Abstract
Previously, we reported that vitamin C facilitates the CpG demethylation of Foxp3 enhancer in CD4+Foxp3+ regulatory T cells (Tregs) by enhancing the activity of a DNA demethylase ten-eleven-translocation (Tet). However, it is not clear whether vitamin C affects other helper T cell lineages like T helper type 17 (Th17) cells which are related with Tregs. Here, we show that the expression of interleukin-17A (IL17) increases with the treatment of vitamin C but not with other antioxidants. Interestingly, the upregulation of IL17 was not accompanied by DNA demethylation in Il17 promoter and was independent of Tet enzymes. Rather, vitamin C reduced the trimethylation of histone H3 lysine 9 (H3K9me3) in the regulatory elements of the Il17 locus, and the effects of vitamin C were abrogated by knockdown of jumonji-C domain-containing protein 2 (jmjd2). These results suggest that vitamin C can affect the expression of IL17 by modulating the histone demethylase activity.
Keywords: H3K9me3, Histone modification, IL17, Jmjd2, Vitamin C
INTRODUCTION
T helper type 17 (Th17) cells are important cellular mediators in diverse immune responses (1). The generation of the Th17 cell is best achieved by stimulation with TCR, CD28 and cytokines like interleukin-6 (IL6) and transforming growth factor-β (TGF-β) (2). TCR/CD28 signals activate pioneering transcription factors to open up and make the target gene chromatin structure permissive to cytokine-associated transcription factor (3), signal transducer and activator of transcription 3 (STAT3). Then, STAT3 acts together with various modifiers to promote the expression of the gene encoding retinoic acid receptor-related orphan receptor γt (RORγt; encoded by Rorc) (4, 5) leading to the lineage specification of Th17 cells. Th17 cells produce a group of proinflammatory cytokines including IL17, which mediate diverse autoimmune and inflammatory diseases (6).
Vitamin C is known to be not only a general anti-oxidant but also a cofactor for a large enzyme family known as the iron-and 2-oxoglutarate-dependent dioxygenases (7). A typical example is the collagen prolyl-4-hydroxylase in collagen maturation and scurvy (8). Recently, we found that ten-eleven-translocation (Tet) DNA dioxygenase also works in a vitamin C dependent manner in regulatory T cells (Tregs). In the absence of vitamin C, the initial hydroxylation of 5-methylcytosine by Tet enzymes proceeded albeit less efficiently; however, the intermediates, 5-hydroxymethylcytosine, accumulated gradually, and a further oxidation reaction was not completed (9). Collectively, vitamin C seems to have a role in reducing inactive iron and maintaining continued enzyme cycling in DNA oxidation followed by demethylation reactions.
In this study, based on our previous report showing the role of vitamin C in Tregs, we investigated Th17 cells after vitamin C treatment and found that vitamin C exploited Jmjd2 histone demethylases to enhance the expression of IL17.
RESULTS AND DISCUSSION
IL17 was upregulated by vitamin C treatment
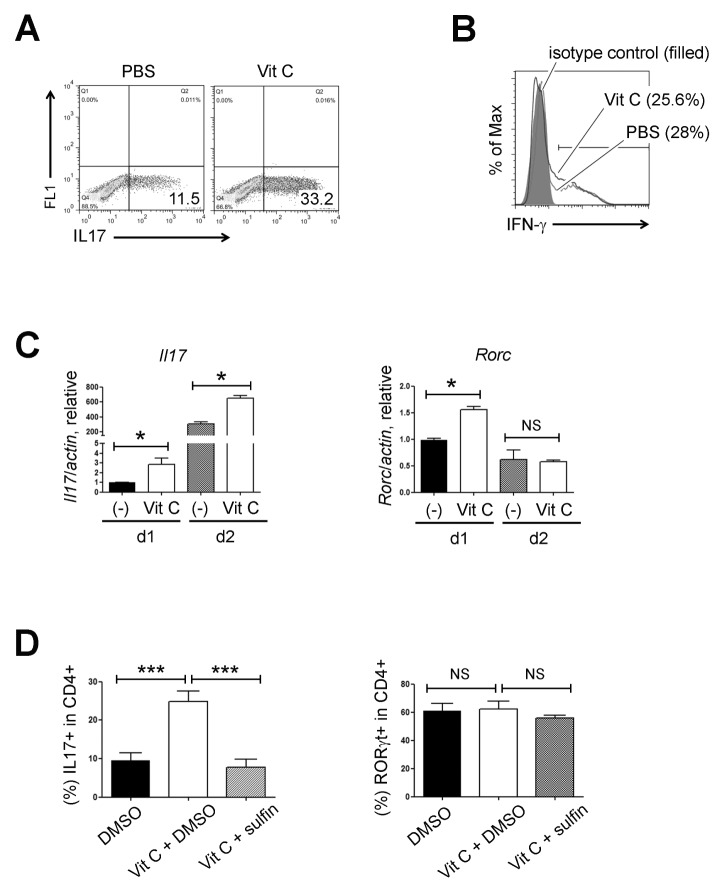
To examine the role of vitamin C in Th17 cell differentiation, we sorted naïve CD4+ T cells from wild type (WT) mice and primed them to differentiate into Th17 in the presence or absence of vitamin C (10 μg/ml). After 3 d, we restimulated the cells with PMA and ionomycin and assessed the expression of cytokines by intracellular staining and flow cytometry. Unexpectedly, in the Th17 polarizing condition (anti-CD3/CD28 mAbs plus rIL6 and TGF-β), some vitamin C–treated cells, but not control cells, differentiated into Foxp3–expressing induced Tregs, which remained up to 5 d (Supplementary Fig. 1). Previously, vitamin C was reported to induce the demethylation of CpG motifs in the Foxp3 enhancer and contribute to the stable expression of Foxp3 in Tregs (9, 10), which led us to hypothesize that vitamin C stabilized the expression of Foxp3 even in the presence of rIL6. Indeed, the CpG motifs in the Foxp3 enhancer regions were demethylated when vitamin C was available. These findings led us to modify the Th17 differentiation condition by adding anti-IL2 blocking mAbs (clone: S4B6), which prevented the Foxp3 expression (Supplemental Fig. 1) and enabled us to analyze the effect of vitamin C on Th17 cell development.
FACS-sorted naïve CD4+ T cells were cultured again under the modified Th17 conditions in the presence or absence of vitamin C. Interestingly, IL17–expressing cells were generated more efficiently from cells treated with vitamin C (Fig. 1A). To check the specificity, we performed Th1 differentiation experiments in parallel and found that vitamin C did not influence the expression of IFN-γ under Th1 conditions (Fig. 1B). The mRNA level of Il17 was also significantly increased in the vitamin C–treated Th17 cells. However, the level of Rorc transcripts was transiently elevated during an early stage (Fig. 1C). We confirmed the effect of vitamin C once again with a sodium-dependent vitamin C transporter inhibitor, sulfinpyrazone (11). Sulfinpyrazone treatment reversed the effect of vitamin C on IL17 expression (Fig. 1D, left) suggesting that the entry of vitamin C is essential. FACS analysis also showed that the frequency of RORγt–expressing cells was not significantly altered by the vitamin C or sulfinpyrazone treatment (Fig. 1D, right). To examine the mechanism of action of vitamin C, we checked whether other anti-oxidants could also increase the expression of IL17. Naïve CD4+ T cells were treated with vitamin C, glutathione (GSH, 5 mM), or dithiothreitol (DTT, 0.3 mM) under the Th17 polarizing conditions for 3 d, and neither GSH nor DTT increased the expression of IL17 (Supplementary Fig. 2A). Altogether, these findings suggest that the effect of vitamin C on IL17 expression was independent of RORγt upregulation and its role as a general reducer.
Fig. 1.
Vitamin C enhances the expression of IL17. (A) FACS-sorted naïve CD4+ T cells were cultured in the presence or absence of vitamin C (10 μg/ml) under Th17 polarizing condition for 3 d and IL17 expression was checked by flow cytometry after PMA/ionomycin restimulation. Data are representative of three independent experiments. (B) FACS-sorted naïve CD4+ T cells were cultured in the presence or absence of vitamin C (10 μg/ml) under Th1 polarizing condition and IFN-γ expression was checked by flow cytometry. Data are representative of two independent experiments. (C) Th17 cells were generated in the presence or absence of vitamin C and then, the expression levels of the indicated gene transcripts were checked by RT-qPCR (mean ± SEM of duplicates, from one experiment representative of three independent experiments). (D) Percentages of IL17 (left) or RORγt (right)–expressing cells cultured under Th17 conditions in the presence of vitamin C ± sulfinpyrazone (sulifin). Bars show the mean ± SEM of the results (n = 6–8) pooled from three independent experiments. FACS plots of (A) and (B) refer to the percentage of each subset. ***P < 0.001; *P < 0.05; NS, not significant.
Tet proteins were not required for the vitamin C–induced IL17 upregulation
Because vitamin C has a role as a cofactor for iron and 2-oxoglutarate dependent dioxygenases including DNA demethylases (Tet family) (12), histone demethylases (jumonji domain-containing proteins, the Jmjd family) (13) and prolyl hydroxylase domain-containing proteins (PHD proteins) (14), we decided to investigate whether these pathways are involved in the vitamin C–induced IL17 upregulation. First, we examined whether PHD proteins and related hypoxia-inducible factor pathways (15) are involved by using N-oxalylglycine (NOG), a potent inhibitor of PHD proteins (16–18). We cultured naïve CD4+ T cells under Th17 conditions in the presence of vitamin C plus various doses of NOG and found that NOG treatment enhances, not inhibits, Th17 differentiation (Supplementary Fig. 2B) consistent with a previous report showing the positive effects of hypoxia-inducible factor on IL17 expression (19).
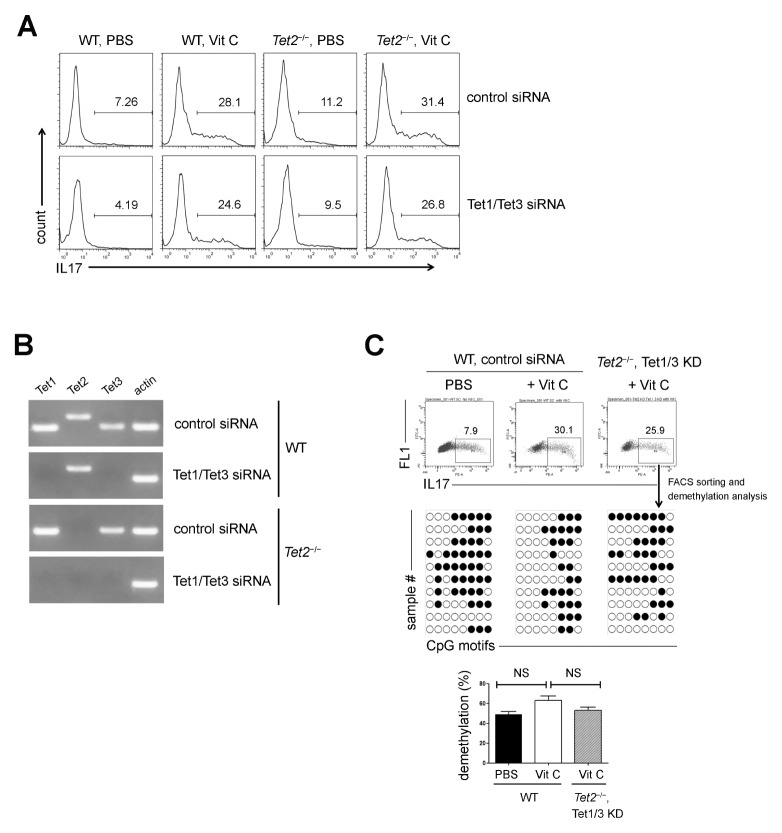
It was reported that the CpG motifs in the IL17 promoter are less methylated in Th17 cells (20), and 5-hydroxymethylcytosine and Tet2 have roles in the generation and functions of Th17 cells (21), which led us to hypothesize that vitamin C might contribute to IL17 expression as a cofactor of Tet2. To address this issue, we cultured WT and Tet2−/− naïve CD4+ T cells with vitamin C under Th17 conditions and examined the effect of vitamin C. Unexpectedly, the expression of IL17 was increased by vitamin C in Tet2−/− cells as well (Fig. 2A). Given the potential redundancy between members of the Tet family (22), we investigated whether Tet1 and Tet3 functioned as substitutes for Tet2. WT and Tet2−/− naïve CD4+ T cells were co-transfected with equal amounts of small interference RNAs (siRNAs) targeting Tet1 and Tet3 and cultured under Th17 conditions in the presence of vitamin C. RT-PCR assays revealed that Tet1 and Tet3 transcripts were specifically downregulated in the Tet1/Tet3 siRNA-transfected cells (Fig. 2B). After 3 d, the expression level of IL17 was analyzed by flow cytometry. IL17 was not altered significantly by the knockdown of Tet1 and Tet3 (Fig. 2A).
Fig. 2.
Tet proteins were not essential for vitamin C–induced IL17 expression. (A) FACS-sorted WT and Tet2−/− naïve CD4+ T cells were transfected with control or Tet1/Tet3 siRNAs and cultured under the indicated conditions. IL17 expression was analyzed by flow cytometry. (B) FACS-sorted WT and Tet2−/− naïve CD4+ T cells were transfected with Tet1/Tet3 siRNAs, and the expression of the indicated gene transcripts was checked by RT-PCR. (C) FACS-sorted WT and Tet2−/− naïve CD4+ T cells were transfected with control or Tet1/Tet3 siRNAs and cultured under the indicated conditions. IL17+ cells were re-sorted and subjected to bisulfate sequencing. Results of statistical analysis are shown as the mean ± SEM of the results (n = 3) pooled from two independent experiments. Numbers in the FACS plots of (A) and (C) refer to the percentage of each subset. Methylation status of individual CpG motif was shown by white (demethylation) or black (methylation) circles.
Next, the methylation status of the CpG motifs in the IL17 promoter was analyzed. FACS-sorted WT and Tet2−/− naïve CD4+ T cells were transfected with control or Tet1/Tet3 siRNAs, cultured under Th17 conditions, and restimulated with PMA and ionomycin. Then, IL17+ cells were re-sorted and subjected to bisulfite sequencing. The extent of CpG demethylation was not significantly changed by vitamin C (Fig. 2C) suggesting that vitamin C facilitated IL17 expression in a Tet–independent manner.
The effects of vitamin C were abrogated by the knockdown of Jmjd2 histone demethylases
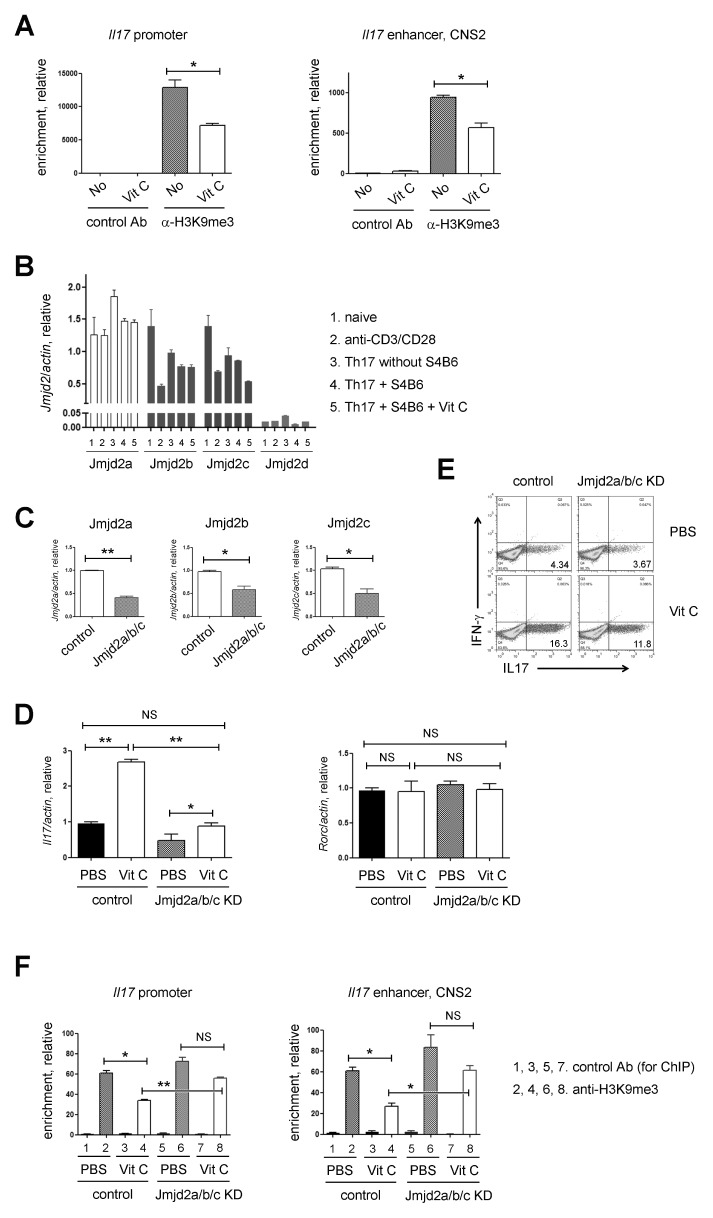
Next, we tested repressive histone modifications like trimethylation of histone H3 lysine 9 (H3K9me3) or histone H3 lysine 27 (H3K27me3) and the related Jmjd histone demethylases. Because the roles of H3K27me3 demethylase, a Jmjd3, in Th17 differentiation were controversial (23, 24) and vitamin C was reported to induce the expression of pluripotent genes in induced stem cells (iPSCs) by erasing the methylation of H3K9 (13), we decided to focus on the changes of H3K9me3 at the IL17 locus (25). As shown in Fig. 3A, vitamin C treatment reduced H3K9me3 in the IL17 promoter and enhancer (CNS2) indicating that H3K9 is one of the sites for chromatin modification in response to vitamin C. Because demethylation at H3K9me3 is catalyzed by the histone demethylase Jmjd2 subfamily (26) and three (Jmjd2a, Jmjd2b, and Jmjd2c) out of four Jmjd2 members are expressed in various types of CD4+ T cells (Fig. 3B), we tried to knock down three Jmjd2 members (referred to as Jmjd2a/b/c hereafter) together using siRNAs to prevent potential redundancy among them (27) and investigated whether vitamin C works in a Jmjd2–dependent manner. Jmjd2a/b/c transcripts were significantly downregulated after siRNA transfection (Fig. 3C). WT naïve CD4+ T cells transfected with control or Jmjd2a/b/c siRNA were cultured under Th17 polarizing conditions in the presence or absence of vitamin C. Subsequent RT-qPCR (Fig. 3D, left) and FACS analysis (Fig. 3E) revealed that the effect of vitamin C on the expression of IL17 was impaired, albeit partially, by the knockdown of Jmjd2a/b/c. In contrast to IL17, the expression of Rorc transcripts was not altered (Fig. 3D, right). Our findings imply that vitamin C enhances IL17 expression by modulating Jmjd2. To examine the H3K9me3 enrichments in the Il17 locus of Th17 cells treated with vitamin C or Jmjd2a/b/c siRNA, we performed the chromatin immunoprecipitation (ChIP) assay. As shown previously (Fig. 3A), the enrichment of H3K9me3 was reduced by vitamin C in the Il17 regulatory elements (promoter and enhancer) in the Th17 cells transfected with control siRNA. However, the effect of vitamin C on H3K9me3 was lost after the treatment with the Jmjd2a/b/c siRNAs (Fig. 3F).
Fig. 3.
Vitamin C worked in a Jmjd2 dependent manner. (A) ChIP-qPCR of H3K9me3 at the IL17 promoter and enhancer (CNS2) locus. FACS-sorted naïve CD4+ T cells were cultured under Th17 conditions in the presence or absence of vitamin C and used for the ChIP analysis. Subsequent qPCR was done on DNA precipitated by control and anti-H3K9me3 Ab (mean ± SEM of duplicates, from one experiment representative of three independent experiments). (B) Quantitative RT-PCR was performed to assess the expression levels of Jmjd2 members in the indicated CD4+ T cells. (C) FACS-sorted naïve CD4+ T cells were transfected with control or Jmjd2a/b/c siRNAs, and then, the expressions of the indicated gene transcripts were checked by RT-qPCR. (D) The expression levels of Il17 (left) and Rorc (right) transcripts were investigated in the indicated Th17 cell populations. (E) IL17 expressions in the indicated Th17 cell populations were checked by flow cytometry. (F) ChIP-qPCR of H3K9me3 at the IL17 promoter and enhancer (CNS2) locus. FACS-sorted naïve CD4+ T cells were cultured under Th17 conditions in the presence or absence of vitamin C and used for the ChIP analysis. Subsequent qPCR was done on DNA precipitated by control and anti-H3K9me3 Ab (mean ± SEM of duplicates, from one experiment representative of three independent experiments). Data are representative of 3 (A, C-E) or 2 (B, F) independent experiments. **P < 0.01; *P < 0.05; NS, not significant.
Altogether, these findings indicate that vitamin C induced the upregulation of IL17 through Jmjd2 histone demethylase enzymes and the related H3K9 histone modifications. Further study on Jmjd2 enzymes and vitamin C will advance our understanding on the regulatory mechanisms of Th17 development and provide new therapeutic targets for Th17–related diseases.
MATERIALS AND METHODS
Mice
WT C57BL/6 mice were purchased from Koatech (Pyeongtaek, Gyeongi-do, Korea). Floxed Tet2 transgenic (B6;129S-Tet2tm1.1Iaai/J, Tet2fl/fl) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). To generate Tet2 deficient mice (referred to as Tet2−/− mice), CD4-Cre transgenic mice were crossed to floxed Tet2 transgenic mice. All animal experimentations were conducted in accordance with guidelines and approval of the International Animal Care and Use Committees of Hallym University (Hallym 2015-63).
Cell isolation and flow cytometry
To sort naïve CD4+ cells, CD44 and CD25 were used (CD4+CD8−CD44lowCD25−). The post-sort purity for each cell type was usually > 97%. Intracellular cytokines were stained using Foxp3 Staining Buffer set (eBioscience, San Diego, CA). For cytokine analysis, cells were cultured for 4 h in the presence of PMA/ionomycin plus monensin (BD biosciences, San Jose, CA) before intracellular cytokine staining (28). Data were acquired through FACS Calibur or FACS Canto-II (BD Biosciences) and were analyzed with FlowJo software (Tree Star, Ashland, OR).
Cell culture
For Th17 cell generation, cells were stimulated by plate–bound anti-CD3 (2C11, 5 μg/ml; eBioscience) plus CD28 (37.51, 1 μg/ml; eBioscience), recombinant murine IL6 (rIL6, 20 ng/ml; Peprotech, Rocky Hill, NJ) and recombinant TGF-β (rTGF-β, 0.5 ng/ml; Peprotech) in the presence or absence of vitamin C (10 μg/ml, Sigma-Aldrich, St. Louis, MO) for 3 d. To prevent the Foxp3 expression, 10 μg/ml of anti-IL2 blocking mAb (clone: S4B6) was always added except some samples in supplementary Fig. 1.
Small interfering RNA transfection
Transfection was performed using the Amaxa Mouse T Cell Nucleofector Kit and the Nucleofector device (X-001 program) according to the previous report (29). Cells were transfected with 300 pmol SMART pool siRNAs (Dharmacon, Lafayette, Colorado) designed against mouse Jmjd2a (M-059020-00-0005), Jmjd2b (M-062955-00-0005) and Jmjd2c (M-051504-00-0005) together. Non-targeting control siRNAs (SN-1001, Bioneer) were also transfected as control treatments. The expression levels of each target molecule were checked 8 h after electro-poration.
Quantitative PCR (qPCR)
PCR reactions were performed on RotorGene 6000 system (Qiagen, Valencia, CA) using AccuPower GreenStar qPCR kit (Bioneer, Seoul, Korea). All data were normalized to actin. Non-specific amplification was checked by using melting curves and agarose gel electorphoresis (30). The sequences of primers are as below.
Mouse Jmjd2a-Forward: 5′-GACCACACTCTGCCCACAC-3′
Mouse Jmjd2a-Reverse: 5′-TCCTGGGGTATTTCCAGACA-3′
Mouse Jmjd2b-Forward: 5′-GGCTTTAACTGCGCTGAGTC-3′
Mouse Jmjd2b-Reverse: 5′-GTGTGGTCCAGCACTGTGAG-3′
Mouse Jmjd2c-Forward: 5′-CACGGAGGACATGGATCTCT-3′
Mouse Jmjd2c-Reverse: 5′-CGAAGGGAATGCCATACTTC-3′
Mouse Actin-Forward: 5′-CATCCGTAAAGACCTCTATGCCAAC-3′
Mouse Actin-Reverse: 5′-ATGGAGCCACCGATCCACA-3′
DNA demethylation analysis
FACS-sorted IL17+ cells were incubated with 300 μl lysis buffer (10 mM Tris-HCl, 100 mM NaCl, 50 mM EDTA, 0.5% SDS, 0.1 μg/ml proteinase K, and 20 μg/ml RNase A) for 24 h at 60°C. Then extracted genomic DNAs were converted by the EZ DNA methylation gold kit (Zymo Research, Irvine, CA) and subjected to PCR (sense: 5′-ACAAATTCATAAACCCCAACAA CTA-3′, antisense: 5′-TGTTGATTTTATTTGAGGATGGAAT-3′). The PCR products were cloned into the pGemT-easy vector (Promega, Madison, WI) and individual clones were sequenced with M13 reverse primer (GAAACAGCTATGACCATG).
Chromatin immunoprecipitation (ChIP)
Nuclear lysate was sonicated to make small DNA fragments ranging from 100–500 base pairs and then incubated with anti-H3K9me3 (MABI 0319, Active motif, Carlsbad, CA) overnight at 4°C. Isotype-matched control Ab was used for the negative control. Immune complexes containing DNA fragments were precipitated using EZ-ChIP kit (Milipore, Darmstadt, Germany). Relative enrichment of the target regions in the precipitated DNA fragments was analyzed by qPCR. The sequences of primers are as follows.
Il17 promoter forward, 5′-GCAGCAGCTTCAGATATGTCC-3′
Il17 promoter reverse, 5′-TGAGGTCAGCACAGAACCAC-3′
Il17 CNS2 enhancer forward, 5′-CCGTTTAGACTTGAAACCCAGTC-3′
Il17 CNS2 enhancer reverse, 5′-GTACCTATGTGTTAGGAGGCGC-3′
Statistical analyses
A two-tailed, unpaired, Student’s t-test was used to calculate the statistical significance of differences between groups. P values are represented as follows: ***P < 0.001; **P < 0.01; *P < 0.05, whereas NS, not significant, is used to denote P values > 0.05. Error bars indicate s.e.m.
Supplementary Information
ACKNOWLEDGEMENTS
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2016R1D 1A1B03934212) and Hallym University (HRF-201612-016).
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting financial interests.
REFERENCES
- 1.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol. 2009;27:485–517. doi: 10.1146/annurev.immunol.021908.132710. [DOI] [PubMed] [Google Scholar]
- 2.Mangan PR, Harrington LE, O’Quinn DB, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 3.Ciofani M, Madar A, Galan C, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durant L, Watford WT, Ramos HL, et al. Diverse targets of the transcription factor STAT3 contribute to T cell pathogenicity and homeostasis. Immunity. 2010;32:605–615. doi: 10.1016/j.immuni.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivanov II, McKenzie BS, Zhou L, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 6.Patel DD, Kuchroo VK. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity. 2015;43:1040–1051. doi: 10.1016/j.immuni.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 7.Young JI, Zuchner S, Wang G. Regulation of the Epigenome by Vitamin C. Annu Rev Nutr. 2015;35:545–564. doi: 10.1146/annurev-nutr-071714-034228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gorres KL, Raines RT. Prolyl 4-hydroxylase. Crit Rev Biochem Mol Biol. 2010;45:106–124. doi: 10.3109/10409231003627991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasidharan Nair V, Song MH, Oh KI. Vitamin C Facilitates Demethylation of the Foxp3 Enhancer in a Tet-Dependent Manner. J Immunol. 2016;196:2119–2131. doi: 10.4049/jimmunol.1502352. [DOI] [PubMed] [Google Scholar]
- 10.Yue X, Trifari S, Aijo T, et al. Control of Foxp3 stability through modulation of TET activity. J Exp Med. 2016;213:377–397. doi: 10.1084/jem.20151438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickson KM, Gustafson CB, Young JI, Zuchner S, Wang G. Ascorbate-induced generation of 5-hydroxymethylcytosine is unaffected by varying levels of iron and 2-oxoglutarate. Biochem Biophys Res Commun. 2013;439:522–527. doi: 10.1016/j.bbrc.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu GL, Walsh CP. Enzymatic DNA oxidation: mechanisms and biological significance. BMB Rep. 2014;47:609–618. doi: 10.5483/BMBRep.2014.47.11.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Liu H, Liu J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet. 2013;45:34–42. doi: 10.1038/ng.2491. [DOI] [PubMed] [Google Scholar]
- 14.Knowles HJ, Raval RR, Harris AL, Ratcliffe PJ. Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 2003;63:1764–1768. [PubMed] [Google Scholar]
- 15.Baek SH, Kim KI. Regulation of HIF-1alpha stability by lysine methylation. BMB Rep. 2016;49:245–246. doi: 10.5483/BMBRep.2016.49.5.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopkinson RJ, Tumber A, Yapp C, et al. 5-Carboxy-8-hydroxyquinoline is a Broad Spectrum 2-Oxoglutarate Oxygenase Inhibitor which Causes Iron Translocation. Chem Sci. 2013;4:3110–3117. doi: 10.1039/c3sc51122g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schofield CJ, Ratcliffe PJ. Oxygen sensing by HIF hydroxylases. Nat Rev Mol Cell Biol. 2004;5:343–354. doi: 10.1038/nrm1366. [DOI] [PubMed] [Google Scholar]
- 18.Chowdhury R, Yeoh KK, Tian YM, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dang EV, Barbi J, Yang HY, et al. Control of T(H)17/T(reg) balance by hypoxia-inducible factor 1. Cell. 2011;146:772–784. doi: 10.1016/j.cell.2011.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang BH, Floess S, Hagemann S, et al. Development of a unique epigenetic signature during in vivo Th17 differentiation. Nucleic Acids Res. 2015;43:1537–1548. doi: 10.1093/nar/gkv014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ichiyama K, Chen T, Wang X, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity. 2015;42:613–626. doi: 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blaschke K, Ebata KT, Karimi MM, et al. Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature. 2013;500:222–226. doi: 10.1038/nature12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Q, Zou J, Wang M, et al. Critical role of histone demethylase Jmjd3 in the regulation of CD4+ T-cell differentiation. Nat Commun. 2014;5:5780. doi: 10.1038/ncomms6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu Z, Cao W, Xu L, et al. The histone H3 lysine-27 demethylase Jmjd3 plays a critical role in specific regulation of Th17 cell differentiation. J Mol Cell Biol. 2015;7:505–516. doi: 10.1093/jmcb/mjv022. [DOI] [PubMed] [Google Scholar]
- 25.Xiao X, Shi X, Fan Y, et al. The Costimulatory Receptor OX40 Inhibits Interleukin-17 Expression through Activation of Repressive Chromatin Remodeling Pathways. Immunity. 2016;44:1271–1283. doi: 10.1016/j.immuni.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hojfeldt JW, Agger K, Helin K. Histone lysine demethylases as targets for anticancer therapy. Nat Rev Drug Discov. 2013;12:917–930. doi: 10.1038/nrd4154. [DOI] [PubMed] [Google Scholar]
- 27.Pedersen MT, Kooistra SM, Radzisheuskaya A, et al. Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 2016;35:1550–1564. doi: 10.15252/embj.201593317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seong MA, Woo JK, Kang JH, et al. Oral administration of fermented wild ginseng ameliorates DSS-induced acute colitis by inhibiting NF-kappaB signaling and protects intestinal epithelial barrier. BMB Rep. 2015;48:419–425. doi: 10.5483/BMBRep.2015.48.7.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantei A, Rutz S, Janke M, et al. siRNA stabilization prolongs gene knockdown in primary T lymphocytes. Eur J Immunol. 2008;38:2616–2625. doi: 10.1002/eji.200738075. [DOI] [PubMed] [Google Scholar]
- 30.Zuo J, Ma H, Cai H, Wu Y, Jiang W, Yu L. An inhibitory role of NEK6 in TGFbeta/Smad signaling pathway. BMB Rep. 2015;48:473–478. doi: 10.5483/BMBRep.2015.48.8.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.