Abstract
Background
We aimed to identify the disease-causing mutations in a consanguineous family featuring intellectual disability and parkinsonism.
Methods
Full phenotypic characterization, followed by genome-wide SNP genotyping and whole genome sequencing, was carried out in all available family members.
Results
The chromosome 2p23.3 was identified as the disease-associated locus, and a homozygous PTRHD1 mutation (c.157C>T) was then established as the disease-causing mutation. The pathogenicity of this PTRHD1 mutation was supported by its segregation with the disease status, its location in a functional domain of the encoding protein, as well as its absence in public databases and ethnicity-matched control chromosomes.
Conclusions
Given the role of 2p23 locus in patients with intellectual disability and the previously reported PTRHD1 mutation (c.155G>A) in patients with parkinsonism and cognitive dysfunction, we concluded that the PTRHD1 mutation identified in this study is likely to be responsible for the phenotypic features of the family under consideration.
Keywords: Intellectual disability, parkinsonism, 2p23.3, PTRHD1 mutation
Introduction
Early-onset parkinsonism (EOP) is a neurodegenerative disease characterized by the young-onset presentations of tremor, muscular rigidity, postural instability, and slowing of movements.1 The most common cause of parkinsonism is Parkinson’s disease (PD), accounting for approximately 80% of the cases with parkinsonism. Atypical juvenile parkinsonism (AJP) usually refers to a complex form of EOP that is inherited in a recessive manner and manifests with diverse neurological and psychiatric manifestations, including pyramidal signs, abnormalities of eye movements, depression, anxiety, psychosis, impulse control disorders, and intellectual disability (ID), among others.2 To date, pathogenic mutations in at least eight different genes have been reported in AJP. These include ATP13A2 [1p36; MIM# 606693], DNAJC6 [1p31.3; MIM# 608375], FBXO7 [22q12.3; MIM# 260300], PLA2G6 [22q12.3; MIM# 612953], SPG11 [15q13-q15; MIM# 610844], SPG15 [14q24.1; MIM# 270700], SYNJ1 [21q22.2; MIM #615530], and VPS13C [15q22.2; MIM# 616840] genes.2–7
ID is characterized by below-average intelligence or mental ability to perform activities of daily living. It is well known that some patients with parkinsonism may manifest ID or some kind of cognitive dysfunction. For instance, patients with null DNAJC6 mutations or large DNAJC6 deletions may develop ID with or without parkinsonism.8, 9 More recently, two other genes have been identified as causatives for ID and PD or motor impairment. These are RAB39B [Xq28; MIM# 300271], mutations of which are reported in ID10 and in patients with ID and parkinsonism,11, 12 and RARB [3p24.2; MIM# 615524], mutations of which have been found in patients with microphthalmia, ID, and progressive motor impairment.13
The goal of this study was to identify the disease-causing mutations in a consanguineous family with two affected siblings featuring ID and parkinsonism without mutations in known genes.
Methods
Subjects
An Iranian consanguineous family featuring early-onset autosomal recessive parkinsonism and ID was clinically examined. The family consisted of healthy parents, two affected sons, and unaffected daughter (Fig. 1A). DNA samples from probands of 15 unrelated families with ID and a movement disorder phenotype and from 504 ethnicity-matched control individuals were as well available. The local ethics committee at Shahid Beheshti University of Medical Sciences approved this study, and informed consent according to the Declaration of Helsinki was obtained from all participants. DNA samples from all participants were isolated from whole blood using standard procedures.
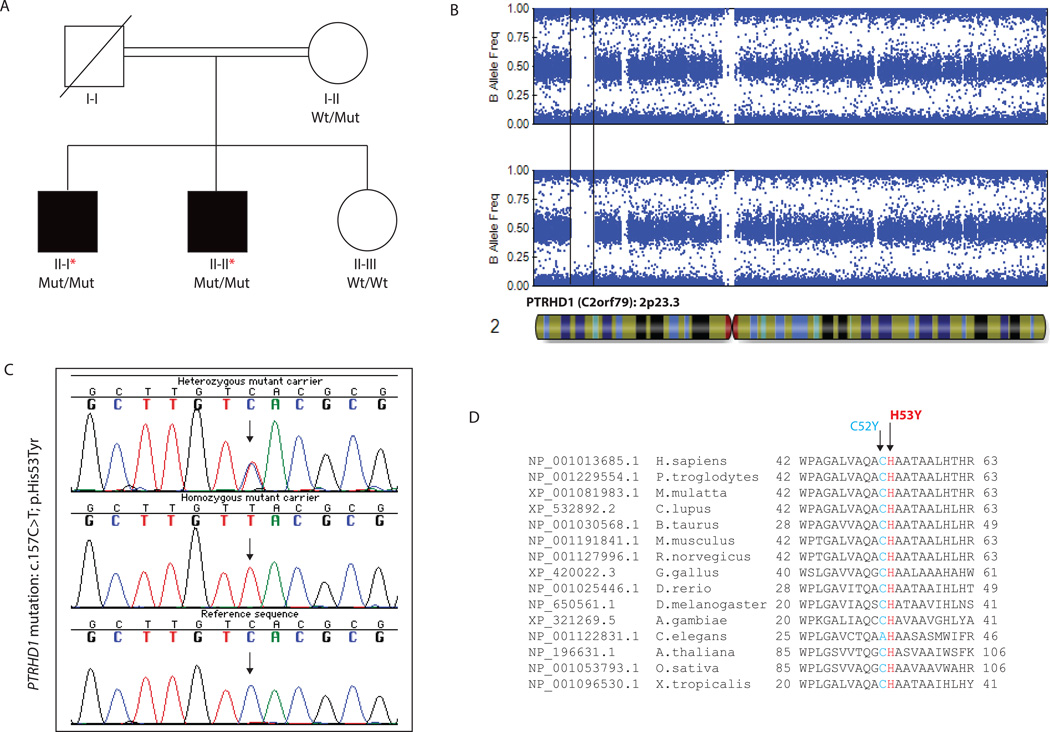
FIG. 1.
(A) Pedigree of the family featuring ID and parkinsonism. Wt/Mut indicates heterozygous carrier for the PTRHD1 c.157C>T mutation; Mut/Mut indicates homozygous carrier; and Wt/Wt indicates non-carrier. Affected siblings are represented with black squares. (B) B allele frequency plots of patients’ chromosome 2 acquired from the Illumina Genome Viewer (IGV) tool within the GS software (Illumina). The final disease-associated homozygous track of 8.2 Mb (2p23.3) is represented between black lines. (C) Sanger chromatogram sequences of the PTRHD1 exon 1 containing the c.157C>T mutation (arrow). (D) Conservation among other orthologous of the two reported PTRHD1 mutations. The mutation reported in this study is highlighted in red while the mutation reported by Jaberi and colleagues is highlighted in blue.
Clinical evaluation
All participants underwent a series of structured questionnaires and a comprehensive neurological and neuropsychological assessment undertaken at the Movement Disorders Unit by three experienced movement disorder specialists. The performed neuropsychological examinations are fully described in the Supplementary Material.
Homozygosity mapping
High-throughput SNP genotyping was carried out in all available family members (Fig. 1A) by using the HumanOmniExpress Exome arrays v1.3 and HiScanSQ system (Illumina Inc., San Diego, CA, USA). Genotyping data was used to perform homozygosity mapping (HM) as previously described (Fig. 1B).3
Whole genome sequencing
WGS was carried out at the New York Genome Center (NYGC) in the two affected family members (II-I, II-II) as described in the Supplementary Material.
Validation and disease-segregating analyses
Primer sequences for the entire coding region of the PTRHD1 gene, which stands for peptidyl-tRNA hydrolase domain containing 1, were designed by using a public primer design website (http://ihg.gsf.de/ihg/ExonPrimer.html). Direct Sanger sequencing using primers flanking the exon 1 of the PTRHD1 (C2orf79) gene was used to validate the c.157C>T mutation (Fig. 1C). Once validated, the mutation was examined in available family members for disease segregation analyses. The entire PTRHD1 gene was examined in 15 familial cases with ID and a movement disorder phenotype, including parkinsonism and/or tremor. The PTRHD1 exon 1 was additionally examined in 104 Iranian control individuals through Sanger sequencing and in 400 Iranian control individuals through allele-specific amplification. For Sanger assays, all purified PCR products were sequenced, resolved, and analyzed as previously described.3 Allele-specific detection was performed using designed primers for the identified mutation and PCR products were analyzed by agarose gel electrophoresis.
Results
Patients’ clinical details
The proband (II-I) presented with ID, muscle stiffness, rest and postural tremor, postural instability, gait disturbances, speech difficulties, as well as psychiatric symptoms such as anxiety, hypersomnia, and hypersexuality. His brother showed similar symptoms but milder, and no additional family members were affected. Full patient’s clinical details are described in the Supplementary Material.
Molecular Analyses
The performed HM identified a total of six potential disease-associated loci shared only by the two affected individuals (Supplementary Table 1). WGS was then performed on the two affected individuals (II-I and II-II). After filtering for common genetic variation, 194 genomic variations, including missense (n = 133), frameshift, splice-site, stop-gained, and start-lost (n = 61) mutations, were found to be present in both patients. Among these, only a homozygous missense mutation was located in one out of the six previously identified disease-associated loci (Fig. 1B). No other homozygous or compound heterozygous mutations were identified outside the disease-associated loci. We then investigated whether genomic deletions to be shared exclusively by affected individuals might as well be causative; however, no genomic deletions were found in the identified disease-associated homozygous segments, and no common genomic deletions were identified within the exome. Due to the presence of parkinsonism in both the affected individuals, PD- and parkinsonism-associated loci were as well examined, but no shared region of homozygosity was observed within the known parkinsonism loci, and no pathogenic mutation was identified within the known PD and parkinsonism genes.
The only homozygous mutation identified in a previously identified disease-associated locus was located in the PTRHD1 gene (C2orf79) at chromosome 2p23.3. It consisted in a C to T transition at nucleotide 157 (c.157C>T), leading to the p.His53Tyr change on the protein level. It did segregate with disease status and was highly conserved among other orthologous (Fig. 1A–C). It was predicted to be pathogenic by various computational methods, was not previously reported, and was not found in any public SNV database, including the NHLBI GO Exome Sequencing Project and the Exome Aggregation Consortium (ExAC; exac.broadinstitute.org) (Table 1). The PTRHD1 c.157C>T mutation was then examined in 504 ethnicity-matched neurologically normal individuals and found to be absent in 1,008 control chromosomes, further supporting its pathogenicity. Fifteen additional families with ID and a movement disorder were selected and screened for PTRHD1 mutations; however, no pathogenic PTRHD1 mutation was identified in these newly recruited cases.
Table 1.
PTRHD1 mutations in Families with ID and parkinsonism
| Gene | Nucleotide Change |
Amino Acid Change |
Protein Domain |
Pathogenicity Predictions | Iranian Control Population |
ExAC Browser |
References | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| SIFT | Polyphen2 | MutPred | SNPs&GO | CADD- phred |
|||||||
| PTRHD1 (C2orf29) |
c.155G>A | p.Cys52Tyr | PTH2 | Deleterious | Probably damaging |
0.749 | 0.604 | 34 | 0/208* | 1 / 119394 | Jaberi at al, 2016 |
|
PTRHD1 (C2orf29) |
c.157C>T | p.His53Tyr | PTH2 | Deleterious | Deleterious | 0.783 | 0.764 | 37 | 0/1008 | Not Found | This study |
The mutation identified in this study is shown in bold. PTH2 stands for peptidyl-tRNA hydrolase 2.
These controls individuals were sequenced in this study as Jaberi and colleagues did not report control data for this specific mutation.
Discussion
Here, we describe the identification of a novel causative gene for a complex form of autosomal recessive ID with parkinsonism. The combination of HM and WGS led us to assign the chromosome 2p23.3 locus as the disease-associated locus, and to identify a homozygous variation in the PTRHD1 gene as the disease-causing mutation (Fig. 1; Table 1). The disease-segregating PTRHD1 mutation, predicted to be pathogenic by various computational methods, was shown to be absent in over 1,000 ethnicity-matched control chromosomes and SNV public databases, likely supporting its pathogenicity.
Despite PTRHD1 mutations having never been reported in parkinsonism or ID, deletions at the 2p23 locus, encompassing the PTRHD1 gene, are known to be associated with several ID syndromes (Supplementary Table 2).14–20 Furthermore, Jaberi et al recently identified a missense PTRHD1 mutation (c.155G>A; p.Cys52Tyr) in a family with early-onset parkinsonism and cognitive dysfunction.21 Although they claimed that the strongest candidate gene for the disease was ADORA1 due to the role of adenosine receptors in brain function and neuronal activity, there were no substantial proofs to discard PTRHD1 as a causative gene, as was well acknowledged in their report.21 The PTRHD1 p.Cys52Tyr mutation was predicted to be pathogenic by various computational methods, was found with extremely low frequency in public SNV databases, and was absent in 208 ethnicity-matched control chromosomes sequenced by us (Table 1). In any case, to exclude ADORA1 as a causative gene, we further investigated the coverage of ADORA1 gene in our SNPs genotyping and WGS data. A total of 33 SNPs covering the ADORA1 locus were genotyped with only 63% of the SNPs being homozygous for Patient 1 (Supplementary Table 3), further excluding ADORA1 as the disease-associated locus. Sequencing of the ADORA1 gene at more than 30x coverage identified no pathogenic mutations, further supporting the role of PTRHD1 as a causative gene in both the reported families.
Although little is known about the function of PTRHD1, both PTRHD1 mutations identified in families with ID and parkinsonism lie within the PTH2 domain (25-139 amino-acids) of the PTRHD1 protein (Table 1). The PTH2 domain is a ubiquitin-like (UBL) domain-binding protein that is known to participate in the ubiquitin-proteasome pathway and has been shown to suppress ubiquitin-mediated degradation.22 In yeast, overexpression of PTH2 has been shown to cause accumulation of polyubiquitinated proteins and to inhibit growth. It is also known that PTH2 binds to the UBL domain of Rad23 and Dsk2 and interacts with polyubiquitinated proteins through their ubiquitin-associated domains.22 The role of ubiquitin-mediated proteolysis in the pathogenesis of ID and neurodegenerative diseases, such as PD and Alzheimer’s disease, is well known and documented.23 Indeed, PD is characterized by the loss of dopaminergic neurons and the progressive neuronal accumulation of protein inclusions containing alpha-synuclein and ubiquitin;24 two different protein ubiquitin ligases (Parkin and FBXO7) are well established as causative genes for early-onset forms of parkinsonism;2, 25–27 and overexpression of mutant alpha-synuclein as well as downregulation of Parkin have been shown to increase sensitivity to proteasome inhibitors, decreasing proteasomal function.28
Moreover, mutations in various ubiquitin enzymes participating in the ubiquitin-proteasome system are known to be associated with the pathophysiology of ID. This is the case of UBE3A (MIM# 105830), mutations of which cause Angelman syndrome29, as well as UBE2A (MIM# 300860), CUL4B (MIM# 300354), and BRWD3 (MIM# 300659), mutations of which are known to cause X-linked ID syndromes.30–32
Therefore, given the role that the ubiquitin proteasome system plays in brain development, synaptic plasticity, and long-term memory formation,33 and the previous association of the chromosome 2p23.3 locus with ID, we speculated that PTRHD1 genetic variability might cause ID and parkinsonism through defects in the ubiquitin-proteasome system, as observed in other inherited forms of parkinsonism and ID.
In conclusion, this study describes for the first time the association of PTRHD1, known to participate in the ubiquitin-proteasome pathway, with the pathogenesis of ID and parkinsonism.
Supplementary Material
Acknowledgments
We thank, patients, relatives and all other participants for their cooperation in this study.
Research project (1): A. Conception (VA, AT, GAS, HD, CPR), B. Organization (HK, LJA, MDO, MG, ST, JJ, VA, AT, GAS, HD, CPR), C. Execution (HK, LJA, MDO, MG, ST, JJ, VA, AT, GAS, HD, CPR). Clinical Assessments (2): A. Design (VA, AT, GAS), B. Execution (VA, AT, GAS), C. Review and Critique (VA, AT, GAS). HM, WGS, Sanger sequencing analyses (3): A. Design (HD, CPR), B. Execution (HK, LJA, MDO, MG, ST, JJ, HD, CPR) C. Review and Critique (HK, LJA, MDO, MG, ST, JJ, HD, CPR). Manuscript Preparation (4): A. Writing of the first draft (CPR), B. Review and Critique (HK, LJA, MDO, MG, ST, JJ, VA, AT, GAS, HD, CPR).
Funding Agencies: This work was supported by the Shahid Beheshti University of Medical Sciences and the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (R01NS079388; CPR).
Footnotes
Relevant conflicts of interests/financial disclosures: The authors declare no conflict of interest and have nothing to report.
References
- 1.Jankovic J. Parkinson's disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry. 2008;79(4):368–376. doi: 10.1136/jnnp.2007.131045. [DOI] [PubMed] [Google Scholar]
- 2.Paisan-Ruiz C, Guevara R, Federoff M, et al. Early-onset L-dopa-responsive parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Mov Disord. 2010;25(12):1791–1800. doi: 10.1002/mds.23221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krebs CE, Karkheiran S, Powell JC, et al. The Sac1 domain of SYNJ1 identified mutated in a family with early-onset progressive Parkinsonism with generalized seizures. Hum Mutat. 2013;34(9):1200–1207. doi: 10.1002/humu.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paisan-Ruiz C, Bhatia KP, Li A, et al. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann Neurol. 2009;65(1):19–23. doi: 10.1002/ana.21415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edvardson S, Cinnamon Y, Ta-Shma A, et al. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS One. 2012;7(5):e36458. doi: 10.1371/journal.pone.0036458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schicks J, Synofzik M, Petursson H, et al. Atypical juvenile parkinsonism in a consanguineous SPG15 family. Mov Disord. 2011;26(3):564–566. doi: 10.1002/mds.23472. [DOI] [PubMed] [Google Scholar]
- 7.Lesage S, Drouet V, Majounie E, et al. Loss of VPS13C Function in Autosomal-Recessive Parkinsonism Causes Mitochondrial Dysfunction and Increases PINK1/Parkin-Dependent Mitophagy. Am J Hum Genet. 2016;98(3):500–513. doi: 10.1016/j.ajhg.2016.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koroglu C, Baysal L, Cetinkaya M, Karasoy H, Tolun A. DNAJC6 is responsible for juvenile parkinsonism with phenotypic variability. Parkinsonism Relat Disord. 2013;19(3):320–324. doi: 10.1016/j.parkreldis.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 9.Vauthier V, Jaillard S, Journel H, Dubourg C, Jockers R, Dam J. Homozygous deletion of an 80 kb region comprising part of DNAJC6 and LEPR genes on chromosome 1P31.3 is associated with early onset obesity, mental retardation and epilepsy. Mol Genet Metab. 2012;106(3):345–350. doi: 10.1016/j.ymgme.2012.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Giannandrea M, Bianchi V, Mignogna ML, et al. Mutations in the small GTPase gene RAB39B are responsible for X-linked mental retardation associated with autism, epilepsy, and macrocephaly. Am J Hum Genet. 2010;86(2):185–195. doi: 10.1016/j.ajhg.2010.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lesage S, Bras J, Cormier-Dequaire F, et al. Loss-of-function mutations in RAB39B are associated with typical early-onset Parkinson disease. Neurol Genet. 2015;1(1):e9. doi: 10.1212/NXG.0000000000000009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson GR, Sim JC, McLean C, et al. Mutations in RAB39B cause X-linked intellectual disability and early-onset Parkinson disease with alpha-synuclein pathology. Am J Hum Genet. 2014;95(6):729–735. doi: 10.1016/j.ajhg.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Srour M, Caron V, Pearson T, et al. Gain-of-Function Mutations in RARB Cause Intellectual Disability with Progressive Motor Impairment. Hum Mutat. 2016 doi: 10.1002/humu.23004. [DOI] [PubMed] [Google Scholar]
- 14.Bloch M, Leonard A, Diplas AA, et al. Further phenotype description, genotype characterization in patients with de novo interstitial deletion on 2p23.2-24.1. Am J Med Genet A. 2014;164A(7):1789–1794. doi: 10.1002/ajmg.a.36516. [DOI] [PubMed] [Google Scholar]
- 15.Okamoto N, Toribe Y, Shimojima K, Yamamoto T. Tatton-Brown-Rahman syndrome due to 2p23 microdeletion. Am J Med Genet A. 2016;170(5):1339–1342. doi: 10.1002/ajmg.a.37588. [DOI] [PubMed] [Google Scholar]
- 16.Rocca MS, Faletra F, Devescovi R, Gasparini P, Pecile V. Molecular cytogenetic characterization of 2p23.2p23.3 deletion in a child with developmental delay, hypotonia and cryptorchism. Eur J Med Genet. 2013;56(1):62–65. doi: 10.1016/j.ejmg.2012.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Shoukier M, Schroder J, Zoll B, et al. A de novo interstitial deletion of 2p23.3-24.3 in a boy presenting with intellectual disability, overgrowth, dysmorphic features, skeletal myopathy, dilated cardiomyopathy. Am J Med Genet A. 2012;158A(2):429–433. doi: 10.1002/ajmg.a.34427. [DOI] [PubMed] [Google Scholar]
- 18.Su PH, Chen JY, Tsao TF, Chen SJ. De novo interstitial deletion of chromosome 2 (p23p24) Pediatr Neonatol. 2011;52(1):46–50. doi: 10.1016/j.pedneo.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 19.Neidich J, Zackai E, Aronson M, Emanuel BS. Deletion of 2p: a cytogenetic and clinical update. Am J Med Genet. 1987;27(3):707–710. doi: 10.1002/ajmg.1320270326. [DOI] [PubMed] [Google Scholar]
- 20.Penchaszadeh VB, Dowling PK, Davis JG, Schmidt R, Wapnir RA. Interstitial deletion of chromosome 2 (p23p25) Am J Med Genet. 1987;27(3):701–706. doi: 10.1002/ajmg.1320270325. [DOI] [PubMed] [Google Scholar]
- 21.Jaberi E, Rohani M, Shahidi GA, et al. Mutation in ADORA1 identified as likely cause of early-onset parkinsonism and cognitive dysfunction. Mov Disord. 2016 doi: 10.1002/mds.26627. [DOI] [PubMed] [Google Scholar]
- 22.Ishii T, Funakoshi M, Kobayashi H. Yeast Pth2 is a UBL domain-binding protein that participates in the ubiquitin-proteasome pathway. EMBO J. 2006;25(23):5492–5503. doi: 10.1038/sj.emboj.7601418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Layfield R, Cavey JR, Lowe J. Role of ubiquitin-mediated proteolysis in the pathogenesis of neurodegenerative disorders. Ageing Res Rev. 2003;2(4):343–356. doi: 10.1016/s1568-1637(03)00025-4. [DOI] [PubMed] [Google Scholar]
- 24.Shimura H, Schlossmacher MG, Hattori N, et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293(5528):263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 25.Shojaee S, Sina F, Banihosseini SS, et al. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500 K SNP arrays. Am J Hum Genet. 2008;82(6):1375–1384. doi: 10.1016/j.ajhg.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karkheiran S, Krebs CE, Darvish H, Asadian M, Shahidi GA, Paisan-Ruiz C. Variable phenotypic expression in families with early-onset Parkinsonism due to PRKN mutations. J Neurol. 2014;261(6):1223–1226. doi: 10.1007/s00415-014-7360-5. [DOI] [PubMed] [Google Scholar]
- 27.Kitada T, Asakawa S, Hattori N, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392(6676):605–608. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 28.Petrucelli L, O'Farrell C, Lockhart PJ, et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36(6):1007–1019. doi: 10.1016/s0896-6273(02)01125-x. [DOI] [PubMed] [Google Scholar]
- 29.Matsuura T, Sutcliffe JS, Fang P, et al. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15(1):74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 30.Nascimento RM, Otto PA, de Brouwer AP, Vianna-Morgante AM. UBE2A, which encodes a ubiquitin-conjugating enzyme, is mutated in a novel X-linked mental retardation syndrome. Am J Hum Genet. 2006;79(3):549–555. doi: 10.1086/507047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Field M, Tarpey PS, Smith R, et al. Mutations in the BRWD3 gene cause X-linked mental retardation associated with macrocephaly. Am J Hum Genet. 2007;81(2):367–374. doi: 10.1086/520677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tarpey PS, Raymond FL, O'Meara S, et al. Mutations in CUL4B, which encodes a ubiquitin E3 ligase subunit, cause an X-linked mental retardation syndrome associated with aggressive outbursts, seizures, relative macrocephaly, central obesity, hypogonadism, pes cavus, and tremor. Am J Hum Genet. 2007;80(2):345–352. doi: 10.1086/511134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jarome TJ, Kwapis JL, Hallengren JJ, Wilson SM, Helmstetter FJ. The ubiquitin-specific protease 14 (USP14) is a critical regulator of long-term memory formation. Learn Mem. 2014;21(1):9–13. doi: 10.1101/lm.032771.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.