Abstract
The two acireductone dioxygenase (ARD) isozymes from the methionine salvage pathway of Klebsiella oxytoca are the only known pair of naturally occurring metalloenzymes with distinct chemical and physical properties determined solely by the identity of the divalent transition metal ion (Fe2+ or Ni2+) in the active site. We now show that this dual chemistry can also occur in mammals. ARD from Mus musculus (MmARD) was studied to relate metal ion identity and three-dimensional structure to enzyme function. The iron-containing isozyme catalyzes the cleavage of 1,2-dihydroxy-3-keto-5-(thiomethyl)pent-1-ene (acireductone) by O2 to formate and the ketoacid precursor of methionine, the penultimate step in methionine salvage. The nickel bound form of ARD catalyzes an off-pathway reaction resulting in formate, carbon monoxide (CO) and 5-(thiomethyl) propionate. Recombinant MmARD was expressed and purified to obtain a homogeneous enzyme with a single transition metal ion bound. The Fe2+ bound protein, which shows about ten-fold higher activity than others, catalyzes on-pathway chemistry, whereas the Ni2+, Co2+ or Mn2+ forms exhibit off-pathway chemistry, as has been seen with ARD from Klebsiella. Thermal stability of the isozymes is strongly affected by metal ion identity, with Ni2+ bound MmARD being the most stable followed by Co2+ and Fe2+, and Mn2+-bound ARD being the least stable. Ni2+ and Co2+ bound MmARD were crystallized and the structures of the two proteins found to be similar. Enzyme-ligand complexes provide insight into substrate binding, metal coordination and catalytic mechanism.
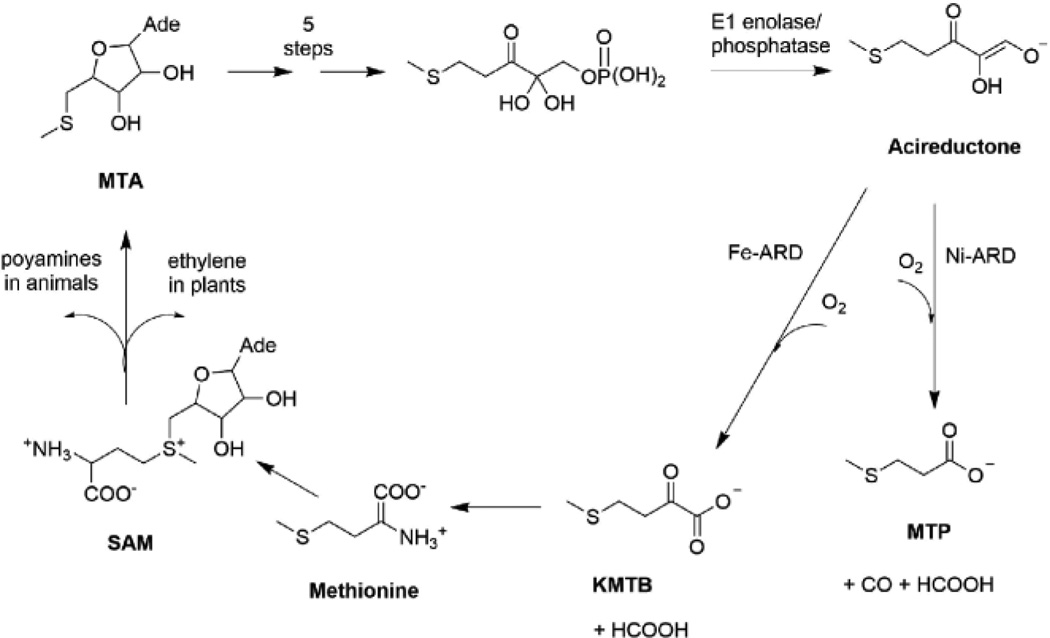
The methionine salvage pathway (MSP) is a ubiquitous pathway found in plants, animals and bacteria. Methylthioadenosine (MTA), is the first intermediate in this pathway and is formed from S-adenosyl methionine (SAM) during polyamine synthesis in animals and ethylene synthesis in plants (Scheme 1).1, 2 Polyamine is required for cell growth and proliferation1 while ethylene is required for ripening of fruits and vegetables.3 MTA is an inhibitor of both polyamine synthesis and transmethylation reactions.1, 4 Inhibition of polyamine synthesis arrests DNA replication,5 and elevated polyamine is associated with tumor formation.6, 7 Hence the concentration of MTA in cells must be tightly regulated. The MSP controls the concentration of MTA by returning it through a series of reactions to methionine, thereby “salvaging” the thiomethyl group of SAM.8 Acireductone dioxygenase (ARD) catalyzes the penultimate step in the pathway, the oxidative decomposition of substrate acireductone (1,2-dihydoxy-3-keto-5-(thiomethyl)pent-1-ene) to formate and 2-keto-4-(thiomethyl)butyrate (KMTB), the keto-acid precursor of methionine.
Scheme 1. Methionine Salvage Pathway.
ARD is a metalloenzyme and requires a metal cofactor for its activity. In the bacterium Klebsiella oxytoca, ARD exhibits dual activity depending on the identity of the metal ion cofactor. ARD with Fe2+-bound catalyzes on-pathway chemistry leading to production of formate and the keto-acid precursor to methionine whereas ARD bound with Ni2+ catalyzes an off-pathway shunt leading to production of carbon monoxide, formate, and methylthiopropionate (MTP).9, 10
Both the Fe2+ and Ni2+ bound forms of the protein have been isolated from Klebsiella,11 and are also obtained upon overexpression in E. coli. The activities of the two enzymes are interconverted by exchanging Fe2+ and Ni2+.12 These represent the only known pair of naturally occurring metalloenzymes with distinct chemical and physical properties determined solely by the identity of the metal ion in the active site. The purpose of the off-pathway reaction catalyzed by Ni2+ bound ARD in Klebsiella oxytoca is unknown. The structures of the two forms of ARD from Klebsiella oxytoca have been solved by NMR spectroscopy. The active site of the Ni2+-bound form was modeled by X-ray Absorption Spectroscopy (XAS) due to paramagnetism of the bound metal ion.13 The structure of the Fe2+-ARD is a model based on the structure of a stable soluble metal-free mutant of H98S ARD.14 An X-ray crystal structure was solved for the mouse homolog by the Joint Center for Structural Genomics (JCSG), but the identity of the metal ion cofactor in the protein and its biochemistry were not determined.15
In addition to its enzymatic function, several studies have shown ARD to serve other functions in mammals. Yeh, et al. have identified an N-terminal truncated version of human ARD (HsARD) called SipL which is implicated in the replication of hepatitis C virus in non-permissive cell lines.16 A separate study showed that HsARD binds the cytoplasmic tail of membrane-type 1 matrix metalloproteinase (MT1-MMP) and acts as a negative regulator of MT1-MMP by inhibiting MT1-MMP-mediated cellular invasiveness.17 A third study has shown that the gene ADI1 encoding ARD is downregulated in rat prostate and human prostate cancer cell lines and its enforced expression was shown to induce apoptosis.18 The same authors also showed that cultured gastric carcinoma cells and fibrosarcoma cells also have downregulated ADI1.17, 19 A recent study has also shown that the gene ADI1 has a direct link with cancer development.20 ARD thus has been shown to perform an inhibitory role in tumor progression and cancer.
To date, no mammalian ARD has been characterized biochemically to relate its chemistry with metal ion identity and protein structure. In this report, we describe purification and characterization of Mus musculus ARD (MmARD) bound to different metal ions to investigate the role of the metal ion center in its catalytic activity. We show that Fe2+-bound MmARD exhibits ~ 10 times higher enzymatic activity than Ni2+, Co2+ or Mn2+ bound MmARD. We were able to crystallize Ni2+ and Co2+-bound forms of the proteins and verify the identity of the metal in the active site using anomalous X-ray diffraction data. Enzymatic product analysis data indicated that MmARD bound to Ni2+, Co2+ or Mn2+ exhibits off-pathway chemistry whereas Fe2+ bound protein exhibited on-pathway chemistry similar to ARD from Klebsiella oxytoca. We thus provide biochemical evidence that a mammalian ARD can also perform dual chemistry in vitro depending on the metal ion cofactor in the active site. The role of ARD in cancer as shown by previous studies makes the possibility of dual chemistry in vivo in mammals interesting since carbon monoxide, one of the products of the off-pathway reaction, is known to be anti-apoptotic and signaling molecule.21–24
MATERIALS AND METHODS
Cloning
6× His tagged construct
A 6× His tagged construct for mouse ARD was obtained from DNASU Plasmid Repository which stores, maintains and distributes protein expression plasmids created by the PSI Protein Structure Initiative (PSI) Biology-Materials (PSI: Biology-MR) centers. This construct (Clone # MmCD00289622) has a 6× His tag on the N-terminus and is in the pMH4 vector and was used for crystal structure determination by JCSG.15
Strep tag II construct
The Strep II tag is a short affinity tag of 8 amino acids (Trp-Ser-His-Pro-Gln-Phe-Glu-Lys) which binds with high affinity to Strep-Tactin which is an engineered Streptavidin protein (IBA Life Sciences). The MmARD gene obtained from DNASU was amplified using the primers 5′-CCATGGCAAGTTGGAGCCACCCGCAGTTCGAGAAG-3′ and 5′-AGTTTTTTGGAAGGAACAGCATAGGAATTC-3′ and cloned into pET28a (Merck Millipore, Darmstadt, Germany) via the restriction sites NcoI and EcoRI. Using the primers, the Strep II tag was placed on the N-terminus of the protein. The sequences were confirmed by sequencing.
Single-metal expression and purification of MmARD
6× His tagged Ni-MmARD and Co-MmARD
Protein samples used for crystallization were prepared by the following method. Cells were transformed on minimal media plates. M9 minimal media was used for expression (6g/L Na2HPO4, 3g/L KH2PO4, 1g/L NH4Cl, 0.5g/L NaCl, 2mM MgSO4, 0.1mM CaCl2) with 0.96% glycerol as the carbon source. Growth was performed at 37°C and the cells were induced at O.D. of 0.5 using L-arabinose (1.2g/L). During induction, the desired metal salt at a final concentration of 20 µM was added. The metal salts used were CoCl2.6H2O and NiCl2.6H2O for getting Co-MmARD and Ni-MmARD respectively. The cells were induced overnight at 37°C. The cell pellet was suspended in 50mM HEPES pH 8.0 with protease inhibitor (Roche protease inhibitor cocktail EDTA-free) and sonicated for 5 min. Ammonium sulfate to a 30% saturation was added to the supernatant at 4°C. The suspension was centrifuged at 20,000 3 g for 20 min. The supernatant was then brought to 70% saturation with ammonium sulfate at 4°C, and then centrifuged at 20,000 3 g for 20 min. The pellet was then suspended in 50mM HEPES pH 7.0 and applied to a Phenyl Sepharose column (BioRad) pre-equilibrated with Buffer A (50mM HEPES, 1.2M (NH4)2SO4 pH 7.0) The column was eluted with a linear gradient increasing from 0–100% buffer B (50mM HEPES pH 7.0). Both Ni and Co-MmARD eluted between 40–70% buffer B. The fractions were pooled, concentrated and passed through a S100 gel filtration column pre-equilibrated with buffer C (50mM HEPES pH 7.0, 100mM NaCl). The protein eluted between 50–70 ml. The pool was concentrated and passed through a MonoQ column pre-equilibrated with buffer D (20mM Tris, pH 7.4). The column was eluted with a linear gradient increasing from 0–20% buffer E (20mM Tris pH 7.4, 1M NaCl). Both Ni and Co-MmARD eluted between 11–15% buffer D. The fractions were pooled and concentrated.
Strep tag II construct
Protein for metal analysis and biochemical assays was prepared by the following method. Cells were transformed on minimal media plates. M9 minimal media was used for expression (6g/L Na2HPO4, 3g/L KH2PO4, 1g/L NH4Cl, 0.5g/L NaCl, 2mM MgSO4, 0.1mM CaCl2) with 0.4% glucose as the carbon source. The growth was performed at 37°C and the cells were induced at O.D. of 0.5 using 0.5mM IPTG. During induction the desired metal salt at a final concentration 20 µM was added. The metal salts used were MnCl2.4H2O, Fe2SO4, CoCl2.6H2O, NiCl2.6H2O, CuCl2.2H2O, ZnCl2 for expressing Mn2+, Fe2+, Co2+, Ni2+, Cu2+ and Zn2+ bound MmARDs respectively. The cells were induced overnight at 37°C. The pellet was suspended in buffer X (100mM HEPES pH 7.0, 100mM NaCl) with protease inhibitor (Roche protease inhibitor cocktail EDTA-free) and sonicated for 5 min. The supernatant was concentrated and passed through Strep-Tactin beads (IBA Life Sciences) pre-equilibrated with buffer X at 4°C. The beads were washed with 5 column volumes (CV) of buffer X and eluted with 2 CV of buffer Y (100 mM HEPES pH 7.0, 10mM D-desthiobiotin). The eluent was buffer-exchanged into the buffer Z (50mM HEPES pH 7.0, 100mM NaCl) using Millipore Amicon concentrators. The pool was concentrated.
E1 protein expression, purification and E1 substrate synthesis
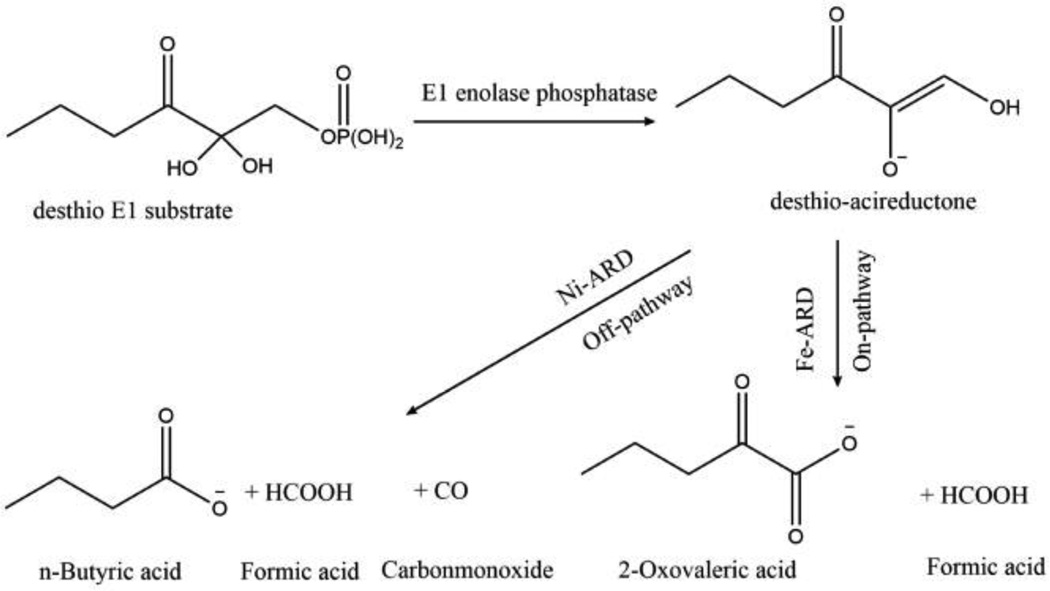
Enzyme E1 enolase phosphatase and the n-propyl desthio-analog of E1 substrate (Scheme 2) for enzymatic assays were prepared according to Zhang et al.25 This desthio analog of E1 substrate will be used for generation of the desthio derivative of acireductone which is also a substrate of ARD.
Scheme 2. ARD reactions using model E1 substrate reaction.
Enzymatic activity and kinetic parameters of single-metal reconstituted MmARD
The enzymatic assay to measure ARD activity was performed according to Zhang et al.25 with some modifications. All experiments were performed in the assay buffer 50mM HEPES pH 7.0, 1mM MgCl2. The assay is a coupled enzyme assay and uses E1 enolase-phosphatase enzyme to produce desthio-acireductone substrate in situ (Scheme 2) where substrate decay is monitored at the absorbance maximum of desthio-acireductone (308 nm) by UV–visible spectroscopy at room temperature. The assay was performed in three consecutive steps. In the first step, E1 substrate (500 µM) was added to E1 enzyme (0.6 µM) in argon saturated assay buffer. The substrate destio-acireductone was allowed to build up to a maximum level after which oxygen saturated assay buffer was added to the assay mixture. The rate of desthio-acireductone consumption due to non-enzymatic decay was monitored at 308 nm. Finally, a controlled amount of ARD was added, and the depletion of desthio-acireductone was monitored for at least 300 s. The initial rates were calculated by selecting the linear portion of the graph and calculating the linear fit in this region after correction for the non-enzymatic reaction rate.
Product quantification assays
The enzymatic reaction products derived from desthio-acireductone substrate from both chemistries are formic acid, 2-oxopentanoic acid, butyric acid, and CO. The organic acids were quantitated by high pressure liquid chromatography (HPLC) using a Biorad Aminex HPX-87 organic acid column (300 × 7.8 mm). The products were eluted using 5 mM H2SO4 at 0.3mL/min. Formic acid and 2-oxopentanoic acid were also monitored by NADH consumption and formation via the formate dehydrogenase (FDH) and lactate dehydrogenase (LDH) assays respectively.11
Metal content analysis
Measurements were performed at Harvard School of Public Health using a Perkin Elmer Elan DRC-II Inductively Coupled Plasma Mass spectrometer (ICP-MS). Samples for analysis were prepared by adding 1 ml of 70% HNO3 to 3 nmol of protein sample. Each sample was then diluted with 2% HNO3 to a final volume of 5 ml. All solutions were made with of deionized water (18 megaohm). A 5 ppb indium solution (in 2% HNO3) was used as internal standard. Isotopes 24Mg, 55Mn, 57Fe, 59Co, and 60Ni were quantitatively monitored using the peak hopping mode with a total of 100 scans per point per isotope (50 milliseconds of dwell time and five replicates). The HNO3 used for the analysis is ultra-high purity acid suitable for quantitative trace metal analysis at the parts-per-trillion level (Aristar Ultra, Part # 87003-658).
Thermal stability
The ThermoFluor assay (differential scanning fluorimetry) used to determine thermal stabilities was performed on a StepOnePlus™ Real-Time PCR System (Applied Biosystems). The samples for analysis were set up in final volumes of 25 µl in a 96-well plate. Each well consisted of 20 µM protein with 10× SYPRO Orange (Invitrogen) and the Strep-Tactin elution buffer 100mM HEPES pH 7.0, 10mM D-desthiobiotin. Thermal melting curves were analyzed using the fluorescence data generated at a ramp rate of 0.3 °C/min and a temperature range of 25°C to 90°C.
Oligomeric state
The MmARD proteins bound to different metal ions were concentrated to 30–100 µM and loaded onto a Superdex 200 5/150 GL column (GE Healthcare Life Sciences) equilibrated with 50 mM HEPES pH 7.0, 100 mM NaCl. A set of low–molecular-weight protein standards (GE Biosciences) dissolved in the same buffer were run on the Superdex-75 column to determine the calibration curve. The standards used were ribonuclease A (13.7 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), conalbumin (75 kDa) and aldolase (158 kDa).
Molecular weights were estimated using a linear regression analysis of Kav and log of molecular weight (MW) where,
Ve is the elution volume of each standard/sample, Vo is the void volume, and Vc is the column volume.
Crystallization
The Ni- and Co- bound proteins were crystallized using the vapor diffusion method with previously reported crystallization condition (19.0% (w/v) polyethylene glycol (PEG) - 4000, 19.0% (w/v) isopropanol, 5.0% glycerol, and 0.095 M Na-citrate pH 4.2 (final pH 5.6)).15 The ligand 2-keto-4-(methylthio)-butyric acid (KMTB) was soaked to a 10 molar excess into already formed Ni and Co-MmARD crystals. The ligands valeric acid (VA) and D-lactic acid (DLA) were soaked to a 10 molar excess into only Ni-MmARD crystals.
X-ray diffraction data collection
Crystals were cryo-cooled and sent to beamline X-29A at the synchrotron at Brookhaven National Laboratory or the 19-ID beamline at Argonne National Laboratory. The data integration, scaling and reduction were done using HKL200026 and refinement was done in Phenix.27 Statistics for these structures are given in Tables S1 to S4. Figures were generated using PyMOL (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC).
Anomalous scattering
The metal ion identities of both Ni-MmARD and Co-MmARD crystals were determined using anomalous X-ray scattering experiments. X-ray diffraction data were collected from the same crystal at two beam energies, 100 eV above and below the absorption edge of the metal expected. The theoretical anomalous edges for Ni (8.3328 keV) and Co (7.7089 keV) were derived using Cromer and Liberman’s approximation28 as implemented on the Biomolecular Structure Center website (http://skuld.bmsc.washington.edu/scatter/).
RESULTS
Purification and characterization of MmARD with specific transition metal ions bound
MmARD was overexpressed and purified in minimal media supplemented with appropriate metal salts during induction to express Mn2+, Fe2+, Co2+, Ni2+, Cu2+ and Zn2+ bound MmARDs. The enzyme overexpressed in all cases and was purified to >99% purity in most cases using Strep-Tactin beads. Fe+2-bound MmARD purification was performed anaerobically, as oxidation to Fe3+ ion leads to metal loss and protein instability. MmARD expressed with Cu+2 and Zn+2 could not be purified to >99% purity using a single affinity column. The metal content of each enzyme was determined using inductively coupled plasma-mass spectroscopy (ICP-MS). The results indicated that each enzyme contained approximately 1 mole of the desired metal ion per mole of the protein, except Cu and Zn which exhibited lower metal occupancy (Table 1).
Table 1.
Metal content of MmARD proteins determined by inductively coupled plasma-mass spectrometry
| Protein | Metal content (mol of metal /mol of protein) | |||||
|---|---|---|---|---|---|---|
| Mn | Fe | Co | Ni | Cu | Zn | |
| Mn-MmARD | 1.09 | 0.03±0.08 | - | 0.05 | - | 0.04 |
| Fe-MmARD | 0.01 | 1.25±0.04 | - | 0.02 | 0.01 | - |
| Co-MmARD | - | 0.01 | 0.96 | 0.02 | - | - |
| Ni-MmARD | - | - | - | 1.16 | 0.12 | - |
| Cu-MmARD | - | 0.01±0.03 | - | 0.09 | 0.74 | 0.09 |
| Zn-MmARD | - | 0.08±0.06 | - | 0.01 | 0.01 | 0.87 |
Errors are represented as standard deviations. All errors are reported within ±0.01 mol of metal/mol of protein except where noted.
Relative thermal stability of MmARD depends on the metal ion cofactor
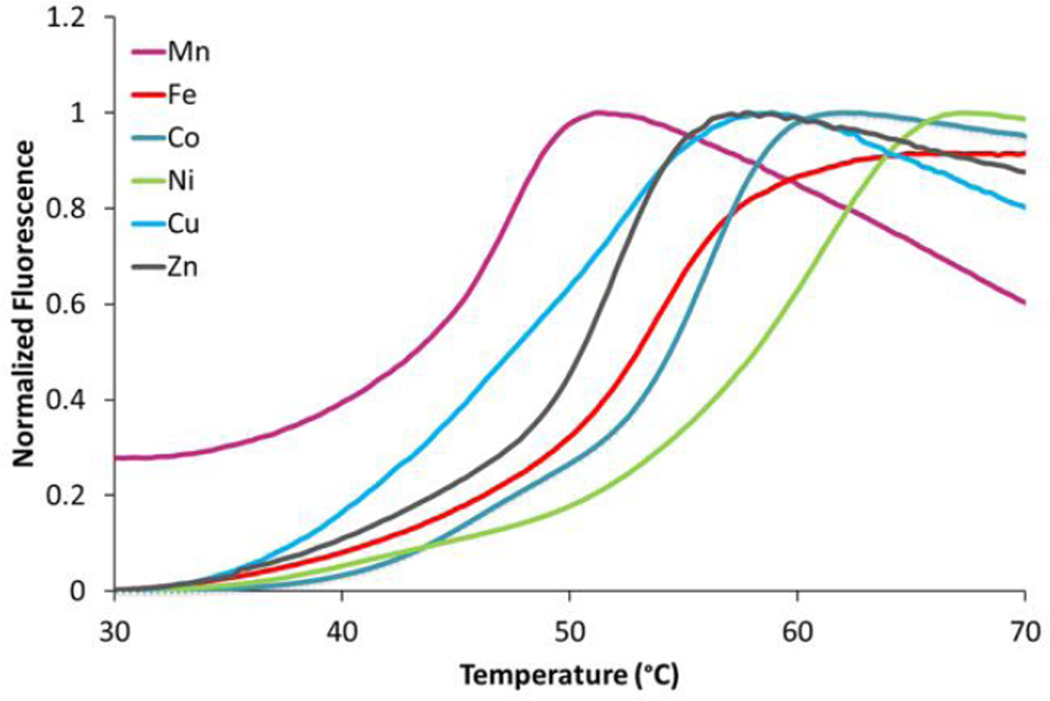
The thermal stability of each metal ion bound MmARD was established using differential scanning fluorimetry (DSF)29. The data (Figure 1) indicated that the Ni2+ bound MmARD is the most thermostable, followed by Co2+ and then Fe2+. Mn2+ bound MmARD exhibited the least thermal stability. These differences in the melting temperatures are likely an indication of the binding affinity of the metal ions to the apo-protein. Since the apo-protein is unfolded, we were unable to perform isothermal titration calorimetry (ITC) experiments to determine absolute dissociation constants.
Figure 1. Thermal stability of MmARD as a function of the bound metal ion.
The data indicate that Ni2+-bound MmARD has the highest melting temperature (58 °C) and Mn2+-bound MmARD has the lowest melting temperature (43 °C).
Kinetic Constants
The Michaelis-Menten kinetic parameters for each of the metal ion bound MmARDs were determined by varying the concentration of substrate desthio-acireductone in the presence of a fixed concentration of O2 (0.25 mM calculated on the basis of solubility of oxygen in water at 25 °C). These constants were obtained using the enzyme activity (substrate depletion) assay discussed in Methods & Materials.
As it was difficult to get 100% occupancy for Mn2+, Cu2+ and Zn2+ bound MmARD proteins, the kinetic analysis was performed only for Fe2+, Co2+ and Ni2+ bound MmARDs. These isoforms showed consistency in stability and metal occupancy from batch to batch. The Fe2+-bound MmARD exhibited ~10× greater turnover number (Table 2) and specific activity (Figure S1) compared to the Ni2+ or Co2+ bound isozymes. This differs from KoARD where the Ni2+ bound form has a higher activity (albeit off-pathway) than Fe2+ bound KoARD.30
Table 2.
Michelis-Menten kinetic constants of MmARD proteins for the substrate desthio-acireductone
| Protein | KM (µM) | kcat (s−1) | kcat/ KM (M−1s−1) |
|---|---|---|---|
| Fe-MmARD | 118.5 | 111.7 | 9.5E5 |
| Co-MmARD | 49.3 | 7.87 | 1.6E5 |
| Ni-MmARD | 287.1 | 17.1 | 6.0E4 |
The kinetic parameters for substrate desthio-acireductone were obtained at a fixed concentration of the second substrate oxygen (0.25 mM calculated on the basis of solubility of oxygen in water at 25 °C).
The identity of bound metal ion does not affect the oligomeric state of MmARD
To investigate whether the metal ion cofactor would alter the oligomeric state of MmARD, analytical size exclusion chromatography was used to estimate the molecular weights of each the MmARD proteins. MmARD eluted as a monomer with a molecular weight ranging from 23 to 26 kDa (Table S1). Figure S2 shows the calibration curve using protein molecular weight standards. All forms remain apparently monomeric regardless of the identity of the bound metal ion. While both Fe- and Ni-ARDs from Klebsiella oxytoca are monomers,10 Fe2+ bound ARD from Oryza sativa L. (OsARD) is a trimer and Ni2+ bound OsARD is a polymer consisting of several types of oligomers.31
Enzymatic Product Analysis
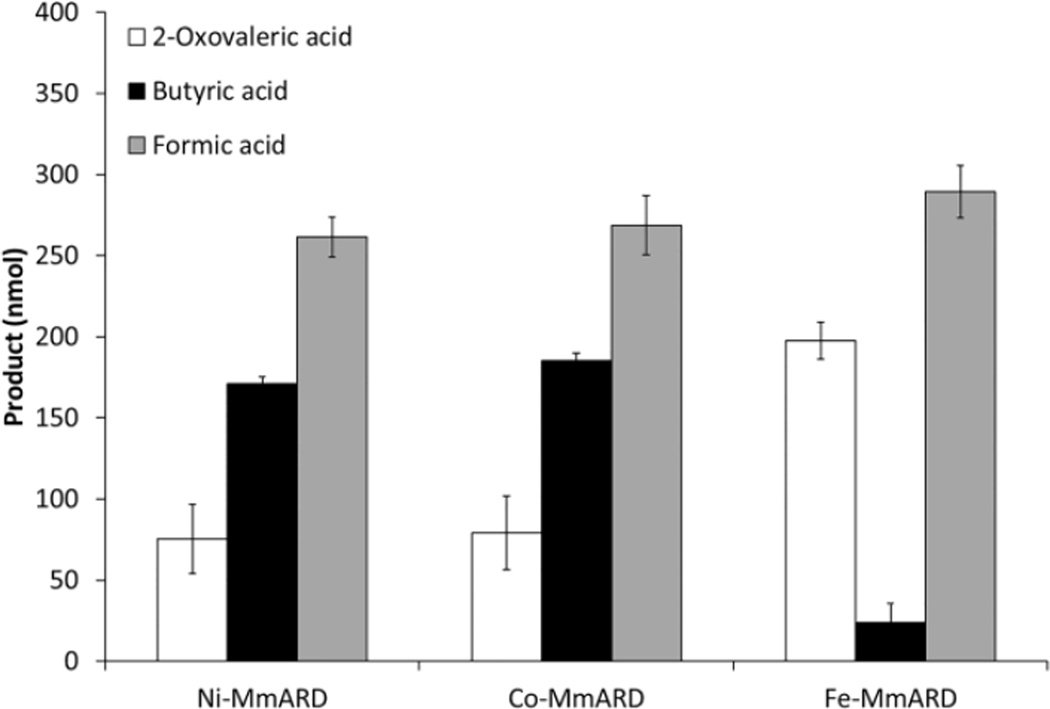
The products from both the on-and off-pathway chemistries were quantified and the results are summarized in Figure 2. Fe-KoARD and Ni-KoARD were used as controls to confirm that the assays are working. The data clearly demonstrate that Fe-MmARD exhibits on-pathway chemistry and Ni-MmARD and Co-MmARD exhibit off-pathway chemistry. The on-pathway product 2-oxovaleric acid seen with both Ni and Co-MmARD is due to the non-enzymatic reaction. Mn-MmARD also exhibits off-pathway chemistry (data not shown). Hence MmARD exhibits the same metal ion-dependent dual chemistry as the bacterial enzyme KoARD.
Figure 2. Quantitation of on-pathway and off-pathway products by MmARD bound to different metal ions.
The quantitation of ARD catalyzed oxidation of desthio-acireductone was done for a reaction mixture volume of 1mL. The error bars are represented as standard deviations.
Structural studies
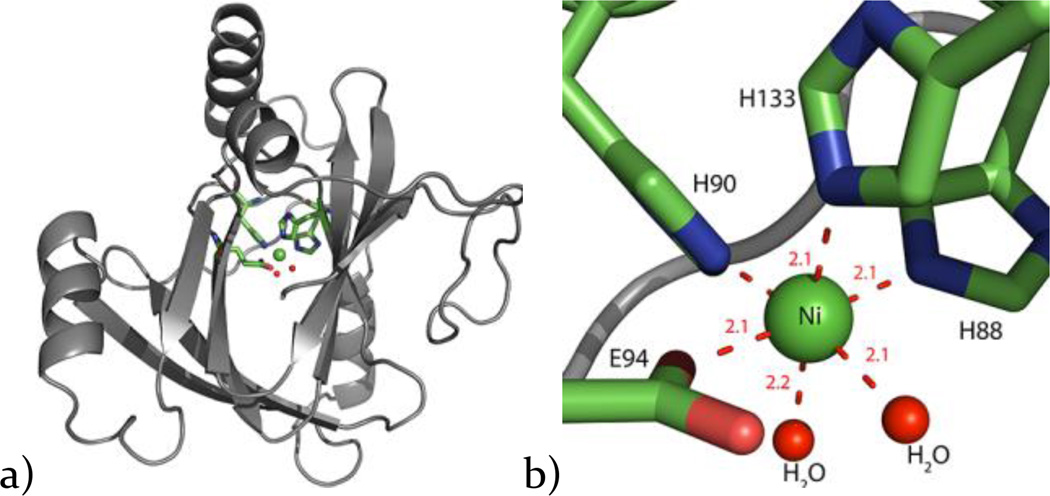
The 6× His tagged construct used for the previously reported structure was used for crystallization.15 This protein construct was expressed in minimal media in the presence of a single metal ion (Co2+ or Ni2+) and purified using traditional purification steps (refer to Methods and Materials) instead of using Ni-NTA affinity resin which was used in the previous study15, to avoid metal cross-contamination. The metal content was verified using ICP-MS metal analysis (Table S2). Both proteins crystallized using the previously reported conditions. Crystal cell parameters were found to be isomorphous with the previously solved structure (1VR3).15 The structures were determined by molecular replacement, using 1VR3 as search model and were refined using PHENIX.27 A summary of the crystallographic statistics is given in Table S3. Consistent with the previously reported structure, both Ni and Co present an octahedral co-ordination geometry in the active site bound to protein ligands H88, H90, H133 and E94 (Figure 3). In addition to the protein ligands, there are two distinct water (or hydroxide) ligands bound to the metal ion center, whereas the previously reported structure showed undefined electron density in the corresponding regions. This is consistent with the NMR solution structure of Ni bound KoARD in which two water molecules were modeled as ligands to fit EXAFS data in addition to the protein-based ligands. The active sites of both Ni-MmARD and Co-MmARD had an extra electron density. This extra electron density in Ni-MmARD could be modeled as 2-oxovaleric acid (not shown) whereas, in the case of Co-MmARD, it could be modeled as the on-pathway product KMTB. While KMTB is the natural on-pathway product of ARD chemistry, 2-oxovaleric acid, an α-ketoacid, is a natural metabolite found in cells32–34.
Figure 3. X-ray crystal structure of Ni-MmARD.
a) The X-ray crystal structure of Ni-MmARD with the Ni atom shown as a green sphere, b) The active site with the Ni atom as a green sphere and its protein ligands H88, H90, H133, E94 represented as sticks and two water molecules shown as red spheres forming an octahedral coordination geometry. The metal co-ordination distances are shown as red dotted lines with distances measured in Å.
Metal identity determination from anomalous scattering
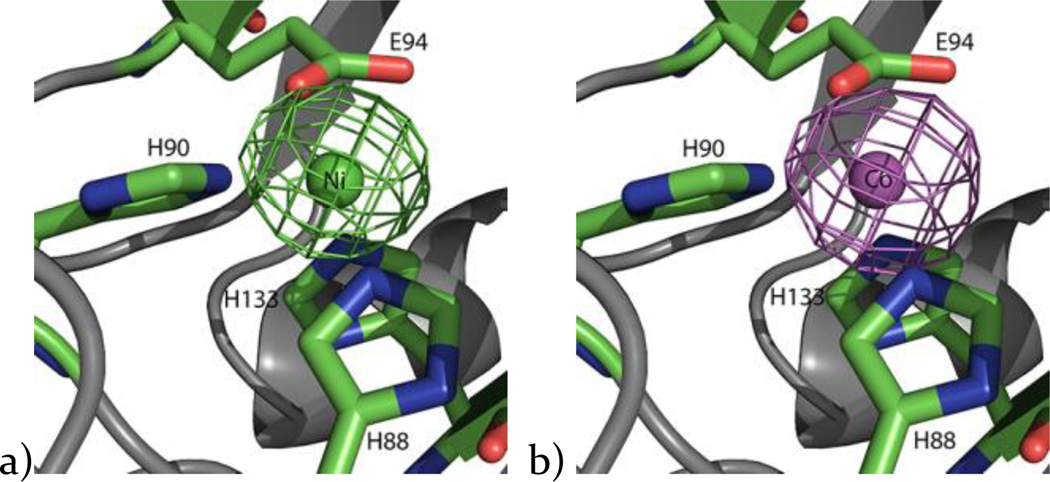
Since each metal has a distinct anomalous scattering wavelength, X-ray diffraction data collected slightly below and above the absorption edge can be used for unambiguous metal identification. To confirm the identity of the active site metals as Ni or Co, X-ray datasets were collected 100 eV below and above the absorption peaks of the K-absorption edges of each of these metals. The X-ray diffraction data collection statistics for Ni-MmARD and Co-MmARD are shown in Tables S4 and S5 respectively. Strong peaks were observed in the expected metal position in the anomalous electron density maps calculated using the above absorption edge X-ray data (Figure 4), whereas much weaker peaks were observed in the corresponding position calculated using the below absorption edge X-ray data. This sharp transition in anomalous scattering upon passing through the absorption edges of Ni or Co clearly confirmed the metal identity in each case of the MmARD protein used in crystallography.
Figure 4. Metal identity established by anomalous scattering experiments.
a) Anomalous electron density map of Ni-MmARD at 8.4328 keV showing the anomalous signal (green) of Ni (green sphere) in the active site; b) anomalous electron density map of Co-MmARD at 7.8089 keV showing the anomalous signal (magenta) of Co (magenta sphere) in the active site.
Product and product analogs bound to MmARD
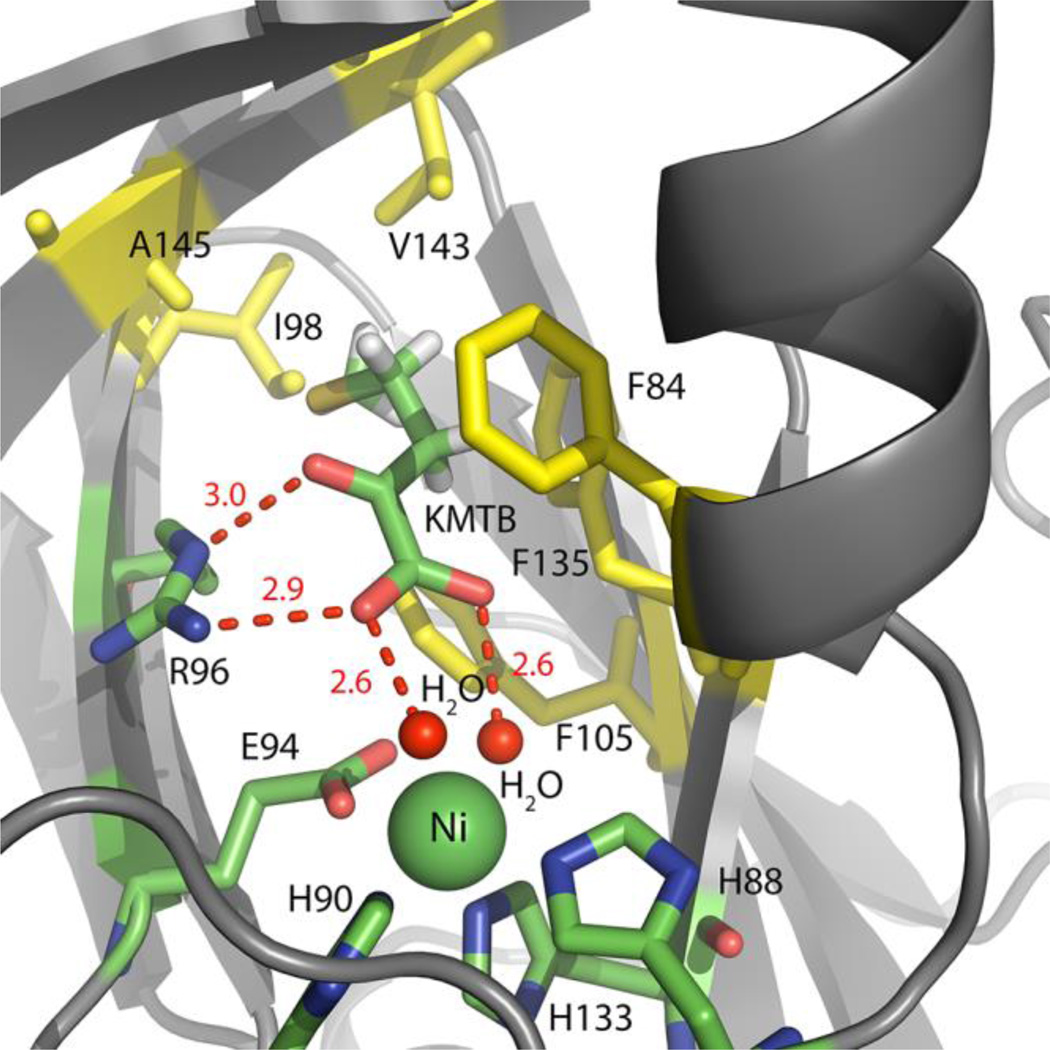
2-keto-4-(methylthio) butyric acid (KMTB) is the on-pathway product of ARD chemistry, and valeric acid (VA) is an analog of the off-pathway product 3-(methylthio) propionic acid (MTP). Thermal melting curves of ARD showed stabilization of both Ni-MmARD and Co-MmARD by the on-pathway product KMTB. Ni-MmARD and Co-MmARD co-crystallized with KMTB using the previously reported conditions15 but KMTB soaked into already formed Ni-MmARD and Co-MmARD crystals gave slightly higher resolution data than the co-crystals. The previously reported structure was solved to 2.06 Å resolution whereas in this study we were able to solve the structures of Ni-MmARD and Co-MmARD to 1.7 Å and 1.9 Å respectively. The data collection and refinement statistics are shown in Table S6. In Ni-MmARD and Co-MmARD, the electron density that could be fit to KMTB was observed in the active site (Figure 5). The structures of Ni-MmARD and Co-MmARD bound to KMTB align with an RMSD of 0.06 Å. The KMTB does not ligate the metal but its two carboxylate oxygens atoms are within hydrogen bonding distance of the two water (or hydroxide) ligands bound to the metal ion center. One of its carboxylate oxygen and the keto oxygen atoms are also within hydrogen bonding distance of one of the Nω atoms (terminal N-atom of the side chain) and Nδ (penultimate N-atom of the side chain) atom of Arg 96, a residue that is strictly conserved in all known ARD sequences. KMTB also forms hydrophobic interactions with F135, F105, F84 and A145, all of which again are strictly conserved. As discussed earlier in the paper, KMTB was also seen present in the active site of Co-MmARD crystals which were not soaked or co-crystallized with KMTB. These two structures (crystallized in the presence and absence of KMTB) were identical. Although KMTB is an on-pathway product, it still binds to both Ni and Co-MmARD both of which exhibit off-pathway chemistry.
Figure 5. The active site of Ni-MmARD showing the on-pathway product KMTB.
The Ni atom is shown as a green sphere. KMTB and active site residues are shown in stick representation. Waters are shown as small red spheres. The residues F84, F105, F135, A145, V143 and I98 which interact with the alkyl group of KMTB are shown in yellow. KMTB is within hydrogen bonding distance of R96 and with the two water molecules bound to Ni. The hydrogen bonding distances are shown as red dotted lines with distances measured in Å.
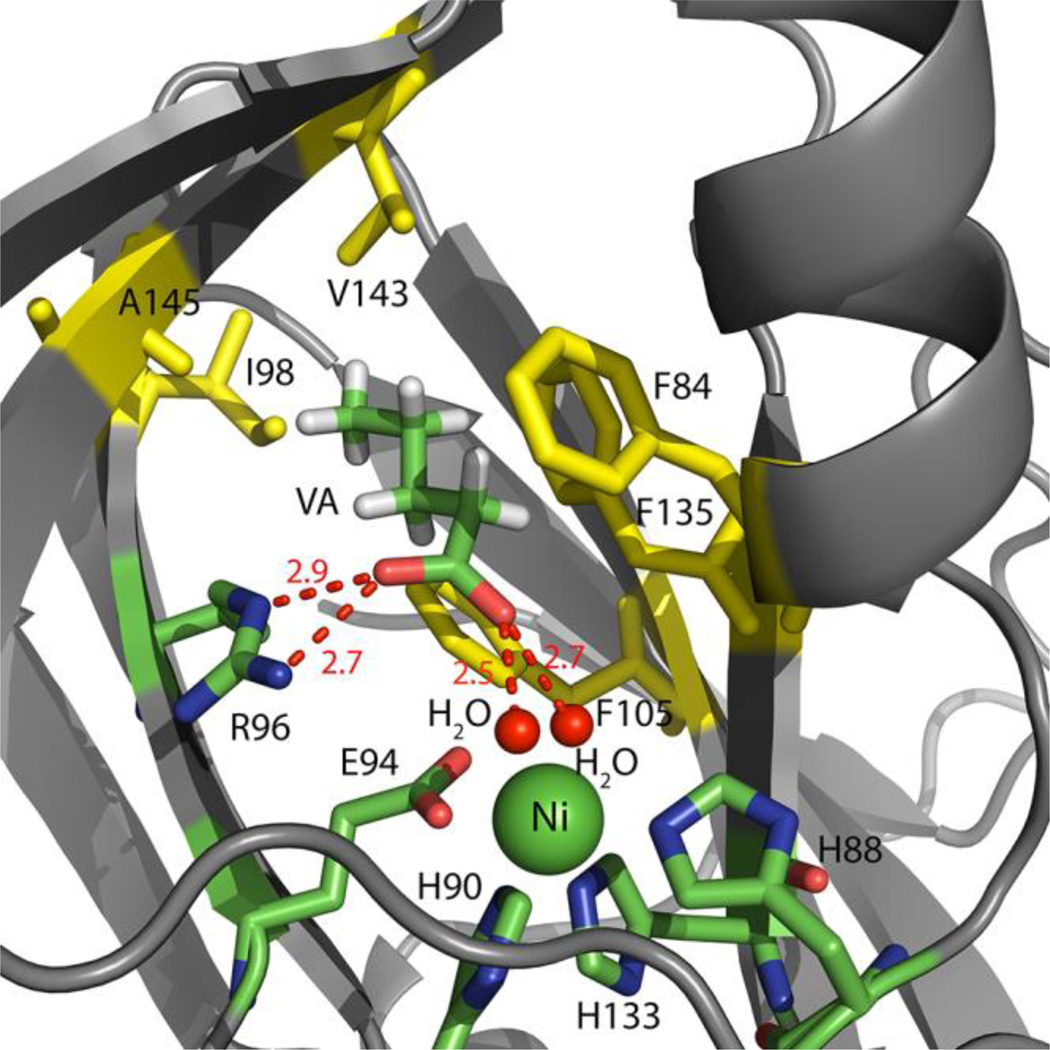
Since VA is an off-pathway product analog, a structure of Ni or Co-MmARD (which perform off-pathway chemistry) bound to VA can provide mechanistic details of the off-pathway chemistry. The structure of Ni-MmARD bound to VA is shown in Figure 6. The data collection and refinement statistics are shown in Table S6. Similar to KMTB, VA was seen to be present in the active site and did not directly interact with the metal ion center. One of its carboxylate oxygen atoms is within hydrogen bonding distance from one of the waters bound to the metal ion. The other carboxylate oxygen atom is within hydrogen bonding distance from Nω and Nδ atoms of R96. Similar to KMTB, The alkyl groups of VA also forms hydrophobic interactions with F135, F105, F84 and A145. It can be seen from the crystallographic data that VA does not form as many hydrogen bonding interactions with MmARD as KMTB since KMTB has an extra keto-oxygen with hydrogen binding capability which is missing in VA. Thermal stability data also indicate that Ni or Co-MmARD bound to KMTB has a higher thermal stability than Ni or Co-MmARD bound to VA.
Figure 6. The active site of Ni-MmARD showing the off-pathway product analog valeric acid (VA).
The Ni atom is shown as a green sphere. VA and active site residues are shown in stick representation. Waters are shown as small red spheres. The residues F84, F105, F135, A145, V143 and I98 which interact with the alkyl group of VA are shown in yellow. VA is within hydrogen bonding distance of R96 and with one of the two water molecules bound to Ni. The hydrogen bonding distances are shown as red dotted lines with distances measured in Å.
Substrate analogs bound to MmARD
In order to get mechanistic details for ARD, a substrate-bound structure in addition to a product-bound structure would be desirable. Desthio-acireductone is highly sensitive to oxidation. In an attempt to obtain a substrate-bound structure, desthio-acireductone was generated in situ in an anaerobic cuvette. It was then added to the drop containing a protein crystal in an anaerobic chamber. The crystals diffracted and the structure was solved, but no ligand was detected in the active site.
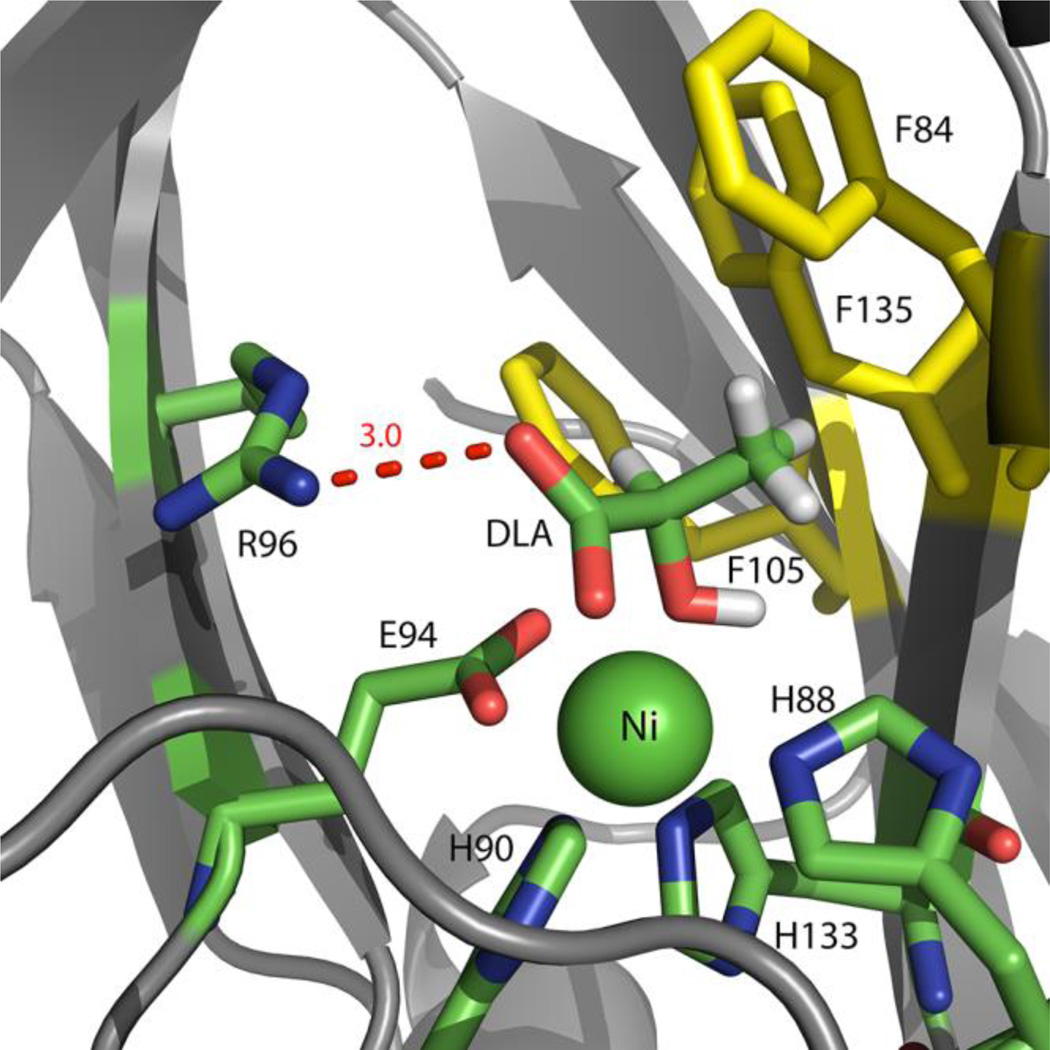
A number of compounds with structural similarity to acireductone were tested for thermal stabilization of the Co/Ni-MmARD (Table S7). The compounds which showed thermal stabilization, were then soaked into Ni-MmARD crystals in order to obtain the structure of MmARD bound to a substrate analog. Of all the compounds tested, only the structure of D-lactic acid (DLA) bound to Ni-MmARD could be solved. It is noteworthy that although DL-lactic acid was soaked into Ni-MmARD crystals, only DLA was seen to bind the protein. The structure of DLA bound to Ni-MmARD was solved to a resolution of 1.7 Å (Table S6). DLA was present in the active site and, unlike the product KMTB and product analog VA, DLA was seen to coordinate directly to the metal ion center (Figure 7). The two water molecules bound to the metal ion in the KMTB-bound or VA-bound protein structures were replaced by one of the carboxylate oxygen atoms and the hydroxyl-oxygen atom of DLA. The carboxylate oxygen atom of DLA not interacting with the metal ion was within hydrogen bonding distance of Nω atom of R96. Lactic acid exhibits a similar sub-structure as acireductone, but lacks the hydrophobic (methylthio) ethylene moiety of the native substrate. The recently deposited structure of Fe-HsARD (4qgn) has L-selenomethionine in the active site and its co-ordination mode to the metal ion is similar to that of D-Lactic acid.
Figure 7. The active site of Ni-MmARD showing D-Lactic acid (DLA).
The Ni atom is shown as a green sphere. DLA and active site residues are shown in stick representation. DLA is coordinating with Ni2+ and is within hydrogen bonding distance of R96. Residues F84, F105 and F135 which interact with the alkyl group of DLA are shown in yellow. The hydrogen bonding distances are shown as red dotted lines with distances measured in Å.
DISCUSSION
Mouse ARD is capable of performing both on- and off-pathway metal-dependent chemistry
ARD functions in the MSP, which is a ubiquitous biochemical pathway found in prokaryotes and eukaryotes.1, 2 ARD from Klebsiella oxytoca (KoARD) is currently the only known enzyme which, as isolated from the native organism, performs two different chemistries based on the metal ion cofactor bound to the protein in the active site.9, 10 The function of the off-pathway chemistry in Klebsiella oxytoca is unknown, although it may be involved in the regulation of the MSP. To date, no mammalian ARD has been characterized biochemically. An X-ray crystallographic structure of MmARD was determined as a part of the Structural Genomics initiative (1VR3), but the identity of the bound metal ion, enzymatic activity and chemistry were not established.15 The biochemical data presented in this report clearly illustrate that a mammalian ARD also exhibits metal ion-dependent on- and off-pathway chemistry in vitro. This is particularly interesting in that carbon monoxide (CO) which is a product of the off-pathway chemistry, is known to be an anti-apoptotic molecule and has been proposed to act as a signaling molecule analogous to NO.21–24 Furthermore, several independent studies have indicated a role for HsARD in cancer. Seiki, et al. have shown that HsARD binds the cytoplasmic tail of MMP14 and inhibits cell migration and invasion activities.17 In another study, Oram, et al., have shown that ADI1 is down-regulated in rat and human prostate cancer cell lines and the enforced expression of ADI1 causes apoptosis.19 This leads to the intriguing possibility that binding of Ni2+, Co2+ or Mn2+ in the ARD active site could lead to inappropriate gain-of-function in carcinogenesis, helping to protect transformed cells from apoptosis by producing CO and/or loss-of-function in regulating MMP activity. Given the increased thermal stability of the Ni2+-bound enzyme relative to the Fe2+-bound form, formation of Ni-ARD could represent a pathological kinetic trap in vivo.
Enzymatic activity of MmARD is metal ion dependent
The enzymatic data indicate that Fe-MmARD has ~10-fold higher activity than Ni-MmARD or Co-MmARD, unlike ARD in Klebsiella oxytoca, where Ni-KoARD has a higher activity than Fe-KoARD.30 On the other hand, Ni2+-bound ARD from Oryza sativa L. (OsARD) polymerizes and has a much reduced activity relative to Fe2+-bound OsARD.31 This suggests that off-pathway chemistry catalyzed by the Ni2+-bound enzyme may be relevant only in pathology in mammals or eukaryotes, unlike in Klebsiella oxytoca where off-pathway chemistry is seen in the native healthy organism. As is KoARD, MmARD is promiscuous in the metal ions it can bind. Co2+ and Mn2+ bound MmARDs perform off-pathway chemistry like Ni-MmARD. We were unable to get 100% occupancy of Mg2+ bound MmARD, since after purification, it was always contaminated with other trace transition metals. Since Fe-MmARD exhibits the maximum on-pathway enzymatic activity and many oxidoreductases use Fe2+ as a metal ion co-factor,35 we suspect that the Fe-MmARD is the native form under normal conditions.
Metal identity
The identity of the metal ion was unknown in the published crystal structure of MmARD (1VR3). Considering the importance of the identity of the metal ion in determining both structure and function of ARD, we deemed it important to identify both the metal(s) that can bind to MmARD as well as the associated enzymatic activity. In this study, we were able to verify the identity of the metal ion both crystallographically and by mass spectrometry. The incorporation of a single metal ion in a recombinantly expressed metalloprotein is experimentally challenging. We tried reconstitution of the desired metal ion by unfolding and refolding Ni-MmARD but, due to compromised protein stability, this procedure was not successful. In this study, we selectively produced a homogeneous single metal ion bound form of the protein by growing E. coli cells in metal-additive free minimal media and introducing the appropriate amount of a single metal during induction. For biochemical studies, in order to unambiguously determine the metal content, we used an N-terminal Strep II affinity tag, avoiding the often-used His tag which has affinity for metals and can complicate metal analysis. The Strep II affinity tag allowed us to use a single affinity column for purification to obtain >99% pure protein, thus avoiding the need for multiple purification steps and concomitant loss of bound metal ion. For the purpose of protein crystallography, we used the same construct as that used for the previously published structure. This was a His tagged construct, but unlike the expression in metal rich media and Ni-NTA purification which was used previously, we used a single metal during expression and purified using traditional purification methods (ammonium sulfate cuts followed by ion exchange and hydrophobic interaction chromatography). The identity of the metal ion in the active site was then confirmed using X-ray anomalous scattering methods.
Implications for the mechanism of ARD-catalyzed dioxygenation reactions
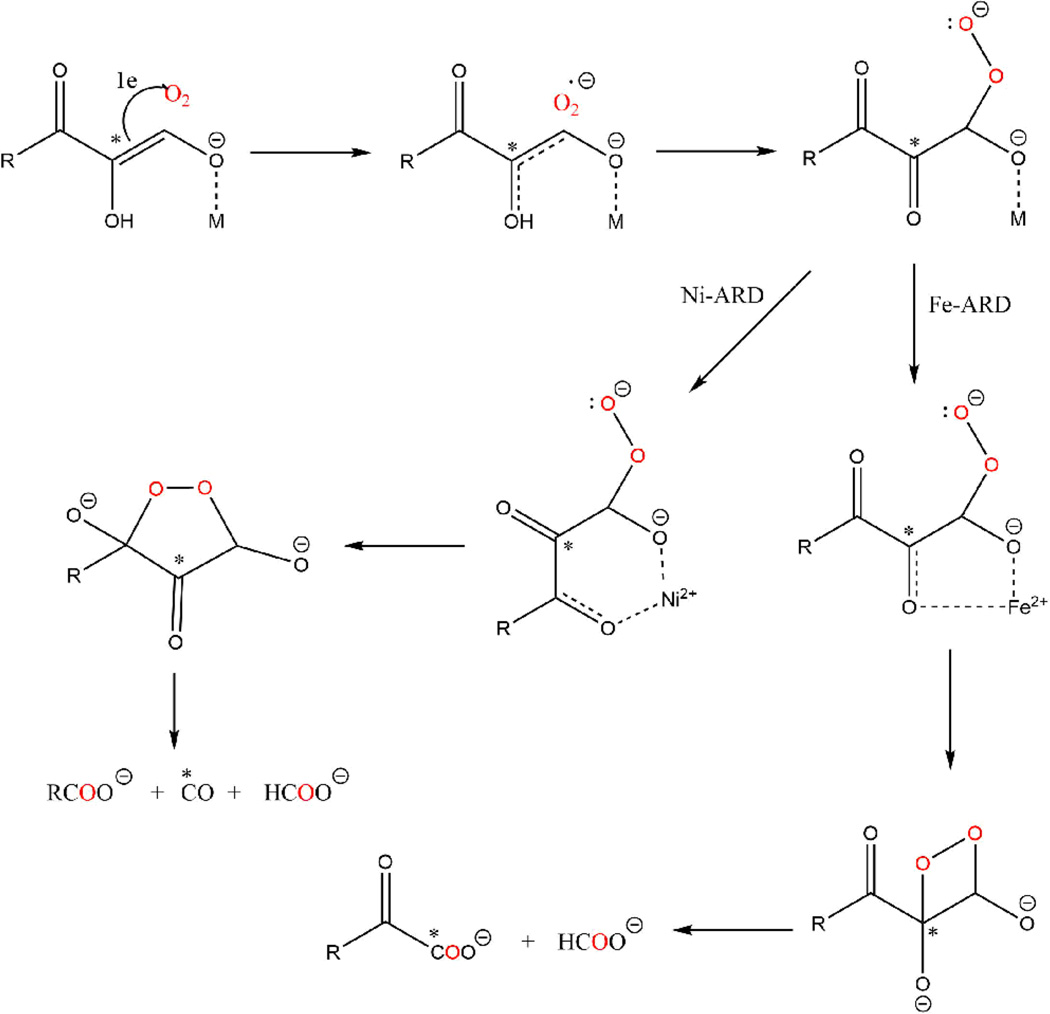
The crystallographic data presented here provide the first detailed structural information regarding the mode of product binding and the likely mode of substrate binding in the active site of Ni2+ or Co2+ bound MmARD. Previous studies have proposed several hypotheses for the dual chemistry of KoARD. The chelate hypothesis postulates that the difference in the chemistry is due to a difference in the coordination modes of substrate to the metal ion center where Fe2+ forms a five-membered ring and Ni2+ forms a six-membered ring leading to different products (Scheme 3). This hypothesis was supported by 18O and 14C labeling studies.13, 14, 30
Scheme 3. Proposed mechanisms of Ni-ARD versus Fe-ARD enzymatic catalysis explained by Chelate hypothesis.
The results of incorporation of 18O and 14C labeling studies are indicated by the blue O atoms and the asterisk. (Figure adapted from ref 39).
Subsequent modeling studies by Bureau et al. challenged the chelate hypothesis, showing that both Ni and Fe can form six-membered ring complexes with substrate homologues and give Ni-like products in anhydrous solvents. The change in the chemistry is seen in the presence of water and can be attributed to the hydration of the tri-ketone reaction intermediate.36–38 These results are further supported by the fact that the NMR structure of Ni-KoARD shows a closed active site compared to the relatively more solvent exposed active site in Fe-KoARD.39 Sparta et al. published computational studies which used mixed quantum-classical molecular dynamics simulations coupled with density functional theoretical calculations to explain the mechanistic differences in the two chemistries. Their energy calculations show that a six membered ring with the substrate has the lowest energy in the cases of both Ni2+ and Fe2+. In these theoretical studies, the difference in the chemistry is attributed to the redox-active nature of Fe2+ relative to Ni2+, allowing Fe-ARD to form an intermediate partially stabilized by charge transfer to the Fe2+.40 However, the DLA-bound Ni-MmARD structure demonstrates that five membered rings can indeed form involving the Ni2+ ion and a substrate analog. In the several ligands we tried (Table S7), we were unable to find a ligand or a substrate analog which formed a six membered ring with the metal ion. While not conclusive evidence for the five-membered ring intermediate in ARD catalysis, these results suggest that the mechanism is still an open question.
In this study we present the crystallographic structures of Ni-MmARD and Co-MmARD. Fe-MmARD requires anaerobic handling due to oxidation from the Fe+2 to Fe+3 state. Although we purified and set crystal trays of Fe-MmARD anaerobically, we were unable to crystallize it. Recently, a low-resolution (3.05 Å) structure of HsARD was deposited in the PDB (4qgn). While complete details of this structure have not been published, the bound metal ion is proposed to be Fe3+ which is surprising to us, as we have found this form of the enzyme to be unstable and inactive. We note, however, that, L-selenomethionine is present in the active site of the 4qgn structure and chelates the metal ion via the amino group and one carboxylate oxygen, generating a five-membered ring in the same fashion as D-lactic acid observed here. The presence of the off-pathway chemistry is particularly interesting in mammals due carbonmonoxide being an anti-apoptotic molecule.22–24 Ni is toxic in mammals, and to date no known native Ni-binding metalloenzyme has been found in mammals. However, since off-pathway chemistry is seen also with Co2+ and Mn2+ in both the MmARD, it will be interesting to test the presence of off-pathway chemistry in vivo to see if ARD performs a switch in cancerous tissue from the on-pathway chemistry to off-pathway chemistry by switching its metal ion.
Supplementary Material
Acknowledgments
The authors thank Nicola Lupoli from Harvard School of Public Health for help with Inductively-coupled Plasma Mass spectrometry (ICP-MS) analysis. A portion of the results in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. The authors would like to acknowledge the assistance of the X29A beam line scientist Howard Robinson at the National Synchrotron Light Source at Brookhaven National Laboratory for help with design and data collection of anomalous scattering experiments. Support for beamline X29A at the National Synchrotron Light Source came from the Offices of Biological and Environmental Research and of Basic Energy Sciences of the US Department of Energy, and from the National Center for Research Resources of the National Institutes of Health. The Schemes were drawn using ChemDraw Professional 15.0.
ABBREVIATIONS
- ARD
Acireductone dioxygenase
- MmARD
Mus musculus ARD
- HsARD
Homo sapiens ARD
- KoARD
Klebsiella oxytoca ARD
- OsARD
Oryza sativa L ARD
- ICP-MS
Inductively coupled plasma mass spectrometry
- KMTB
2-keto-4-(methylthio)-butyric acid
- MTP
3-(methylthio) propionic acid
- DLA
D-lactic acid
- VA
Valeric acid
- DSF
Differential scanning fluorimetry
Footnotes
ASSOCIATED CONTENT
Supporting Information: The crystallographic data and refinement statistics, specific activity data, oligomerization state data and tables of compounds tested for thermal stabilization are included in the supplementary information. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. / All authors have given approval to the final version of the manuscript.
The atomic coordinates and the structure factors will be deposited in the Research Collaboratory for Structural Bioinformatics Protein data Bank, www.rcsb.org [ID codes XXX (Ni-MmARD bound to KMTB, Co-MmARD bound to KMTB, Ni-MmARD bound to VA, Ni-MmARD bound to DLA, Ni-MmARD bound to 2-oxovaleric acid]
REFERENCES
- 1.William-Ashman HG, Pegg AE. Polyamines in Biology and Medicine. New York: Marcel Dekker; 1981. pp. 43–73. [Google Scholar]
- 2.Williams-Ashman HG, Seidenfeld J, Galletti P. Trends in the biochemical pharmacology of 5'-deoxy-5'-methylthioadenosine. Biochem Pharmacol. 1982;31:277–288. doi: 10.1016/0006-2952(82)90171-x. [DOI] [PubMed] [Google Scholar]
- 3.Taiz L. Plant Physiology. Redwood City, CA: Benjamin/Cummins Publishin Co.; 1991. [Google Scholar]
- 4.Schlenk F. Methylthioadenosine. Adv Enzymol Relat Areas Mol Biol. 1983;54:195–265. doi: 10.1002/9780470122990.ch4. [DOI] [PubMed] [Google Scholar]
- 5.Oredsson SM. Polyamine dependence of normal cell-cycle progression. Biochem Soc Trans. 2003;31:366–370. doi: 10.1042/bst0310366. [DOI] [PubMed] [Google Scholar]
- 6.Marton LJ, Pegg AE. Polyamines as targets for therapeutic intervention. Annu Rev Pharmacol Toxicol. 1995;35:55–91. doi: 10.1146/annurev.pa.35.040195.000415. [DOI] [PubMed] [Google Scholar]
- 7.Pegg AE. Polyamine metabolism and its importance in neoplastic growth and a target for chemotherapy, Cancer Res. 1988;48:759–774. [PubMed] [Google Scholar]
- 8.Shapiro SK, Barrett A. 5-Methylthioribose as a precursor of the carbon chain of methionine. Biochem Biophys Res Commun. 1981;102:302–307. doi: 10.1016/0006-291x(81)91521-7. [DOI] [PubMed] [Google Scholar]
- 9.Wray JW, Abeles RH. The methionine salvage pathway in Klebsiella pneumoniae and rat liver. Identification and characterization of two novel dioxygenases. J Biol Chem. 1995;270:3147–3153. doi: 10.1074/jbc.270.7.3147. [DOI] [PubMed] [Google Scholar]
- 10.Dai Y, Wensink PC, Abeles RH. One protein, two enzymes. J Biol Chem. 1999;274:1193–1195. doi: 10.1074/jbc.274.3.1193. [DOI] [PubMed] [Google Scholar]
- 11.Myers RW, Wray JW, Fish S, Abeles RH. Purification and characterization of an enzyme involved in oxidative carbon-carbon bond cleavage reactions in the methionine salvage pathway of Klebsiella pneumoniae. J Biol Chem. 1993;268:24785–24791. [PubMed] [Google Scholar]
- 12.Wray JW, Abeles RH. A bacterial enzyme that catalyzes formation of carbon monoxide. J Biol Chem. 1993;268:21466–21469. [PubMed] [Google Scholar]
- 13.Pochapsky TC, Pochapsky SS, Ju T, Mo H, Al-Mjeni F, Maroney MJ. Modeling and experiment yields the structure of acireductone dioxygenase from Klebsiella pneumoniae. Nat Struct Biol. 2002;9:966–972. doi: 10.1038/nsb863. [DOI] [PubMed] [Google Scholar]
- 14.Ju T, Goldsmith RB, Chai SC, Maroney MJ, Pochapsky SS, Pochapsky TC. One protein, two enzymes revisited: a structural entropy switch interconverts the two isoforms of acireductone dioxygenase. J Mol Biol. 2006;363:823–834. doi: 10.1016/j.jmb.2006.08.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Q, Schwarzenbacher R, Krishna SS, McMullan D, Agarwalla S, Quijano K, Abdubek P, Ambing E, Axelrod H, Biorac T, Canaves JM, Chiu HJ, Elsliger MA, Grittini C, Grzechnik SK, DiDonato M, Hale J, Hampton E, Han GW, Haugen J, Hornsby M, Jaroszewski L, Klock HE, Knuth MW, Koesema E, Kreusch A, Kuhn P, Miller MD, Moy K, Nigoghossian E, Paulsen J, Reyes R, Rife C, Spraggon G, Stevens RC, van den Bedem H, Velasquez J, White A, Wolf G, Hodgson KO, Wooley J, Deacon AM, Godzik A, Lesley SA, Wilson IA. Crystal structure of acireductone dioxygenase (ARD) from Mus musculus at 2.06 angstrom resolution. Proteins. 2006;64:808–813. doi: 10.1002/prot.20947. [DOI] [PubMed] [Google Scholar]
- 16.Yeh CT, Lai HY, Chen TC, Chu CM, Liaw YF. Identification of a hepatic factor capable of supporting hepatitis C virus replication in a nonpermissive cell line. J Virol. 2001;75:11017–11024. doi: 10.1128/JVI.75.22.11017-11024.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uekita T, Gotoh I, Kinoshita T, Itoh Y, Sato H, Shiomi T, Okada Y, Seiki M. Membrane-type 1 matrix metalloproteinase cytoplasmic tail-binding protein-1 is a new member of the Cupin superfamily. A possible multifunctional protein acting as an invasion suppressor down-regulated in tumors. J Biol Chem. 2004;279:12734–12743. doi: 10.1074/jbc.M309957200. [DOI] [PubMed] [Google Scholar]
- 18.Oram SW, Ai J, Pagani GM, Hitchens MR, Stern JA, Eggener S, Pins M, Xiao W, Cai X, Haleem R, Jiang F, Pochapsky TC, Hedstrom L, Wang Z. Expression, Function of the Human Androgen-Responsive Gene AD11 in Prostate Cancer. Neoplasia. 2007;9:643–651. doi: 10.1593/neo.07415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oram S, Jiang F, Cai X, Haleem R, Dincer Z, Wang Z. Identification and characterization of an androgen-responsive gene encoding an aci-reductone dioxygenase-like protein in the rat prostate. Endocrinology. 2004;145:1933–1942. doi: 10.1210/en.2003-0947. [DOI] [PubMed] [Google Scholar]
- 20.Espin-Perez A, de Kok TM, Jennen DG, Hendrickx DM, De Coster S, Schoeters G, Baeyens W, van Larebeke N, Kleinjans JC. Distinct genotype-dependent differences in transcriptome responses in humans exposed to environmental carcinogens. Carcinogenesis. 2015;36:1154–1161. doi: 10.1093/carcin/bgv111. [DOI] [PubMed] [Google Scholar]
- 21.Verma A, Hirsch DJ, Glatt CE, Ronnett GV, Snyder SH. Carbon monoxide: a putative neural messenger. Science. 1993;259:381–384. doi: 10.1126/science.7678352. [DOI] [PubMed] [Google Scholar]
- 22.Gunther L, Berberat PO, Haga M, Brouard S, Smith RN, Soares MP, Bach FH, Tobiasch E. Carbon monoxide protects pancreatic beta-cells from apoptosis and improves islet function/survival after transplantation. Diabetes. 2002;51:994–999. doi: 10.2337/diabetes.51.4.994. [DOI] [PubMed] [Google Scholar]
- 23.Liu XM, Chapman GB, Wang H, Durante W. Adenovirus-mediated heme oxygenase-1 gene expression stimulates apoptosis in vascular smooth muscle cells. Circulation. 2002;105:79–84. doi: 10.1161/hc0102.101369. [DOI] [PubMed] [Google Scholar]
- 24.Thom SR, Fisher D, Xu YA, Notarfrancesco K, Ischiropoulos H. Adaptive responses and apoptosis in endothelial cells exposed to carbon monoxide. Proc Natl Acad Sci U S A. 2000;97:1305–1310. doi: 10.1073/pnas.97.3.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Heinsen MH, Kostic M, Pagani GM, Riera TV, Perovic I, Hedstrom L, Snider BB, Pochapsky TC. Analogs of 1-phosphonooxy-2,2-dihydroxy-3-oxo-5-(methylthio)pentane, an acyclic intermediate in the methionine salvage pathway: a new preparation and characterization of activity with E1 enolase/phosphatase from Klebsiella oxytoca. Bioorg Med Chem. 2004;12:3847–3855. doi: 10.1016/j.bmc.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 26.Otwinowski Z, Minor W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. In: Carter CWJ, Sweet RM, editors. Methods in Enzymology. New York: Academic Press; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 27.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 28.Brennan S, Cowan PL. A suite of programs for calculating x-ray absorption, reflection, and diffraction performance for a variety of materials at arbitrary wavelengths. Rev. Sci. Instrum. 1992;63:850–853. [Google Scholar]
- 29.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 30.Dai Y, Pochapsky TC, Abeles RH. Mechanistic studies of two dioxygenases in the methionine salvage pathway of Klebsiella pneumoniae. Biochemistry. 2001;40:6379–6387. doi: 10.1021/bi010110y. [DOI] [PubMed] [Google Scholar]
- 31.Sauter M, Lorbiecke R, Ouyang B, Pochapsky TC, Rzewuski G. The immediate-early ethylene response gene OsARD1 encodes an acireductone dioxygenase involved in recycling of the ethylene precursor S-adenosylmethionine. Plant J. 2005;44:718–729. doi: 10.1111/j.1365-313X.2005.02564.x. [DOI] [PubMed] [Google Scholar]
- 32.Wang ZJ, Zaitsu K, Ohkura Y. High-performance liquid chromatographic determination of alpha-keto acids in human serum and urine using 1,2-diamino-4,5-methylenedioxybenzene as a precolumn fluorescence derivatization reagent. J Chromatogr. 1988;430:223–231. doi: 10.1016/s0378-4347(00)83157-6. [DOI] [PubMed] [Google Scholar]
- 33.Lee SH, Kim SO, Chung BC. Gas chromatographic-mass spectrometric determination of urinary oxoacids using O-(2,3,4,5,6-pentafluorobenzyl)oxime-trimethylsilyl ester derivatization and cation-exchange chromatography. J Chromatogr B Biomed Sci Appl. 1998;719:1–7. doi: 10.1016/s0378-4347(98)00388-0. [DOI] [PubMed] [Google Scholar]
- 34.Fu X, Kimura M, Iga M, Yamaguchi S. Gas chromatographic-mass spectrometric screening for organic acidemias using dried urine filter paper: determination of alpha-ketoacids. J Chromatogr B Biomed Sci Appl. 2001;758:87–94. doi: 10.1016/s0378-4347(01)00101-3. [DOI] [PubMed] [Google Scholar]
- 35.Waldron KJ, Rutherford JC, Ford D, Robinson NJ. Metalloproteins and metal sensing. Nature. 2009;460:823–830. doi: 10.1038/nature08300. [DOI] [PubMed] [Google Scholar]
- 36.Allpress CJ, Grubel K, Szajna-Fuller E, Arif AM, Berreau LM. Regioselective aliphatic carbon-carbon bond cleavage by a model system of relevance to iron-containing acireductone dioxygenase. J Am Chem Soc. 2013;135:659–668. doi: 10.1021/ja3038189. [DOI] [PubMed] [Google Scholar]
- 37.Szajna E, Arif AM, Berreau LM. Aliphatic carbon-carbon bond cleavage reactivity of a mononuclear Ni(II) cis-beta-keto-enolate complex in the presence of base and O2: a model reaction for acireductone dioxygenase (ARD) J Am Chem Soc. 2005;127:17186–17187. doi: 10.1021/ja056346x. [DOI] [PubMed] [Google Scholar]
- 38.Berreau LM, Borowski T, Grubel K, Allpress CJ, Wikstrom JP, Germain ME, Rybak-Akimova EV, Tierney DL. Mechanistic studies of the O2-dependent aliphatic carbon-carbon bond cleavage reaction of a nickel enolate complex. Inorg Chem. 2011;50:1047–1057. doi: 10.1021/ic1017888. [DOI] [PubMed] [Google Scholar]
- 39.Maroney MJ, Ciurli S. Nonredox nickel enzymes. Chem Rev. 2014;114:4206–4228. doi: 10.1021/cr4004488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sparta M, Valdez CE, Alexandrova AN. Metal-dependent activity of Fe and Ni acireductone dioxygenases: how two electrons reroute the catalytic pathway. J Mol Biol. 2013;425:3007–3018. doi: 10.1016/j.jmb.2013.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.