Abstract
Meiosis, the mechanism of creating haploid gametes, is a complex cellular process observed across sexually reproducing organisms. Fundamental to meiosis is the process of homologous recombination, whereby DNA double-strand breaks are introduced into the genome and are subsequently repaired to generate either noncrossovers or crossovers. Although homologous recombination is essential for chromosome pairing during prophase I, the resulting crossovers are critical for maintaining homolog interactions and enabling accurate segregation at the first meiotic division. Thus, the placement, timing, and frequency of crossover formation must be exquisitely controlled. In this review, we discuss the proteins involved in crossover formation, the process of their formation and designation, and the rules governing crossovers, all within the context of the important landmarks of prophase I. We draw together crossover designation data across organisms, analyze their evolutionary divergence, and propose a universal model for crossover regulation.
Keywords: meiosis, homologous recombination, crossover designation
INTRODUCTION
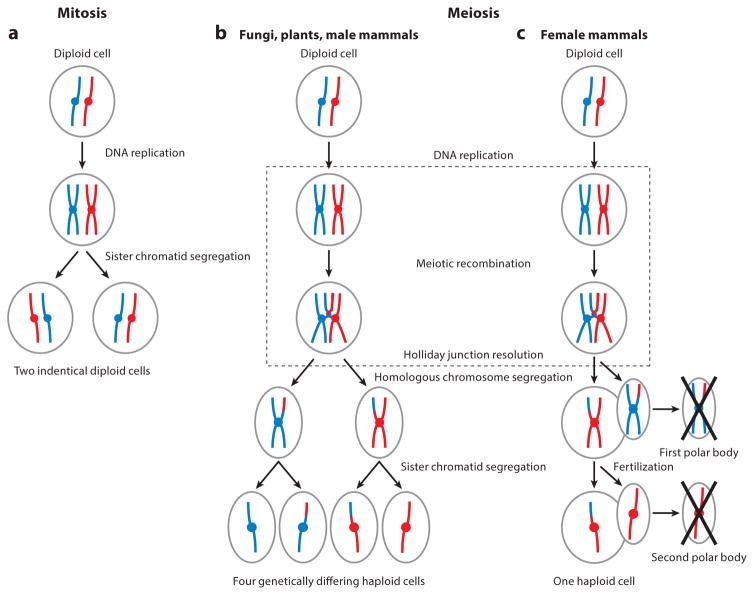
Meiosis is a unique, specialized cellular division process fundamental to the production of viable gametes and the propagation of sexually reproducing species. The resulting cells can, for example, be in the form of spermatozoa and oocytes (mammals), pollen and ovule (flowering plants), and spores (fungi). Mitosis produces genetically identical diploid daughter cells from a diploid progenitor cell through one round of DNA replication and one ensuing chromosome segregation event that involves equal sister chromatid separation (Figure 1a). This is facilitated by the fact that the sister chromatids are intricately linked through the cohesin rings that keep them tethered along their entire lengths. By contrast, meiosis produces haploid gametes from a diploid parental cell by means of two rounds of chromosome segregation preceded by a single round of DNA replication (Figure 1b). The second meiotic division (MII) is highly reminiscent of a mitotic division, in which sister chromatids divide equally into their respective daughter cells. However, during the first meiotic division (MI), the requirement to faithfully halve the genome requires that homologous (maternal and paternal) chromosomes must find each other, pair, and then become tethered to each other until the appropriate time. This tethering process, involving both pro-tein:protein interactions (synapsis) and DNA:DNA interactions (meiotic recombination leading to crossover formation), defines prophase I of meiosis. Thus, meiosis I, and specifically prophase I, involves highly complex, coordinated processes that are essential for successful meiosis. Although these events are highly conserved across sexually reproducing organisms, there are fundamental differences in specific components of prophase I across species that make this one of the most fascinating aspects of biology and an area of intense research interest.
Figure 1.
Mitosis and meiosis. (a) During mitosis, a diploid cell containing a pair of homologous chromosomes (one red and one blue) undergoes DNA replication to generate sister chromatids. In the mitotic division, sister chromatids are segregated to separate daughter cells. The final products of mitosis are two genetically identical cells. As in mitosis, (b) in meiosis, diploid cells containing two homologous chromosomes undergo DNA replication to generate sister chromatids. During prophase I (dotted box), however, meiotic cells then undergo recombination, forming physical links between homologs and resulting in genetic exchange. In the first meiotic division, homologous chromosomes are segregated, followed by sister chromatids in the second division. The products of meiosis in fungi, plants, and male mammals are four genetically differing haploid gametes from one starting diploid cell. (c) In female meiosis in mammals, cells arrest prior to the first meiotic division (dictyate arrest). Upon ovulation, the first meiotic division occurs, segregating homologous chromosomes and forming a polar body, which is subsequently discarded. Upon fertilization, segregation of the sister chromatids occurs during the second meiotic division, creating a second polar body, which is also subsequently discarded. The product of female meiosis is one genetically differing haploid cell.
Vive la Différence: Studying Meiosis in Different Species
The meiotic process has been studied in a number of organisms across different kingdoms, each providing important insight into conserved mechanisms, while at the same time possessing unique features that help to shape our understanding. Simple unicellular organisms, such as the budding yeast Saccharomyces cerevisiae, allow for examination of large-scale and synchronous cultures [>70% (28)], enabling temporal-/stage-specific analysis at genomic and biochemical scales. Such organisms are easily modified genetically and allow analysis of meiotic products by spore dissection and viability. In addition genetic variations between strains enable offspring DNA sequences to be compared with that of their parental strains, identifying where genetic exchange has taken place [e.g., (43, 146)]. Arabidopsis thaliana, a species from the Brassicaceae family, is an excellent model for plants containing many genes similar to other organisms yet also containing genes specific to plants (6). A. thaliana offers additional value as a model organism: It is an organism in which mutant alleles are easily generated, its genome is small and sequenced unlike other plants, and its life cycle is relatively short (reviewed in 179). The unique properties of the nematode Caenorhabditis elegans and the exquisite spatial organization of meiotic cells allows visualization of multiple stages of meiosis within the same organism, and extensive shared mutant banks allow complex genetic analysis of meiosis in worms (reviewed in 138). In addition, C. elegans exist as hermaphrodites (as well as males), allowing the understanding of the impact of meiotic mutations on both spermatogenesis and oogenesis within the same organism.
Mammals, such as the lab mouse Mus musculus, provide an additional level of complexity not observed in the aforementioned single cell and simple organisms. For example, the endocrine control required to coordinate the meiotic program with sexual receptivity and the general hormonal milieu of the individual offers another layer to an already complex process. Moreover, mammals and other organisms with two distinct sexes offer systems in which male and female meiosis can vary widely. Such sexual dimorphism is most notable in, for example, the fruit fly Drosophila melanogaster, in which females undertake a typical meiotic program for oogenesis and males do not undergo recombination and synaptonemal complex (SC) formation, which are the canonical methods for ensuring homologous chromosome segregation (83, 149). In mammals, male meiosis and female meiosis are more similar than in male and female flies, utilizing similar genetic pathways in each sex. However, the control of these pathways and their temporal coordination exhibit distinct sexual dimorphism (157).
Meiosis in female mammals initiates during embryonic development, when retinoic acid from the adjoining mesonephros acts on the oogonial cells of the primitive gonad to induce their entry into prophase I of meiosis (30, 116). Retinoic acid promotes prophase I entry through the induction of several retinoic acid genes, including Stra8, with the entire population of oogonia entering prophase I in synchrony or near synchrony (5, 240). In embryonic males, retinoic acid production is no less abundant; however, the somatic cells of the male gonad express Cyp26B1, whose protein product rapidly and efficiently degrades retinoic acid (30, 116). Thus, in males, degradation of retinoic acid prevents the induction of retinoic acid–responsive genes and thereby prevents meiotic entry. In neonatal mice, expression of Cyp26B1 no longer occurs, and retinoic acid, this time from the Sertoli cells of the seminiferous tubules, can once again act on spermatogonia to induce meiotic entry (30). Indeed, it is believed that the localized production of retinoic acid along the seminiferous tubules is responsible for inducing the waves of meiotic entry that underlie the spermatogenic wave in male mammals (206).
In males, the canonical meiotic program results in the creation of four haploid spermatozoa from each diploid spermatocyte precursor (Figure 1b). Meiosis in male mammals initiates within the seminiferous tubules of the testis, whose spermatogonial stem cell progenitors line the tubular periphery close to the basement membrane. Following several rounds of proliferation, spermatogonia enter meiosis as spermatocytes, proceed through both cellular divisions before exiting meiosis as spermatids, and then undergo a protracted differentiation process to become mature spermatozoa. Populations of spermatogonia initiate meiosis sequentially along the tubule, resulting in a phenomenon known as the spermatogenic wave, resulting in continual production of sperm throughout the life of the individual. By contrast, in females, all oocytes enter the initial stages of meiosis I in utero but then enter an arrested state known as dictyate arrest at around the time of birth, depending on the species. Dictyate arrest persists until after puberty when, upon stimulation by the gonadotropin luteinizing hormone (LH) during the estrous/menstrual cycle, the oocyte is stimulated to resume meiosis, completing the first meiotic division and releasing the first polar body. Polar body extrusion coincides with ovulation, both events being stimulated by LH, and thus synchronizing meiotic events with the events of mating. The first polar body contains half the genomic content of the cell but is less than a fraction of the size of the final MII oocyte. The oocyte then commences meiosis II but arrests once again at metaphase II. Upon fertilization, calcium waves initiated by sperm-derived factors result in resumption of MII and the second division takes place, creating a second polar body. As with the first division, the second division results in a halving of genomic material but in contrast to the first polar body, the second polar body contains a quarter of the overall genomic material of the progenitor oogonial cell. Thus, unlike males who produce four sperm from a meiotic progenitor, females produce just one oocyte and two discarded polar bodies (Figure 1c).
At the experimental level, studying meiosis in mouse males is complicated because of the total population of cells consisting of subpopulations having initiated meiosis at different times. This therefore makes it difficult to easily access stage-specific meiotic cells. Techniques have been developed to isolate stage-specific populations of mouse testis extract by either differential sedimentation velocity (STA-put) (122) or FACS isolation using markers of spermatogenesis (148), or by collecting the initial wave of meiosis that occurs postnatally (13). In contrast, and because meiosis in females initiates in the embryo, it is necessary to observe fertilization by the presence of vaginal plugs and then to harvest embryos at specific times in gestational development. However, the number of meiocytes that can be obtained through this method is considerably smaller than for males.
Aneuploidy
It is critical that gametes formed during meiosis have the correct genetic complement, as these provide the basis for the propagation of the organism. Thus, the need for chromosome segregation to occur correctly is of paramount importance. Inaccurate chromosome segregation can lead to gametes that are aneuploid, containing either too many or too few copies of one (or more) chromosomes. Subsequent fertilization with one such gamete can lead to offspring that bear the wrong number of chromosomes, most commonly in the form of trisomies, which have three copies of a specific chromosome. Interestingly, trisomies within the human population occur far more frequently than in other species (80) and originate from defects that occur prior to, or during, the meiotic divisions.
Many of the aneuploidy events that occur in humans appear to arise most commonly during meiosis in females rather than males, with the oocyte thought to account for between 90% and 95% of autosomal aneuploidy events (81). The most frequent of these is Down syndrome, resulting from trisomy of chromosome 21, which accounts for 1 in 700 live births in the United States (173). Reduced/altered meiotic recombination has been associated with a higher incidence of trisomy 21, as well as with trisomy of other chromosomes, including 15, 16, and 18 (82, 200). Moreover, many chromosomal aneuploidies are lethal to the developing embryo, with estimates of chromosomal abnormalities occurring within aborted fetuses being around 35% (79, 80).
The high incidence of aneuploidies in human gametes highlights the importance of understanding how and where these abnormalities arise. Indeed, accumulating evidence points to a high degree of variability among human germ cells undergoing meiotic prophase I. For example, although maternal age appears to be the only factor that has been definitively associated with increased risk of aneuploidy in women and a number of molecular pathways have been proposed to account for such age-related phenomena (70), several reports have identified significant variation in recombination frequencies among women independent of age (32). In line with this, studies of human fetal oocytes have demonstrated similarly high levels of variation in the events that give rise to crossovers at prophase I (125), events that would be extremely stringently regulated in other meiotic species. Such observations suggest that mechanisms of crossover regulation during prophase I, and not relating to extended periods of dictyate arrest, must be important for the etiology of human aneuploidy events.
Scope of this Review
Prophase I encapsulates the unique and defining events of meiosis that are essential for sexual reproduction. Of particular importance are the crossovers that form between homologous chromosomes and that serve to maintain a strong connection between homologs, thus enabling them to be accurately segregated during MI. Crossovers must be tightly regulated, both temporally and spatially, and yet often arise from an excessive number of initiating DNA double-strand break (DSB) events. These additional DSBs serve to facilitate the equally fascinating processes of homology searching and pairing but must be repaired with equally exquisite precision as those designated to become crossovers.
In mouse, as in most other organisms studied, the number of DSBs outweighs the final number of crossovers by at least tenfold. This raises many questions, not least of which is how crossovers are selected from this pool of DSBs, how the timing of this selection (or designation) process is regulated with respect to other cytogenetic events, whether crossover designation is a multistep or single-step process (licensing and designation, for example), and what the consequences are for altered/aberrant crossover designation. Are there alternative pathways that can compensate/modulate crossover designation if critical events fail?
This review focuses on these and other questions, utilizing new data arising from studies across the eukaryotic spectrum. Although we attempt to synthesize an overarching view across organisms, it is not possible to explicitly describe all processes employed by all organisms. Thus, we illustrate areas of shared conservation and unique differences where possible. By necessity, we first begin with a brief description of the events of synapsis and DSB formation and repair, as well as the highly correlated process of SC formation. For more complete reviews of these two hallmark features of prophase I, the reader is directed to many excellent recent reviews on these topics (101, 123, 210, 242). We then discuss the intrinsic/cis-regulatory mechanisms of crossover formation in various species, comparing and contrasting mechanisms of crossover formation where possible. Finally, we incorporate recent data concerning the roles of various extrinsic/trans-regulatory mechanisms that oversee crossover licensing and designation, culminating with an integrated model to explain how different organisms achieve the appropriate placement and frequency of crossovers during prophase I.
INITIATION OF SYNAPSIS AND RECOMBINATION
Disjunction of sister chromatids during mitosis and meiosis II is accomplished accurately as a result of the cohesin complexes that are laid down during DNA replication. Cohesin complexes encircle both sister chromatids, thereby allowing tension to form when each sister is pulled in opposite directions by the cellular division machinery (reviewed by 34). In meiosis I, however, homologous chromosomes, each of which has been derived from a different parent, do not have any such pre-established attachments. Thus, the chromosomes must find and connect with their homologous partner to allow them to then be correctly segregated from each other at the first meiotic division. These events all occur during prophase I of meiosis and include pairing (where homologous chromosomes move and locate each other), meiotic recombination (the process of DSB formation and repair), and synapsis (the formation of a proteinaceous structure, the SC) between homologs.
Pairing
Early prophase I chromosome movements are essential for pairing of homologs prior to the initiation of synapsis and recombination. These movements are often preceded by initial coupling or contact, and are concomitant, in most cases, with the tethering of telomeres to the nuclear envelope in a manner that is dependent on the SUN/KASH family of proteins (reviewed by 92, 114). In certain cases, such as in the mouse, telomere clustering also appears to be dependent on certain proteins of the SC and the DSB formation protein SPO11, independent of its DSB function (19, 130, 193). In addition, the telomeres of many species appear to cluster within a limited domain of the nuclear envelope, resulting in a chromosome configuration known as the telomere bouquet (241). The classic example of chromosome movements and telomere clustering is provided by Schizosaccharomyces pombe, in which the telomeres cluster at the spindle pole body and the entire nucleus oscillates between the cell poles, a movement often described as the horsetail (46, 114). Centromere association appears to be important also, at least in certain species, including S. cerevisiae, S. pombe, A. thaliana, and D. melanogaster (reviewed by 58).
In C. elegans, pairing occurs independently of recombination through the use of pairing centers (PCs), which are regions of DNA sequence that are highly correlated with initial interactions between homologous chromosomes. These PCs are located at the ends of chromosomes and are composed of highly repetitive DNA sequences that recruit and bind the C2H2 zinc-finger proteins, ZIM-1, ZIM-2, ZIM-3, and HIM-8 (reviewed by 138). Although HIM-8 binds specifically to the X-chromosome, the three ZIM proteins facilitate pairing of the five autosomes. Pairing of chromosomes II and III are mediated by ZIM-1, whereas pairing of chromosomes I and IV are mediated by ZIM-3, suggesting additional mechanisms to specify the correct chromosomal pairing (177).
Pairing centers do not appear to be confined to worms and are a feature of meiosis in both D. melanogaster males and in S. pombe. In the former, homologous chromosomes enter meiosis in a paired state, with the exception of the sex chromosomes, which pair via ribosomal DNA (rDNA) repeats of the nucleolus organizing region (NOR) (reviewed in 149). Deletion of the NOR abolishes XY pairing and leads to nondisjunction at the first meiotic division, whereas exogenous rDNA sequences can restore this pairing, indicating that the NOR serves as a pairing center for the sex chromosomes in Drosophila males (149).
The pairing center defined in S. pombe is mechanistically distinct from those mentioned above. Ding et al. (62) have identified the sme2 locus of S. pombe to encode two meiosis-restricted long noncoding RNAs (lncRNA), meiRNA-S and meiRNA-L, that mediate homolog recognition. The lncRNA transcripts accumulate at the sme2 locus on chromosome II during the horsetail stage in early prophase I and thereby facilitate robust and sustained pairing. Deletion of the putative sme2 promoter leads to reduced lncRNA and a resulting reduction in pairing, whereas transfer of the sme2 coding sequence to nonhomologous chromosomes facilitates their ectopic pairing in early prophase I (62). Thus, localization of this lncRNA to its locus of origin acts as a cis-acting pairing factor but is unlikely to be the sole contributing factor that ensures pairing in S. pombe.
Meiotic Recombination
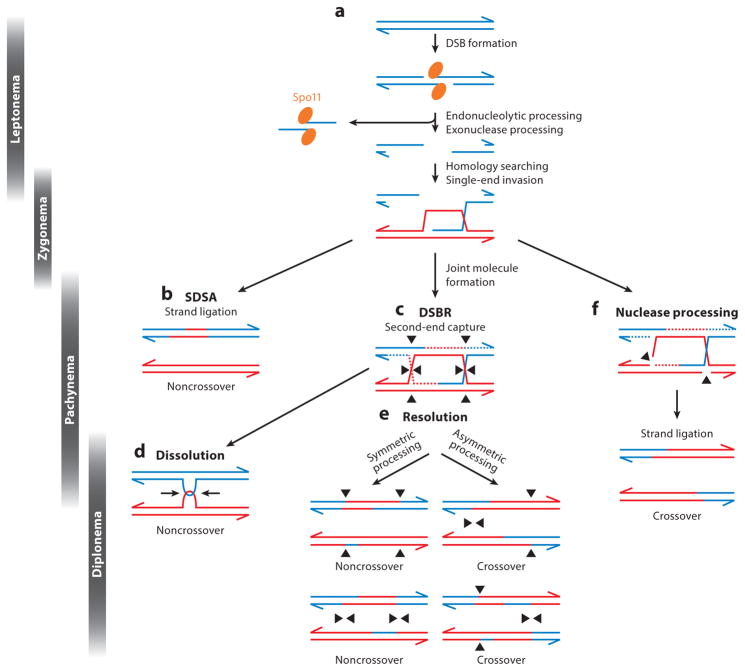
Meiotic recombination, the process by which programmed DSBs are introduced throughout the genome and subsequently repaired, is a mechanism used by the majority of eukaryotes to enable accurate homologous chromosome segregation. In this process, multiple (often hundreds of) DSBs are introduced throughout the genome by the topoisomerase type II–like protein Spo11 (111). These DSBs must subsequently be repaired to form either crossovers or noncrossovers. A crossover is a structure resulting from the repair of a DSB using a homologous chromosome template in a manner that creates a physical connection between the two distinct DNA molecules. Resolution of a crossover results in a DNA molecule that is genetically distinct from either parental molecule—a recombinant chromosome. By contrast, noncrossovers arise when the DSB is repaired from either the sister chromatid or homologous chromosome and results in localized genetic exchange, referred to as gene conversion, immediately in the vicinity of the initiating DSB but does not result in a physical link between homologs. Thus, the DNA sequence flanking the site of a noncrossover does not vary from its original parental sequence (see Figure 2).
Figure 2.
Summary of meiotic recombination based on data from Saccharomyces cerevisiae. (a) Meiotic recombination initiated by the formation of DNA double-strand breaks (DSBs) by the protein Spo11 (orange ellipse) and other accessory proteins. Following DNA cleavage downstream of the DSB event, Spo11 is liberated, forming a Spo11-oligonucleotide complex. Exonuclease activity generates single-stranded DNA (ssDNA) that, through the action of the RecA homologs Rad51 and Dmc1, coat the ssDNA, invade homologous DNA templates on the homolog, and create a displacement (D)-loop. (b) In the following extension of the invading DNA strand, the strand can become disrupted and displaced, reanneal with the opposite side of the DSB, and be repaired as a noncrossover event in a process known as synthesis-dependent strand annealing (SDSA). Alternatively, the invading strand can continue to become extended creating a larger D-loop. (c) Upon second-end capture of the other side of the DSB and subsequent DNA synthesis, a double Holliday junction (dHJ) is formed. (d) By the actions of a helicase and a topoisomerase, dHJs can be migrated toward each other and dissolved (in a process called dissolution) into a noncrossover event. (e) Alternatively, dHJs can be resolved in a symmetrical manner creating noncrossover events or asymmetrically creating crossover events. (f) Following strand invasion, structure-specific nucleases can process joint molecule intermediates to generate a crossover. Many of these events are drawn largely from studies in S. cerevisiae and can be extrapolated to many meiotic species. Abbreviation: DSRB, double-strand break repair.
Initiation of recombination via DNA double-strand break induction
The initiating event of meiotic recombination is the formation of the DSB. The location and numbers of DSBs formed are controlled by factors detailed below and define the location of recombination events.
Double-strand break timing, placement, and modulation
In S. cerevisiae, induction of meiotic DSBs is temporally and mechanistically linked with the completion of DNA replication, ensuring that one event does not precede the other (26, 161). S. pombe and the barley Hordeum vulgare also show a correlation between completion of replication and DSB formation (90). Meiotic DSB formation in S. cerevisiae requires multiple proteins (discussed below; reviewed in 109), but one component in particular, Mer2, is targeted for phosphorylation by the cell cycle kinases CDK (Cdc28 with either Clb5 or Clb6) and DDK (Cdc7-Dbf4 complex) (85, 226). Phosphorylation of Mer2 results in the initiation of DSB formation (85, 226). No equivalent DSB-initiating phos-phorylation event has yet been defined in other organisms, but a homolog of Mer2, Rec15, has been identified in S. pombe (63, 155).
DSB formation does not occur uniformly across the genome (12, 56, 71). Instead, certain regions appear to be much more favorable for DSB formation and these regions, termed hot spots, tend to be intergenic and/or promoter regions where nucleosomes are more widely dispersed (176). High-resolution DSB maps have been obtained by ChIPseq using antibodies raised against known DSB repair proteins (31, 38, 112, 178, 204). In addition, following DSB formation, the initial repair stage produces a protein-DNA complex (Spo11-oligonucleotide) that specifically marks DSB location (166). Using antibodies against Spo11, these Spo11-oligonucleotides can be enriched and sequenced to provide a high-resolution DSB map (171). Mapping of DSB hot spots through ChIPseq correlates extremely well with Spo11-oligo maps, attesting to the specificity and utility of both approaches (38, 129, 171).
In budding yeast, DSB hot spots tend to reside adjacent to transcriptional start sites and are devoid of any obvious DNA sequence motif, whereas in humans and mice, they are located far away from such regions and appear to contain a common sequence motif (162, 163). In many organisms, including yeast, mouse, and human, hot spots correlate, at least in part, with regions enriched for trimethylation of histone H3 on lysine 4 (H3K4me3) (27, 37). A role for chromatin modification in driving DSB position/frequency has also been indicated in C. elegans, in which the condensin DPY-28 negatively regulates DSB frequency, whereas the chromatin-associated proteins HIM-5, HIM-17, and XND-1 have all been implicated in regulating the timing and frequency of DSB induction (219).
Studies of specific hot spots in the mouse, comparative studies of recombination rates between mouse strains, and differences in recombination rates across human subpopulations all led to the identification of the methyltransferase PRDM9 (PR domain encoding protein 9) as a major determinant of recombination activity in human and mouse (10, 174). The role of PRDM9 in driving hot spot utilization in the mouse has been reviewed extensively elsewhere (11, 25, 128, 165).
Given that DSBs form in the absence of PRDM9 in mouse meiocytes, it is plausible that PRDM9 is not the sole determinant of hot spot activity in the mouse. Indeed, promoter regions that are enriched for H3K4me3 signals do not display high DSB rates (204). Moreover, whereas DSB induction at the pseudoautosomal region (PAR) of the mouse XY bivalent is associated with H3K4me3, this histone modification at this region is not PRDM9 dependent (31). Together, these observations suggest that non-PRDM9 mechanisms are involved in the recruitment of SPO11 complexes to DNA, a finding that is supported by recent observations of canine meiotic recombination, in which the Prdm9 allele is nonfunctional, and yet SPO11-invoked recombination events still occur during meiosis (8). In canids, the recombination landscape is dramatically different from that seen in other mammals, with a high accumulation of DSBs around promoter-associated CpG islands, reminiscent of the high correspondence of DSBs with promoter-associated H3K4me3 marks seen in Prdm9−/− mice (31) and vaguely reminiscent of the pattern of recombination reported for budding yeast (171).
Factors influencing double-strand break frequency: negative and positive regulation
Critical to the correct repair of meiotic DSBs is the action of checkpoint proteins. Checkpoints function to inhibit progression of the cell program until sufficient repair of the DNA damage event has occurred. In addition, checkpoints also function to recruit repair factors and help bias the repair toward the homologous chromosome—a key difference compared with mitotic DSB repair (reviewed in 141). Interestingly, two related checkpoint kinases also play a role in meiotic DSB formation. In fly and yeast, Tel1/ATM inhibits DSB formation (41, 107), whereas in yeast Mec1/ATR functions to promote DSB formation in situations in which DSB catalysis is defective (7, 73). In addition to modulating DSB frequency, these checkpoint proteins also function to define DSB distribution across chromosomes and chromatids (68, 238). Thus, these two kinases appear to function as homeostatic mechanisms, at least in yeast, ensuring the correct number and placement of DSBs (reviewed in 53).
Contradictory evidence in mouse suggests that the role of ATM is less clear. For example, the number of DSBs, as measured by SPO11-oligonucleotide complexes, is elevated tenfold in Atm mutant spermatocytes, whereas the meiotic defects observed in these mutant animals can be suppressed by heterozygosity of the Spo11 gene (9, 124). However, confounding these findings is the observation that mouse mutants that result in elevated ATM protein levels during prophase I do not apparently alter DSB frequency and distribution (156).
The mechanics of double-strand break formation and repair
Much of our biochemical knowledge of DSB formation and repair comes from organisms that we can easily genetically modify to observe or assay for repair intermediates (18, 39, 40, 59, 102, 111, 146, 166, 211, 234). However, studies in other organisms, such as mouse, have enabled us to confirm DSB formation and repair pathways while identifying critical differences in DSB processing across species (49, 51, 74).
Spo11 was identified by Scott Keeney and colleagues as the enzyme responsible for creating DSBs during meiotic prophase I in S. cerevisiae (111). Homologous proteins have since been discovered in plants (SPO11-1,2,3), worm (SPO-11), fly (mei-w68), and mouse (SPO11α and SPO11β), to name just a few (60, 78, 110, 150, 215). This topoisomerase II–like protein has similarities to the archaeal TopoVI A subunit. Archaeal TopoVI requires both A and B subunits to carry out its function, and recent investigations in plant and mouse have uncovered equivalent subunits that work with SPO11β during meiosis, named MTOPVIB and TOPVIBL, respectively (184, 225).
Two Spo11 monomers are required to induce a single DSB (Figure 2a), with each protein breaking one DNA strand (111). In the process of forming the DSB, Spo11 becomes covalently attached to the DNA and must then be removed for subsequent processing. In addition, Spo11 action requires additional proteins, most notably in S. cerevisiae, at least nine additional proteins are required: Ski8, Mer2, Mei4, Rec102, Rec104, Rec114, Mre11, Rad50, and Xrs2 (reviewed in 109). Of these, only Mei4 is known to be functionally conserved in mouse and A. thaliana (PRD2) (118). Plant, mouse, fly, and worm all require additional proteins not described in S. cerevisiae. A. thaliana requires the proteins DFO, PRD1, and SWI1 (153), mouse requires MEI1 (equivalent to A. thaliana PRD1) (127), worm requires DSB-1 (207), and fly requires Mei-P22, Trem, and Vilya (121, 133).
Initial double-strand break repair gives rise to structures that can be repaired via multiple distinct pathways
Following DSB induction in S. cerevisiae and S. pombe, the Mre11-Rad50-Xrs2/Nbs1 complex, in conjunction with the endonuclease Sae2/Ctp1, cleaves the DNA strand downstream from the Spo11 attachment, thereby liberating the Spo11-oligonucleotide complex (77, 166). Similar processing of Spo11-induced breaks is observed in C. elegans (138). Spo11 does not cleave the DNA at the same nucleotide position on each strand, resulting in a two-nucleotide overhang, thus preventing simple gap filling from the opposite strand (134). The resulting double-stranded DNA molecule now has a short single-stranded DNA (ssDNA) tail that is further extended by the 5′–3′ exonuclease Exo1 (69, 234). This extends the ssDNA tail by an average of 800 nt (in S. cerevisiae) and thereby facilitates the next step in the recombination process: strand invasion (234).
The ssDNA tail is initially coated by the replication protein A (RPA) (reviewed in 183), protecting the potentially fragile ssDNA molecule and impairing secondary structure formation. RPA is gradually replaced by the RecA family members Rad51 and Dmc1, the latter being a meiosis-specific protein (16, 197, 201). Recent results from the Bishop lab have indicated that in S. cerevisiae, both Rad51 and Dmc1 form filaments at both sides of the DSB when observed using super-resolution microscopy (35). This appears to be in contrast to the observations made in A. thaliana, where RAD51 and DMC1 localize to opposite sides of a DSB (119). In conjunction with chromatin remodeling factors (154), the RecA-DNA complexes can invade DNA templates in search of the correct homology in a process known as single-end invasion (SEI) (102). The ultimate result of this RecA-led ssDNA invasion of an opposing DNA helix is the displacement of a complementary DNA strand and the formation of a displacement loop (D-loop), representing the last intermediate stage that is common to all repair outcomes (215) (Figure 2).
Crossover/noncrossover designation
Following D-loop formation, DNA polymerases can undertake DNA synthesis, extending the single-stranded molecule using the invaded strand as a template. At this point, if the invading strand is disrupted and displaced, it can reanneal with the other side of the DSB. Following further DNA synthesis and ligation, the DSB is repaired. This repair pathway, named synthesis-dependent strand annealing (SDSA) (Figure 2b) (164), leads only to noncrossover events, with any region of potential heteroduplex DNA being resolved by the mismatch repair machinery (2, 151). These SDSA-derived noncrossovers are temporally distinct from crossovers in yeast and mouse, leading to the idea that the two DSB outcomes arise from mechanistically distinct events (3, 49). Importantly, however, SDSA represents the major, but not the sole, source of noncrossovers, at least in budding yeast (146).
If SDSA is not utilized, the extended D-loop becomes captured by the other side of the DSB in a process known as second-end capture, creating a joint molecule (JM) between two opposing chromatids (215). At this stage, structure-specific endonucleases can cleave the junctions, allowing for further elongation and ligation to create a crossover (Figure 2f) (reviewed in 196). If nucleases do not act on the JM, a double Holliday junction (dHJ) forms (Figure 2c), and these appear to be the predominant product of D-loop extension during meiosis (39, 93). dHJs can be repaired in two different ways, either by the resolution (Figure 2e) or dissolution (Figure 2d), the former resulting in either a noncrossover or crossover event. For reasons that are not altogether clear, but which may involve members of the DNA mismatch repair machinery or other repair proteins, dHJ repair in meiosis is heavily biased toward the formation of crossovers (3). Alternatively, dHJs can undergo dissolution through the action of helicases and topoisomerases to result in a noncrossover (reviewed in 17).
As described, these complex pathways, requiring different proteins and acting on different structures, are defined separately; however, multiple cross talk between these pathways exist. In addition, unlike DSBs arising within other environments, DSB repair during meiosis takes place within the confines of the proteinaceous structure of the SC. In this way, the SC serves to promote mechanisms of DSB repair that are fundamental to, and defining of, the meiotic process.
Synaptonemal Complex Formation
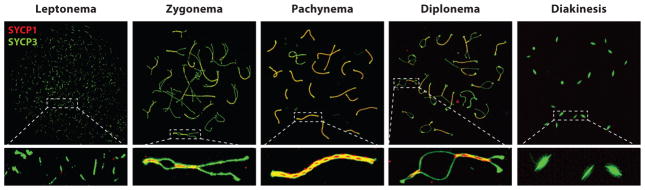
Following homolog recognition and initial pairing (alignment), homologous chromosomes then become tethered together in a process termed synapsis. The process of synapsis is a sequential event that culminates in the completion of a tripartite structure known as the SC, which maintains homologous chromosomes within 100 nm of each other (169). First identified independently by Fawcett (64) and Moses in 1956 (158), the SC defines the individual substages of prophase I according to the physical state of the SC (Figure 3). SC assembly begins in leptonema (from the Greek for thin threads) with the accumulation of the axial element (AE) along each chromosome. The AEs serve as docking sites for numerous proteins involved in cell cycle control and recombination, and they also interact with the cohesin rings that help to facilitate both sister chromatid cohesion and homologous chromosome interactions. During zygonema (paired threads), the two AEs, which are now known as lateral elements (LE), become closely apposed at ~400 nm, and become progressively connected via a structure known as the central region (CR), which comprises two transverse filaments (TFs) that intersect with the central element (CE) region. By pachynema (thick threads), the two LEs are now completely zippered together along their lengths by the CR, and the intact tripartite structure persists until diplonema when the SC begins to disassemble. Diakinesis is the final stage of prophase I, when the majority of the SC has broken down and chromosomes are now held together through their chiasmata, the physical manifestation of crossing over (reviewed by 66, 169).
Figure 3.
Synaptonemal complex (SC) formation and synapsis during prophase I in mouse. Meiotic chromosome spread preparations from male mouse spermatocytes are immunofluorescently stained and imaged at different stages of prophase I using 3D structured illumination microscopy (3D-SIM). During prophase I, axial elements [stained with antibodies against SYCP3 (green)] start to form along the homologous chromosomes during leptonema. In zygonema, axial elements have formed along the entire axis of the homologous chromosome, and as synapsis occurs proteins of the central/transverse filament [stained with antibodies against SYCP1 (red)] begin to zipper the homologous chromosomes together. In pachynema, full synapsis has occurred along all autosomes and within the pseudoautosomal region of the sex chromosomes (in males). During diplonema, the SC starts to disassemble and homologous chromosomes repel each other, except for at regions of crossovers. Finally, at diakinesis, the axial elements are visible only at the centromeres.
In most cases, SC formation is dependent on the initiation of recombination and is, in turn, essential for the processing of recombination events and for prophase I progression. However, some exceptions exist: For example, S. pombe and Aspergillus nidulans both undergo recombination but without an SC, whereas D. melanogaster females and C. elegans undergo pairing and synapsis independently of recombination (reviewed by 120, 138, 242). Moreover, in Drosophila males, alternative systems for homolog engagement have been established (149).
Components of the synaptonemal complex: organizational and functional conservation without sequence conservation
Although SC structural organization is conserved across many eukaryotic species, the constituent proteins that make up this complex structure vary markedly between organisms. Multiple AE/LE proteins are usually present, such as the mouse SC-2 and SC-3 proteins (SYCP2, SYCP3) (229) and the C. elegans four HORMA-domain (Hop1, Rev7, Mad2) proteins HIM-3, HTP-1, HTP-2, and HTP-3 (reviewed by 138). AE/LE components in several species are known to interact with cohesin components, such as Drosophila C(2)M (120, 143) and C. elegans HTP-3 (72, 145, 199), and these interactions are usually essential for normal SC formation.
TF proteins share very little sequence homology; however, their overall structures are similar, with N-terminal and C-terminal globular domains separated by a coiled-coil central domain (84). Within the SC structure, TF proteins form parallel coiled-coil homodimers with their globular C-terminal ends embedded in the LE and their N-terminal ends interacting with proteins of the CE (reviewed by 88). Many organisms appear to have only a single TF protein, exemplified by Zip1 of S. cerevisiae (213) and SYCP1 of mouse (61). By contrast, Drosophila appears to have two TF proteins: C(3)G and Corolla (52, 168).
The central element proteins vary greatly among SC-bearing organisms, with mouse having four CE proteins, SC central element proteins 1–3 (SYCE1, SYCE2, and SYCE3) (22, 23, 55, 76, 195), and TEX12 (76). Drosophila females have only a single known CE protein, Corona, which is also known as Cona (170). In S. cerevisiae, appropriate installation of the TF protein, Zip1, is dependent on the presence of the CE proteins Ecm11 and Gmc2 (100, 237) and, more specifically, requires the SUMOylation of Ecm11 in a Gmc2-dependent manner (224, 236). Indeed, SUMO appears to be an integral component of the SC in many organisms and is required for initial loading of Zip1 (reviewed by 242). As is discussed below, post-translational modification events such as this one appear to be central for events in prophase I.
The entire CR in C. elegans consists of four proteins, SYP-1–4 (48, 140). Studies by Schild-Prüfert et al. (194) have demonstrated a complex interaction between these four proteins, in which a homodimer of SYP-1 connects SYP-2 in the central core regions and SYP-3 is tethered to the AE. SYP-3 also interacts with SYP-4, which is thereby retained in the core of the CE. Thus, for worms at least, the CR structure is more complex and less defined into TF and CE subcompartments.
Initiation of synapsis
In budding yeast, Zip2, Zip3, and Zip4 are essential components of the synapsis initiation complex (SIC). These proteins initially arise at sites called axial associations (AAs), representing sites where opposing AEs become very closely aligned. The appearance of AAs are dependent on DSB formation, whereas proteins required for synapsis localize to these sites initially, indicating that AAs are the precursors to SICs (1, 47, 67, 175, 221). Both the number of AAs and the accumulation of Zip3 are dependent on and can be altered by varying the dose of Spo11, and they become loaded with RAD51 and DMC1, indicating that these structures represent the sites of DSB induction (86, 87, 185).
Prior to DSB induction, when the homologs are aligned and oppose each other at a distance of ~400 nm, Zip3 loading initiates at the centromeres independently of Spo11 activity but dependent on Zip1 preloading at this site (198, 220). Zip3 then associates with chromosome axes and recruits Zip2 to a subset of AAs, acting in a complex with Zip4 (Spo22) to convert the AA into an SIC at the leptotene-to-zygotene transition, bringing the homologs to within 100 nm of each other. Zip2 also interacts with key recombination proteins, including Rad51, Mre11, and the MutS homologs Msh4/5 (components of the ZMM pathway, see below) (1, 47, 221). SIC formation is concomitant with recruitment of Zip1 along the cores to form the TF of the central element.
Importantly, as is discussed below, Zip3 association with the chromosomes requires its SUMO E3 ligase activity as well as phosphorylation of Zip3 by Mec1/Tel1, and this leads to accumulation of the TF protein Zip1 (198). ChIP analysis demonstrates that further Zip3 recruitment is dependent on Zip1 accumulation and, specifically, on the stabilization of SUMO chains by Zip1. This leads to a second wave of Zip3 recruitment through the Zip3 SUMO binding motif, but this is confined to those DSB sites that have transitioned into dHJs (45, 198). A similar two-stage accumulation/specification is observed for the worm ortholog of Zip3, ZHP-3, although the worm protein does not appear to be critical for SC formation (15, 103). Thus, although SUMOylation may be critical for early synapsis in some organisms, it appears to be of more widespread importance for DSB induction, as well as for the conversion of a subset of DSBs into dHJs. This process of DSB selection to become a dHJ lies at the heart of crossover fate designation, and these data implicate proteins of the SC, as well as the DSB repair machinery, in the process of crossover designation.
CROSSOVER FATE DESIGNATION
Class I Crossovers and the ZMM Pathway
One of the major arguments in favor of temporally distinct crossover/noncrossover processing during meiotic prophase I is the emergence of a group of genes whose deletion or mutation results in a severe reduction in crossover, but not noncrossover, frequency (reviewed by 139). Termed the ZMM family for the yeast genes that originally defined the group, members include proteins involved in DNA metabolism, including Mer3 helicase, which extends the D-loop, and the meiosis-specific MutS homologs of the DNA mismatch repair (MMR) family, Msh4 and Msh5 (59, 94), which stabilize dHJs. The ZMM family also includes members of the Zip2, Zip3, and Zip4 families of SUMO and/or ubiquitin E3 ligases (1, 29, 221), and the SC protein Zip1 (139, 213), described above. Orthologs for most, if not all, of the ZMM proteins have been identified across eukaryotes. The majority of crossovers in yeast, some plants, and mice, and all crossovers in C. elegans, are processed through the ZMM-regulated crossover pathway, also known as the class I crossover pathway (74). Thus, the ZMM proteins are also classified as pro-crossover factors. However, recent studies in a variety of organisms have revealed many more pro-crossover factors involved in class I events than were originally defined by the ZMM classification, implying that the ZMM proteins represent only a minor fraction of those proteins that are specifically essential for class I crossovers.
In most organisms, the class I crossover pathway is characterized by the sequential accumulation of homologs of the bacterial MutS and MutL proteins. In the canonical eukaryotic MMR system, a heterodimer of MutS homologs binds to a specific DNA substrate and then recruits a heterodimer of MutL homologs as the signaling and effector components of the pathway (reviewed by 126). The MSH4/MSH5 heterodimer, known collectively as MutSγ, is first recruited to specific sites of ongoing DSB repair and is unique among the MutS heterodimers in that it does not itself engage in mismatch correction (20, 94, 172, 187, 192). Instead, in vitro studies of human MutSγ indicate that the complex forms a sliding clamp that encircles dHJ substrates and can translocate along the DNA in an ATP-dependent fashion, allowing the accumulation of further MutSγ clamps at that DSB repair site (205). Other studies have suggested that the preferred binding substrate for MutSγ is the SEI structure where the heterodimer promotes second-end capture (reviewed in 142).
In mouse, MSH4 and MSH5 associate with meiotic chromosome cores in zygonema, with the number of foci formed by these proteins matching only about half of the initiating DSBs (approximately 100–150 of the 250–300 DSBs) (115). Accordingly, loss of either gene in mouse results in meiotic disruption prior to the pachytene stage, with incomplete synapsis, persistence of RAD51, and a failure to complete DSB repair (115).
Like all MutS heterodimers, MutSγ recruits and associates with a heterodimer of MutL homologs that, in the case of meiotic DSB repair, involves the MutLγ heterodimer, which consists of MLH1 and MLH3 (reviewed by 142). In mouse and plants, MLH1 and MLH3 associate with chromosomes at pachytene of prophase I, somewhat later than the appearance of their DNA-binding partners, MSH4 and MSH5 (132). Importantly, the number of MutLγ foci is a subset of those observed for MutSγ foci, and these later foci exhibit interference in that the placement of one focus prevents the nearby localization of another focus (discussed below). The meiotic phenotype of mice lacking either Mlh1 or Mlh3 is manifested somewhat later than that of Msh4 or Msh5, although in all cases spermatocytes fail to complete prophase I. This temporal difference in phenotype is surprising given the fact that these proteins are presumed to act together, as is the case for other MutS and MutL heterodimer combinations, and points to additional roles for MutSγ in the selection of DSB repair pathways and/or stabilization of DSB repair intermediates. Indeed, MSH4 and MSH5 are thought to prevent dissolution of dHJ structures, and so it is conceivable that an early function of MutSγ is to promote dHJ stability at all JMs and not just those that are substrates of the class I machinery.
The precise role of MutLγ is unclear in the context of crossing over, but the presence of a functional endonuclease activity within yeast Mlh3 points to a role for this protein complex in resolution of dHJ intermediates (186). Thus, MutLγ is considered to be the all-important class I Holliday junction resolvase, capable of making the cuts in the dHJ that result in the formation of two intact recombinant DNA molecules. Evidence in support of this resolvase function has emerged in budding yeast, where MutLγ functions together with Exo1 and the helicase Sgs1 to resolve class crossovers (235). Importantly, although functional MutSγ orthologs exist in C. elegans (48), MutLγ proteins do not, suggesting that the resolvase activity of MutLγ must be fulfilled by other proteins in worms.
Although MLH1 and MLH3 are not traditionally defined as ZMM proteins, they function exclusively in the class I crossover pathway in the context of meiotic DSB repair, and, most importantly, these proteins are considered to be the final markers of class I crossover events in late prophase I in yeast, mouse, humans, and plants (125, 132). Conversely, in light of the early meiotic phenotype of mice harboring mutations in the MutSγ components, it is plausible that MSH4 and MSH5 may actually function outside of the class I pathways, at least in mammals, again suggesting that ZMM classification may not be appropriate for all (or limited to) class I–type crossover events.
Class II Crossovers and MUS81
Although the class I pathway accounts for the majority of crossovers, genetic evidence from yeast, plants, and mouse points to the presence of a minor class II crossover pathway, orchestrated by the MUS81-EME1 structure-specific endonuclease (Mus81-Mms4 in S. cerevisiae) (21). This pathway accounts for 5–10% of crossovers in the mouse (95), approximately 5% of crossovers in A. thaliana (89), and 15–35% of crossovers in budding yeast (59). Class II crossovers do not form in C. elegans, although worm MUS-81 is required for processing additional DSBs that arise from irradiation or from mutation of the helicase Rtel-1 (reviewed by 138).
In S. cerevisiae, Mus81 acts in concert with the RecQ helicase Sgs1 (BLM helicase in mammals) to promote the resolution of complex multichromatid JM intermediates that arise as a result of various events downstream of DSB resection, including secondary-strand invasion events (104). Moreover, formation of class II–regulated crossovers is not constrained by interference in the way that class I crossovers are controlled. Thus, organisms, such as S. pombe, in which the class I pathway is absent, do not exhibit crossover interference during meiosis (57), although it should be noted that S. pombe also does not have a classically defined SC structure (137). Moreover, in mammals at least, it appears that there are functional links between the two classes, given that loss of the class II crossover pathway in Mus81 homozygous mutant male mice results in a compensatory rise in the frequency of MutLγ-marked class I crossover events, without any overall increase in crossover frequency (95). This suggests that crossover numbers are maintained in the absence of class II events in a homeostatic mechanism that results in upregulation of class I events. Whether these additional class I crossovers represent those sites that would have been class II crossovers or whether additional class I crossovers are drawn from the pool of excess MutSγ sites that exist during zygonema remains to be seen.
Interactions Between Class I and Class II Crossovers Pathways
The increase in class I crossovers that occurs in mice lacking a functional class II pathway (Mus81 mutant mice) suggests that there must be some degree of interaction between class I and class II events. Supporting evidence of this comes from studies in multiple organisms and, in particular, implicates the RecQ helicases in this cross talk. The yeast RecQ helicase Sgs1 and its orthologs in mouse (BLM) and plants (RECQ4A) all appear to play vital roles in meiotic DSB repair processes (97). Loss of Sgs1 in yeast leads to a significant rise in complex JMs that have the potential to become substrates for Mus81-Mms4 action. In addition, the appearance of crossover and non-crossover events become temporally linked in Sgs1 mutants, suggesting that the normal SDSA pathway is suppressed by loss of Sgs1, whereas the class I crossover pathway also becomes suppressed (104). As a result, both crossovers and noncrossovers now arise out of repair pathways that involve other resolvases, including Mus81-Mms4 and, at least in yeast, two other resolvases more commonly associated with mitotic DNA repair: Slx1-Slx4 and Yen1 (reviewed by 75). Thus, taken together, these observations suggest that Sgs1 functions to regulate DSB repair by modifying the existing repair intermediate such that they can now become substrates for the appropriate repair pathway. Under normal circumstances, Sgs1 would facilitate distribution of DSB repair intermediates between the class I and class II pathways, as appropriate. In addition, Sgs1 is critical for the resolvase activity of MutLγ (together with Exo1) in class I crossovers (235).
In mammals, much less is known about the function of BLM in meiosis. Loss of Blm in at least a subset of spermatocytes results in impaired meiotic progression, which is associated with increased crossing over (97). This increased crossing over is not related to class I crossover events, however, because MLH1 appearance and frequency are normal in Blm mutant spermatocytes. Instead, this observation suggests that class II crossovers are increased, a possibility that has yet to be confirmed as a result of the lack of available antibodies against MUS81 or EME1. Similar to the situation in mice, C. elegans MUS-81 appears to function in conjunction with the worm ortholog of BLM, HIM-6, because mus-81 him-6 double mutants exhibit synthetic lethality (191).
SLX4 is a scaffold protein that interacts with multiple structure-specific endonucleases to orchestrate DNA repair in many organisms (reviewed by 113). Most prominently, SLX4 interacts with SLX1, MUS81, and XPF (65, 160, 212). During meiosis, SLX4 appears to play roles in crossover formation in Drosophila, C. elegans, and mouse but does not play any appreciable role in budding yeast (96, 191). In C. elegans, the SLX4 ortholog HIM-18 is responsible for a subset of Holliday junctions because him-18 mutant worms exhibit a 30–50% reduction in crossing over (191). This meiotic function of HIM-18 does not appear to involve interactions with SLX-1, because slx-1 mutant worms do not exhibit similarly reduced crossovers, although him-18 slx-1 double mutant worms show a slightly exacerbated crossover phenotype compared to him-18 mutation alone (190). Additionally, there is mounting evidence that SLX-1 functions separately from HIM-18 and works with other structure-specific endonucleases, including MUS-81 and XPF-1, to promote crossover formation in worms (reviewed by 188). Conversely, in Drosophila, the function of SLX4 in meiotic crossing over appears to require interaction with XPF (230).
In the mouse, SLX1 does not appear to function in meiotic prophase I (K.J. Grive, K. Campbell-Peterson, P.E. Cohen, unpublished data), whereas SLX4 appears to function to promote crossing over via the class II pathway (96). Interestingly, the loss of Slx4 in mice results in elevated MLH1 focus frequency at the pachytene stage of prophase I, indicating increased class I crossover utilization. However, chiasmata counts show normal levels of crossing over in Slx4 homozygous mutant animals, indicating that class II crossovers must be downregulated in these animals (96). SLX4 interacts with components of both class I and class II machineries in the mouse (189), making it well placed to orchestrate crossover decisions between the two crossover classes.
MECHANISMS GOVERNING CROSSOVERS
Given the requirement for crossover formation to ensure accurate chromosome segregation at MI, it is not surprising that the precise timing, frequency, and distribution of crossovers must adhere to strict rules and be tightly regulated. Three basic principles have been described for governing crossover distribution: (a) obligate crossover formation, whereby at least one crossover forms between homologous chromosomes (105); (b) crossover interference, whereby the presence of one crossover prevents the formation of another crossover forming nearby (159, 209); and (c) crossover homeostasis, whereby crossover levels are maintained when DSB formation is altered, presumably at the expense of noncrossovers (147).
The Obligate Crossover
The obligate crossover rule, also known as crossover assurance, ensures that at least one crossover occurs between any given homolog pair, thus ensuring accurate segregation of those chromosomes. The presence of at least one crossover per homologous pair has been observed in most organisms (33, 105, 106), although on rare occasions crossovers do not occur between homologs, leading to nonexchange chromosomes (reviewed in 228).
Crossover Interference
Coincident with the need for the obligate crossover is the process of crossover interference, which was first described in the early 1900s and has been observed in multiple organisms, including S. cerevisiae, C. elegans, D. melanogaster, A. thaliana, mouse, and humans (33, 54, 152, 159, 168, 181, 209, 214). Of the two classes of crossovers formed, only class I crossovers are subject to interference (14, 59, 95). Interestingly, however, in mice lacking a functional Mus81 allele and therefore deficient in the class II crossover pathway, the additional MLH1 foci/class I crossovers observed are not constrained by interference (95). This suggests that, at least in mouse, the loss of class II crossovers and/or the acquisition of additional class I crossovers occurs somewhat later than the signal to impose interference, because these newly defined crossovers are not constrained in the same way as the normal cohort of class I events.
Crossover Homeostasis
Crossover homeostasis was first described in S. cerevisiae and has since been described in S. pombe, C. elegans, maize, and mouse (50, 108, 147, 202, 231, 232). Crossover homeostasis, as initially defined, is the maintenance of crossover frequency even when precursor DSB numbers are decreased (147). There must also be a mechanism for establishing the upper limit of crossover formation, as not all DSBs are repaired as crossovers. This upper limit may be defined by crossover interference or another mechanism; either way, it contributes to the homeostatic mechanism. Given that the phenomena of the obligate crossover and crossover interference exist, these two mechanisms may already ensure a minimum, one per homolog pair, and maximum amount of crossovers form due to crossover interference. Thus, it is often difficult to distinguish crossover homeostasis as a separate mechanism.
In the original yeast experiments that defined crossover homeostasis (147), altered DSB numbers were achieved using hypomorphic mutants in Spo11 and were measured in a rad50S mutant background, a mutation that results in the accumulation of DSBs with persistent attachment of Spo11 protein (111). However, in other studies, where researchers instead utilized later stage mutations (such as Rad51 or Dmc1), the number of calculated DSBs on the same Spo11 hypomorph background was shown to be different (73). Moreover, additional data suggest that the rad50S mutants can themselves be limiting in DSB formation, possibly due to Rad50 being required for both DSB formation and subsequent Spo11 processing in S. cerevisiae (38). Given this, it is difficult to accurately determine the DSB levels in the Spo11 hypomorph mutants on the rad50S, or indeed any, background. These observations therefore question the extent to which crossover homeosta-sis can truly be determined, because true DSB levels are often difficult to measure. Despite these concerns over true DSB counts, given that yeast produces approximately 80 crossovers and has 16 chromosomes, there are more crossovers than can be governed by the obligate crossover, thereby supporting the idea of crossover homeostasis. Similar mutations in S. pombe Spo11 ortholog Rec12 also lead to a decrease in measured DSBs but a maintained crossover level (108).
Several questions remain regarding crossover homeostasis, not least of which is whether crossover homeostasis establishes a minimal threshold level or, instead, whether the homeostatic mechanism keeps crossovers levels within a particular ideal range? Thus, would an increase in DSBs be prevented from a concurrent increase in crossovers by the homeostatic mechanism? In mouse, such questions were tested in a study in which DSB numbers were altered by changing the copy number of the Spo11 gene (50). Accordingly, DSB levels (as determined by RAD51 and DMC1 foci) decreased or increased with respect to wild type, depending on Spo11 dosage, yet crossovers (determined by foci of the class I–specific crossover component MLH1) did not (50). Mouse produces fewer crossovers than S. cerevisiae, 26 in male mice (approximately 90% from class I and 10% from class II) compared with 80 in yeast (74, 95). Similar to yeast, in the experimental situation in which DSBs are decreased, the number of crossovers formed is still more than required for the obligate crossover on each chromosome (with 20 chromosomes in mouse), thereby supporting the concept of crossover homeostasis (50). Interestingly, when DSBs are increased by introducing additional copies of the Spo11 gene by BAC (bacterial artificial chromosome) transgenesis, the observed number of MLH1 foci is similarly unchanged relative to wild type, indicating that class I crossover numbers are maintained and supporting the concept of the homeostatic mechanism in the mouse (50). However, the caveat to these data is that final crossovers, or chiasmata, were not quantified, and thus class II crossover events were not accounted for, leaving open the question as to whether increased DSBs can affect class II crossovers and/or whether the class II pathway is regulated by crossover homeostasis. Given that class II events are not subject to interference, it is possible that these class II–mediated crossovers are not constrained by homeostatic mechanisms. Indeed, in other mouse mutants discussed below, increased crossover counts are observed with normal MLH1 focus frequency, attesting to the fact that class II events can be differentially and independently regulated.
In support of the idea that class II crossovers are differentially affected by altered DSB frequency compared to class I crossovers, as described earlier, recent investigations in mouse, fly, and yeast have identified that mutation of Tel1/ATM leads to an increase in DSB levels (41, 68, 107, 124, 238). At least for yeast, this increased DSB frequency results specifically in increased crossovers of the class II variety (4). Mutation of Atm in mouse causes an approximately tenfold increase in DSBs (as measured by SPO11-oligonucleotides) compared with wild type but also causes a prophase I arrest, such that crossover levels are unable to be measured because the cells die prior to diplonema (124). However, by reducing the number of DSBs in the Atm−/− mouse by SPO11 heterozygosity, DSBs (as measured by SPO11-oligonucleotides) remain 4.5–7.8-fold above wild-type levels because of the loss of ATM-mediated negative feedback; however, they bypass the meiotic arrest caused by loss of Atm (9, 124). Interestingly, in this Spo11+/− Atm−/− mouse, class I crossovers, as measured by MLH1 focus frequency, are increased above wild-type levels (9), suggesting that either the homeostatic mechanism or crossover interference is not maintained in the absence of ATM in mouse. However, in this scenario, it remains to be seen what effect this genotype has on class II crossovers. It is possible that the increased MLH1 foci, and hence the increase in flux through the class I pathway, is counter-balanced by a decrease in class II crossovers. Interestingly, a mouse mutant that exhibits a twofold increase in the ATM protein during meiosis fails to show altered DSB numbers or chiasmata counts compared to wild-type littermates (156). Taken together, these observations in mouse suggest that ATM may have different effects on DSB numbers and that the relationship between DSB numbers and final crossovers in the mouse may be more complex than previously assumed.
Crossover homeostasis has also been described in C. elegans, an organism with arguably the most stringent crossover control, which results in precisely six crossovers, one per homologous pair, with little variation (15, 103, 231). All crossovers formed in C. elegans are class I crossovers and show complete interference. This defined number of crossovers could be a consequence of crossover homeostasis or of requirements determined by both the obligate crossover and crossover interference. Interestingly, dpy-28 and rtel-1 mutants increase both the number of DSBs and the number of crossovers (219, 233). Whether the increase in crossovers in these mutants is due to a defect in either crossover interference or a homeostatic mechanism is unclear.
Across species, there are currently no examples in which DSB levels have increased and total crossovers have remained the same. In maize, for example, Sidhu et al. (202) analyzed strains with varying numbers of meiotic DSBs, as measured by RAD51 foci. Interestingly, they found that crossover numbers increased accordingly, suggesting that perhaps homeostasis mechanisms are different in plants. Whether these crossovers are class I or class II events is unknown. Given these observations, it appears that crossover homeostasis functions as a mechanism to establish crossovers at a particular level (the lower limit), but it remains unclear as to whether a homeostatic mechanism, other than crossover interference, exists to limit excess crossover numbers (the upper limit). Further experiments will provide clarification for crossover homeostasis, particularly as it pertains to class I and class II interactions, or may suggest that the phenomenon of a homeostatic capping mechanism is indistinguishable from crossover interference.
Crossover Selecting/Deselecting Factors
Crossover designation relates to the process of DSB selection to become crossovers. This process is not a one step process; it involves multiple proteins at intermediary stages. A key transition/designation step in this process is the paring down of high-number, early MutSγ to low-number, late MutLγ complexes.
In mouse, the overabundance of MutSγ sites in zygonema suggests that specific mechanisms must be in place to allow MutLγ to bind to only a subset of these sites (approximately 150 foci versus 22–24). Loss of Mlh3 in the mouse does not prevent the reduction in MutSγ focus frequency observed in late pachytene, suggesting that it is not simply loading of MutLγ that stabilizes some sites and allows loss of the extraneous sites (X. Sun & P.E. Cohen, unpublished work). Instead, it has been proposed that regulated loss of MutSγ may occur via post-translational modification of target proteins through ubiquitylation and SUMOylation, which lead to degradation and stabilization of target proteins, respectively (reviewed in 91). Therefore, identification of proteins that may target MutSγ for modification is important for gaining mechanistic insight into the early to late pro-crossover factors.
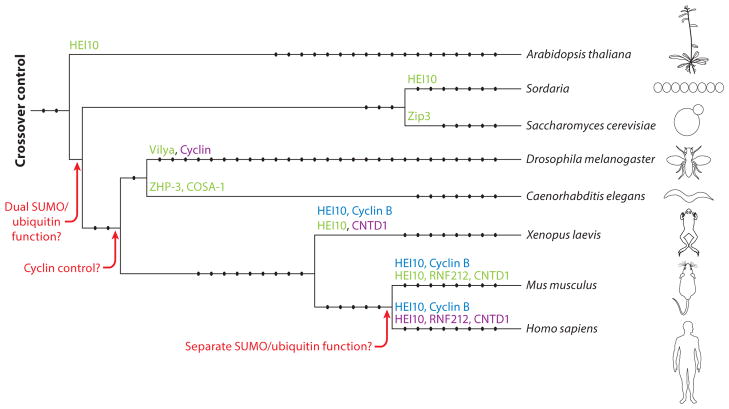
In yeast, computational analysis of the synapsis initiation protein Zip3 revealed an N-terminal RING-finger domain, likely allowing the protein to function as an E3 SUMO and/or ubiquitin ligase (175). Later studies further clarified the function of Zip3 to be that of a SUMO E3 ligase. In addition, these studies revealed the presence of a Smt3(SUMO) binding domain (SBD) on Zip3 (45). Although global ubiquitin levels analyzed by Western blot do not change significantly in zip3 mutants, it is possible that subtle changes in ubiquitin levels induced by Zip3 at the point of synapsis initiation are not readily detectable. Proteins with structural similarity exist within other organisms, namely ZHP-3 in worm (15), Vilya in fly (121), and RNF212 in mouse (182). Mutation of Zip3, ZHP-3, Vilya, or RNF212 in the respective organisms leads to defects in crossover formation, subsequently leading to a reduction in spore viability (yeast), a reduction in fertility (worm), high levels of nondisjunction (fly), and infertility (mouse) (1, 29, 103, 121, 182). In yeast, a Zip3 mutation causes defective SC formation (29); however, in worm and mouse, the equivalent mutation leads to normal SC formation, albeit potentially delayed (15, 103, 182). Interestingly, in worms, the phenotype observed with a separation-of-function allele of zhp-3 produces a similar phenotype to a smo-1 (SUMO) mutant with the defect exacerbated in double mutants (15), supporting the idea that ZHP-3 and this family of proteins function as SUMO E3 ligases. Similar localization of these four proteins within their respective organisms during prophase I highlights potential similarities and differences in their meiotic function.
In yeast, Zip3 colocalizes with another SC protein, Zip2, at synapsis initiation sites early in prophase and colocalizes and interacts with the MutSγ complex (1). Additional evidence focused on reducing DSBs while maintaining crossovers shows that Zip3 foci do not decrease with the same severity as DSB reduction, implicating a role of Zip3 in crossover formation (86). ZHP-3, Vilya, and RNF212 have many similarities in localization; initially forming foci/filaments at early prophase correlating with early synapsis sites, and observed as foci at late prophase correlating with crossover sites (15, 121, 182). Subtle differences exist: ZHP-3 appears to undergo relocalization forming nonuniform protein filaments along the SC, until at late prophase it forms discrete foci marking crossover events (15); Vilya is observed as both discrete foci and stretches at early pachytene (121); and RNF212 is excluded from unsynapsed regions (182). Such subtle differences may be changes in protein function but may also be due to the ability to detect or visualize proteins given the technologies at the time.
Interestingly in budding yeast, worm, and mouse, components of MutSγ interact with the SUMO E3 ligase orthologs Zip3, ZHP-3, and RNF212, respectively (1, 182, 231). Although neither MSH4 nor MSH5 has been demonstrated to be a SUMOylation target, other components of the MMR family are proven SUMO targets (24). Moreover, another MutS homolog family member, MSH2, has been shown to have a genetic interaction with the SUMO proteases Ulp2 and SENP6 in multiple yeast species and in humans, respectively (218). Thus, MutSγ may be a target of SUMO and/or may promote SUMO removal through its interaction with SUMO proteases.
In addition to the SUMO E3 ligases that have been described, there is a second set of RING-finger–domain family proteins that function in crossover formation. Human enhancer of invasion 10 (HEI10) has been identified as interacting with cyclin B in Xenopus, yeast ortholog Clb2, and the E2 ubiquitin conjugating protein UbcH7 (217). HEI10 shares amino acid similarity with other E3 ubiquitin ligases. During mitosis, HEI10 is thought to cause degradation of cyclin B in the G2/M transition and also inhibits migration and invasion of cancer cells (203, 217). Other HEI10 orthologs have been identified in rice, A. thaliana, Sordaria, and a type of scallop (42, 135, 222, 239). Characterization of the HEI10 mutant allele Hei10mei4/mei4 in mouse [not to be confused with the meiotic DSB protein MEI4 (118)] showed sterility in both males and females, along with a lack of localization of late-stage crossover class I–specific protein complex MutLγ and a severe decrease in chiasmata, with only residual class II chiasmata remaining (180, 208, 227). Mutation of both rice and Arabidopsis HEI10 causes a loss of class I chiasmata (42, 239). Localization of HEI10 in these three species correlates with a higher number of foci in early prophase down to a discrete number of foci correlating with class I, MutLγ-marked crossovers at late pachytene (42, 180, 227, 239). In addition, rice HEI10 also colocalizes with the ZMM protein MER3 helicase at early prophase and then forms filaments following synapsis (239). Given the defects in transition from early to late pro-crossover factors, and reduction in crossover of these E3 ligase mutants, this highlights the important role of SUMO/ubiquitin modification in crossover designation.
Recent characterization of worm COSA-1 (crossover associated-1) and its mouse ortholog, CNTD1 (cyclin N-terminal domain-containing-1), have demonstrated an essential and conserved role in crossover formation (98, 231). These proteins have sequence similarity with cyclin B but also predict structural differences from the canonical cyclin family (231). In C. elegans, COSA-1 functions to ensure crossover homeostasis when DSB levels are limiting and for the process of maturing DSBs to interhomolog crossovers (231). Both COSA-1 and CNTD1 mutants have meiotic defects that lead to a lack of crossovers, drastically reduced viability in worm, and infertility in mouse (98, 231). Specifically, cosa-1 mutants form normal DSBs and SC but fail to undergo SC remodeling consistent with crossover formation (231). In mouse, Cntd1 mutant males are defective in the transition from early pro-crossover factors MutSγ to the late pro-crossover MutLγ, leading to a prolonged persistence of MutSγ (98). In addition, RNF212 persists through pachynema, and HEI10 fails to load in Cntd1 mutant mice (98). Interestingly, the phenotype of Hei10 mutant mice phenocopies that of Cntd1 mutant mice almost identically, with persistent RNF212 and MutSγ foci. This, together with the observation that HEI10 contains a cyclin-interacting motif, suggests that CNTD1 is essential for recruiting and/or facilitating HEI10 activity or that CNTD1 is a ubiquitin substrate of HEI10.
EMERGING THEMES OF CROSSOVER CONTROL
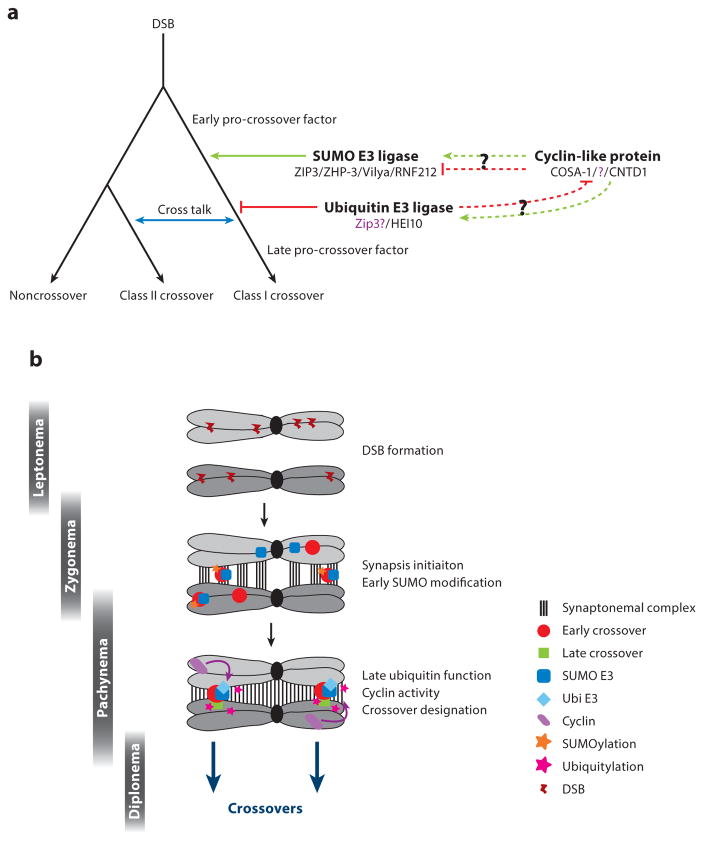
In the study of crossover formation during meiosis, various themes have been touched upon during this review and additional themes are emerging in the field. The following are areas of focus in which much has been learned in recent years and where ongoing studies are likely to yield important information. These themes are summarized in Figure 4.
Figure 4.
Model for crossover control. (a) The interaction between double-strand break (DSB) repair pathways is orchestrated by SUMO and ubiquitin E3 ligases and overseen by cyclin-like proteins such as COSA-1/CNTD1. DSBs can repair as either noncrossovers or crossovers, with cross talk occurring between the crossover classes (blue arrow). Three groups of proteins have been described as affecting crossover control: SUMO E3 ligases, ubiquitin E3 ligases, and cyclin-like proteins. All three proteins function in the process of paring down early crossover promoting factors to late crossover promoting factors. SUMO E3 ligases likely function to select/determine/designate some pro-crossover sites. Given that the mitotic role of HEI10 is inhibition of related cyclin-like proteins, it is likely that the ubiquitin E3 ligases inhibit cyclin-like proteins in meiosis and target some pro-crossover sites. It is unknown how the cyclin-like proteins function in relation to the SUMO E3 ligases. (b) Spatiotemporal relationship between SUMO and ubiquitin E3 ligases. The role of synapsis initiation sites and RING-finger proteins are depicted.
Crossover Classes
Studies have revealed at least two crossover pathways, each orchestrated by different repair machineries and each having unique regulatory networks governing their usage. In species in which both class I and class II crossovers exist, we have yet to fully determine the requirement/preferences for both the classes or for the interactions between pathways. Indeed, in organisms such as mouse, where a high percentage of crossovers form from the class I pathway, it will be interesting to learn what the function of the class II crossovers are. The Class I crossover mechanism generates enough crossovers to ensure the obligate crossover per homolog and crossover interference ensures that these crossovers are distributed across the genome. Could the class II crossovers be a mechanism established to increase genetic diversity? Given that they are not subject to interference, are they historical products of a preexisting crossover mechanism? Or are they simply repair pathways of last resort to deal with aberrant JMs that if left unrepaired would be toxic to the cell? In mouse, removal of proteins that function in class II crossover formation causes compensation by the class I crossover machinery (95). This compensation highlights the interesting cross talk that occurs between the repair pathways (Figure 4a). How exactly this cross talk between the two classes of crossovers is achieved remains to be understood.
MutSγ to MutLγ Designation
A key stage in crossover designation is the recruitment of MutLγ to MutSγ complexes—going from a high number of MutSγ to a refined number of MutLγ crossover designating foci. Two interesting points arise from this research: First, it is important to note that as certain DSB repair sites gradually lose MutSγ, these sites must be processed for repair via another set of proteins. Second, in mouse, Msh4/Msh5 mutant spermatocytes die in zygonema before recruitment of MLH1/3. Given this arrest, it may be that MutSγ plays a key role in cross talk or control of all crossovers. In addition, preliminary observations of MSH5 foci during prophase I have revealed a heterogeneous size population of MSH5 foci (J.K. Holloway & P.E. Cohen, unpublished results). These distinct populations of MSH5 foci may indicate different classes of DSB repair intermediates, one in which the amount of MutSγ is sufficient to drive class I crossover resolution and one in which a threshold of MutSγ requirement is not reached. Super-resolution microscopy technologies provide increased detail and quantitative information regarding foci. Future investigations may therefore provide mechanistic insight into this crossover designation stage and reveal differences in early to late designated sites and/or between different classes of MutSγ-marked sites.
Post-Translational Modifications
Investigations into crossover formation have previously focused on the phenotypic consequences of complete gene mutations. More recent investigations have instead focused on the effect of specific post-translational modifications of proteins of interest, as well as on the mechanisms of feedback control of crossover formation during meiosis (44, 198). Checkpoint kinases are already known to play a feedback role in the DSB formation (7, 41, 73, 107, 124), so it is unsurprising to learn that these play a similar role in crossover formation, as already observed in S. cerevisiae Tel1 mutants (4). Meiotic DSB formation is a complex regulated process with feedback mechanisms to coordinate appropriate levels of formation. Given the emergence of post-translational modifi-cations regulating crossover formation, it may be difficult to define a particular temporal/repair stage for crossover designation.
Recent studies have pointed to the idea that post-translational modification of key pro-crossover factors may regulate crossover designation/placement. In S. cerevisiae, for example, the ZMM protein Zip3 is phosphorylated by Cdc28 (CDK) and by the checkpoint kinases Mec1 and Tel1 (198). Although Cdc28 phosphorylation appears to be unimportant for crossover levels, mutation of the Mec1/Tel1 phosphorylation sites on Zip3 leads to decreased loading of Zip3 onto the chromosomes and subsequent reduction in crossover numbers (198). In addition to Zip3, Zip1 phosphorylation by the meiotic kinase Mek1 is important for the formation of class I crossovers (44). Interestingly, these phosphorylation events occur by action of kinases involved in checkpoint signaling and meiotic progression, thereby tying multiple meiotic events together in a coordinated fashion.
Currently, there is direct evidence only for kinase-driven phosphorylation events playing a role in crossover formation events. SUMOylation, however, has been shown to be important for multiple meiotic events across multiple species (reviewed in 136). Specifically related to crossover events, SUMO modification has currently been observed to be important for correct SC formation in S. cerevisiae, C. elegans, mouse, and human (15, 36, 45, 99, 100, 117, 131, 223, 224). With knowledge of SUMO-modifying and ubiquitin-modifying proteins functioning in crossover formation (such as Zip3/ZHP-3, HEI10, and RNF212) across multiple species (Figure 5), it will be interesting to investigate the direct targets of these RING-finger proteins and thereby to gain mechanistic understanding.
Figure 5.
Divergence of crossover control mechanisms. Phylogenetic tree of species divergence indicates known meiotic crossover designation proteins (green) with mitotic equivalents (if known; blue). Proteins in purple are predicted or currently unknown proteins. Predicted crossover control mechanisms and their divergences are marked in red.
Similarly, exciting avenues of investigation have been opened with the discovery of a cyclin-like protein in C. elegans and mouse (COSA-1 and CNTD1, respectively). Given the cyclin domains contained within these proteins, it is likely that they function with a CDK and may act as a regulatory mechanism during crossover designation. Understanding the CDK partner(s) and their subsequent targets may help to uncover further proteins involved in crossover formation and how the crossover designation pathway functions.
It is evident from species that have an SC that this structure plays an important role in crossover formation. Work on yeast Zip3 and fly Vilya proteins has revealed secondary roles for these proteins in DSB formation. Zip3 mutants, in particular, continue to form meiotic DSBs because of a lack of homolog engagement (216). These additional DSBs have been argued as being important in chromosome dynamics. Given the importance of crossover formation, however, and that crossover levels are decreased in Zip3 mutants, it will be interesting to determine whether the persistent DSBs forming are targeted to establishing crossovers.
Proposed Model for Crossover Designation
Across species there appear to be three different groupings of protein modifiers involved in crossover designation: ZIP3/ZHP-3/Vilya/RNF212 SUMO E3 ligases, HEI10 ubiquitin ligases, and the cyclin domain-containing proteins COSA-1/CNTD1. Prior to COSA-1/CNTD1 discovery, Qiao et al. (180) presented a model whereby the process of crossover designation occurs initially by action of a SUMO ligase (RNF212) recruitment to a number of early crossover sites, which are then targeted by a ubiquitin ligase (HEI10) working antagonistically against RNF212 to select crossover sites. There is little evidence to support this as yet, but elucidation of the substrates for both of these RING-finger proteins will help to support or refute this model in time. Given the action of HEI10 inhibiting cyclin B in mitotic cells (203), we hypothesize that in mouse, HEI10 inhibits CNTD1, which functions to restrain/inhibit RNF212 recruitment/function (Figure 4). Alternatively, HEI10 may be a target of CNTD1/CDK activity as part of a cell cycle network that seeks to tie crossover designation to cell cycle progression.
Meiotic SUMO and/or ubiquitin E3 ligases have been discovered in multiple organisms. Given the proposed function of these proteins, the different species may require mechanisms to stimulate, dampen, or modulate a particular Goldilocks zone of crossover levels: not too few and not too many. Analysis of the divergence of species in which these proteins are known has revealed that there are potential points at which crossover designation proteins gain or split function or become controlled by cyclin-like proteins (Figure 5). Given this, we hypothesize that both Xenopus and D. melanogaster may have a meiotic cyclin-like protein similar to that of COSA-1 and CNTD1. However, this hypothesis is based on the current proteins already known in these species and other discoveries may refute or clarify this.
The concept of a protein functioning as either a SUMO or ubiquitin E3 ligase is complicated by the fact that some proteins are known to function as both SUMO and ubiquitin E3 ligases, and distinguishing these enzymatic functions is often difficult (144, 167). Given the phenotypes of mouse mutants lacking the aforementioned RING and cyclin-like genes, it is evident that they play a role in the sequential paring-down process of MutSγ to MutLγ sites and/or the crossover designation process inherent within this paring-down process. How these proteins and this general mechanism function in regard to the class II crossovers and the cross talk between both crossovers and noncrossovers are of fascinating interest. It is likely that future discoveries will help define both intimate and overarching details of crossover control.
Acknowledgments
We are thankful to Monica Colaiácovo for helpful discussion and to Mathilde Grelon and Bernard de Massy for sharing unpublished information. We are also thankful to Miguel Brieño-Enriquez, Elizabeth Crate, Kathryn Grive, Carolyn Milano, Wojtek Pawlowski, John Schimenti, Melissa Toledo, and Adele Zhou for providing critical feedback for the manuscript. Imaging data (Figure 3) were acquired through the Cornell University Biotechnology Resource Center, with NSF 14,28922 funding for the shared Zeiss Elyra super-resolution microscope. Work in the Co-hen Lab is supported through funding from the NICHD (HD041012) and NIGMS (GM097263) to P.E.C.
Glossary
- SC
synaptonemal complex
- DSB
DNA double-strand break
- PCs
pairing centers
- rDNA
ribosomal DNA
- NOR
nucleolus organizing region
- CDK
cyclin-dependent kinase
- DDK
Dbf4-dependent kinase
- PAR
pseudoautosomal region
- SEI
single-end invasion
- D-loop
displacement loop
- SDSA
synthesis-dependent strand annealing
- JM
joint molecule
- dHJ
double Holliday junction
- AE
axial element
- CR
central region
- TF
transverse filament
- CE
central element
- SIC
synapsis initiation complex
- AA
axial association
- BAC
bacterial artificial chromosome
- RING
really interesting new gene
Footnotes
Errata
An online log of corrections to Annual Review of Genetics articles may be found at http://www.annualreviews.org/errata/genet
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Agarwal S, Roeder GS. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell. 2000;102(2):245–55. doi: 10.1016/s0092-8674(00)00029-5. [DOI] [PubMed] [Google Scholar]
- 2.Allers T, Lichten M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell. 2001;8(1):225–31. doi: 10.1016/s1097-2765(01)00280-5. [DOI] [PubMed] [Google Scholar]
- 3.Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001;106(1):47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- 4.Anderson CM, Oke A, Yam P, Zhuge T, Fung JC. Reduced crossover interference and increased zmm-independent recombination in the absence of Tel1/ATM. PLOS Genet. 2015;11(8):e1005478. doi: 10.1371/journal.pgen.1005478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson EL, Baltus AE, Roepers-Gajadien HL, Hassold TJ, de Rooij DG, et al. Stra8 and its inducer, retinoic acid, regulate meiotic initiation in both spermatogenesis and oogenesis in mice. PNAS. 2008;105(39):14976–80. doi: 10.1073/pnas.0807297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arabidopsis Genome Initiat. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408(6814):796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 7.Argunhan B, Farmer S, Leung WK, Terentyev Y, Humphryes N, et al. Direct and indirect control of the initiation of meiotic recombination by DNA damage checkpoint mechanisms in budding yeast. PLOS ONE. 2013;8(6):e65875. doi: 10.1371/journal.pone.0065875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Auton A, Rui Li Y, Kidd J, Oliveira K, Nadel J, et al. Genetic recombination is targeted towards gene promoter regions in dogs. PLOS Genet. 2013;9(12):e1003984. doi: 10.1371/journal.pgen.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barchi M, Roig I, Di Giacomo M, de Rooij DG, Keeney S, Jasin M. ATM promotes the obligate XY crossover and both crossover control and chromosome axis integrity on autosomes. PLOS Genet. 2008;4(5):e1000076. doi: 10.1371/journal.pgen.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, et al. Prdm9 is a major determinant of meiotic recombination hotspots in humans and mice. Science. 2010;327(5967):836–40. doi: 10.1126/science.1183439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baudat F, Imai Y, de Massy B. Meiotic recombination in mammals: localization and regulation. Nat Rev Genet. 2013;14(11):794–806. doi: 10.1038/nrg3573. [DOI] [PubMed] [Google Scholar]
- 12.Baudat F, Nicolas A. Clustering of meiotic double-strand breaks on yeast chromosome III. PNAS. 1997;94(10):5213–18. doi: 10.1073/pnas.94.10.5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bellvé AR, Cavicchia JC, Millette CF, O’Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol. 1977;74(1):68–85. doi: 10.1083/jcb.74.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berchowitz LE, Francis KE, Bey AL, Copenhaver GP. The role of ATMus81 in interference-insensitive crossovers in A. thaliana. PLOS Genet. 2007;3(8):e132. doi: 10.1371/journal.pgen.0030132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bhalla N, Wynne DJ, Jantsch V, Dernburg AF. Zhp-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLOS Genet. 2008;4(10):e1000235. doi: 10.1371/journal.pgen.1000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bishop DK, Park D, Xu L, Kleckner N. Dmc1: a meiosis-specific yeast homolog of E. coli RecA required for recombination, synaptonemal complex formation, and cell cycle progression. Cell. 1992;69(3):439–56. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- 17.Bizard AH, Hickson ID. The dissolution of double Holliday junctions. Cold Spring Harb Perspect Biol. 2014;6(7):a016477. doi: 10.1101/cshperspect.a016477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blat Y, Protacio RU, Hunter N, Kleckner N. Physical and functional interactions among basic chromosome organizational features govern early steps of meiotic chiasma formation. Cell. 2002;111(6):791–802. doi: 10.1016/s0092-8674(02)01167-4. [DOI] [PubMed] [Google Scholar]
- 19.Boateng KA, Bellani MA, Gregoretti IV, Pratto F, Camerini-Otero RD. Homologous pairing preceding spo11-mediated double-strand breaks in mice. Dev Cell. 2013;24(2):196–205. doi: 10.1016/j.devcel.2012.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocker T, Barusevicius A, Snowden T, Rasio D, Guerrette S, et al. Hmsh5: A human MutS homologue that forms a novel heterodimer with hMSH4 and is expressed during spermatogenesis. Cancer Res. 1999;59(4):816–22. [PubMed] [Google Scholar]
- 21.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107(4):537–48. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 22.Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R, et al. Syce2 is required for synap-tonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol. 2007;176(6):741–47. doi: 10.1083/jcb.200610027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bolcun-Filas E, Hall E, Speed R, Taggart M, Grey C, et al. Mutation of the mouse syce1 gene disrupts synapsis and suggests a link between synaptonemal complex structural components and DNA repair. PLOS Genet. 2009;5(2):e1000393. doi: 10.1371/journal.pgen.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bologna S, Altmannova V, Valtorta E, Koenig C, Liberali P, et al. Sumoylation regulates EXO1 stability and processing of DNA damage. Cell Cycle. 2015;14(15):2439–50. doi: 10.1080/15384101.2015.1060381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Borde V, de Massy B. Programmed induction of DNA double strand breaks during meiosis: setting up communication between DNA and the chromosome structure. Curr Opin Genet Dev. 2013;23(2):147–55. doi: 10.1016/j.gde.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 26.Borde V, Goldman AS, Lichten M. Direct coupling between meiotic DNA replication and recombination initiation. Science. 2000;290(5492):806–9. doi: 10.1126/science.290.5492.806. [DOI] [PubMed] [Google Scholar]
- 27.Börde V, Robine N, Lin W, Bonfils S, Géli V, Nicolas A. Histone H3 lysine 4 trimethylation marks meiotic recombination initiation sites. EMBO J. 2009;28(2):99–111. doi: 10.1038/emboj.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Börner GV, Cha RS. Induction and analysis of synchronous meiotic yeast cultures. Cold Spring Harb Protoc. 2015;2015(10):908–13. doi: 10.1101/pdb.prot085035. [DOI] [PubMed] [Google Scholar]
- 29.Börner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117(1):29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- 30.Bowles J, Koopman P. Retinoic acid, meiosis and germ cell fate in mammals. Development. 2007;134(19):3401–11. doi: 10.1242/dev.001107. [DOI] [PubMed] [Google Scholar]
- 31.Brick K, Smagulova F, Khil P, Camerini-Otero RD, Petukhova GV. Genetic recombination is directed away from functional genomic elements in mice. Nature. 2012;485(7400):642–45. doi: 10.1038/nature11089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Broman KW, Murray JC, Sheffield VC, White RL, Weber JL. Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet. 1998;63(3):861–69. doi: 10.1086/302011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Broman KW, Rowe LB, Churchill GA, Paigen K. Crossover interference in the mouse. Genetics. 2002;160(3):1123–31. doi: 10.1093/genetics/160.3.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooker AS, Berkowitz KM. The roles of cohesins in mitosis, meiosis, and human health and disease. Methods Mol Biol. 2014;1170:229–66. doi: 10.1007/978-1-4939-0888-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brown MS, Grubb J, Zhang A, Rust MJ, Bishop DK. Small Rad51 and Dmc1 complexes often co-occupy both ends of a meiotic DNA double strand break. PLOS Genet. 2015;11(12):e1005653. doi: 10.1371/journal.pgen.1005653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brown PW, Hwang K, Schlegel PN, Morris PL. Small ubiquitin-related modifier (SUMO)-1, SUMO-2/3 and sumoylation are involved with centromeric heterochromatin of chromosomes 9 and 1 and proteins of the synaptonemal complex during meiosis in men. Hum Reprod. 2008;23(12):2850–57. doi: 10.1093/humrep/den300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buard J, Barthés P, Grey C, de Massy B. Distinct histone modifications define initiation and repair of meiotic recombination in the mouse. EMBO J. 2009;28(17):2616–24. doi: 10.1038/emboj.2009.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Buhler C, Borde V, Lichten M. Mapping meiotic single-strand DNA reveals a new landscape of DNA double-strand breaks in Saccharomyces cerevisiae. PLOS Biol. 2007;5(12):e324. doi: 10.1371/journal.pbio.0050324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464(7290):937–41. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61(6):1089–101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- 41.Carballo JA, Panizza S, Serrentino ME, Johnson AL, Geymonat M, et al. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLOS Genet. 2013;9(6):e1003545. doi: 10.1371/journal.pgen.1003545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chelysheva L, Vezon D, Chambon A, Gendrot G, Pereira L, et al. The Arabidopsis HEI10 is a new zmm protein related to Zip3. PLOS Genet. 2012;8(7):e1002799. doi: 10.1371/journal.pgen.1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen SY, Tsubouchi T, Rockmill B, Sandler JS, Richards DR, et al. Global analysis of the meiotic crossover landscape. Dev Cell. 2008;15(3):401–15. doi: 10.1016/j.devcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen X, Suhandynata RT, Sandhu R, Rockmill B, Mohibullah N, et al. Phosphorylation of the synaptonemal complex protein Zip1 regulates the crossover/noncrossover decision during yeast meiosis. PLOS Biol. 2015;13(12):e1002329. doi: 10.1371/journal.pbio.1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cheng CH, Lo YH, Liang SS, Ti SC, Lin FM, et al. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006;20(15):2067–81. doi: 10.1101/gad.1430406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chikashige Y, Yamane M, Okamasa K, Mori C, Fukuta N, et al. Chromosomes rein back the spindle pole body during horsetail movement in fission yeast meiosis. Cell Struct Funct. 2014;39(2):93–100. doi: 10.1247/csf.14007. [DOI] [PubMed] [Google Scholar]
- 47.Chua PR, Roeder GS. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell. 1998;93(3):349–59. doi: 10.1016/s0092-8674(00)81164-2. [DOI] [PubMed] [Google Scholar]
- 48.Colaiácovo MP, MacQueen AJ, Martinez-Perez E, McDonald K, Adamo A, et al. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev Cell. 2003;5(3):463–74. doi: 10.1016/s1534-5807(03)00232-6. [DOI] [PubMed] [Google Scholar]
- 49.Cole F, Baudat F, Grey C, Keeney S, de Massy B, Jasin M. Mouse tetrad analysis provides insights into recombination mechanisms and hotspot evolutionary dynamics. Nat Genet. 2014;46(10):1072–80. doi: 10.1038/ng.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cole F, Kauppi L, Lange J, Roig I, Wang R, et al. Homeostatic control of recombination is implemented progressively in mouse meiosis. Nat Cell Biol. 2012;14(4):424–30. doi: 10.1038/ncb2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cole F, Keeney S, Jasin M. Comprehensive, fine-scale dissection of homologous recombination outcomes at a hot spot in mouse meiosis. Mol Cell. 2010;39(5):700–10. doi: 10.1016/j.molcel.2010.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Collins KA, Unruh JR, Slaughter BD, Yu Z, Lake CM, et al. Corolla is a novel protein that contributes to the architecture of the synaptonemal complex of Drosophila. Genetics. 2014;198(1):219–28. doi: 10.1534/genetics.114.165290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cooper TJ, Wardell K, Garcia V, Neale MJ. Homeostatic regulation of meiotic DSB formation by ATM/ATR. Exp Cell Res. 2014;329(1):124–31. doi: 10.1016/j.yexcr.2014.07.016. [DOI] [PubMed] [Google Scholar]
- 54.Copenhaver GP, Housworth EA, Stahl FW. Crossover interference in Arabidopsis. Genetics. 2002;160(4):1631–39. doi: 10.1093/genetics/160.4.1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Costa Y, Speed R, Ollinger R, Alsheimer M, Semple CA, et al. Two novel proteins recruited by synaptonemal complex protein 1 (SYCP1) are at the centre of meiosis. J Cell Sci. 2005;118(Pt. 12):2755–62. doi: 10.1242/jcs.02402. [DOI] [PubMed] [Google Scholar]
- 56.Cromie GA, Hyppa RW, Cam HP, Farah JA, Grewal SI, Smith GR. A discrete class of intergenic DNA dictates meiotic DNA break hotspots in fission yeast. PLOS Genet. 2007;3(8):e141. doi: 10.1371/journal.pgen.0030141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday junctions are intermediates of meiotic recombination. Cell. 2006;127(6):1167–78. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Da Ines O, White CI. Centromere associations in meiotic chromosome pairing. Annu Rev Genet. 2015;49:95–114. doi: 10.1146/annurev-genet-112414-055107. [DOI] [PubMed] [Google Scholar]
- 59.De los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164(1):81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dernburg AF, McDonald K, Moulder G, Barstead R, Dresser M, Villeneuve AM. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell. 1998;94(3):387–98. doi: 10.1016/s0092-8674(00)81481-6. [DOI] [PubMed] [Google Scholar]
- 61.De Vries FA, de Boer E, van den Bosch M, Baarends WM, Ooms M, et al. Mouse Sycp1 functions in synaptonemal complex assembly, meiotic recombination, and XY body formation. Genes Dev. 2005;19(11):1376–89. doi: 10.1101/gad.329705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ding DQ, Okamasa K, Yamane M, Tsutsumi C, Haraguchi T, et al. Meiosis-specific noncoding RNA mediates robust pairing of homologous chromosomes in meiosis. Science. 2012;336(6082):732–36. doi: 10.1126/science.1219518. [DOI] [PubMed] [Google Scholar]
- 63.Doll E, Molnar M, Hiraoka Y, Kohli J. Characterization of rec15, an early meiotic recombination gene in Schizosaccharomyces pombe. Curr Genet. 2005;48(5):323–33. doi: 10.1007/s00294-005-0024-3. [DOI] [PubMed] [Google Scholar]
- 64.Fawcett DW. The fine structure of chromosomes in the meiotic prophase of vertebrate spermatocytes. J Biophys Biochem Cytol. 1956;2(4):403–6. doi: 10.1083/jcb.2.4.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138(1):78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fraune J, Schramm S, Alsheimer M, Benavente R. The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp Cell Res. 2012;318(12):1340–46. doi: 10.1016/j.yexcr.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 67.Fung JC, Rockmill B, Odell M, Roeder GS. Imposition of crossover interference through the nonrandom distribution of synapsis initiation complexes. Cell. 2004;116(6):795–802. doi: 10.1016/s0092-8674(04)00249-1. [DOI] [PubMed] [Google Scholar]
- 68.Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature. 2015;520(7545):114–18. doi: 10.1038/nature13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garcia V, Phelps SE, Gray S, Neale MJ. Bidirectional resection of DNA double-strand breaks by Mre11 and Exo1. Nature. 2011;479(7372):241–44. doi: 10.1038/nature10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Garcia-Cruz R, Brieño MA, Roig I, Grossmann M, Velilla E, et al. Dynamics of cohesin proteins REC8, STAG3, SMC1β and SMC3 are consistent with a role in sister chromatid cohesion during meiosis in human oocytes. Hum Reprod. 2010;25(9):2316–27. doi: 10.1093/humrep/deq180. [DOI] [PubMed] [Google Scholar]
- 71.Gerton JL, DeRisi J, Shroff R, Lichten M, Brown PO, Petes TD. Global mapping of meiotic recombination hotspots and coldspots in the yeast Saccharomyces cerevisiae. PNAS. 2000;97(21):11383–90. doi: 10.1073/pnas.97.21.11383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Goodyer W, Kaitna S, Couteau F, Ward JD, Boulton SJ, Zetka M. HTP-3 links DSB formation with homolog pairing and crossing over during C. elegans meiosis. Dev Cell. 2008;14(2):263–74. doi: 10.1016/j.devcel.2007.11.016. [DOI] [PubMed] [Google Scholar]
- 73.Gray S, Allison RM, Garcia V, Goldman AS, Neale MJ. Positive regulation of meiotic DNA double-strand break formation by activation of the DNA damage checkpoint kinase Mec1(ATR) Open Biol. 2013;3(7):130019. doi: 10.1098/rsob.130019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guillon H, Baudat F, Grey C, Liskay RM, de Massy B. Crossover and noncrossover pathways in mouse meiosis. Mol Cell. 2005;20(4):563–73. doi: 10.1016/j.molcel.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 75.Haber JE. Topping off meiosis. Mol Cell. 2015;57(4):577–81. doi: 10.1016/j.molcel.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hamer G, Gell K, Kouznetsova A, Novak I, Benavente R, Höög C. Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J Cell Sci. 2006;119(Pt. 19):4025–32. doi: 10.1242/jcs.03182. [DOI] [PubMed] [Google Scholar]
- 77.Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM. Ctp1CtiP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009;29(7):1671–81. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hartung F, Puchta H. Molecular characterisation of two paralogous SPO11 homologues in Ara-bidopsis thaliana. Nucleic Acids Res. 2000;28(7):1548–54. doi: 10.1093/nar/28.7.1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hassold T, Abruzzo M, Adkins K, Griffin D, Merrill M, et al. Human aneuploidy: incidence, origin, and etiology. Environ Mol Mutagen. 1996;28:167–75. doi: 10.1002/(SICI)1098-2280(1996)28:3<167::AID-EM2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 80.Hassold T, Hall H, Hunt P. The origin of human aneuploidy: Where we have been, where we are going. Hum Mol Genet. 2007;16(Spec No 2):R203–8. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- 81.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–91. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
- 82.Hassold T, Sherman S. Down syndrome: genetic recombination and the origin of the extra chromosome 21. Clin Genet. 2000;57(2):95–100. doi: 10.1034/j.1399-0004.2000.570201.x. [DOI] [PubMed] [Google Scholar]
- 83.Hawley RS. Meiosis: how male flies do meiosis. Curr Biol. 2002;12(19):R660–62. doi: 10.1016/s0960-9822(02)01161-2. [DOI] [PubMed] [Google Scholar]
- 84.Hawley RS. Solving a meiotic lego puzzle: transverse filaments and the assembly of the synaptone-mal complex in Caenorhabditis elegans. Genetics. 2011;189(2):405–9. doi: 10.1534/genetics.111.134197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125(7):1321–32. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Henderson KA, Keeney S. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. PNAS. 2004;101(13):4519–24. doi: 10.1073/pnas.0400843101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Henderson KA, Keeney S. Synaptonemal complex formation: Where does it start? BioEssays. 2005;27(10):995–98. doi: 10.1002/bies.20310. [DOI] [PubMed] [Google Scholar]
- 88.Heyting C. Meiotic transverse filament proteins: essential for crossing over. Transgenic Res. 2005;14(5):547–50. doi: 10.1007/s11248-005-8925-y. [DOI] [PubMed] [Google Scholar]
- 89.Higgins JD, Buckling EF, Franklin FC, Jones GH. Expression and functional analysis of ATMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 2008;54(1):152–62. doi: 10.1111/j.1365-313X.2008.03403.x. [DOI] [PubMed] [Google Scholar]
- 90.Higgins JD, Perry RM, Barakate A, Ramsay L, Waugh R, et al. Spatiotemporal asymmetry of the meiotic program underlies the predominantly distal distribution of meiotic crossovers in barley. Plant Cell. 2012;24(10):4096–109. doi: 10.1105/tpc.112.102483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hilgarth RS, Murphy LA, Skaggs HS, Wilkerson DC, Xing H, Sarge KD. Regulation and function of SUMO modification. J Biol Chem. 2004;279(52):53899–902. doi: 10.1074/jbc.R400021200. [DOI] [PubMed] [Google Scholar]
- 92.Hiraoka Y, Dernburg AF. The sun rises on meiotic chromosome dynamics. Dev Cell. 2009;17(5):598–605. doi: 10.1016/j.devcel.2009.10.014. [DOI] [PubMed] [Google Scholar]
- 93.Holliday R. A mechanism for gene conversion in fungi. Genet Res. 1964;5(2):282. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 94.Hollingsworth NM, Ponte L, Halsey C. MSH5, a novel MutS homolog, facilitates meiotic reciprocal recombination between homologs in Saccharomyces cerevisiae but not mismatch repair. Genes Dev. 1995;9(14):1728–39. doi: 10.1101/gad.9.14.1728. [DOI] [PubMed] [Google Scholar]
- 95.Holloway JK, Booth J, Edelmann W, McGowan CH, Cohen PE. MUS81 generates a subset of MLH1-MLH3-independent crossovers in mammalian meiosis. PLOS Genet. 2008;4(9):e1000186. doi: 10.1371/journal.pgen.1000186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Holloway JK, Mohan S, Balmus G, Sun X, Modzelewski A, et al. Mammalian BTBD12 (SLX4) protects against genomic instability during mammalian spermatogenesis. PLOS Genet. 2011;7(6):e1002094. doi: 10.1371/journal.pgen.1002094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Holloway JK, Morelli MA, Borst PL, Cohen PE. Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination. J Cell Biol. 2010;188(6):779–89. doi: 10.1083/jcb.200909048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Holloway JK, Sun X, Yokoo R, Villeneuve AM, Cohen PE. Mammalian CNTD1 is critical for meiotic crossover maturation and deselection of excess precrossover sites. J Cell Biol. 2014;205(5):633–41. doi: 10.1083/jcb.201401122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hooker GW, Roeder GS. A role for SUMO in meiotic chromosome synapsis. Curr Biol. 2006;16(12):1238–43. doi: 10.1016/j.cub.2006.04.045. [DOI] [PubMed] [Google Scholar]
- 100.Humphryes N, Leung WK, Argunhan B, Terentyev Y, Dvorackova M, Tsubouchi H. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLOS Genet. 2013;9(1):e1003194. doi: 10.1371/journal.pgen.1003194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hunter N. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol. 2015;7(12):a016618. doi: 10.1101/cshperspect.a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell. 2001;106(1):59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- 103.Jantsch V, Pasierbek P, Mueller MM, Schweizer D, Jantsch M, Loidl J. Targeted gene knockout reveals a role in meiotic recombination for ZHP-3, a Zip3-related protein in Caenorhabditis elegans. Mol Cell Biol. 2004;24(18):7998–8006. doi: 10.1128/MCB.24.18.7998-8006.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008;31(3):313–23. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jones GH. The control of chiasma distribution. Symp Soc Exp Biol. 1984;38:293–320. [PubMed] [Google Scholar]
- 106.Jones GH, Franklin FC. Meiotic crossing-over: obligation and interference. Cell. 2006;126(2):246–48. doi: 10.1016/j.cell.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 107.Joyce EF, Pedersen M, Tiong S, White-Brown SK, Paul A, et al. Drosophila ATM and ATR have distinct activities in the regulation of meiotic DNA damage and repair. J Cell Biol. 2011;195(3):359–67. doi: 10.1083/jcb.201104121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kan F, Davidson MK, Wahls WP. Meiotic recombination protein Rec12: functional conservation, crossover homeostasis and early crossover/non-crossover decision. Nucleic Acids Res. 2011;39(4):1460–72. doi: 10.1093/nar/gkq993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Keeney S. Mechanism and control of meiotic recombination initiation. Curr Top Dev Biol. 2001;52:1–53. doi: 10.1016/s0070-2153(01)52008-6. [DOI] [PubMed] [Google Scholar]
- 110.Keeney S, Baudat F, Angeles M, Zhou ZH, Copeland NG, et al. A mouse homolog of the Saccha-romyces cerevisiae meiotic recombination DNA transesterase Spo11p. Genomics. 1999;61(2):170–82. doi: 10.1006/geno.1999.5956. [DOI] [PubMed] [Google Scholar]
- 111.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88(3):375–84. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 112.Khil PP, Smagulova F, Brick KM, Camerini-Otero RD, Petukhova GV. Sensitive mapping of recombination hotspots using sequencing-based detection of ssDNA. Genome Res. 2012;22(5):957–65. doi: 10.1101/gr.130583.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Kim Y. Nuclease delivery: versatile functions of SLX4/FANCP in genome maintenance. Mol Cells. 2014;37(8):569–74. doi: 10.14348/molcells.2014.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Klutstein M, Cooper JP. The chromosomal courtship dance-homolog pairing in early meiosis. Curr Opin Cell Biol. 2014;26:123–31. doi: 10.1016/j.ceb.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kneitz B, Cohen PE, Avdievich E, Zhu L, Kane MF, et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14(9):1085–97. [PMC free article] [PubMed] [Google Scholar]
- 116.Koubova J, Menke DB, Zhou Q, Capel B, Griswold MD, Page DC. Retinoic acid regulates sex-specific timing of meiotic initiation in mice. PNAS. 2006;103(8):2474–79. doi: 10.1073/pnas.0510813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kovalenko OV, Plug AW, Haaf T, Gonda DK, Ashley T, et al. Mammalian ubiquitin-conjugating enzyme Ubc9 interacts with Rad51 recombination protein and localizes in synaptonemal complexes. PNAS. 1996;93(7):2958–63. doi: 10.1073/pnas.93.7.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kumar R, Bourbon HM, de Massy B. Functional conservation of Mei4 for meiotic DNA double-strand break formation from yeasts to mice. Genes Dev. 2010;24(12):1266–80. doi: 10.1101/gad.571710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kurzbauer MT, Uanschou C, Chen D, Schlögelhofer P. The recombinases DMC1 and RAD51 are functionally and spatially separated during meiosis in Arabidopsis. Plant Cell. 2012;24(5):2058–70. doi: 10.1105/tpc.112.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Lake CM, Hawley RS. The molecular control of meiotic chromosomal behavior: events in early meiotic prophase in Drosophila oocytes. Annu Rev Physiol. 2012;74:425–51. doi: 10.1146/annurev-physiol-020911-153342. [DOI] [PubMed] [Google Scholar]
- 121.Lake CM, Nielsen RJ, Guo F, Unruh JR, Slaughter BD, Hawley RS. Vilya, a component of the recombination nodule, is required for meiotic double-strand break formation in Drosophila. eLife. 2015;4:e08287. doi: 10.7554/eLife.08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Lam DM, Furrer R, Bruce WR. The separation, physical characterization, and differentiation kinetics of spermatogonial cells of the mouse. PNAS. 1970;65(1):192–99. doi: 10.1073/pnas.65.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lam I, Keeney S. Mechanism and regulation of meiotic recombination initiation. Cold Spring Harb Perspect Biol. 2015;7(1):a016634. doi: 10.1101/cshperspect.a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lange J, Pan J, Cole F, Thelen MP, Jasin M, Keeney S. ATM controls meiotic double-strand-break formation. Nature. 2011;479(7372):237–40. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lenzi ML, Smith J, Snowden T, Kim M, Fishel R, et al. Extreme heterogeneity in the molecular events leading to the establishment of chiasmata during meiosis I in human oocytes. Am J Hum Genet. 2005;76(1):112–27. doi: 10.1086/427268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18(1):85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 127.Libby BJ, Reinholdt LG, Schimenti JC. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. PNAS. 2003;100(26):15706–11. doi: 10.1073/pnas.2432067100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lichten M. Molecular biology. Putting the breaks on meiosis. Science. 2015;350(6263):913. doi: 10.1126/science.aad5404. [DOI] [PubMed] [Google Scholar]
- 129.Lichten M, de Massy B. The impressionistic landscape of meiotic recombination. Cell. 2011;147(2):267–70. doi: 10.1016/j.cell.2011.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Liebe B, Alsheimer M, Höög C, Benavente R, Scherthan H. Telomere attachment, meiotic chromosome condensation, pairing, and bouquet stage duration are modified in spermatocytes lacking axial elements. Mol Biol Cell. 2004;15(2):827–37. doi: 10.1091/mbc.E03-07-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Lin FM, Lai YJ, Shen HJ, Cheng YH, Wang TF. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 2010;29(3):586–96. doi: 10.1038/emboj.2009.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Lipkin SM, Moens PB, Wang V, Lenzi M, Shanmugarajah D, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31(4):385–90. doi: 10.1038/ng931. [DOI] [PubMed] [Google Scholar]
- 133.Liu H, Jang JK, Kato N, McKim KS. mei-P22 encodes a chromosome-associated protein required for the initiation of meiotic recombination in Drosophila melanogaster. Genetics. 2002;162(1):245–58. doi: 10.1093/genetics/162.1.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu J, Wu TC, Lichten M. The location and structure of double-strand DNA breaks induced during yeast meiosis: evidence for a covalently linked DNA-protein intermediate. EMBO J. 1995;14(18):4599–4608. doi: 10.1002/j.1460-2075.1995.tb00139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Llera-Herrera R, García-Gasca A, Abreu-Goodger C, Huvet A, Ibarra AM. Identification of male gametogenesis expressed genes from the scallop Nodipecten subnodosus by suppressive subtraction hybridization and pyrosequencing. PLOS ONE. 2013;8(9):e73176. doi: 10.1371/journal.pone.0073176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lomelí H, Vázquez M. Emerging roles of the SUMO pathway in development. Cell Mol Life Sci. 2011;68(24):4045–64. doi: 10.1007/s00018-011-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lorenz A, Wells JL, Pryce DW, Novatchkova M, Eisenhaber F, et al. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J Cell Sci. 2004;117(Pt. 15):3343–51. doi: 10.1242/jcs.01203. [DOI] [PubMed] [Google Scholar]
- 138.Lui DY, Colaiácovo MP. Meiotic development in Caenorhabditis elegans. Adv Exp Med Biol. 2013;757:133–70. doi: 10.1007/978-1-4614-4015-4_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Lynn A, Soucek R, Börner GV. ZMM proteins during meiosis: crossover artists at work. Chromo-some Res. 2007;15(5):591–605. doi: 10.1007/s10577-007-1150-1. [DOI] [PubMed] [Google Scholar]
- 140.MacQueen AJ, Colaiácovo MP, McDonald K, Villeneuve AM. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes Dev. 2002;16(18):2428–42. doi: 10.1101/gad.1011602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.MacQueen AJ, Hochwagen A. Checkpoint mechanisms: the puppet masters of meiotic prophase. Trends Cell Biol. 2011;21(7):393–400. doi: 10.1016/j.tcb.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 142.Manhart CM, Alani E. Roles for mismatch repair family proteins in promoting meiotic crossing over. DNA Repair. 2016;38:84–93. doi: 10.1016/j.dnarep.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Manheim EA, McKim KS. The synaptonemal complex component C(2)M regulates meiotic crossing over in Drosophila. Curr Biol. 2003;13(4):276–85. doi: 10.1016/s0960-9822(03)00050-2. [DOI] [PubMed] [Google Scholar]
- 144.Marshall H, Bhaumik M, Aviv H, Moore D, Yao M, et al. Deficiency of the dual ubiquitin/SUMO ligase topors results in genetic instability and an increased rate of malignancy in mice. BMC Mol Biol. 2010;11:31. doi: 10.1186/1471-2199-11-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Martinez-Perez E, Villeneuve AM. Htp-1-dependent constraints coordinate homolog pairing and synapsis and promote chiasma formation during C. elegans meiosis. Genes Dev. 2005;19(22):2727–43. doi: 10.1101/gad.1338505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Martini E, Borde V, Legendre M, Audic S, Regnault B, et al. Genome-wide analysis of heterodu-plex DNA in mismatch repair–deficient yeast cells reveals novel properties of meiotic recombination pathways. PLOS Genet. 2011;7(9):e1002305. doi: 10.1371/journal.pgen.1002305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Martini E, Diaz RL, Hunter N, Keeney S. Crossover homeostasis in yeast meiosis. Cell. 2006;126(2):285–95. doi: 10.1016/j.cell.2006.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mays-Hoopes LL, Bolen J, Riggs AD, Singer-Sam J. Preparation of spermatogonia, spermatocytes, and round spermatids for analysis of gene expression using fluorescence-activated cell sorting. Biol Reprod. 1995;53(5):1003–11. doi: 10.1095/biolreprod53.5.1003. [DOI] [PubMed] [Google Scholar]
- 149.McKee BD, Yan R, Tsai JH. Meiosis in male Drosophila. Spermatogenesis. 2012;2(3):167–84. doi: 10.4161/spmg.21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McKim KS, Hayashi-Hagihara A. mei-W68 in Drosophila melanogaster encodes a Spo11 homolog: evidence that the mechanism for initiating meiotic recombination is conserved. Genes Dev. 1998;12(18):2932–42. doi: 10.1101/gad.12.18.2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLOS Biol. 2007;5(11):e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Meneely PM, Farago AF, Kauffman TM. Crossover distribution and high interference for both the X chromosome and an autosome during oogenesis and spermatogenesis in Caenorhabditis elegans. Genetics. 2002;162(3):1169–77. doi: 10.1093/genetics/162.3.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Mercier R, Armstrong SJ, Horlow C, Jackson NP, Makaroff CA, et al. The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development. 2003;130(14):3309–18. doi: 10.1242/dev.00550. [DOI] [PubMed] [Google Scholar]
- 154.Mets DG, Meyer BJ. Condensins regulate meiotic DNA break distribution, thus crossover frequency, by controlling chromosome structure. Cell. 2009;139(1):73–86. doi: 10.1016/j.cell.2009.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Miyoshi T, Ito M, Kugou K, Yamada S, Furuichi M, et al. A central coupler for recombination initiation linking chromosome architecture to S phase checkpoint. Mol Cell. 2012;47(5):722–33. doi: 10.1016/j.molcel.2012.06.023. [DOI] [PubMed] [Google Scholar]
- 156.Modzelewski AJ, Hilz S, Crate EA, Schweidenback CT, Fogarty EA, et al. Dgcr8 and Dicer are essential for sex chromosome integrity during meiosis in males. J Cell Sci. 2015;128(12):2314–27. doi: 10.1242/jcs.167148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Morelli MA, Cohen PE. Not all germ cells are created equal: aspects of sexual dimorphism in mammalian meiosis. Reproduction. 2005;130(6):761–81. doi: 10.1530/rep.1.00865. [DOI] [PubMed] [Google Scholar]
- 158.Moses MJ. Chromosomal structures in crayfish spermatocytes. J Biophys Biochem Cytol. 1956;2(2):215–18. doi: 10.1083/jcb.2.2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Muller H. The mechanism of crossing-over. Am Nat. 1916;50(592):193–221. [Google Scholar]
- 160.Muñoz IM, Hain K, Déclais AC, Gardiner M, Toh GW, et al. Coordination of structure-specific nucleases by human SLX4/BTBD12 is required for DNA repair. Mol Cell. 2009;35(1):116–27. doi: 10.1016/j.molcel.2009.06.020. [DOI] [PubMed] [Google Scholar]
- 161.Murakami H, Keeney S. Temporospatial coordination of meiotic DNA replication and recombination via DDK recruitment to replisomes. Cell. 2014;158(4):861–73. doi: 10.1016/j.cell.2014.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Myers S, Freeman C, Auton A, Donnelly P, McVean G. A common sequence motif associated with recombination hot spots and genome instability in humans. Nat Genet. 2008;40:1124–29. doi: 10.1038/ng.213. [DOI] [PubMed] [Google Scholar]
- 163.Myers S, Spencer CC, Auton A, Bottolo L, Freeman C, et al. The distribution and causes of meiotic recombination in the human genome. Biochem Soc Trans. 2006;34(Pt. 4):526–30. doi: 10.1042/BST0340526. [DOI] [PubMed] [Google Scholar]
- 164.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14(3):1613–25. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Neale MJ. PRDM9 points the zinc finger at meiotic recombination hotspots. Genome Biol. 2010;11(2):104. doi: 10.1186/gb-2010-11-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436(7053):1053–57. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Oh Y, Chung KC. UHRF2, a ubiquitin E3 ligase, acts as a small ubiquitin-like modifier E3 ligase for zinc finger protein 131. J Biol Chem. 2013;288(13):9102–11. doi: 10.1074/jbc.M112.438234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Page SL, Hawley RS. c(3)G encodes a Drosophila synaptonemal complex protein. Genes Dev. 2001;15(23):3130–43. doi: 10.1101/gad.935001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Page SL, Hawley RS. The genetics and molecular biology of the synaptonemal complex. Annu Rev Cell Dev Biol. 2004;20:525–58. doi: 10.1146/annurev.cellbio.19.111301.155141. [DOI] [PubMed] [Google Scholar]
- 170.Page SL, Khetani RS, Lake CM, Nielsen RJ, Jeffress JK, et al. Corona is required for higher-order assembly of transverse filaments into full-length synaptonemal complex in Drosophila oocytes. PLOS Genet. 2008;4(9):e1000194. doi: 10.1371/journal.pgen.1000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Pan J, Sasaki M, Kniewel R, Murakami H, Blitzblau HG, et al. A hierarchical combination of factors shapes the genome-wide topography of yeast meiotic recombination initiation. Cell. 2011;144(5):719–31. doi: 10.1016/j.cell.2011.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Paquis-Flucklinger V, Santucci-Darmanin S, Paul R, Sauniéres A, Turc-Carel C, Desnuelle C. Cloning and expression analysis of a meiosis-specific MutS homolog: the human MSH4 gene. Genomics. 1997;44(2):188–94. doi: 10.1006/geno.1997.4857. [DOI] [PubMed] [Google Scholar]
- 173.Parker SE, Mai CT, Canfield MA, Rickard R, Wang Y, et al. Updated national birth prevalence estimates for selected birth defects in the United States, 2004–2006. Birth Defects Res Part A Clin Mol Teratol. 2010;88(12):1008–16. doi: 10.1002/bdra.20735. [DOI] [PubMed] [Google Scholar]
- 174.Parvanov ED, Petkov PM, Paigen K. Prdm9 controls activation of mammalian recombination hotspots. Science. 2010;327(5967):835. doi: 10.1126/science.1181495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Perry J, Kleckner N, Börner GV. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. PNAS. 2005;102(49):17594–99. doi: 10.1073/pnas.0508581102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Petes TD. Meiotic recombination hot spots and cold spots. Nat Rev Genet. 2001;2(5):360–69. doi: 10.1038/35072078. [DOI] [PubMed] [Google Scholar]
- 177.Phillips CM, Meng X, Zhang L, Chretien JH, Urnov FD, Dernburg AF. Identification of chromosome sequence motifs that mediate meiotic pairing and synapsis in C. elegans. Nat Cell Biol. 2009;11(8):934–42. doi: 10.1038/ncb1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Pratto F, Brick K, Khil P, Smagulova F, Petukhova GV, Camerini-Otero RD. DNA recombination. Recombination initiation maps of individual human genomes. Science. 2014;346(6211):1256442. doi: 10.1126/science.1256442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Provart NJ, Alonso J, Assmann SM, Bergmann D, Brady SM, et al. 50 years of Arabidopsis research: highlights and future directions. New Phytol. 2016;209(3):921–44. doi: 10.1111/nph.13687. [DOI] [PubMed] [Google Scholar]
- 180.Qiao H, Prasada Rao HB, Yang Y, Fong JH, Cloutier JM, et al. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet. 2014;46(2):194–99. doi: 10.1038/ng.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Rasmussen SW, Holm PB. The synaptonemal complex, recombination nodules and chiasmata in human spermatocytes. Symp Soc Exp Biol. 1984;38:271–92. [PubMed] [Google Scholar]
- 182.Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet. 2013;45(3):269–78. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Ribeiro J, Abby E, Livera G, Martini E. RPA homologs and ssDNA processing during meiotic recombination. Chromosoma. 2015;125:265–76. doi: 10.1007/s00412-015-0552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Robert T, Nore A, Brun C, Maffre C, Crimi B, et al. The TopoVIB-like protein family is required for meiotic DNA double-strand break formation. Science. 2016;351(6276):943–49. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- 185.Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995;9(21):2684–95. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- 186.Rogacheva MV, Manhart CM, Chen C, Guarne A, Surtees J, Alani E. Mlh1-Mlh3, a meiotic crossover and DNA mismatch repair factor, is a Msh2-Msh3-stimulated endonuclease. J Biol Chem. 2014;289(9):5664–73. doi: 10.1074/jbc.M113.534644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Ross-Macdonald P, Roeder GS. Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell. 1994;79(6):1069–80. doi: 10.1016/0092-8674(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 188.Saito TT, Colaiácovo MP. Crossover recombination mediated by HIM-18/SLX4-associated nucleases. Worm. 2014;3:e28233. doi: 10.4161/worm.28233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Saito TT, Lui DY, Kim HM, Meyer K, Colaiácovo MP. Interplay between structure-specific endonucleases for crossover control during Caenorhabditis elegans meiosis. PLOS Genet. 2013;9(7):e1003586. doi: 10.1371/journal.pgen.1003586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Saito TT, Mohideen F, Meyer K, Harper JW, Colaiácovo MP. SLX-1 is required for maintaining genomic integrity and promoting meiotic noncrossovers in the Caenorhabditis elegans germline. PLOS Genet. 2012;8(8):e1002888. doi: 10.1371/journal.pgen.1002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Saito TT, Youds JL, Boulton SJ, Colaiácovo MP. Caenorhabditis elegans HIM-18/SLX-4 interacts with SLX-1 and XPF-1 and maintains genomic integrity in the germline by processing recombination intermediates. PLOS Genet. 2009;5(11):e1000735. doi: 10.1371/journal.pgen.1000735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Santucci-Darmanin S, Walpita D, Lespinasse F, Desnuelle C, Ashley T, Paquis-Flucklinger V. MSH4 acts in conjunction with MLH1 during mammalian meiosis. FASEB J. 2000;14(11):1539–47. doi: 10.1096/fj.14.11.1539. [DOI] [PubMed] [Google Scholar]
- 193.Scherthan H. Telomere attachment and clustering during meiosis. Cell Mol Life Sci. 2007;64(2):117–24. doi: 10.1007/s00018-006-6463-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schild-Prüfert K, Saito TT, Smolikov S, Gu Y, Hincapie M, et al. Organization of the synaptonemal complex during meiosis in Caenorhabditis elegans. Genetics. 2011;189(2):411–21. doi: 10.1534/genetics.111.132431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Höög C, et al. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLOS Genet. 2011;7(5):e1002088. doi: 10.1371/journal.pgen.1002088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Schwartz EK, Heyer WD. Processing of joint molecule intermediates by structure-selective en-donucleases during homologous recombination in eukaryotes. Chromosoma. 2011;120(2):109–27. doi: 10.1007/s00412-010-0304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Sehorn MG, Sigurdsson S, Bussen W, Unger VM, Sung P. Human meiotic recombinase Dmc1 promotes ATP-dependent homologous DNA strand exchange. Nature. 2004;429(6990):433–37. doi: 10.1038/nature02563. [DOI] [PubMed] [Google Scholar]
- 198.Serrentino ME, Chaplais E, Sommermeyer V, Borde V. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLOS Genet. 2013;9(4):e1003416. doi: 10.1371/journal.pgen.1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Severson AF, Ling L, van Zuylen V, Meyer BJ. The axial element protein HTP-3 promotes cohesin loading and meiotic axis assembly in C. elegans to implement the meiotic program of chromosome segregation. Genes Dev. 2009;23(15):1763–78. doi: 10.1101/gad.1808809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Sherman SL, Takaesu N, Freeman SB, Grantham M, Phillips C, et al. Trisomy 21: association between reduced recombination and nondisjunction. Am J Hum Genet. 1991;49(3):608–20. [PMC free article] [PubMed] [Google Scholar]
- 201.Shinohara A, Ogawa H, Ogawa T. Rad51 protein involved in repair and recombination in S. cerevisiae is a RecA-like protein. Cell. 1992;69(3):457–70. doi: 10.1016/0092-8674(92)90447-k. [DOI] [PubMed] [Google Scholar]
- 202.Sidhu GK, Fang C, Olson MA, Falque M, Martin OC, Pawlowski WP. Recombination patterns in maize reveal limits to crossover homeostasis. PNAS. 2015;112(52):15982–87. doi: 10.1073/pnas.1514265112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Singh MK, Nicolas E, Gherraby W, Dadke D, Lessin S, Golemis EA. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene. 2007;26(33):4825–32. doi: 10.1038/sj.onc.1210282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Smagulova F, Gregoretti IV, Brick K, Khil P, Camerini-Otero RD, Petukhova GV. Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature. 2011;472(7343):375–78. doi: 10.1038/nature09869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15(3):437–51. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 206.Snyder EM, Small C, Griswold MD. Retinoic acid availability drives the asynchronous initiation of spermatogonial differentiation in the mouse. Biol Reprod. 2010;83(5):783–90. doi: 10.1095/biolreprod.110.085811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Stamper EL, Rodenbusch SE, Rosu S, Ahringer J, Villeneuve AM, Dernburg AF. Identification of DSB-1, a protein required for initiation of meiotic recombination in Caenorhabditis elegans, illuminates a crossover assurance checkpoint. PLOS Genet. 2013;9(8):e1003679. doi: 10.1371/journal.pgen.1003679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Strong ER, Schimenti JC. Evidence implicating CCNB1IP1, a ring domain-containing protein required for meiotic crossing over in mice, as an E3 SUMO ligase. Genes. 2010;1(3):440–51. doi: 10.3390/genes1030440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Sturtevant AH. The behavior of the chromosomes as studied through linkage. Z Indukt Abstamm Vererb. 1915;13(1):234–87. [Google Scholar]
- 210.Subramanian VV, Hochwagen A. The meiotic checkpoint network: step-by-step through meiotic prophase. Cold Spring Harb Perspect Biol. 2014;6(10):a016675. doi: 10.1101/cshperspect.a016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Sun H, Treco D, Schultes NP, Szostak JW. Double-strand breaks at an initiation site for meiotic gene conversion. Nature. 1989;338(6210):87–90. doi: 10.1038/338087a0. [DOI] [PubMed] [Google Scholar]
- 212.Svendsen JM, Smogorzewska A, Sowa ME, O’Connell BC, Gygi SP, et al. Mammalian BTBD12/SLX4 assembles a Holliday junction resolvase and is required for DNA repair. Cell. 2009;138(1):63–77. doi: 10.1016/j.cell.2009.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sym M, Engebrecht JA, Roeder GS. Zip1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72(3):365–78. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- 214.Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79(2):283–92. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- 215.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33(1):25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 216.Thacker D, Mohibullah N, Zhu X, Keeney S. Homologue engagement controls meiotic DNA break number and distribution. Nature. 2014;510(7504):241–46. doi: 10.1038/nature13120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Toby GG, Gherraby W, Coleman TR, Golemis EA. A novel ring finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol Cell Biol. 2003;23(6):2109–22. doi: 10.1128/MCB.23.6.2109-2122.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Tosti E, Katakowski JA, Schaetzlein S, Kim HS, Ryan CJ, et al. Evolutionarily conserved genetic interactions with budding and fission yeast MutS identify orthologous relationships in mismatch repair-deficient cancer cells. Genome Med. 2014;6(9):68. doi: 10.1186/s13073-014-0068-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Tsai CJ, Mets DG, Albrecht MR, Nix P, Chan A, Meyer BJ. Meiotic crossover number and distribution are regulated by a dosage compensation protein that resembles a condensin subunit. Genes Dev. 2008;22(2):194–211. doi: 10.1101/gad.1618508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Tsubouchi T, Macqueen AJ, Roeder GS. Initiation of meiotic chromosome synapsis at centromeres in budding yeast. Genes Dev. 2008;22(22):3217–26. doi: 10.1101/gad.1709408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tsubouchi T, Zhao H, Roeder GS. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev Cell. 2006;10(6):809–19. doi: 10.1016/j.devcel.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 222.Vasnier C, de Muyt A, Zhang L, Tessé S, Kleckner NE, et al. Absence of SUN-domain protein Slp1 blocks karyogamy and switches meiotic recombination and synapsis from homologs to sister chromatids. PNAS. 2014;111(38):E4015–23. doi: 10.1073/pnas.1415758111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Voelkel-Meiman K, Johnston C, Thappeta Y, Subramanian VV, Hochwagen A, MacQueen AJ. Separable crossover-promoting and crossover-constraining aspects of Zip1 activity during budding yeast meiosis. PLOS Genet. 2015;11(6):e1005335. doi: 10.1371/journal.pgen.1005335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Voelkel-Meiman K, Taylor LF, Mukherjee P, Humphryes N, Tsubouchi H, Macqueen AJ. SUMO localizes to the central element of synaptonemal complex and is required for the full synapsis of meiotic chromosomes in budding yeast. PLOS Genet. 2013;9(10):e1003837. doi: 10.1371/journal.pgen.1003837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Vrielynck N, Chambon A, Vezon D, Pereira L, Chelysheva L, et al. A DNA topoisomerase VI-like complex initiates meiotic recombination. Science. 2016;351(6276):939–43. doi: 10.1126/science.aad5196. [DOI] [PubMed] [Google Scholar]
- 226.Wan L, Niu H, Futcher B, Zhang C, Shokat KM, et al. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 2008;22(3):386–97. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Ward JO, Reinholdt LG, Motley WW, Niswander LM, Deacon DC, et al. Mutation in mouse HEI10, an E3 ubiquitin ligase, disrupts meiotic crossing over. PLOS Genet. 2007;3(8):e139. doi: 10.1371/journal.pgen.0030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Wolf KW. How meiotic cells deal with non-exchange chromosomes. BioEssays. 1994;16(2):107–14. doi: 10.1002/bies.950160207. [DOI] [PubMed] [Google Scholar]
- 229.Yang F, De La Fuente R, Leu NA, Baumann C, McLaughlin KJ, Wang PJ. Mouse SYCP2 is required for synaptonemal complex assembly and chromosomal synapsis during male meiosis. J Cell Biol. 2006;173(4):497–507. doi: 10.1083/jcb.200603063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Yildiz O, Majumder S, Kramer B, Sekelsky JJ. Drosophila MUS312 interacts with the nucleotide excision repair endonuclease MEI-9 to generate meiotic crossovers. Mol Cell. 2002;10(6):1503–9. doi: 10.1016/s1097-2765(02)00782-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yokoo R, Zawadzki KA, Nabeshima K, Drake M, Arur S, Villeneuve AM. COSA-1 reveals robust homeostasis and separable licensing and reinforcement steps governing meiotic crossovers. Cell. 2012;149(1):75–87. doi: 10.1016/j.cell.2012.01.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Youds JL, Boulton SJ. The choice in meiosis: defining the factors that influence crossover or non-crossover formation. J Cell Sci. 2011;124(Pt. 4):501–13. doi: 10.1242/jcs.074427. [DOI] [PubMed] [Google Scholar]
- 233.Youds JL, Mets DG, McIlwraith MJ, Martin JS, Ward JD, et al. RTEL-1 enforces meiotic crossover interference and homeostasis. Science. 2010;327(5970):1254–58. doi: 10.1126/science.1183112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Zakharyevich K, Ma Y, Tang S, Hwang PY, Boiteux S, Hunter N. Temporally and biochemically distinct activities of Exo1 during meiosis: double-strand break resection and resolution of double Holliday junctions. Mol Cell. 2010;40(6):1001–15. doi: 10.1016/j.molcel.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149(2):334–47. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Zavec AB, Comino A, Lenassi M, Komel R. Ecm11 protein of yeast Saccharomyces cerevisiae is regulated by SUMOylation during meiosis. FEMS Yeast Res. 2008;8(1):64–70. doi: 10.1111/j.1567-1364.2007.00307.x. [DOI] [PubMed] [Google Scholar]
- 237.Zavec AB, Lesnik U, Komel R, Comino A. The Saccharomyces cerevisiae gene ECM11 is a positive effector of meiosis. FEMS Microbiol Lett. 2004;241(2):193–99. doi: 10.1016/j.femsle.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 238.Zhang L, Kim KP, Kleckner NE, Storlazzi A. Meiotic double-strand breaks occur once per pair of (sister) chromatids and, via Mec1/ATR and Tel1/ATM, once per quartet of chromatids. PNAS. 2011;108(50):20036–41. doi: 10.1073/pnas.1117937108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Zhang L, Tang D, Luo Q, Chen X, Wang H, et al. Crossover formation during rice meiosis relies on interaction of OsMSH4 and OsMSH5. Genetics. 2014;198(4):1447–56. doi: 10.1534/genetics.114.168732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhou Q, Nie R, Li Y, Friel P, Mitchell D, et al. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A–sufficient postnatal murine testes. Biol Reprod. 2008;79(1):35–42. doi: 10.1095/biolreprod.107.066795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Zickler D, Kleckner N. The leptotene-zygotene transition of meiosis. Annu Rev Genet. 1998;32:619–97. doi: 10.1146/annurev.genet.32.1.619. [DOI] [PubMed] [Google Scholar]
- 242.Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 2015;7(6):a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]