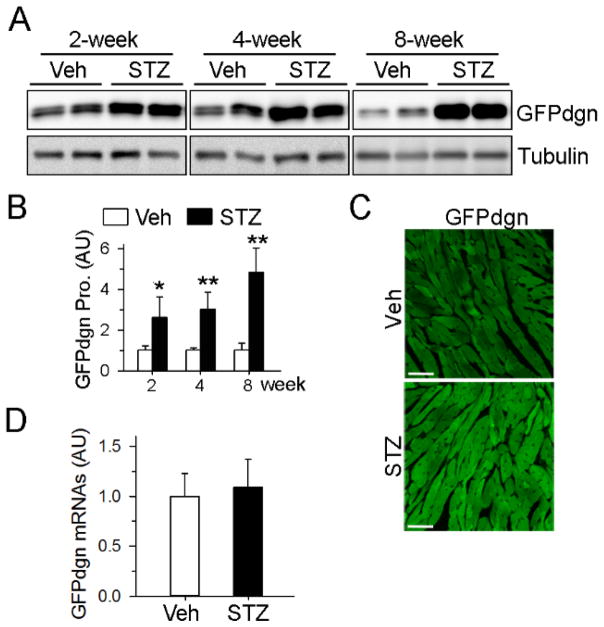
Various short-lived reporter proteins have been widely employed as sensitive and reliable tools to monitor the functional status of the UPS[24]. We have previously created a transgenic mouse model with ubiquitous expression of a UPS functional reporter GFPdgn (GFP fused with CL1 degron signal), in which GFPdgn protein levels inversely reflect UPS function. We and others have used this model to successfully measure UPS function in vivo under various experimental disease settings[23, 25, 26]. To assess cardiac UPS function following the onset of diabetes, we injected STZ into 8-week-old GFPdgn transgenic mice. Diabetes was successfully established, as indicated by prompt and sustained elevation of blood glucose levels (Supplemental Fig. 1A). As expected, the diabetic mice displayed loss of body weight and heart weight after STZ injections (Supplemental Fig. 1B–1C). Western blot analysis revealed a 2-fold increase in cardiac GFPdgn protein levels at 2 weeks post STZ injection (Fig. 1A–1B), which increased further to 3- and 4-fold at 4 and 8 weeks after STZ injections, respectively, compared with the non-diabetic mice. The increase in GFPdgn proteins in the cardiomyocytes of diabetic mice was further confirmed by confocal fluorescent microscopy (Fig. 1C). We did not detect any change in GFPdgn transcripts in diabetic hearts by quantitative real-time PCR (Fig. 1D), suggesting that the increase in GFPdgn proteins was not due to upregulation of GFPdgn transcription.
Figure 1.
Accumulation of GFPdgn proteins in hearts of diabetic mice. A, Western blots showed progressively increased cardiac GFPdgn levels after diabetes induction. B, Quantification of (A), n=4 per group. *P<0.05, ** P<0.01 versus vehicle (Veh). C, Confocal microscopy analysis revealed increased direct GFP fluorescence (green) in myocardial sections 4 weeks after STZ injections. Bar, 50 μm. D, Quantitative real-time PCR analysis showed no change in levels of GFPdgn transcripts in diabetic hearts 2 weeks after STZ treatment.
3.2 Accumulation of ubiquitinated proteins in diabetic hearts
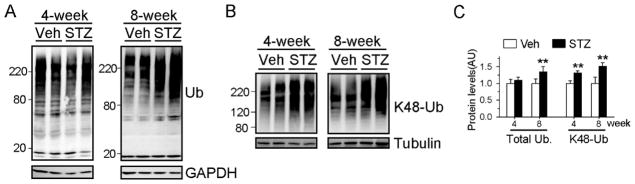
Insufficient UPS function can be associated with accumulation of ubiquitinated proteins, especially those tagged by a lysine 48 (K48)-linked ubiquitin chain[27]. Consistent with GFPdgn reporter protein data, levels of K48-linked ubiquitinated proteins in the heart were significantly increased 4 weeks after STZ injection (Fig. 2A–2C). Notably, an increase in total ubiquitinated proteins in the heart was not detected until 8 weeks post STZ injection (Fig. 2B–2C).
Figure 2.
Accumulation of ubiquitinated proteins in hearts of diabetic mice. Diabetes was induced in a cohort of non-transgenic FVB/N mice with STZ injections. A–B, Western blot analysis of total (A) and lysine 48 (K48)-linked (B) ubiquitinated proteins (Ub) in myocardial homogenates at the indicated times after STZ treatment. C, Quantification of (A) and (B). ** P<0.01 versus Veh at the respective time points.
3.3 Temporal changes of proteasomal activities in diabetic hearts
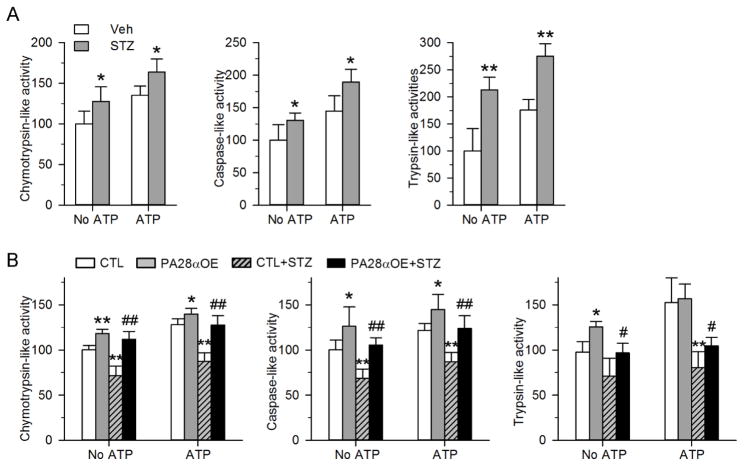
To understand how diabetes influences cardiac UPS function, we measured cardiac proteasome peptidase activity using small fluorogenic peptide substrates. Since ATP was found to stimulate peptidase activity[28], we performed the assay in both the absence and presence of ATP. Interestingly, we observed significant upregulation of chymotrypsin-like, caspase-like and trypsin-like activities in the hearts of STZ-treated mice at one month after diabetes induction, which was maintained in the presence of ATP (Fig. 3A). However, at two months after diabetes induction, all three peptidase activities in diabetic hearts showed 30–40% reduction when compared with those in non-diabetic hearts (Fig. 3B). The temporal kinetics suggest that cardiac proteasome activity is induced in the early stage of diabetes but subsequently becomes impaired with diabetes progression.
Figure 3.
Evaluation of proteasome chymotrypsin-like, caspase-like and trypsin-like activities in myocardium homogenates from the indicated mice at 1 month (A) and 2 months (B) post STZ treatment, in the absence or presence of ATP. The activity in individual groups was normalized to that of non-diabetic hearts in the absence of ATP. N=4–6 mice per group. * P<0.05, ** P<0.01 versus Veh or CTL. # P<0.05, ## P<0.01 versus CTL+STZ.
Cumulatively, the data demonstrate progressive and severe impairment of UPS proteolytic function in diabetic hearts, which is partially ascribed to the diminished proteasome function. Importantly, since cardiac dysfunction in STZ-induced diabetic mice is typically not detected until 2 to 3 months after diabetes induction (Supplemental Fig. 1D), the accumulation of GFPdgn and polyubiquitinated proteins at an early stage of diabetes indicates that insufficient UPS function precedes the onset of diabetic cardiomyopathy and thus potentially could play a pathogenic role in the disease.
3.4 PA28a overexpression enhances cardiac proteasome activities
The temporal changes of proteasome activity in diabetic hearts prompted us to hypothesize that the initial upregulation of proteasome function is an adaptive response to the onset of diabetic cardiomyopathy, while the subsequent reduction is maladaptive. To determine if UPS malfunction is causal in the development of diabetic cardiomyopathy, we employed a mouse model with cardiomyocyte-restricted proteasome function enhancement induced by overexpression of PA28α (PA28αOE). PA28αOE is achieved by crossing Tet trans-activator (tTA) transgenic mice with a responder transgenic line carrying PA28α transgene [17]. We have previously demonstrated that PA28αOE increased the 11S proteasome complex, elevated proteasome activity, enhanced UPS proteolytic function, and attenuated desmin-related cardiomyopathy and ischemia/reperfusion injury [17]. Given the diminished proteasome function in diabetic hearts (Fig. 3B), we first determined whether PA28αOE can restore cardiac proteasome function. Consistent to our previous findings in cultured cardiomyocytes [22], PA28αOE mouse hearts exhibited enhanced proteasome function at baseline as demonstrated by ~20–30% increase in all three proteasomal peptidase activities when compared with tTA transgenic (control) mice (Fig. 3B). Moreover, PA28αOE diabetic hearts displayed significantly improved proteasomal peptidase activities compared to diabetic tTA transgenic hearts, at levels comparable to non-diabetic tTA transgenic hearts and non-diabetic PA28αOE hearts (Fig. 3B). These data suggest PA28αOE enhances cardiac proteasome activity at baseline and preserves its function under diabetic conditions.
3.5 PA28α overexpression restores cardiac UPS function of diabetic mice
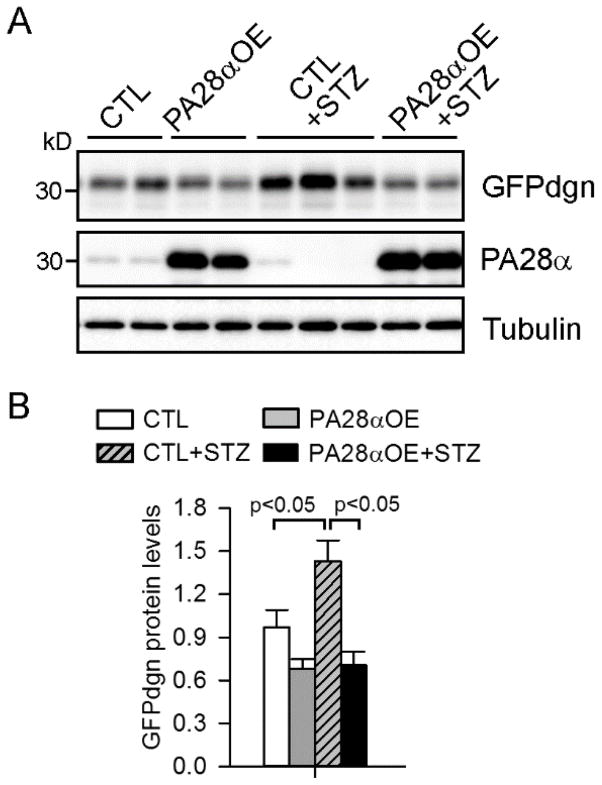
We next asked whether enhancement of proteasome activity in cardiomyocytes by PA28αOE suffices to normalize diabetes-impaired UPS function. We first introduced the GFPdgn transgene into tTA or PA28αOE mice and induced diabetes in the resultant tTA/GFPdgn double transgenic mice and tTA/PA28α/GFPdgn triple transgenic mice by STZ injections. PA28αOE diabetic mice showed significantly attenuated GFPdgn accumulation in the heart compared with control diabetic mice (Fig. 4A–4B), despite the comparable levels of GFPdgn transcripts (data not shown). We also induced diabetes in a separate cohort of tTA (controls, CTL) and PA28αOE transgenic mice and observed a significant reduction of both K48-linked ubiquitinated proteins and total polyubiquitinated proteins in diabetic PA28αOE mouse hearts compared with controls (Fig. 5A–5B). Moreover, immunostaining of myocardium sections with ubiquitin antibodies identified fewer protein aggregates in the hearts of PA28αOE diabetic mice (Fig. 5C). Taken together, these findings suggest that PA28αOE recovers the loss of UPS function in the hearts of diabetic mice.
Figure 4.
PA28αOE decreased GFPdgn accumulation in the hearts of diabetic mice. GFPdgn transgene was introduced into CTL and PA28αOE mice. Diabetes was then induced by STZ injection into the resultant CTL/GFPdgn and PA28αOE/GFPdgn mice 8 weeks after STZ injection. A–B, Western blots (A) of the indicated proteins in myocardial homogenates and the quantification (B) of GFPdgn proteins. n=3–4 mice per group.
Figure 5.
PA28αOE reduced ubiquitinated proteins in the hearts of diabetic mice. A–B, Western blots (A) and the quantification (B) of total ubiquitinated and K48-linked ubiquitinated proteins in myocardial homogenates 8 weeks after STZ injection. C, Representative confocal micrographs of myocardial sections stained with ubiquitin (green) antibody. Ubiquitin-positive protein aggregates (arrowheads) were evident in STZ+CTL hearts but were markedly reduced in STZ+PA28αOE hearts. Bar, 20 μm.
3.6 Overexpression of PA28α ameliorates diabetes-induced pathological cardiac remodeling
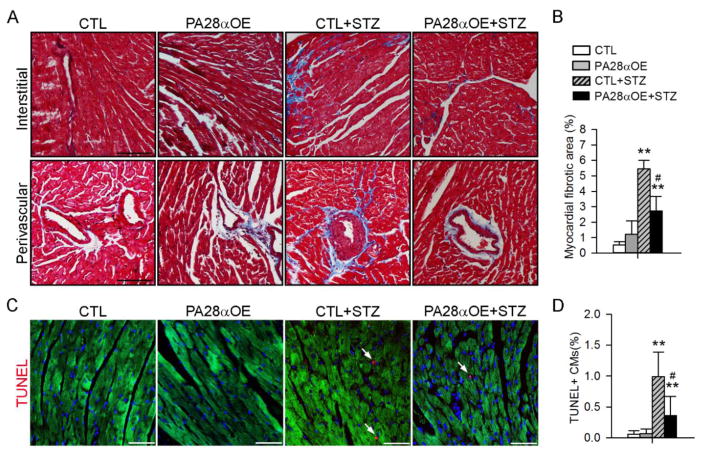
Having shown that PA28αOE restores UPS function in diabetic hearts, we next determined the impact of PA28αOE on the development of diabetic cardiomyopathy, which is characterized by fibrosis, reactivation of fetal genes, increased cardiomyocyte cell death and impaired left ventricle contractile and relaxing function in STZ-induced diabetes[6]. Serial STZ injections resulted in comparable increases in fasting blood glucose between PA28αOE mice and control mice (Supplemental Fig. 2A). As expected, diabetes induction caused body weight loss and heart weight loss, and a decrease in heart weight to tibial length ratio, all of which were unaffected by PA28αOE (Supplemental Fig. 2B–2D). Moreover, cardiac interstitial and perivascular fibrosis was markedly increased in CTL+STZ hearts but this increase was significantly attenuated in PA28αOE+STZ hearts (Fig. 6A–6B). Cardiomyocyte apoptosis was evident in CTL+STZ hearts significantly blunted in PA28αOE+STZ hearts (Fig. 6C–6D). These data suggest that PA28αOE attenuates diabetes-induced pathological cardiac remodeling.
Figure 6.
PA28αOE attenuated diabetes-induced cardiac fibrosis and apoptosis. Diabetes was induced in CTL and PA28αOE mice by STZ injection. A–B, Representative microscopic images (A) of myocardial sections subjected to Masson’s Trichrome staining and the quantification (B) of fibrotic area relative to myocardium tissue area 8 weeks after STZ injection. Bar, 100 μm. C, Representative confocal micrographs (C) of myocardial sections stained with TUNEL (red) to identify apoptotic cardiomyocytes. The sections were counterstained with Alexa Flour 488-conjugated phalloidin (green) and DAPI to label cardiomyocytes and nuclei, respectively. D, Quantitative analysis of TUNEL+ cardiomyocytes (CMs). Bar, 50 μm. **P<0.01 versus CTL; #P<0.05 versus CTL+STZ.
3.7 Overexpression of PA28α improves cardiac function in diabetic mice
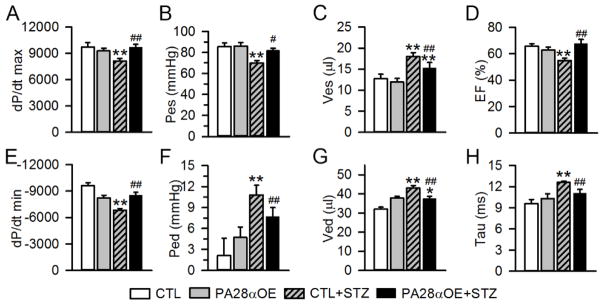
We next performed left ventricle (LV) pressure-volume relationship analysis to determine the impact of PA28αOE on cardiac function in diabetic mice. LV systolic function was impaired at 8 weeks after diabetes induction in the CTL+STZ group compared to the CTL group, with a significant decrease in dP/dt max (−16%), Pes (−18%) and EF (−15%), and an increase in Ves (+41%). PA28αOE led to a significant improvement of systolic function as compared to CTL+STZ, with an increase in dP/dt max (+19%), Pes (+32%) and EF (+24%), and a decrease in Ves (−15%) (Fig. 7A–7D). LV diastolic function was impaired 8 weeks after STZ treatment, as demonstrated by a significant decrease in dP/dt min (−29%) compared to CTL group. This was accompanied by an increase in Ped (+414%) and Ved (+35%), and a prolongation of Tau (+31%). PA28a OE significantly restored diastolic function as compared to CTL+STZ, with an increase in dP/dt min (+25%), and a decrease in Ped (−29%), Ved (−13%) and Tau (−12%). (Fig. 7E–7H).
Figure 7.
PA28αOE improved cardiac function in diabetic mice. Invasive hemodynamic measurements were obtained in CTL and PA28aOE mice 8 weeks after STZ injection. Results of pressure-volume (PV) loop analyses showing measures of systolic (A–D) and diastolic (E–H) function. dP/dt max, maximal dP/dt; Pes, end-systolic pressure; Ves, end-systolic volume; EF, ejection fraction; dP/dt min, minimal dP/dt; Ped, end-diastolic pressure; Ved, end-diastolic volume; Tau, the load-independent isovolumic relaxation constant. N=7–9 mice per group. Data are expressed as Mean+SEM. **P<0.01 versus CTL; #P<0.05, ##P<0.01 versus CTL+STZ, respectively.
Collectively, these data demonstrate that PA28αOE improves both systolic and diastolic function in STZ-induced diabetic mice.
4. Discussion
We report that UPS function is progressively impaired in the heart of STZ-induced diabetic mice, preceding the onset of cardiomyopathy. Using a mouse model with enhanced proteasome function by cardiac-specific overexpression of PA28α, we demonstrate that restoration of UPS function attenuates cardiac dysfunction. These novel findings suggest that insufficient proteasome function contributes to the pathogenesis of diabetic cardiomyopathy.
UPS-mediated proteolysis involves the ubiquitination of a substrate, the recognition of ubiquitinated proteins and their delivery to the proteasome by ubiquitin receptors, and finally proteasomal degradation of the ubiquitinated substrates[29]. By assessing the cleavage of small fluorogenic peptide substrates in vitro, previous studies have reported either upregulation or downregulation of cardiac proteasome activities in murine models of type I diabetes[18–20], yet global function of the UPS has not been systematically probed. The discrepant results regarding cardiac proteasome activity in diabetes likely reflects differences in STZ dosing, stage of disease, and methods to assess UPS function and underscores the importance of assessment of the temporal changes in overall cardiac UPS function. To circumvent these limitations, our study employed a surrogate UPS substrate GFPdgn as a reporter, whose degradation requires coordinated function of the whole UPS[25, 30], to longitudinally evaluate cardiac UPS function in vivo. Our data demonstrate that GFPdgn accumulates in the heart shortly after diabetes induction (Fig. 1), indicating that insufficient cardiac UPS function is an early event in diabetes. In support of this conclusion, we also observed subsequent accumulation of ubiquitinated proteins in the hearts of diabetic mice, despite at a later stage (Fig. 2), confirming the sensitivity of GFPdgn as a functional UPS reporter. In concordance with our findings, other studies also reported increased ubiquitinated and oxidized proteins in the hearts of STZ-induced diabetic mice[18–20]. Together, our data and others suggest an increasing demand for the clearance of abnormal proteins consequent to insufficient UPS function in the hearts of type I diabetic animals.
By temporally assessing proteasome function in diabetic hearts, we detected initial upregulation followed by suppression of activity (Fig. 3). Given that the temporal changes of proteasome function inversely correlated with the progression of diabetic cardiomyopathy (Supplemental Fig. 1D), we reasoned the initial upregulation of proteasome function is a compensatory response to increasing proteotoxic stress, which is subsequently lost with persisting diabetic stress. In general, proteasome function can be regulated by the expression and posttranslational modification of proteasome subunits, the composition of the proteasome and its binding partners [31, 32]. For instance, Nrf1 and Nrf2 were shown to induce the expression of 20S and 11S proteasomes and are required for adaption to oxidative stress, which is present in diabetic hearts [33, 34]. Interestingly, Nrf2 is downregulated in diabetic hearts [35], which could potentially contribute to diabetes-induced impairment of proteasome function. The functions of cardiac proteasomes are also regulated by diverse posttranslational modifications including phosphorylation, nitrosylation and glycosylation, etc. [31], which could be altered by diabetes-induced metabolic, oxidative and nitrative stress. The precise mechanisms underlying the temporal changes of proteasome function in diabetic hearts remains to be investigated. [36, 37]
Regarding mechanisms whereby PA28αOE improves UPS function in diabetic hearts, we previously reported that PA28αOE upregulates 11S proteasome abundance by stabilizing PA28β, leading to increased association of 11S proteasomes with 20S proteasomes[17, 22]. The 20S proteasomes are capable of degrading denatured, non-ubiquitinated proteins in an ATP-independent manner[38, 39] and the 11S proteasomes upregulate 20S proteolytic activity by binding to the 20S α-ring subunits and facilitating gate opening of the 20S proteasome[37]. Therefore, it is plausible that under pathological conditions such as diabetes, cells may resort to the 11S–20S proteasomes for bulk degradation of damaged proteins, and as a consequence of PA28αOE, the increased 11S proteasomes protect cardiomyocytes against diabetes-induced proteotoxicity. In support of this notion, the 11S proteasomes were reported to be strongly induced by oxidative stress, in turn promoting the degradation of oxidized proteins[22, 40, 41]. Whether PA28αOE alleviates oxidative stress in diabetic hearts, however, remains to be determined.
Another important finding of our study is the protective effect of overexpression of PA28α on diabetic cardiomyopathy. The improvement of cardiac hemodynamics in diabetic PA28aOE mice may be attributable to the attenuation of proteotoxic stress (now Fig. 4–5) and pathological cardiac remodeling (now Fig. 6). Indeed, emerging studies have established the pathogenic role of proteotoxicity in various forms of cardiac diseases. Disruption of protein quality control by either excessive production of misfolded proteins or inhibition of proteasome or autophagy, results in impairment of cardiac contractility and heart failure in mouse models [9, 12, 42]. In a high fat diet-induced obesity mouse model, adiponectin deficiency was shown to diminish autophagy function and aggravate cardiac dysfunction [43]. Conversely, restoration of protein quality control by improvement of proteasome or autophagic function has been shown to preserve cardiac hemodynamics against various pathological insults [17, 44]. These prior findings, along with our findings from the present study, demonstrate that protein homeostasis is essential for cardiomyocyte function and survival and call for the development of therapeutic measures to curtail cardiac proteotoxicity. Notably, activation of proteasome function can be achieved by pharmacological manipulation of the cAMP-protein kinase A (PKA) pathway, protein kinase G (PKG) signaling and ubiquitin-specific protease 14 (USP14)[45–48], warranting future studies to determine the effects of these pharmacological agents in diabetic cardiomyopathy.
We showed that diabetic PA28αOE hearts displayed significant reduction in diabetes-induced cardiac fibrosis and apoptosis, which, however, remained higher than in non-diabetic hearts (now Fig. 6). These data indicate that insufficient UPS function is only one of the major factors that trigger pathological remodeling in diabetic hearts. Since diabetes-induced cardiac damage is known to be caused by a combination of cellular stresses including increased oxidative stress, altered substrate metabolism, mitochondrial dysfunction and inflammation [5, 6, 49], it is conceivable that PA28αOE may not be able to antagonize all these stresses and that the fibrosis and apoptosis may be further augmented by persisting diabetic stress, eventually dampening the cardiac function of PA28αOE diabetic mice. Interestingly, both attenuation of inflammation by pharmacological proteasome inhibition and maintenance of mitochondrial function by overexpressing mitochondrial aldehyde dehydrogenase (ALDH2) have been shown to benefit diabetic hearts[20, 50, 51]. Our findings suggest that enhancement of proteasome function in cardiomyocytes, in combination with other strategies (such as elevation of autophagy, anti-oxidant and anti-inflammatory), could be more effective in limiting diabetic cardiac injury.
Supplementary Material
Highlights.
Cardiac UPS function is impaired prior to the onset of diabetic cardiomyopathy.
Diabetes diminishes cardiac proteasome function.
PA28a overexpression restores cardiac UPS function after diabetes.
Enhancement of cardiac proteasome function ameliorates diabetic cardiomyopathy.
Acknowledgments
Funding Sources
This work was in part supported by the National Institute of Health grants R01HL124248 (to H.S.), R01HL072166 (to X.W.), R01HL124251 (to I.K.), R01HL112640, R01HL126949, R01HL134354 and R01AR070029 (to N.L.W.), and the American Heart Association grants 11SDG6960011 (to H.S.), 16SDG30940002 (to J.L.) and 14SDG18970040 (to I.K.).
Footnotes
Disclosures
None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Danaei G, Finucane MM, Lu Y, Singh GM, Cowan MJ, Paciorek CJ, et al. National, regional, and global trends in fasting plasma glucose and diabetes prevalence since 1980: systematic analysis of health examination surveys and epidemiological studies with 370 country-years and 2.7 million participants. Lancet. 2011;378(9785):31–40. doi: 10.1016/S0140-6736(11)60679-X. [DOI] [PubMed] [Google Scholar]
- 2.Falcao-Pires I, Leite-Moreira AF. Diabetic cardiomyopathy: understanding the molecular and cellular basis to progress in diagnosis and treatment. Heart Fail Rev. 2012;17(3):325–44. doi: 10.1007/s10741-011-9257-z. [DOI] [PubMed] [Google Scholar]
- 3.Jia G, DeMarco VG, Sowers JR. Insulin resistance and hyperinsulinaemia in diabetic cardiomyopathy. Nat Rev Endocrinol. 2016;12(3):144–53. doi: 10.1038/nrendo.2015.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Song Y, Wang Q, Kralik PM, Epstein PN. Causes and characteristics of diabetic cardiomyopathy. Rev Diabet Stud. 2006;3(3):108–17. doi: 10.1900/RDS.2006.3.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poornima IG, Parikh P, Shannon RP. Diabetic cardiomyopathy: the search for a unifying hypothesis. Circulation research. 2006;98(5):596–605. doi: 10.1161/01.RES.0000207406.94146.c2. [DOI] [PubMed] [Google Scholar]
- 6.Boudina S, Abel ED. Diabetic cardiomyopathy revisited. Circulation. 2007;115(25):3213–23. doi: 10.1161/CIRCULATIONAHA.106.679597. [DOI] [PubMed] [Google Scholar]
- 7.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82(2):373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, Su H, Ranek MJ. Protein quality control and degradation in cardiomyocytes. Journal of molecular and cellular cardiology. 2008;45(1):11–27. doi: 10.1016/j.yjmcc.2008.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Q, Liu JB, Horak KM, Zheng H, Kumarapeli AR, Li J, et al. Intrasarcoplasmic amyloidosis impairs proteolytic function of proteasomes in cardiomyocytes by compromising substrate uptake. Circulation research. 2005;97(10):1018–26. doi: 10.1161/01.RES.0000189262.92896.0b. [DOI] [PubMed] [Google Scholar]
- 10.Liu J, Chen Q, Huang W, Horak KM, Zheng H, Mestril R, et al. Impairment of the ubiquitin-proteasome system in desminopathy mouse hearts. FASEB J. 2006;20(2):362–4. doi: 10.1096/fj.05-4869fje. [DOI] [PubMed] [Google Scholar]
- 11.Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, et al. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121(8):997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang M, Li J, Huang W, Su H, Liang Q, Tian Z, et al. Proteasome functional insufficiency activates the calcineurin-NFAT pathway in cardiomyocytes and promotes maladaptive remodelling of stressed mouse hearts. Cardiovasc Res. 2010;88(3):424–33. doi: 10.1093/cvr/cvq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Z, Zheng H, Li J, Li Y, Su H, Wang X. Genetically induced moderate inhibition of the proteasome in cardiomyocytes exacerbates myocardial ischemia-reperfusion injury in mice. Circulation research. 2012;111(5):532–42. doi: 10.1161/CIRCRESAHA.112.270983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann J, Wohlert C, Saguner AM, Flores A, Nesbitt LL, Chade A, et al. Primary proteasome inhibition results in cardiac dysfunction. Eur J Heart Fail. 2013;15(6):614–23. doi: 10.1093/eurjhf/hft034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranek MJ, Zheng H, Huang W, Kumarapeli AR, Li J, Liu J, et al. Genetically induced moderate inhibition of 20S proteasomes in cardiomyocytes facilitates heart failure in mice during systolic overload. Journal of molecular and cellular cardiology. 2015;85:273–81. doi: 10.1016/j.yjmcc.2015.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rajagopalan V, Zhao M, Reddy S, Fajardo G, Wang X, Dewey S, et al. Altered ubiquitin-proteasome signaling in right ventricular hypertrophy and failure. American journal of physiology Heart and circulatory physiology. 2013;305(4):H551–62. doi: 10.1152/ajpheart.00771.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Horak KM, Su H, Sanbe A, Robbins J, Wang X. Enhancement of proteasomal function protects against cardiac proteinopathy and ischemia/reperfusion injury in mice. The Journal of clinical investigation. 2011;121(9):3689–700. doi: 10.1172/JCI45709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu J, Klein JD, Du J, Wang XH. Cardiac muscle protein catabolism in diabetes mellitus: activation of the ubiquitin-proteasome system by insulin deficiency. Endocrinology. 2008;149(11):5384–90. doi: 10.1210/en.2008-0132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell SR, Samuel SM, Wang P, Divald A, Thirunavukkarasu M, Koneru S, et al. Upregulation of myocardial 11S-activated proteasome in experimental hyperglycemia. Journal of molecular and cellular cardiology. 2008;44(3):618–21. doi: 10.1016/j.yjmcc.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 20.Marfella R, Di Filippo C, Portoghese M, Siniscalchi M, Martis S, Ferraraccio F, et al. The ubiquitin-proteasome system contributes to the inflammatory injury in ischemic diabetic myocardium: the role of glycemic control. Cardiovascular pathology: the official journal of the Society for Cardiovascular Pathology. 2009;18(6):332–45. doi: 10.1016/j.carpath.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 21.Li J, Ma W, Li H, Hou N, Wang X, Kim IM, et al. NEDD8 Ultimate Buster 1 Long (NUB1L) Protein Suppresses Atypical Neddylation and Promotes the Proteasomal Degradation of Misfolded Proteins. J Biol Chem. 2015;290(39):23850–62. doi: 10.1074/jbc.M115.664375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li J, Powell SR, Wang X. Enhancement of proteasome function by PA28α overexpression protects against oxidative stress. FASEB J. 2011;25(3):883–93. doi: 10.1096/fj.10-160895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Su H, Li J, Menon S, Liu J, Kumarapeli AR, Wei N, et al. Perturbation of cullin deneddylation via conditional Csn8 ablation impairs the ubiquitin-proteasome system and causes cardiomyocyte necrosis and dilated cardiomyopathy in mice. Circulation research. 2011;108(1):40–50. doi: 10.1161/CIRCRESAHA.110.230607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bence NF, Bennett EJ, Kopito RR. Application and analysis of the GFPu family of ubiquitin-proteasome system reporters. Methods Enzymol. 2005;399:481–90. doi: 10.1016/S0076-6879(05)99033-2. [DOI] [PubMed] [Google Scholar]
- 25.Kumarapeli AR, Horak KM, Glasford JW, Li J, Chen Q, Liu J, et al. A novel transgenic mouse model reveals deregulation of the ubiquitin-proteasome system in the heart by doxorubicin. FASEB J. 2005;19(14):2051–3. doi: 10.1096/fj.05-3973fje. [DOI] [PubMed] [Google Scholar]
- 26.Liu Y, Hettinger CL, Zhang D, Rezvani K, Wang X, Wang H. Sulforaphane enhances proteasomal and autophagic activities in mice and is a potential therapeutic reagent for Huntington’s disease. Journal of neurochemistry. 2014;129(3):539–47. doi: 10.1111/jnc.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu P, Duong DM, Seyfried NT, Cheng D, Xie Y, Robert J, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137(1):133–45. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Powell SR, Davies KJ, Divald A. Optimal determination of heart tissue 26S-proteasome activity requires maximal stimulating ATP concentrations. Journal of molecular and cellular cardiology. 2007;42(1):265–9. doi: 10.1016/j.yjmcc.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang X, Terpstra EJ. Ubiquitin receptors and protein quality control. Journal of molecular and cellular cardiology. 2013;55:73–84. doi: 10.1016/j.yjmcc.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dong X, Liu J, Zheng H, Glasford JW, Huang W, Chen QH, et al. In situ dynamically monitoring the proteolytic function of the ubiquitin-proteasome system in cultured cardiac myocytes. American journal of physiology Heart and circulatory physiology. 2004;287(3):H1417–25. doi: 10.1152/ajpheart.01233.2003. [DOI] [PubMed] [Google Scholar]
- 31.Cui Z, Scruggs SB, Gilda JE, Ping P, Gomes AV. Regulation of cardiac proteasomes by ubiquitination, SUMOylation, and beyond. Journal of molecular and cellular cardiology. 2014;71:32–42. doi: 10.1016/j.yjmcc.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsukamoto O, Minamino T, Kitakaze M. Functional alterations of cardiac proteasomes under physiological and pathological conditions. Cardiovasc Res. 2010;85(2):339–46. doi: 10.1093/cvr/cvp282. [DOI] [PubMed] [Google Scholar]
- 33.Pickering AM, Linder RA, Zhang H, Forman HJ, Davies KJ. Nrf2-dependent induction of proteasome and Pa28alphabeta regulator are required for adaptation to oxidative stress. J Biol Chem. 2012;287(13):10021–31. doi: 10.1074/jbc.M111.277145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radhakrishnan SK, Lee CS, Young P, Beskow A, Chan JY, Deshaies RJ. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol Cell. 2010;38(1):17–28. doi: 10.1016/j.molcel.2010.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan Y, Ichikawa T, Li J, Si Q, Yang H, Chen X, et al. Diabetic downregulation of Nrf2 activity via ERK contributes to oxidative stress-induced insulin resistance in cardiac cells in vitro and in vivo. Diabetes. 2011;60(2):625–33. doi: 10.2337/db10-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi S, Mao K, Zheng H, Wang X, Patterson C, O’Connell TD, et al. Diminished GATA4 protein levels contribute to hyperglycemia-induced cardiomyocyte injury. J Biol Chem. 2007;282(30):21945–52. doi: 10.1074/jbc.M703048200. [DOI] [PubMed] [Google Scholar]
- 37.Odiete O, Konik EA, Sawyer DB, Hill MF. Type 1 diabetes mellitus abrogates compensatory augmentation of myocardial neuregulin-1beta/ErbB in response to myocardial infarction resulting in worsening heart failure. Cardiovascular diabetology. 2013;12:52. doi: 10.1186/1475-2840-12-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shringarpure R, Grune T, Mehlhase J, Davies KJ. Ubiquitin conjugation is not required for the degradation of oxidized proteins by proteasome. J Biol Chem. 2003;278(1):311–8. doi: 10.1074/jbc.M206279200. [DOI] [PubMed] [Google Scholar]
- 39.Davies KJ. Degradation of oxidized proteins by the 20S proteasome. Biochimie. 2001;83(3–4):301–10. doi: 10.1016/s0300-9084(01)01250-0. [DOI] [PubMed] [Google Scholar]
- 40.Seifert U, Bialy LP, Ebstein F, Bech-Otschir D, Voigt A, Schroter F, et al. Immunoproteasomes preserve protein homeostasis upon interferon-induced oxidative stress. Cell. 2010;142(4):613–24. doi: 10.1016/j.cell.2010.07.036. [DOI] [PubMed] [Google Scholar]
- 41.Pickering AM, Koop AL, Teoh CY, Ermak G, Grune T, Davies KJ. The immunoproteasome, the 20S proteasome and the PA28alphabeta proteasome regulator are oxidative-stress-adaptive proteolytic complexes. Biochem J. 2010;432(3):585–94. doi: 10.1042/BJ20100878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13(5):619–24. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 43.Guo R, Zhang Y, Turdi S, Ren J. Adiponectin knockout accentuates high fat diet-induced obesity and cardiac dysfunction: role of autophagy. Biochim Biophys Acta. 2013;1832(8):1136–48. doi: 10.1016/j.bbadis.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bhuiyan MS, Pattison JS, Osinska H, James J, Gulick J, McLendon PM, et al. Enhanced autophagy ameliorates cardiac proteinopathy. The Journal of clinical investigation. 2013;123(12):5284–97. doi: 10.1172/JCI70877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myeku N, Clelland CL, Emrani S, Kukushkin NV, Yu WH, Goldberg AL, et al. Tau-driven 26S proteasome impairment and cognitive dysfunction can be prevented early in disease by activating cAMP-PKA signaling. Nat Med. 2016;22(1):46–53. doi: 10.1038/nm.4011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lokireddy S, Kukushkin NV, Goldberg AL. cAMP-induced phosphorylation of 26S proteasomes on Rpn6/PSMD11 enhances their activity and the degradation of misfolded proteins. Proc Natl Acad Sci U S A. 2015;112(52):E7176–85. doi: 10.1073/pnas.1522332112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranek MJ, Terpstra EJ, Li J, Kass DA, Wang X. Protein kinase g positively regulates proteasome-mediated degradation of misfolded proteins. Circulation. 2013;128(4):365–76. doi: 10.1161/CIRCULATIONAHA.113.001971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee BH, Lee MJ, Park S, Oh DC, Elsasser S, Chen PC, et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature. 2010;467(7312):179–84. doi: 10.1038/nature09299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palomer X, Salvado L, Barroso E, Vazquez-Carrera M. An overview of the crosstalk between inflammatory processes and metabolic dysregulation during diabetic cardiomyopathy. Int J Cardiol. 2013;168(4):3160–72. doi: 10.1016/j.ijcard.2013.07.150. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Sun W, Du B, Miao X, Bai Y, Xin Y, et al. Therapeutic effect of MG-132 on diabetic cardiomyopathy is associated with its suppression of proteasomal activities: roles of Nrf2 and NF-kappaB. American journal of physiology Heart and circulatory physiology. 2013;304(4):H567–78. doi: 10.1152/ajpheart.00650.2012. [DOI] [PubMed] [Google Scholar]
- 51.Zhang Y, Babcock SA, Hu N, Maris JR, Wang H, Ren J. Mitochondrial aldehyde dehydrogenase (ALDH2) protects against streptozotocin-induced diabetic cardiomyopathy: role of GSK3beta and mitochondrial function. BMC Med. 2012;10:40. doi: 10.1186/1741-7015-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.