Abstract
The trans-Atlantic slave trade brought millions of Africans to the New World. Advances in genomics are providing novel insights into the history and health of Africans and the diasporan populations. Recent examples reviewed here include the unraveling of substantial hunter-gatherer and “Eurasian” admixtures across sub-Saharan Africa, expanding our understanding of ancestral African genetics; the global ubiquity of mixed ancestry; the revealing of African ancestry in Latin Americans that likely derived from the slave trade; and understanding of the ancestral backgrounds of APOL1 and LPL found to influence kidney disease and lipid levels, respectively, providing specific insights into disease etiology and health disparities.
Introduction
Anatomically modern humans originated in Africa before migrating to populate the rest of the world in the last 100,000 years, hence the expression “we are all Africans beneath our skin” [1,2]. This generally accepted consensus begs the question of why is the global dispersion of some human populations out of Africa referred to as the “African Diaspora” and others are not? Who and from where are these groups that constitute the African Diaspora and how has their history shaped patterns of genomic variation, the distribution of fitness influencing mutations, and health? Here, we review recent data on these questions and explore how these data, especially the accelerated cataloging of global human genetic variation, are informing our understanding of the identities and health of these populations in their current homelands. We illustrate opportunities offered by the African Diaspora to study interactions of old genes with modern environments, thereby lending novel insights into disease etiology, ancestry-based disease gene mapping, and health disparities.
The African Diaspora – History and Definition
The term “African Diaspora” first appeared in the literature in the 1950s and has been broadly defined to include all global communities descended from the historic migrations of peoples from Africa since the 15th century [3,4]. This delineates it from the pre-historic Out-of-Africa migrations that led to the peopling of the world. The African Diaspora has also been more narrowly defined to include only the trans-Atlantic slave trade. This narrower definition, which emphasizes the important roles that blackness, slavery, colonialism, racism, and geography played in sustaining the trans-Atlantic slave trade, is the reason why some refer to the “African Diaspora” as the “Black Diaspora” [3,5]. The trans-Atlantic slave trade was the single largest immigration of Africans from the Old World to the New World. The first leg of the triangular trade involved ships from Europe carrying goods (e.g., iron, brandy, weapons, and gunpowder) that were traded for slaves in Africa. The second leg, termed the Middle Passage, involved the shipment of between 12 and 14 million enslaved Africans across the Atlantic Ocean to the Americas. The last leg was the transportation of goods (e.g., sugar, cotton, tobacco, rum, and molasses) from the Americas to Europe.
Most enslaved Africans were brought to European colonies in Latin America, while 3–5% were brought to the United States of America (USA) [6]. These enslaved Africans and their descendants in the USA represent the group “African Americans”. As with other diasporan populations and indeed most of the world populations [7], African Americans have multiple ancestries with lineages from Africa, Europe, Asia, and Native America among others. Hence, the term “African American” is not a genetically homogeneous entity as reflected in the fact that self-identified African Americans include individuals ranging from almost no African ancestry to almost no European ancestry [8,9] (Figure 1). In addition, cultural diversity abounds in the descendants of the African Diaspora as exemplified by Brazilians. With over 4 million slaves, Brazil has long been a melting pot of ancestries and cultures, as evidenced by the blending of African and European religions (Candomble, Catholicism), foods (Feijoada), music, and dance (Samba). A recent census indicates that about 50% of the ~200 million Brazilians self-identify as persons of African ancestry or mixed ancestry [10]. These mixtures of ancestries, acquired over hundreds of years, have serious implications for national and global biomedical initiatives such as precision medicine because phenotypic appearances and self and group identification are unlikely to adequately capture the ancestral backgrounds of individuals that make up the African Diaspora.
Figure 1.
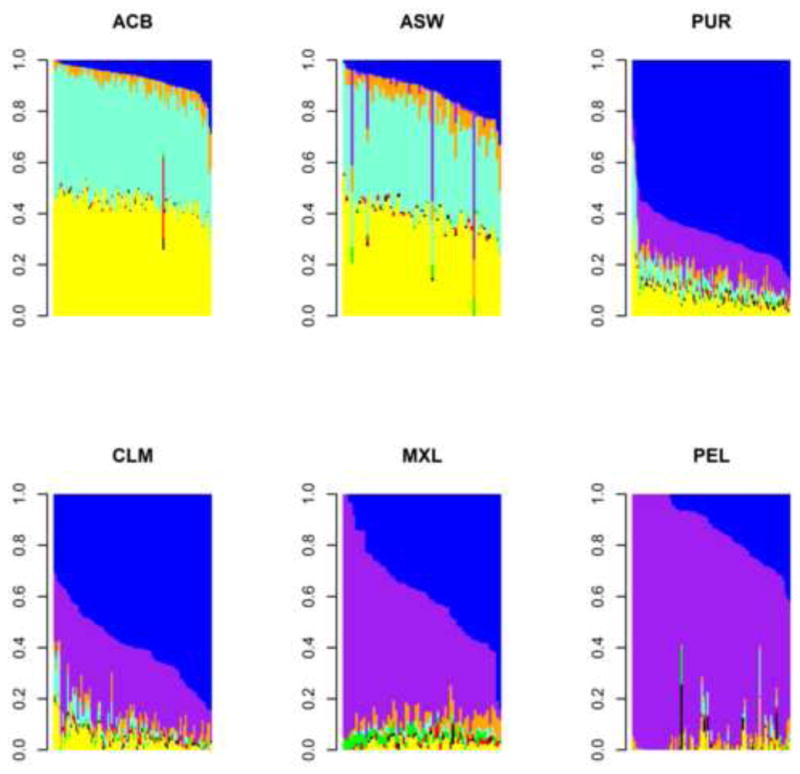
Individual admixture proportions from New World samples [12]. The six samples are African Caribbean in Barbados (ACB), People with African Ancestry in Southwest USA (ASW), Puerto Ricans in Puerto Rico (PUR), Colombians in Medellín, Colombia (CLM), People with Mexican Ancestry in Los Angeles, California (MXL), and Peruvians in Lima, Peru (PEL). Conditional on eight ancestries, yellow corresponds to Western African ancestry, aquamarine corresponds to West-Central African ancestry, blue corresponds to Southern European ancestry, orange corresponds to Northern European ancestry, purple corresponds to Native American ancestry, red corresponds to South Asian ancestry, green corresponds to East Asian ancestry, and black corresponds to Southeastern Asian ancestry. Summing the Western and West-Central African ancestries, the range of individual admixture proportion across these six samples goes from 0% to 97.8%. Similarly, the range of individual admixture proportion for Native American ancestry goes from 0% to 100%.
Population Structure and Genetic Diversity among African Populations
The landscape of population structure and genetic diversity of sub-Saharan Africans (SSA) was recently illuminated by two large international projects: the African Genome Variation Project (AGVP) [11] and the 1000 Genomes Project [12]. The AGVP data, generated from 20 African ethno-linguistic groups, revealed previously unappreciated population structure, including regionally distinct patterns of admixture. Using principal components analysis, unsupervised cluster analysis, and the f3 test for admixture, evidence for substantial “Eurasian” and hunter-gatherer admixture was observed across SSA [11]. The timing and sources of admixture were regionally distinct, with admixture in West Africans dating to ~9000 years ago and a source similar to present-day Khoe-San populations and with admixture in East Africans dating to ~3000 years ago and a source similar to Mbuti rainforest hunter-gatherer populations [11].
African populations are highly subdivided, with population structure across Africa currently recognized as 11 ancestries that correspond to a combination of geographic and linguistic separation: Khoisan in southern Africa; Central African, predominant in Pygmies; Hadza in Tanzania; Western African, predominant in Mande-speaking peoples; West-Central African, predominant in both Bantu-speaking and non-Bantu-speaking peoples in the area from Ghana to Cameroon; and ancestries corresponding to speakers of Berber, Cushitic, Eastern Bantu, Omotic, Nilo-Saharan, and Southern Bantu languages [7,11,13]. By comparison, 12 ancestries have been detected in the rest of world, including two that define north-to-south differentiation in Europe and one that encompasses Native American ancestry [7]. Genetic differentiation, measured by FST, between SSA ancestries can exceed that between pairs of non-African ancestries. For example, FST is 0.054 between Khoisan and Omotic ancestries, compared to 0.024 between Southern and Northern European ancestries or 0.042 between Arabian and Indian ancestries [7].
Higher levels of genetic diversity are observed among SSA. The number of variant sites per individual of SSA ancestry is ~5 million, compared to ~4.0–4.2 million variants per individual of East Asian, European, or South Asian ancestry [12]. The average rate of nucleotide differences of 1.2 per kilobase between a pair of Khoe-San individuals exceeds that of 1.0 per kilobase between an Asian individual and a European individual [14]. As a function of physical distance, linkage disequilibrium decays faster in SSA populations than in non-African populations [12], such that haplotypes are shorter in SSA. Lower burdens of runs of homozygosity tend to be observed in SSA ancestry populations, as well as in admixed populations [15], leading to lower risk of autosomal recessive diseases.
Genomic Profile of Contemporary Populations of the African Diaspora
In admixed African Americans, continental-level differences between Africans and Europeans can explain up to 8% of phenotypic variance across a range of anthropometric and cardio-metabolic traits [16]. Given the progress in delineating population structure at the sub-continental level described above, a deeper understanding of the fine-scale genetic structure in African Diaspora populations is needed. Western African ancestry is the predominant ancestry in the Mende people from Sierra Leone and Jola, Mandinka, and Wolof peoples from The Gambia [11,12]. West-Central African ancestry is the predominant ancestry in non-Bantu-speaking peoples such as the Ga-Adangbe peoples from Ghana and the Esan, Igbo, and Yoruba peoples from Nigeria [11,12]. Among Bantu-speaking peoples, genetic differentiation following the Bantu expansion gave rise to distinct ancestries in West-Central Africa (found in Bamum and M’fang peoples in Cameroon and Kongo people from the Democratic Republic of the Congo), Eastern Africa (found in Baganda, Barundi, and Banyarwanda peoples from Uganda and Luhya and Kikuyu peoples from Kenya), and Southern Africa (found in Sotho and Zulu peoples from South Africa) [7,8,11]. At the Y DNA level, Western Africans have comparatively more E1b1a1a1f, whereas West-Central Africans have comparatively more E1b1a1a1g [12]. These findings of regional ancestry within continental Africa indicate that some degree of localization of African origin for African Americans and other diasporan Africans is possible.
The genomic profile of contemporary populations of the Americas reflects admixture that occurred among Europeans, Native Americans, and enslaved Africans in the New World. For several reasons, including where slaves disembarked and socio-cultural practices such as ancestry-positive assortative mating, the average percentage of African ancestry in the Americas varies widely. Recent estimates range from 87% in African Caribbeans from Barbados, 75% in African Americans in Southwest USA, 19% in Puerto Ricans, 12% in Colombians in Medellín, 7% in people with Mexican ancestry in Los Angeles, and 4% in both Peruvians in Lima and Argentinians sampled from across Argentina [12,17] (Figure 1). African ancestry in the Caribbean appears consistent with two waves, the first from Western Africa followed by the second from West-Central Africa [18]. In Spanish-speaking South America, African ancestry averages 5% [19]. In Brazil, African ancestry varies from 51% in Salvador in the Northeast to 15% in Bambuí in the Southeast and 16% in Pelotas in the South [20]. Western African and West-Central African ancestries are more prevalent in Northeastern Brazil due to a larger proportion of disembarkation from Western and West-Central Africa in Salvador whereas Eastern African ancestry is more prevalent in the Southeast and South due to a larger proportion of disembarkation from Mozambique in Rio de Janeiro [20]. Population structure in the Americas displays further complexity by the widely varied average percentage of Native American ancestry in these populations, with about 1% in African Caribbeans from Barbados, 4% in African Americans in Southwest USA, 15% in Puerto Ricans, 28% in Colombians in Medellín, 49% in people with Mexican ancestry in Los Angeles, and 78% in Peruvians in Lima [12] (Figure 1).
Genetic Signals of Natural Selection and Implications for Health and Disease
Genetic adaptations that took place across Africa, particularly against fatal pathogens and ecological forces, have resulted in elevated frequencies of alleles conferring survival advantages detectable in present-day African ancestry individuals on the continent and in the Diaspora (Table 1) [21–26]. Unfortunately, some of these alleles are maladaptive in modern-day environments. The discordance between ancestral genetic background and modern-day environmental exposures became pronounced in the African Diaspora, contributing to the disproportionately high burden of some chronic diseases and health disparities in these groups.
Table 1.
Examples of natural selection with implications for human health
| Adapted Genes | Beneficial Trait | Negative Outcome | References |
|---|---|---|---|
| APOL1 | Protection against Human African Trypanosomiasis (HAT) | Kidney disease | [27–36,49] |
| ATP1A1, AQP2, CSK | Climate adaptation | Hypertension and osmoregulation | [11,37,38] |
| PPARA | Energy metabolism during prolonged food deficiency | None known to date | [50] |
| CIC, PAFAH1B3, LIPE, BHLHE41 | High altitude adaptation | None known to date | [25,26] |
| LARGE, IL21 | Confers protection against the Lassa virus | None known to date | [51] |
| DMD | Confers protection against the Lassa virus | 1) Increased replication of vaccinia virus. 2) Duchenne and Becker muscular dystrophy | [52] |
| DARC | Confers resistance to P. vivax | Benign ethnic neutropenia | [53–59] |
| HBB | Confer incomplete resistance to lethal forms of malaria | Sickle Cell Trait and Sickle Cell Disease | [60–65] |
| Regulatory deficiencies of HBA and HBB | Confers incomplete resistance to lethal forms of malaria | α and β thalassemia | [66,67] |
| CCR5 Δ32 deletion | Confers HIV protection | 1) CCR5 Δ32 are at higher risk for tick-borne encephalitis. 2) Fully functional CCR5 reduces symptoms from infection with West Nile virus | [68–71] |
| LCT | Lactase persistence | None known to date | [21,72,73] |
| AMY1 | Increased copies of the gene in areas where starch is consumed | None known to date | [74] |
A recent striking example of the evolutionary importance of genetic variants in populations of the African Diaspora, with implications for health disparities, is the link between kidney disease, African sleeping sickness, and two missense haplotypes called G1 (consisting of S342G and I384M) and G2 (delN388/Y389) in the gene Apolipoprotein L1 (APOL1) [27,28]. The initial studies that linked this genomic region to kidney diseases took advantage of the demographic history of African Americans by conducting admixture mapping [29,30]. The two renal disease risk haplotypes are present at relatively high frequency in individuals of recent African ancestry (especially from West-Central Africa where the trypanosome parasite is endemic) but are absent in Europeans and Asians. These observations, and the known trypanolytic activity of the variants against Trypanosoma brucei rhodesiense, have led to the hypothesis that these variants evolved in SSA and have risen to high frequency because they confer protection against a deadly form of African sleeping sickness [27]. Interestingly, these African-specific renal risk variants are also seen in the Americas as a result of admixture since the trans-Atlantic slave trade [28,31] (Figure 2). G1 and G2 haplotypes recapitulate the molecular characteristics of APOL1 in Old World monkeys, and innate immune activity afforded by APOL1 may extend beyond trypanosomes [32]. S342G has the highest frequency in West-Central Africa, whereas G2 is more evenly distributed and at lower frequency throughout SSA (Figure 2). S342G was the most differentiated locus (P=5.11×10−7) in a genome-wide comparison of the Western African Yoruba and the Eastern African Luhya ethnic groups, suggesting the role of positive selection in the evolution of the locus [32]. Clinically, kidney transplant failure is higher in recipients of donor kidneys from African ancestry individuals carrying APOL1 renal risk variants [33]. APOL1 high-risk genotypes (i.e., two risk alleles) explain an estimated 7–37% variance and 52–68% population attributable risk for different forms of end-stage kidney disease [34]. The APOL1 variants have also been implicated in cardiovascular diseases (CVD) [35]. Individuals of African ancestry with the risk genotype display less protection against kidney disease from HDL compared to Europeans and Asians [36].
Figure 2.
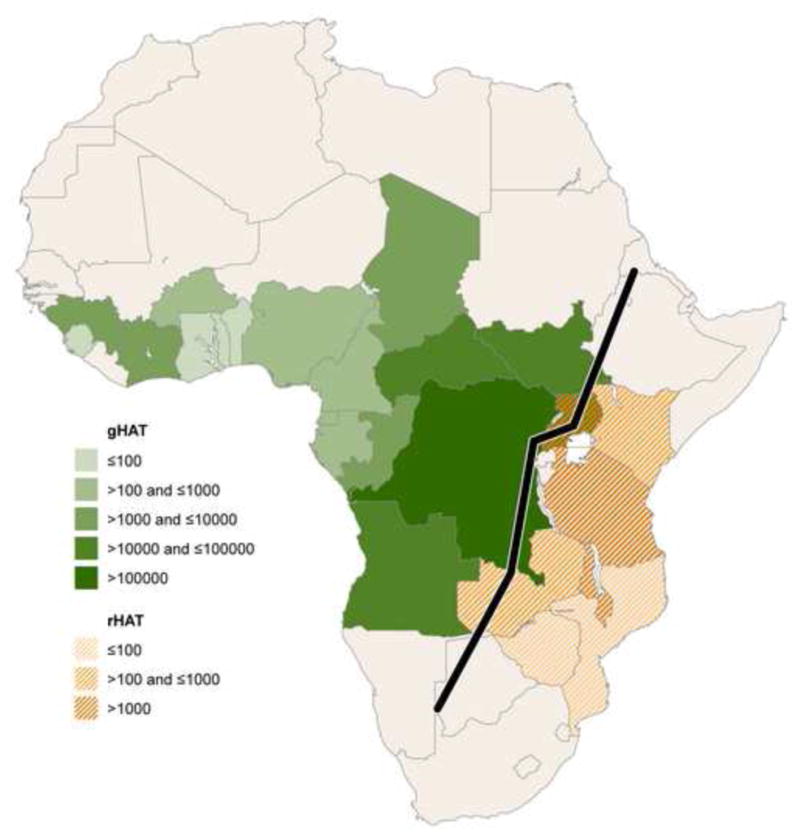
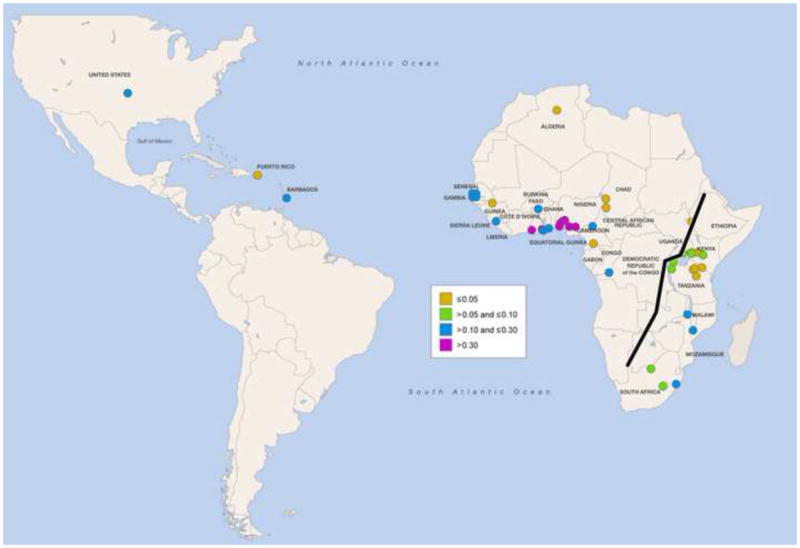
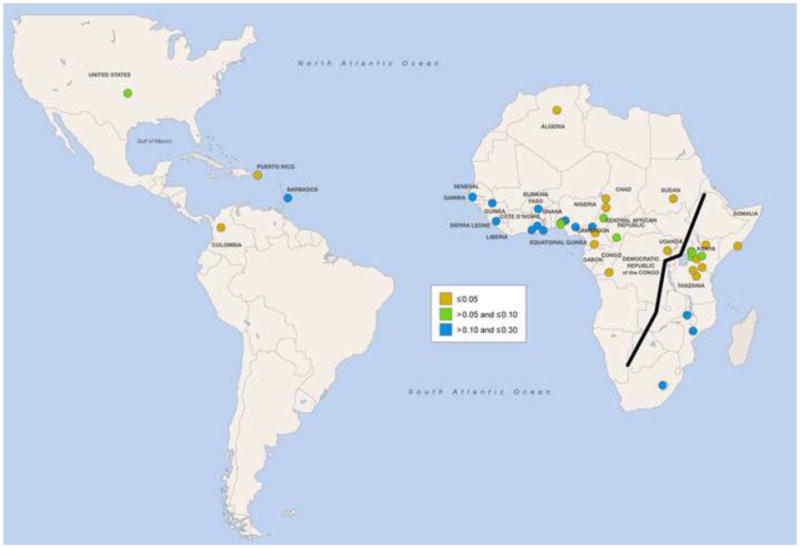
Geographic distribution of Trypansoma brucei and APOL1 risk variants. (A) Cumulative incidence from 1990 to 2014 of human African trypanosomiasis due to Trypansoma brucei gambiense (gHAT) and Trypanosoma brucei rhodesiense (rHAT) [75]. Uganda is the only country reporting trypanosomiasis due to both subspecies, with a higher cumulative incidence of gHAT. (B) Global frequency distribution of the G allele of the S342G mutation [11,12,28,49,76–78]. The Esan in Nigeria have the highest frequency (49.5%). (C) Global frequency distribution of the G2 deletion [12,28,32,49,77]. Bantu-speaking people in South Africa (Herero, Ovambo, Pedi, Sotho, Tswana, and Zulu) have the highest frequency (21.4%).
Ecological adaptations to tropical climate have also shaped the genetic structure of Africans. Local temperature-induced adaptive genetic changes may be one mechanism involved in hypertension and differences in salt sensitivity in humans [37]. Heat-adapted people, particularly members of the African Diaspora, have greater risk of hypertension due to exposures of the modern world (e.g., increased salt intake) interacting with ancestral susceptibility [37]. Consistent with this hypothesis, signals of selection have been detected in ATP1A1, AQP2, and CSK genes previously implicated in hypertension and osmoregulation [11]. The ancestral allele of the CSK locus displaying high differentiation among African populations is strongly correlated with hypertension risk and its frequency is inversely correlated with latitude [11]. These findings support the hypothesis that adaptation to climate produced a latitudinal cline in hypertension susceptibility. If validated with more mechanistic studies, these observations could provide significant insight into the pathogenesis of hypertension. However, the ubiquity of exposure to environmental risk factors for hypertension at the global level is likely to make validating this hypothesis difficult; some of the highest rates of hypertension have been observed in non-African Diaspora populations [38].
Climatic adaptations in low latitude and high ultraviolet radiation (UVR) regions of the tropics are thought to have maintained dark skin pigmentation to protect against UVR-induced DNA damage and folate photolysis [39]. After modern humans migrated out of Africa, the challenges of producing vitamin D in the skin from the low UVR outside of the tropics were met by natural selection acting on mutations producing skin depigmentation more than 30,000 years ago [40]. Dark skin pigmentation is maladaptive in low UVR environments, potentially contributing to health disparities in diseases associated with vitamin D deficiency including several forms of cancer and cardiometabolic diseases [41]. Future mechanistic studies are needed to disentangle the effects of the ancestral alleles of genes selected for skin pigmentation from those of dietary and lifestyle changes leading to vitamin D deficiency and related co-morbidities.
Novel insights into disease etiology have also been gained by comparing diasporan populations to their ancestral populations in SSA and by characterizing local admixture at disease risk loci. A recent study found that the association between the LPL SNP rs328 and lipid levels was stronger and the levels of HDL cholesterol were higher among African Americans with predominantly European ancestry than among those with African ancestry at this locus [42]. Lipid levels and their association with the LPL variant in African Americans with two African ancestry alleles at this locus were similar to those of West Africans despite widely different lifestyles and diets [42]. Another example of disease-associated alleles introduced to the Americas during the trans-Atlantic slave trade is the LEPRE1 c.1080+1G>T allele that causes type VIII osteogenesis imperfecta, so far found only in African Americans and West Africans [43]. This LEPRE1 mutation arose in West Africa more than 650 years ago and was transported to the Americas during the trans-Atlantic slave trade. Approximately 0.4% of African Americans and 1.5% of Nigerians and Ghanaians are heterozygous carriers [43].
Call for Large-Scale Deep Sequencing across the African Continent and the Diaspora
Earlier genotyping arrays were less efficient for interrogating the genomes of African ancestry populations [44] due to the fact that greater genetic variation is seen in present-day Africa populations than in populations outside of the continent, resulting in an increased number of haplotypes, lower levels of linkage disequilibrium (LD), more divergent patterns of LD, and more complex patterns of population substructure [11–13]. For example, our recent whole-genome sequencing effort in 320 SSA identified about 30 million variants [11], of which up to a quarter were unobserved in other populations from the 1000 Genomes sequencing project [12], indicating a need for large-scale deep sequencing of diverse populations across SSA and a need for a better genotyping array. Although some efforts have been made to address these concerns, including the recent development of the Infinium Multi-Ethnic Genotyping Array (MEGA) and the Affymetrix® Axiom® Genome-Wide Pan-African (PanAFR) Array, there remains a need for the development of a pan-African genotyping array that captures a larger proportion of common genetic variation across diverse African populations [11]. To achieve this goal, the H3Africa consortium has joined with the Wellcome Trust Sanger Institute, Illumina, and others to develop a genotyping array by interrogating whole-genome sequences from over 4,000 Africans sampled across the continent. There is also a need for improvements in algorithms for ancestry inference from whole-genome sequence data. Methodological issues include accounting for uncertain genotypes [45,46], linkage disequilibrium and phase [47], low-coverage sequence reads [48], and inadequate or missing source populations.
Conclusions
The history of the African Diaspora as defined in this opinion piece is complex, dynamic and continuous. With Africa at its root, members of the Diaspora developed multifaceted religious, cultural, and socio-political characteristics to adapt and survive in their new environments. As demonstrated above, genomics is beginning to facilitate better understanding of these multilayered stories and their implications for human history and health. However, more comprehensive sampling and genetic characterization of the populations of the African Diaspora and their ancestral homelands is urgently needed if these groups are to benefit from genomic medicine.
Acknowledgments
The study was supported by the Intramural Research Program of the National Institutes of Health (NIH) in the Center for Research on Genomics and Global Health (CRGGH). The CRGGH is supported by the National Human Genome Research Institute (NHGRI), the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), the Center for Information Technology (CIT), and the Office of the Director at the NIH (1ZIAHG200362).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
*of special interest
**of outstanding interest
- 1.Barry SL. All Africans under the Skin. In: Wolzak E, editor. National Geographic Program: The Genographic Project. [Google Scholar]
- 2.Morris K. Charles Rotimi: engaging Africa in human genomic research. Lancet. 2010;376:1383. doi: 10.1016/S0140-6736(10)61943-5. [DOI] [PubMed] [Google Scholar]
- 3.Palmer C. Defining and Studying the Modern African Diaspora. Perspectives: American Historical Association Newsletter. 1998;36:22–25. [Google Scholar]
- 4.Shepperson G. African Diaspora: Concept and Context. In: Harris J, editor. The Global Dimensions of the African Diaspora. Howard University Press; 1993. [Google Scholar]
- 5.Segal R. The Black Diaspora: Five Centuries of the Black Experience outside Africa. New York: The Noonday Press: Farrar, Straus and Giroux; 1995. [Google Scholar]
- 6.Lovejoy P. The Abolition of the Slave Trade. The New York Public Library; 2007. U.S. Slave Trade. [Google Scholar]
- 7**.Shriner D, Tekola-Ayele F, Adeyemo A, Rotimi CN. Genome-wide genotype and sequence-based reconstruction of the 140,000 year history of modern human ancestry. Sci Rep. 2014;4:6055. doi: 10.1038/srep06055. Using 163 global samples, the study showed that 94.4% of global populations showed mixed ancestry. Ubiquity of mixed ancestry emphasizes the importance of accounting for ancestry in history, forensics, and health (including drug labeling) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryc K, Auton A, Nelson MR, Oksenberg JR, Hauser SL, Williams S, Froment A, Bodo JM, Wambebe C, Tishkoff SA, et al. Genome-wide patterns of population structure and admixture in West Africans and African Americans. Proc Natl Acad Sci U S A. 2010;107:786–791. doi: 10.1073/pnas.0909559107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shriner D, Adeyemo A, Ramos E, Chen G, Rotimi CN. Mapping of disease-associated variants in admixed populations. Genome Biol. 2011;12:223. doi: 10.1186/gb-2011-12-5-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips T. US, editor. The Guardian. 2011. Nov 17, Brazil census shows African-Brazilians in the majority for the first time. [Google Scholar]
- 11**.Gurdasani D, Carstensen T, Tekola-Ayele F, Pagani L, Tachmazidou I, Hatzikotoulas K, Karthikeyan S, Iles L, Pollard MO, Choudhury A, et al. The African Genome Variation Project shapes medical genetics in Africa. Nature. 2015;517:327–332. doi: 10.1038/nature13997. This study, which has substantially expanded our understanding of African genome variation, provides novel clues about ancient populations in Africa that pre-dated the Bantu expansion, provides the first practical framework for genetic research in Africa, and developed an invaluable resource for researchers across the world. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12**.Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. Using a combination of low-coverage whole-genome sequencing, deep exome sequencing, and dense microarray genotyping, the 1000 Genomes Project reconstructed the genomes of 2,504 individuals from 26 populations. It characterized a broad spectrum of genetic variation, described the distribution of these variations across the global sample, and discussed the implications for common disease studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schuster SC, Miller W, Ratan A, Tomsho LP, Giardine B, Kasson LR, Harris RS, Petersen DC, Zhao F, Qi J, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joshi PK, Esko T, Mattsson H, Eklund N, Gandin I, Nutile T, Jackson AU, Schurmann C, Smith AV, Zhang W, et al. Directional dominance on stature and cognition in diverse human populations. Nature. 2015;523:459–462. doi: 10.1038/nature14618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shriner D, Bentley AR, Doumatey AP, Chen G, Zhou J, Adeyemo A, Rotimi CN. Phenotypic variance explained by local ancestry in admixed African Americans. Front Genet. 2015;6:324. doi: 10.3389/fgene.2015.00324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avena S, Via M, Ziv E, Perez-Stable EJ, Gignoux CR, Dejean C, Huntsman S, Torres-Mejia G, Dutil J, Matta JL, et al. Heterogeneity in genetic admixture across different regions of Argentina. PLoS One. 2012;7:e34695. doi: 10.1371/journal.pone.0034695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moreno-Estrada A, Gravel S, Zakharia F, McCauley JL, Byrnes JK, Gignoux CR, Ortiz-Tello PA, Martinez RJ, Hedges DJ, Morris RW, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19*.Homburger JR, Moreno-Estrada A, Gignoux CR, Nelson D, Sanchez E, Ortiz-Tello P, Pons-Estel BA, Acevedo-Vasquez E, Miranda P, Langefeld CD, et al. Genomic Insights into the Ancestry and Demographic History of South America. PLoS Genet. 2015;11:e1005602. doi: 10.1371/journal.pgen.1005602. This study used genome-wide SNP data for 437 admixed individuals from 5 countries (Colombia, Ecuador, Peru, Chile, and Argentina) to explore the population structure and demographic history of South American Latinos. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20*.Kehdy FS, Gouveia MH, Machado M, Magalhaes WC, Horimoto AR, Horta BL, Moreira RG, Leal TP, Scliar MO, Soares-Souza GB, et al. Origin and dynamics of admixture in Brazilians and its effect on the pattern of deleterious mutations. Proc Natl Acad Sci U S A. 2015;112:8696–8701. doi: 10.1073/pnas.1504447112. The EPIGEN Brazil Project studied the genomic diversity of admixed 6,487 Brazilians. Findings broadened understanding of the African diaspora by revealing an African ancestry component that likely derives from the slave trade. It showed that continental admixture is the main and complex determinant of the amount of deleterious genotypes in admixed individuals. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanchard N, Elzein A, Trafford C, Rockett K, Pinder M, Jallow M, Harding R, Kwiatkowski D, McKenzie C. Classical sickle beta-globin haplotypes exhibit a high degree of long-range haplotype similarity in African and Afro-Caribbean populations. BMC Genet. 2007;8:52. doi: 10.1186/1471-2156-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tishkoff SA, Varkonyi R, Cahinhinan N, Abbes S, Argyropoulos G, Destro-Bisol G, Drousiotou A, Dangerfield B, Lefranc G, Loiselet J, et al. Haplotype diversity and linkage disequilibrium at human G6PD: recent origin of alleles that confer malarial resistance. Science. 2001;293:455–462. doi: 10.1126/science.1061573. [DOI] [PubMed] [Google Scholar]
- 24.Scheinfeldt LB, Soi S, Thompson S, Ranciaro A, Woldemeskel D, Beggs W, Lambert C, Jarvis JP, Abate D, Belay G, et al. Genetic adaptation to high altitude in the Ethiopian highlands. Genome Biol. 2012;13:R1. doi: 10.1186/gb-2012-13-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huerta-Sanchez E, Degiorgio M, Pagani L, Tarekegn A, Ekong R, Antao T, Cardona A, Montgomery HE, Cavalleri GL, Robbins PA, et al. Genetic signatures reveal high-altitude adaptation in a set of ethiopian populations. Mol Biol Evol. 2013;30:1877–1888. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Udpa N, Ronen R, Zhou D, Liang J, Stobdan T, Appenzeller O, Yin Y, Du Y, Guo L, Cao R, et al. Whole genome sequencing of Ethiopian highlanders reveals conserved hypoxia tolerance genes. Genome Biol. 2014;15:R36. doi: 10.1186/gb-2014-15-2-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, et al. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, et al. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, et al. MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet. 2008;40:1185–1192. doi: 10.1038/ng.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopp JB, Smith MW, Nelson GW, Johnson RC, Freedman BI, Bowden DW, Oleksyk T, McKenzie LM, Kajiyama H, Ahuja TS, et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosset S, Tzur S, Behar DM, Wasser WG, Skorecki K. The population genetics of chronic kidney disease: insights from the MYH9-APOL1 locus. Nat Rev Nephrol. 2011;7:313–326. doi: 10.1038/nrneph.2011.52. [DOI] [PubMed] [Google Scholar]
- 32**.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, Winkler CA, Kopp J, Rotimi C, Adeyemo A, et al. Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci U S A. 2014;111:E2130–2139. doi: 10.1073/pnas.1400699111. This paper combined genetic, physiological, and biochemical studies to explore coevolution between the APOL1 gene and trypanosomes by analyzing the APOL1 sequences in modern and archaic humans and baboons along with geographic distribution in present day Africa to understand how the APOL1 kidney risk variants evolved. It showed that APOL1 variants that protect against T. brucei rhodesiense have recapitulated molecular signatures found in Old World monkeys. This study raises the possibility that APOL1 has innate immune activity that extends beyond trypanosomes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kofman T, Audard V, Narjoz C, Gribouval O, Matignon M, Leibler C, Desvaux D, Lang P, Grimbert P. APOL1 polymorphisms and development of CKD in an identical twin donor and recipient pair. Am J Kidney Dis. 2014;63:816–819. doi: 10.1053/j.ajkd.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 34.Dummer PD, Limou S, Rosenberg AZ, Heymann J, Nelson G, Winkler CA, Kopp JB. APOL1 Kidney Disease Risk Variants: An Evolving Landscape. Semin Nephrol. 2015;35:222–236. doi: 10.1016/j.semnephrol.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukamal KJ, Tremaglio J, Friedman DJ, Ix JH, Kuller LH, Tracy RP, Pollak MR. APOL1 Genotype, Kidney and Cardiovascular Disease, and Death in Older Adults. Arterioscler Thromb Vasc Biol. 2016;36:398–403. doi: 10.1161/ATVBAHA.115.305970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36**.Bentley AR, Divers J, Shriner D, Doumatey AP, Gutierrez OM, Adeyemo AA, Freedman BI, Rotimi CN. APOL1 G1 genotype modifies the association between HDLC and kidney function in African Americans. BMC Genomics. 2015;16:421. doi: 10.1186/s12864-015-1645-7. This study discovered a lipids locus (rs328 in LPL) in African Americans for which the association of the minor allele was different depending on the ancestral background at that locus. African Americans with only West African ancestry at that locus had lipid levels similar to West Africans whereas African Americans with only European ancestry at this locus had levels more similar to that of Europeans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young JH, Chang YP, Kim JD, Chretien JP, Klag MJ, Levine MA, Ruff CB, Wang NY, Chakravarti A. Differential susceptibility to hypertension is due to selection during the out-of-Africa expansion. PLoS Genet. 2005;1:e82. doi: 10.1371/journal.pgen.0010082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooper RS, Wolf-Maier K, Luke A, Adeyemo A, Banegas JR, Forrester T, Giampaoli S, Joffres M, Kastarinen M, Primatesta P, et al. An international comparative study of blood pressure in populations of European vs. African descent BMC Med. 2005;3:2. doi: 10.1186/1741-7015-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jablonski NG, Chaplin G. Colloquium paper: human skin pigmentation as an adaptation to UV radiation. Proc Natl Acad Sci U S A. 2010;107(Suppl 2):8962–8968. doi: 10.1073/pnas.0914628107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beleza S, Santos AM, McEvoy B, Alves I, Martinho C, Cameron E, Shriver MD, Parra EJ, Rocha J. The timing of pigmentation lightening in Europeans. Mol Biol Evol. 2013;30:24–35. doi: 10.1093/molbev/mss207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Batai K, Murphy AB, Shah E, Ruden M, Newsome J, Agate S, Dixon MA, Chen HY, Deane LA, Hollowell CM, et al. Common vitamin D pathway gene variants reveal contrasting effects on serum vitamin D levels in African Americans and European Americans. Hum Genet. 2014;133:1395–1405. doi: 10.1007/s00439-014-1472-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bentley AR, Chen G, Shriner D, Doumatey AP, Zhou J, Huang H, Mullikin JC, Blakesley RW, Hansen NF, Bouffard GG, et al. Gene-based sequencing identifies lipid-influencing variants with ethnicity-specific effects in African Americans. PLoS Genet. 2014;10:e1004190. doi: 10.1371/journal.pgen.1004190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cabral WA, Barnes AM, Adeyemo A, Cushing K, Chitayat D, Porter FD, Panny SR, Gulamali-Majid F, Tishkoff SA, Rebbeck TR, et al. A founder mutation in LEPRE1 carried by 1.5% of West Africans and 0. 4% of African Americans causes lethal recessive osteogenesis imperfecta. Genet Med. 2012;14:543–551. doi: 10.1038/gim.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3. 1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bansal V, Libiger O. Fast individual ancestry inference from DNA sequence data leveraging allele frequencies for multiple populations. BMC Bioinformatics. 2015;16:4. doi: 10.1186/s12859-014-0418-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skotte L, Korneliussen TS, Albrechtsen A. Estimating individual admixture proportions from next generation sequencing data. Genetics. 2013;195:693–702. doi: 10.1534/genetics.113.154138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Guan Y. Detecting structure of haplotypes and local ancestry. Genetics. 2014;196:625–642. doi: 10.1534/genetics.113.160697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang C, Zhan X, Liang L, Abecasis GR, Lin X. Improved ancestry estimation for both genotyping and sequencing data using projection procrustes analysis and genotype imputation. American Journal of Human Genetics. 2015;96:926–937. doi: 10.1016/j.ajhg.2015.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, et al. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tekola-Ayele F, Adeyemo A, Chen G, Hailu E, Aseffa A, Davey G, Newport MJ, Rotimi CN. Novel genomic signals of recent selection in an Ethiopian population. Eur J Hum Genet. 2015;23:1085–1092. doi: 10.1038/ejhg.2014.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Andersen KG, Shylakhter I, Tabrizi S, Grossman SR, Happi CT, Sabeti PC. Genome-wide scans provide evidence for positive selection of genes implicated in Lassa fever. Philos Trans R Soc Lond B Biol Sci. 2012;367:868–877. doi: 10.1098/rstb.2011.0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kunz S, Rojek JM, Kanagawa M, Spiropoulou CF, Barresi R, Campbell KP, Oldstone MB. Posttranslational modification of alpha-dystroglycan, the cellular receptor for arenaviruses, by the glycosyltransferase LARGE is critical for virus binding. J Virol. 2005;79:14282–14296. doi: 10.1128/JVI.79.22.14282-14296.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gething PW, Elyazar IR, Moyes CL, Smith DL, Battle KE, Guerra CA, Patil AP, Tatem AJ, Howes RE, Myers MF, et al. A long neglected world malaria map: Plasmodium vivax endemicity in 2010. PLoS Negl Trop Dis. 2012;6:e1814. doi: 10.1371/journal.pntd.0001814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Howes RE, Patil AP, Piel FB, Nyangiri OA, Kabaria CW, Gething PW, Zimmerman PA, Barnadas C, Beall CM, Gebremedhin A, et al. The global distribution of the Duffy blood group. Nat Commun. 2011;2:266. doi: 10.1038/ncomms1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Howes RE, Reiner RC, Jr, Battle KE, Longbottom J, Mappin B, Ordanovich D, Tatem AJ, Drakeley C, Gething PW, Zimmerman PA, et al. Plasmodium vivax Transmission in Africa. PLoS Negl Trop Dis. 2015;9:e0004222. doi: 10.1371/journal.pntd.0004222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Langhi DM, Jr, Bordin JO. Duffy blood group and malaria. Hematology. 2006;11:389–398. doi: 10.1080/10245330500469841. [DOI] [PubMed] [Google Scholar]
- 57.Mendis K, Sina BJ, Marchesini P, Carter R. The neglected burden of Plasmodium vivax malaria. Am J Trop Med Hyg. 2001;64:97–106. doi: 10.4269/ajtmh.2001.64.97. [DOI] [PubMed] [Google Scholar]
- 58.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 59.Reich D, Nalls MA, Kao WH, Akylbekova EL, Tandon A, Patterson N, Mullikin J, Hsueh WC, Cheng CY, Coresh J, et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 2009;5:e1000360. doi: 10.1371/journal.pgen.1000360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aidoo M, Terlouw DJ, Kolczak MS, McElroy PD, ter Kuile FO, Kariuki S, Nahlen BL, Lal AA, Udhayakumar V. Protective effects of the sickle cell gene against malaria morbidity and mortality. Lancet. 2002;359:1311–1312. doi: 10.1016/S0140-6736(02)08273-9. [DOI] [PubMed] [Google Scholar]
- 61.Allison AC. Protection afforded by sickle-cell trait against subtertian malareal infection. Br Med J. 1954;1:290–294. doi: 10.1136/bmj.1.4857.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fumagalli M, Cagliani R, Pozzoli U, Riva S, Comi GP, Menozzi G, Bresolin N, Sironi M. Widespread balancing selection and pathogen-driven selection at blood group antigen genes. Genome Res. 2009;19:199–212. doi: 10.1101/gr.082768.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, Bennett S, Brewster D, McMichael AJ, Greenwood BM. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 64.Pasvol G, Weatherall DJ, Wilson RJ. Cellular mechanism for the protective effect of haemoglobin S against P. falciparum malaria. Nature. 1978;274:701–703. doi: 10.1038/274701a0. [DOI] [PubMed] [Google Scholar]
- 65.Piel FB, Patil AP, Howes RE, Nyangiri OA, Gething PW, Dewi M, Temperley WH, Williams TN, Weatherall DJ, Hay SI. Global epidemiology of sickle haemoglobin in neonates: a contemporary geostatistical model-based map and population estimates. Lancet. 2013;381:142–151. doi: 10.1016/S0140-6736(12)61229-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Enevold A, Alifrangis M, Sanchez JJ, Carneiro I, Roper C, Borsting C, Lusingu J, Vestergaard LS, Lemnge MM, Morling N, et al. Associations between alpha+-thalassemia and Plasmodium falciparum malarial infection in northeastern Tanzania. J Infect Dis. 2007;196:451–459. doi: 10.1086/519390. [DOI] [PubMed] [Google Scholar]
- 67.Muncie HL, Jr, Campbell J. Alpha and beta thalassemia. Am Fam Physician. 2009;80:339–344. [PubMed] [Google Scholar]
- 68.Eugen-Olsen J, Iversen AK, Garred P, Koppelhus U, Pedersen C, Benfield TL, Sorensen AM, Katzenstein T, Dickmeiss E, Gerstoft J, et al. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. Aids. 1997;11:305–310. doi: 10.1097/00002030-199703110-00007. [DOI] [PubMed] [Google Scholar]
- 69.Kindberg E, Mickiene A, Ax C, Akerlind B, Vene S, Lindquist L, Lundkvist A, Svensson L. A deletion in the chemokine receptor 5 (CCR5) gene is associated with tickborne encephalitis. J Infect Dis. 2008;197:266–269. doi: 10.1086/524709. [DOI] [PubMed] [Google Scholar]
- 70.Lim JK, Louie CY, Glaser C, Jean C, Johnson B, Johnson H, McDermott DH, Murphy PM. Genetic deficiency of chemokine receptor CCR5 is a strong risk factor for symptomatic West Nile virus infection: a meta-analysis of 4 cohorts in the US epidemic. J Infect Dis. 2008;197:262–265. doi: 10.1086/524691. [DOI] [PubMed] [Google Scholar]
- 71.Michael NL, Chang G, Louie LG, Mascola JR, Dondero D, Birx DL, Sheppard HW. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat Med. 1997;3:338–340. doi: 10.1038/nm0397-338. [DOI] [PubMed] [Google Scholar]
- 72.Ingram CJ, Mulcare CA, Itan Y, Thomas MG, Swallow DM. Lactose digestion and the evolutionary genetics of lactase persistence. Hum Genet. 2009;124:579–591. doi: 10.1007/s00439-008-0593-6. [DOI] [PubMed] [Google Scholar]
- 73.Itan Y, Jones BL, Ingram CJ, Swallow DM, Thomas MG. A worldwide correlation of lactase persistence phenotype and genotypes. BMC Evol Biol. 2010;10:36. doi: 10.1186/1471-2148-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–1260. doi: 10.1038/ng2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.World Health Organization. Global Health Observatory. 2016 Accessed from: http://www.who.int/topics/trypanosomiasis_african/en/
- 76.Rotimi CN, Dunston GM, Berg K, Akinsete O, Amoah A, Owusu S, Acheampong J, Boateng K, Oli J, Okafor G, et al. In search of susceptibility genes for type 2 diabetes in West Africa: the design and results of the first phase of the AADM study. Ann Epidemiol. 2001;11:51–58. doi: 10.1016/s1047-2797(00)00180-0. [DOI] [PubMed] [Google Scholar]
- 77.Ko WY, Rajan P, Gomez F, Scheinfeldt L, An P, Winkler CA, Froment A, Nyambo TB, Omar SA, Wambebe C, et al. Identifying Darwinian selection acting on different human APOL1 variants among diverse African populations. Am J Hum Genet. 2013;93:54–66. doi: 10.1016/j.ajhg.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Behar DM, Kedem E, Rosset S, Haileselassie Y, Tzur S, Kra-Oz Z, Wasser WG, Shenhar Y, Shahar E, Hassoun G, et al. Absence of APOL1 risk variants protects against HIV-associated nephropathy in the Ethiopian population. Am J Nephrol. 2011;34:452–459. doi: 10.1159/000332378. [DOI] [PubMed] [Google Scholar]


