Abstract
DNA methylation is one of the most common epigenetic alterations, providing important information regarding cancer risk and prognosis. A case-control study (423 breast cancer cases, 509 controls) and a case-only study (326 cases) were conducted to evaluate the association of DUSP1 promoter methylation with breast cancer risk and clinicopathological characteristics. No significant association between DUSP1 methylation in peripheral blood leukocyte (PBL) DNA and breast cancer risk was observed. DUSP1 methylation was significantly associated with ER/PR-negative status; in particular, triple-negative breast cancer patients showed the highest frequency of DUSP1 methylation in both tumour DNA and PBL DNA. Soybean intake was significantly correlated with methylated DUSP1 only in ER-negative (OR 2.978; 95% CI 1.245–7.124) and PR negative (OR 2.735; 95% CI 1.315–5.692) patients. Irregular menstruation was significantly associated with methylated DUSP1 only in ER-positive (OR 3.564; 95% CI 1.691–7.511) and PR-positive (OR 3.902, 95% CI 1.656–9.194) patients. Thus, DUSP1 methylation is a cancer-associated hypermethylation event that is closely linked with triple-negative status. Further investigations are warranted to confirm the association of environmental factors, including fruit and soybean intake, irregular menstruation, and ER/PR status, with DUSP1 methylation in breast tumour DNA.
Breast cancer is the most common cancer among women worldwide. The World Health Organization reported that there were 1.67 million new breast cancer cases and 0.52 million deaths attributed to breast cancer worldwide in 2012, while in the same year in China, newly diagnosed cases and deaths totalled 187,000 and 48,000, respectively1. According to latest estimates, 246,660 new female breast cancer cases and 40, 450 cancer deaths are projected to occur in the United States in 20162.
Among many signalling pathways, the mitogen-activated protein kinase (MAPK) cascades are central to cell proliferation and apoptosis. The first MAPK phosphatase to be identified was mitogen-activated protein kinase phosphatase-1 (MKP-1), which is encoded by the dual-specificity phosphatase 1 (DUSP1) gene and mediates the dephosphorylation of MAPKs3. MKP-1 is an endogenous inhibitor of the mitogen-activated protein kinase (MAPK) pathway through inhibiting the activation of ERK4,5. Although the mechanisms of MAPK signalling pathways in breast cancer development, progression, and tamoxifen resistance have been well-documented6,7,8,9, very little is known about the role of MKP-1 in breast carcinogenesis. Accumulating evidence has shown reduced MKP-1 mRNA or protein expression in several types of cancers including prostate10, epithelial11, renal12 and urothelial13 cancers. Chen et al.14 suggested there was a significant reduction in DUSP1 mRNA expression in five breast cancer cell lines compared with a normal control.
Carcinogenesis is a multi-stage process driven by the accumulation of genetic and epigenetic abnormalities15. DNA methylation is a critical mechanism of epigenetic modification involved in gene expression programming. Abnormal DNA methylation occurs primarily in CpG islands within gene promoters, resulting in transcriptional inactivation and gene silencing, and contributes to the tumorigenesis of several cancers16,17,18. It has been proposed that the methylation status of some CpG sites could be passed on from previous generations as an inherited marker19. Several studies have been conducted on the changes in DNA methylation in blood leukocyte DNA, and suggested a link of blood leukocyte DNA methylation with cancer susceptibility20,21,22,23. Ji-Yeob et al. found that leukocyte DNA hypomethylation is independently associated with the development of breast cancer24. Thus, peripheral blood leukocyte (PBL) DNA might be a potential surrogate biomarker for cancer risk assessment. In addition, epigenetic variation can arise as a consequence of environmental, dietary, and aging factors25,26,27,28. Different tissues may exhibit different responses to environmental factors, and methylation status in leukocytes may not fully reflect the changes in the target tissue29. Methylation–environment interactions may provide further explanations for the complexity of cancer development.
Genome-wide differing methylated regions have been detected by comparing breast tumour tissue DNA and adjacent normal tissue DNA using next-generation sequencing techniques30. Several promising methylated biomarkers have been identified from circulating cell-free DNA between breast cancer cases and controls31,32,33,34. Specific methylation patterns were proposed to correlate with distinct clinicopathological characteristics35,36 and assist in identifying individuals who will respond to therapy and survive longer. Hence, with suitable assays and validation in large populations, such associations can be exploited in non-invasive diagnosis and personalized treatment decisions.
Given the lack of research on DUSP1 methylation in breast cancer in epidemiological studies, we first investigated the association between DUSP1 methylation in PBL DNA, interactions with environmental factors, and breast cancer risk. We also explored the correlation between clinicopathological characteristics and DUSP1 methylation in both tumour DNA and PBL DNA, as well as the effect of environmental factors on DUSP1 methylation in tumour tissue DNA.
Results
Association between DUSP1 methylation in PBL DNA and breast cancer risk
PBL DNA was extracted from 423 patients and 509 controls. Supplemental Table 1 shows the distribution of demographic characteristics in cases and controls. No significant difference was found for age (P = 0.276) and BMI (P = 0.154). However, there were significant differences for the distribution of marital status (P = 0.023), educational level (P = 0.002), occupation (P = 0.001), and family history of cancer (P = 0.000) between cases and controls. Hence, these four variables were adjusted in the subsequent multivariate analyses.
DUSP1 methylation was detected in 5.2% (22/423) breast cancer cases and 4.9% (25/509) controls in PBL DNA (Table 1). After adjusting for marital status, educational level, occupation, and family history of cancer, no significant difference in DUSP1 methylation was observed between cases and controls. Therefore, we cannot conclude any association between DUSP1 methylation in PBL DNA and breast cancer risk (OR 0.79, 95% CI 0.414–1.504, P = 0.472).
Table 1. Association of DUSP1 methylation in peripheral blood leukocyte DNA and environmental factors on breast cancer risk.
| Factors | Cases No. (%) | Controls No. (%) | ORcrude (95%CI) | P-value | ORadj (95%CI) | P-value | Interaction | |
|---|---|---|---|---|---|---|---|---|
| ORi (95%CI) | P-value | |||||||
| 423 | 509 | |||||||
| DUSP1 methylation | ||||||||
| Methylated | 22 (5.2) | 25 (4.9) | 1.000 | 1.000 | ||||
| Unmethylated | 401 (94.8) | 484 (95.1) | 0.790 (0.414–1.504) | 0.472 | 1.247 (0.540–2.878) | 0.605 | ||
| Refined grain (g/day) | ||||||||
| <100 | 260 (57.4) | 290 (42.6) | 1.000 | 1.000 | 1.000 | |||
| ≥100 | 90 (25.7) | 215 (97.2) | 0.434 (0.308–0.610) | 0.000 | 0.424 (0.275–0.652) | 0.000 | 2.169 (0.463–10.163) | 0.326 |
| Vegetable (g/day) | ||||||||
| <500 | 283 (68.2) | 261 (52.0) | 1.000 | 1.000 | 1.000 | |||
| ≥500 | 132 (31.8) | 241 (48.0) | 0.657 (0.487–0.888) | 0.006 | 0.374 (0.241–0.582) | 0.000 | 0.581 (0.156–2.167) | 0.419 |
| Fruit (g/week) | ||||||||
| <1500 | 216 (52.4) | 291 (58.3) | 1.000 | 1.000 | 1.000 | |||
| ≥1500 | 196 (47.6) | 208 (41.7) | 1.569 (1.164–2.114) | 0.003 | 1.775 (1.172–2.687) | 0.007 | 2.030 (0.491–8.403) | 0.328 |
| Garlic (times/week) | ||||||||
| <4 | 50 (11.8) | 288 (56.6) | 1.000 | 1.000 | 1.000 | |||
| ≥4 | 173 (41.7) | 221 (43.4) | 0.228 (0.157–0.332) | 0.000 | 0.233 (0.143–0.380) | 0.000 | 0.589 (0.056–6.151) | 0.659 |
| Seafood (times/month) | ||||||||
| <1 | 378 (91.3) | 421 (83.2) | 1.000 | 1.000 | 1.000 | |||
| ≥1 | 36 (8.7) | 85 (16.8) | 0.528 (0.325–0.857) | 0.010 | 0.431 (0.223–0.832) | 0.012 | 1.260 (0.185–8.602) | 0.814 |
| Milk (times/week) | ||||||||
| <3 | 310 (74.0) | 300 (59.8) | 1.000 | 1.000 | 1.000 | |||
| ≥3 | 109 (26.0) | 202 (40.2) | 0.487 (0.349–0.679) | 0.000 | 0.402 (0.252–0.641) | 0.000 | 1.439 (0.350–5.910) | 0.614 |
| Health care products | ||||||||
| No | 244 (58.5) | 35 (72.3) | 1.000 | 1.000 | 1.000 | |||
| Yes | 173 (41.5) | 13 (27.7) | 1.685 (1.224–2.320) | 0.001 | 2.356 (1.505–3.689) | 0.000 | 0.455 (0.113–1.837) | 0.269 |
| Overnight Food (times/week) | ||||||||
| ≤3 | 239 (57.2) | 350 (70.4) | 1.000 | 1.000 | 1.000 | |||
| >3 | 179 (42.8) | 147 (29.6) | 1.934 (1.422–2.632) | 0.000 | 1.745 (1.152–2.643) | 0.009 | 1.172 (0.298–4.603) | 0.821 |
| Sports | ||||||||
| No | 270 (64.4) | 270 (53.7) | 1.000 | 1.000 | 1.000 | |||
| Yes | 149 (35.6) | 233 (46.3) | 0.490 (0.362–0.665) | 0.000 | 0.447 (0.294–0.681) | 0.000 | 1.552 (0.415–5.807) | 0.514 |
| Menstrual cycle | ||||||||
| regular | 342 (82.2) | 45 (90.7) | 1.000 | 1.000 | 1.000 | |||
| irregular | 74 (17.8) | 47 (9.3) | 1.891 (1.218–2.936) | 0.005 | 2.510 (1.429–4.410) | 0.001 | 0.332 (0.046–2.420) | 0.277 |
| Breast massage | ||||||||
| No | 185 (46.7) | 36 (74.0) | 1.000 | 1.000 | 1.000 | |||
| Yes | 211 (53.3) | 127 (26.0) | 3.746 (2.720–5.159) | 0.000 | 3.961 (2.594–6.047) | 0.000 | 0.502 (0.136–1.858) | 0.302 |
| Mammary gland hyperplasia medication history | ||||||||
| No | 332 (79.4) | 488 (96.3) | 1.000 | 1.000 | 1.000 | |||
| Yes | 86 (20.6) | 19 (3.7) | 6.013 (3.416–10.582) | 0.000 | 3.692 (1.811–7.526) | 0.000 | 0.296 (0.040–2.200) | 0.234 |
| Contraceptive ring | ||||||||
| No | 105 (25.1) | 157 (30.9) | 1.000 | 1.000 | 1.000 | |||
| Yes | 313 (74.9) | 351 (69.1) | 1.443 (1.040–2.002) | 0.028 | 1.563 (1.001–2.442) | 0.050 | 0.697 (0.181–2.678) | 0.599 |
ORcrude, odds ratio generated by univariate logistic regression; ORadj, odds ratio generated by multivariate logistic regression; ORi, odds ratio generated by multivariate logistic regression for the interaction of DUSP1 methylation and environmental factors; 95%CI, 95% confidence interval.
Association of DUSP1 methylation in PBL DNA and environmental factors on breast cancer risk
Supplemental Table 3 shows the univariate and multivariate logistic regression analyses for all associations between environmental factors and breast cancer risk. Several environmental factors, including the consumption of refined grains, vegetables, fruit, seafood, milk, smoked food, etc., were found to be associated with the development of breast cancer following adjustment for educational level, occupation, marital status, and family history of cancer. We analysed the interactions of DUSP1 methylation with all of the above significant environmental factors. However, no significant interaction was observed (as shown in Table 1). Therefore, we concluded that there was not enough evidence for DUSP1 methylation in PBL DNA as a biomarker for breast cancer risk assessment.
Differences in DUSP1 methylation frequency between tumour DNA and PBL DNA in breast cancer patients
Genomic DNA from 326 breast tumour tissue samples was detected for DUSP1 methylation: the positive frequency of DUSP1 methylation was 59.2% (193/326). We successfully detected DUSP1 methylation in both PBL DNA and tumour DNA from 155 breast cancer patients. As shown in Table 2, a total of 83 tumour DNA were methylated in 155 tumour samples with a methylation frequency of 53.55%; in contrast, only five (3.23%) PBL DNA was methylated among the same patients. The P-value (0.000) from the McNemar Test indicates that there was a significant difference for the DUSP1 methylation frequency between the DNA samples from these two tissue types.
Table 2. Differences in DUSP1 methylation frequency between tumour DNA and PBLa DNA in breast cancer patients.
| PBL DNAa | Tumour tissue DNA | Total No. (%) | P-valueb | |
|---|---|---|---|---|
| Methylated No. (%) | Unmethylated No. (%) | |||
| Methylated No. (%) | 3 (1.94) | 2 (1.29) | 5 (3.23) | 0.000 |
| Unmethylated No. (%) | 80 (51.61) | 70 (45.16) | 150 (96.77) | |
| Total No. (%) | 83 (53.55) | 72 (46.45) | 155 (100) | |
aPBL, peripheral blood leukocytes. bP-value was generated by McNemar Test.
Correlation between clinicopathological characteristics and DUSP1 methylation in breast tumour DNA and PBL DNA
As show in Table 3, similar significant associations of ER and PR status and molecular subtypes with DUSP1 methylation in tumour DNA and PBL DNA were observed. Aberrant methylation of DUSP1 occurred more frequently in tumour DNA (OR = 2.278, 95% CI 1.389–3.735, P = 0.001) and PBL DNA (OR = 2.534, 95% CI 1.062–6.044, P = 0.036) with oestrogen receptor (ER)-negativity, as well as for progesterone receptor (PR)-negativity in tumour DNA (OR = 2.016, 95% CI 1.275–3.186, P < 0.01) and PBL DNA (OR = 3.034, 95% CI 1.264–7.282, P = 0.013). In particular, in a molecular subtype analysis, compared with luminal A breast cancer (ER and/or PR + , HER2-), patients with HER2-enriched (ER and PR-, HER2 + ) and basal-like (ER-, PR-, HER2-) subtypes showed significantly higher DUSP1 methylation frequencies with ORs of 2.661 (95% CI 1.345–5.267, P = 0.005) and 5.636 (95% CI 2.205–14.406, P = 0.000), respectively, in tumour DNA; patients with the basal-like (ER-, PR-, HER2-) subtype showed significantly higher DUSP1 methylation frequency with an OR of 5.238 (95% CI 1.108–24.763, P = 0.000) in PBL DNA, which indicated that DUSP1 methylation is linked with ER/PR-negative status and is a significant characteristic of triple-negative breast cancer.
Table 3. Correlation between clinicopathological characteristics and DUSP1 methylation in breast tumour DNA and PBL DNAa.
| Clinicopathologic characteristics |
DUSP1 methylation (tumour tissue DNA) |
DUSP1 methylation (PBL DNAa) |
||||||
|---|---|---|---|---|---|---|---|---|
| Methylated No. (%) | Unmethylated No. (%) | ORcrude (95% CI)b | P-value | Methylated No. (%) | Unmethylated No. (%) | ORcrude (95% CI)b | P-value | |
| TNM Stages | 0.886 | 0.327 | ||||||
| I | 38 (60.3) | 25 (39.7) | 1.000 | 8 (8.0) | 92 (92.0) | 1.000 | ||
| II | 113 (59.8) | 76 (40.2) | 0.978 (0.546–1.751) | 0.941 | 11 (4.7) | 221 (95.3) | 0.572 (0.223–1.469) | 0.246 |
| III & IV | 42 (56.8) | 32 (43.2) | 0.863 (0.436–1.709) | 0.674 | 3 (3.3) | 87 (96.7) | 0.397 (0.102–1.543) | 0.182 |
| Tumour invasion | 0.566 | 0.110 | ||||||
| T0–T1 | 86 (61.0) | 55 (39.0) | 1.000 | 13 (7.3) | 166 (92.7) | 1.000 | ||
| T2–T4 | 107 (57.8) | 78 (42.2) | 1.140 (0.729–1.782) | 9 (3.7) | 234 (96.3) | 0.491 (0.205–1.176) | ||
| Lymphnodes involved | 0.506 | |||||||
| N0 | 103 (60.9) | 66 (39.1) | 1.000 | 11 (5.2) | 201 (94.8) | 1.000 | ||
| N1/N3 | 90 (57.3) | 67 (42.7) | 1.162 (0.747–1.808) | 11 (5.3) | 197 (94.7) | 1.020 (0.432–2.408) | ||
| Metastasis status | 0.950 | 0.999 | ||||||
| M0 | 186 (59.2) | 128 (40.8) | 1.000 | 22 (5.4) | 389 (94.6) | |||
| M1 | 7 (58.3) | 5 (41.7) | 1.038 (0.322–3.343) | 0 (0.0) | 11 (100.0) | |||
| Histological type | 0.243 | 0.871 | ||||||
| Invasive | 143 (57.4) | 106 (42.6) | 1.000 | 17 (5.3) | 303 (94.7) | 1.000 | ||
| Noninvasive | 50 (64.9) | 27 (35.1) | 1.373 (0.807–2.335) | 5 (4.9) | 97 (95.1) | 0.919 (0.330–2.556) | ||
| ER status | 0.001 | 0.036 | ||||||
| Positive | 113 (52.8) | 101 (47.2) | 1.000 | 12 (3.8) | 301 (96.2) | 1.000 | ||
| Negative | 79 (71.8) | 31 (28.1) | 2.278 (1.389–3.7335) | 10 (9.2) | 99 (90.8) | 2.534 (1.062–6.044) | ||
| PR status | 0.003 | 0.013 | ||||||
| Positive | 94 (51.9) | 87 (48.1) | 1.000 | 9 (3.2) | 271 (96.8) | 1.000 | ||
| Negative | 98 (68.5) | 45 (31.5) | 2.016 (1.275–3.186) | 13 (9.2) | 129 (90.8) | 3.034 (1.264–7.282) | ||
| HER2 expression | 0.503 | 0.779 | ||||||
| Positive | 127 (60.5) | 83 (39.5) | 1.000 | 15 (5.4) | 261 (94.6) | 1.000 | ||
| Negative | 64 (56.6) | 49 (43.4) | 0.854 (0.537–1.357) | 7 (4.8) | 139 (95.2) | 1.141 (0.455–2.865) | ||
| Molecular subtypec | 0.001 | 0.108 | ||||||
| Luminal A | 33 (44.0) | 42 (56.0) | 1.000 | 3 (2.7) | 110 (97.3) | 1.000 | ||
| Luminal B | 81 (57.0) | 61 (43.0) | 1.690 (0.961–2.971) | 0.068 | 9 (4.4) | 197 (95.6) | 1.675 (0.444–6.317) | 0.446 |
| HER-2 enriched | 46 (67.6) | 22 (32.4) | 2.661 (1.345–5.267) | 0.005 | 6 (8.5) | 65 (91.5) | 3.385 (0.819–13.944) | 0.092 |
| Basal-like | 31 (81.6) | 7 (18.4) | 5.636 (2.205–14.406) | 0.000 | 4 (12.5) | 28 (87.5) | 5.238 (1.108–24.763) | 0.037 |
| P53 | 0.649 | 0.151 | ||||||
| Positive | 49 (61.2) | 31 (38.8) | 1.000 | 8 (8.1) | 91 (91.9) | 1.000 | ||
| Negative | 143 (58.4) | 102 (41.6) | 0.887 (0.529–1.487) | 14 (4.3) | 308 (95.7) | 0.517 (0.210–1.271) | ||
aPBL, peripheral blood leukocytes; bORcrude, odds ratio generated by univariate logistic regression; 95%CI, 95% confidence interval. cSubtypes were classified by immunohistochemical surrogates as basal-like (ER-, PR-, HER-2−, triple-negative), luminal A (ER and/or PR+, HER-2−), luminal B (ER and/or PR+, HER-2+), or HER-2 enriched (ER and PR−, HER-2+).
No significant association of DUSP1 methylation in tumour DNA with TNM stage, tumour invasion, lymph node involvement, metastasis status, histological type, or TP53 mutation status was observed.
Effect of exposure to environmental factors on DUSP1 methylation in tumour tissue DNA
Supplemental Table 2 shows the distribution of demographic characteristics between patients with methylated and unmethylated tumour DNA. No statistically significant difference was observed for any demographic characteristic (all P > 0.05).
We analysed the associations between environmental factors and DUSP1 methylation in tumour DNA in a multivariate analysis. Among all the environmental factors, there was no significant effect on DUSP1 methylation status from menopause, breast massage, breast hyperplasia, breast disease, smoking, alcohol etc. (Supplemental Table 4). However, the consumption of fruit and soybean, and irregular menstruation significantly correlated with the DUSP1 methylation status of tumour DNA in both univariate and multivariate analyses (Table 4). Individuals with a lower fruit intake (≤ 1000 g/week) had a higher proportion (67.9%) of methylated DUSP1. Individuals with a higher soybean intake (>1 times/week) and irregular menstruation had higher proportions of methylated DUSP1 with OR of 1.955 (P = 0.006) and 2.000 (P = 0.020), respectively.
Table 4. Effect of exposure to environmental factors on DUSP1 methylation in tumour DNA.
| Environmental factors (%) |
DUSP1 methylation |
ORcrude (95% CI) | P-value | ORadj (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| Methylated | Un-methylated | |||||
| Fruit (g/week) | 0.021 | 0.023 | ||||
| ≤1000 | 89 (67.9) | 42 (32.1) | 1.000 | 1.000 | ||
| >1000 | 100 (54.9) | 82 (45.1) | 0.576 (0.360–0.920) | 0.567 (0.348–0.924) | ||
| Soybean (times/week) | 0.007 | 0.006 | ||||
| ≤1 | 72 (51.8) | 67 (48.2) | 1.000 | 1.000 | ||
| >1 | 119 (66.9) | 59 (33.1) | 1.877 (1.189–2.962) | 1.955 (1.211–3.154) | ||
| Menstrual regularity | 0.014 | 0.020 | ||||
| regular | 134 (55.8) | 106 (44.2) | 1.000 | 1.000 | ||
| irregular | 54 (72.0) | 21 (28.0) | 2.034 (1.156–3.578) | 2.000 (1.113–3.593) | ||
ORcrude, odds ratio generated by univariate logistic regression; ORadj, odds ratio generated by multivariate logistic regression; 95%CI, 95% confidence interval.
Since soybean intake and irregular menstruation are two hormone-related factors, the associations between soybean intake, menstrual cycle and DUSP1 methylation were further examined by dividing the subjects according to ER and PR status. As show in Table 5, we found significant data for soybean intake and irregular menstruation amongst the subgroups. Soybean intake (>1 times/week) was significantly correlated with increased DUSP1 methylation only in patients with ER-negative (OR 2.978, 95% CI 1.245–7.124) and PR-negative (OR 2.735, 95% CI 1.315–5.692) breast cancer. Meanwhile, irregular menstruation was significantly linked with increased DUSP1 methylation only in patients with ER-positive (OR 3.564, 95% CI 1.691–7.511) and PR-positive (OR 3.902, 95% CI 1.656–9.194) breast cancer.
Table 5. Association of environmental exposures and DUSP1 methylation in tumour DNA by ER and PR status.
| Environmental factors (%) | ER− | ER+ | PR− | PR+ | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Metha | Unmethb | ORadjc (95%CI) P-value | Metha | Unmethb | ORadjc (95%CI) P-value | Metha | Unmethb | ORadjc (95%CI) P-value | Metha | Unmethb | ORadjc (95%CI) P-value | |
| Soybean (times/week) | ||||||||||||
| ≤1 | 29 | 19 | 1.000 | 43 | 47 | 1.000 | 36 | 27 | 1.000 | 36 | 39 | 1.000 |
| >1 | 50 | 11 | 2.978 (1.245–7.124) 0.017 | 68 | 48 | 1.548 (0.889–2.696) 0.159 | 62 | 17 | 2.735 (1.315–5.692) 0.010 | 56 | 42 | 1.444 (0.789–2.643) 0.282 |
| Menstrual regularity | ||||||||||||
| Yes | 58 | 21 | 1.000 | 75 | 84 | 1.000 | 69 | 31 | 1.000 | 64 | 74 | 1.000 |
| No | 19 | 10 | 0.688 (0.276–1.716) 0.475 | 35 | 11 | 3.564 (1.691–7.511) 0.001 | 27 | 13 | 0.933 (0.425–2.047) 0.863 | 27 | 8 | 3.902 (1.656–9.194) 0.001 |
aMeth, methylated; bUnmeth, unmethylated; cORadj, odds ratio generated by multivariate logistic regression; 95%CI, 95% confidence interval.
Discussion
Several studies have suggested that individual variation in the epigenome in blood is associated with aging and environmental factors encountered throughout life37,38, with consequent risk of breast39, ovarian40, bladder20, and small-cell lung cancer41. The potential utility of specific methylation biomarkers in PBL DNA as novel markers of cancer susceptibility has been proposed. In this study, we first explored the value of DUSP1 methylation in PBL DNA for the risk assessment of breast cancer, but we failed to find any association between DUSP1 methylation in PBL DNA with breast cancer risk, or for the interactive effects of DUSP1 methylation and environmental factors. However, we did find significant correlations of triple-negative status with DUSP1 methylation in both tumour DNA and PBL DNA, and significant associations among soybean intake, irregular menstruation, ER/PR status, and DUSP1 methylation in tumour DNA.
Increasing numbers of studies have identified tissue-specific differential methylation42, which will provide important novel insights into normal and pathogenic mechanisms, as well as help in identifying markers of carcinogenesis and future epigenetic therapies. In this study, we analysed differences in DUSP1 promoter methylation between PBL DNA and tumour DNA. A significantly higher DUSP1 methylation frequency was observed in tumour DNA than in PBL DNA. In the research of Chen et al., they identified a normal breast cell line (M10) that was completely unmethylated while several breast cancer cell lines (MCF7, MDA-MB-231, SKBR3, and BT474) exhibited 100% methylation; unmethylation was dominant (86.2%) in benign breast tumours whereas methylation was dominant (57.2%) in invasive breast tumours14. Together, we concluded that DUSP1 methylation is a cancer-associated hypermethylation event. The biological significance of DUSP1 hypermethylation in breast cancer should be addressed in future in vitro studies. Given the very low (3.23–5.2%) methylation frequency in PBL DNA, DUSP1 methylation has potential as a biomarker in non-invasive breast cancer diagnosis if we can detect DUSP1 methylation from circulating cell-free DNA in plasma that is released by breast tumour cells.
Some genes that exhibit special methylation status in tumours are correlated with ER/PR status. ER-positive tumours were found to be more frequently methylated on RASSF1A than ER-negative tumours43. PR had a positive correlation with DUSP1 expression in 30 human breast cancer cell lines by binding to two progesterone response elements downstream of the DUSP1 transcriptional start site to upregulate DUSP1 promoter activity44. A critical novel finding of this study was the linkage of ER/PR-negativity with methylated DUSP1 in both breast tumour DNA and PBL DNA, which might account for the lower MKP-1 expression.
It is well known that negative results for oestrogen receptor (ER-), progesterone receptor (PR-), and HER2 (HER2-) expression in breast cancer cells signifies that the cancer is triple-negative cancer. These negative results indicate that the growth of the cancer is not supported by the oestrogen and progesterone hormones, or by the presence of too many HER2 receptors. Therefore, triple-negative breast cancer does not respond to hormonal therapy (such as tamoxifen or aromatase inhibitors) or therapies that target HER2 receptors. In contrast, ER-positive and PR-positive tumours are associated with improved response to hormonal therapy and with a longer disease-free interval and improved survival45,46. Doctors and researchers have an intense interest in developing their understanding of triple-negative breast cancer pathogenesis and finding new medications that can treat this breast cancer type. Holm et al. determined the methylation status of 807 breast cancer-related genes according to molecular subtype and found that basal-like, luminal A and luminal B tumours have different methylation profiles47. In this study, we found triple-negative breast tumour showed the highest frequency of DUSP1 methylation. Hence, DUSP1 methylation might be considered as a distinctive subtype-specific marker of triple-negative patients.
Most established risk factors for female breast cancer are thought to influence the susceptibility to cancer through hormone-related pathways48. Epidemiological experimental evidence implicated that increased concentrations of endogenous oestrogen level or exogenous oestrogen intake may induce aberrant DNA methylation49. In this study, we found high intake of fruit was correlated with decreased DUSP1 methylation, while high intake of soybean and irregular menstruation were correlated with increased DUSP1 methylation.
Soybean is a unique food because it contains large amounts of isoflavones50,51. Isoflavones have a chemical structure that is very similar to the hormone oestrogen52. To the best of knowledge, breast cancer is a heterogeneous disease and biological differences in subtypes depend on the expression of receptors, including ER, PR, and HER2. Because of the ability of isoflavones to bind oestrogen receptors, the varied associations between soybean intake and breast cancer risk by the hormone receptor status of tumours have been suggested in eight published epidemiological studies53,54,55,56,57,58,59,60. However, few studies have researched the modified effect of soybean intake on DNA methylation, which may have pivotal functions in relation to tumour suppression, apoptosis, etc. in breast cancer. Harlid et al. used an Illumina Human Methylation450 BeadChip to evaluate epigenome-wide DNA methylation in vaginal cells from soy formula-fed and cow formula-fed girls; the results indicated that girls fed soy formula had altered DNA methylation in their vaginal cell DNA61. Only one study has been published that provides evidence on the potential effects of two naturally occurring isoflavones, genistein and daidzein, on the methylation of BRCA1 and BRCA2 tumour suppressor genes in breast cancer cell lines (MCF-7, MDA-MB 231, and MCF10a)62. Our study is the first to explore the putative effects of soybean consumption on DUSP1 promotor methylation in breast cancer. We found that soybean intake was significantly associated with methylated DUSP1 in tumour DNA in ER/PR-negative patients. Messina and colleagues reviewed substantial epidemiological data from observational studies and cell culture data, and noted that the current limited knowledge regarding the effect of soybean on breast cancer-related issues suggested that clinicians should be careful of what they prescribe for patients63,64. Although information regarding soybean consumption was provided as retrospective data for the patients’ dietary habit prior to cancer diagnosis, given the significant correlation between soybean intake and DUSP1 methylation we observed, prolonged or excessive consumption of soybean in ER/PR-negative patients is not recommended.
In addition, progesterone balances oestrogen and in doing so minimises the negative effects of oestrogen–progesterone imbalances65. When these hormones become unbalanced it is usually because of oestrogen dominance, which means too much oestrogen compared with the levels of progesterone. Irregular menstruation flow is an oestrogen-dominant symptom. In the subgroup analyses in our study, irregular menstruation was significantly correlated with increased DUSP1 methylation in ER/PR-positive patients, which suggested that irregular menstruation may correlate with DUSP1 methylation through the indirect effect of oestrogen. Further in vitro studies are needed to validate this inference.
There are some limitations with the interpretation of the present results. First, some studies have suggested cell-specific variation in DNA methylation66,67; in our case-control study design, we only focused on the differential methylation of DUSP1 in leukocyte DNA between cases and controls, and we did not further explore the cell type composition difference of PBL between the cases and controls. Second, our conclusion for the association between ER/PR status and DUSP1 methylation in breast tumour DNA was generated based on a population study without an experimental validation in vitro study. Third, the small number of subjects in the stratified analysis limited the statistical power to evidence the conclusion; further studies with larger sample sizes are encouraged to verify the relationship between environmental factors, ER/PR status, and DUSP1 methylation.
Conclusions
The results of this study indicated that DUSP1 methylation in PBL DNA and its interaction with environmental factors was not associated with breast cancer risk. This is the first study to report the modified effects of soybean consumption on DNA methylation in tumour by ER/PR status, and we provide preliminary evidence on potential epigenetic changes through DUSP1 methylation in triple-negative patients. Further validation of the association of environmental factors, including fruit and soybean intake, and irregular menstruation, with DUSP1 methylation by hormone receptor status in breast cancer should be undertaken.
Materials and Methods
Study subjects
We carried out this study after obtaining informed written consent from study subjects and approval from the Human Research and Ethics Committee of Harbin Medical University. All experiments including all relevant details were performed in accordance with relevant guidelines and regulations.
A case-control study was designed to assess the role of DUSP1 methylation and interactions with environmental factors on breast cancer risk. All breast cancer patients were newly diagnosed cases recruited from the Third Affiliated Clinical Hospital of Harbin Medical University from 2010 to 2014. Controls were recruited from patients admitted to the Orthopaedic and Ophthalmology of the Second Affiliated Hospital of Harbin Medical University, and volunteers from the Xiangfang community of Harbin City within the same time period. Any individual with a history of benign breast disease or any other cancer was excluded from the control group. Approximately 5 ml of peripheral venous blood was obtained from all cases either before surgery for the patients and at enrolment for the controls.
A case-only study was designed to explore the difference in DUSP1 methylation between breast tumour DNA and PBL DNA. Tumour tissues specimen were collected during surgery and rapidly frozen in liquid nitrogen after removal, then returned to the lab and stored at −80 °C immediately. We analysed the correlation between clinicopathological characteristics and DUSP1 methylation in tumour DNA and PBL DNA, as well as the effect of exposure to environmental factors on DUSP1 methylation in tumour DNA.
Data collection
All subjects were interviewed face-to-face by well-trained interviewers using the same questionnaires, which included questions on demographic information (age, marital status, education, occupation, family cancer history, height and weight), behaviours (smoking, drinking, physical activity), dietary status (intake of milk, vegetables, fruits, soy bean etc.) during the 12 months prior to cancer diagnosis, menstruation and reproductive history, and any other disease history. The clinical and pathological information of cancer patients was extracted from medical records, including TNM stage, histological, and pathological results.
Genomic DNA extraction
PBL DNA was extracted from blood samples using a commercial DNA extraction kit (QIAamp DNA Blood Mini Kit, Hilden, Germany) according to the manufacturer’s protocol and then stored at −80 °C. Less than 25 mg of minced tumour tissue was used for DNA extraction. Tumour tissues were removed from the deep freeze and ground into small pieces immediately by a tissue grinder. DNA was extracted from tumour tissues using a DNA extraction kit (PureLinkTM Genomic DNA Kit, Carlsbad, USA) according to the manufacturer’s protocol and then stored at −80 °C. DNA quantity was measured using the Nanodrop 2000 Spectrophotometer (Thermo Scientific).
Sodium bisulphite modification
Bisulphite conversion was performed using 2 μg DNA and an EpiTect Bisulfite Kit (Qiagen, Hilden, Germany) according to the manufacturer’s guidelines. DNA yield after bisulphite conversion was in the range of 50–100 ng/μl; DNA was stored at −80 °C.
Analysis of the methylation status of DUSP1
Methylation-sensitive high-resolution melting analysis (MS-HRM) was performed on a LightCycler 480 (Roche Applied Science, Mannheim, Germany) equipped with Gene Scanning software (version 2.0) to detect and analyse the methylation status of DUSP168. Universal methylated and unmethylated DNA standards (ZYMO, USA) were used as the positive and negative controls. To create the range of methylated and unmethylated allele dilutions, the above two standards were mixed at 1, 5, 10, and 20% ratios.
Primers were designed for MS-HRM analysis using Primer Premier 5.0 software as follows: forward primer, 5′-TGGTTTGGTAGGGCGGGTGA-3′, and reverse primer, 5′–GTCGCACACACAACCCAAATA-3′. The PCR product (range = chr5:172198165–172198336, 171 bp) was located at CpG island IV located on the border of the promoter and exon 1 of DUSP1. There is an Illumina 450 K probe within this region (cg11757894 = chr5:172197877), as shown in Supp. Fig. 1. PCR reactions were performed using LightCycler 480 ResoLight Dye (Roche Applied Science), primers at 200 nmol/L final concentration, 3 nmol/L MgCl2 for DUSP1, and 5 ng of bisulphite-converted DNA sample in 10 μl final volume.
The PCR amplification protocol consisted of denaturation for 10 min at 95 °C for one cycle, denaturation for 10 s at 95 °C, annealing with a touchdown (65–55 °C, 30 s, in tumour DNA; 70–64 °C, 40 s, in PBL DNA) of each primer annealing temperature and extension for 10 s at 72 °C for 58 cycles. The HRM melting protocol then consisted of 95 °C for 1 min, cool down to 40 °C for 1 min, 70 °C for 5 s and continuous acquisition to 90 °C at 20 acquisitions per 1 °C (LightCycler480, Roche, Mannheim, Germany). We repeated the MS-HRM assay for the DNA samples without a good application curve.
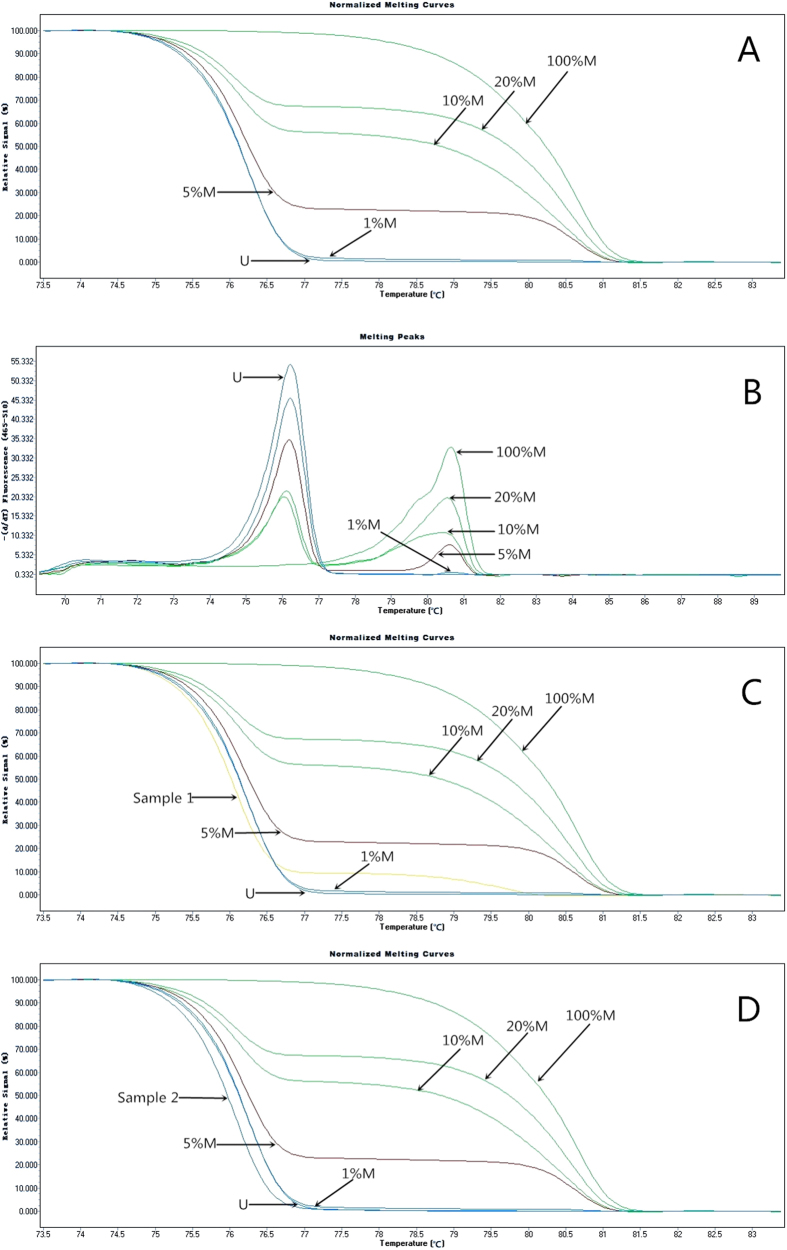
Then, 0% M (universal unmethylated DNA standards) served as the cut-off value to distinguish methylation and non-methylation of DUSP1. We analysed methylation as a qualitative variable, methylated (any methylated status with methylation level higher than 0%M) and unmethylated. We duplicated sample DNA, two blank controls, and gradient methylated DNA standards in each plate. Figure 1(a,b) showed the profile of fluorescence obtained at the melting temperature for serial dilutions of methylated DNA (100%, 20%, 10%, 5%, 1%, and 0%). Figure 1(c,d) showed the melting profiles of a methylated breast tumour DNA and a unmethylated sample.
Figure 1. MS-HRM of the DUSP1 promoter methylation for serials standards and samples.
(A) Normalized HRM curves. The DNA methylation standards of 0 (universal unmethylated DNA), 1, 5, 20, and 100% methylation (universal methylated DNA) are indicated. (B) Tm plot (negative first derivative of the HRM curves) of serials standards. (C) The melting profile of a methylated breast tumour DNA sample (sample 1 with methylation level of 1–5%). (D) The melting profile of an unmethylated tumour DNA sample (sample 2).
Immunohistochemical assay
The presence of oestrogen receptor (ER), progesterone receptor (PR), and HER2 in breast tumour tissue was tested by immunohistochemical (IHC) assay; further verification using fluorescence in situ hybridization (FISH) was needed if the results of IHC assays showed HER2-positivity.
Statistical analysis
Categorical and continuous variables were tested by chi-square test and two-sample t-test, respectively. Univariate and multivariate logistic-regression analyses were used to calculate the crude and adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the association of environmental factors, DUSP1 methylation in PBL DNA, and their association with breast cancer risk.
Correlation between clinicopathological characteristics and DUSP1 methylation status in tumour DNA and PBL DNA was evaluated using odds ratios (ORs) and 95% CIs derived from unconditional logistic regression. The effect of environment factors on DUSP1 methylation in tumour DNA was calculated using unconditional univariate and multivariate logistic regression.
All statistical analyses were performed using SAS version 9.2, with P-values of < 0.05 considered statistically significant.
Additional Information
How to cite this article: Li, J. et al. DUSP1 promoter methylation in peripheral blood leukocyte is associated with triple-negative breast cancer risk. Sci. Rep. 7, 43011; doi: 10.1038/srep43011 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Material
Acknowledgments
This work is funded by the grant from the National Nature Science Foundation of China (81202262), Postdoctoral Science Foundation of China (2013T60390), National Nature Science Foundation of China (81172743), and Educational Commission of Heilongjiang Province (12541340) and Dr. Wu Lien-the Science Foundation of Harbin Medical University (WLD-QN1106).
Footnotes
The authors declare no competing financial interests.
Author Contributions F.W. and Y.Z. contributed to study design, data interpretation, manuscript draft, and the acquisition of funding. D.P. contributed to the study design and supervision. J.L. and Y.C. contributed to primer design and data analysis. J.L. and H.Y. contributed to DNA extraction, bisulfate conversion, and MS-HRM detection. F.Y. and J.F. contributed to sample collection and questionnaire. L.Z. contributed to data management. J.L. and J.T. contributed to manuscript draft. All authors contributed to review and revision of the manuscript.
References
- GLOBOCAN2012:Estimated Cancer Incidence, Mortality and Prevalence Worldwide in 2012., http://globocan.iarc.fr/Pages/fact_sheets_cancer.aspx. (2012).
- Siegel R. L., Miller K. D. & Jemal A. Cancer statistics, 2016. CA Cancer J Clin 66, 7–30, doi: 10.3322/caac.21332 (2016). [DOI] [PubMed] [Google Scholar]
- Tsujita E. et al. Suppressed MKP-1 is an independent predictor of outcome in patients with hepatocellular carcinoma. Oncology 69, 342–347, doi: 10.1159/000089766 (2005). [DOI] [PubMed] [Google Scholar]
- Wang X. & Liu Y. Regulation of innate immune response by MAP kinase phosphatase-1. Cellular signalling 19, 1372–1382, doi: 10.1016/j.cellsig.2007.03.013 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. W. & Yang J. L. Cooperation of ERK and SCFSkp2 for MKP-1 destruction provides a positive feedback regulation of proliferating signaling. The Journal of biological chemistry 281, 915–926, doi: 10.1074/jbc.M508720200 (2006). [DOI] [PubMed] [Google Scholar]
- Adeyinka A. et al. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clinical cancer research: an official journal of the American Association for Cancer Research 8, 1747–1753 (2002). [PubMed] [Google Scholar]
- Fu X. et al. Overcoming endocrine resistance due to reduced PTEN levels in estrogen receptor-positive breast cancer by co-targeting mammalian target of rapamycin, protein kinase B, or mitogen-activated protein kinase kinase. Breast cancer research: BCR 16, 430, doi: 10.1186/s13058-014-0430-x (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M. C. et al. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 23, 2469–2476, doi: 10.1200/jco.2005.01.172 (2005). [DOI] [PubMed] [Google Scholar]
- Yang F., Tang X. Y., Liu H. & Jiang Z. W. Inhibition of mitogen-activated protein kinase signaling pathway sensitizes breast cancer cells to endoplasmic reticulum stress-induced apoptosis. Oncology reports 35, 2113–2120, doi: 10.3892/or.2016.4580 (2016). [DOI] [PubMed] [Google Scholar]
- Rauhala H. E. et al. Dual-specificity phosphatase 1 and serum/glucocorticoid-regulated kinase are downregulated in prostate cancer. International journal of cancer 117, 738–745, doi: 10.1002/ijc.21270 (2005). [DOI] [PubMed] [Google Scholar]
- Loda M. et al. Expression of mitogen-activated protein kinase phosphatase-1 in the early phases of human epithelial carcinogenesis. The American journal of pathology 149, 1553–1564 (1996). [PMC free article] [PubMed] [Google Scholar]
- Mizuno R. et al. Inhibition of MKP-1 expression potentiates JNK related apoptosis in renal cancer cells. The Journal of urology 172, 723–727, doi: 10.1097/01.ju.0000124990.37563.00 (2004). [DOI] [PubMed] [Google Scholar]
- Shimada K. et al. c-Jun NH2 terminal kinase activation and decreased expression of mitogen-activated protein kinase phosphatase-1 play important roles in invasion and angiogenesis of urothelial carcinomas. The American journal of pathology 171, 1003–1012, doi: 10.2353/ajpath.2007.070010 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. M. et al. The mitogen-activated protein kinase phosphatase-1 (MKP-1) gene is a potential methylation biomarker for malignancy of breast cancer. Experimental & molecular medicine 44, 356–362, doi: 10.3858/emm.2012.44.5.040 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin S. B. & Jones P. A. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews. Cancer 11, 726–734, doi: 10.1038/nrc3130 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayar N. et al. Transgelin gene is frequently downregulated by promoter DNA hypermethylation in breast cancer. Clinical epigenetics 7, 104, doi: 10.1186/s13148-015-0138-5 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond C. E. et al. Methylation and expression of the tumour suppressor, PRDM5, in colorectal cancer and polyp subgroups. BMC cancer 15, 20, doi: 10.1186/s12885-015-1011-9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutkowska A. et al. RARbeta Promoter Methylation as an Epigenetic Mechanism of Gene Silencing in Non-small Cell Lung Cancer. Advances in experimental medicine and biology 878, 29–38, doi: 10.1007/5584_2015_159 (2016). [DOI] [PubMed] [Google Scholar]
- Kile M. L. et al. Correlation of global and gene-specific DNA methylation in maternal-infant pairs. PloS one 5, e13730, doi: 10.1371/journal.pone.0013730 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsit C. J. et al. DNA methylation array analysis identifies profiles of blood-derived DNA methylation associated with bladder cancer. Journal of clinical oncology: official journal of the American Society of Clinical Oncology 29, 1133–1139, doi: 10.1200/jco.2010.31.3577 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim U. et al. Genomic methylation of leukocyte DNA in relation to colorectal adenoma among asymptomatic women. Gastroenterology 134, 47–55, doi: 10.1053/j.gastro.2007.10.013 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. E. et al. Genomic DNA hypomethylation as a biomarker for bladder cancer susceptibility in the Spanish Bladder Cancer Study: a case-control study. The Lancet. Oncology 9, 359–366, doi: 10.1016/s1470-2045(08)70038-x (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiung D. T. et al. Global DNA methylation level in whole blood as a biomarker in head and neck squamous cell carcinoma. Cancer epidemiology, biomarkers & prevention: a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology 16, 108–114, doi: 10.1158/1055-9965.epi-06-0636 (2007). [DOI] [PubMed] [Google Scholar]
- Choi J. Y. et al. Association between global DNA hypomethylation in leukocytes and risk of breast cancer. Carcinogenesis 30, 1889–1897, doi: 10.1093/carcin/bgp143 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. et al. HIF3A DNA Methylation Is Associated with Childhood Obesity and ALT. PloS one 10, e0145944, doi: 10.1371/journal.pone.0145944 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsli-Ceppioglu S. et al. The Role of Soy Phytoestrogens on Genetic and Epigenetic Mechanisms of Prostate Cancer. The Enzymes 37, 193–221, doi: 10.1016/bs.enz.2015.05.004 (2015). [DOI] [PubMed] [Google Scholar]
- Dadon Bar-El S. & Reifen R. Vitamin A and the Epigenome. Critical reviews in food science and nutrition, 0, doi: 10.1080/10408398.2015.1060940 (2015). [DOI] [PubMed] [Google Scholar]
- Issa J. P. CpG-island methylation in aging and cancer. Current topics in microbiology and immunology 249, 101–118 (2000). [DOI] [PubMed] [Google Scholar]
- McKay J. A. et al. Blood as a surrogate marker for tissue-specific DNA methylation and changes due to folate depletion in post-partum female mice. Molecular nutrition & food research 55, 1026–1035, doi: 10.1002/mnfr.201100008 (2011). [DOI] [PubMed] [Google Scholar]
- Li Z. et al. Methylation profiling of 48 candidate genes in tumor and matched normal tissues from breast cancer patients. Breast Cancer Res Treat 149, 767–779, doi: 10.1007/s10549-015-3276-8 (2015). [DOI] [PubMed] [Google Scholar]
- Radpour R. et al. Hypermethylation of tumor suppressor genes involved in critical regulatory pathways for developing a blood-based test in breast cancer. PloS one 6, e16080, doi: 10.1371/journal.pone.0016080 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Preston R. et al. Differential promoter methylation of kinesin family member 1a in plasma is associated with breast cancer and DNA repair capacity. Oncology reports 32, 505–512, doi: 10.3892/or.2014.3262 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimonidou M. et al. CST6 promoter methylation in circulating cell-free DNA of breast cancer patients. Clinical biochemistry 46, 235–240, doi: 10.1016/j.clinbiochem.2012.09.015 (2013). [DOI] [PubMed] [Google Scholar]
- Chimonidou M., Strati A., Malamos N., Georgoulias V. & Lianidou E. S. SOX17 promoter methylation in circulating tumor cells and matched cell-free DNA isolated from plasma of patients with breast cancer. Clinical chemistry 59, 270–279, doi: 10.1373/clinchem.2012.191551 (2013). [DOI] [PubMed] [Google Scholar]
- Wei M. et al. Estrogen receptor alpha, BRCA1, and FANCF promoter methylation occur in distinct subsets of sporadic breast cancers. Breast Cancer Res Treat 111, 113–120, doi: 10.1007/s10549-007-9766-6 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift-Scanlan T., Vang R., Blackford A., Fackler M. J. & Sukumar S. Methylated genes in breast cancer: associations with clinical and histopathological features in a familial breast cancer cohort. Cancer biology & therapy 11, 853–865 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E. et al. Age-dependent DNA methylation of genes that are suppressed in stem cells is a hallmark of cancer. Genome research 20, 440–446, doi: 10.1101/gr.103606.109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen B. C. et al. Aging and environmental exposures alter tissue-specific DNA methylation dependent upon CpG island context. PLoS genetics 5, e1000602, doi: 10.1371/journal.pgen.1000602 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M. et al. Epigenotyping in peripheral blood cell DNA and breast cancer risk: a proof of principle study. PloS one 3, e2656, doi: 10.1371/journal.pone.0002656 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teschendorff A. E. et al. An epigenetic signature in peripheral blood predicts active ovarian cancer. PloS one 4, e8274, doi: 10.1371/journal.pone.0008274 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. et al. Methylation markers for small cell lung cancer in peripheral blood leukocyte DNA. Journal of thoracic oncology: official publication of the International Association for the Study of Lung Cancer 5, 778–785, doi: 10.1097/JTO.0b013e3181d6e0b3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokk K. et al. DNA methylome profiling of human tissues identifies global and tissue-specific methylation patterns. Genome biology 15, r54, doi: 10.1186/gb-2014-15-4-r54 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajabova V. et al. RASSF1A Promoter Methylation Levels Positively Correlate with Estrogen Receptor Expression in Breast Cancer Patients. Translational oncology 6, 297–304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. C., Hardy D. B. & Mendelson C. R. Progesterone receptor inhibits proliferation of human breast cancer cells via induction of MAPK phosphatase 1 (MKP-1/DUSP1). The Journal of biological chemistry 286, 43091–43102, doi: 10.1074/jbc.M111.295865 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunnwald L. K., Rossing M. A. & Li C. I. Hormone receptor status, tumor characteristics, and prognosis: a prospective cohort of breast cancer patients. Breast cancer research: BCR 9, R6, doi: 10.1186/bcr1639 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzdar A. U. Role of biologic therapy and chemotherapy in hormone receptor- and HER2-positive breast cancer. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO 20, 993–999, doi: 10.1093/annonc/mdn739 (2009). [DOI] [PubMed] [Google Scholar]
- Holm K. et al. Molecular subtypes of breast cancer are associated with characteristic DNA methylation patterns. Breast cancer research: BCR 12, R36, doi: 10.1186/bcr2590 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddad S. A. et al. Hormone-related pathways and risk of breast cancer subtypes in African American women. Breast Cancer Res Treat 154, 145–154, doi: 10.1007/s10549-015-3594-x (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez S. V. & Russo J. Estrogen and xenoestrogens in breast cancer. Toxicologic pathology 38, 110–122, doi: 10.1177/0192623309354108 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshima A., Mine W., Nakada M. & Yanase E. Analysis of isoflavones and coumestrol in soybean sprouts. Bioscience, biotechnology, and biochemistry, 1–3, doi: 10.1080/09168451.2016.1196577 (2016). [DOI] [PubMed] [Google Scholar]
- Lee S. J. et al. Analysis of isoflavones and phenolic compounds in Korean soybean [Glycine max (L.) Merrill] seeds of different seed weights. Journal of agricultural and food chemistry 56, 2751–2758, doi: 10.1021/jf073153f (2008). [DOI] [PubMed] [Google Scholar]
- Han K. K., Soares J. M. Jr., Haidar M. A., de Lima G. R. & Baracat E. C. Benefits of soy isoflavone therapeutic regimen on menopausal symptoms. Obstetrics and gynecology 99, 389–394 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang C. et al. Soy product and isoflavone intake and breast cancer risk defined by hormone receptor status. Cancer science 101, 501–507, doi: 10.1111/j.1349-7006.2009.01376.x (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y. A. et al. Effect of dietary soy intake on breast cancer risk according to menopause and hormone receptor status. Eur J Clin Nutr 64, 924–932, doi: 10.1038/ejcn.2010.95 (2010). [DOI] [PubMed] [Google Scholar]
- Iwasaki M. et al. Dietary isoflavone intake and breast cancer risk in case-control studies in Japanese, Japanese Brazilians, and non-Japanese Brazilians. Breast Cancer Res Treat 116, 401–411, doi: 10.1007/s10549-008-0168-1 (2009). [DOI] [PubMed] [Google Scholar]
- Touillaud M. S. et al. Effect of dietary intake of phytoestrogens on estrogen receptor status in premenopausal women with breast cancer. Nutrition and cancer 51, 162–169, doi: 10.1207/s15327914nc5102_6 (2005). [DOI] [PubMed] [Google Scholar]
- Dai Q. et al. Population-based case-control study of soyfood intake and breast cancer risk in Shanghai. Br J Cancer 85, 372–378, doi: 10.1054/bjoc.2001.1873 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson L. N., Cotterchio M., Boucher B. A. & Kreiger N. Phytoestrogen intake from foods, during adolescence and adulthood, and risk of breast cancer by estrogen and progesterone receptor tumor subgroup among Ontario women. International journal of cancer 132, 1683–1692, doi: 10.1002/ijc.27788 (2013). [DOI] [PubMed] [Google Scholar]
- Zhang M., Yang H. & Holman C. D. Dietary intake of isoflavones and breast cancer risk by estrogen and progesterone receptor status. Breast Cancer Res Treat 118, 553–563, doi: 10.1007/s10549-009-0354-9 (2009). [DOI] [PubMed] [Google Scholar]
- Suzuki T. et al. Effect of soybean on breast cancer according to receptor status: a case-control study in Japan. International journal of cancer 123, 1674–1680, doi: 10.1002/ijc.23644 (2008). [DOI] [PubMed] [Google Scholar]
- Harlid S. et al. Soy Formula and Epigenetic Modifications: Analysis of Vaginal Epithelial Cells from Infant Girls in the IFED Study. Environ Health Perspect, doi: 10.1289/EHP428 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosviel R., Dumollard E., Dechelotte P., Bignon Y. J. & Bernard-Gallon D. Can soy phytoestrogens decrease DNA methylation in BRCA1 and BRCA2 oncosuppressor genes in breast cancer? OMICS 16, 235–244, doi: 10.1089/omi.2011.0105 (2012). [DOI] [PubMed] [Google Scholar]
- Messina M. A brief historical overview of the past two decades of soy and isoflavone research. The Journal of nutrition 140, 1350s–1354s, doi: 10.3945/jn.109.118315 (2010). [DOI] [PubMed] [Google Scholar]
- Peethambaram P., Olson J. & Loprinzi C. L. Soyfood consumption in breast cancer survivors: don’t overstate the facts! Oncology (Williston Park, N.Y.) 27, 442, 448, 450 (2013). [PubMed] [Google Scholar]
- Maybin J. A. & Critchley H. O. Progesterone: a pivotal hormone at menstruation. Annals of the New York Academy of Sciences 1221, 88–97, doi: 10.1111/j.1749-6632.2011.05953.x (2011). [DOI] [PubMed] [Google Scholar]
- Montano C. M. et al. Measuring cell-type specific differential methylation in human brain tissue. Genome biology 14, R94, doi: 10.1186/gb-2013-14-8-r94 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Baker-Andresen D., Zhao Q., Marshall V. & Bredy T. W. Methyl CpG binding domain ultra-sequencing: a novel method for identifying inter-individual and cell-type-specific variation in DNA methylation. Genes, brain, and behavior 13, 721–731, doi: 10.1111/gbb.12150 (2014). [DOI] [PubMed] [Google Scholar]
- Wojdacz T. K., Dobrovic A. & Hansen L. L. Methylation-sensitive high-resolution melting. Nature protocols 3, 1903–1908, doi: 10.1038/nprot.2008.191 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.